Introduction
Acute myocardial infarction (AMI) is one of the top
three causes of mortality and disability in the world (1). The pathological process of AMI is
myocardial necrosis due to persistent myocardial ischemia and
hypoxia, caused by coronary plaque rupture or paralysis. At
present, biomarkers reflecting myocardial injury, such as cardiac
troponins, creatine kinase (CK)-MB and lactate dehydrogenase (LDH),
and electrocardiogram (ECG) findings, are the most common
diagnostic methods for AMI in clinical practice. However, the
aforementioned biomarkers are not sensitive enough for the early
diagnosis of AMI in the emergency room, thereby increasing the risk
of complications and mortality (2). The low specificity of elevated
cardiac troponins also affects the diagnosis of AMI, as these
elevated levels may result from other non-cardiac issues (3). In order to develop practical
surveillance tools, there is a clear need to identify new
biomarkers for the diagnosis of AMI.
Noncoding RNAs (ncRNAs) are classified into small
ncRNAs and long ncRNAs (lncRNAs) based on size, and play a variety
of roles in cell cycle regulation, gene expression regulation,
cellular differentiation, transcription, translation and chromatin
modification (4,5). Emerging evidence has indicated that
lncRNAs participate in multiple physiological and pathological
processes of cardiovascular disease (6). Circulating RNAs have been described
as being relatively stable in different human bodily fluids, such
as serum, plasma and urine (7–9),
making them suitable for the clinical assessment and monitoring of
pathological conditions. Circulating mitochondrial lncRNA
uc022bqs.1 has been proven to be a novel biomarker of cardiac
remodeling and predicts future mortality in patients with heart
failure (10). lncRNA urothelial
carcinoma-associated 1 was found to be aberrantly expressed in AMI
patients (11).
A previous study demonstrated that the lncRNA
myocardial infarction associated transcript (MIAT) is associated
with myocardial infarction through single-nucleotide polymorphism
(SNP) association experiments (12). The present study aimed to detect
the plasma level of MIAT in patients with AMI to determine whether
it can be used as a potential biomarker to monitor myocardial
pathological changes, and to explore its function at the cellular
level.
Materials and methods
Patients
Between August 2016 and December 2017, 260 patients
aged 40–70 years treated in the emergency department of The First
Affiliated Hospital of Xinjiang Medical University within 3 h of
the onset of chest pain were recruited. In the present study, 58
patients diagnosed with ST-segment elevated myocardial infarction
(STEMI) were selected as the observer group and 50 patients with
unstable angina (UA) as the control group. A total of 72 patients
were excluded due to a lack of four serial time samples, or the
presence of cardiomyopathy, myocarditis, heart failure, chronic
renal failure, pulmonary infection or psychiatric problems. The
other 80 patients diagnosed with non-ST segment elevation
myocardial infarction were not included in the study. A total of
180 patients were included in the present study (49 female and 59
male) All of the patients received a clinical assessment by an
experienced cardiologist, which included medical history, physical
examination, renal function assessment, and ECG and cardiac enzyme
monitoring at 0, 3, 6, 12 and 24 h after the onset of chest pain.
The inclusion criteria for STEMI were based on the 2017 European
Society of Cardiology (ESC) guidelines (13) and the diagnostic parameters
included ischemic symptoms, an elevated ST-segment on ECG, and
evidently increased cardiac troponin T (cTnT) and CK-MB. The
inclusion criteria for UA were also based on the 2017 ESC
guidelines, including recent episodes of angina, and onset of or
new angina at rest lasting >20 min, with or without ECG ST-T
changes at onset. Written informed consent was obtained from all
enrolled subjects and the study protocol was approved by the ethics
committee of The First Affiliated Hospital of Xinjiang Medical
University.
Plasma collection and determination of
myocardial enzymes
Plasma was collected at 0, 3, 6, 12 and 24 h after
the chest pain episode, and the concentrations of myocardial
enzymes (cTnT; cat. no. MAB18742; and CK-MB; cat. no. MAB9076;
R&D Systems, Inc.) at the different times were detected using
ELISA. Fasting venous blood was collected from all subjects on the
morning following admission. Samples were placed in heparin-coated
anticoagulant tubes and centrifuged at 3,000 × g for 15 min at room
temperature to separate the plasma. The supernatant was obtained
and stored at −80°C.
Animals and establishment of the rat
AMI model
All animal procedures were approved by the
Experimental Animal Ethics Committee of The First Affiliated
Hospital of Xinjiang Medical University. Male 8 week old Wistar
rats (n=40; 200–250 g) were purchased from the laboratory animal
center of Xinjiang Medical University. All animals were fed a
standard rat diet and subjected to 1 week of adaptive feeding. The
animals were allowed free access to drinking water and feed at a
temperature of 23–25°C and a humidity of 55–70% with a 12 h
light/dark cycle. In total, 10 normal rats were sacrificed after
1.0% isoflurane anesthesia and the heart, liver, spleen, lungs and
kidneys were removed. In total, 50 g of tissue homogenate was
obtained and the expression of MIAT was detected by reverse
transcription-quantitative (RT-q)PCR.
The remaining 30 rats were divided into a sham group
(n=15) and an AMI group (n=15). AMI surgery was performed according
to a previously published procedure (14). Briefly, rats were deeply
anaesthetized with 1.0% isoflurane using a rodent ventilator, fixed
onto the operating table, and connected to a standard limb lead II
ECG. Thoracotomy and pericardiotomy in the 3rd to 4th ribs were
performed to expose the heart, and then a segment of saline-soaked
7/0 suture was looped around the left anterior descending (LAD)
coronary artery. When the left ventricular myocardium turned white,
and the ECG ST-segment was elevated >0.1 mv, the model was
considered to have been successfully established. In the sham
group, the LAD was encircled without ligation. Post-operative blood
samples were collected at 0, 3, 6, 12 and 24 h.
Neonatal rat cardiomyocyte culture and
treatment
Neonatal Wistar rats (1–2 days old) were purchased
from the laboratory animal center of Xinjiang Medical University.
The rats were deeply anesthetized with 1.0% isoflurane, and the
ventricles were cut into small pieces and transferred into a
digestion solution containing 0.1% collagenase and 0.25% trypsin at
37°C for 30 min. The cells were then cultured in Dulbecco's
Modified Eagle's medium/F12 (Gibco; Thermo Fisher Scientific, Inc.)
with 10% fetal bovine serum (Thermo Fisher Scientific, Inc., MA,
USA), 100 U/ml penicillin and 100 mg/ml streptomycin in an
incubator at 37°C. Cells were cultured in a culture flask and
exposed to normoxic conditions in a humidified atmosphere of 5%
CO2 and 95% O2, or hypoxic conditions in 5%
CO2 and 95% N2 in a hypoxic incubator chamber
for 24 h and then subjected to further experiments.
Cell transfection
lncRNA MIAT small interfering (si)RNA (si-MIAT-1 and
si-MIAT-2) and si-negative control (NC) were synthesized by
Shanghai GenePharma Co., Ltd. Cardiomyocytes were transfected with
100 nM of si-NC, si-MIAT-1 or si-MIAT-2 when the cells reached ~80%
confluence in 24-well plates using Lipofectamine® 2000
reagent (Life Technologies; Thermo Fisher Scientific, Inc.). After
48 h, cells were harvested for further experiments. The sequences
of the siRNAs are as follows: si-NC, 5′-CCCACGCACTTCCTGCAA-3′;
si-MIAT-1, 5′-CCTCTCATCTTTCATTCCAATCCTTA-3′; and si-MIAT-2,
5′-UCCUCCGAACCUGGCACGU-3′.
TUNEL assay
The TUNEL assay was performed followed instructions
of the in situ apoptosis detection kit (Roche Diagnostics).
The detection procedure was in accordance with a previous study
(15). Cells were exposed to
normoxic or hypoxic conditions, and fixed with 4% paraformaldehyde
at room temperature for 20 min. After permeabilization with 0.1%
Triton-X 100 for 2 min, cells were added to the TUNEL reaction
mixture (including TdT and fluorescein-dUTP) and incubated at 37°C
for 1 h. After washing with PBS, cells were incubated with 10 µg/ml
DAPI (Solarbio, Beijing) for 10 min at room temperature. The rate
of apoptosis was expressed as the ratio of TUNEL-positive
cardiomyocyte nuclei to the total number of cardiomyocyte
nuclei.
RNA extraction and reverse
transcription-quantitative PCR (RT-qPCR)
Total RNA was extracted using TRIzol®
reagent (Invitrogen; Thermo Fisher Scientific, Inc.). RT was
performed using a Prime Script™ RT reagent kit (Takara Bio,
Inc.)with the following conditions: 42°C for 15 min and 95°C for 3
min. The specific primers were synthesized by Sangon Biotech Co.,
Ltd., and the sequences of the primers were as follows: MIAT
forward, 5′-TAGCTCGAGTCTTTTTAGCTACTTCGACTACGGC-3′ and reverse,
5′-TCAAGAATGCGGACGCGACAGGATAGGCCACTTTGTC-3′; and GAPDH forward,
5′-TGTGTCCGTCGTGGATCTGA-3′ and reverse,
5′-CCTGCTTCACCACCTTCTTGA-3′. The lncRNA levels were quantified via
a standard RT-qPCR protocol with SYBR Premix Ex Taq (Takara Bio,
Inc.). The thermal cycling conditions were as follows: 95°C for 15
min, followed by 40 cycles of 95°C for 10 sec, 60°C for 30 sec and
72°C for 60 sec. The lncRNA levels were calculated based on the Cq
values and were normalized to GAPDH in each sample. The data were
analyzed using the 2−ΔΔCq method (16).
Western blotting
Total protein was extracted from tissues or cells
using RIPA lysis buffer containing protease inhibitor and PMSF
(Nanjing KeyGen Biotech Co., Ltd.). The supernatant protein
concentration was detected using a standard BCA assay (Nanjing
KeyGen Biotech Co., Ltd.). Protein samples (60 µg) were loaded into
the wells of 10 or 15% SDS-PAGE gels for electrophoresis, and
transferred to PVDF membranes. The membranes were blocked with 5%
nonfat milk at room temperature for 2 h and then incubated with
primary antibodies at 4°C overnight. The primary antibodies used
were as follows: Anti-caspase 3 (1:1,000; Cell Signaling
Technology, Inc.; cat. no. 9662), anti-cleaved caspase 3 (1:1,000;
Cell Signaling Technology, Inc.; cat. no. 9661), anti-Bax (1:1,000;
Cell Signaling Technology, Inc.; cat. no. 2772), anti-Bcl2
(1:1,000; Abcam; cat. no. ab196495) and anti-GAPDH (1:5,000; Abcam;
cat. no. ab9484) were used in this study. After washing with PBS
three times, the membranes were incubated with HRP-conjugated
secondary antibody (1:8,000; Abcam; cat. no. ab7090) for 2 h at
room temperature. ECL (EMD Millipore) was used to detect the
protein bands. The western blot bands were captured using a
ChemiDoc MP Imager (ChemiDoc™ MP imaging system; Bio-Rad,
Laboratories, Inc.) and analyzed with ImageJ v14.0 software
(National Institutes of Health).
Statistical analysis
All data are presented as the mean ± SD of three
independent experiments. Differences between groups were analyzed
using SPSS 19.0 (IBM Corp.) and GraphPad 6.0 (GraphPad Software,
Inc.) software with a Student's t-test or one-way ANOVA.
Comparisons between groups were performed using Tukey's or
Dunnett's tests, based on whether the variances were consistent.
Spearman's rho correlation coefficient was used to assess the
relationships among biomarkers. The sensitivity and specificity of
biomarkers were assessed using a receiver operating characteristic
(ROC) curve. P<0.05 was considered to indicate a statistically
significant difference.
Results
Pattern of plasma lncRNA MIAT levels
in the patients with AMI
A total of 58 patients with AMI and 50 subjects with
UA were included for the detection of circulating MIAT levels. The
baseline characteristics of the subjects are shown in Table I. There were significant
differences in smoking status, coronary artery disease, left
ventricular ejection fraction, CK-MB and cTnT (P<0.05). As shown
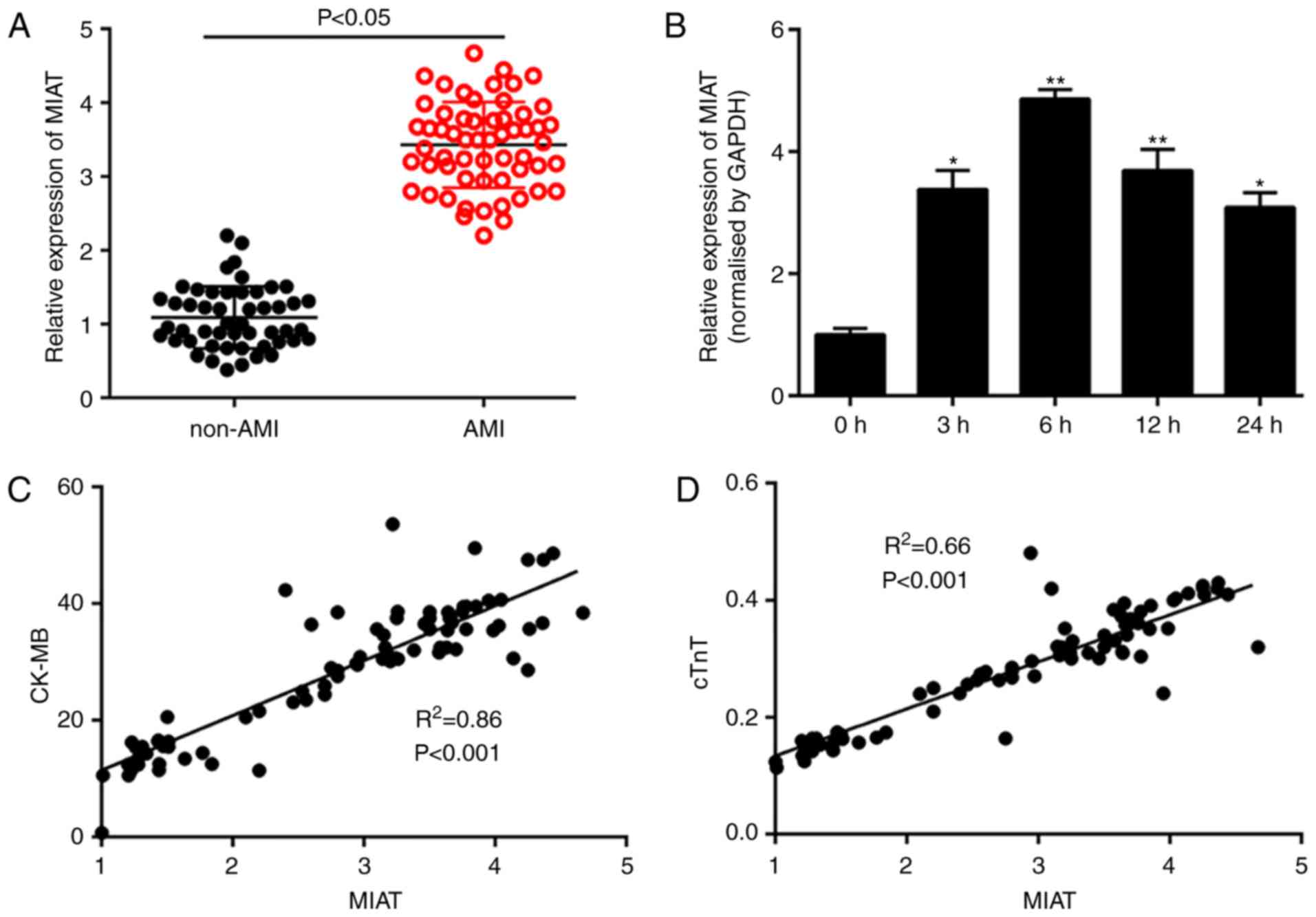
in Fig. 1A, the plasma level of
MIAT in the AMI group was significantly upregulated by
approximately three times compared with that of the UA group. In
addition, dynamic monitoring showed that MIAT was significantly
upregulated within 3 h of the onset of ischemic symptoms, and
reached its highest level at 6 h, making it more sensitive than
cTnT or CK-MB (Fig. 1B) (17). Moreover, correlation analysis
indicated that the expression trend of MIAT was positively
correlated to those of CK-MB and cTnT, particularly with CK-MB
(r=0.86; P<0.01; Fig. 1C and
D). These results suggested that the expression level of MIAT
is closely related to myocardial injury, and that circulating MIAT
may be detected within hours of the onset of chest pain.
 | Table I.Demographic and clinical baseline
characteristics. |
Table I.
Demographic and clinical baseline
characteristics.
| Variable | UA group (n=50) | AMI group (n=58) | P-value |
|---|
| Age, years | 60.4 | 61.64 | 0.41 |
| Male/female, n/n | 26/24 | 33/25 | 0.69 |
| Currently smoking, n
(%) | 18 (36) | 33
(56.8)a | 0.02 |
| Heart rate,
beats/min | 72.2 (67.3,
76.9) | 75.4 (71, 83) | 0.37 |
| SBP, mmHg | 120 (115, 125) | 118 (110, 127) | 0.61 |
| DBP, mmHg | 73.64±4.39 | 72.2±11.36 | 0.53 |
| Hypertension, n
(%) | 12 (24) | 16 (27.3) | 0.14 |
| Diabetes mellitus, n
(%) | 8 (16) | 22 (37.9) | 0.03 |
| Coronary artery
disease, n (%) | 25 (50) | 48
(82.5)a | <0.01 |
| LVEF, % | 60.72 | 52.44a | <0.01 |
| CK-MB (U/l) | 3.2 | 38.65a | <0.01 |
| cTnT, ng/ml | 0.12 | 0.49a | <0.01 |
| eGFR, ml/min/1.73
m2 | 91.5 (83.5,
102.8) | 96.7 (86.3,
104) | 0.35 |
lncRNA MIAT is a sensitive biomarker
that reflects the extent of myocardial injury
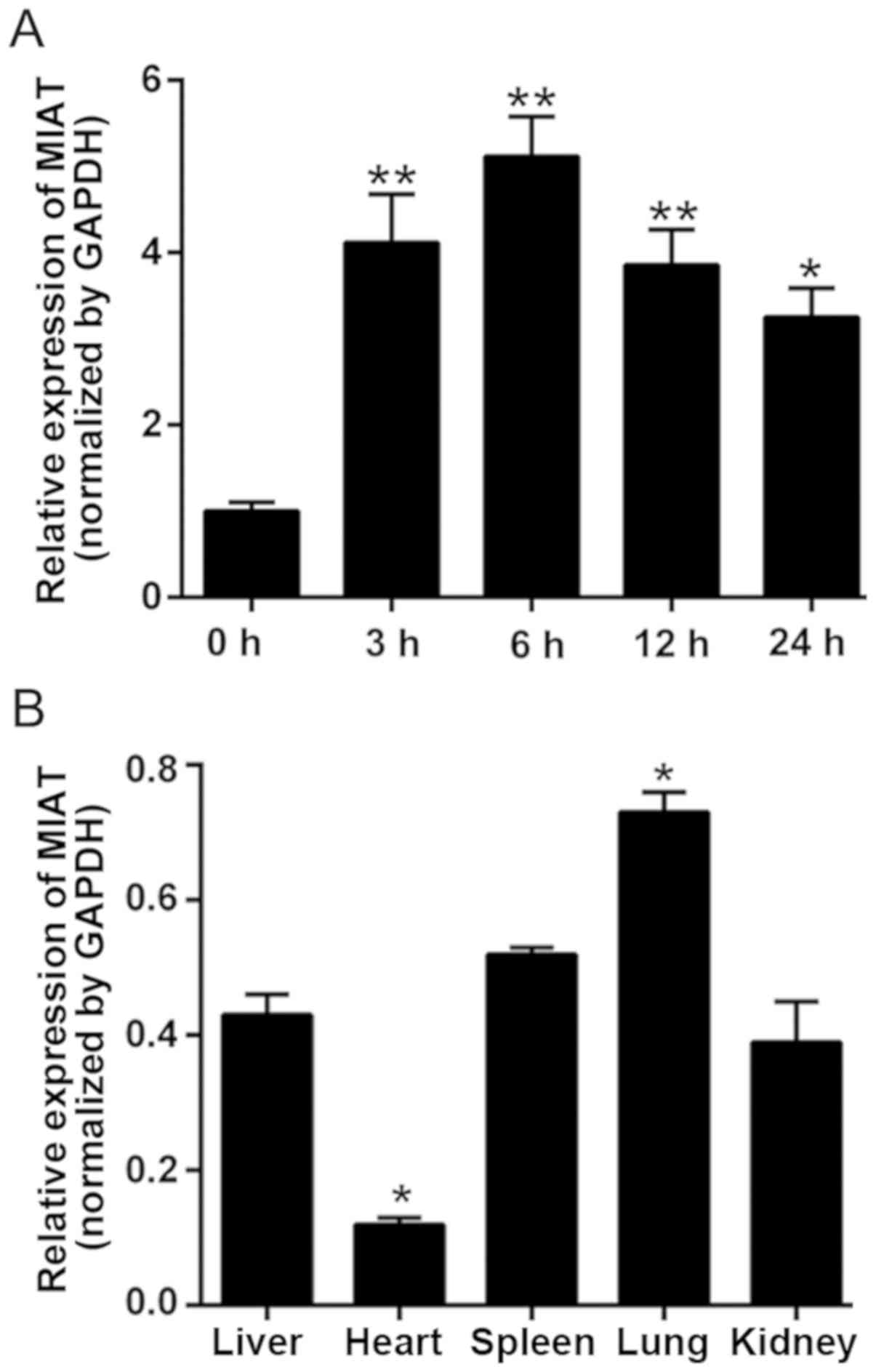
To validate the expression trend of MIAT, a rat AMI
model was established. As presented in Fig. 2A, the expression level of MIAT was
significantly increased within 3 h, indicating that the dynamic
trend of MIAT expression in rat plasma is similar to that in
humans. In addition, the expression levels of MIAT were detected in
different organs to further understand its potential as a biomarker
of myocardial damage. As expected, MIAT was expressed at the lowest
level in the normal myocardium, which was obviously opposite in AMI
tissues (Fig. 2B). The results
indicated that the expression level of MIAT reflects the state of
myocardial injury. Monitoring the level of MIAT may be beneficial
for understanding the condition of patients with myocardial
infarction.
Diagnostic value of lncRNA MIAT in
patients with AMI
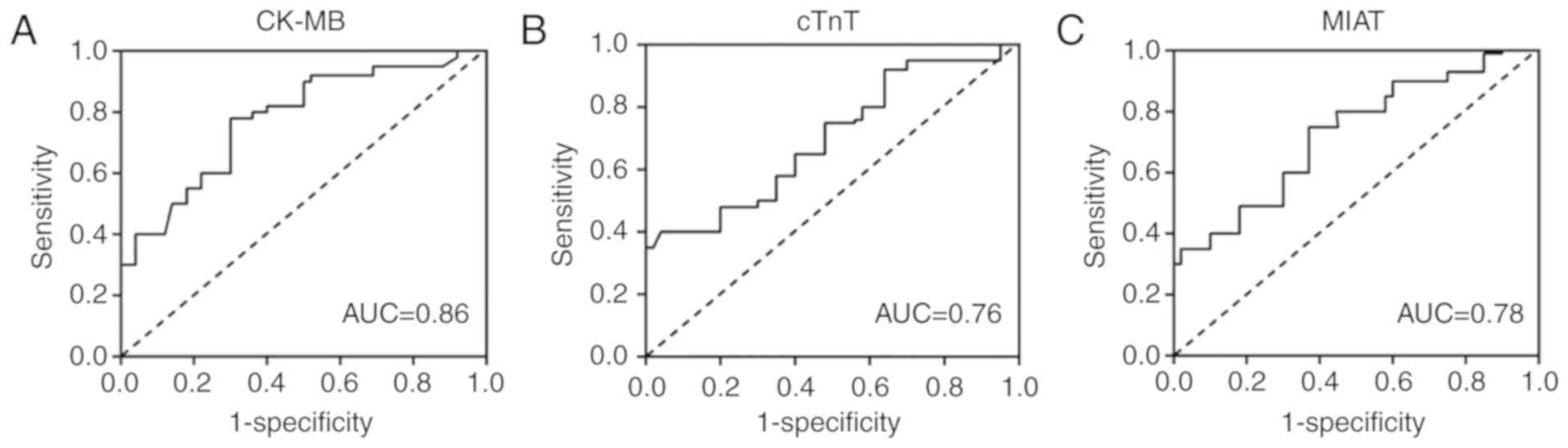
ROC curve analysis was performed to test the
reliability of MIAT as a biomarker for diagnosing AMI. As shown in
Fig. 3, CK-MB provided the
greatest diagnostic value [area under the curve (AUC)=0.86; 95% CI,
0.814–0.0.92], while cTnT and MIAT obtained AUC values of 0.76 and
0.78, respectively. These results indicated that MIAT has the same
value as cTnT in the diagnosis of AMI.
Knockdown of MIAT alleviates
cardiomyocyte apoptosis
To characterize the functional role of MIAT in
myocardial injury, a loss-of-function approach was used in neonatal
rat cardiomyocytes. As shown in Fig.
4A, the expression of MIAT was significantly increased after
exposure to hypoxia for 24 h, and this result was consistent with
the results in the plasma of patients with AMI and AMI rats.
Subsequently, MIAT siRNAs were transfected into cell and the
expression of MIAT was significantly decreased in the si-MIAT-1 and
si-MIAT-2 groups (Fig. 4B).
Moreover, the TUNEL assay showed that MIAT knockdown significantly
repressed cardiomyocyte apoptosis (Fig. 4C and D). Caspases are a family of
cysteine proteases that play essential roles in cell apoptosis.
Cleaved caspase 3 was increased when cells were exposed to hypoxia,
and MIAT knockdown significantly decreased caspase 3 activity.
Besides, the pro-apoptotic protein Bax and the anti-apoptotic
protein Bcl2 were detected using western blotting. The results
indicated that knockdown of MIAT significantly decreased the
expression of Bax and increased the expression of Bcl2 (Fig. 4E and F). These results indicated
that MIAT knockdown may prevent the apoptosis of cardiomyocytes by
regulating the expression of Bcl2 family proteins.
 | Figure 4.Knockdown of MIAT alleviates
cardiomyocyte apoptosis. (A) Cardiomyocytes were exposed to hypoxia
for 24 h, and the expression of MIAT was measured using RT-qPCR.
(B) Cells were transfected with MIAT siRNAs, and relative
expression of MIAT was detected using RT-qPCR. Comparisons between
groups were performed using Tukey's test. (C) Cells were incubated
with TUNEL reaction mixture and DAPI stain, and representative
images indicated that condensed nuclei were present (magnification,
×200). (D) The relative apoptotic cell percentages were calculated
at least three times. Comparisons between groups were performed
using Tukey's test. (E) Cells were collected and expression levels
of cleaved caspase 3, caspase 3, Bax and Bcl2 were detected using
western blotting. (F) Relative protein quantitative differences
were calculated in the different treatment groups. Comparisons
between groups were performed using Tukey's test. The data are
presented as the mean ± SD. *P<0.05, **P<0.01 vs. respective
control group; #P<0.05, ##P<0.01 vs.
respective si-NC group. RT-qPCR, reverse transcription-quantitative
PCR; siRNA, small interfering RNA; NC, negative control; MIAT,
myocardial infarction associated transcript. |
Discussion
AMI has become a major public health problem, owing
to its high mortality and morbidity. Therefore, there is an urgent
need to discover new biomarkers for better and faster diagnosis of
AMI. The present study found enough advantages of MIAT as a novel
biomarker to diagnose AMI. Firstly, MIAT was rapidly upregulated by
two times within 3 h of ischemic symptom onset, and reached a peak
at 6 h. Secondly, correlation analysis and ROC analysis
demonstrated that MIAT had the same diagnostic value as cTnT.
Thirdly, MIAT is specifically expressed at low levels in the heart,
and rapidly increases during the onset of MI, which may be
beneficial for monitoring the condition of the myocardium in
patients with AMI. A previous study reported that the expression
level of MIAT was correlated with a high risk of mortality from
AMI, suggesting that MIAT may play a role in promoting
cardiomyocyte apoptosis and even mortality (12).
Several lncRNAs have been implicated as regulators
of the cardiovascular system. lncRNA-p21 is induced by p53 to
inhibit smooth muscle cell proliferation and apoptosis (18). lncRNA Mhrt, as a cardiac-specific
lncRNA, plays an important role in preventing cardiac remodeling
and hypertrophy by regulating the chromatin-binding protein
transcription activator BRG1 (19). MIAT, also known as RNCR2, AK028326
or GOMAFU, was first reported to be expressed in mitotic
progenitors and post-mitotic retinal precursor cells (20). Subsequent studies have confirmed
that MIAT is correlated with the progression of multiple diseases,
including tumor proliferation and apoptosis (21), a high risk of mortality from AMI
(12,22), microvascular dysfunction (23) and neuronal activity (24).
A large clinical trial (25) compared MIAT levels in peripheral
blood cells between patients with MI and healthy subjects, and the
results indicated that there was no statistical difference in MIAT
levels between the two groups. However, the difference in
expression of MIAT appears between patients with STEMI and those
with non-ST-segment elevated myocardial infarction. Ishii et
al (12) identified that the
aberrant expression of MIAT with SNP rs2301523 was related to the
pathogenesis of MI, which is consistent with the present results.
Yan et al (23)
demonstrated that the upregulation of MIAT induced by high glucose
leads to diabetic microvascular dysfunction. In the present study,
the proportion of patients with concomitant diabetes was higher in
the AMI group than in the non-AMI group, which may be a reason for
the difference in MIAT expression between the two groups.
Early rescue of apoptosis in cardiomyocytes induced
by ischemia and hypoxia is the key to the treatment of AMI. With
more in-depth study of lncRNAs, their influence on cardiomyocyte
apoptosis and pathological mechanisms may be further revealed,
which will provide a theoretical basis for the role of lncRNAs in
clinical cardiovascular disease and anti-atherosclerosis therapies.
The pro-apoptotic effect of MIAT in cardiomyocytes, by affecting
the expression of death-associated protein kinase 2 (DAPK2), has
been demonstrated in a rat diabetic cardiomyopathy model (26). Specifically, it may act as a
competitive endogenous RNA, upregulating DAPK2 expression by
sponging miR-22-3p, which leads to cardiomyocyte apoptosis.
There are some limitations to the present study. A
larger sample size would allow for investigating the possibility of
using MIAT as an AMI biomarker. Animal experiments are needed to
further verify the role of MIAT in promoting cardiomyocyte
apoptosis, and its regulatory mechanism at the transcriptional
level.
The present study evaluated the plasma levels and
functional role of lncRNA MIAT in the process of AMI to investigate
the potential of MIAT as a biomarker for diagnosing AMI. Moreover,
in vivo and in vitro experimental results showed that
MIAT was able to sensitively reflect the degree of myocardial
injury and that MIAT knockdown markedly suppressed cardiomyocyte
apoptosis. These findings provide a new insight into the potential
role of MIAT in the diagnosis of AMI, and its functional role in
AMI pathogenesis.
Acknowledgements
Not applicable.
Funding
No funding was received.
Availability of data and materials
The datasets used and/or analyzed during the present
study are available from the corresponding author on reasonable
request.
Authors' contributions
MA was responsible for drafting the manuscript,
analysis and interpretation of data. AA and XH were responsible for
acquisition of data. RG was responsible for searching documents and
acquisition of data. PP was responsible for the design of the study
and revising it critically for important intellectual content.
Ethics approval and consent to
participate
Written informed consent was obtained from all
enrolled subjects and the study protocol was approved by the Ethics
Committee of The First Affiliated Hospital of Xinjiang Medical
University. All animal procedures were approved by the Experimental
Animal Ethics Committee of The First Affiliated Hospital of
Xinjiang Medical University.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
Glossary
Abbreviations
Abbreviations:
|
lncRNAs
|
long noncoding RNAs
|
|
AMI
|
acute myocardial infarction
|
|
MIAT
|
myocardial infarction associated
transcript
|
|
LDH
|
lactate dehydrogenase
|
|
ECG
|
electrocardiogram
|
|
cTnT
|
cardiac troponin T
|
|
CK-MB
|
creatine kinase-MB
|
References
|
1
|
Puaschitz NG, Assmus J, Strand E, Karlsson
T, Vinknes KJ, Lysne V, Drevon CA, Tell GS, Dierkes J and Nygård O:
Adherence to the Healthy Nordic Food Index and the incidence of
acute myocardial infarction and mortality among patients with
stable angina pectoris. J Hum Nutr Diet. 32:86–97. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Khan DA, Sharif MS and Khan FA: Diagnostic
performance of high-sensitivity troponin T, myeloperoxidase, and
pregnancy-associated plasma protein A assays for triage of patients
with acute myocardial infarction. Korean J Lab Med. 31:172–178.
2011. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Omran MM, Zahran FM, Kadry M, Belal AAM
and Emran TM: Role of myeloperoxidase in early diagnosis of acute
myocardial infarction in patients admitted with chest pain. J
Immunoassay Immunochem. 39:337–347. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Tan J and Yang L: Long noncoding RNA
VPS9D1-AS1 overexpression predicts a poor prognosis in non-small
cell lung cancer. Biomed Pharmacother. 106:1600–1606. 2018.
View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Yang Z, Li H, Li J, Lv X, Gao M, Bi Y,
Zhang Z, Wang S, Li S, Li N, et al: Association between long
noncoding RNA MEG3 polymorphisms and lung cancer susceptibility in
chinese northeast population. DNA Cell Biol. 37:812–820. 2018.
View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Viereck J and Thum T: Circulating
noncoding RNAs as biomarkers of cardiovascular disease and injury.
Circ Res. 120:381–399. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
El-Hefnawy T, Raja S, Kelly L, Bigbee WL,
Kirkwood JM, Luketich JD and Godfrey TE: Characterization of
amplifiable, circulating RNA in plasma and its potential as a tool
for cancer diagnostics. Clin Chem. 50:564–573. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Lorenzen JM, Schauerte C, Kölling M,
Hübner A, Knapp M, Haller H and Thum T: Long noncoding RNAs in
urine are detectable and may enable early detection of acute T
Cell-mediated rejection of renal allografts. Clin Chem.
61:1505–1514. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Bär C, Chatterjee S and Thum T: Long
noncoding RNAs in cardiovascular pathology, diagnosis, and therapy.
Circulation. 134:1484–1499. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Kumarswamy R, Bauters C, Volkmann I, Maury
F, Fetisch J, Holzmann A, Lemesle G, de Groote P, Pinet F and Thum
T: Circulating long noncoding RNA, LIPCAR, predicts survival in
patients with heart failure. Circ Res. 114:1569–1575. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Yan Y, Zhang B, Liu N, Qi C, Xiao Y, Tian
X, Li T and Liu B: Circulating long noncoding RNA UCA1 as a novel
biomarker of acute myocardial infarction. Biomed Res Int.
2016:80793722016. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Ishii N, Ozaki K, Sato H, Mizuno H, Saito
S, Takahashi A, Miyamoto Y, Ikegawa S, Kamatani N, Hori M, et al:
Identification of a novel non-coding RNA, MIAT, that confers risk
of myocardial infarction. J Hum Genet. 51:1087–1099. 2006.
View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Arslan F, Bongartz L, Ten Berg JM, Jukema
JW, Appelman Y, Liem AH, de Winter RJ, van't Hof AWJ and Damman P:
2017 ESC guidelines for the management of acute myocardial
infarction in patients presenting with ST-segment elevation:
Comments from the Dutch ACS working group. Neth Heart J.
26:417–421. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Diao H, Kang Z, Han F and Jiang W:
Astilbin protects diabetic rat heart against ischemia-reperfusion
injury via blockade of HMGB1-dependent NF-kB signaling pathway.
Food Chem Toxicol. 63:104–110. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Lv X, Yu X, Wang Y, Wang F, Li H, Wang Y,
Lu D, Qi R and Wang H: Berberine inhibits doxorubicin-triggered
cardiomyocyte apoptosis via attenuating mitochondrial dysfunction
and increasing Bcl-2 expression. PLoS One. 7:e473512012. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(-Delta Delta C(T)) method. Methods. 25:402–408. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Ndrepepa G, Colleran R, Braun S, Xhepa E,
Hieber J, Cassese S, Fusaro M, Kufner S, Laugwitz KL, Schunkert H
and Kastrati A: Comparative prognostic value of postprocedural
creatine kinase myocardial band and high-sensitivity troponin T in
patients with non-ST-segment elevation myocardial infarction
undergoing percutaneous coronary intervention. Catheter Cardiovasc
Interv. 91:215–223. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Wu G, Cai J, Han Y, Chen J, Huang ZP, Chen
C, Cai Y, Huang H, Yang Y, Liu Y, et al: LincRNA-p21 regulates
neointima formation, vascular smooth muscle cell proliferation,
apoptosis, and atherosclerosis by enhancing p53 activity.
Circulation. 130:1452–1465. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Han P, Li W, Lin CH, Yang J, Shang C,
Nuernberg ST, Jin KK, Xu W, Lin CY, Lin CJ, et al: A long noncoding
RNA protects the heart from pathological hypertrophy. Nature.
514:102–106. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Qu Y, Xiao H, Xiao W, Xiong Z, Hu W, Gao
Y, Ru Z, Wang C, Bao L, Wang K, et al: Upregulation of MIAT
regulates LOXL2 expression by competitively binding MiR-29c in
clear cell renal cell carcinoma. Cell Physiol Biochem.
48:1075–1087. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Luan T, Zhang X, Wang S, Song Y, Zhou S,
Lin J, An W, Yuan W, Yang Y, Cai H, et al: Long non-coding RNA MIAT
promotes breast cancer progression and functions as ceRNA to
regulate DUSP7 expression by sponging miR-155-5p. Oncotarget.
8:76153–76164. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Li Y, Wang J, Sun L and Zhu S: LncRNA
myocardial infarction-associated transcript (MIAT) contributed to
cardiac hypertrophy by regulating TLR4 via miR-93. Eur J Pharmacol.
818:508–517. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Yan B, Yao J, Liu JY, Li XM, Wang XQ, Li
YJ, Tao ZF, Song YC, Chen Q and Jiang Q: lncRNA-MIAT regulates
microvascular dysfunction by functioning as a competing endogenous
RNA. Circ Res. 116:1143–1156. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Barry G, Briggs JA, Vanichkina DP, Poth
EM, Beveridge NJ, Ratnu VS, Nayler SP, Nones K, Hu J, Bredy TW, et
al: The long non-coding RNA Gomafu is acutely regulated in response
to neuronal activation and involved in schizophrenia-associated
alternative splicing. Mol Psychiatry. 19:486–494. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Vausort M, Wagner DR and Devaux Y: Long
noncoding RNAs in patients with acute myocardial infarction. Circ
Res. 115:668–677. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Zhou X, Zhang W, Jin M, Chen J, Xu W and
Kong X: lncRNA MIAT functions as a competing endogenous RNA to
upregulate DAPK2 by sponging miR-22-3p in diabetic cardiomyopathy.
Cell Death Dis. 8:e29292017. View Article : Google Scholar : PubMed/NCBI
|