Introduction
THP-1 human acute monocytic leukemia cells can
differentiate into macrophages (1), osteoclasts (2) and dendritic cells (3). THP-1 cells can also be differentiated
from macrophages to osteoclasts using cytokines and exogenous
hormones, such as macrophage colony-stimulating factor (M-CSF) and
receptor activator of nuclear factor-κB ligand (RANKL) (2,4).
M-CSF and RANKL are expressed in osteoblasts or activated T cells
and are involved in osteoclastogenesis (5,6). The
expression of nuclear factor of activated T-cells, cytoplasmic 1
(NFATc1), a key transcription factor involved in
osteoclastogenesis, is induced by RANKL (7). Tartrate-resistant acid phosphatase
(TRAP) and cathepsin K are osteoclast-specific markers and are
involved in the NFATc1 pathway (8). In addition, microRNA (miR)-218
negatively regulates osteoclast differentiation and bone remodeling
by blocking the RANKL-induced p38 mitogen activated protein
kinase/c-Fos/NFATc1 pathway (9).
MicroRNAs (miRNAs) are small non-coding RNAs, ~22
nucleotides in length, that are involved in post-transcriptional
regulations (10). miRNAs regulate
cell growth and differentiation of osteoblasts and osteoclasts from
embryonic bone to skeletal maturation (11). Members of the miR-17-92 family,
including miR-20a, are associated with monocytic differentiation
and maturation by regulating the expression levels of acute myeloid
leukemia-1 protein and M-CSF receptor (12). However, the expression and function
of miR-20a during osteoclast differentiation remain elusive.
miR-20a downregulates autophagy by targeting the 3′ untranslated
region (3′-UTR) of autophagy related 16-like 1 during
hypoxia-induced osteoclast differentiation (13). miR-20a overexpression was also
previously reported to suppress osteoclast differentiation and bone
resorption by downregulating RANKL expression (14). Therefore, elucidation of the
potential mechanism involving miR-20a during osteoclastogenesis is
crucial.
The present study investigated the potential
mechanisms of miR-20a during the differentiation of THP-1 cells
into osteoclasts. The results may provide a new perspective to the
understanding of miR-20a in osteoclast differentiation.
Materials and methods
Cell culture and differentiation
induction
THP-1 human acute monocytic leukemia cells (cat. no.
TIB-202; American Type Culture Collection, Manassas, VA, USA) were
incubated in RPMI-1640 medium (Sigma-Aldrich; Merck KGaA,
Darmstadt, Germany) containing 10% fetal bovine serum (Gibco;
Thermo Fisher Scientific, Inc., Waltham, MA, USA) at 37°C under 5%
CO2 and saturation humidity. THP-1 cells in the
logarithmic stage of growth were seeded (2×105
cells/well) into 6-well plates and co-cultured with 200 ng/ml
phorbol-12-myristate-13-acetate (PMA; cat. no. 79346;
Sigma-Aldrich; Merck KGaA), 25 ng/ml M-CSF (PeproTech, Inc., Rocky
Hill NJ, USA) and 30 ng/ml RANKL (PeproTech, Inc.) to induce their
differentiation into osteoclast-like cells, as previously described
(2). Cells were cultured for 0,
24, 72 and 120 h, collected and stained by using an Acid
Phosphatase, Leukocyte (TRAP) kit (cat. no. 387; Sigma-Aldrich;
Merck KGaA) according to the manufacturer's protocol. Subsequently,
the TRAP-positive stained cells were photographed using a light
microscopy (Nikon Corporation, Tokyo, Japan), and the number of
TRAP-positive staining cells were counted and analyzed.
Subsequently, the miR-20a expression levels were detected by
reverse transcription-quantitative polymerase chain reaction
(RT-qPCR) at 0, 24, 72 and 120 h, as described below.
Cell treatment and miR-20a mimics
transfection
To investigate the influences of miR-20a on the
process of monocyte differentiate into osteoclasts, THP-1 cells
were co-cultured with 10 ng/ml PMA to induce their differentiation
into macrophages (osteoclast precursors) at 37°C for three days
(1). Subsequently, the THP-1
cell-derived macrophages were seeded (1×104 cells/well)
into 96-well plates and transfected with 100 nM miR-20a mimics or
100 nM miRNA negative controls (miR-NC; GenePharma, Shanghai,
China) at 37°C for 6 h using Lipofectamine® 2000
(Invitrogen; Thermo Fisher Scientific, Inc., Waltham, MA, USA),
according to the manufacturer's guidelines. Untransfected THP-1
cell-derived macrophages were used as a Blank group. At 24 h
post-transfection, the transfection efficiency of miRNAs was
assessed by determining the miR-20a expression levels by RT-qPCR,
using the following primers: Forward, 5′-CCGCTCGTGAAATGTTTAGG-3′,
reverse primer, 5′-ATGGAGCCTGGGACGAGA-3′. U6 (forward primer
5′-ATTGGAACGATACAGAGAAGATT-3′; reverse primer,
5′-GGAACGCTTCACGAATTTG-3′) was used as an internal control for
normalization based on the study of Zhou et al (15). For RT-PCR, miRNAs was extracted
from 2×106 cells by using a mirVana miRNA isolation kit
(Applied Biosystems, Foster City, CA, USA) as per the
manufacturer's protocol. The cDNA was synthesized by using the
Prime Script RT-PCR kit (Takara Biotechnology Co., Ltd., Dalian,
China). The RT-PCR was performed using SYBR®
PrimeScript® miRNA RT-PCR kit (Takara Biotechnology Co.,
Ltd.) on FTC-3000™ Real-time PCR Cycler (Funglyn Biotech Corp.
Ltd., Toronto, ON, Canada). The parameters for RT-PCR were as
follows: 95°C for 2 min, followed by 40 cycles of 95°C for 30 sec
and 60°C for 45 sec.
Cell proliferation assay
Following miR-20a mimics transfection,
2×105 cells were seeded into 6-well plates. Cell
proliferation was examined at 0, 24, 48 and 72 h using the Cell
Counting Kit-8 (Dojindo Molecular Technologies, Inc., Kumamoto,
Japan), following the manufacturer's protocol. Optical density (OD)
was detected at 450 nm using a Benchmark microplate reader (Bio-Rad
Laboratories, Inc., Hercules, CA, USA). The rate of cell
proliferation was calculated using the following formula:
(ODtest-ODblank)/(ODcontrol-ODblank)
×100. All assays were performed in triplicates.
Cell cycle analysis
Following miR-20a mimic treatment for 48 h, cells
were fixed overnight in ice-cold ethanol (at 4°C). The cells were
washed with PBS and incubated with a 1 ml solution comprising 20
mg/ml propidium iodide (PI) and 10 U/ml RNaseA (cat. no. KGA214;
Nanjing KeyGen Biotech Co., Ltd., Nanjing, China) for 30 min at
room temperature. Cells were counted by Accuri C6
Fluorescence-activated cell sorting (BD Biosciences, Franklin
Lakes, NJ, USA). Progression of the cell cycle was assayed using
the ModFit LT 3.0 software (Verity Software House, Topsham, ME,
USA).
Assessment of apoptosis
Following miR-20a mimic treatment, cells were
collected at 48 h. Cell suspensions were prepared by centrifuging
cell suspensions at 1,000 × g for 5 min at room temperature. Cells
from pellets were re-suspended in PBS and centrifuged at 1,000 × g
for 5 min under room temperature, and the supernatant was removed.
Subsequently, cells were incubated with Annexin V-Fluorescein
isothiocyanate (FITC)/PI, from the Annexin V-FITC/PI
Double-staining Apoptosis Detection kit (cat. no. M3021-3;
Molecular Biology and Chemical, Shanghai, China) for 15 min away
from light, according to the manufacturer' protocol. Cells were
analyzed immediately on a BD Accuri C6 flow cytometer equipped with
BD Accuri C6 software (BD Biosciences; Becton-Dickinson and
Company, Franklin Lakes, NJ, USA). Experiments were performed three
times.
RT-qPCR
Transfected cells were further incubated with M-CSF
(25 ng/ml) and RANKL (30 ng/ml) for 3 days (2). To assess the changes of TRAP, NFATc1
and peroxisome proliferator-activated receptor γ (PPARγ) mRNA
expression levels, 2×106 cells were collected and
treated with TRIzol reagent (Invitrogen; Thermo Fisher Scientific,
Inc.) according to the manufacturer's protocol. Total RNA was used
for RT-qPCR. The cDNA was synthesized by the Prime Script RT-PCR
kit. SYBR-Green-based real time RT-PCR was performed using
SYBR® Premix Ex Taq™ II (Takara Biotechnology Co., Ltd)
on FTC-3000™ Real-time PCR Cycler. The parameters for RT-PCR were
as follows: 95°C for 3 min, followed by 40 cycles of 95°C for 30
sec and 60°C for 30 sec, and finally 72°C for 90 sec. Primers
specific for TRAP, NFATc1, PPARγ and GAPDH were synthesized as
previously described (16) and as
follows: TRAP forward, 5′-GGAGATCAGCTCCAAAGAGATG-3′ and reverse
primer, 5′-GGGCAGTCATGGGAGTTCAG-3′; NFATc1 forward,
5′-AGACCGTGTCCACCACCAGC-3′ and reverse primer,
5′-CAGGATTCCGGCACAGTCAAT-3′; PPARγ forward,
5′-TGGCCTCCTTGATGAATA-3′ and reverse primer,
5′-GGCTTGTAGCAGGTTGTC-3′; GAPDH forward,
5′-CATGAGAAGTATGACAACAGCCT-3′ and reverse primer,
5′-AGTCCTTCCACGATACCAAAGT-3′. GAPDH was used as an internal control
and for the normalization of TRAP, NFATc1 and PPARγ mRNA levels.
Relative mRNA levels were calculated using the 2−ΔΔCq
method (17).
Western blotting
Cells in the Blank and miR-transfected groups were
treated with M-CSF and RANKL for three days. Subsequently, the
cells (5×106) were lysed using M-PER Mammalian Protein
Extraction Reagent (Thermo Fisher Scientific, Inc.). Cell lysates
were centrifuged at 12,000 × g for 1 min and 30 µg total proteins,
the concentration determined by BCA kit (cat. no. BCA1;
Sigma-Aldrich; Merck KGaA) were separated by 8% SDS-PAGE, followed
by transfer of separated proteins to polyvinylidene difluoride
membranes. Membranes were blocked with Blocking Buffer (1X PBS;
0.1% Tween-20; 5% w/v bovine serum albumin (Sigma-Aldrich; Merck
KGaA) for 1 h at room temperature and incubated with antibodies
against TRAP (1:100; cat. no. sc-28204; Santa Cruz Biotechnology,
Inc., Dallas, TX, USA), NFATc1 (1:100; cat. no. sc-13033; Santa
Cruz Biotechnology, Inc.) or PPARγ (1:100; cat. no. sc-7273; Santa
Cruz Biotechnology, Inc.) at room temperature for 1.5 h.
Subsequently, the goat anti-mouse IgG secondary antibody (1:10,000;
cat. no. 115-035-003; Jackson ImmunoResearch Laboratories Inc.,
West Grove, PA, USA), goat anti-Rabbit IgG secondary antibody
(1:10,000; cat. no. 115-035-003; Jackson ImmunoResearch
Laboratories Inc.) and rabbit anti-Goat IgG secondary antibody
(1:10,000; cat. no. 33701ES60; Yeasen Corporation, Shanghai, China)
was added correspondingly for incubation at 4°C overnight. Protein
levels were normalized to those of GAPDH (1:200; cat. no. sc-47724;
Santa Cruz Biotechnology, Inc.). Expression levels of protein in
each sample were calculated using the Gel-Pro analyzer 3.0 software
(Media Cybernetics, Inc., Rockville, MD).
Statistical analysis
All experiments were performed in triplicate. Data
are expressed as mean ± standard deviation, and were analyzed using
SPSS 19.0 software (IBM Corp., Armonk, NY, USA). Student's t-test
was used to analyze the statistical significance between two
groups. Data from more >3 groups were analyzed by one-way or
two-way analysis of variance, followed by Bonferroni's post hoc
test. P<0.05 was considered to indicate a statistically
significant difference.
Results
miR-20a expression during THP-1 cell
osteoblast differentiation
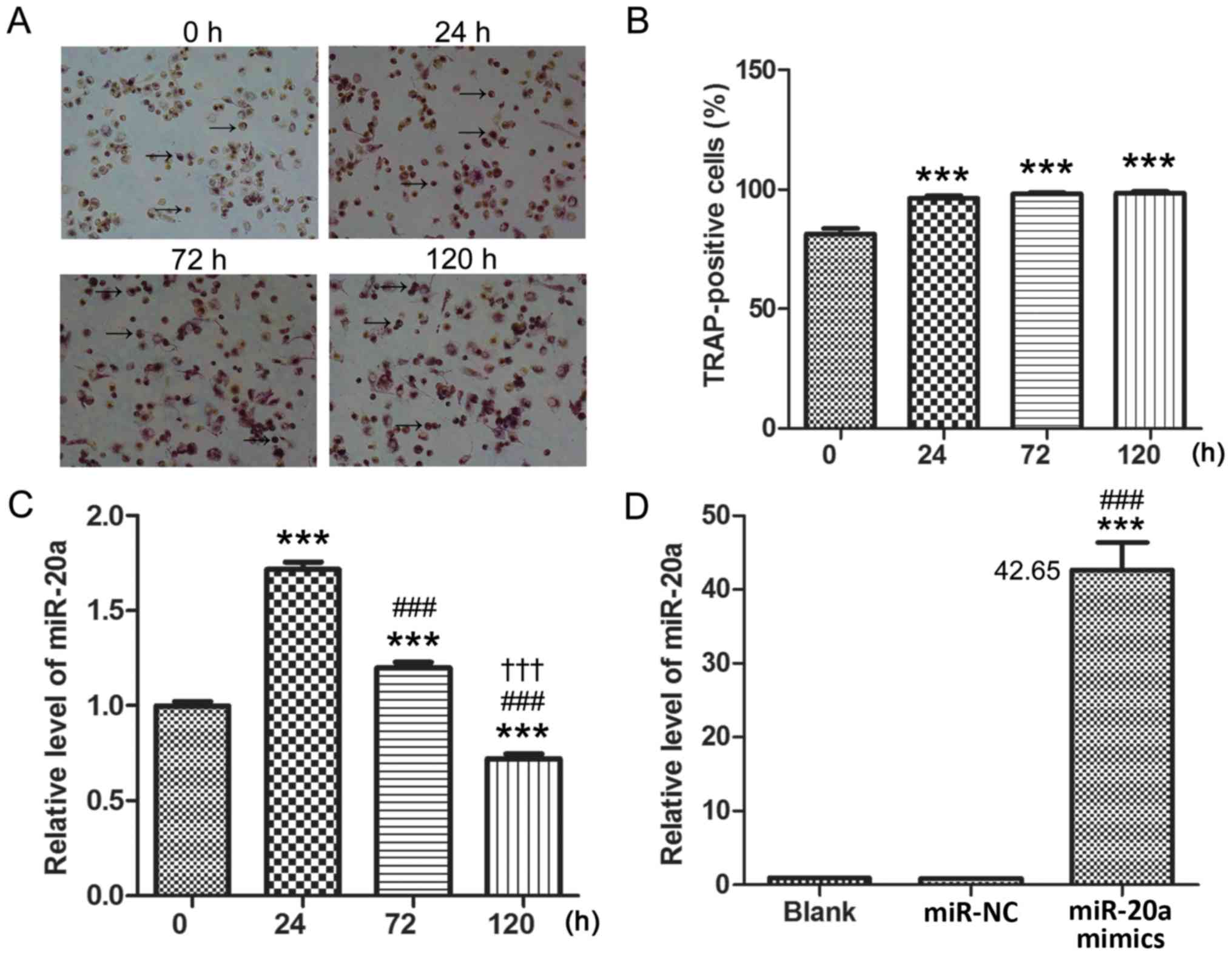
It was observed that the number of TRAP-positive
staining cells was significantly increased and some of the THP-1
cells were induced into osteoblasts (P<0.001; Fig. 1A and B). The TRAP-positive cells
included both macrophages (osteoclast precursors) and osteoclasts.
In addition, miR-20a expression levels were significantly increased
at 24 h (P<0.001) and 72 h (P<0.001), but significantly
decreased at 120 h (P<0.001) compared with expression levels at
0 h (Fig. 1C). In addition, there
was a time-dependent decrease in miR-20a expression levels from 24
to 120 h. These data demonstrated that miR-20a expression was
decreased post-differentiation induction. Following transfection
with miR-20a mimics into PMA treated THP-1 cells, miR-20a
expression levels were significantly increased in cells of by
~40-fold compared with the Blank and miR-NC-transfected groups
(P<0.001; Fig. 1D). Since the
result in Fig. 1D proved that
there was no significant difference between the Blank and miR-NC
group, therefore, only the Blank group was used in Figs. 2 and 3.
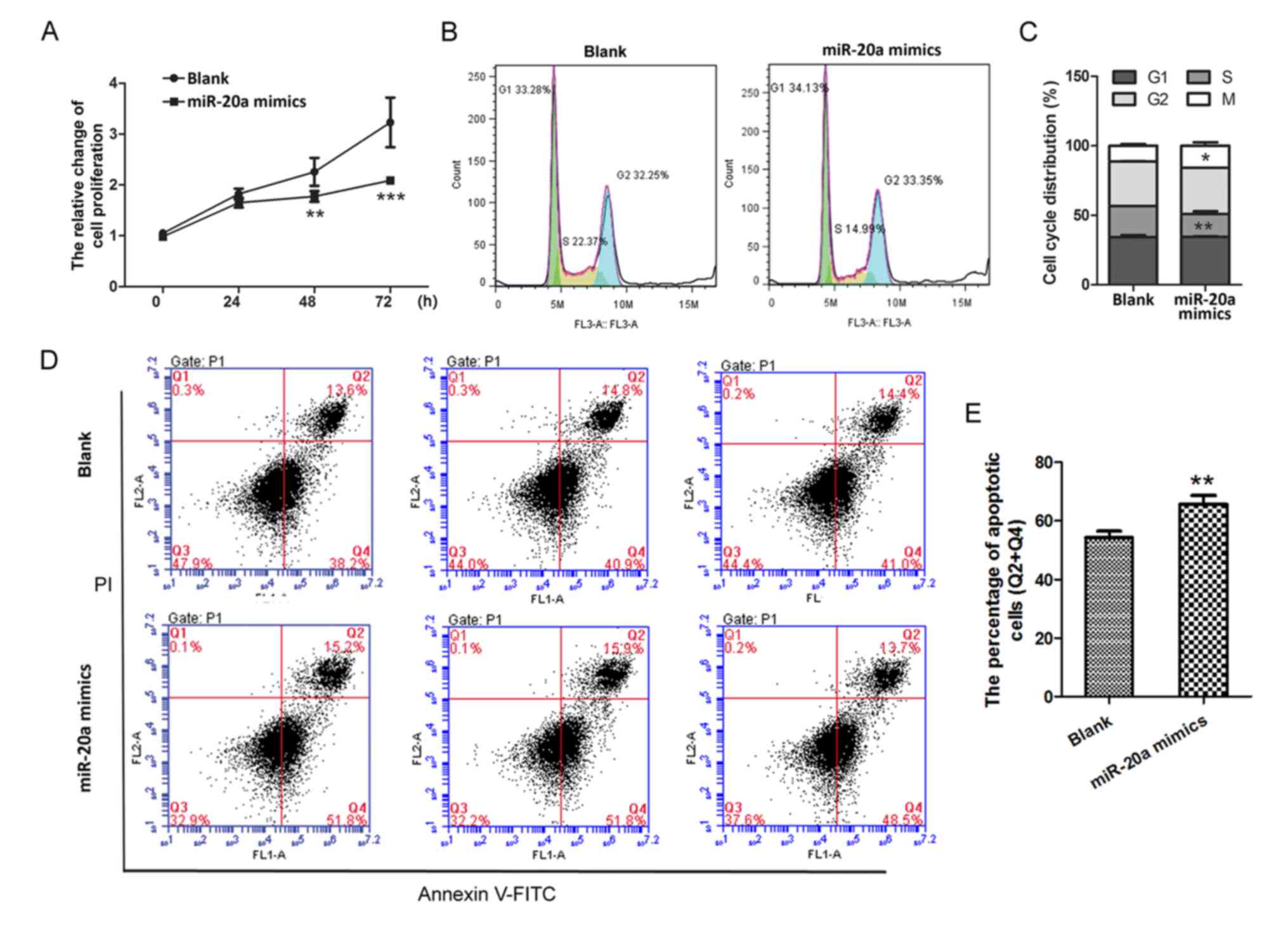
miR-20a inhibits the proliferation of
PMA-treated THP-1 cells
Results of cell proliferation assays demonstrated
that cell proliferation was markedly inhibited in the miR-20a
mimics group compared with proliferation in the Blank group at 48
and 72 h (P<0.01 and P<0.001, respectively; Fig. 2A). Furthermore, the percentage of
cells in the S phase of the cell cycle was significantly decreased
in the miR-20a mimics group compared with the Blank group
(P<0.01; Fig. 2B and C), which
indicated that cell division was inactive following miR-20a mimics
transfection. In addition, the total number of early and late
apoptotic cells was significantly higher in the miR-20a mimics
group (65.63±2.99%) compared with apoptotic cells in the Blank
group (54.3±2.17%; P<0.01, Fig. 2D
and E). These results demonstrated that miR-20a may inhibit the
growth of PMA-treated THP-1 cells.
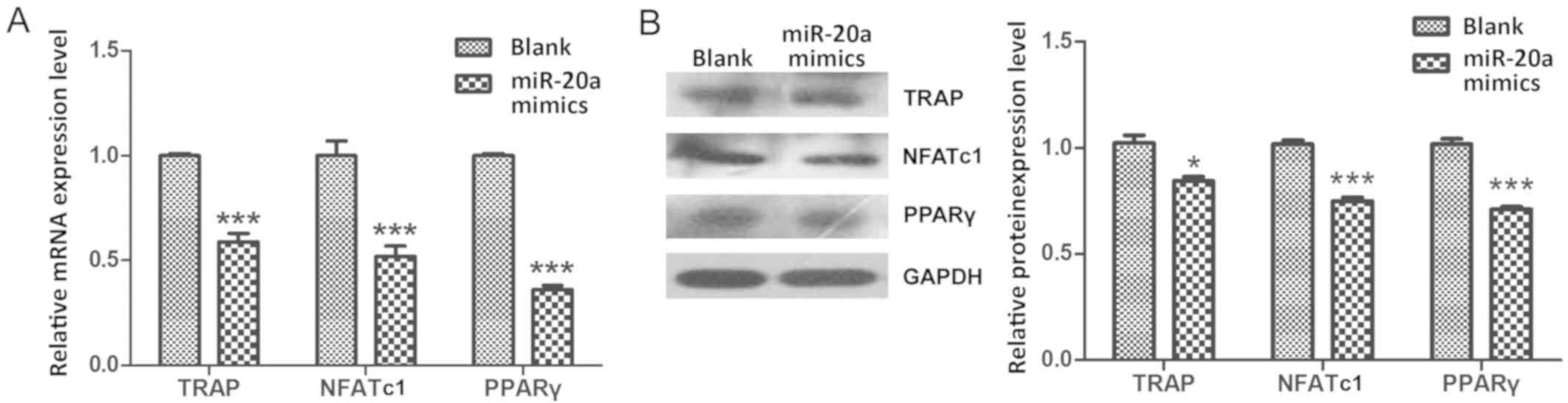
miR-20a negatively regulates
osteoclastogenesis by decreasing PPARγ during THP-1 cell
differentiation process
As the master regulators of osteoclastogenesis, the
mRNA expression levels of TRAP (18), NFATc1 (19) and PPARγ (20) were significantly decreased during
the THP-1 cell differentiation process in cells transfected with
miR-20a mimics compare with cells in the Blank group (P<0.001;
Fig. 3A). In addition, results of
western blot analysis confirmed that the protein levels of TRAP
(P<0.05), NFATc1 (P<0.001) and PPARγ (P<0.001) were also
significantly decreased in the miR-20a mimics group compare with
the Blank group (Fig. 3B). These
results indicated that miR-20a may regulate osteoclastogenesis by
decreasing PPARγ expression during THP-1 cell differentiation
process.
Discussion
Results from the present study revealed that the
expression levels of miR-20a in THP-1 cells decreased during PMA,
M-CSF and RANKL-induced osteoclastogenesis. Following miR-20a
mimics transfections, PMA-treated THP-1 cells exhibited weakened
cell growth ability. In addition, TRAP, NFATc1 and PPARγ levels
were significantly decreased following upregulated miR-20a
expression during the THP-1 cell differentiation progress. In
summary, increased miR-20a expression negatively regulated the
growth and osteoclastogenesis of THP-1 cells during its osteoclast
differentiation progress by downregulating PPARγ.
miRNAs not only positively regulate osteoblast
differentiation, but also negatively regulate osteoclastogenesis
(21–23). It has been reported that miR-20a
promotes osteoblast differentiation through the activated bone
morphogenetic protein (BMP)/Runx2 pathway in vitro by
directly binding to the 3′-UTR of BMP-2 (24,25).
However, another study demonstrated that miR-20a inhibited
dexamethasone-induced osteoclast differentiation through the
downregulation of RANKL expression (14), and miR-20a was reported to be
downregulated during osteoclastogenesis owing to its decreased
levels in mature osteoblasts (26). Therefore, the role of miR-20a
regulation in osteoclastogenesis is controversial. In the present
study, miR-20a was demonstrated to be downregulated in a time
dependent manner during PMA, M-CSF and RANKL-induced
osteoclastogenesis. Notably, miR-20a expression levels were
significantly increased at 24 and 72 h when compared to 0 h, but
significantly decreased at 120 h when compared to 0 h, which seemed
to exist contradict but was true results. It was hypothesized that
this may be associated with the differentiation state and degree of
monocytes, which need to be investigated in the authors' future
studies.
A previous study confirmed that miR-20a
overexpression reduced the levels of osteoclast differentiation
markers TRAP and NFATc1 in hypoxia-induced osteoclast
differentiation (13). In
addition, it was previously reported that miR-20a inhibited NFATc1
activity (27).
The nuclear transcription factor PPARγ is a subtype
of the PPAR family and is involved in cell differentiation, growth
and apoptosis (28). Human PPARγ
was reported to be a target gene of miR-20a and a component of the
BMP/Runx2 pathway, and is involved in osteoblast differentiation in
human mesenchymal stem cells (25). Additionally, mouse miR-20a
suppressed the differentiation of adipocyte progenitor cells by
inducing the expression of PPARγ in bone marrow stem cells
(29). During the differentiation
of bone marrow stem cells to osteoblasts, naringin promoted the
differentiation of osteoblasts through the overexpression of
miR-20a, which negatively regulated expression levels of PPARγ
(30). In the present study, the
upregulation of miR-20a negatively regulated osteoclast
differentiation of THP-1 cells by decreasing PPARγ. However,
whether miR-20a directly targeted PPARγ during THP-1 osteoblast
differentiation is still unclear and requires further
investigation.
A previous study demonstrated that the regulation of
miR-181a induces osteoclast apoptosis (31), and the overexpression of miR-20a
was reported to inhibit hepatocellular carcinoma cell proliferation
by regulating cell cycle and apoptosis (32). miR-20a downregulated
proliferation-associated target genes to modulate cell cycle
progression and to downregulate E2F transcription factors in order
to control the G1/S transition as well as WEE1 involved in the G2/M
transition (33). In the present
study, miR-20a overexpression inhibited cell proliferation, and
induced S phase cell cycle arrest and cell apoptosis of PMA treated
THP-1 cells.
In conclusion, miR-20a may negatively regulate the
growth and osteoclastogenesis of THP-1 cells during osteoclast
differentiation by downregulating PPARγ expression, which indicated
a role for miR-20a in osteoclast differentiation and also provided
a new direction for the investigation of pathological processes
involved in osteoclast differentiation.
Acknowledgements
Not applicable.
Funding
No funding was received.
Availability of data and materials
The datasets used/and or analyzed during the current
study are available from the corresponding author on reasonable
request.
Authors' contributions
HW conceived and designed the research, acquired the
data and drafted the manuscript. YS performed the statistical
analysis and revised the manuscript for important intellectual
contents.
Ethics approval and consent to
participate
Not applicable.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Cheng J, Liang H, Li Q, Peng C, Li Z, Shi
S, Yang L, Tian Z, Tian Y, Zhang Z and Cao W: Hematoporphyrin
monomethyl ether-mediated photodynamic effects on THP-1
cell-derived macrophages. J Photochem Photobiol B. 101:9–15. 2010.
View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Sengupta S, Park SH, Seok GE, Patel A,
Numata K, Lu CL and Kaplan DL: Quantifying osteogenic cell
degradation of silk biomaterials. Biomacromolecules. 11:3592–3599.
2010. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Geijtenbeek TB, Torensma R, van Vliet SJ,
van Duijnhoven GC, Adema GJ, van Kooyk Y and Figdor CG:
Identification of DC-SIGN, a novel dendritic cell-specific ICAM-3
receptor that supports primary immune responses. Cell. 100:575–585.
2000. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Jakob F, Siggelkow H, Homann D, Köhrle J,
Adamski J and Schütze N: Local estradiol metabolism in osteoblast-
and osteoclast-like cells. J Steroid Biochem Mol Biol. 61:167–174.
1997. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Hattersley G, Owens J, Flanagan AM and
Chambers TJ: Macrophage colony stimulating factor (M-CSF) is
essential for osteoclast formation in vitro. Biochem Biophys Res
Commun. 177:526–531. 1991. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Takahashi N, Udagawa N and Suda T: A new
member of tumor necrosis factor ligand family,
ODF/OPGL/TRANCE/RANKL, regulates osteoclast differentiation and
function. Biochem Biophys Res Commun. 256:449–455. 1999. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Takayanagi H, Kim S, Koga T, Nishina H,
Isshiki M, Yoshida H, Saiura A, Isobe M, Yokochi T, Inoue J, et al:
Induction and activation of the transcription factor NFATc1 (NFAT2)
integrate RANKL signaling in terminal differentiation of
osteoclasts. Dev Cell. 3:889–901. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Matsumoto M, Kogawa M, Wada S, Takayanagi
H, Tsujimoto M, Katayama S, Hisatake K and Nogi Y: Essential role
of p38 mitogen-activated protein kinase in cathepsin K gene
expression during osteoclastogenesis through association of NFATc1
and PU.1. J Biol Chem. 279:45969–45979. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Qu B, Xia X, Yan M, Gong K, Deng S, Huang
G, Ma Z and Pan X: miR-218 is involved in the negative regulation
of osteoclastogenesis and bone resorption by partial suppression of
p38MAPK-c-Fos-NFATc1 signaling: Potential role for osteopenic
diseases. Exp Cell Res. 338:89–96. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Esteller M: Non-coding RNAs in human
disease. Nat Rev Genet. 12:861–874. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Kapinas K and Delany AM: MicroRNA
biogenesis and regulation of bone remodeling. Arthritis Res Ther.
13:2202011. View
Article : Google Scholar : PubMed/NCBI
|
|
12
|
Fontana L, Pelosi E, Greco P, Racanicchi
S, Testa U, Liuzzi F, Croce CM, Brunetti E, Grignani F and Peschle
C: MicroRNAs 17-5p-20a-106a control monocytopoiesis through AML1
targeting and M-CSF receptor upregulation. Nat Cell Biol.
9:775–787. 2007. View
Article : Google Scholar : PubMed/NCBI
|
|
13
|
Sun KT, Chen MY, Tu MG, Wang IK, Chang SS
and Li CY: MicroRNA-20a regulates autophagy related protein-ATG16L1
in hypoxia-induced osteoclast differentiation. Bone. 73:145–153.
2015. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Shi C, Qi J, Huang P, Jiang M, Zhou Q,
Zhou H, Kang H, Qian N, Yang Q, Guo L and Deng L: MicroRNA-17/20a
inhibits glucocorticoid-induced osteoclast differentiation and
function through targeting RANKL expression in osteoblast cells.
Bone. 68:67–75. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Zhou BR, Guo XF, Zhang JA, Xu Y, Li W, Wu
D, Yin ZQ, Permatasari F and Luo D: Elevated miR-34c-5p mediates
dermal fibroblast senescence by ultraviolet irradiation. Int J Biol
Sci. 9:743–752. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Kim MH, Ryu SY, Choi JS, Min YK and Kim
SH: Saurolactam inhibits osteoclast differentiation and stimulates
apoptosis of mature osteoclasts. J Cell Physiol. 221:618–628. 2009.
View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(-Delta Delta C(T)) method. Methods. 25:402–408. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
18
|
M'Baya-Moutoula E, Louvet L, Metzinger-Le
Meuth V, Massy ZA and Metzinger L: High inorganic phosphate
concentration inhibits osteoclastogenesis by modulating miR-223.
Biochim Biophys Acta. 1852:2202–2212. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Yuan FL, Xu RS, Jiang DL, He XL, Su Q, Jin
C and Li X: Leonurine hydrochloride inhibits osteoclastogenesis and
prevents osteoporosis associated with estrogen deficiency by
inhibiting the NF-κB and PI3K/Akt signaling pathways. Bone.
75:128–137. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Wan Y, Chong LW and Evans RM: PPAR-gamma
regulates osteoclastogenesis in mice. Nat Med. 13:1496–1503. 2007.
View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Li H, Li T, Fan J, Li T, Fan L, Wang S,
Weng X, Han Q and Zhao RC: miR-216a rescues dexamethasone
suppression of osteogenesis, promotes osteoblast differentiation
and enhances bone formation, by regulating c-Cbl-mediated PI3K/AKT
pathway. Cell Death Differ. 22:1935–1945. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Guo DW, Han YX, Cong L, Liang D and Tu GJ:
Resveratrol prevents osteoporosis in ovariectomized rats by
regulating microRNA-338-3p. Mol Med Report. 12:2098–2106. 2015.
View Article : Google Scholar
|
|
23
|
Sun T, Leung F and Lu WW: miR-9-5p,
miR-675-5p and miR-138-5p damages the strontium and LRP5-mediated
skeletal cell proliferation, differentiation, and adhesion. Int J
Mol Sci. 17:2362016. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Tiago DM, Marques CL, Roberto VP, Cancela
ML and Laizé V: Mir-20a regulates in vitro mineralization and BMP
signaling pathway by targeting BMP-2 transcript in fish. Arch
Biochem Biophys. 543:23–30. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Zhang JF, Fu WM, He ML, Xie WD, Lv Q, Wan
G, Li G, Wang H, Lu G, Hu X, et al: MiRNA-20a promotes osteogenic
differentiation of human mesenchymal stem cells by co-regulating
BMP signaling. RNA Biol. 8:829–838. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Zhou M, Ma J, Chen S, Chen X and Yu X:
MicroRNA-17-92 cluster regulates osteoblast proliferation and
differentiation. Endocrine. 45:302–310. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Zhou G, Chen T and Raj JU: MicroRNAs in
pulmonary arterial hypertension. Am J Respir Cell Mol Biol.
52:139–151. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Krishnan A, Nair SA and Pillai MR: Biology
of PPAR gamma in cancer: A critical review on existing lacunae.
Curr Mol Med. 7:532–540. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Zhou J, Guo F, Wang G, Wang J, Zheng F,
Guan X, Chang A, Zhang X, Dai C, Li S, et al: miR-20a regulates
adipocyte differentiation by targeting lysine-specific demethylase
6b and transforming growth factor-β signaling. Int J Obes (Lond).
39:1282–1291. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Fan J, Li J and Fan Q: Naringin promotes
differentiation of bone marrow stem cells into osteoblasts by
upregulating the expression levels of microRNA-20a and
downregulating the expression levels of PPARγ. Mol Med Report.
12:4759–4765. 2015. View Article : Google Scholar
|
|
31
|
Shao B, Liao L, Yu Y, Shuai Y, Su X, Jing
H, Yang D and Jin Y: Estrogen preserves Fas ligand levels by
inhibiting microRNA-181a in bone marrow-derived mesenchymal stem
cells to maintain bone remodeling balance. FASEB J. 29:3935–3944.
2015. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Fan MQ, Huang CB, Gu Y, Xiao Y, Sheng JX
and Zhong L: Decrease expression of microRNA-20a promotes cancer
cell proliferation and predicts poor survival of hepatocellular
carcinoma. J Exp Clin Canc Res. 32:212013. View Article : Google Scholar
|
|
33
|
Trompeter HI, Abbad H, Iwaniuk KM, Hafner
M, Renwick N, Tuschl T, Schira J, Müller HW and Wernet P: MicroRNAs
MiR-17, MiR-20a, and MiR-106b act in concert to modulate E2F
activity on cell cycle arrest during neuronal lineage
differentiation of USSC. PLoS One. 6:e161382011. View Article : Google Scholar : PubMed/NCBI
|