Introduction
Pulmonary arterial hypertension (PAH) is an
intractable cardiovascular disease, a key feature of which is
pulmonary vascular remodeling (PVR) (1). Hypoxia is considered to be the
predominant factor in the pathogenesis of PAH (2,3).
During the hypoxic exposure, the dysfunction of pulmonary artery
smooth muscle cells (PASMCs), including both proliferation and
migration, is the major reason for medial hypertrophy in PVR
(4). To limit morbidity and
mortality, attention has been focused on identifying the cellular
and molecular mechanisms underlying aberrant proliferation and
migration in human (H)PASMCs. However, the mechanism of PASMCs
proliferation and migration at the molecular level is still not
very clear, and further research is needed.
Long non-coding RNAs (lncRNAs), with transcripts
>200 nucleotides in length, were once considered as irrelevant
transcriptional ‘noise’ without biological function. Previous
studies, however, have shown that lncRNAs play a significant role
in diverse biological and disease processes (5). LncRNAs can be used as a ‘scaffold’ to
bind a variety of proteins to exert biological functions (6). Also, lncRNAs can form an anchor point
to regulate different bioactivities by collecting or sequestering
certain protein factors and participating in the synthesis and
reconstruction of nucleic acid sequences (7).
On chromosome 9p21, there is a lncRNA named
antisense noncoding RNA in the INK4 locus (ANRIL) with ~126,000 bps
(8). LncRNA ANRIL was first
identified and named in the investigation of a melanoma-neural
system tumor syndrome family (9).
A previous study identified that ANRIL is associated with ~40% of
all tumor types (10). Moreover,
ANRIL also plays an important role in the development and
progression of various diseases to a certain degree, including
coronary heart disease (11,12).
Knocking down the expression of ANRIL in human aortic vascular
smooth muscle cells, the results obtained suggested that ANRIL
splicing variants played a role in coordinating tissue remodeling
(13). Based on the above
evidence, it is possible to hypothesize that ANRIL may regulate the
process of PAH and that this deserves further investigation. Based
on the potential roles of ANRIL in maintaining cellular functions,
the present study speculated that ANRIL participates in
hypoxia-induced proliferation and migration of HPASMCs in PAH. To
this end, the expression of ANRIL was studied in HPASMCs where PAH
had been induced by hypoxia.
Materials and methods
Materials
Antibodies against proliferating cell nuclear
antigen (PCNA), Cyclin A, Cyclin D and Cyclin E were purchased from
Boster Biological Technology. Antibody against Ki67 was purchased
from Abcam. Bromodeoxyuridine (BrdU) proliferation assay kit was
purchased from EMD Millipore. DMEM was purchased from Hyclone; GE
Healthcare Life Sciences. The remaining chemical reagents were
domestic analytical pure and biochemical reagents.
Cell culture
HPASMCs were obtained from the Center Laboratory of
Harbin Medical University Daqing Campus. The cells were secondary
cultured in 20% Clark serum (CLARK Bioscience)-DMEM at 37°C in a 5%
CO2 humidified incubator. HPASMCs were cultured with a
gas mixture containing 92% N2, 5% CO2 and 3%
O2 to create hypoxic conditions.
Small interfering (si)RNA and
transfection
The sequence of siRNA against lncRNA ANRIL and
scrambled negative control siRNA (si-NC) were synthesized by
Shanghai GenePharma Co., Ltd. The target sequences of these siRNAs
were follows: Si-lncRNA ANRIL,
5′-GCCCAAGCAUAUAGAUCAATTUUGAUCUAUAUGCUUGGGCTT-3′ and si-NC,
5′-UUCUCCGAACGUGUCACGUTTACGUGACACGUUCGGAGAATT-3′. HPASMCs cultured
on six-well plates were transfected with the si-lncRNA ANRIL or
si-NC (3.75 µl per 1.1 ml DMEM) using X-tremeGENE (Sigma-Aldrich;
Merck KGaA) (14). A total of 24 h
post transfection, cells were harvested for quantitative (q)PCR and
western blotting analyses.
Revers transcription-qPCR
Total RNA was extracted and isolated from HPASMCs
using TRIzol reagent (Invitrogen; Thermo Fisher Scientific, Inc.).
For the qPCR assay, the total RNA was reverse transcribed (15 min
at 37°C, 5 min at 50°C, 5 min at 98°C and holding at 4°C.) into
cDNA using One-step cDNA First Strand Reverse Transcription kit
(HaiGene) according to the manufacturer's protocol. The expression
levels were normalized against the expression of GAPDH. The PCR
primers were as follows: ANRIL forward, 5′-TCCTGCCACTTCCTCTACTGA-3′
and reverse, 5′-TGGTATGGAAGGTGCTATGGA-3′; GAPDH forward,
5′-CAATGACCCCTTCATTGACC-3′ and reverse, 5′-TGGAAGATGGTGATGGGATT-3′.
The amplification reaction using SYBR® Green Realtime
PCRMaster Mix was performed for 1 min at 95°C, 15 min at 95°C,
followed by 40 cycles at 95°C for 15 sec and 52°C for 15 sec. The
relative quantification of target genes expression was relative to
the GAPDH levels. The 2−ΔΔCq method was used to
calculate the expression of lncRNA ANRIL (15). All qPCR reactions were performed in
duplicate.
Western blotting
Proteins were solubilized and extracted with lysis
buffer (Tris 50 mM, pH 7.4, NaCl 150 mM, Triton X-100 1%, EDTA 1
mM, and PMSF 2 mM) and incubated for 30 min on ice. The lysates
were sonicated for 30 sec at 25 kHz in an ice bath and centrifuged
at 15,000 × g for 15 min at 4°C and the insoluble fractions were
discarded. Protein concentrations were determined by the Bradford
assay using bovine serum albumin (Sigma-Aldrich; Merck KGaA) as
standard. Cell samples containing 20 µg of protein were subjected
to electrophoresis on a 10% SDS-polyacrylamide gel. Following
electrophoresis, proteins were transferred to nitrocellulose
membranes. These membranes were blocked at room temperature for 1 h
in blocking buffer (Tris 20 mM, pH 7.6, NaCl 150 mM and Tween 20
0.1%) containing 5% nonfat dry milk and incubated with PCNA (Boster
Biological Technology; cat. no. BM3888; anti-PCNA antibody; 1:400),
cyclin A (Boster Biological Technology; cat. no. BM4673;
anti-Cyclin A2 antibody; 1:200), cyclin D (Boster Biological
Technology; cat. no. BM4272; anti-Cyclin D1 antibody; 1:200),
cyclin E (Boster Biological Technology; cat. no. BM4658;
anti-Cyclin E2 antibody; 1:100), β-actin (Boster Biological
Technology; cat. no. BM0627; anti-β-actin antibody; 1:2,000) at 4°C
overnight. Secondary antibodies [Beyotime institute of
Biotechnology; cat. no. A0208; horseradish peroxidase (HRP)-labeled
Goat Anti-Rabbit IgG(H+L); 1:10,000; cat. no. A0216; HRP-labeled
Goat Anti-Mouse IgG(H+L); 1:12,000] were prepared according to the
corresponding proportion and species, and incubated at room
temperature for 1 h. The proteins were visualized with enhanced
chemiluminescence reagents (Super Signal; Pierce; Thermo Fisher
Scientific, Inc.). Grayscale values were calculated using Quantity
One v4.6.2 (Bio-Rad Laboratories, Inc.) and statistical data were
obtained using Microsoft Excel and Origin 7.5 (OriginLab).
Immunofluorescence assay
The pre-configured primary antibody against Ki67
(Santa Cruz Biotechnology, Inc.; cat. no. sc-23900, Ki-67 Antibody;
1:50) was added to the cells of different components and incubated
at 4°C overnight. Then the cells were incubated with the secondary
antibody [Invitrogen; Thermo Fisher Scientific, Inc.; cat. no.
R-6393; Goat anti-Mouse IgG (H+L)] (1:100) conjugated with
rhodamine for 1 h in the dark at 37°C and washed with PBS. The
nuclei were stained with 4,6-diamidino-2-phenylindole (DAPI) at
room temperature for 5 min. The slides were examined using NA1.4
inverted fluorescence microscope (Leica DMI6000; Leica
Microsystems, Inc.), images were visualized by Hamamatsu ORCA-R2
camera (Hamamatsu Photonics K.K.) and recorded by LAS AF 2.6.0
software (Leica Microsystems, Inc.). The experiments were conducted
in triplicate.
Scratch-wound assay
The confluent HPASMCs cultured in 6-well plates were
wounded by pipette tips and the detached cells were washed out by
PBS (16). After that, the cells
were treated with vehicle or the chemicals of interest in 5%
FBS-DMEM. The cells in all study groups were imaged in the same
area of the culture plate immediately after 24 h after the wound
was inflicted.
Transwell assay
Cell migration was measured using a
Matrigel®-coated modified Boyden chamber with a
polycarbonate filter with a pore size of 8 µm. A total of
5×104 HPASMCs was added to each upper chamber in a
serum-free medium. The lower chambers contained 20% Clark serum
(CLARK Bioscience)-DMEM. After 24 h of incubation at 37°C in a 5%
CO2 humidified incubator, the nonmigrating cells in the
upper chamber removed. The cells on the underside of the membrane
were incubated in 4% formaldehyde solution for 10 min at room
temperature, followed by incubation in 0.4% Crystal Violet for 5
min at room temperature. The number of migrated cells was measured
by counting the number of stained nuclei per high power field using
a light microscope (Olympus Corporation). Each sample was counted
randomly at nine separate locations in the center of the membrane
and HPASMCs migration activity was reported as the number of cells
migrating per field of view. The experiments were performed at
least three times in quadruplicate.
BrdU incorporation
HPASMCs were plated in 96-well plates at a density
of 1×104 cells/well and then transfected by siRNA after
growth arrest for 24 h. BrdU was added to a final concentration of
30 µM and incubated for 30–60 min. BrdU was removed and washed 3
times with PBS. 4% paraformaldehyde was used to fix cells at room
temperature for 30 min. Then cells were washed three times with
PBS. The following steps were done according to the manufacturer's
protocol. Finally, the plate was read using a spectrophotometer
microplate reader at dual wavelengths of 450/550 nm.
3-(4,5-Dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide (MTT)
assay
After the model was established, the cells were
drained of the original culture solution and 90 µM culture solution
and 10 µM MTT (5 mg·ml−1) were added to each well
(17). The cultures were incubated
at 37°C for 4 h, then drained of liquid and 150 µl DMSO was added
to each well. The absorbance was read at 490 nm using a
spectrophotometer, to determine the cell viability.
Cell cycle analysis
The proportions of cells in the G0/G1, S and G2/M
phases were detected using a flow cytometer. The cell cycle
analysis kit (Beyotime institute of Biotechnology) was used. The
cells were digested by trypsin then centrifuged at 1,000 × g for 5
min at room temperature to remove the supernatant. A total of 1 ml
ice bath precooled PBS buffer was added to suspend the cells and
centrifuged again (1 000 × g, 5 min, room temperature). A total of
1 ml 70% ethanol precooled by ice bath was added and mixed. Cells
were fixed at 4°C for 2 h or longer then centrifuged at 1,000 × g
for 3–5 min at room temperature. A total of 1 ml ice bath precooled
PBS buffer was added then the mixture was centrifuged again (1,000
× g, 5 min, room temperature) and discard the supernatant. The
prepared staining solution was added according to the protocol of
the kit and the cell precipitation was resuspended. The cells were
then placed in a temperature bath at 37°C for half an hour.
Finally, the cells were filtered once through 400-mesh sieves and
detected by flow cytometry (CytoFLEX, Beckman Coulter, Inc.). The
results were analyzed by CytExpert 2.0 (Beckman Coulter, Inc.).
Statistical analysis
Each experiment was repeated ≥3 times. The composite
data were expressed as the mean ± standard error of the mean.
Statistical analysis was performed with analysis of variance
followed by Dunnett's test or Student's t-test or Pearson
correlation test. Quantity One-4.6.2 (Bio-Rad Laboratories, Inc.),
Microsoft Office Excel 2007 (Microsoft Corporation), Origin 7.5
(OriginLab) and IPP 6.0 (Media Cybernetics, Inc.) software were
used to analyze data. P<0.05 was considered to indicate a
statistically significant difference.
Results
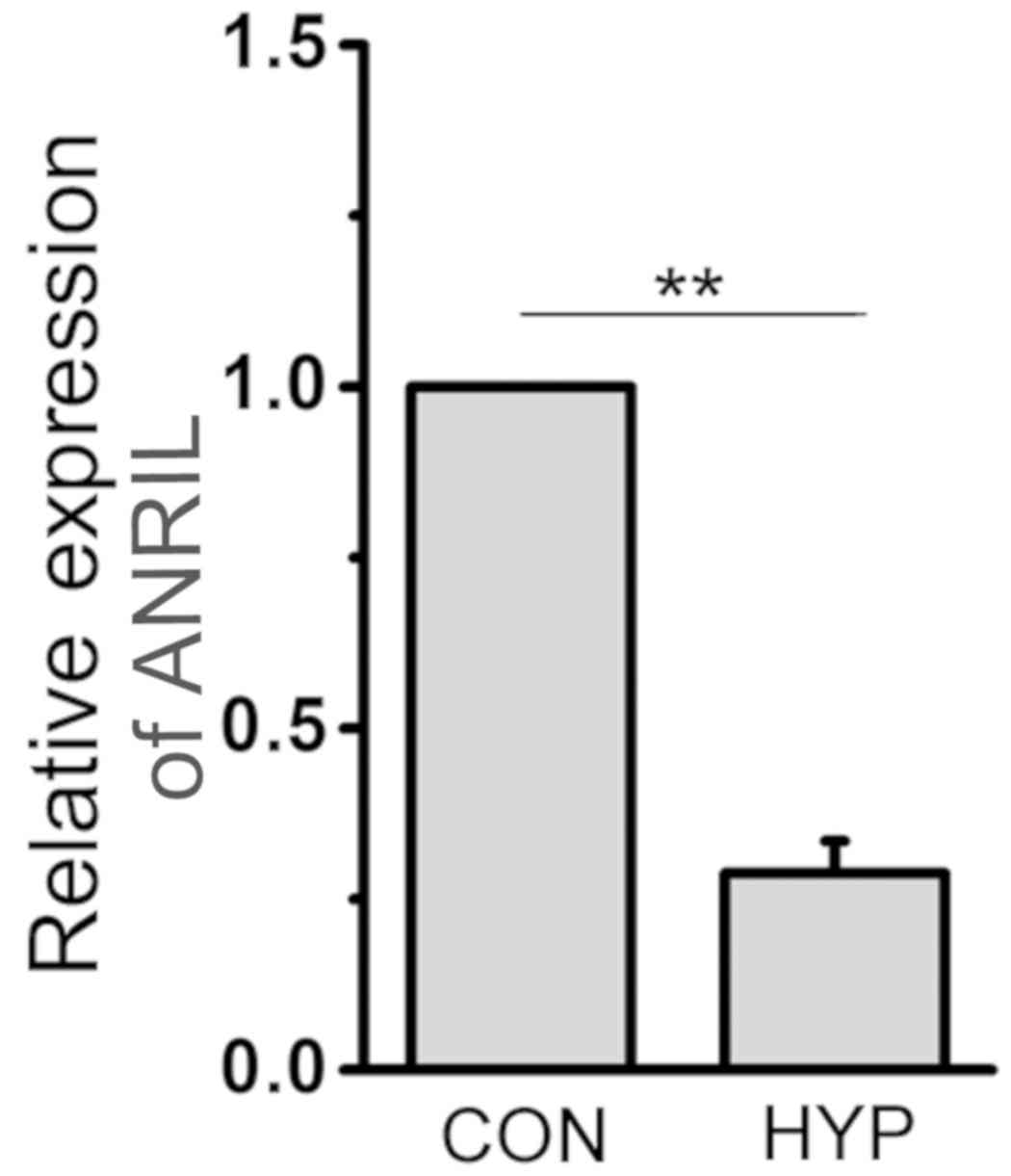
ANRIL expression is downregulated in
hypoxic HPASMCs
In order to prove that lncRNA ANRIL was related to
hypoxic PAH, the expression of ANRIL in HPASMCs exposed to both
normal and hypoxic conditions was detected by qPCR. The results
revealed that the expression of ANRIL in hypoxic HPASMCs was
significantly downregulated compared with that in normoxic cells
(P<0.01; Fig. 1).
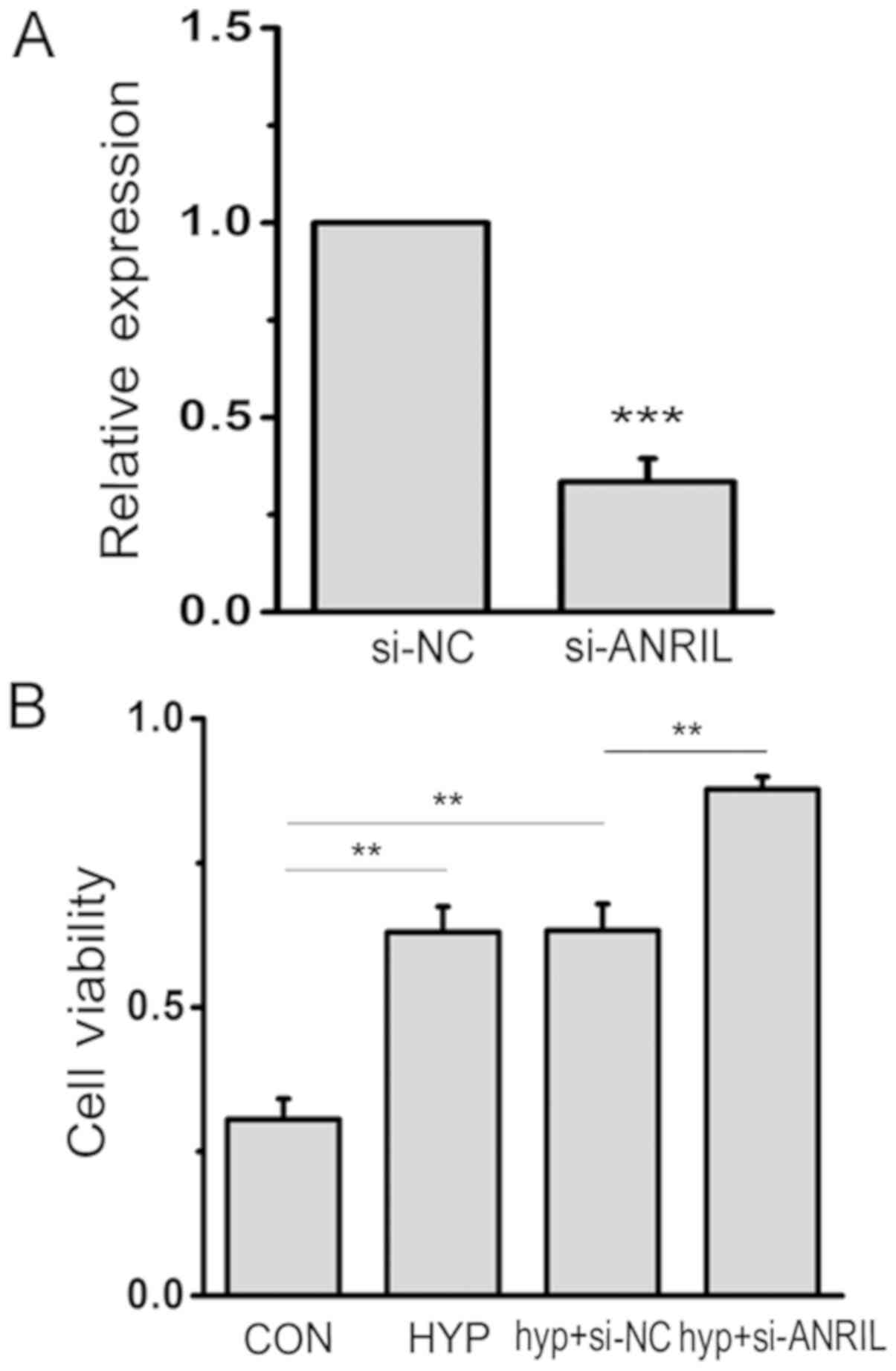
ANRIL regulates the viability of
HPASMCs
To determine the biological function of ANRIL in
HPASMCs under hypoxic conditions, siRNA targeting lncRNA ANRIL were
designed to inhibit the expression. The efficiency and specificity
of siRNA transfection and the downregulation of endogenous lncRNA
ANRIL were confirmed by qPCR (Fig.
2A). Meanwhile, to explore the effects of ANRIL on cell
survival ability, the viability of HPASMCs was examined by MTT
assay. The result revealed the significantly increased viability of
hypoxic HPASMCs compared with those under conditions of normoxia,
which was strengthened by si-ANRIL under hypoxic conditions
(P<0.05; Fig. 2B).
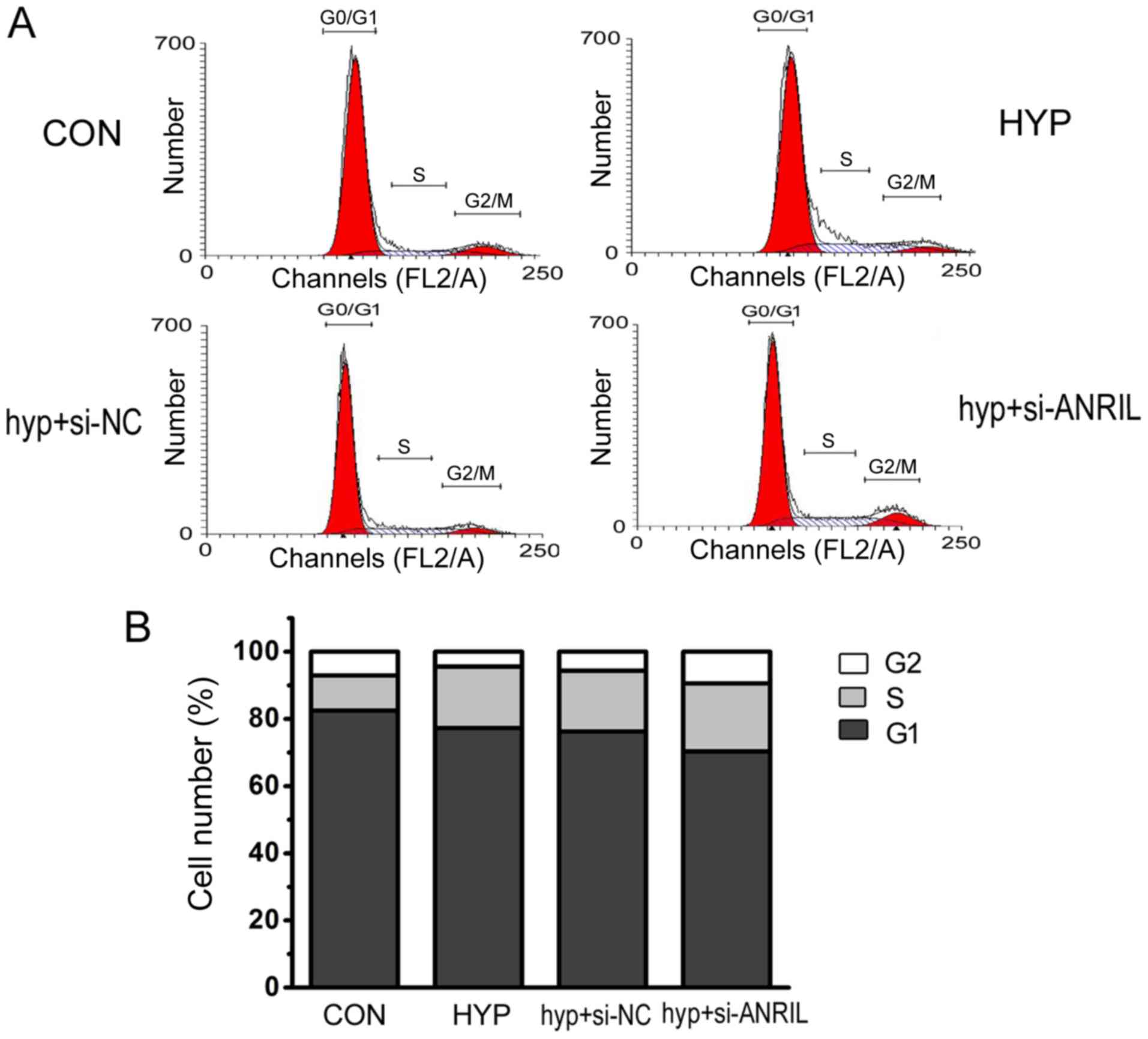
ANRIL regulates the cell cycle
progression of HPASMCs
In order to investigate whether hypoxia affected
cell cycle progression through the lncRNA ANRIL pathway, flow
cytometry was used to detect the number of cells at different cell
cycle phases. The results showed that hypoxia increased the
percentage of cells in the G2/M+S phase and the downregulated ANRIL
strengthened the increase in the percentage of cells in the G2/M+S
phase under hypoxic conditions (Fig.
3).
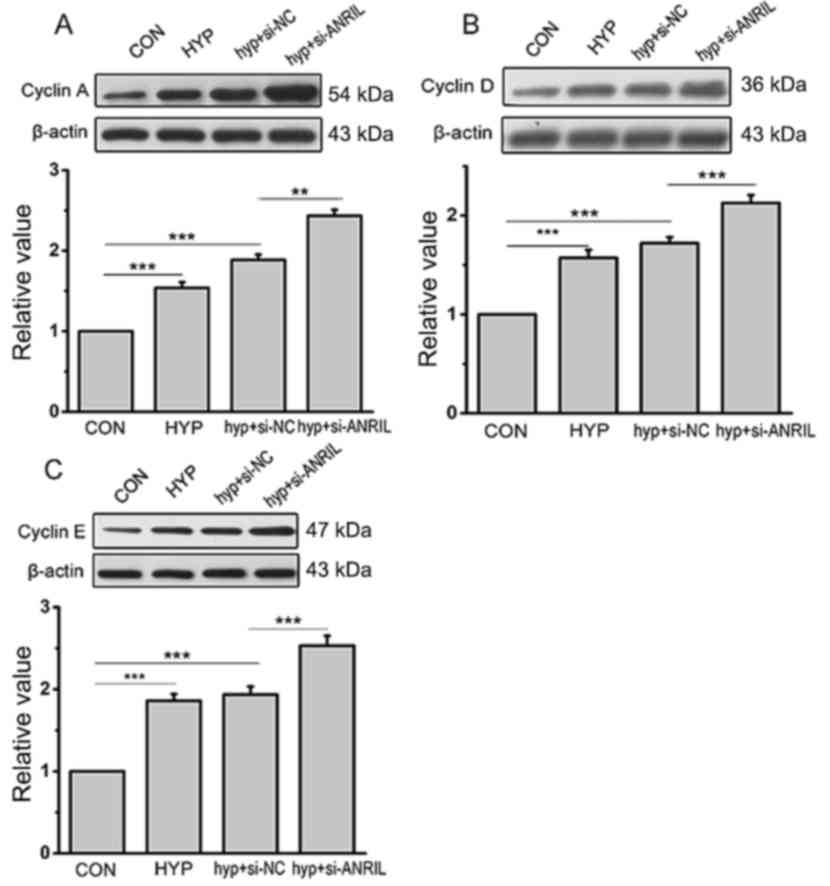
In addition, cyclins, proteins that regulate the
cell cycle and play important roles in both the S phase and G2/M
phase of cell cycle, have been generally considered as markers of
cellular proliferation. In order to understand the molecular
mechanism by which ANRIL regulates the cell cycle, the expression
of cell cycle-associated proteins, Cyclin A, Cyclin D and Cyclin E,
were detected by western blotting. The results revealed that the
expression of cyclins in hypoxic HPASMCs was significantly
increased compared with normoxia, which was further promoted by
ANRIL siRNA (P<0.001; Fig.
4).
ANRIL affects the proliferation of
HPASMCs
Subsequently, the effect of ANRIL on HPASMC
proliferation under hypoxic conditions was further investigated.
The results of western blotting displayed that the expression of
PCNA was significantly elevated under hypoxic conditions contrasted
with normoxia and si-ANRIL increased the trend further (P<0.05;
Fig. 5A). To evaluate the number
of cells that were actively synthesizing DNA, the BrdU
incorporation assay was carried out. The present results revealed
that the ratio of proliferating cells in HPASMCs under hypoxic
conditions was significantly increased compared with under normoxic
conditions (P<0.001), which was enhanced by si-ANRIL (Fig. 5B). The Ki67 protein is a cell
proliferation marker that is closely related to cell proliferation
and the cell cycle. The result of immunofluorescence assay showed
that the expression of Ki67 increased in hypoxia compared with
under normal conditions. When the expression of ANRIL was
downregulated by siRNA, Ki67 fluorescence staining was found to be
increased (Fig. 5C). Consistently,
these results demonstrated that the proportion of proliferating
cells with a low ANRIL content was higher.
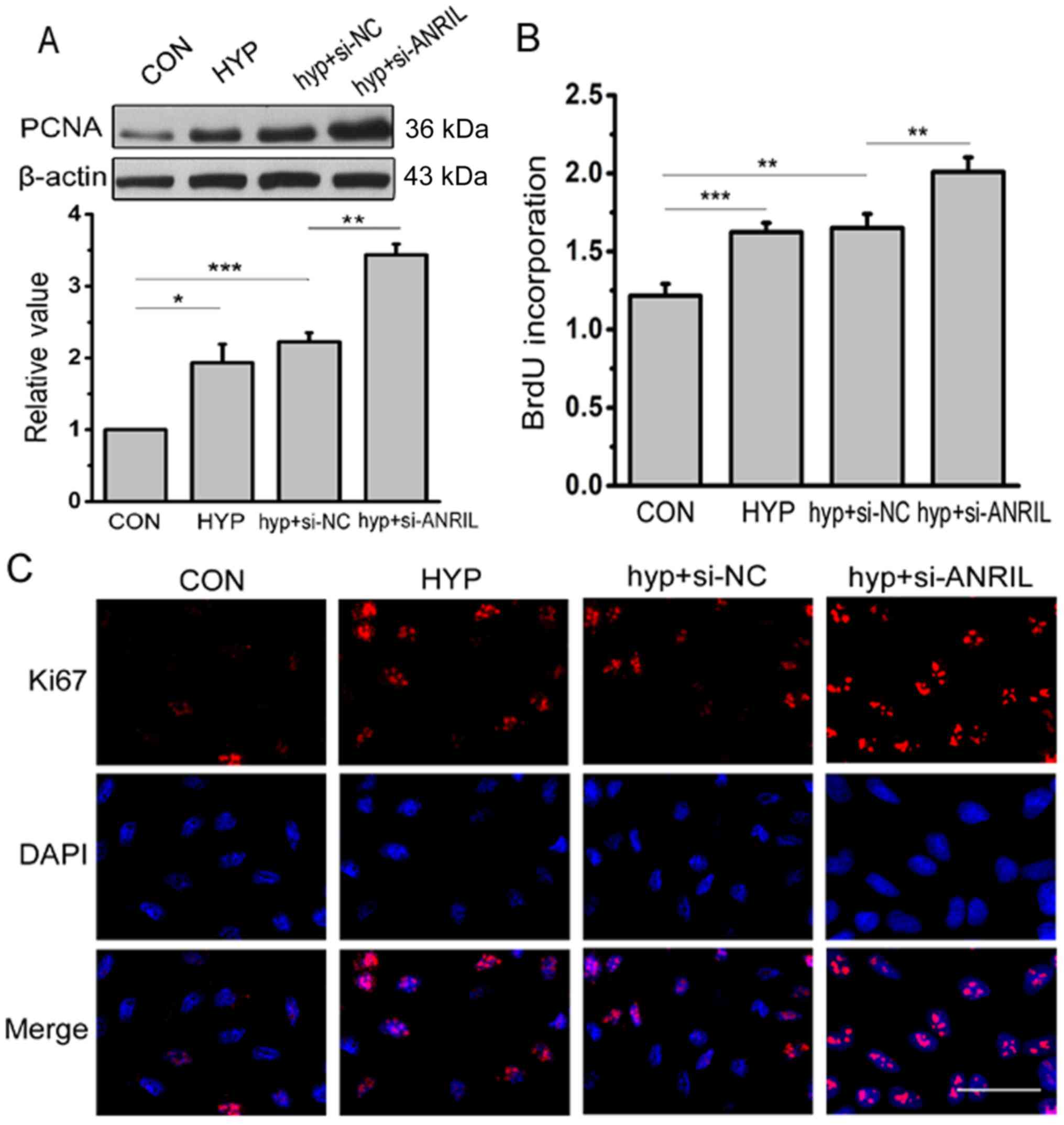 | Figure 5.ANRIL induces higher levels of
proliferation in HPASMCs. (A) The protein level of PCNA was
analyzed by western blotting after transfection with si-ANRIL;
densitometric quantification of protein bands showed a significant
increase in PCNA following 24 h of transfection with si-ANRIL
(n=3). (B) 5-bromodeoxyuridine incorporation assay was performed to
determine the proliferation of HPASMCs (n=5). (C)
Immunofluorescence assay was used to detect the expression of Ki67.
The red color denotes Ki67 staining by rhodamine, whereas the blue
color denotes nucleus staining by DAPI (n=3). Scale bar=50 µm.
*P<0.05, **P<0.01 and ***P<0.001. CON, normoxia control;
HYP, hypoxia; hyp+si-NC, cells treated with negative control under
hypoxic conditions; hyp+si-ANRIL, cells treated with si-ANRIL under
hypoxic conditions; HPASMCS, human pulmonary artery smooth muscle
cells; PCNA, proliferating cell nuclear antigen; ANRIL, antisense
noncoding RNA in the INK4 locus; DAPI,
4,6-diamidino-2-phenylindole. |
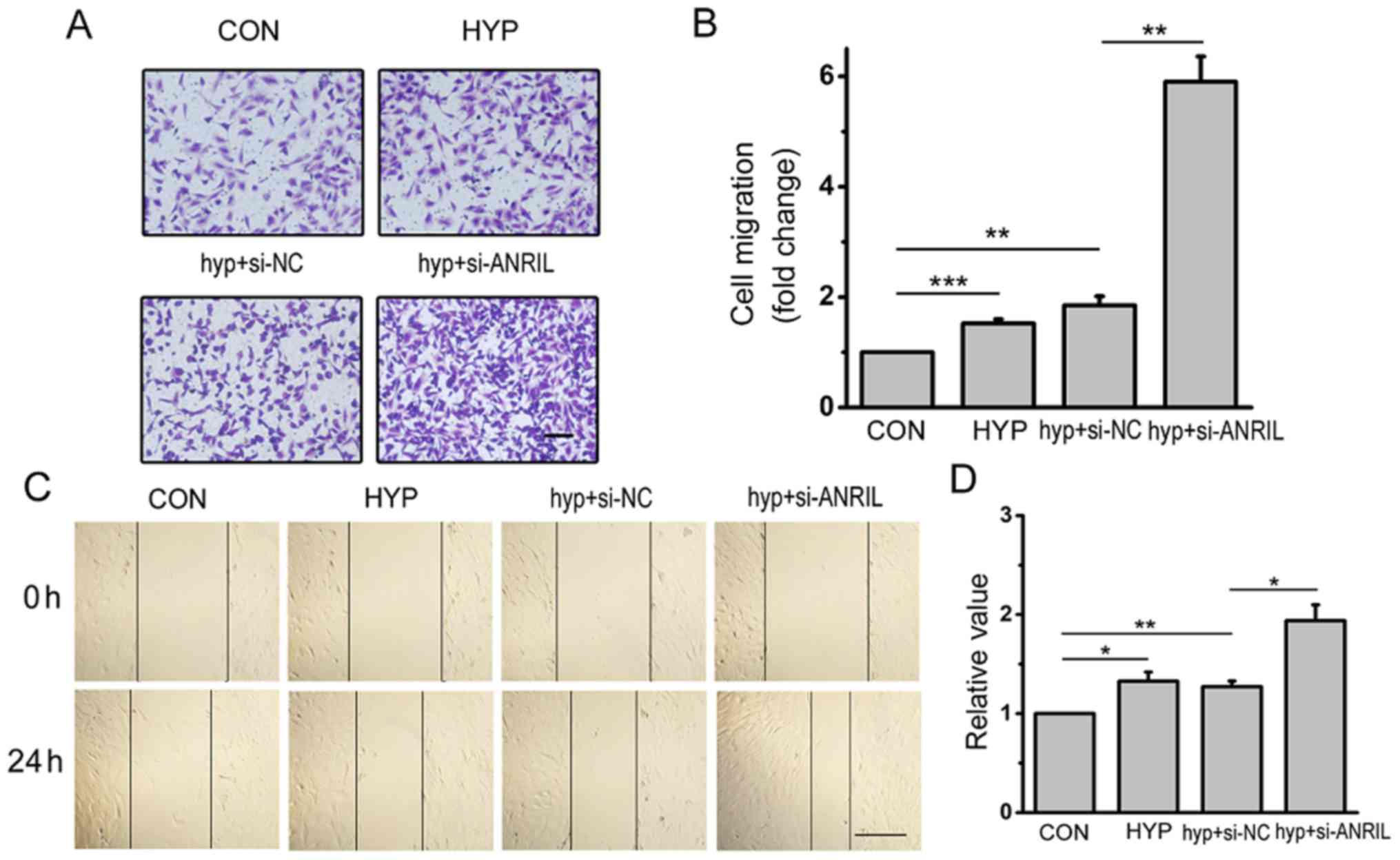
ANRIL promotes migration of hypoxic
HPASMCs
The effects of ANRIL on cell migration was examined
by Transwell migration assay and scratch-wound assay. As shown in
Fig. 6A, the number of migrated
cells under hypoxic conditions was more than that under normoxia.
Downregulated ANRIL significantly increased the cell migration
according to the Transwell migration assay (P<0.01; Fig. 6B). The scratch-wound assay results
showed that the migration distance of hypoxic HPASMCs was longer
than that in normoxia, which was strengthened by ANRIL siRNA under
hypoxic condition in Fig. 6C.
Histograms showing the results of the Transwell migration and
scratch-wound assays are shown Fig. 6B
and D, respectively. Taken together, these data indicated that
the downregulation of ANRIL induced by hypoxia enhanced the
capacity of migration in HPASMCs.
Discussion
Previous studies have indicated that, in numerous
diseases, lncRNA ANRIL regulates cell proliferation, migration and
cell cycle progression including cardiovascular diseases, tumors
and diabetes (18,19). In this study, it was found that the
expression of ANRIL was downregulated in hypoxic HPASMCs. Hypoxia
accelerated the cell cycle progression and induced cell
proliferation of HPASMCs through the downregulation of the lncRNA
ANRIL pathway. Moreover, downregulated ANRIL in the hypoxic HPASMCs
promoted cell migration. Therefore, solid evidence demonstrated
that ANRIL is a critical regulator in HPASMCs induced by
hypoxia.
It is well established that hypoxia is the main
cause of PAH. Recent reports have highlighted the relationship
between lncRNAs and hypoxia in PAH. The downregulated lncRNA,
maternally expressed 3, in hypoxic PASMCs was shown to trigger cell
proliferation and migration via the p53 signaling pathway (1). The lncRNA Hoxa cluster antisense RNA
3 (Hoxaas3) was upregulated in both pulmonary vessels in hypoxic
mice and PASMCs under hypoxic conditions, while a high expression
level of Hoxaas3 was associated with cell proliferation and
modulated cell cycle distribution by upregulating Homeobox a3 at
the mRNA and protein levels (20).
The results from the present study showed that the decrease in the
expression level of ANRIL when downregulated in hypoxic HPASMCs was
twice that which occurred under normoxic conditions by qPCR,
indicating that ANRIL was able to regulate hypoxic PAH.
The proliferation of PASMCs is one of the main
causes of medial hypertrophy of PVR in PAH (4). A previous study showed that in
epithelial ovarian cancer, ANRIL promoted proliferation and cell
cycle progression (21). The
present study demonstrated that si-ANRIL may accelerate cell cycle
progression by increasing the percentage of cells in the G2/M+S
phase and also elevated the expression level of cyclins under
hypoxic conditions. Downregulated ANRIL promoted the expression of
PCNA in HPASMCs and increased the expression of Ki-67 under
hypoxia. The above results indicated that ANRIL could regulate the
cell cycle progression and the proliferation of HPASMCs.
A recently published study reported that the
expression of ANRIL in the pancreatic cancer cells was higher than
that in normal pancreatic duct epithelial cells and after ANRIL is
silenced, the migratory and invasive abilities of the cells were
decreased (22). In the present
study, Transwell and scratch-wound assays were used to examine the
effects of ANRIL on cell migration. The results showed that
downregulated ANRIL increased the migration of hypoxic HPASMCs.
These data indicated that the downregulation of ANRIL could enhance
the capacity of migration in HPASMCs.
At present, the importance of cell biology and the
role of ANRIL in the pathogenesis of various diseases have been
major topics for study. To the best of our knowledge, this is the
first study that has reported on the involvement of ANRIL in PAH,
however, there remain several limitations that need to be resolved
in future studies. Although the downregulation of ANRIL was
detected in HPASMCs, it is still unclear how the decrease of ANRIL
is caused by hypoxia. In addition, it is worth noting that one
lncRNA may target multiple microRNAs and exert different effects.
Whether ANRIL can effectively function as a competing endogenous
RNA by binding with microRNAs remains to be further studied.
Further experiments will be performed in the future to explore the
role of ANRIL in PAH.
In conclusion, the present study has demonstrated
that the expression of lncRNA ANRIL in HPASMCs of PAH was reduced.
The decreased expression of lncRNA ANRIL contributed to the
proliferation and migration of HPASMCs in the pathogenesis of PAH.
Therefore, the authors' proposal is that ANRIL-mediated PASMCs are
involved in the pathogenesis of vascular remodeling in PAH. These
findings provide a novel potential target for the treatment of PAH
and may help to improve the therapeutic efforts in the future.
Acknowledgements
The authors would like to thank Professor Daling
Zhu, Dr Ying Liu and Dr Hongyue Zhang (Harbin Medical University)
for lending their expertise for the studies.
Funding
The present study was supported by The Key Project
of Natural Science Foundation of Heilongjiang province, project no.
2D201416.
Availability of data and materials
The datasets used and/or analyzed during the current
study are available from the corresponding author on reasonable
request.
Authors' contributions
SW, CZ and XZ made substantial contributions towards
the conception and design of the experiments. SW completed all the
studies and aggregated the figures and discussed the results. SW
and CZ drafted the manuscript and critically revised and added
important intellectual content.
Ethics approval and consent to
participate
Not applicable.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Sun Z, Nie X, Sun S, Dong S, Yuan C, Li Y,
Xiao B, Jie D and Liu Y: Long non-coding RNA MEG3 downregulation
triggers human pulmonary artery smooth muscle cell proliferation
and migration via the p53 signaling pathway. Cell Physiol Biochem.
42:2569–2581. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Stenmark KR, Fagan KA and Frid MG:
Hypoxia-induced pulmonary vascular remodeling: Cellular and
molecular mechanisms. Circ Res. 99:675–691. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Howell K, Preston RJ and McLoughlin P:
Chronic hypoxia causes angiogenesis in addition to remodelling in
the adult rat pulmonary circulation. J Physiol. 547:133–145. 2003.
View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Pietra GG, Capron F, Stewart S, Leone O,
Humbert M, Robbins IM, Reid LM and Tuder RM: Pathologic assessment
of vasculopathies in pulmonary hypertension. J Am Coll Cardiol. 43
(Suppl S):25S–32S. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Harries LW: Long non-coding RNAs and human
disease. Biochem Soc Trans. 40:902–906. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Park JY, Lee JE, Park JB, Yoo H, Lee SH
and Kim JH: Roles of long non-coding RNAs on tumorigenesis and
glioma development. Brain Tumor Res Treat. 2:1–6. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Li T, Mo X, Fu L, Xiao B and Guo J:
Molecular mechanisms of long noncoding RNAs on gastric cancer.
Oncotarget. 7:8601–8612. 2016.PubMed/NCBI
|
|
8
|
Roberts R: Genetics of coronary artery
disease: An update. Methodist Debakey Cardiovasc J. 10:7–12. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Pasmant E, Laurendeau I, Héron D, Vidaud
M, Vidaud D and Bièche I: Characterization of a germ-line deletion,
including the entire INK4/ARF locus, in a melanoma-neural system
tumor family: Identification of ANRIL, an antisense noncoding RNA
whose expression coclusters with ARF. Cancer Res. 67:3963–3969.
2007. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Tano K and Akimitsu N: Long non-coding
RNAs in cancer progression. Front Genet. 3:2192012. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Schaefer AS, Bochenek G, Manke T,
Nothnagel M, Graetz C, Thien A, Jockel-Schneider Y, Harks I,
Staufenbiel I, Wijmenga C, et al: Validation of reported genetic
risk factors for periodontitis in a large-scale replication study.
J Clin Periodontol. 40:563–572. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Masharawi YM, Kjaer P, Bendix T, Manniche
C, May H, Mirovsky Y, Anekshtein Y, Jensen TS and Hershkovitz I:
Lumbar facet and interfacet shape variation during growth in
children from the general population: A three-year follow-up MRI
study. Spine (Phila Pa 1976). 34:408–412. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Congrains A, Kamide K, Katsuya T, Yasuda
O, Oguro R, Yamamoto K, Ohishi M and Rakugi H: CVD-associated
non-coding RNA, ANRIL, modulates expression of atherogenic pathways
in VSMC. Biochem Biophys Res Commun. 419:612–616. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Zhou X, Han X, Wittfeldt A, Sun J, Liu C,
Wang X, Gan LM, Cao H and Liang Z: Long non-coding RNA ANRIL
regulates inflammatory responses as a novel component of NF-κB
pathway. RNA Biol. 13:98–108. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(-Delta Delta C(T)) method. Methods. 25:402–408. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Yu X, Li T, Liu X, Yu H, Hao Z, Chen Y,
Zhang C, Liu Y, Li Q, Mao M and Zhu D: Modulation of pulmonary
vascular remodeling in hypoxia: Role of 15-LOX-2/15-HETE-MAPKs
Pathway. Cell Physiol Biochem. 35:2079–2097. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Siegel R, Ma J, Zou Z and Jemal A: Cancer
statistics, 2014. CA Cancer J Clin. 64:9–29. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Aguilo F, Di Cecilia S and Walsh MJ: Long
Non-coding RNA ANRIL and Polycomb in human cancers and
cardiovascular disease. Curr Top Microbiol Immunol. 394:29–39.
2016.PubMed/NCBI
|
|
19
|
Zhang B, Wang D, Ji TF, Shi L and Yu JL:
Overexpression of lncRNA ANRIL up-regulates VEGF expression and
promotes angiogenesis of diabetes mellitus combined with cerebral
infarction by activating NF-κB signaling pathway in a rat model.
Oncotarget. 8:17347–17359. 2017.PubMed/NCBI
|
|
20
|
Zhang H, Liu Y, Yan L, Wang S, Zhang M, Ma
C, Zheng X, Chen H and Zhu D: Long noncoding RNA Hoxaas3
contributes to hypoxia-induced pulmonary artery smooth muscle cell
proliferation. Cardiovasc Res. 115:647–657. 2018. View Article : Google Scholar
|
|
21
|
Qiu JJ, Wang Y, Liu YL, Zhang Y, Ding JX
and Hua KQ: The long non-coding RNA ANRIL promotes proliferation
and cell cycle progression and inhibits apoptosis and senescence in
epithelial ovarian cancer. Oncotarget. 7:32478–32492. 2016.
View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Chen S, Zhang JQ, Chen JZ, Chen HX, Qiu
FN, Yan ML, Chen YL, Peng CH, Tian YF and Wang YD: The over
expression of long non-coding RNA ANRIL promotes
epithelial-mesenchymal transition by activating the ATM-E2F1
signaling pathway in pancreatic cancer: An in vivo and in vitro
study. Int J Biol Macromol. 102:718–728. 2017. View Article : Google Scholar : PubMed/NCBI
|