Introduction
Bone remodeling is finely regulated by the balance
between bone formation and bone resorption (1). In adult organisms, bone formation is
mediated by the recruitment of bone marrow mesenchymal stem cells
(BMSCs), which can differentiate into osteoblast cells (2,3). A
previous study identified that inhibition of osteogenic
differentiation of BMSCs leads to impaired bone formation and
contributes to osteoporosis (4).
Therefore, to treat bone loss, it is necessary to understand the
regulatory mechanisms underlying osteogenic differentiation of
BMSCs in order to identify novel potential therapeutic targets.
MicroRNAs (miRNAs) are single-stranded small RNA
molecules of ~22 nucleotides in length that regulate translation of
target mRNAs by binding to their 3′untranslated region (UTR) and
mediating translational inhibition or degradation (5). Previous studies have demonstrated
that miRNAs have essential roles in a variety of biological
processes, including cellular proliferation, apoptosis,
differentiation and immunity (6–8).
Numerous previous studies identified various miRNAs able to
regulate osteogenic differentiation by targeting important
osteogenic factors, such as histone deacetylase 9 (HDAC9), RPTOR
independent companion of MTOR complex 2, HDAC5 and runt-related
transcription factor 2 (RUNX2) (4,9,10,11).
However, the roles and the number of miRNAs involved in osteogenic
differentiation remain unclear.
In the present study, miR-483-3p was identified to
be significantly upregulated during osteogenic differentiation of
BMSCs. Overexpression of miR-438-3p using agomiR-438-3p markedly
promoted osteogenic differentiation in vitro, whereas
inhibition of miR-438-3p using antagomiR-438-3p reversed these
effects. Moreover, BMSC-targeted aptamer-agomiR-483-3p enhanced
bone formation in vivo. Collectively, the present study
identified a novel role for miR-483-3p and its underlying mechanism
in promoting osteogenic differentiation of BMSCs.
Materials and methods
Animals and reagents
In total, 20 female C57BL/6J mice of 12 months of
age and weighing 35–40 g were purchased from Hunan Slaccas Jingda
Laboratory Animal Co., Ltd. All mice were maintained in the
specific pathogen-free facility of The Laboratory Animal Research
Center at Central South University. Animals were housed under a
controlled temperature (25±1°C) and humidity (50%), with a 12-h
light/dark cycle and free access to food and water. All animal
experimental procedures were approved by The Animal Care and Use
Committee of The Xiangya Hospital of Central South University.
AgomiR-negative control (NC), agomiR-438-3p, antagomiR-NC and
antagomiR-483-3P were purchased from Shanghai GenePharma Co., Ltd.
The BMSC-specific aptamer
(5′-GAATTCAGTCGGACAGCGACGACGGTGATATGTCAAGGTCGTATGCACGAGTCAGAGGGATGGACGAATATCGTCTCCC-3′)
was synthesized by GenScript Corporation.
Cell transfection
BMSCs were seeded into 6-well plates
(1×106 cells/well) and cultured at 37°C for 24 h. After
24 h, BMSCs were transfected with 100 nM agomiR-NC
(5′-UUUGUACUACACAAAAGUACUG-3′), agomiR-438-3p
(5′-UCACUCCUCCCCUCCCGUCUU-3′), antagomiR-NC
(5′-CAGUACUUUUGUGUAGUACAAA-3′) or antagomiR-483-3P
(5′-AAGACGGGAGGGGAGGAGUGA-3′) using Lipofectamine® 2000
(Invitrogen; Thermo Fisher Scientific, Inc.) 48 h prior to further
experimentation, according to the manufacturer's protocol.
BMSC culture and osteogenic
differentiation
Mouse BMSCs were isolated from bone marrow of femurs
and tibia of C57BL/6J mice (age, 6–8 weeks) and cultured in α-MEM
medium containing 10% FBS (Gibco; Thermo Fisher Scientific, Inc.),
1% penicillin and streptomycin (Gibco; Thermo Fisher Scientific,
Inc.) at 37°C with 5% CO2, as previously described
(4). Human MSCs (hMSCs) were
isolated from the bone marrow of 30 healthy donors (female; age,
25.0±4.0 years) and the procedure was approved by The Ethics
Committee of The Xiangya Hospital of Central South University. All
patients provided written informed consent. Osteogenesis induction
medium containing 10% FBS, 300 ng/ml bone morphogenetic protein 2
(Sigma-Aldrich; Merck KGaA), 50 µg/ml ascorbic acid and 5 mM
β-glycerol phosphate (Sigma-Aldrich; Merck KGaA) was used to induce
the osteogenic differentiation of BMSCs and hMSCs for 0, 3, 7, 14
and 21 days.
miRNA microarray assay and miRNA
targets prediction
Total RNA was extracted from 3×106 BMSCs
using TRIzol®reagent (Invitrogen; Thermo Fisher
Scientific, Inc.) after treatment with osteogenic medium for 7
days. Total RNA was quantified using the NanoDrop ND-2000 (Thermo
Fisher Scientific, Inc.) and the RNA integrity was assessed using
the Agilent Bioanalyzer 2100 (Agilent Technologies, Inc.). Then,
total RNA was dephosphorylated, denatured and labeled with
Cyanine-3-CTP using the miRNA Complete Labeling and Hub kit
(Agilent Technologies, Inc.), according to the manufacturer's
protocol. The labeled RNAs were hybridized onto a mouse miRNA 8×15K
chip (Agilent Technologies, Inc.). After washing, all slides were
scanned with the Agilent Scanner G2505C (Agilent Technologies,
Inc.). Feature Extraction software (version 10.7.1.1; Agilent
Technologies, Inc.) and GeneSpring software (version 13.1; Agilent
Technologies, Inc.) were used to analyze the microarray results.
Differentially expressed miRNAs were then identified based on the
fold change and P<0.05, calculated using Student's t-test, as
previously described (4). In
total, 4 experimental groups (control group and osteogenic
differentiation group) were analyzed, n=5/group. The target genes
of miRNAs were predicted using TargetScan Human version 7.2
(http://www.targetscan.org) and miRanda
(http://www.microrna.org/microrna/home.do).
Alkaline phosphatase (ALP) activity,
osteocalcin (OCN) secretion assay and Alizarin Red S Staining
ALP activity was determined using an enzymatic
colorimetric ALP kit (Roche Diagnostics), according to the
manufacturer's protocol. Secreted levels of OCN were measured using
a specific OCN immunoassay kit (cat. no. 310950; DiaSorin SpA),
according to the manufacturer's protocol. Following culture with
osteogenesis induction medium for 21 days, BMSCs were fixed with 4%
paraformaldehyde at 4°C for 10 min and then stained with 2%
Alizarin Red S (Sigma-Aldrich; Merck KGaA) at room temperature for
5 min to evaluate cell matrix mineralization. Staining results were
visualized using a Diaphot Inverted Microscope and Camera System
(magnification, ×100; Leica Microsystems GmbH), as previously
described (12). n=5 in each
group.
RNA isolation and reverse
transcription quantitative (RT-q) PCR
Total RNA was extracted from 3×106 BMSCs
using TRIzol reagent (Invitrogen; Thermo Fisher Scientific, Inc.).
The mRNAs were reverse-transcribed into cDNA using PrimeScript RT
reagent kit with gDNA Eraser (Takara Bio, Inc) by incubating at
37°C for 30 min. The qPCR was performed using SYBR®
Premix Ex Taq™ (Takara Bio, Inc.) with an ABI 7900 thermocycler
(Thermo Fisher Scientific, MA, USA) using 40 cycles of 95°C for 5
sec and 60°C for 30 sec. Nucleotide sequences of primers for STAT1,
RUNX2, osterix, Type 1 collagen, ALP, OCN and β-actin are listed in
Table I. miRNAs were
reverse-transcribed at 37°C for 60 min using One Step
PrimeScript® miRNA cDNA Synthesis kit (Takara Bio,
Inc.). Primers for HsnRNA U6 (cat. no. HmiRQP9001), MsnRNA U6(cat.
no. MmiRQP9002), hsa-miR-483 (cat. no. HmiRQP0517), mmu-miR-483
(cat. no. MmiRQP1052) were purchased from GeneCopoeia, Inc. The
qPCR was performed using SYBR® Premix Ex Taq™ (Takara
Bio, Inc.) with an ABI 7900 thermocycler (Thermo Fisher Scientific,
Inc) using the following thermocycling conditions: 40 cycles of
95°C for 5 sec and 60°C for 30 sec. The expression levels of the
target genes were calculated using the 2−∆∆Cq method
(13). β-actin and U6 were used as
an internal control for normalizing the expression of mRNA and
miRNA, respectively.
 | Table I.Nucleotide sequences of primers used
for quantitative PCR. |
Table I.
Nucleotide sequences of primers used
for quantitative PCR.
| Gene | Genbank accession
no. | Primer sequence
(5′-3′) |
|---|
| RUNX2 | NM_001146038 | F:
AGAGTCAGATTACAGATCCCAGG |
|
|
| R:
TGGCTCTTCTTACTGAGAGAGG |
| Osterix | NM_130458.4 | F:
ACCAGGTCCAGGCAACAC |
|
|
| R:
GCAAAGTCAGATGGGTAAGTAG |
| STAT1 | XM_017319463.1 | F:
TCACAGTGGTTCGAGCTTCAG |
|
|
|
R:GCAAACGAGACATCATAGGCA |
| Type 1
collagen | NM_007742 | F:
GCTCCTCTTAGGGGCCACT |
|
|
| R:
CCACGTCTCACCATTGGGG |
| ALP | XM_017319924 |
F:CCCCATGTGATGGCGTAT |
|
|
|
R:CGGTAGGGAGAGCACAGC |
| OCN | NM_001032298 |
F:AAGCAGGAGGGCAATAAGGT |
|
|
|
R:ATGCGTTTGTAGGCGGTCTT |
| β-actin | NM_007393.5 | F:
GGCTGTATTCCCCTCCATCG |
|
|
| R:
CCAGTTGGTAACAATGCCATGT |
Plasmid construction and luciferase
reporter assay
The 3′-UTR of STAT1, containing the predicted
miR-483-3p binding sites, was amplified by PCR using the Phanta
Super-Fidelity DNA Polymerase (Vazyme) from complementary DNA,
using the STAT1 forward primer, 5′-GGGGTACCAGCCATAAACTTGGAGAA-3′
and STAT1 reverse primer, 5′-CCCAAGCTTTCTTGCCCTGACTTGGAT-3′. The
PCR thermocycling conditions were as follows: 95°C for 2 min
followed by 30 cycles of 95°C for 30 sec, 55°C for 30 sec and 72°C
for 60 sec, and a final extension at 72°C for 10 min. The PCR
products were digested using KpnI and HindIII, and
inserted into apGL3-basic vector (Promega Corporation) to generate
the reporter vector wild-type-pGL3-STAT1 3′-UTR. A vector
containing a mutant version of the STAT1 3′-UTR was constructed
using STAT1-mutant forward primer,
5′-GGGGTACCCTTCCCATGTCTCCAGTTAAGTT-3′ and STAT1 reverse primer,
5′-CCCAAGCTTTCTTGCCCTGACTTGGAT-3′. Reporter plasmid containing the
promoter of OCN was constructed as previously described (14). The RUNX2 or STAT1 coding sequence
was cloned into the pCMV-Myc vector (Sino Biological, Inc.) using
EcoRI and KpnI restriction enzymes, generatingRUNX2
or STAT1 overexpression plasmids. BMSCs cells were transfected with
2 µg of the RUNX2 plasmid or the STAT1 plasmids along with 100 nM
of agomiR-483-3P or antagomiR-483-3 Pusing
Lipofectamine® 2000. Relative luciferase activities were
determined by a dual-luciferase reporter assay system (Promega
Corporation). Luciferase activity was determined 48 h after
transfection. Renilla activity was used to normalize Firefly
luciferase activity.
Western blot analysis
BMSCs were collected and lysed in RIPA buffer
(Beyotime Institute of Biotechnology) with protease inhibitors
(Roche Diagnostics) for 30 min on ice. Cells were then sonicated at
a power level of 55% on a microtip-equipped Sonic Dismembrator for
5 sec, 3 times, on ice. After sonication, the cell lysates were
cleared by centrifugation at 14,000 × g for 10 min at 4°C. Nuclear
protein was isolated using a Nuclear and Cytoplasmic Protein
Extraction kit (Beyotime Institute of Biotechnology), according to
the manufacturer's instructions (15). The protein concentration was
measured using the Coomassie-blue method, and equal amounts of
total protein (15 µg/lane) were separated on a 12% SDS-PAGE, and
transferred to PVDF membranes. PVDF membranes were then blocked
with 5% BSA for 1 h at room temperature and incubated with primary
antibodies overnight at 4°C: Mouse-anti-Runx2 (1:1,000; cat. no.
ab76956; Abcam), rabbit-anti-STAT1 (1:1,000; cat. no. 14994; Cell
Signaling Technology, Inc.), rabbit-anti-proliferating cell nuclear
antigen (1:1,000; cat. no. 13110; Cell Signaling Technology, Inc.)
and mouse-anti-β-actin (1:2,000; cat. no. BM5422; Boster Biological
Technology). The following day, membranes were washed with 0.1%
TBS-Tween 20 (TBST) and incubated with the appropriate horseradish
peroxidase-conjugated secondary antibody (1:2,000; cat. nos.
sc-2004 and sc-2005; Santa Cruz Biotechnology, Inc.) for 1 h at
room temperature. After washing, protein bands were visualized
using SuperSignal West Dura Extended Duration Substrate (Pierce;
Thermo Fisher Scientific, Inc.). Western blot analysis was
performed in triplicate.
Microcomputed tomography analysis
Female mice (age, 12 months) were randomly divided
into three groups (n=6 per group). The BMSC-specific
aptamer-agomiR-483-3p was synthesized by GenScript Corporation.
Aptamer-agomiR-NC, Aptamer-agomiR-483-3P or PBS was periosteally
injected into the medullary cavity of the femur twice per month for
3 months. After 3 months of treatment, microcomputed tomography
analysis, three-point bending test and bone histomorphometrically
analysis were performed as previously reported (4,16).
Statistical analysis
Statistical analysis was performed using SPSS
(version 13.0; SPSS, Inc.). Data are presented as the mean ± SD.
One-way ANOVA followed by Dunnett's post hoc test was used to
analyze differences among multiple groups. All experiments were
repeated >3 times. P<0.05 was considered to indicate a
statistically significant difference.
Results
miR-483-3p is increased during
osteogenic differentiation
To investigate changes in miRNA expression during
osteogenic differentiation, BMSCs were cultured with osteogenic
medium for 7 days. Changes in miRNA expression were identified by
performing miRNA microarray analysis of differentiated BMSCs
compared with undifferentiated control samples. The z-score
illustrated the relative expression level of a specific miRNA. Each
row represents an miRNA and each column represents a sample. From
the analysis of the microarray data based on the fold change and
probability values, 10 miRNAs were found to be downregulated and 11
were upregulated significantly (fold change, >2; P<0.05).
Certain miRNAs that have previously been reported to regulate
osteogenic differentiation, such as miR-129, miR-188, miR-195 and
miR-497 (4,17,18),
were detected in the present microarray analysis (Fig. 1A). Among the differentially
expressed miRNAs identified, miR-483-3p was significantly
upregulated during osteogenic differentiation (Fig. 1A). Previous studies have shown that
miR-483-3p is involved in cell proliferation, cell cycle and
apoptosis (19,20), but its role in osteogenic
differentiation remains uninvestigated. To examine the expression
level of miR-483 during osteogenic differentiation, BMSCs were
treated with osteogenic differentiation medium for 0, 3, 7, 14, and
21 days, and the expression level of miR-483-3p was assessed by
RT-qPCR. The qPCR results suggested that miR-483-3p levels
gradually increased during osteogenic differentiation, in line with
the miRNA microarray analysis results (Fig. 1B). Additionally, the expression
levels of osteogenic gene markers such as ALP and OCN gradually
increased during osteogenic differentiation (Fig. 1C and D). Furthermore, the change in
miRNA-483-3p expression also gradually increased in hMSCs during
osteogenic differentiation (Fig.
1E). The present results suggested that miRNA-483-3p may play
an important role in the process of osteogenic differentiation of
BMSCs.
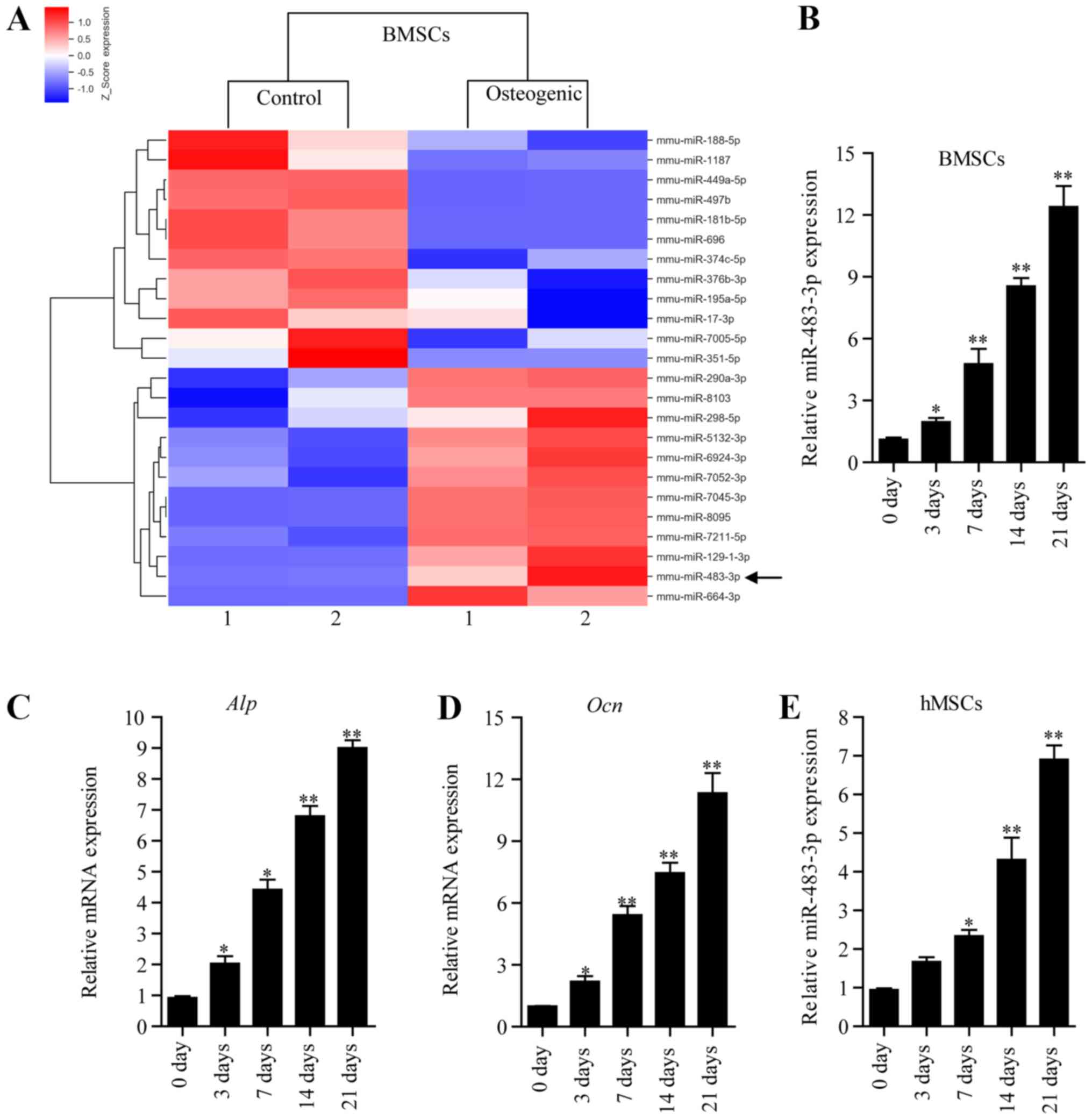 | Figure 1.Expression level of miR-483-3p during
osteogenic differentiation of BMSCs. (A) Changes in miRNA
expression were analyzed by miRNA microarray using BMSCs cultured
in osteogenic differentiation medium or in α-MEM control medium for
7 days. Red indicates increased expression and blue indicates
decreased expression compared with the control group. (B) RT-qPCR
analysis of the expression level of miR-483-3p in BMSCs cultured in
osteogenic differentiation medium for 0, 3, 7, 14 and 21 days.
RT-qPCR analysis of the mRNA levels of (C) OCN and (D) ALP at 0, 3,
7, 14 and 21 days. (E) RT-qPCR analysis of the expression level of
miR-483-3p in hMSCs cultured in osteogenic differentiation medium
for 0, 3, 7, 14 and 21 days. Data are presented as the mean ± SD.
n=5 per group. *P<0.05, **P<0.01 vs. 0 day. BMSCs, bone
marrow mesenchymal stem cells; miRNA, microRNA; RT-qPCR, reverse
transcription-quantitative PCR; ALP, alkaline phosphatase; OCN,
osteocalcin; hMSCs, human mesenchymal stem cells; miR,
microRNA. |
miR-483-3p promotes osteogenic
differentiation of BMSCs
The expression level of miR-483-3p increased during
osteogenic differentiation, which suggested that miR-483 may have a
role in promoting osteogenic differentiation. To test this
hypothesis, BMSCs were transfected with agomiR-483-3p,
antagomiR-483-3p or control, and cultured in osteogenic
differentiation medium for 48 h. The expression of miR-483-3p was
assessed by qPCR (Fig. 2A). ALP
activity and OCN secretion, two of the most important osteogenic
differentiation markers (4), were
identified to be increased in agomiR-483-3p-transfected BMSCs
compared with the control group (Fig.
2B and C). By contrast, these effects were reversed following
antagomiR-483-3p transfection (Fig. 2B
and C). In addition, overexpression of miR-483-3p increased
mineralized nodule formation following a21-day incubation in
osteogenesis induction medium, as assessed by Alizarin Red staining
(Fig. 2D). By contrast, knockdown
of miR-483-3p decreased the mineralized nodule formation (Fig. 2D). Moreover, the expression of
osteogenic gene markers such as ALP, OCN, Type I collagen, RUNX2
and osterix were also increased following transfection with
agomiR-483-3p (Fig. 2E-I).
However, the transfection of antagomiR-483-3p decreased the
expression levels of these genes (Fig.
2E-I). Collectively, the present results suggested that
miR-483-3p promoted osteogenic differentiation of BMSCs.
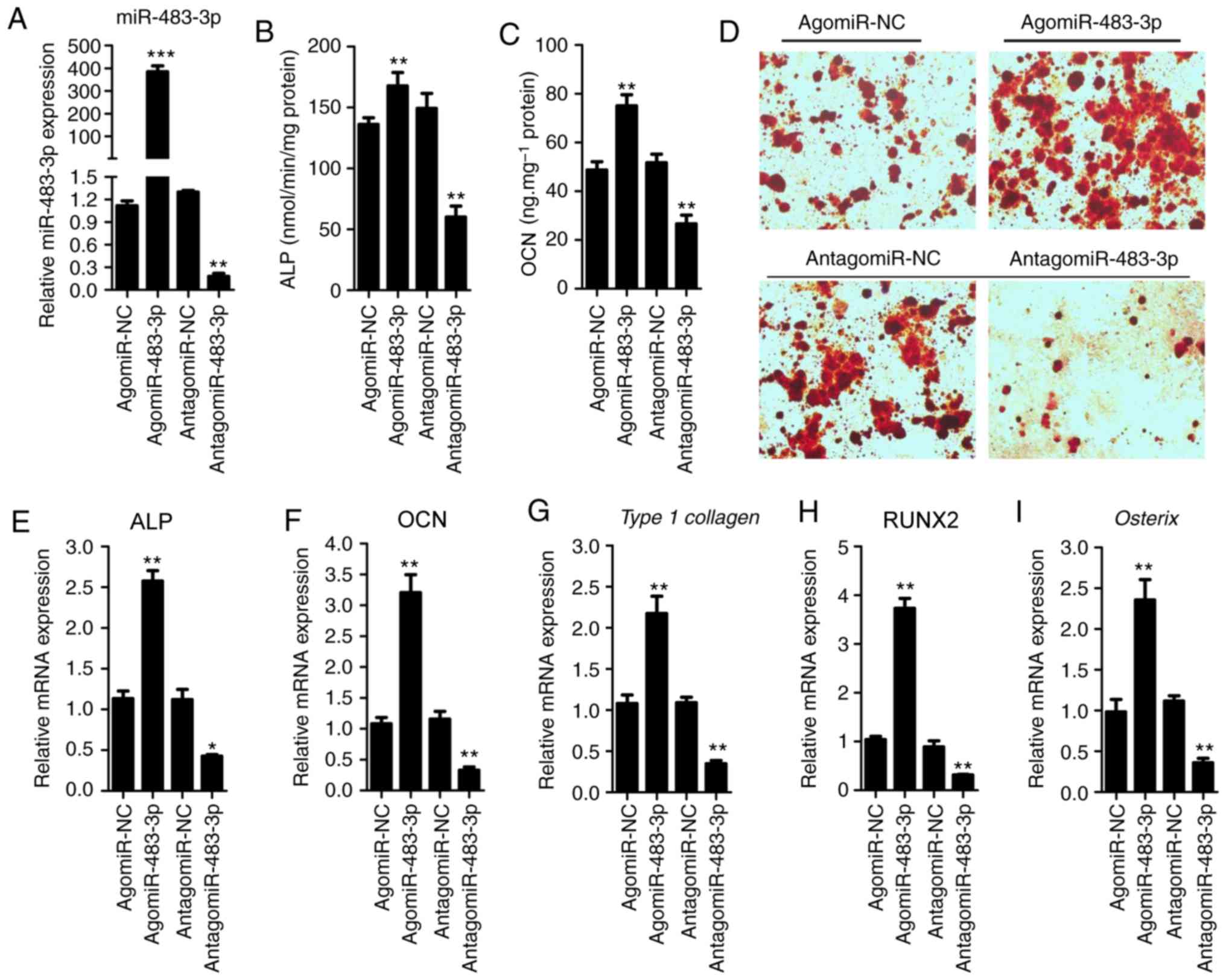 | Figure 2.miR-483-3p promotes osteogenic
differentiation of BMSCs. (A) RT-qPCR analysis was performed to
assess the expression level of miR-483-3p in BMSCs after
transfection with agomiR-NC, agomiR-483-3p, antagomiR-NC or
antagomiR-483-3p for 48 h. (B) ALP activity and (C) OCN secretion
were measured after transfection with agomiR-483-3p,
antagomiR-483-3p or NC for 48 h. (D) Alizarin Red staining was used
to quantify matrix mineralization in BMSCs after induction of
osteoblast differentiation for 21 days. Magnification, ×200.
RT-qPCR analysis of the mRNA levels of (E) ALP, (F) OCN, (G) Type 1
collagen, (H) RUNX2 and (I) osterix in BMSCs cultured in osteogenic
differentiation medium for 48 h. Data are presented as the mean ±
SD. n=5 in each group. *P<0.05, **P<0.01, ***P<0.001 vs.
corresponding NC. BMSCs, bone marrow mesenchymal stem cells; miR,
microRNA; ALP, alkaline phosphatase; OCN, osteocalcin; RUNX2,
runt-related transcription factor 2; NC, negative control; RT-qPCR,
reverse transcription-quantitative PCR. |
miR-483-3p directly targets STAT1 to
regulate osteogenic differentiation
To understand the molecular mechanisms underlying
miR-483-3p promotion of osteogenic differentiation, the potential
target genes of miR-483-3p were investigated using the miRNA target
prediction tools Targets can and miRanda. Among the putative target
genes identified, the present study examined STAT1, which was
predicted to be a potential target gene in both databases, and has
previously been reported to regulate osteogenic differentiation
(21). Sequence analysis suggested
that miR-483-3p may target STAT1 by binding to its 3′-UTR in the
position 1511–1517 (Fig. 3A). To
examine whether miR-483-3p can directly bind the 3′-UTR of STAT1, a
luciferase reporter plasmid containing the 3′-UTR of STAT1 and a
mutated luciferase reporter plasmid were constructed (Fig. 3A). Wild-type and mutant STAT1
luciferase expression vectors were then transfected into BMSCS
cells along with agomiR-483-3p. Transfection with agomiR-483-3p
suppressed the luciferase activity of the wild-type STAT1 3′-UTR,
but exhibited no effects on the mutated version of STAT1 3′-UTR
(Fig. 3B). Moreover,
overexpression of miR-483-3p downregulated endogenous STAT1 protein
expression, whereas inhibition of miR-483-3p increased STAT1
protein expression levels (Fig.
3C). However, STAT1 mRNA levels were not affected by
transfection with agomiR-483-3p or antagomiR-483-3p (Fig. 3D). The present results suggested
that miR-483-3p targetedSTAT1 by binding to its 3′-UTR and
affectedSTAT1 expression at the post-transcriptional level.
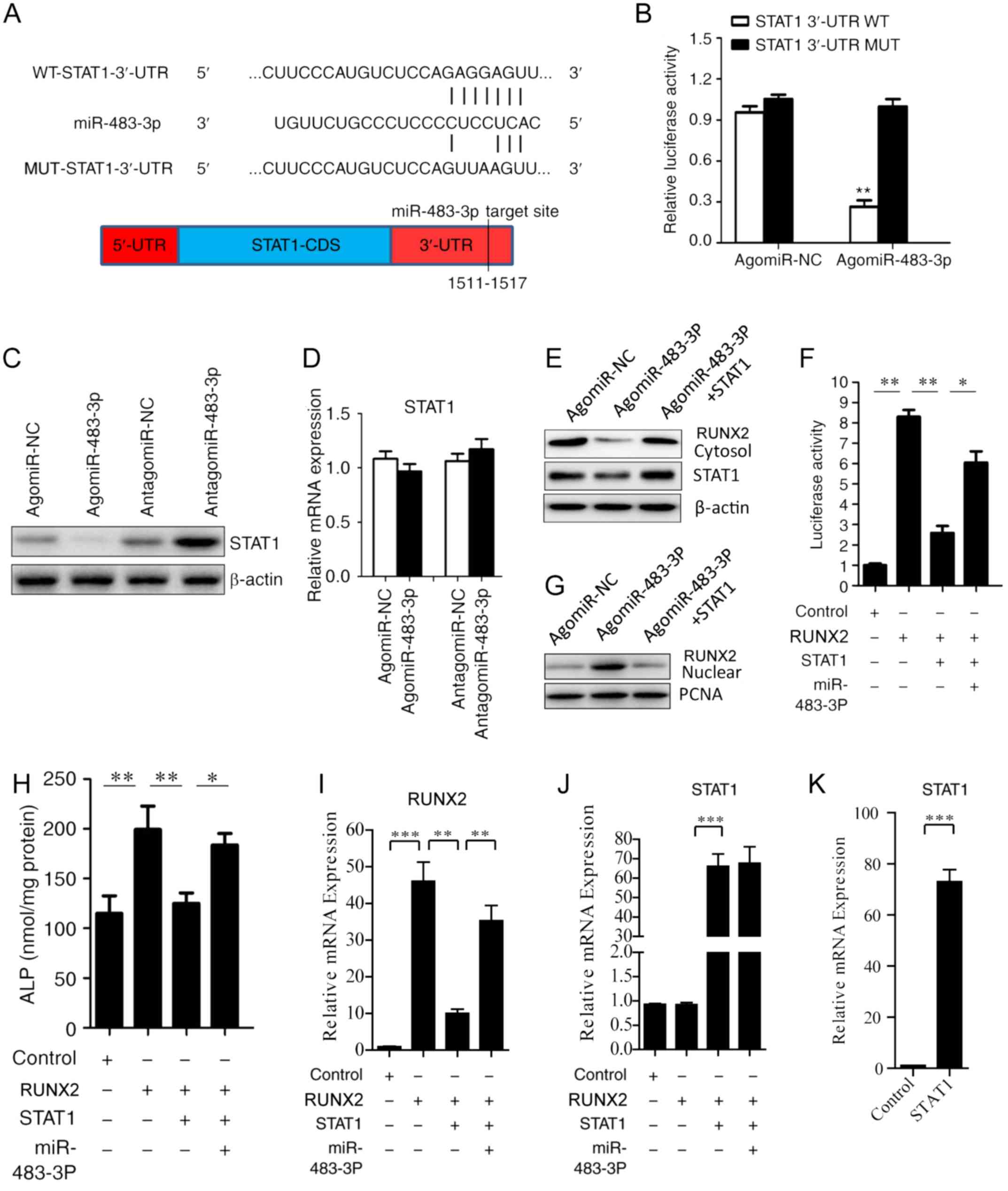 | Figure 3.miR-483-3p targets STAT1 to
functionally promote osteogenic differentiation. (A) Base pairing
of miR-483-3p with STAT1 3′-UTR. Luciferase reporters with the
WT-STAT13′-UTR and the MUT-STAT13′-UTR. (B) AgomiR-NC or
agomiR-483-3p were co-transfected with WT-STAT13′-UTR or MUT-STAT1
3′-UTRinto BMSCs, and luciferase activity was determined 48 h after
transfection. Renilla activity was used to normalize Firefly
luciferase activity. (C) Western blotting and (D) RT-qPCR analysis
of the relative expression level of STAT1 after transfection with
agomiR-483-3p or antagomiR-483-3p into BMSCs. (E) Western blot
analysis of STAT1 and RUNX2 protein expression in the cytoplasm.
β-actin was used as loading control. (F) Luciferase reporter assay
of OCN promoter activity in BMSCs after co-transfection with a
dual-luciferase reporter plasmid containing the OCN promoter along
with RUNX2 plasmid or RUNX2 and STAT1 overexpression vectors. (G)
Western blot analysis of RUNX2 protein expression in the nucleus.
PCNA was used as loading control. (H) ALP activity in BMSCs was
analyzed after transfection with RUNX2 plasmid or RUNX2 and STAT1
plasmids. (I) RUNX2 and (J) STAT1 expression levels were analyzed
in the four experimental groups. (K) STAT1 expression level was
analyzed following transfection of STAT1 plasmid. Data are
presented as the mean ± SD. n=5 in each group. *P<0.05,
**P<0.01, ***P<0.001. miR, microRNA; RUNX2, runt-related
transcription factor 2; OCN, osteocalcin; RT-qPCR, reverse
transcription-quantitative PCR; UTR, untranslated region; BMSCs,
bone marrow mesenchymal stem cells; CDS, coding sequence; MUT,
mutant; WT, wild-type; PCNA, proliferating cell nuclear antigen;
ALP, alkaline phosphatase; NC, negative control. |
A previous study reported that STAT1 inhibits
osteoblast differentiation by inhibiting RUNX2 transcriptional
activity and RUNX2 nuclear translocation, which has been reported
to have a key role in osteogenic differentiation and bone formation
(21). In the present study,
miR-483-3p was identified to downregulate STAT1 expression, and it
was hypothesized that miR-483-3p may promote osteogenic
differentiation of BMSCs by downregulating STAT1 and enhancing
RUNX2 transcriptional activity and nuclear translocation.
Interestingly, a previous study reported that overexpression of
STAT1 significantly inhibited RUNX2 nuclear translocation and
osteoblast differentiation mediated by miR-194 (15). In the present study, overexpression
of miR-483-3p was identified to increase the nuclear translocation
of RUNX2, but this effect was suppressed by transfection with STAT1
overexpression plasmid (Fig. 3E and
G). Next, to study the effects of miR-483-3p on RUNX2
transcriptional activity, a reporter plasmid containing the OCN
promoter was transfected into BMSCs along with RUNX2 vector, STAT1
plasmid, and/or agomiR-483-3p. Overexpression of RUNX2
significantly increased RUNX2-mediated luciferase activity in a
plasmid containing the OCN promoter, whereas co-transfection with
STAT1 overexpression plasmid suppressed this effect. Interestingly,
RUNX2-mediated luciferase activity was rescued by co-transfection
with agomiR-483-3p (Fig. 3F). The
present results suggested that miR-483-3p promoted osteogenic
differentiation of BMSCs by increasing RUNX2 nuclear translocation
and RUNX2 transcriptional activity via themiR-483-3p/STAT1/RUNX2
axis.
To further investigate this hypothesis, RUNX2, STAT1
or agomiR-483-3p were transfected into BMSCs to investigate their
effects on ALP activity. The present results suggested that ALP
activity was significantly increased in the RUNX2 overexpression
plasmid transfection group, whereas this effect was inhibited
following STAT1 overexpression. Notably, ALP activity was rescued
following agomiR-483-3p co-transfection (Fig. 3H). Additionally, the transfection
efficiencies of RUNX2 and STAT1 overexpression plasmids were
examined (Fig. 3I-K).
Collectively, the present results suggested that miR-483-3p
regulated osteogenic differentiation via themiR-483-3p/STAT1/RUNX2
axis.
miR-483-3p regulates bone formation in
vivo
In order to examine the effects of miR-483-3p in
vivo, BMSC-specific aptamer-agomiR-NC, BMSC-specific
aptamer-agomiR-483-3P or PBS was injected into the femoral bone
marrow cavity of 12-month-old mice twice per month for 3 months
(n=6 in each group). After 3 months, miR-483-3p mRNA levels were
measured by RT-qPCR in mice bone tissues. The results showed that
the levels of miR-483-3p were significantly increased in bone after
injection of BMSC-targeted aptamer-agomiR-483-3p (Fig. 4B). Moreover, these mice exhibited
significantly higher bone density, trabecular bone volume per
tissue volume, trabecular thickness, and tibia maximum load and
stiffness, and lower trabecular separation compared with
vehicle-treated mice (Fig. 4).
Furthermore, the number of osteoblasts on the bone surface was
significantly increased in mice treated with agomiR-483-3p compared
with controls (Fig. 4G). The
present results suggested that overexpression of miR-483-3p
increased bone formation in aged mice.
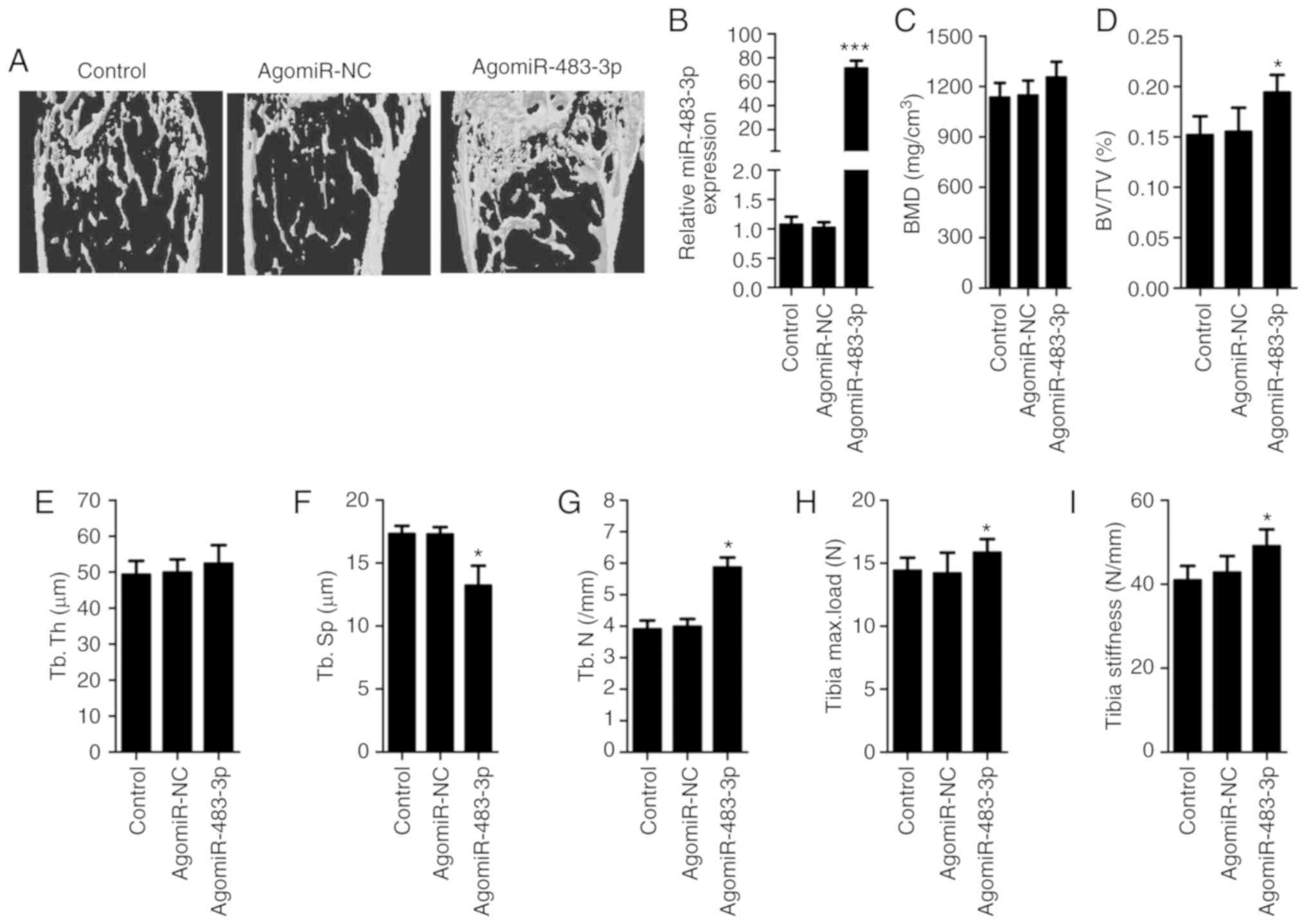 | Figure 4.miR-483-3p regulates bone formation
in vivo. BMSC-specific aptamer-agomiR-483-3p,
aptamer-agomiR-NC or PBS was periosteally injected into
12-month-old mice twice per month for 3 months. (A) Effect of
BMSC-specific aptamer-agomiR-483-3p in aged mice as determined by
microCT examination. (B) RT-qPCR analysis of the expression level
of miR-483-3P in bone. Quantitative microCT analysis of femoral
histomorphometric parameters, including (C) BMD, (D) BV/TV, (E)
Tb.Th, (F) Tb.Sp and (G) Tb.N. Three-point bending measurement of
tibia (H) maximum load and (I) stiffness. Data are presented as the
mean ± SD. n=6 in each group. *P<0.05, ***P<0.001 vs.
control. NC, negative control; miR, microRNA; BMSCs, bone marrow
mesenchymal stem cells; BMD, bone mineral density; BV/TV, bone
volume/tissue volume; Tb.Th, trabecular thickness; Tb.Sp,
trabecular spacing; Tb.N, trabecular number; microCT, microcomputed
tomography. |
Discussion
Numerous previous studies identified that miRNAs
serve an important regulatory role in bone biology, including
osteogenic differentiation and bone formation (4,5). In
the present study, miR-438-3p expression was identified to
gradually increase during osteogenic differentiation. In addition,
miR-438-3p promoted osteogenic differentiation of BMSCs by
downregulating STAT1 and enhancing the transcriptional activity of
RUNX2. Present in vivo experiments suggested that increasing
miR-483-3p levels using a BMSC-specific delivery system promoted
bone formation in aged mice.
miR-483-3p has been widely investigated in various
cellular processes and cell types. Specifically, miR-483-3p has
been reported to be involved in regulating apoptosis,
proliferation, diabetes-associated heart diseases and inhibition of
adipocyte differentiation (22,23).
However, the role of miR-483-3p in regulating osteogenic
differentiation of BMSCs remains unclear. In the present study, a
novel role of miR-483-3p and its underlying mechanism were
identified, and miR-483-3p was found to be involved in regulating
the osteogenic differentiation of BMSCs by targeting STAT1.
STAT1 was first identified as a signaling molecule
in the interferon signaling pathway (24,25).
A previous study identified that STAT1 is also involved in
osteogenic differentiation and bone formation (26). STAT1 mutant mice exhibit increased
bone formation and bone mass, although they also present increased
osteoclasts and bone resorption (21). In addition, a previous study
demonstrated that STAT1 directly interacts and inhibits RUNX2, a
transcription factor with a key role in osteoblast differentiation
and bone formation, suppressing the nuclear localization and
transcriptional activity of RUNX2 (15,27).
These previous findings suggested that STAT1 functions as a potent
osteogenic inhibitor able to suppress bone formation in vivo
(15,27). A previous study suggested that
downregulation of STAT1 could be used as a potential strategy to
improve bone formation or osteogenic differentiation (21). The present results suggested that
the change in miR-483-3p levels resulted in an alteration in the
amount of STAT1 protein. In addition, overexpression of STAT1 can
abolish the positive effect of RUNX2 on ALP activity in BMSCs,
which is rescued by agomiR-483-3p transfection. Collectively, the
present results suggested that STAT1 was a functional target of
miR-483-3p and that increasing the expression level ofmiR-483-3p
may be a potential strategy for promoting osteogenic
differentiation.
Due to the increase in the aging population,
osteoporosis has become a major public health problem (28). Intermittent injections of
parathyroid hormone 1–38 is a common treatment approach for
osteoporosis, but this approach is limited to a 2-year time period
and has some potential side effects, such as excessive bone
resorption (29,30). Novel safe anabolic drugs that
increase bone formation are thus required for treating this
disease. Previous studies demonstrated that intra-bone marrow
injection of agomiR or antagomiR with a BMSC-specific aptamer
increased BMSC differentiation and bone formation, suggesting that
this approach may represent a potential a strategy to treat
osteoporosis (4,31). In the present study, a
BMSC-specific aptamer was used to deliver agomiR-483-3p into mouse
BMSCs via intra-bone marrow injections. The present results
suggested that overexpression of miR-483-3p increased bone
formation in aged mice. The present results support the
aptamer-mediated modulation of miRNA expression levels in BMSCs as
a new strategy to treat senile osteoporosis.
Collectively, the present findings suggested that
miR-483-3p promoted osteogenic differentiation of BMSCs by
targeting STAT1 and enhancing the transcriptional activity of
RUNX2, identifying a new role for miR-483-3p and suggesting a novel
potential therapeutic target to treat osteoporosis.
Acknowledgements
Not applicable.
Funding
The present study was supported by The National
Natural Science Foundation of China (grant nos. 81700785, 81500686,
81770877 and 81670809).
Availability of data and materials
The datasets used and/or analyzed during the current
study are available from the corresponding author on reasonable
request.
Authors' contributions
YX performed the experiments. WX designed the study.
QG and TJJ analyzed and interpreted the data. YY, LY and GWW
performed the statistical analysis and drafted the manuscript. All
authors read and approved the final manuscript.
Ethics approval and consent to
participate
Human mesenchymal stem cells were isolated from the
bone marrow of healthy donors. The procedure was approved by The
Ethics Committee of the Xiangya Hospital of Central South
University. All animal experimental procedures were approved by The
Animal Care and Use Committee of the Xiangya Hospital of Central
South University.
Patient consent for publication
All patients provided written informed consent.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Krane SM: Identifying genes that regulate
bone remodeling as potential therapeutic targets. J Exp Med.
201:841–843. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Pittenger MF, Mackay AM, Beck SC, Jaiswal
RK, Douglas R, Mosca JD, Moorman MA, Simonetti DW, Craig S and
Marshak DR: Multilineage potential of adult human mesenchymal stem
cells. Science. 284:143–147. 1999. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Crane JL and Cao X: Bone marrow
mesenchymal stem cells and TGF-β signaling in bone remodeling. J
Clin Invest. 124:466–472. 2014. View
Article : Google Scholar : PubMed/NCBI
|
|
4
|
Li CJ, Cheng P, Liang MK, Chen YS, Lu Q,
Wang JY, Xia ZY, Zhou HD, Cao X, Xie H, et al: MicroRNA-188
regulates age-related switch between osteoblast and adipocyte
differentiation. J Clin Invest. 125:1509–1522. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Khalaj M, Woolthuis CM, Hu W, Durham BH,
Chu SH, Qamar S, Armstrong SA and Park CY: miR-99 regulates normal
and malignant hematopoietic stem cell self-renewal. J Exp Med. Jul
21–2017.(Epub ahead of print). jem.20161595, 2017. doi:
10.1084/jem.20161595. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Edmonds MD, Boyd KL, Moyo T, Mitra R,
Duszynski R, Arrate MP, Chen X, Zhao Z, Blackwell TS, Andl T and
Eischen CM: MicroRNA-31 initiates lung tumorigenesis and promotes
mutant KRAS-driven lung cancer. J Clin Invest. 126:349–364. 2016.
View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Mori MA, Thomou T, Boucher J, Lee KY,
Lallukka S, Kim JK, Torriani M, Yki-Järvinen H, Grinspoon SK,
Cypess AM and Kahn CR: Altered miRNA processing disrupts
brown/white adipocyte determination and associates with
lipodystrophy. J Clin Invest. 124:3339–3351. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Zhou X, Jeker LT, Fife BT, Zhu S, Anderson
MS, McManus MT and Bluestone JA: Selective miRNA disruption in T
reg cells leads to uncontrolled autoimmunity. J Exp Med.
205:1983–1991. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Li H, Xie H, Liu W, Hu R, Huang B, Tan YF,
Xu K, Sheng ZF, Zhou HD, Wu XP and Luo XH: A novel microRNA
targeting HDAC5 regulates osteoblast differentiation in mice and
contributes to primary osteoporosis in humans. J Clin Invest.
119:3666–3677. 2009. View
Article : Google Scholar : PubMed/NCBI
|
|
10
|
Li Z, Hassan MQ, Volinia S, van Wijnen AJ,
Stein JL, Croce CM, Lian JB and Stein GS: A microRNA signature for
a BMP2-induced osteoblast lineage commitment program. Proc Natl
Acad Sci USA. 105:13906–13911. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Gu H, Xu J, Huang Z, Wu L, Zhou K, Zhang
Y, Chen J, Xia J and Yin X: Identification and differential
expression of microRNAs in 1, 25-dihydroxyvitamin D3-induced
osteogenic differentiation of human adipose-derived mesenchymal
stem cells. Am J Transl Res. 11:4856–4871. 2017.
|
|
12
|
Maruyama K, Uematsu S, Kondo T, Takeuchi
O, Martino MM, Kawasaki T and Akira S: Strawberry notch homologue 2
regulates osteoclast fusion by enhancing the expression of
DC-STAMP. J Exp Med. 210:1947–1960. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(-Delta Delta C(T)) method. Methods. 25:402–408. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Javed A, Gutierrez S, Montecino M, van
Wijnen AJ, Stein JL, Stein GS and Lian JB: Multiple Cbfa/AML sites
in the rat osteocalcin promoter are required for basal and vitamin
D-responsive transcription and contribute to chromatin
organization. Mol Cell Biol. 19:7491–7500. 1999. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Li J, He X, Wei W and Zhou X: MicroRNA-194
promotes osteoblast differentiation via downregulating STAT1.
Biochem Biophys Res Commun. 460:482–488. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Zou W, Greenblatt MB, Brady N, Lotinun S,
Zhai B, de Rivera H, Singh A, Sun J, Gygi SP, Baron R, et al: The
microtubule-associated protein DCAMKL1 regulates osteoblast
function via repression of RUNX2. J Exp Med. 210:1793–1806. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Xiao WZ, Gu XC, Hu B, Liu XW, Zi Y and Li
M: Role of microRNA-129-5p in osteoblast differentiation from bone
marrow mesenchymal stem cells. Cell Mol Biol (Noisy-le-grand).
62:95–99. 2016.PubMed/NCBI
|
|
18
|
Grünhagen J, Bhushan R, Degenkolbe E,
Jäger M, Knaus P, Mundlos S, Robinson PN and Ott CE: miR-497
approximately 195 cluster microRNAs regulate osteoblast
differentiation by targeting BMP signaling. J Bone Miner Res.
30:796–808. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Bertero T, Gastaldi C, Bourget-Ponzio I,
Imbert V, Loubat A, Selva E, Busca R, Mari B, Hofman P, Barbry P,
et al: miR-483-3p controls proliferation in wounded epithelial
cells. FASEB J. 25:3092–3105. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Lupini L, Pepe F, Ferracin M, Braconi C,
Callegari E, Pagotto S, Spizzo R, Zagatti B, Lanuti P, Fornari F,
et al: Over-expression of the miR-483-3p overcomes the miR-145/TP53
pro-apoptotic loop in hepatocellular carcinoma. Oncotarget.
7:31361–31371. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Kim S, Koga T, Isobe M, Kern BE, Yokochi
T, Chin YE, Karsenty G, Taniguchi T and Takayanagi H: Stat1
functions as a cytoplasmic attenuator of RUNX2 in the
transcriptional program of osteoblast differentiation. Genes Dev.
17:1979–1991. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Bertero T, Bourget-Ponzio I, Puissant A,
Loubat A, Mari B, Meneguzzi G, Auberger P, Barbry P, Ponzio G and
Rezzonico R: Tumor suppressor function of miR-483-3p on squamous
cell carcinomas due to its pro-apoptotic properties. Cell Cycle.
12:2183–2193. 2013. View
Article : Google Scholar : PubMed/NCBI
|
|
23
|
Ferland-McCollough D, Fernandez-Twinn DS,
Cannell IG, David H, Warner M, Vaag AA, Bork-Jensen J, Brøns C,
Gant TW, Willis AE, et al: Programming of adipose tissue miR-483-3p
and GDF-3 expression by maternal diet in type 2 diabetes. Cell
Death Differ. 19:1003–1012. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Meraz MA, White JM, Sheehan KC, Bach EA,
Rodig SJ, Dighe AS, Kaplan DH, Riley JK, Greenlund AC, Campbell D,
et al: Targeted disruption of the Stat1 gene in mice reveals
unexpected physiologic specificity in the JAK-STAT signaling
pathway. Cell. 84:431–442. 1996. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Huang Z, Richmond TD, Muntean AG, Barber
DL, Weiss MJ and Crispino JD: STAT1 promotes megakaryopoiesis
downstream of GATA-1 in mice. J Clin Invest. 117:3890–3899. 2007.
View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Takayanagi H, Ogasawara K, Hida S, Chiba
T, Murata S, Sato K, Takaoka A, Yokochi T, Oda H, Tanaka K, et al:
T-cell-mediated regulation of osteoclastogenesis by signalling
cross-talk between RANKL and IFN-gamma. Nature. 408:600–605. 2000.
View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Tajima K, Takaishi H, Takito J, Tohmonda
T, Yoda M, Ota N, Kosaki N, Matsumoto M, Ikegami H, Nakamura T, et
al: Inhibition of STAT1 accelerates bone fracture healing. J Orthop
Res. 28:937–941. 2010.PubMed/NCBI
|
|
28
|
Slemenda C, Longcope C, Peacock M, Hui S
and Johnston CC: Sex steroids, bone mass, and bone loss. A
prospective study of pre-, peri-, and postmenopausal women. J Clin
Invest. 97:14–21. 1996. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Lane NE, Sanchez S, Modin GW, Genant HK,
Pierini E and Arnaud CD: Parathyroid hormone treatment can reverse
corticosteroid-induced osteoporosis. Results of a randomized
controlled clinical trial. J Clin Invest. 102:1627–1633. 1998.
View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Balani DH, Ono N and Kronenberg HM:
Parathyroid hormone regulates fates of murine osteoblast precursors
in vivo. J Clin Invest. 127:3327–3338. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Wang X, Guo B, Li Q, Peng J, Yang Z, Wang
A, Li D, Hou Z, Lv K, Kan G, et al: miR-214 targets ATF4 to inhibit
bone formation. Nat Med. 19:93–100. 2013. View Article : Google Scholar : PubMed/NCBI
|


















