Introduction
Atopic dermatitis (AD) is a chronic, relapsing
inflammatory skin disease that commonly occurs in children and
teenagers. It is characterized by recurrent severe itching and
eczema lesions in an atopic population (1). The morbidity of AD is increasing
annually, particularly in developed countries and regions of rapid
urbanization, and affects 15–30% of children and 2–10% of adults
worldwide (2). This trend has also
been observed in China; the AD morbidity rate of children aged 3–6
years was 8.3% in Shanghai in 2012 (3) and 12.94% in 1–7 year-olds in China in
2016 (4). The burden and
recurrence of AD greatly reduces patient quality of life and places
a significant economic burden on their households, creating
potentially severe social impacts. The exact cause of AD is
unclear, but it may result from a combination of genetic
predispositions and environmental factors. Currently, it has been
considered that a predominant T-helper (Th)2 immune response
arising from an imbalance of Th1/Th2 cells (the two major types of
CD4+ T helper cell) is a classical pathway of the disease (5).
MicroRNAs (miRNA/miR) are a type of non-coding small
RNA molecule that is 21–25 nucleotides in length. They are involved
the post-transcriptional regulation of endogenous gene expression
by binding to specific mRNA sequences via base pairing, resulting
in the subsequent recruitment of a silencing complex that degrades
target mRNA or prohibit its translation (6,7).
Such regulation can lead to abnormalities in protein synthesis
(8,9), which may be associated with various
human diseases (10,11). miR-146a is a known
anti-inflammatory and NF-κB pathway-dependent miRNA that is
expressed in a variety of immune cells, including B cells, T cells,
monocytes and dendritic cells (12–15).
It serves as a crucial regulatory factor of innate and adaptive
immunity, and controls immune cell differentiation, antibody
production and inflammatory factor secretion (16). A previous study revealed an
increased miR-146a expression in the keratin and lesions of skin in
patients with AD (17); however,
its expression status in patient serum remains unclear.
The aim of the present study was to determine the
expression of miR-146a in the serum of patients with AD and to
establish a 2,4-dinitrofluorobenzene (DNFB)-induced AD-like skin
lesion mouse model to investigate whether an imbalance in the
Th1/Th2 ratio of murine spleen cells is consistent with that
demonstrated in patients with AD. The present study also aimed to
establish the expression status of miR-146a in skin lesions in a
mouse model.
Materials and methods
Ethics statement
The use of human samples was approved by the Ethics
Committee of Guangdong Provincial Hospital of Chinese Medicine
(approval no. B2015-017-01). Written informed consent was obtained
from each participant prior to enrollment.
Patients
A total of 25 patients with moderate-to-severe AD
(19 males, 6 females; age range, 5–33 years; mean age 17.8 years;
disease span range, 1–22 years; average length of disease, 9.64
years) participated in the current study. All patients were
eligible for assessment using the Hanifin and Rajka criteria
(18) (5 years old or older with a
disease span >1 year), and Investigator Global Assessment
criteria (scores ≥3) (19). No
patients had received systematic hormone or immunosuppressor
treatment within 6 months, or partial treatment of hormones and
calcineurin inhibitors within 2 weeks prior to study commencement.
No patients exhibited any psychological, tumor, cardiovascular,
liver, kidney, brain or hematopoietic system disorders. The healthy
control group comprised 16 individuals (9 males, 7 females; age
range, 10–35 years; average age, 18.2 years). No individual in the
control group possessed the aforementioned diseases or AD, asthma,
allergic rhinitis or other allergic disorders.
Animals
Specific pathogen-free (SPF) C57BL/6 male mice (age,
6–8 weeks; weight, 16–22 g) were purchased from Guangdong Medical
Science Experiment Center (batch no. 44007200026387). Mice were
raised in an SPF grade animal room in the animal center of
Guangdong Traditional Chinese Medicine Academy of Sciences. Animal
care and experiments were conducted in accordance with the
Laboratory Animal Research Ethical Committee Guidelines of
Guangdong Provincial Hospital of Chinese Medicine, and was approved
by the Laboratory Animal Ethics Committee of Guangdong Provincial
Hospital of Chinese Medicine, approval no. 2015016). The room
temperature was maintained at 25±2°C with 50–60% relative humidity.
The circadian rhythm was set to 12 h, and food and water were
supplied freely. All experiments followed national regulations
(Guangdong Provincial Hospital of Chinese Medicine, Guangdong
Provincial Academy of Chinese Medical Sciences, Public Laboratory
Animal Experiment Center, Standard Operating Procedure PH-C032 to
PH-C036) on the usage, welfare and ethics of experimental
animals.
Reagents
Human peripheral blood lymphocyte separation liquid
(Tianjin Haoyang Biological), the serum miRNA extraction kit
(Qiagen GmbH), 2,4-dinitrofluorobenzene (DNFB, Sigma-Aldrich; Merck
KGaA), the human and mouse Th1/Th2 cytokine kit (BD Biosciences),
TRIzol® reagent (Thermo Fisher Scientific, Inc.),
reverse transcription kit (Thermo Fisher Scientific, Inc.),
real-time qPCR kit (Roche Diagnostics), miR-146a primer and miR-39
primer (Guangzhou Ruibo Biological Technology, Co., Ltd.) were used
in the following experiments. The 3′-untranslated region (UTR) of
small ubiquitin-related modifier 1 (SUMO1) luciferase vectors
(GV272/SUMO1 3′-UTR wild type (wt) and GV272/SUMO1 3′-UTR mutant
(mu)] was synthesized and cloned into GV272 (SV40-Luc-MCS, Fig. S1) firefly luciferase reporter
vector by Shanghai GeneChem Co., Ltd. The wt miR-146a
(GV268/miR-146a wt) and miRNA-NC (GV268 empty vector) were
constructed, packed and purified by Shanghai GeneChem Co., Ltd.
GV268 (CMV-MCS-SV40-Neomycin) plasmid information was presented in
Fig. S2. The sequence of
hsa-miR-146a was (5′→3′): CCUCUGAAAUUCAGUUCUUCAG.
AD-like model
A total of 12 C57BL/6 male mice were randomly and
evenly divided into the control and model groups. AD mice were
modelled based on a previously described method (20–22).
Mice were shaved with a razor and back hair was removed with
depilatory paste 24 h prior to model establishment. The size of the
shaved region was ~2×2 cm. On the first day of experimentation,
0.5% DNFB solution made with 50 µl acetone and olive oil
(acetone:olive oil, 4:1) was applied to the back skin of the model
group, once, on day 1. On the 5th day of experimentation, an
allergic reaction was induced on the right ear and back using 20
and 50 µl 0.2% DNFB solution, respectively. This procedure was
repeated every 3 days (on day 8, 11 and 14). The same quantity of
acetone-olive-oil solution was administered to the control group
without the induction of an allergic reaction (only acetone/olive
oil matrix).
Mouse ear thickness measurement
To investigate AD-like the change in ear thickness,
ear swelling was observed on the last day of the experiment (day
15). Mouse ear thickness was also measured and recorded using an
electronic digital caliper (Guilin Guanglu Measuring Instrument
Co., Ltd.).
Histological examination
The dorsal skin samples of mice were harvested and
fixed with 10% neutral-buffered formalin for 48 h at 4°C,
dehydrated and embedded in paraffin. Then, 4 µm sections were
stained with hematoxylin for 5 min and eosin for 1–2 min, then
dehydrated in 95% and absolute alcohols at room temperature. Tissue
sections were examined in three different areas (magnification,
×200) using an Olympus BX53 light microscope (Olympus
Corporation).
Western blotting
The protein samples derived from 293T cells were
detected by western blotting. Briefly, the sample was lysed in RIPA
buffer (Cell Signaling Technology, Inc.) with protease inhibitor
cocktail (Roche Applied Science) and centrifuged at 12,000 × g for
10 min at 4°C. The concentration of protein samples was detected
using the BCA protein assay kit (Thermo Fisher Scientific, Inc.).
Equal amounts of cellular protein (40 µg/lane) were separated by
10% tris-glycine sodium dodecyl sulfate-polyacrylamide gel
electrophoresis, and then transferred onto nitrocellulose filter
membranes. Following transfer, the membranes were blocked with
Tris-buffered saline (TBS) containing 5% non-fat milk, and 0.1%
tween (TBST) for 1 h at room temperature. The membranes were washed
three times for 5 min each with 10 ml of TBST at room temperature,
and sequentially incubated with various primary antibodies: GAPDH
(diluted in 5% BSA at 1:1,000, Cell Signaling Technology, Inc.,
catalog no. 2118), SUMO1 (diluted in 5% BSA at 1:1,000, Cell
Signaling Technology, Inc., catalog no. 4940) overnight at 4°C. The
washing was repeated 3 times, followed by incubation with
HRP-conjugated anti-rabbit secondary antibody (diluted in 5% BSA at
1:5,000, Cell Signaling Technology, Inc., catalog no. 7074) for 1 h
at room temperature. The bands were detected with ECL reagents
(Merck KGaA). All data are representatives of three independent
experiments. Bands were quantified with densitometry analysis using
Gel-Pro Analyzer version 4.0 software (Media Cybernetics,
Inc.).
Flow cytometry analysis of Th1
[CD4+interferon (IFN)-γ+] and Th2
[CD4+ interleukin (IL)-4+]
For blood sampling, ethylenediamine tetraacetic acid
anticoagulant tubes were used to collect left arm venous blood from
AD patients. Peripheral blood mononuclear cells (PBMCs) were
isolated via discontinuous density gradient centrifugation with
Lymphoprep™ (STEMCELL Technologies) for 20 min at 800 × g at room
temperature (20–25°C). PBMCs were placed in a 37°C water bath
immediately after removal from a liquid nitrogen tank and slowly
rotated until samples were fully thawed. According to the
manufacturer's protocols of the Th1/Th2 phenotyping kit (BD
Biosciences), PBMCs were examined on a cell sorter after cells were
resuspended in RPMI1640 medium (Thermo Fisher Scientific, Inc.)
supplemented with 10% fetal bovine serum (Thermo Fisher Scientific,
Inc.), then 1 ml of cold fixation buffer (provided in the kit) was
added and the cells incubated for 20 min at room temperature. The
cells were permeabilized in 1X Perm/Wash™ buffer (provided in the
kit) and incubated at room temperature for 15 min then sample
antibodies (20 µl) added and incubated for 30 min in the dark at
room temperature. The number of CD4+T cellular gates
were measured using a PerCP-Cy5.5-conjugated anti-CD4 antibody
(clone SK3, BD Pharmingen; BD Biosciences, catalog no. 560751),
with 100,000 CD4+T cells for each sample incubated at
room temperature for 30 min in the dark. In addition, samples were
stained with fluorescein isothiocyanate (FITC)-labeled anti-IFNγ
(clone B27, BD Pharmingen; BD Biosciences, catalog no. 560751),
allophycocyanin (APC)-anti-IL-4 (clone MP4-25D2, BD Pharmingen; BD
Biosciences, catalog no. 560751) and incubated at room temperature
for 30 min in the dark. The percentage of Th1
(CD4+IFN-γ+) and Th2
(CD4+IL-4+) cells was subsequently
calculated. Analysis was conducted with a flow cytometer and
FACSDiva 7.0 software (BD Biosciences).
Flow cytometry analysis of
splenocytes
Murine spleens were acquired under aseptic
conditions and ground in a Petri dish. Samples were then
centrifuged at 1,500 × g for 30 min at room temperature and the
supernatant discarded. Red blood cell lysis buffer (5 ml, BD
Biosciences, catalog no. 555899) was added to samples and the
mixture was incubated in the dark for 15 min at 37°C. The mixture
was subsequently centrifuged at 200 × g for 5 min at room
temperature a second time and the supernatant discarded. Similar to
flow cytometry analysis of PBMCs, spleen cells were examined for
expression of Th1 and Th2 according to the manufacturer's
instructions (BD Biosciences, catalog no. 560758). The number of
CD4+T cellular gates was measured using FITC-conjugated
anti-CD4 antibodies (clone RM4-5, provided in the kit), with
100,000 CD4+T cells for each sample. They were stained with
PerCP-Cy5.5-labeled anti-IFNγ (clone XMG1.2, provided in the kit)
and APC-anti-IL-4 (clone 11B11, provided in the kit) incubated at
room temperature for 30 min in the dark. The percentage of Th1
(CD4+IFN-γ+) and Th2
(CD4+IL-4+) cells was then calculated. Flow
cytometry data was analyzed with FACSDiva 7.0 software.
Reverse transcription quantitative
(RT-q)PCR analysis of miR-146a human serum expression
miR-146a extracted from serum of patients with AD
and healthy controls and total RNA was extracted using the QIAGEN
miRNeasy serum/plasma kit (Qiagen GmbH, catalog no. 217184) and the
First strand cDNA synthesis kit (Thermo Fisher Scientific, Inc.),
respectively. Serum (220 µl) was obtained after transferring the
sample from −80°C storage and thawing on ice. miRNA was then
extracted from the serum using aforementioned methods. Reagents
were added during the RT process according to the manufacturer's
protocols (Qiagen GmbH, catalog no. 217184); qPCR was performed
with an ABI Prism 7500 PCR system (Applied Biosystems; Thermo
Fisher Scientific, Inc.). The miR-146a and miR-39 primers were
synthesized by Guangzhou RiboBio Co., Ltd. and due to the company's
patents, specific primer sequences cannot be given. Samples were
amplified under the following conditions: 95°C for 10 min, followed
by 45 cycles of 95°C for 15 sec and 60°C for 1 min. To calculate
the expression of the target miR-146a (Bulge-Loop miRNA-146a
reverse transcription-primers, Guangzhou RiboBio Co., Ltd.)
relative to suitable reference gene (Bulge-Loop miRNA-39 reverse
transcription-primers, Guangzhou RiboBio Co., Ltd.). The expression
of miR-146a was calculated using the 2−∆∆Cq method
(23).
RT-qPCR analysis of miR-146a
expression in mouse skin
Back skin samples from mice were collected and
transferred to a refining tube. TRIzol (1 ml) was added and total
RNA was extracted. RNA concentrations were quantified using a UV
spectrophotometer and RT was performed with 1,000 ng RNA in
accordance with the manufacturer's protocols. qPCR was performed
based on the corresponding kit procedures (Guangzhou RiboBio Co.,
Ltd.) and the expression of miR-146a was calculated using the
2−∆∆Cq method (23).
The amplification program consisted of 1 cycle of 95°C for 10 min,
followed by 45 cycles of 95°C for 15 sec and 60°C for 1 min. The
expression of miR-146a (Bulge-Loop miRNA-146a reverse
transcription-primers, Guangzhou RiboBio Co., Ltd.) relative to the
expression of the miR-39 (Bulge-Loop miRNA-39 reverse
transcription-primers, Guangzhou RiboBio Co., Ltd.) used as a
reference gene.
miRNA sequencing and target
prediction
miRNA analysis was performed using the Illumina
HiSeq 4000 sequencing platform (Illumina, Inc.). TargetScan
(version 7.2, http://www.targetscan.org/vert_72/) (24) were utilized to predict the target
gene of miR-146a. In accordance with a recent study (25), it was predicted that SUMO1 was a
target gene of miR-146a (25).
Transfection and luciferase
assays
293T cells (Cell Bank of Type Culture Collection of
Chinese Academy of Sciences) were transfected using the
X-tremeGENE™ HP DNA transfection reagent (Roche Diagnostics) in
accordance with the manufacturer's protocols. Dual-luciferase
assays were performed with 0.1 µg 3′UTR luciferase plasmids, 0.4 µg
miRNA expression plasmids and 0.02 µg Renilla plasmids in a
24-well plate at a density of 1×105 cells/well.
Dual-luciferase assays were conducted at 48 h post-transfection
using the Dual-Glo® Luciferase assay system according to
the manufacturer's protocols (Promega Corporation). Luciferase
readings were corrected for background, and Firefly luciferase
values were used to normalized Renilla luciferase activity.
We constructed 3′UTR-NC (3′-UTR GV272 empty vector), 3′UTR-MU
(3′-UTR GV272/SUMO1 mu), 3′UTR (3′-UTR GV272/SUMO1 wt), as well as
miRNA-NC (GV268 empty vector) and miRNA (GV268/miR-146a wt)
plasmids. RT-qPCR analysis (as aforementioned) was also performed
on 293T cells transfected with GV268/miR-146a and GV268 empty
vector (negative control), which demonstrated significant
upregulation of miR-146a in the cells transfected with
GV268/miR-146a compared with the non-transfected control cells as a
negative control group (Fig.
S3).
Statistical analysis
Statistical analyses were performed using SPSS 17.0
software (SPSS, Inc.) and data are presented as the mean + standard
error of the mean. If group data were normally distributed, an
independent t-test was performed. Otherwise data were analyzed
using a Mann-Whitney U test. One-way analysis of variance was used
for comparing the data of three or more groups; a Dunnett's
post-hoc test was also conducted. P<0.05 was considered to
indicate a statistically significant difference.
Results
Clinical characteristics of healthy
controls and patients with AD
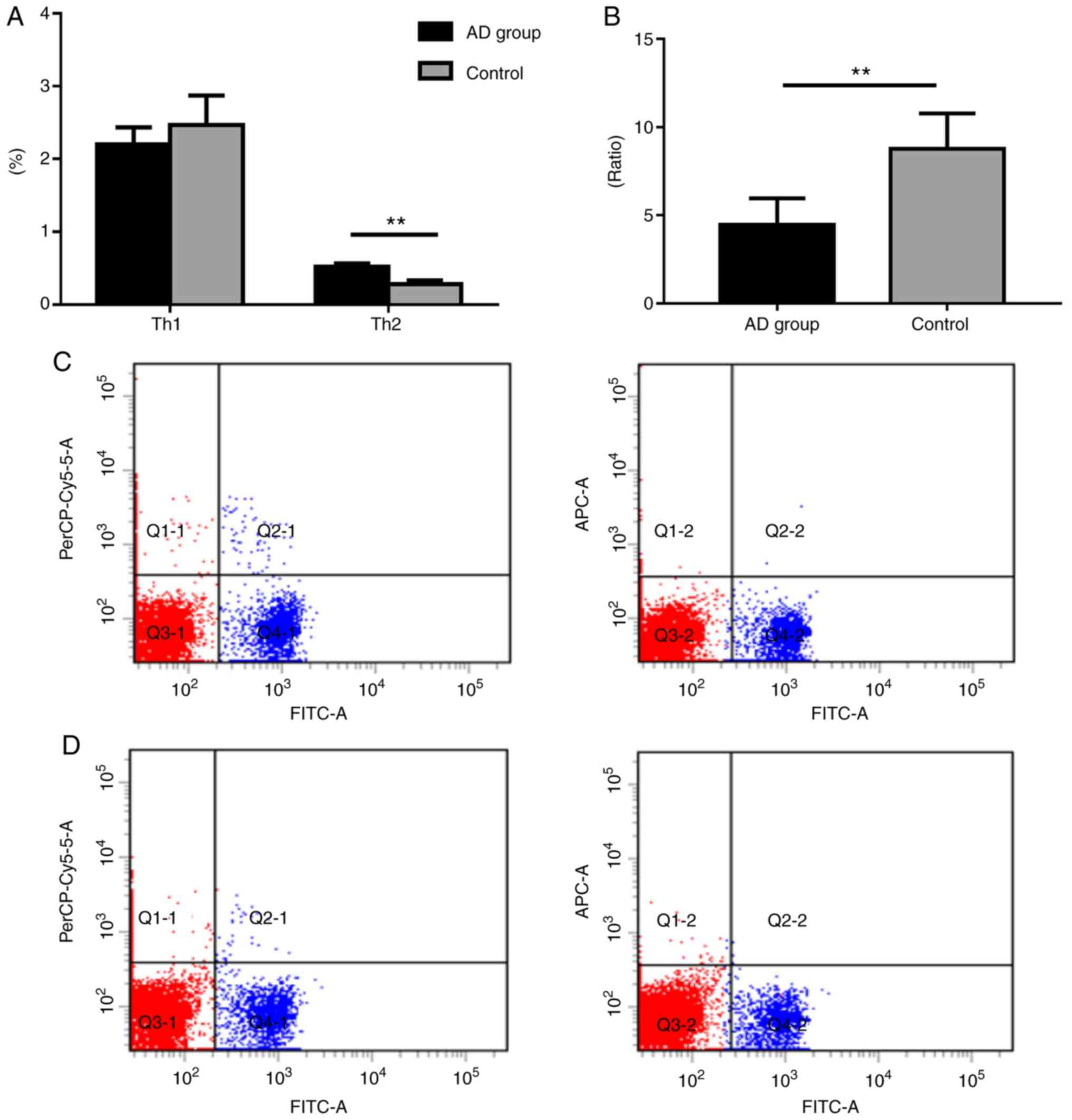
To assess the role of miR-146a in the serum of
patients with AD, Th1/Th2 ratio expression profiling was performed
in healthy individuals and AD patients. The results revealed that
the AD group exhibited significantly decreased Th1 levels compared
with the control group (P<0.01). Patients with AD also exhibited
increased Th2 levels, although the difference was not statistically
significant (P>0.05). Additionally, our results demonstrated
that the AD group exhibited a significantly lower Th1/Th2 ratio
compared with the control (P<0.05; Fig. 1).
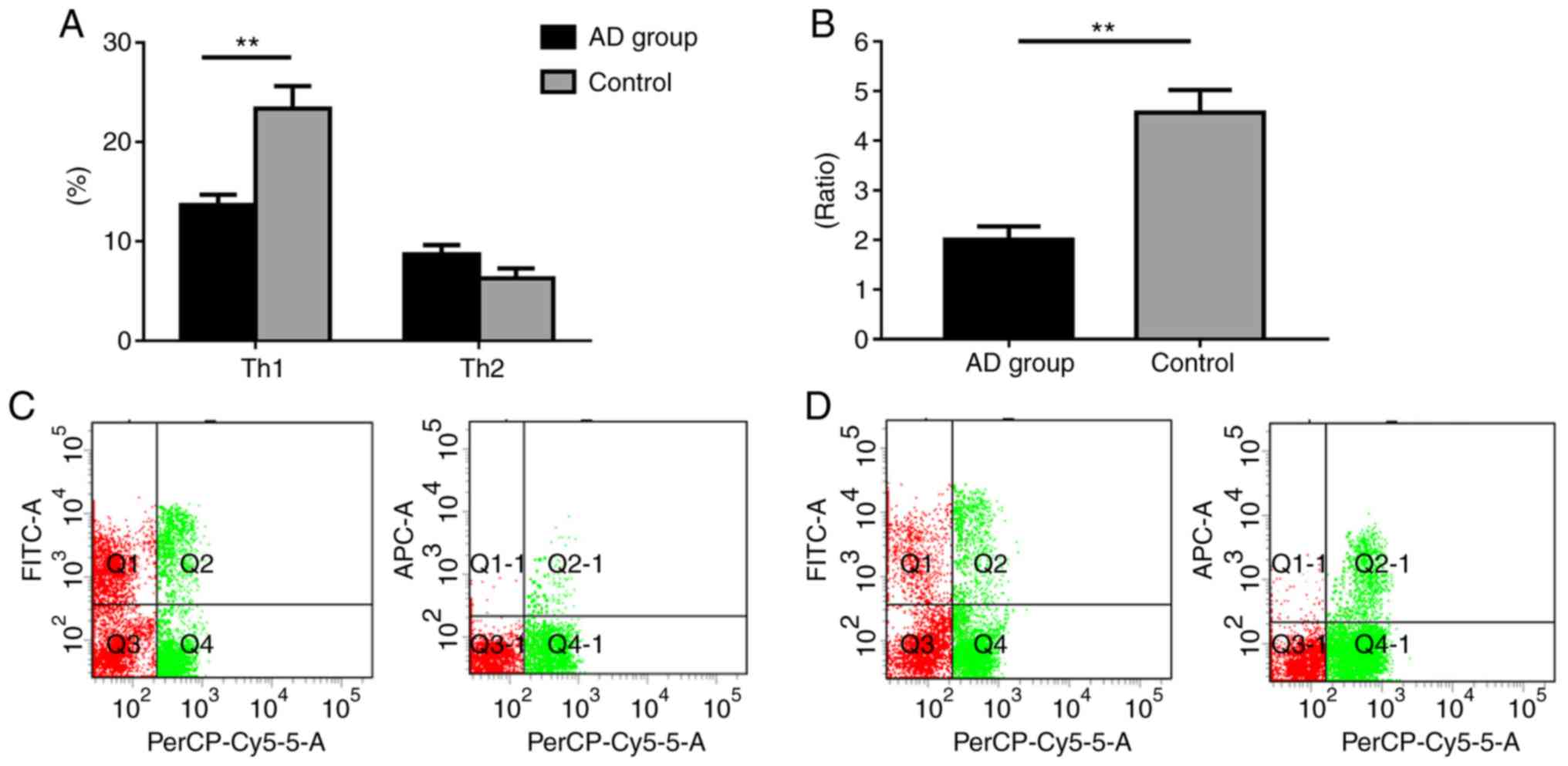 | Figure 1.Th1/Th2 ratio expression profiling of
patients with AD and healthy controls. (A) Percentage of Th1 and
Th2 cells. (B) Th1/Th2 ratio. (C) Control group flow cytometry
analysis. (D) Flow cytometry analysis of patients with AD.
PerCP-Cy5.5, FITC, and APC denote CD4+, Th1
(CD4+IFN-γ+), and Th2
(CD4+IL-4+), respectively. Data are presented
as the mean ± SEM (AD group, n=25; Control group, n=16).
**P<0.01. Th, T-helper; IFN, interferon; IL, interleukin; FITC,
fluorescein isothiocyanate; AD, atopic dermatitis. |
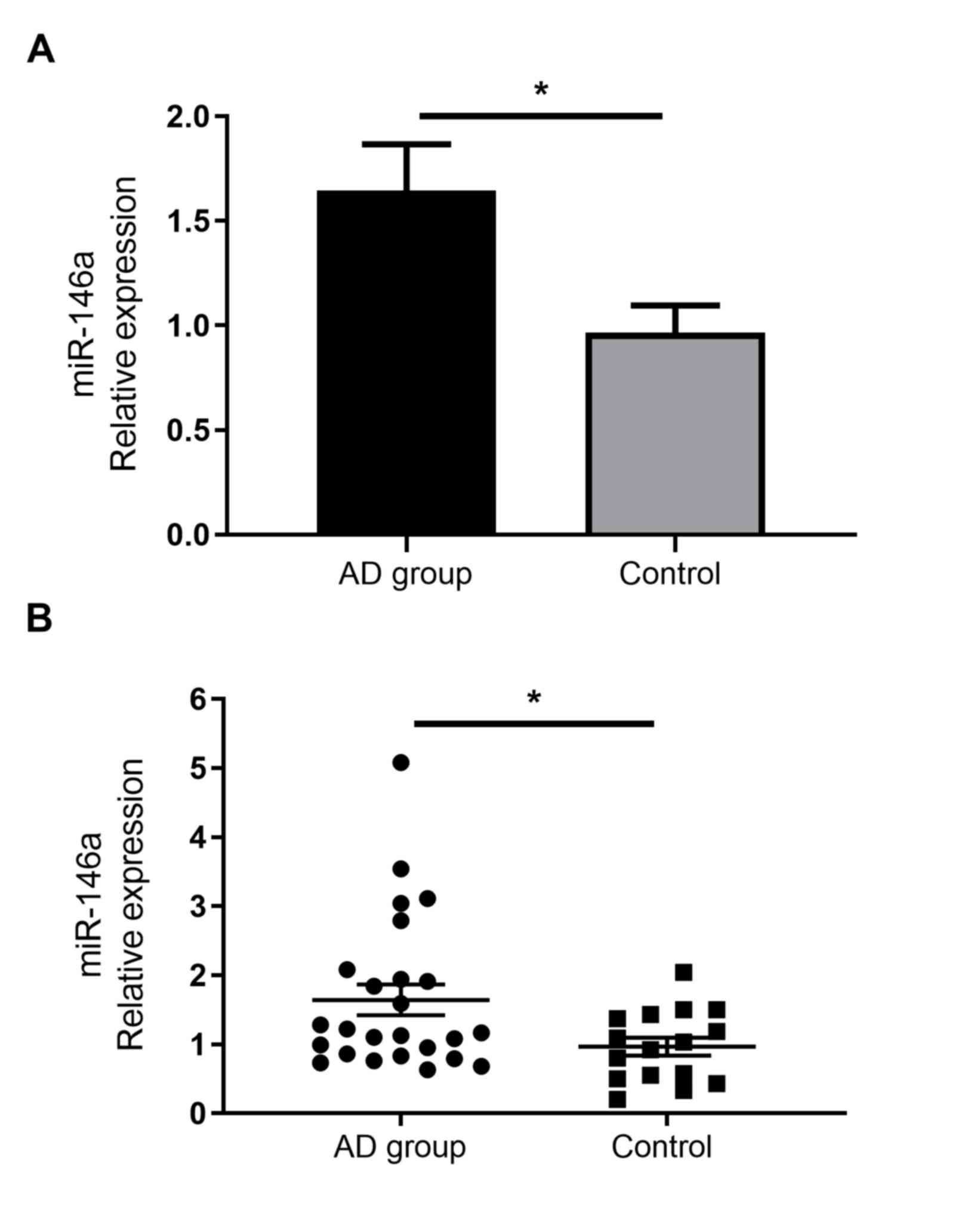
miR-146a is upregulated in the serum
of patients with AD
As miR-146a has been demonstrated to serve immune
regulatory functions, previous reports have confirmed its increased
expression in the non-lesional and chronic lesional skin of
patients with AD (14). However,
to the best of our knowledge, no studies have assessed the
expression of miR-146a in the serum of patients with AD. The
current study demonstrated that miR-146a expression was
significantly increased in the serum of patients with AD than in
healthy individuals (Fig. 2).
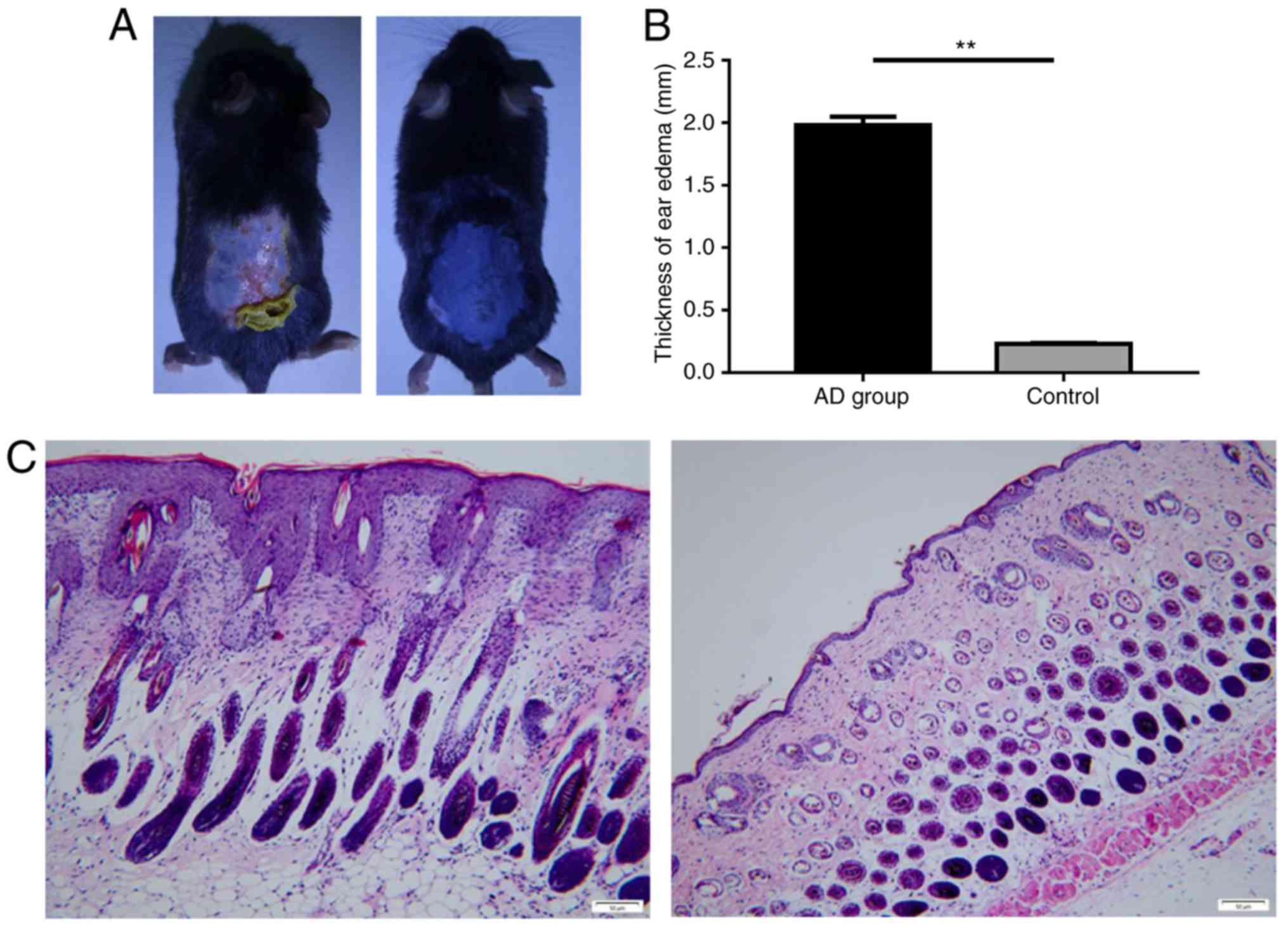
DNFB-induces AD-like skin inflammation
in C57BL/6 mice
To determine whether miR-146a expression serves a
role in AD animal models, AD-like skin inflammation was induced via
DNFB in a mouse model. The results revealed that the skin of the
control mice appeared to be normal, while the skin of the model
group was thicker, roughened and scarred. Cutaneous ulcers,
erythema and desquamation also developed (Fig. 3A). Furthermore, the degree of ear
swelling induced by DNBF in C57BL/6 mice was significantly higher
in the AD group than in the control (Fig. 3B). In addition, the skin tissue
pathology of the control group indicated no apparent abnormality,
yet the model group exhibited notable pathological changes,
including epidermal parakeratosis, hyperkeratosis, spongiosis,
acanthosis and excessive lymphocyte infiltration in the dermis
(Fig. 3C).
Shift of the Th1/Th2 ratio in murine
spleens
The Th1 (CD4+IFN-γ+) levels of
the model group were markedly decreased compared with the control
group; however, the count of Th2 (CD4+IL-4+)
cells in the model group was significantly higher when compared
with the control group (Fig. 4).
Furthermore, the ratio of Th1/Th2 was significantly lower in the
model group compared with control group (P<0.01; Fig. 4).
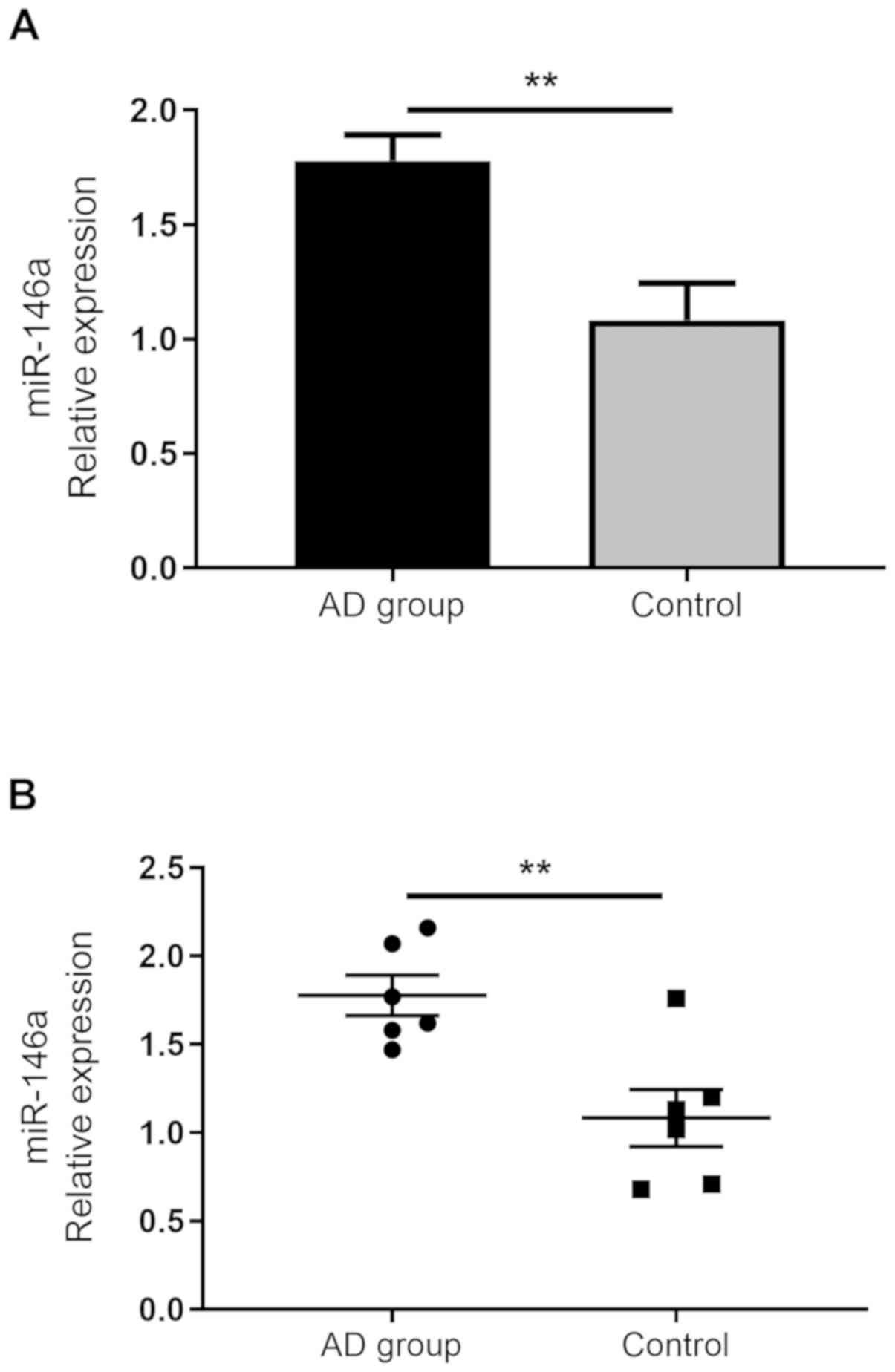
miR-146a expression in mouse skin
To investigate the expression of miR-146a during
skin inflammation, the expression of miR-146a was determined in
mouse skin using a DNFB-induced AD-like model. The results revealed
significantly upregulated miR-146a expression in the model group
compared with the control group (Fig.
5). This result was consistent with the expression of miR-146a
in the serum of the patients with AD.
miR-146a affects SUMO1 expression
Second-generation sequencing and a combined
literature analysis was performed in the current study. The results
revealed that the potential molecular target of miR-146a was SUMO1
(21). To functionally determine
whether miR-146a has a direct effect on SUMO1 activation, vectors
encoding a partial sequence of the 3′UTR of SUMO1 mRNA were
utilized, where the predicted miR-146a target sites were located
(Fig. 6A). The results revealed
that luciferase activity significantly decreased following
co-transfection with miR-146a and the vector carrying the 3′UTR of
SUMO1 compared with the negative control (Fig. 6B). Then, we evaluated the protein
expression levels of SUMO1 in 293T cells transfected with miR-146a
and negative control. As presented in Fig. 6C, the quantification of band
density of SUMO1 expression was not statistically different in each
group.
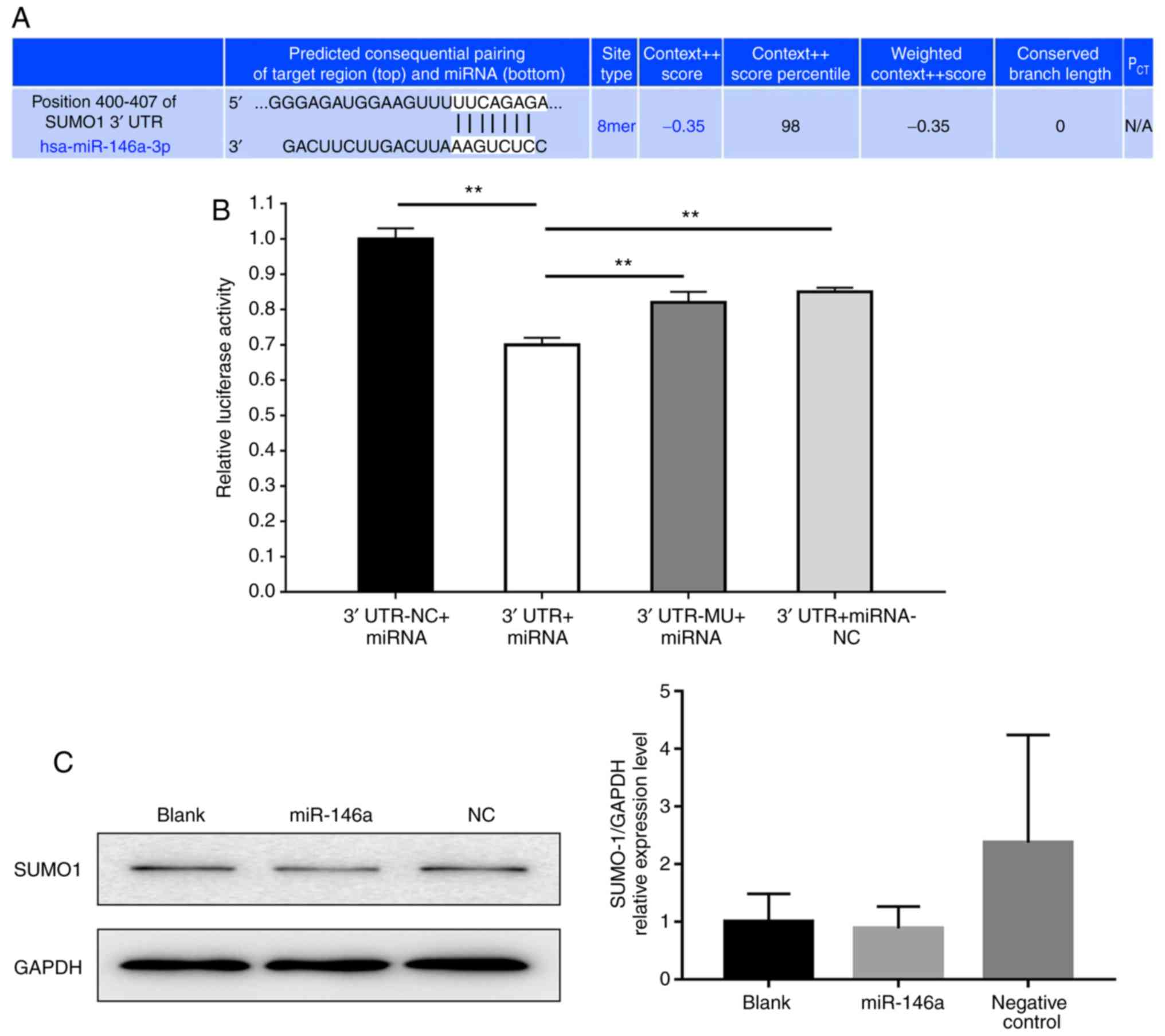 | Figure 6.miR-146a affects SUMO1 expression.
(A) Bioinformatic analysis indicated the prediction of the
complementary site of miR-146a in the SUMO1 3′-UTR (TargetScan).
(B) Firefly luciferase activity was used to normalize the
luciferase activity of Renilla following co-transfection,
and subsequently normalized for SUMO1 expression in the presence of
miR-146a. Values are presented as the relative luciferase activity
± the standard deviation of three independent experiments. For each
3′UTR construct, normalized luciferase activity in the absence of
miR-146a was set to 1. **P<0.01. (C) Western blot analysis the
protein expression of SUMO1 in 293T cells transfected with miR-146a
and negative control. Data are representative of three independent
experiments. 3′UTR-NC + miRNA, 3′-UTR GV272 empty vector +
GV268/miR-146a wt group; 3′UTR + miRNA, 3′-UTR GV272/SUMO1 wt +
GV268/miR-146a wt group; 3′UTR-MU + miRNA, 3′-UTR GV272/SUMO1 mu +
GV268/miR-146a wt group; 3′UTR + miRNA-NC, 3′-UTR GV272/SUMO1 wt +
GV268 empty vector group; miRNA/miR, microRNA; SUMO1, small
ubiquitin-related modifier 1; 3′-UTR, 3′-untranslated region; NC,
negative control; mu, mutant; wt, wild type. |
Discussion
AD is a common skin disease that causes pruritic and
chronically relapsing inflammation in children and adults (26,27).
miRNAs are relatively novel molecules that have been widely studied
in recent years to determine their exact function in the human body
(28). However, information
regarding the role of miRNAs in AD is limited. Recent studies have
described an altered expression of miRNAs in the skin or serum of
patients with AD (29,30). Psoriatic AD patients have been
characterized by the reduced expression of miR-122a, miR-133a-133b,
miR-133b, miR-215 and miR-326, whereas the downregulated expression
of miR-33, miR-483, miR-515-5p and miR-519d has occurred only in
patients with AD (31). In
addition, 10 upregulated and 34 downregulated miRNAs have been
identified in the affected skin of patients with AD (32). It has been indicated that miRNAs
control an early phase of inflammation by delaying the expression
of cytotoxic T-lymphocyte-associated protein 4 (33). Specifically, multiple
CD4+ helper cells, including Th1, Th2, Th9, Th17, Th22
and T regulatory (Treg) cells, as well as their associated
cytokines, participate in the onset of disease. These cells
interact with, control and affect each other, forming a complex
cellular and cytokine network that alters immunoregulation
(34). The classic pathway of AD
development states that the imbalance of the two major types of
CD4+ helper T cells, Th1 and Th2, causes a dominant
Th2-type immune response. The key feature is the bidirectional
polarization of T cells (35,36).
In the acute phase, IL-4-expressing Th2 cells dominate and in the
chronic phase, IFN-γ-expressing Th1 cells serve a major role
(37,38). The current study revealed a
decrease in Th1, an increase in Th2 and an imbalance in the Th1/Th2
ratio in the PBMCs of patients with AD, which is in agreement with
the classic pathway of AD development. In addition, flow cytometry
analysis of spleen cells in a mouse AD model demonstrated a
statistically significant decrease in Th1 cells, an increase in Th2
cells and a decrease in the Th1/Th2 ratio of mice in the model
group compared with the control group. This model is in accordance
with the classic pathway in human AD, in which an imbalance in
Th1/Th2 was reported to cause a dominant Th2-type immune response
(36). The pathological
characteristics and skin lesions of mice may therefore similar to
that of AD in humans. Hence, effective animal models could be used
for studying the mechanisms of AD and developing novel therapeutic
strategies.
A previous study demonstrated that miR-146a serves a
major role in certain physiological and pathological processes,
including the onset and development of immunity, tumors and
inflammation (39). The abnormal
expression of miR-146a is closely associated with systemic lupus
erythematosus, psoriasis, rheumatoid arthritis, tumors and asthma
(40–43). High expression of miR-146a in Treg
cells greatly contributes to the maintenance of normal cell
immunoregulatory functions (44).
Through regulating its target genes, including tumor necrosis
factor receptor associated 6, IL1 receptor associated kinase 1, IFN
regulatory factor 5, and signal transducer and activator of
transcription 1, miR-146a participates in the development of many
diseases (45,46). A recent study indicated that
miR-146a negatively regulates osteogenesis and bone regeneration
from adipose-derived mesenchymal stem cells (47). Furthermore, it has been suggested
that miR-146a alleviates chronic skin inflammation in AD by
controlling nuclear factor-κB-dependent signaling inflammatory
responses in keratinocytes (17).
Rebane et al (17) revealed
that certain molecules and cytokines are regulated by miR-146a, in
which miR-146a-deficient mice were used. It was demonstrated that
stronger inflammatory features were exhibited in these
miR-146a-deficient mice (17).
Compared with the aforementioned study, we reported a potential
target gene of miR-146a as SUMO1. Furthermore, miR-146a may be
considered as a potential regulator involved in the pathogenesis of
AD. It has also been revealed that miR-146a induced cardiac
dysfunction in maladaptive hypertrophy and suppressed SUMO1
expression in heart failure (25).
In the present study, increased expression of miR-146a was observed
in the serum of patients with AD and in the skin of model group
mice, indicating that miR-146a may participate in the development
of AD; however, further studies are required to reveal its
underlying mechanisms.
The current study analyzed the expression of
miR-146a in patients with AD and in an animal model of AD. The
limitations of the study include the few patients with AD that were
enrolled. A larger sample size may better reflect whether miR-146a
is a novel target for drug intervention. Our results demonstrated
that the C57BL/6 mouse AD model may reflect the classic process of
human AD (48), in which an
imbalance in Th1/Th2 causes a dominant Th2-type immune response.
The results of luciferase assay in the present study indicated that
the potential direct target of miR-146a may be SUMO1. Therefore, it
could be considered to be an appropriate model for AD studies.
Increased expression of miR-146a in the serum of patients with AD
and the skin of model group mice indicates that miR-146a may be
involved in the development of AD and that SUMO1 could be a
potential direct mRNA target of miR-146a. Further investigation
into its regulatory mechanisms is required, particularly in the
immune regulation of AD, such as T cell regulation and Th1/Th2
balance. In conclusion, miR-146a may be a novel target for drug
intervention in the treatment of patients with AD.
Supplementary Material
Supporting Data
Acknowledgements
Not applicable.
Funding
The current study was supported by the National
Natural Science Foundation of China (grant no. 81774307), the
Specific Research Fund for TCM Science and Technology of Guangdong
Provincial Hospital of Chinese Medicine (grant nos. YN2015MS06,
YN2015QN09 and YN2016QJ11), the Traditional Chinese Medicine Bureau
of Guangdong Province (grant no. 20183004) and the Department of
Science and Technology of Guangdong Province [grant nos. (2017)105
and 2017A030310122)].
Availability of data and materials
The datasets used and/or analyzed during the current
study are available from the corresponding author on reasonable
request.
Authors' contributions
DC and YL designed the experiments, supervised all
research and revised the manuscript. FY and WM conducted the
experiments and wrote the manuscript. SY, XZ, XM and JL conducted
the experiments and contributed to the interpretation of data. All
authors read and approved the final version.
Ethics approval and consent to
participate
The use of human samples was approved by the Ethics
Committee of Guangdong Provincial Hospital of Chinese Medicine
(approval no. B2015-017-01). Written informed consent was obtained
from each participant prior to enrollment. The care and use of
laboratory animals was performed in accordance to the Laboratory
Animal Research Ethical Committee Guidelines of Guangdong
Provincial Hospital of Chinese Medicine. The present study was
approved by the Laboratory Animal Ethics Committee of Guangdong
Provincial Hospital of Chinese Medicine, approval number:
2015016).
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Weidinger S, Beck LA, Bieber T, Kabashima
K and Irvine AD: Atopic dermatitis. Nat Rev Dis Primers. 4:12018.
View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Bieber T: Atopic dermatitis. N Engl J Med.
358:1483–1494. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Xu F, Yan S, Li F, Cai M, Chai W, Wu M, Fu
C, Zhao Z, Kan H, Kang K and Xu J: Prevalence of childhood atopic
dermatitis: An urban and rural community-based study in shanghai,
China. PLoS One. 7:e361742012. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Guo Y, Li P, Tang J, Han X, Zou X, Xu G,
Xu Z, Wei F, Liu Q, Wang M, et al: Prevalence of atopic dermatitis
in Chinese children aged 1–7 ys. Sci Rep. 6:297512016. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Wang AX and Xu Landén N: New insights into
T cells and their signature cytokines in atopic dermatitis. IUBMB
Life. 67:601–610. 2015. View
Article : Google Scholar : PubMed/NCBI
|
|
6
|
Li S, Liu L, Zhuang X, Yu Y, Liu X, Cui X,
Ji L, Pan Z, Cao X, Mo B, et al: MicroRNAs inhibit the translation
of target mRNAs on the endoplasmic reticulum in arabidopsis. Cell.
153:562–574. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Wahid F, Shehzad A, Khan T and Kim YY:
MicroRNAs: Synthesis, mechanism, function, and recent clinical
trials. Biochim Biophys Acta. 1803:1231–1243. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Lau PW and MacRae IJ: The molecular
machines that mediate microRNA maturation. J Cell Mol Med.
13:54–60. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Sonkoly E and Pivarcsi A: Advances in
microRNAs: Implications for immunity and inflammatory diseases. J
Cell Mol Med. 13:24–38. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Yoshizawa JM and Wong DT: Salivary
microRNAs and oral cancer detection. Methods Mol Biol. 936:313–324.
2013. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Tili E, Michaille JJ, Wernicke D, Alder H,
Costinean S, Volinia S and Croce CM: Mutator activity induced by
microRNA-155 (miR-155) links inflammation and cancer. Proc Natl
Acad Sci USA. 108:4908–4913. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
West C and McDermott MF: Effects of
microRNA-146a on the proliferation and apoptosis of human
osteochondrocytes by targeting TRAF6 through the NF-κB signalling
pathway. Biosci Rep. 37(pii): BSR201701802017. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Park H, Huang X, Lu C, Cairo MS and Zhou
X: MicroRNA-146a and microRNA-146b regulate human dendritic cell
apoptosis and cytokine production by targeting TRAF6 and IRAK1
proteins. J Biol Chem. 290:2831–2841. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Lindner JM, Kayo H, Hedlund S, Fukuda Y,
Fukao T and Nielsen PJ: Cutting Edge: The transcription factor Bob1
counteracts B cell activation and regulates miR-146a in B cells. J
Immunol. 192:4483–4486. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Wang S, Zhang X, Ju Y, Zhao B, Yan X, Hu
J, Shi L, Yang L, Ma Z, Chen L, et al: MicroRNA-146a feedback
suppresses T cell immune function by targeting Stat1 in patients
with chronic Hepatitis B. J Immunol. 191:293–301. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Williams AE, Perry MM, Moschos SA,
Larner-Svensson HM and Lindsay MA: Role of miRNA-146a in the
regulation of the innate immune response and cancer. Biochem Soc
Trans. 36:1211–1215. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Rebane A, Runnel T, Aab A, Maslovskaja J,
Rückert B, Zimmermann M, Plaas M, Kärner J, Treis A, Pihlap M, et
al: MicroRNA-146a alleviates chronic skin inflammation in atopic
dermatitis through suppression of innate immune responses in
keratinocytes. J Allergy Clin Immunol. 134:836–847.e11. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
18
|
M HJ and Georg R: Diagnostic features of
atopic dermatitis. Acta Dermatovener. 60:44–47. 1980.
|
|
19
|
Eichenfield LF, Lucky AW, Boguniewicz M,
Langley RG, Cherill R, Marshall K, Bush C and Graeber M: Safety and
efficacy of pimecrolimus (ASM 981) cream 1% in the treatment of
mild and moderate atopic dermatitis in children and adolescents. J
Am Acad Dermatol. 46:495–504. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Heo JC, Nam DY, Seo MS and Lee SH:
Alleviation of atopic dermatitis-related symptoms by Perilla
frutescens Britton. Int J Mol Med. 28:733–737. 2011.PubMed/NCBI
|
|
21
|
Heo JC, Son HU, Kim SL and Lee SH: A
derivative of L-allo threonine alleviates
2,4-dinitrofluorobenzene-induced atopic dermatitis indications.
Biosci Biotechnol Biochem. 76:2021–2025. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Nam DY, Lee JM, Heo JC and Lee SH:
Mitigation of 2,4-dinitrofluorobenzene-induced atopic
dermatitis-related symptoms by Terminalia chebula Retzius. Int J
Mol Med. 28:1013–1018. 2011.PubMed/NCBI
|
|
23
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(-Delta Delta C(T)) method. Methods. 25:402–408. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Agarwal V, Bell GW, Nam JW and Bartel DP:
Predicting effective microRNA target sites in mammalian mRNAs.
Elife. 42015.doi: 10.7554/eLife.05005.
|
|
25
|
Oh JG, Watanabe S, Lee A, Gorski PA, Lee
P, Jeong D, Liang L, Liang Y, Baccarini A, Sahoo S, et al: miR-146a
suppresses SUMO1 expression and induces cardiac dysfunction in
maladaptive hypertrophy. Circ Res. 123:673–685. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Deleanu D and Nedelea I: Biological
therapies for atopic dermatitis: An update. Exp Ther Med.
17:1061–1067. 2019.PubMed/NCBI
|
|
27
|
Thomsen SF: Atopic dermatitis: Natural
history, diagnosis, and treatment. ISRN Allergy. 2014:3542502014.
View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Rożalski M, Rudnicka L and Samochocki Z:
MiRNA in atopic dermatitis. Postepy Dermatol Alergol. 33:157–162.
2016. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Specjalski K and Jassem E: MicroRNAs:
Potential biomarkers and targets of therapy in allergic diseases?
Arch Immunol Ther Exp (Warsz). 67:213–223. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Bin L and Leung DY: Genetic and epigenetic
studies of atopic dermatitis. Allergy Asthma Clin Immunol.
12:522016. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Sonkoly E, Wei T, Janson PC, Sääf A,
Lundeberg L, Tengvall-Linder M, Norstedt G, Alenius H, Homey B,
Scheynius A, et al: MicroRNAs: Novel regulators involved in the
pathogenesis of psoriasis? PLoS One. 2:e6102007. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Bhardwaj N: MicroRNAs in atopic
dermatitis: A review. J Transl Genet Genom. 1:15–22. 2017.
View Article : Google Scholar
|
|
33
|
Sonkoly E, Janson P, Majuri ML, Savinko T,
Fyhrquist N, Eidsmo L, Xu N, Meisgen F, Wei T, Bradley M, et al:
MiR-155 is overexpressed in patients with atopic dermatitis and
modulates T-cell proliferative responses by targeting cytotoxic T
lymphocyte-associated antigen 4. J Allergy Clin Immunol.
126:581–589.e1-e20. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Auriemma M, Vianale G, Amerio P and Reale
M: Cytokines and T cells in atopic dermatitis. Eur Cytokine Netw.
24:37–44. 2013.PubMed/NCBI
|
|
35
|
Lipozencić J, Pastar Z, Kulisić SM and
Pavić I: Immunologic aspects of atopic dermatitis. Acta
Dermatovenerol Croat. 17:226–234. 2009.PubMed/NCBI
|
|
36
|
Agrawal R, Wisniewski JA and Woodfolk JA:
The role of regulatory T cells in atopic dermatitis. Curr Probl
Dermatol. 41:112–124. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Miraglia del Giudice M, Decimo F, Leonardi
S, Maioello N, Amelio R, Capasso A, Capristo C and Capristo AF:
Immune dysregulation in atopic dermatitis. Allergy Asthma Proc.
27:451–455. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Grewe M, Bruijnzeel-Koomen CA, Schöpf E,
Maioello N, Amelio R, Capasso A, Capristo C and Capristo AF: A role
for Th1 and Th2 cells in the immunopathogenesis of atopic
dermatitis. Immunol Today. 19:359–361. 1998. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Li L, Chen XP and Li YJ: MicroRNA-146a and
human disease. Scand J Immunol. 71:227–231. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Ammari M, Jorgensen C and Apparailly F:
Impact of microRNAs on the understanding and treatment of
rheumatoid arthritis. Curr Opin Rheumatol. 25:225–233. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Dai Y, Huang YS, Tang M, Lv TY, Hu CX, Tan
YH, Xu ZM and Yin YB: Microarray analysis of microRNA expression in
peripheral blood cells of systemic lupus erythematosus patients.
Lupus. 16:939–946. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Bartel DP: MicroRNAs: Genomics,
biogenesis, mechanism, and function. Cell. 116:281–297. 2004.
View Article : Google Scholar : PubMed/NCBI
|
|
43
|
Jiménez-Morales S, Gamboa-Becerra R, Baca
V, Del Río-Navarro BE, López-Ley DY, Velázquez-Cruz R,
Saldaña-Alvarez Y, Salas-Martínez G and Orozco L: MiR-146a
polymorphism is associated with asthma but not with systemic lupus
erythematosus and juvenile rheumatoid arthritis in Mexican
patients. Tissue Antigens. 80:317–321. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
44
|
Lu LF, Boldin MP, Chaudhry A, Lin LL,
Taganov KD, Hanada T, Yoshimura A, Baltimore D and Rudensky AY:
Function of miR-146a in controlling Treg cell-mediated regulation
of Th1 responses. Cell. 142:914–929. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
45
|
Tang Y, Luo X, Cui H, Ni X, Yuan M, Guo Y,
Huang X, Zhou H, de Vries N, Tak PP, et al: MicroRNA-146A
contributes to abnormal activation of the type I interferon pathway
in human lupus by targeting the key signaling proteins. Arthritis
Rheum. 60:1065–1075. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
46
|
Kamali K, Korjan ES, Eftekhar E,
Malekzadeh K and Soufi FG: The role of miR-146a on NF-κB expression
level in human umbilical vein endothelial cells under hyperglycemic
condition. Bratisl Lek Listy. 117:376–380. 2016.PubMed/NCBI
|
|
47
|
Xie Q, Wei W, Ruan J, Ding Y, Zhuang A, Bi
X, Sun H, Gu P, Wang Z and Fan X: Effects of miR-146a on the
osteogenesis of adipose-derived mesenchymal stem cells and bone
regeneration. Sci Rep. 7:428402017. View Article : Google Scholar : PubMed/NCBI
|
|
48
|
Lyons JJ and Milner JD: Primary atopic
disorders. J Exp Med. 215:1009–1022. 2018. View Article : Google Scholar : PubMed/NCBI
|