Introduction
Hepatocellular carcinoma (HCC), the most common
malignant tumor of the liver, is the fourth most prevalent cancer
and the third leading cause of cancer-associated mortality in the
world (1). It is estimated that
there is ~792,000 incident cases and 818,000 mortalities due to HCC
each year worldwide (2). Despite
the continuous progresses made in diagnosis and therapy, the
long-term survival of patients with HCC remains poor, with a 5-year
survival rate of ~30% (3,4). The poor prognosis of patients with
HCC is primarily due to high rate of intrahepatic and distal
metastasis, recurrence and the lack of effective therapeutic
methods for those patients diagnosed at advanced stages (5,6).
Several well-documented factors have been identified to be closely
associated with HCC initiation and progression, including chronic
infection with hepatitis B or C virus, alcoholism, aflatoxin and
excessive obesity caused by high-fat diets (7,8);
however, the detailed mechanisms underlying the pathogenesis of HCC
remain poorly characterized. Therefore, it is imperative to improve
the understanding of the mechanisms involved in HCC occurrence and
development, which may contribute to the identification of
potential effective therapeutic options for the treatment of
patients with this disease.
MicroRNAs (miRNAs) are a group of endogenous,
non-coding and short regulatory RNA molecules that are 18–24
nucleotides in length (9). miRNAs
bind to the 3′-untranslated regions (UTRs) of their target genes to
decrease their translation and/or promote mRNAs degradation
(10). miRNAs are differently
expressed in a number of types of human cancer, including HCC
(11), and ovarian (12), thyroid (13) and gastric cancer (14). Dysregulated miRNAs serve a pivotal
role in numerous biological behaviors, including cell
proliferation, cell cycle, apoptosis, metastasis and angiogenesis
(15–17). miRNAs are able to serve
tumor-suppressing or oncogenic roles in the formation and
progression of HCC, which primarily depends on the characteristic
of their target genes (18,19).
For example, miR-29a inhibits HCC cell proliferation, colony
formation and inhibits cell cycle progression by directly targeting
Sirtuin 1 (15). miR-1468 is
upregulated in HCC and promotes aggressive tumor behaviors by
activating peroxisome proliferator-activated receptor
gamma-mediated protein B kinase signaling (20). Considering their crucial roles in
HCC, miRNAs have the potential to be developed as novel biomarkers
for diagnosis and therapeutic targets for treating patients with
HCC.
Abnormal miR-663b expression has been previously
described in different types of human cancer, including
nasopharyngeal carcinoma (21),
bladder cancer (22), osteosarcoma
(23) and endometrial cancer
(24). However, the expression
pattern, potential biological roles and underlying mechanisms of
miR-663b in HCC remain largely unknown. Therefore, the present
study aimed to detect miR-663b expression in HCC tissues and cell
lines, determine its effects and investigate the molecular
regulatory mechanism of miR-663b in HCC progression. To the best of
our knowledge, these data indicate for the first time a fundamental
role for miR-663b in the pathogenesis of HCC.
Materials and methods
Patients and tissue specimens
In total, 34 pairs of HCC tissues and adjacent
normal tissues were obtained from patients (23 males, 11 females;
age range 43–72 years; median age, 61 years) who underwent surgical
resection at the Affiliated Hospital of Yan'an University (Yan'an,
China) between May 2015 and February 2017. Adjacent normal tissues
were obtained 2 cm away from HCC tissues. The clinicopathological
features of these patients are summarized in Table I. Patients were diagnosed using the
tumor-node-metastasis (TNM) system (25). Patients treated with radiotherapy,
chemotherapy or other treatments prior to surgery were excluded
from the study cohort.
 | Table I.Clinicopathological features of
patients with hepatocellular carcinoma. |
Table I.
Clinicopathological features of
patients with hepatocellular carcinoma.
| Patient no. | Sex | Age | TNM stage | Patient no. | Sex | Age | TNM stage |
|---|
| 1 | F | 52 | T1N0M0 | 18 | F | 63 | T1N0M0 |
| 2 | M | 45 | T2N1M0 | 19 | F | 69 | T2N1M0 |
| 3 | F | 62 | T2N0M0 | 20 | M | 54 | T2N0M0 |
| 4 | F | 68 | T1N0M0 | 21 | F | 45 | T1N0M0 |
| 5 | M | 58 | T1N0M0 | 22 | M | 60 | T3aN1M0 |
| 6 | M | 69 | T1N0M0 | 23 | M | 66 | T3bN1M0 |
| 7 | M | 58 | T1N0M0 | 24 | M | 71 | TaN1M0 |
| 8 | M | 67 | T2N0M0 | 25 | M | 59 | T2N1M0 |
| 9 | F | 53 | T1N0M0 | 26 | F | 43 | T1N0M0 |
| 10 | F | 70 | T2N1M0 | 27 | M | 67 | T2N0M0 |
| 11 | M | 72 | T1N0M0 | 28 | M | 68 | T1N0M0 |
| 12 | M | 48 |
T3aN1M0 | 29 | M | 53 | T3aN1M0 |
| 13 | F | 56 | T2N1M0 | 30 | F | 64 | TaN0M0 |
| 14 | M | 71 | T1N0M0 | 31 | M | 58 | T3aN0M0 |
| 15 | M | 63 | T1N0M0 | 32 | M | 49 | T1N1M0 |
| 16 | M | 49 | T1N0M0 | 33 | M | 47 | T3bN1M0 |
| 17 | M | 55 | T1N0M0 | 34 | M | 63 | T1N0M0 |
The present study was approved by the Ethics
Committee of Affiliated Hospital of Yan'an University. Written
informed consent was obtained from all participants.
Cell lines and culture conditions
A total of 2 human HCC Huh7 and Hep3B cell lines and
the immortalized normal human liver epithelial L-O2 cell line were
purchased from Institute of Biochemistry and Cell Biology, Shanghai
Institutes for Biological Sciences, Chinese Academy of Science
(Shanghai, China). All cells were grown at 37°C in a humidified
atmosphere containing 5% CO2 and 95% air, and cultured
in Dulbecco's modified Eagle medium (DMEM) containing 10% fetal
bovine serum (FBS), 100 µ/ml penicillin and 100 µg/ml streptomycin
(all from Gibco; Thermo Fisher Scientific, Inc., Waltham, MA,
USA).
Oligonucleotide transfection
Cells were plated into 6-well plates with a density
of 6×105 cells/well. Then, 12 h after inoculation, cells
were transfected with miRNA negative control mimics (miR-NC) or
miR-663b mimics (both from Guangzhou Ribobio Co., Ltd., Guangzhou,
China) using Lipofectamine® 2000 (Invitrogen; Thermo
Fisher Scientific, Inc.), as per the manufacturer's protocol. To
restore Grb2-associated binding 2 (GAB2) expression, the full
length sequence of GAB2 was chemically synthesized by Guangzhou
Fueneng Gene Co., Ltd. (Guangzhou, China), cloned into pcDNA3.1
vector and named as pcDNA3.1-GAB2. The transfection efficiencies of
miR-663b mimics and pcDNA3.1-GAB2 were determined using reverse
transcription-quantitative polymerase chain reaction (RT-qPCR) and
western blot analysis, respectively. RT-qPCR and western blot
analysis were performed at 48 and 72 h after transfection, and
carried out as described in the following paragraphs. Cell Counting
kit-8 (CCK-8) and Transwell invasion assays were conducted at 24
and 48 h post-transfection, respectively.
RNA extraction and RT-qPCR
TRIzol® reagent (Thermo Fisher
Scientific, Inc.) was used to isolate total RNA from tissue samples
or cells. The concentration of total RNA was measured using a
NanoDrop 1000 spectrophotometer (NanoDrop Technologies; Thermo
Fisher Scientific, Inc., Wilmington, DE, USA). For the detection of
miR-663b expression, total RNA was subjected to cDNA synthesis
using a TaqMan™ MicroRNA Reverse Transcription kit (Applied
Biosystems; Thermo Fisher Scientific, Inc.), and then qPCR was
conducted using a TaqMan™ MicroRNA Assay kit (Applied Biosystems;
Thermo Fisher Scientific, Inc.). The cycling conditions were: 50°C
for 2 min, 95°C for 10 min, 40 cycles of denaturation at 95°C for
15 sec and annealing/extension at 60°C for 60 sec. To analyze GAB2
mRNA expression, RT was performed to produce cDNA using a
PrimeScript RT Reagent kit (Takara Biotechnology Co., Ltd., Dalian,
China), followed by qPCR with a SYBR Premix Ex Taq™ kit (Takara
Biotechnology Co., Ltd.). The thermocycling conditions were: 5 min
at 95°C, 40 cycles of 95°C for 30 sec and 65°C for 45 sec. U6 small
nuclear RNA and GAPDH were used as internal references for miR-663b
and GAB2 expression, respectively. All data was calculated by the
2−∆∆Cq method (26).
The primers were designed as follows: miR-663b,
5′-CGCTAACAGTCTCCAGTC-3′ (forward) and 5′-GCGACACCAAACTGGATGA-3′
(reverse); U6, 5′-CTCGCTTCGGCAGCACATATACT-3′ (forward) and
5′-ACGCTTCACGAATTTGCGTGTC-3′ (reverse); GAB2,
5′-CTGAGACTGATAACGGAGAT-3′ (forward) and 5′-GAGGTGTTTCTGCTTGAC-3′
(reverse); and GAPDH, 5′-CGGAGTCAACGGATTTGGTCGTAT-3′ (forward) and
5′-AGCCTTCTCCATGGTGGTGAAGAC-3′ (reverse).
CCK-8 assay
Transfected cells were incubated at 37°C with 5%
CO2 for 24 h. Next, transfected cells were collected and
plated into 96-well plates with a density of 3×103 cells
per well. CCK-8 assays were performed at 0, 1, 2 and 3 days after
inoculation to detect cell proliferation, in accordance with the
manufacturer's protocol. In detail, 10 µl CCK-8 solution (Dojindo
Molecular Technologies, Inc., Kumamoto, Japan) was added into each
well. Following incubation at 37°C for 2 h, the absorbance was
measured at a wavelength of 450 nm using a microplate reader
(Bio-Rad Laboratories, Inc., Hercules, CA, USA).
Transwell invasion assay
Transwell plate cell culture chambers (Corning Life
Sciences, Cambridge, MA, USA) coated with Matrigel (BD Biosciences,
San Jose, CA, USA) were utilized to evaluate invasion capacities of
HCC cells. Matrigel coating was performed at 37°C for 2 h.
Following transfection for 48 h, cells were harvested, washed and
diluted into FBS-free DMEM. A total of 1×105 transfected
cells were placed into the upper compartment of the Transwell plate
cell culture chambers, and DMEM containing 20% FBS was added into
the lower compartments. The chambers were then incubated at 37°C at
5% CO2 for 24 h. Non-invasive cells remaining on the
upper chamber surface were gently removed using a cotton swab. The
invasive cells that had passed through the membranes were fixed
with 4% paraformaldehyde at room temperature for 20 min and stained
with 0.5% crystal violet at room temperature for 20 min. The number
of invasive cells was counted in 5 randomly chosen visuals per
chamber under an inverted microscope (IX73; magnification, ×200;
Olympus Corporation, Tokyo, Japan).
Target prediction for miR-663b
Bioinformatics analysis was applied to predict the
putative targets of miR-663b using TargetScan (http://www.targetscan.org/) and microRNA.org (http://www.microrna.org/). GAB2 was predicted as a
major target of miR-663b and was considered for subsequent
experimental analysis.
Luciferase reporter assay
The 3′-UTR fragments of GAB2 containing the
wild-type (wt) or mutant (mut) miR-663b-binding sequences were
synthesized by Shanghai GenePharma Co., Ltd. (Shanghai, China),
inserted into the pGL3 luciferase reporter vector (Promega
Corporation, Madison, WI, USA) and named as pGL3-wt-GAB2-3′-UTR and
pGL3-mut-GAB2-3′-UTR, respectively. Cells were plated into 24-well
plates at a density of 60–70% confluence 12 h prior to
transfection. Cells were co-transfected with miR-663b mimics (50
pmol) or miR-NC (50 pmol), and pGL3-wt-GAB2-3′-UTR or
pGL3-mut-GAB2-3′-UTR, using Lipofectamine® 2000
(Invitrogen; Thermo Fisher Scientific, Inc.) based on the
manufacturer's instructions. Luciferase activity was measured at 48
h post-transfection using the Dual-Luciferase reporter system
(Promega Corporation). Renilla luciferase activity was used
as an internal reference.
Western blot analysis
Western blot analysis was performed to detect GAB2
protein expression. Total protein was isolated from homogenized
tissues or cultured cells using cold radioimmunoprecipitation assay
buffer, and then was subjected to the detection of protein
concentration using a BCA Assay kit (both Nanjing KeyGen Biotech
Co., Nanjing, Ltd, China). Protein samples (20 µg) were separated
on a 10% SDS-PAGE and transferred onto polyvinylidene difluoride
membranes (EMD Millipore, Billerica, MA, USA). The membranes were
then blocked at room temperature for 2 h with 5% skimmed milk
diluted in TBS/0.1% Tween (TBST) and incubated overnight at 4°C
with primary antibodies. Following extensive washing with TBST, the
membranes were additionally probed with the goat anti-rabbit
horseradish peroxidase-conjugated secondary antibody (cat. no.
ab205718; 1:5,000 dilution; Abcam, Cambridge, UK) for 2 h at room
temperature, followed by visualization of the protein bands using
an enhanced chemiluminescence reagent (Bio-Rad Laboratories, Inc.).
The primary antibodies used in the present study included rabbit
anti-human GAB2 antibody (cat. no. ab108423; 1:1,000 dilution;
Abcam) and rabbit anti-human GAPDH antibody (cat. no. ab128915;
1:1,000 dilution; Abcam). GAPDH was used as a loading control.
Statistical analysis
All data are presented as the means ± standard
deviation, and were analyzed using the SPSS 21.0 software (IBM
Corp., Armonk, NY, USA). The differences between two groups were
examined using Student's t-test or Wilcoxon signed-rank test, and
differences between three or more groups were analyzed using
one-way analysis of variance (ANOVA). The Student-Newman-Keuls test
was employed as a post-hoc test following ANOVA. The association
between miR-663b and GAB2 mRNA levels in HCC tissues was determined
using Spearman's correlation analysis. P<0.05 was considered to
indicate a statistically significant difference.
Results
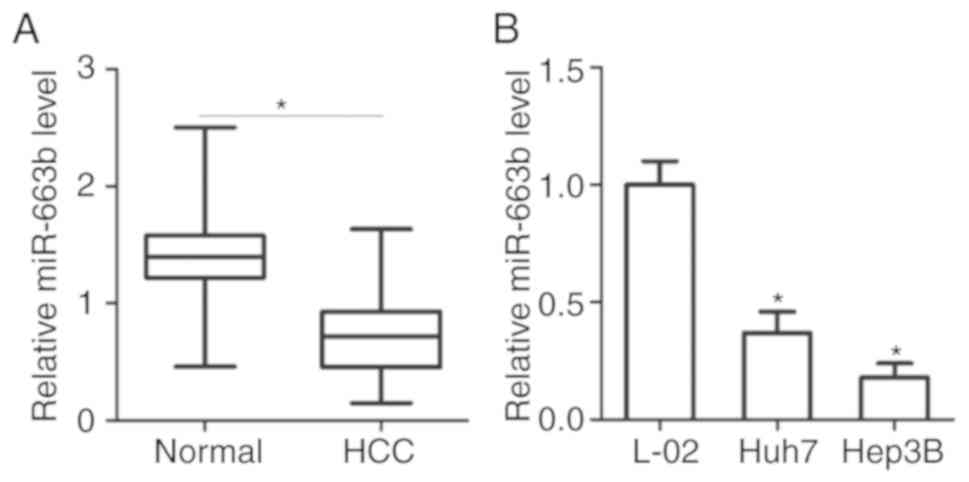
miR-663b is decreased in HCC tissues
and cell lines
To detect the expression status of miR-663b in HCC,
miR-663b expression in HCC was first determined by initially
comparing the levels of miR-663b in 34 pairs of HCC tissues and
adjacent normal tissues. Results from RT-qPCR analysis revealed
that the expression levels of miR-663b were decreased in HCC
tissues compared with that in adjacent normal tissues (Fig. 1A; P<0.05). miR-663b expression
was then additionally measured in 2 human HCC Huh7 and Hep3B cell
lines, and an immortalized normal human liver epithelial L-O2 cell
line. Significantly decreased expression levels of miR-663b were
observed in the 2 HCC cell lines compared with that in L-O2
(Fig. 1B; P<0.05). These
results suggest that the decreased miR-663b expression may be
associated with the progression and development of HCC.
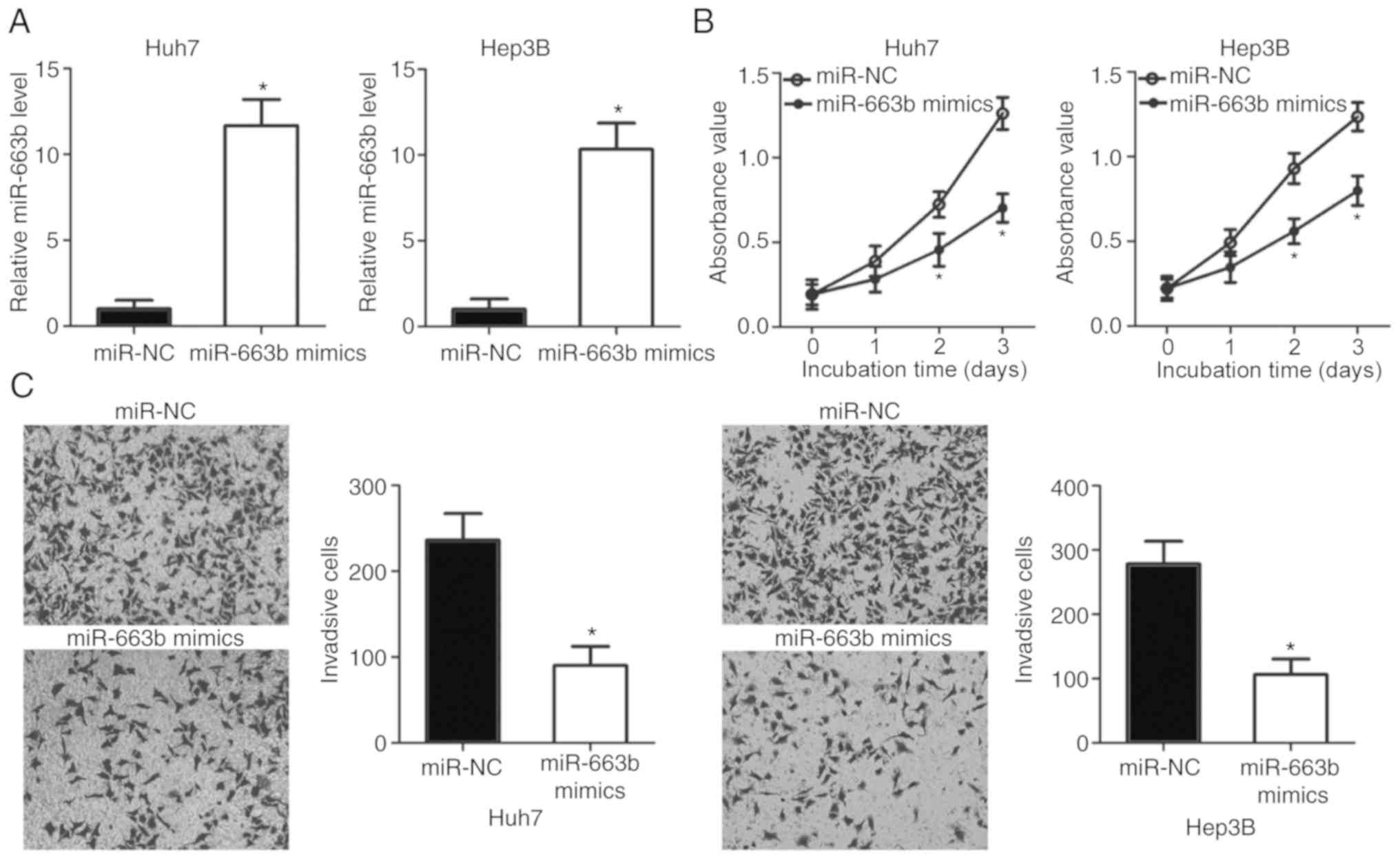
miR-663b overexpression suppresses HCC
cell proliferation and invasion
To examine the biological roles of miR-663b in HCC,
Huh7 and Hep3B cells were treated with miR-663 mimics to increase
its endogenous expression (Fig.
2A; P<0.05). The effect of miR-663b overexpression on HCC
cell proliferation was determined by CCK-8 assay. The results
indicated that ectopic miR-663b expression significantly inhibited
the proliferation of Huh7 and Hep3B cells (Fig. 2B; P<0.05). In addition, a
Transwell invasion assay was applied to detect the invasion ability
of Huh7 and Hep3B cells transfected with miR-663b mimics or miR-NC.
The invasion ability of Huh7 and Hep3B cells was markedly decreased
following treatment with miR-663b mimics (Fig. 2C; P<0.05). Taken together, these
results suggested that miR-663b may serve as a tumor suppressor in
HCC.
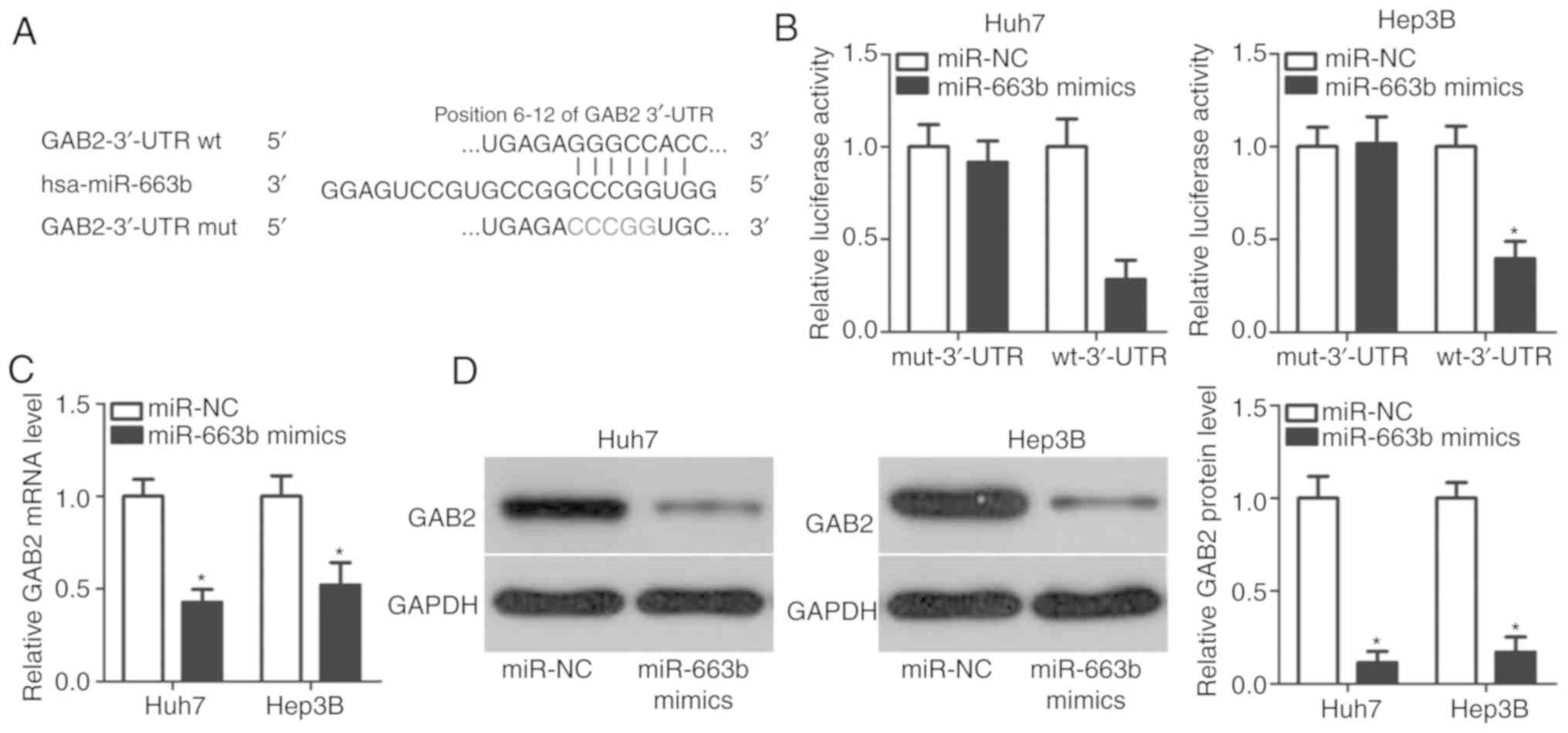
miR-663b inhibits GAB2 expression in
HCC cells by binding to its 3′-UTR
To explore the fundamental mechanisms of miR-663b in
HCC cells, bioinformatics analysis was used to predict the
potential targets of miR-663b. A total of 135 genes were predicted
as candidates of miR-663b, including GAB2, tumor suppressor
candidate 2 (TUSC2), sex-determining region Y-box 12,
chromodomain-helicase-DNA-binding protein 5, cyclin D2 and
transcription factor forkhead box A2. GAB2 was predicted as a major
target of miR-663b (Fig. 3A) and
was considered for additional experimental analysis as GAB2 has
been demonstrated to be closely associated with HCC oncogenesis and
development (27–29). To affirm this prediction, a
luciferase reporter assay was performed to clarify whether miR-663b
may directly interact with the 3′-UTR of GAB2. Fig. 3B indicated that miR-663b
overexpression suppressed the luciferase activity of the reporter
plasmid with wt 3′-UTR of GAB2 in Huh7 and Hep3B cells (P<0.05);
however, the luciferase activity of the reporter plasmid carrying
the mut 3′-UTR of GAB2 was unaffected. In order to additionally
confirm that GAB2 was a direct target for miR-663b, RT-qPCR and
western blot analysis were performed to detect GAB2 mRNA and
protein levels in Huh7 and Hep3B cells following transfection with
miR-663b or miR-NC. The results demonstrated that mRNA (Fig. 3C; P<0.05) and protein (Fig. 3D; P<0.05) levels of GAB2 in Huh7
and Hep3B cells were markedly downregulated by miR-663b
overexpression. Therefore, GAB2 is a direct target of miR-663b in
HCC cells.
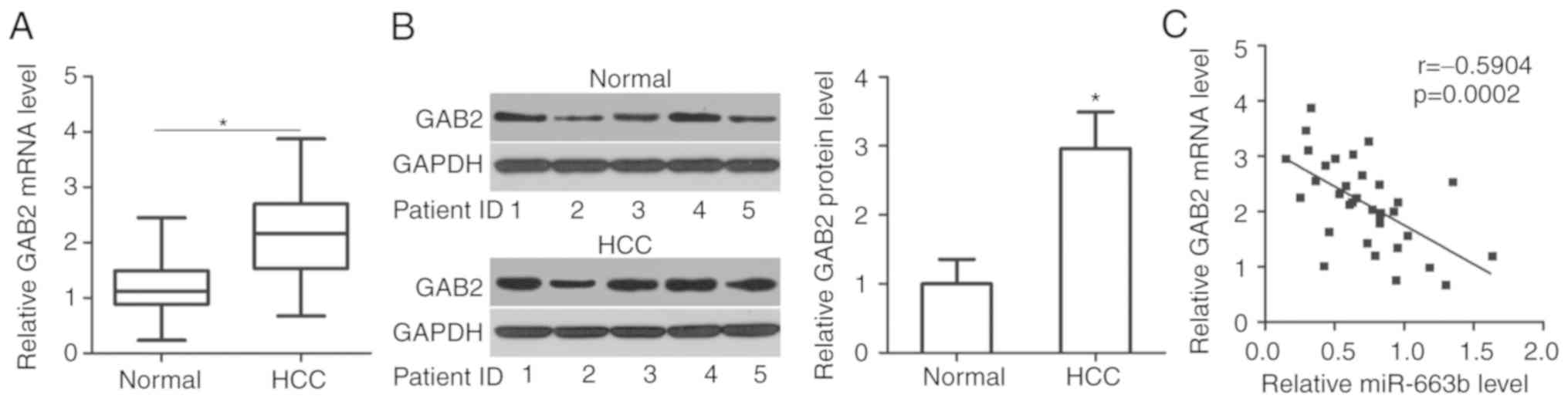
miR-663b expression is inversely
associated with GAB2 in HCC tissues
To additionally investigate the association between
miR-663b and GAB2 in HCC, GAB2 expression was measured in 34 pairs
of HCC tissues and adjacent normal tissues. The data from the
RT-qPCR analysis indicated that GAB2 mRNA levels were upregulated
in HCC tissues compared with that in adjacent normal tissues
(Fig. 4A; P<0.05).
Concomitantly, western blot analysis revealed that the expression
level of GAB2 protein was significantly increased in HCC tissues
compared with that in adjacent normal tissues (Fig. 4B; P<0.05). Furthermore,
Spearman's correlation analysis revealed an inverse correlation
between miR-663b and GAB2 mRNA expression in HCC tissues (Fig. 4C; r=−0.5904; P=0.0002). These
results suggest that the upregulation of GAB2 in HCC tissues was,
at least partly, caused by miR-663b downregulation.
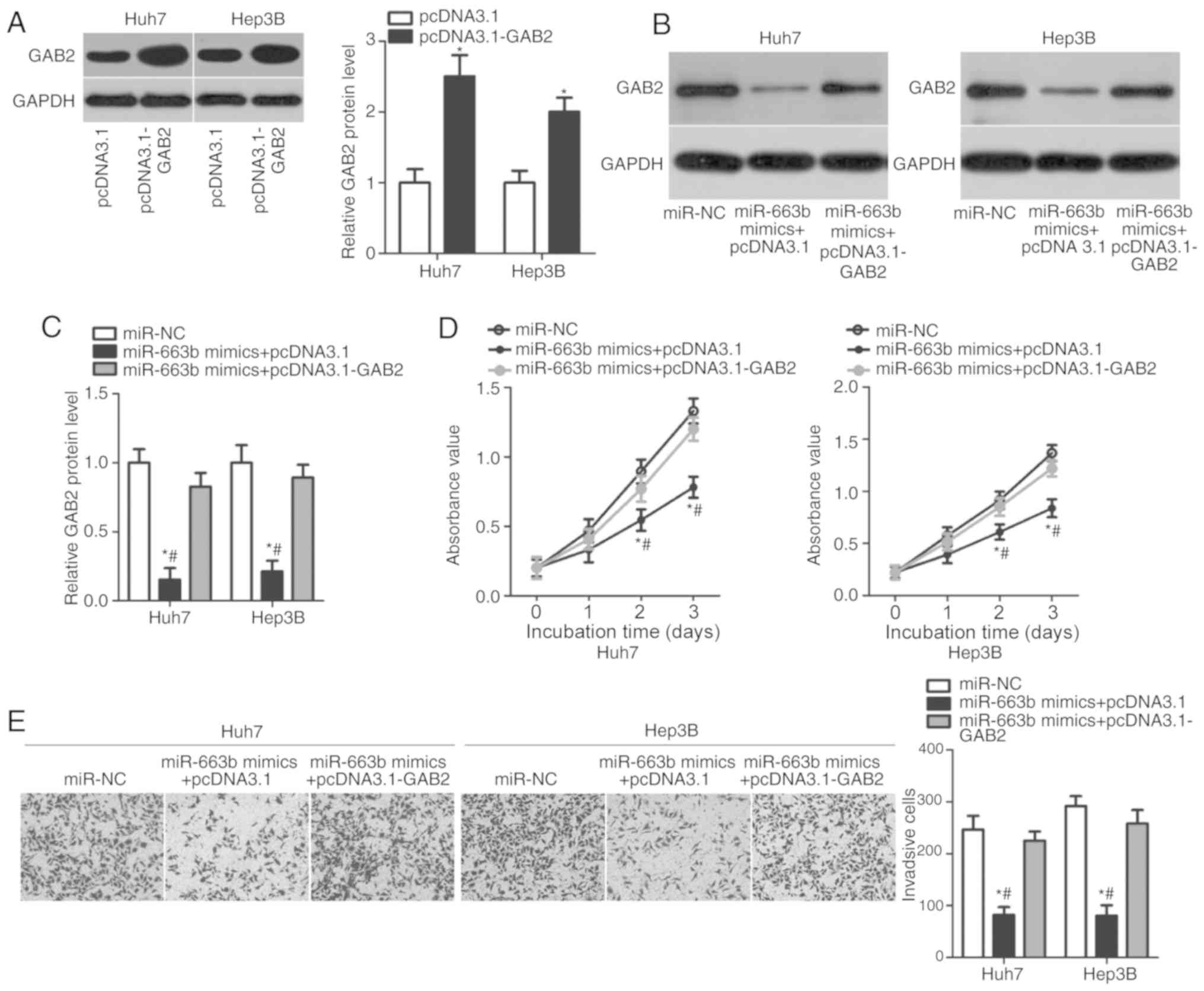
GAB2 overexpression rescues the
inhibitory effects of miR-663b overexpression on HCC cell
proliferation and invasion
A series of rescue experiments were performed to
determine whether GAB2 overexpression may reverse cell
proliferation and invasion suppression caused by miR-663b
upregulation in Huh7 and Hep3B cells. Therefore, pcDNA3.1-GAB2 or
empty pcDNA3.1 plasmids were transfected into Huh7 and Hep3B cells,
and western blot analysis was performed to detect GAB2 protein
expression. GAB2 protein levels were significantly increased in
pcDNA3.1-GAB2-transfected Huh7 and Hep3B cells compared with that
in cells transfected with pcDNA3.1 (Fig. 5A; P<0.05). Western blot analysis
also identified that the downregulation of GAB2 protein induced by
miR-663b overexpression was restored in Huh7 and Hep3B cells by
co-transfecting with pcDNA3.1-GAB2 (Fig. 5B and C; P<0.05). Furthermore,
CCK-8 and Transwell invasion assays demonstrated that GAB2
overexpression partially rescued the inhibitory effects of miR-663b
on the proliferation (Fig. 5D;
P<0.05) and invasion (Fig. 5E;
P<0.05) of Huh7 and Hep3B cells. These data confirmed that
miR-663b may serve inhibitory roles in HCC, at least partly, by
decreasing GAB2 expression.
Discussion
Previous studies have demonstrated that numerous
tumor-specific miRNAs are upregulated or downregulated in HCC, and
that their dysregulation is implicated in the HCC occurrence and
development (30–32). Therefore, investigation of crucial
miRNAs involved in HCC oncogenesis and progression may provide
novel insights into the therapy of patients with this malignant
tumor. The present study demonstrated that miR-663b expression was
significantly downregulated in HCC tissues and cell lines.
Exogenous overexpression of miR-663b impeded the proliferation and
invasion of HCC cells. Additionally, GAB2 was demonstrated to be a
direct target gene of miR-663b in HCC cells. Furthermore, it was
identified that miR-663b expression was negatively correlated with
GAB2 expression in HCC tissues. Finally, restoration of GAB2
expression partially abrogated the tumor suppressive roles of
miR-663b in HCC cells. All these results demonstrated that miR-663b
is downregulated in HCC, and inhibits the proliferation and
invasion of HCC cells by directly targeting GAB2.
miR-663b dysregulation is observed in several types
of human malignancies. For example, miR-663b is upregulated in
nasopharyngeal carcinoma tissues and cell lines (21). High miR-663b expression is markedly
associated with clinical stage and lymph node metastasis of
patients with nasopharyngeal carcinoma (21). Expression of miR-663b was increased
in the plasma of patients with bladder cancer (22). miR-663b level was suggested to be
an effective circulating biomarker for the diagnosis of bladder
cancer (22). miR-663b is also
overexpressed in osteosarcoma (23) and endometrial cancer (24) tissues. Patients with endometrial
cancer with high miR-663b levels exhibited poorer prognosis
compared with those patients with low miR-663b levels (24). However, miR-663b is downregulated
in pancreatic cancer tissues and cell lines (33). These contrary data indicated that
the expression pattern of miR-663b in human cancer is
tissue-specific. Therefore, miR-663b may be a promising biomarker
for the diagnosis of these human cancer types.
Aberrantly expressed miR-663b serves oncogenic roles
in the progression of multiple human cancer types. For example,
miR-663b overexpression promotes cell growth and metastasis of
nasopharyngeal carcinoma (21).
Shu et al (23)
demonstrated that inhibition of miR-663b suppresses the
proliferation and induced apoptosis of osteosarcoma cells. Wang
et al (24) identified that
miR-663b re-expression promotes cell viability and repressed cell
apoptosis in endometrial cancer. Nevertheless, miR-663b serves
tumor-suppressing roles in pancreatic cancer via affecting cell
proliferation, colony formation, migration, invasion and apoptosis
in vitro and tumor growth in vivo (33). These conflicting results
demonstrated that the biological roles of miR-663b in the
tumorigenesis and tumor development exhibit tissue specificity. In
general, miR-663b may be developed as a prospective therapeutic
cancer target for the treatment of patients with these specific
cancer types.
Multiple genes have been confirmed to be the direct
target genes of miR-663b, including TUSC2 in nasopharyngeal
carcinoma (21), tumor protein P73
in osteosarcoma (23), apoptosis
facilitator Bcl-2-like protein 14 in endometrial cancer (24) and insulin-like growth factor 2 in
pancreatic cancer (33). In the
present study, GAB2, a member of the mammalian Gab
scaffolding/adapter family, was validated as a direct and
functional downstream target of miR-663b in HCC; its expression was
upregulated in HCC tissues and a subset of HCC cell lines compared
with normal tissues and cell lines. High GAB2 expression exhibits a
significant correlation with histological grade and tumor size in
patients with HCC (27,28). Patients with HCC with high GAB2
expression exhibit poorer prognosis compared with those patients
with low GAB2 levels (27).
Kaplan-Meier survival curves identified GAB2 expression level as an
independent prognostic marker for predicting overall survival of
patients with HCC (28). In
addition, GAB2 serves as an oncogene in HCC carcinogenesis and
cancer progression through regulating a variety of biological
behaviors, including cell proliferation, cycle, apoptosis,
migration and invasion in vitro and tumor growth in
vivo (28,29). On the basis of the results of the
present study, miR-663b and its target GAB2 may be potential
therapeutic targets for patients with HCC.
In conclusion, the results of the present study
demonstrated that miR-663b is downregulated in HCC tissues and cell
lines. The restoration of miR-663b expression inhibited cell
proliferation and invasion in HCC through directly targeting and
inhibiting GAB2. These results suggested the key roles of miR-663b
in HCC progression, in the diagnosis and therapy of patients with
HCC. However, the present study did not investigate the effects of
miR-663b in HCC cell apoptosis in vitro, or in vivo
tumor growth and metastasis. In addition, the association between
GAB2 expression and histological grades in patients with HCC was
not described. These are limitations of the present study, and will
be resolved in the future.
Acknowledgements
Not applicable.
Funding
The present study was supported by the Special
Research Funds for Discipline Construction of High Level University
Construction (grant no. 2013SXTS02).
Availability of data and materials
The datasets used and/or analyzed during the present
study are available from the corresponding author on reasonable
request.
Authors' contributions
LG and JY designed the study. LG and BL performed
the CCK-8 assay and reverse transcription-quantitative polymerase
chain reaction. MM conducted the Transwell invasion and luciferase
reporter assays. Western blot analysis and statistical analysis
were performed by JY and JJ. All authors read and approved the
final draft.
Ethics approval and consent to
participate
The present study was approved by the Research
Ethics Committee of Affiliated Hospital of Yan'an University, and
was performed in accordance with the Declaration of Helsinki and
the guidelines of the Ethics Committee of Affiliated Hospital of
Yan'an University. Written informed consent was obtained from all
patients for the use of their clinical tissues.
Patient consent for publication
All patients enrolled in this study provided written
informed consent for the publication of any associated data.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Siegel RL, Miller KD and Jemal A: Cancer
statistics, 2016. CA Cancer J Clin. 66:7–30. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Global Burden of Disease Cancer
Collaboration, ; Fitzmaurice C, Dicker D, Pain A, Hamavid H,
Moradi-Lakeh M, MacIntyre MF, Allen C, Hansen G, Woodbrook R, et
al: The global burden of cancer 2013. JAMA Oncol. 1:505–527. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Cabrera R and Nelson DR: Review article:
The management of hepatocellular carcinoma. Aliment Pharmacol Ther.
31:461–476. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Marrero JA and Welling T: Modern diagnosis
and management of hepatocellular carcinoma. Clin Liver Dis.
13:233–247. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Forner A, Hessheimer AJ, Isabel Real M and
Bruix J: Treatment of hepatocellular carcinoma. Crit Rev Oncol
Hematol. 60:89–98. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Ishizawa T, Hasegawa K, Aoki T, Takahashi
M, Inoue Y, Sano K, Imamura H, Sugawara Y, Kokudo N and Makuuchi M:
Neither multiple tumors nor portal hypertension are surgical
contraindications for hepatocellular carcinoma. Gastroenterology.
134:1908–1916. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Forner A, Llovet JM and Bruix J:
Hepatocellular carcinoma. Lancet. 379:1245–1255. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Yu MC and Yuan JM: Environmental factors
and risk for hepatocellular carcinoma. Gastroenterology 127 (5
Suppl 1). S72–S78. 2004.
|
|
9
|
Borchert GM, Lanier W and Davidson BL: RNA
polymerase III transcribes human microRNAs. Nat Struct Mol Biol.
13:1097–1101. 2006. View
Article : Google Scholar : PubMed/NCBI
|
|
10
|
Bartel DP: MicroRNAs: Genomics,
biogenesis, mechanism, and function. Cell. 116:281–297. 2004.
View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Klingenberg M, Matsuda A, Diederichs S and
Patel T: Non-coding RNA in hepatocellular carcinoma: Mechanisms,
biomarkers and therapeutic targets. J Hepatol. 67:603–618. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Weidle UH, Birzele F, Kollmorgen G and
Nopora A: Potential microRNA-related targets for therapeutic
intervention with ovarian cancer metastasis. Cancer Genomics
Proteomics. 15:1–15. 2018.PubMed/NCBI
|
|
13
|
Celano M, Rosignolo F, Maggisano V, Pecce
V, Iannone M, Russo D and Bulotta S: MicroRNAs as Biomarkers in
Thyroid Carcinoma. Int J Genomics. 2017:64965702017. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Irmak-Yazicioglu MB: Mechanisms of
microRNA deregulation and microRNA targets in gastric cancer. Oncol
Res Treat. 39:136–139. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Zhang Y, Yang L, Wang S, Liu Z and Xiu M:
MiR-29a suppresses cell proliferation by targeting SIRT1 in
hepatocellular carcinoma. Cancer Biomark. 22:151–159. 2018.
View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Li H, Wang H and Ren Z: MicroRNA-214-5p
inhibits the invasion and migration of hepatocellular carcinoma
cells by targeting Wiskott-Aldrich syndrome like. Cell Physiol
Biochem. 46:757–764. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Wu M, Huang C, Huang X, Liang R, Feng Y
and Luo X: MicroRNA-144-3p suppresses tumor growth and angiogenesis
by targeting SGK3 in hepatocellular carcinoma. Oncol Rep.
38:2173–2181. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Hayes CN and Chayama K: MicroRNAs as
biomarkers for liver disease and hepatocellular carcinoma. Int J
Mol Sci. 17:2802016. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Anwar SL and Lehmann U: MicroRNAs:
Emerging Novel Clinical Biomarkers for Hepatocellular Carcinomas. J
Clin Med. 4:1631–1650. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Liu Z, Wang Y, Dou C, Sun L, Li Q, Wang L,
Xu Q, Yang W, Liu Q and Tu K: MicroRNA-1468 promotes tumor
progression by activating PPAR-gamma-mediated AKT signaling in
human hepatocellular carcinoma. J Exp Clin Cancer Res. 37:492018.
View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Liang S, Zhang N, Deng Y, Chen L, Zhang Y,
Zheng Z, Luo W, Lv Z, Li S and Xu T: miR-663b promotes tumor cell
proliferation, migration and invasion in nasopharyngeal carcinoma
through targeting TUSC2. Exp Ther Med. 14:1095–1103. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Du M, Shi D, Yuan L, Li P, Chu H, Qin C,
Yin C, Zhang Z and Wang M: Circulating miR-497 and miR-663b in
plasma are potential novel biomarkers for bladder cancer. Sci Rep.
5:104372015. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Shu Y, Ye W, Gu YL and Sun P: Blockade of
miR-663b inhibits cell proliferation and induces apoptosis in
osteosarcoma via regulating TP73 expression. Bratisl Lek Listy.
119:41–46. 2018.PubMed/NCBI
|
|
24
|
Wang YL, Shen Y, Xu JP, Han K, Zhou Y,
Yang S, Yin JY, Min DL and Hu HY: Pterostilbene suppresses human
endometrial cancer cells in vitro by down-regulating miR-663b. Acta
Pharmacol Sin. 38:1394–1400. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Faria SC, Szklaruk J, Kaseb AO, Hassabo HM
and Elsayes KM: TNM/Okuda/Barcelona/UNOS/clip international
multidisciplinary classification of hepatocellular carcinoma:
Concepts, perspectives, and radiologic implications. Abdom Imaging.
39:1070–1087. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(-Delta Delta C(T)) method. Methods. 25:402–408. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Hu X, He B, Zhou L, Xie H and Zheng S:
Expression pattern and clinical significance of Gab2 protein in
hepatocellular carcinoma. Clin Lab. 62:1087–1092. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Chen Y, Liu Q, Wu M, Li M, Ding H, Shan X,
Liu J, Tao T, Ni R and Chen X: GAB2 promotes cell proliferation by
activating the ERK signaling pathway in hepatocellular carcinoma.
Tumour Biol. 37:11763–11773. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Cheng J, Zhong Y, Chen S, Sun Y, Huang L,
Kang Y, Chen B, Chen G, Wang F, Tian Y, et al: Gab2 mediates
hepatocellular carcinogenesis by integrating multiple signaling
pathways. FASEB J. 31:5530–5542. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Zhang T, Liu W, Meng W, Zhao H, Yang Q, Gu
SJ, Xiao CC, Jia CC and Fu BS: Downregulation of miR-542-3p
promotes cancer metastasis through activating TGF-beta/Smad
signaling in hepatocellular carcinoma. Onco Targets Ther.
11:1929–1939. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Cui H, Song R, Wu J, Wang W, Chen X and
Yin J: MicroRNA-337 regulates the PI3K/AKT and Wnt/β-catenin
signaling pathways to inhibit hepatocellular carcinoma progression
by targeting high-mobility group AT-hook 2. Am J Cancer Res.
8:405–421. 2018.PubMed/NCBI
|
|
32
|
Morishita A and Tsutomu M: MicroRNAs as
possible biomarkers for hepatocellular carcinoma. Hepatol Res.
48:499–501. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Cai H, An Y, Chen X, Sun D, Chen T, Peng
Y, Zhu F, Jiang Y and He X: Epigenetic inhibition of miR-663b by
long non-coding RNA HOTAIR promotes pancreatic cancer cell
proliferation via up-regulation of insulin-like growth factor 2.
Oncotarget. 7:86857–86870. 2016. View Article : Google Scholar : PubMed/NCBI
|