Introduction
Gestational diabetes mellitus (GDM) is a type of
diabetes mellitus (DM) that develops in pregnant women as the
pregnancy progresses, and can be caused by maternal hyperglycemia
and decreased glucose intolerance (1). A high glucose concentration in the
internal environment caused by maternal hyperglycemia is dangerous
to the fetus, as glucose can be easily transferred via the
fetus-placenta circulation. However, maternal insulin cannot be
transferred across the placental barrier, and therefore under high
glucose conditions the fetus experiences fetal hyperinsulinemia,
leading to fetal disorders (2).
Previous studies have demonstrated that despite recovery following
pregnancy, mothers with GDM are more likely to have type 2 DM and
other cardiovascular diseases (3).
In addition, phenotypic changes induced by GDM can persist in
infants carried by mothers with GDM, and these infants are more
likely to have DM or obesity in later life (4). Therefore, epigenetic modifications
may be involved in the GDM-induced high glucose environment and
further accentuating the physical consequence of GDM (5).
MicroRNAs (miRNAs) are small non-coding RNAs that
can modulate gene expressions at the post-transcriptional level.
miRNAs (miR) have been widely studied in tumor research, as they
are closely associated with tumorigenesis and the development of
numerous cancer types. In cervical cancer, miR-125a serves as a
tumor suppressor by inhibiting tumor growth, invasive ability and
metastasis via sponging STAT3 (6).
Furthermore, miR-193a inhibits cell proliferation and metastasis in
breast cancer by targeting WT1 transcription factor (7). However, to the best of our knowledge,
there are few studies investigating the association between miRNAs
and GDM. In total, 6 miRNAs: miR-155-5p; −21-3p; −146b-5p; −223-3p;
−517-5p; and miR-29a-3p, may have the potential to increase the
likelihood of GDM occurrence in pregnant women (8). In addition, a miRNA profiling
analysis in patients with GDM revealed that miR-195-5p exhibited
the highest fold upregulation in GDM and was involved in metabolism
(9). However, the function and
mechanism of miR-195-5p in human umbilical vein endothelial cells
(HUVECs) with GDM remains unknown.
It has been hypothesized that epigenetic changes
induce gene expression deregulation in response to perturbations of
the utero environmental (10).
Enhancer of zeste homolog 2 (EZH2) is a component of the polycomb
repressor complex 2 (PRC2). Moreover, PRC2 is an epigenetic
regulator that initiates and maintains the trimethylation of
histone H3 on lysine 27 (H3K27me3), which is an epigenetic marker
associated with heterochromatin formation and transcription
silencing (11,12). A previous study demonstrated that
EZH2 is a target of GDM and mediates the damaging effects of GDM on
HUVECs (10). However, the
function of EZH2 on cell survival of GDM-HUVECs and its association
with miR-195-5p are not fully understood.
Therefore, the aim of the present study was to
investigate the effects of miR-195-5p and EZH2, a hypothesized
target gene for miR-195-5p, on HUVECs.
Materials and methods
Patient recruitment and ethical
approval
Human umbilical cords were obtained from 25
full-term healthy women (age, 35–23) and 25 women with GDM (age,
45–25) in Jingmen No. 1 People's Hospital from February 2017 to
March 2018. The diagnosis of GDM was based on the 2010 GDM
diagnosis criteria introduced by International Association of The
Diabetes and Pregnancy Study Group (13). The study was approved by Jingmen
No. 1 People's Hospital Ethics Committee (approval no.,
NJ201702463) and was performed in accordance with The Declaration
of Helsinki (1964) and its later amendments. All patients provided
written informed consent.
HUVECs
HUVECs derived from umbilical cords were prepared
(14). Umbilical cord veins were
rinsed in PBS, and HUVECs were incubated with 0.05% collagenase
type II derived from Clostridium histolyticum;
(Sigma-Aldrich; Merck KGaA) in M199 medium (Invitrogen; Thermo
Fisher Scientific Inc.) containing 100 U/ml penicillin G sodium
salt and 100 µg/ml streptomycin sulfate (Sigma-Aldrich; Merck KGaA)
at 37°C with 5% CO2 for 10 min. Cells were centrifugated
at 1,000 × g for 10 min at room temperature, and then resuspended
in M199 medium containing 10% FBS (Invitrogen; Thermo Fisher
Scientific, Inc.) and 10% newborn calf serum (Invitrogen; Thermo
Fisher Scientific, Inc.), 2 mM glutamine and antibiotics as
mentioned above at 37°C with 5% CO2. For detecting cell
endothelial properties, cells were cultured in endothelial cell
basal medium (Lonza Group AG) containing 10% FBS at 37°C for 24 h
and a EGM-2 Epithelial SingleQuots™ kit (cat. no. CC-4176; Lonza
Group AG) was used. The cells were fixed with 1% paraformaldehyde
at 37°C for 30 min. The cells were then analyzed using a flow
cytometer (FACS Canto II; BD Biosciences) with cellQuest software
(version 7.5.3; BD Biosciences), with mouse anti-human
phycoerythrin-platelet endothelial cell adhesion molecule (CD31)
antibody (cat. no. MHCD3104; eBioscience; Thermo Fisher Scientific,
Inc.; 1:100) at 4°C for 20 min used as an endothelial cell
marker.
Cell Counting Kit-8 assay (CCK-8)
After 24 h cell cultivation, a cell solution with a
density of 3×104 cells/ml was prepared and cultured in
96-well plates at 37°C for 24 h. Then, cell viability was
determined using a CCK-8 assay (Dojindo Molecular Technologies,
Inc.) according to the manufacturer's protocol, and optical density
value was measured at the wavelength of 450 nm.
Colony formation assay
HUVECs were lysed using 0.25% trypsin-0.02% EDTA
(Gibco; Thermo Fisher Scientific, Inc.), and the cell suspension
was prepared and seeded (1×104) into 6-well plate for 10
days at 37°C. HUVECs were washed with PBS 3 times, and cells were
fixed with 10% methanol under room temperature for 15 min and
stained with 0.1% crystal violet solution at room temperature for
30 min. Images of the cells were captured (light microscope;
magnification, ×1), The number of colonies were counted using a
light microscope (Olympus CKX41; Olympus Corporation).
Flow cytometry assay
Cell suspension was prepared in 500 µl binding
buffer (Gibco; Thermo Fisher Scientific, Inc.). Cell staining was
performed with Annexin V-fluorescein isothiocyanate and propidium
iodide (20 µg/ml; Biovision, Inc.) to determine the apoptotic rate
of HUVECs in the dark for 15 min at room temperature. Cells were
analyzed by flow cytometry (FACSCalibur; BD Biosciences) with BD
CellQuest Pro Software version 1.2 (BD Biosciences) in the
dark.
Cell transfection
pCMV6-XL5-EZH2 (cat. no. SC101257) and its empty
control vector were purchased from OriGene Technologies, Inc.
miR-195-5p mimics (cat. no. miR10000461-1-5,
5′-CGGUUAUAAAGACACGACGAU-3′) and its negative control RNA were
obtained from Guangzhou RiboBio Co., Ltd. HUVECs were incubated to
70–80% confluence at 37°C for 24 h. A total of 20 µM miR-195-5p
mimics, negative control (NC), miR-195-5p mimic, mimic + EZH2 or
EZH2 were used and the transfection procedure was conducted using
Lipofectamine 2000 (Invitrogen; Thermo Fisher Scientific, Inc.).
After 24 h, the cells were used to perform the subsequent
experiments.
Dual-luciferase reporter assay
The 3′-untranslated regions (UTR) of EZH2 contained
a predicted binding-site for miR-195-5p, as identified using
Targetscan7.2 (http://www.targetscan.org/vert_72/) analysis. To
assess whether miR-195-5p specifically targets EZH2, this was
examined using the luciferase pGL3-Basic vector (Promega
Corporation). Firstly, wild-type (WT) and mutant (MUT) EZH2-3′-UTR
were purchased from Shanghai GenePharma Co., Ltd. and inserted into
the luciferase vector. Then, the miR-195-5p mimic was
co-transfected with WT or MUT luciferase vector into 293T cells
(American Type Culture Collection) using Lipofectamine®
2000 (Invitrogen; Thermo Fisher Scientific, Inc.). After 48 h of
transfection, luciferase activity was measured by the
Dual-Luciferase Reporter Assay System (Promega Corporation).
Renilla luciferase activity was detected in the same
method.
Reverse transcription-quantitative
polymerase chain reaction (RT-qPCR)
miR-195-5p and EZH2 mRNA expression levels in HUVECs
were determined by RT-qPCR. Total RNA of EZH2 was harvested using
QIAzol Lysis reagent (Qiagen, Inc.), and total RNAs of miR-195-5p
was isolated using miRVana miRNA Isolation kit (Thermo Fisher
Scientific, Inc.). RT of EZH2 and miR-195-5p from RNAs to cDNAs was
performed using a QuantiTect RT kit (Qiagen, Inc.) and TaqMan miRNA
RT kit (Thermo Fisher Scientific, Inc.) at 37°C for 45 min and then
at 80°C for 5 min. The Step One Plus RT PCR system (Applied
Biosystems; Thermo Fisher Scientific, Inc.) was used to perform
RT-qPCR. The PCR reaction was as follows: Initital denaturation at
95°C for 5 min, followed by 40 cycles at 94°C for 30 sec, 55°C for
30 sec, 72°C for 45 sec and 72°C extension for 10 min. Data were
analyzed using 2−ΔΔCq method (15). GAPDH and U6 served as internal
references for EZH2 and miR-195-5p, respectively. The primers
sequences used are shown in Table
I.
 | Table I.Primers sequence used for reverse
transcription- quantitative PCR. |
Table I.
Primers sequence used for reverse
transcription- quantitative PCR.
| Primer | Sequence |
|---|
| EZH2 |
| Forward |
5′-CCTGAAGTATGTCGGCATCGAAAGAG-3′ |
| Reverse |
5′-TGCAAAAATTCACTGGTACAAAACACT-3′ |
| miR-195-5p |
| Forward |
5′-GTCGTATCCAGTGCAGGGTCCGAGGT-3′ |
| Reverse |
5′-ATTCGCACTGGATACGACTATAACCG-3′ |
| U6 |
| Forward |
5′-CTCGCTTCGGCAGCACA-3′ |
| Reverse |
5′-AACGCTTCACGAATTTGCGT-3′ |
| GAPDH |
| Forward |
5′-CGGAGTCAACGGATTTGGTCGTAT-3′ |
| Reverse |
5′-AGCCTTCTCCATGGTGGTGAAGAC-3′ |
Western blot analysis
Following rinsing the cells in cold PBS 3 times,
HUVECs were lysed in lysis buffer containing 10 mM HEPES (pH 7.5),
10 mM KCl, 1.5 mM MgCl2, 0.34 M sucrose, 10% glycerol
and 0.1% TritonX-100 with protease inhibitors at 4°C. Total
proteins were extracted and the concentration was determined with a
bicinchoninic acid assay kit (Thermo Fisher Scientific, Inc.).
Proteins (20 µg/lane) were isolated on 10% SDS-PAGE and then
transferred into PVDF membranes, which were blocked with 5% skimmed
milk powder at room temperature for 2 h. Blots were incubated
overnight at 4°C with the primary antibody against EZH2 (cat. no.
GTX110384; 1:500; GeneTex, Inc.) and GAPDH (cat. no. GTX100118;
1:5,000; GeneTex, Inc.). Following incubation, the membranes were
washed three times with TBST (0.05% Tween20) and cultured with
horseradish peroxidase-conjugated goat anti-rabbit immunoglobulin G
(cat. no., GTX213110-01; 1:1,000; GeneTex, Inc.) for 2 h at room
temperature. The bands were detected by chemiluminescence (ECL™
Prime; GE Healthcare Life Sciences), imaged on X-ray film (GE
Healthcare Lifesciences) and quantified using ImageJ (version
1.8.0; National Institutes of Health).
Statistical analysis
All experiments were repeated in triplicate.
Statistical analyses were performed using SPSS v.19.0 software (IBM
Corp.), and GraphPad Prism v.5.02 software (GraphPad Prism
Software, Inc.) was used to create the graphs. Data are presented
as the mean ± standard deviation, and were evaluated using unpaired
Student's t-test in luciferase activity assay, independent samples
t-test in comparison with Healthy and GDM groups or analysis of
variance followed by Tukey's post-hoc test. P<0.05 was
considered to indicate a statistically significant difference.
Results
GDM-induced phenotypic alterations in
HUVECs
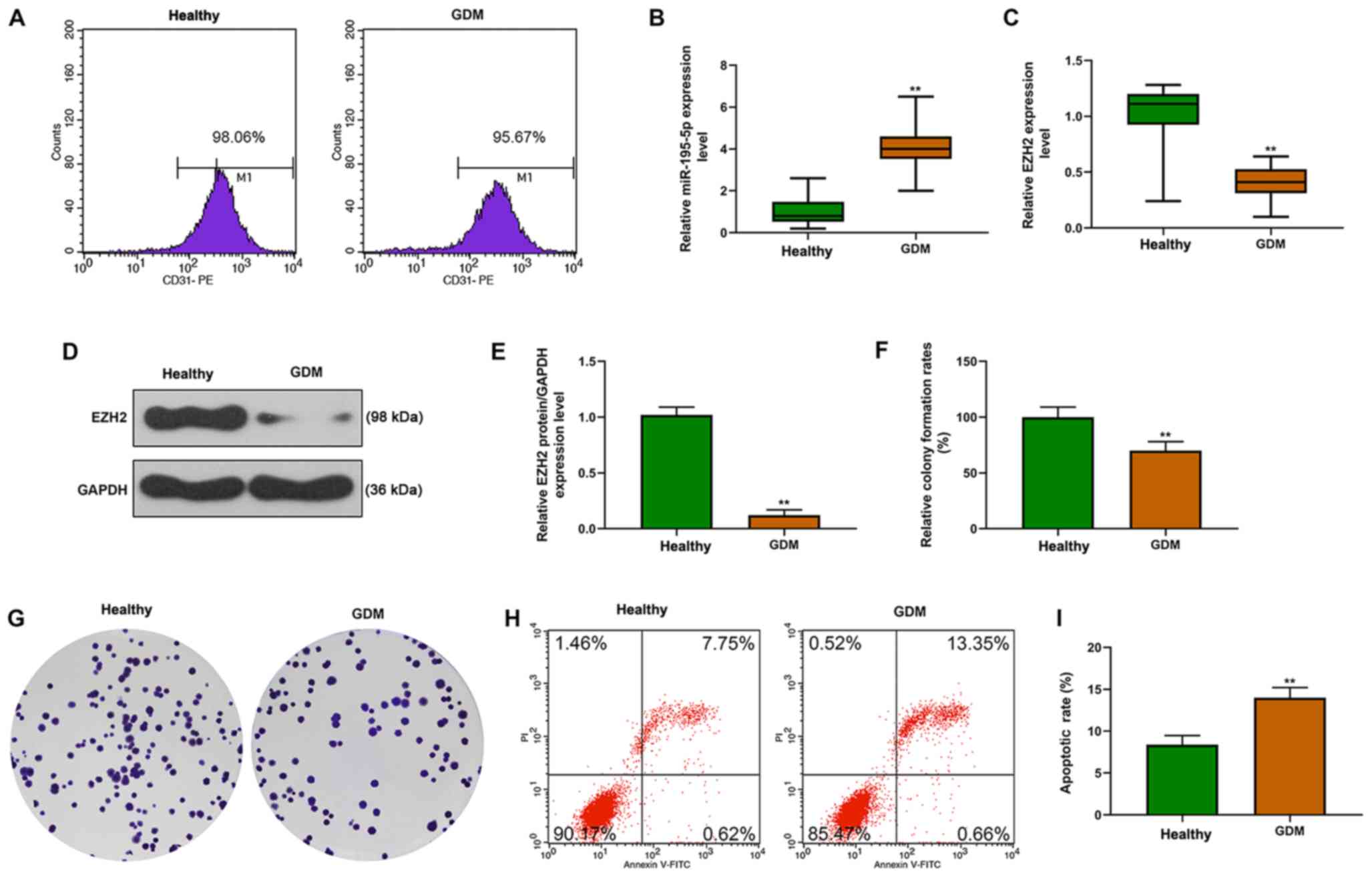
The endothelial phenotype of HUVECs indicated by
CD31 was detected by flow cytometry, and >95% of GDM-HUVECs and
healthy HUVECs were identified as CD31-positive (Fig. 1A). Moreover, compared with the
healthy controls, it was identified that miR-195-5p mRNA expression
in GDM-HUVECs was significantly increased, but EZH2 mRNA and
protein expression levels were significantly decreased (Fig. 1B-E). In addition, decreased cell
proliferation (Fig. 1F and G) and
elevated apoptosis were identified in GDM-HUVECs (Fig. 1H and I).
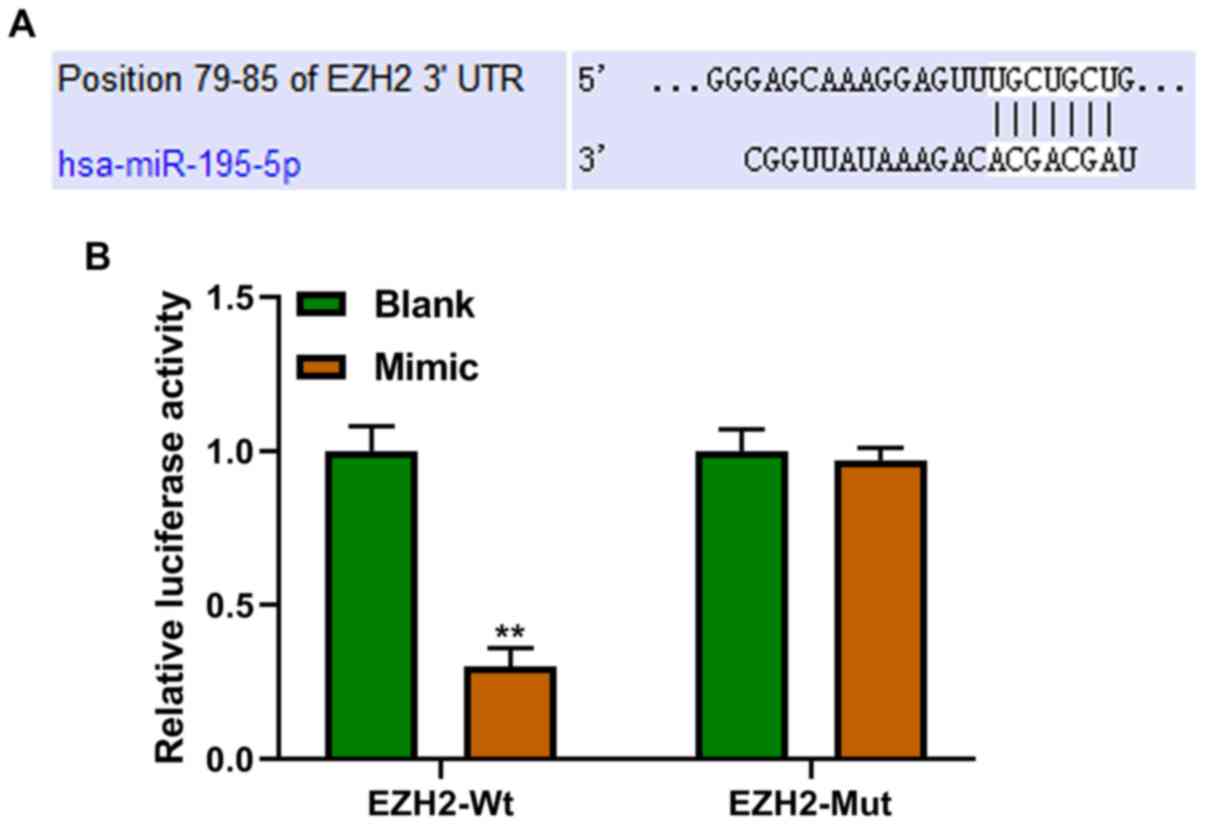
miR-195-5p targets EZH2
EZH2 was predicted to be the target gene of
miR-195-5p (Fig. 2A), which was
confirmed using a dual-luciferase reporter assay. Furthermore, it
was identified that cells transfected with miR-195-5p mimics
exhibited significantly decreased luciferase activity compared with
the blank group in the EZH2-WT group (Fig. 2B).
Effects of miR-195-5p and EZH2 on
functional capacities of control HUVECs
Cell viability, proliferation and apoptosis were
detected following overexpression of miR-195-5p, EZH2 or the
combination of both in control HUVECs (Fig. 3A-E). It was demonstrated that
miR-195-5p overexpression suppressed cell viability and
proliferation, but increased the apoptotic rate. However,
overexpression of EZH2 promoted cell viability and proliferation,
and inhibited apoptosis. In addition, the combination of the
overexpression of miR-195-5p and EZH2 was identified to partially
alleviate the effects caused by miR-195-5p overexpression on cell
viability, proliferation and apoptosis.
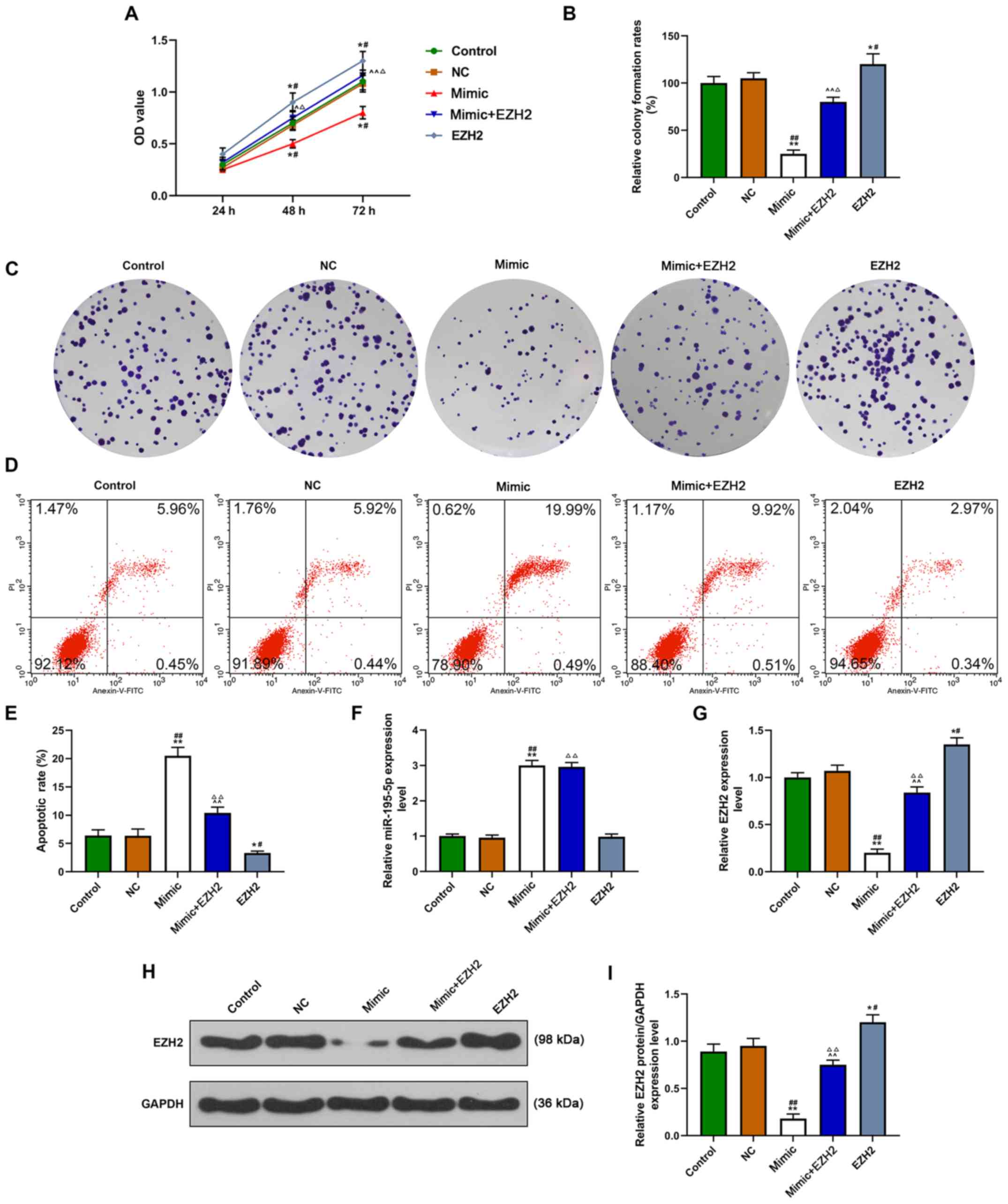 | Figure 3.Effects of miR-195-5p and EZH2 on cell
viability, proliferation and apoptosis in healthy HUVECs. Groups
were divided into control (cells treated with PBS), NC, miR-195-5p
mimic, mimic + EZH2 and EZH2 groups. (A) Cell viability in HUVECs
determined using a Cell Counting Kit-8 assay. (B) Cell
proliferation in HUVECs determined by colony formation assays. (C)
Representative images of cell colonies (D) Flow cytometry was used
to assess cell apoptosis in HUVECs. (E) Quantification of the flow
cytometry data. (F) miR-195-5p mRNA expression in HUVECs was
determined by RT-qPCR. (G) RT-qPCR and (H) western blot analysis
were used to determine EZH2 mRNA and protein expression levels in
HUVECs. (I) Quantification of the western blot analysis
densitometric data. *P<0.05, **P<0.01 vs. Control.
#P<0.05, ##P<0.01 vs. NC.
^P<0.05, ^^P<0.01 vs. Mimic;
ΔP<0.05, ΔΔP<0.01 vs. EZH2. miR,
microRNA; EZH2, enhancer of zeste homolog 2; GDM, gestational
diabetes mellitus; NC, negative control; HUVECs, human umbilical
vein endothelial cells; RT-qPCR, reverse transcription-quantitative
polymerase chain reaction; FITC, fluorescein isothiocyanate; PI,
propidium iodide; OD, optical density. |
Effects of miR-195-5p and EZH2 on the
expression levels of miR-195-5p and EZH2 in control HUVECs
The present results suggested that the expression of
miR-195-5p was upregulated by miR-195-5p overexpression, while EZH2
overexpression had no significant effect on miR-195-5p (Fig. 3F). Furthermore, EZH2 expression was
affected by overexpression of both miR-195-5p and EZH2, as
upregulation of EZH2 promoted EZH2 expression, while miR-195-5p
overexpression downregulated EZH2 (Fig. 3G-I).
Discussion
In pregnant women with DM, exposure to hyperglycemia
will result in long-lasting effects to the vascular cells, which
can cause vascular complications (16,17).
The underlying epigenetic mechanisms, involving DNA modification
and histone marks, affect the activation of gene transcription
(10). These epigenetic
modifications enable the cells to respond to changing internal and
external environments and adjust to these environmental
stimulations. Initially, epigenetics was referred to as to
chromatin changes that were inherited; however, this definition has
been expanded, and epigenetics are now known to have long-term
effects, from pregnancy through to every stage of human growth
(18). In addition, epigenetic
modification can be altered (strengthened, alleviated or even
erased) over time due to the effects of drug use, change in
lifestyles or other environment stimuli (10). DM is closely associated with the
genome (19), and miR-216 may be a
potential therapeutic agent for DM due to its role in angiogenesis
and its protective effects on vascular integrity in preventing the
pathogenesis and complications of DM (20). Furthermore, miR-155, miR-146a and
long non-coding RNA homeodomain interacting protein kinase 3 have
been demonstrated to serve important roles in the dysfunction of
type 1 or type 2 DM (21–23). Unlike DM, the changes to glucose
metabolism caused by GDM during pregnancy can be reverted following
childbirth (24). However, GDM is
associated with increased cardiovascular risk and type 2 DM in both
the mothers and their children in later life (24).
Endothelial dysfunction is a common feature in
numerous pregnancy-associated diseases, as well as in the different
forms of DM (25). HUVECs from
human umbilical cords are widely used as a cellular model for the
analysis of the effects of uterine environmental changes on fetal
endothelium. Previous GDM data have demonstrated that trophoblasts
from human term placenta exhibit high apoptotic rates, and that
maternal GDM is associated with alternations in apoptotic and
inflammatory gene expression levels (26). Furthermore, proliferation of
pancreatic β cells is inhibited by decreases in placental growth
factor in GDM (27).
The association between GDM and miRNAs has also been
previously investigated: miRNA expression levels in HUVECs were
associated with the occurrence of GDM, and alternations of miRNAs
in HUVECs are speculated to affect metabolic processes of GDM
(28). A previous study revealed
that miR-195-5p was significantly upregulated in patients with deep
vein thrombosis, which is one of the most common cardiovascular
diseases (29). In addition, it
has been demonstrated that the expression of miR-195-5p in GDM is
significantly increased in comparison with the healthy group
(9). Similar to these data
concerning deep vein thrombosis, the results of the present study
identified that miR-195-5p was also significantly upregulated in
GDM. In addition, the downstream target gene of miR-195-5p was
identified to be EZH2, which elicited the opposite effects on
cellular processes compared with miR-195-5p: It was identified that
high expression of miR-195-5p decreased cell viability and
proliferation, and promoted apoptosis, while EZH2 overexpression
resulted in the opposite effects. Furthermore, the combination of
both factors partially attenuated the previous inhibition of cell
growth and promotion of cell apoptosis induced by the upregulation
of miR-195-5p.
Previous studies have indicated that EZH2 enhances
the proliferative capacity of gastric cancer cells by suppressing
cyclin-dependent kinase inhibitor 1 (p21) expression (30,31).
Furthermore, in laryngeal carcinoma, EZH2 promotes cell
proliferation by regulating Runt-related transcription factor 3
(RUNX3) (32). It has also been
demonstrated that EZH2 promotes the epigenetic silencing of miR-205
and miR-31 to suppresses apoptosis (1). Therefore, EZH2 can regulate
proliferation and apoptosis in multiple cell types. In concordance
with these previous studies, the results of the present study
indicated that EZH2 was downregulated in GDM, and that
overexpression of EZH2 enhanced proliferation and inhibited
apoptosis of HUVECs. Moreover, we hypothesized that EZH2 may
regulate proliferation and apoptosis by directly modulating the
cell cycle, proliferation, and apoptotic-associated genes or
miRNAs, including p21, RUNX3, miR-205 and miR-31.
Previous studies have focused on the role of
miR-195-5p in tumorigenesis: In hepatocellular carcinoma, miR-195
can act as a tumor suppressor via the inhibition of cell
proliferation by targeting astrocyte elevated gene-1 (33). In papillary thyroid carcinoma, it
was demonstrated that miR-195 inhibited tumor growth and prevented
metastasis via sponging cyclin D1 and fibroblast growth factor 2,
suppressing tumor development (34). Furthermore, EZH2 was identified to
be closely associated with cancer, with regards to its therapeutic
effects on cell proliferation, invasion and metastasis (35). Moreover, by suppressing the
expression of EZH2, which promotes the phosphorylation process in
breast cancer cells, the progression of breast cancer is markedly
inhibited (36). In GDM-HUVECs,
EZH2 can affect the regulation of β-cells via the interaction with
prolactin receptor genes (37). A
previous study has also revealed that EZH2 is downregulated in
GDM-HUVECs, and that the overexpression of EZH2 inhibits apoptosis
and promotes migration by targeting the miR-101 promoter for
inhibition of HUVECs (10).
Therefore, based on previous data and the results from the present
study, we hypothesized that miR-195-5p and EZH2 may participate in
cellular processes of HUVECs, and the interaction of these 2
factors affects the progression of GDM. EZH2 was demonstrated to
suppress the expression of miR-195-5p in cervical cancer cells via
increasing H3K27me3 in the promoter region of miR-195 (31), which is different from the present
results. Therefore, it is hypothesized that the association between
miR-195-5p and EZH2 may be complicated, and vary between different
cell types. Therefore, further studies are required to investigate
the mechanism underlying the association between miR-195-5p and
EZH2. Furthermore, additional indicators associated with GDM-HUVECs
should be assessed in future experiments, such as changes in
apoptotic and inflammatory gene expression levels, metabolic
product levels and angiogenesis. Moreover, the function of
miR-195-5p and EZH2 in GDM should be further examined in an animal
model.
In conclusion, the present study investigated the
role of miR-195-5p in HUVECs with GDM, and identified that
miR-195-5p may be associated with cell proliferation and apoptosis
of HUVECs by sponging EZH2. The present results provide evidence of
a novel underlying mechanism involving miRNA and epigenetic
regulation in GDM, which may facilitate the development of
potential therapeutic strategies for GDM.
Acknowledgements
Not applicable.
Funding
No funding was received.
Availability of data and materials
The datasets used and/or analyzed during the current
study are available from the corresponding author on reasonable
request.
Authors' contributions
XL made substantial contributions to conception and
design of the project. ZZ, XZ and XL were responsible for data
acquisition, data analysis and interpretation. XL was involved in
drafting the article and critically revising it for important
intellectual content. All authors read and approved the final
version of the manuscript. All authors agreed to be accountable for
all aspects of the work in ensuring that questions related to the
accuracy or integrity of the work are appropriately investigated
and resolved.
Ethics approval and consent to
participate
The present study was approved by Jingmen No. 1
People's Hospital Ethics Committee (approval no. NJ201702463) and
was performed in accordance with The Declaration of Helsinki (1964)
and its later amendments. All patients provided written informed
consent.
Patient consent for publication
All patients provided written informed consent.
Competing interests
The authors declare that they have no competing
interests.
Glossary
Abbreviations
Abbreviations:
|
GDM
|
gestational diabetes mellitus
|
|
DM
|
diabetes mellitus
|
|
HUVECs
|
human umbilical vein endothelial
cells
|
|
miRNAs
|
microRNAs
|
References
|
1
|
Zhang Q, Padi SK, Tindall DJ and Guo B:
Polycomb protein EZH2 suppresses apoptosis by silencing the
proapoptotic miR-31. Cell Death Dis. 5:e14862014. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Desoye G and Nolan CJ: The fetal glucose
steal: An underappreciated phenomenon in diabetic pregnancy.
Diabetologia. 59:1089–1094. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Leach L: Placental vascular dysfunction in
diabetic pregnancies: Intimations of fetal cardiovascular disease?
Microcirculation. 18:263–269. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Greene MF and Solomon CG: Gestational
diabetes mellitus-time to treat. N Engl J Med. 352:2544–2546. 2005.
View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Elliott HR, Sharp GC, Relton CL and Lawlor
DA: Epigenetics and gestational diabetes: A review of epigenetic
epidemiology studies and their use to explore epigenetic mediation
and improve prediction. Diabetologia. 62:2171–2178. 2019.
View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Fan Z, Cui H, Xu X, Lin Z, Zhang X, Kang
L, Han B, Meng J, Yan Z, Yan X and Jiao S: MiR-125a suppresses
tumor growth, invasion and metastasis in cervical cancer by
targeting STAT3. Oncotarget. 6:25266–25280. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Xie F, Hosany S, Zhong S, Jiang Y, Zhang
F, Lin L, Wang X, Gao S and Hu X: MicroRNA-193a inhibits breast
cancer proliferation and metastasis by downregulating WT1. PLoS
One. 12:e01855652017. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Wander PL, Boyko EJ, Hevner K, Parikh VJ,
Tadesse MG, Sorensen TK, Williams MA and Enquobahrie DA:
Circulating early- and mid-pregnancy microRNAs and risk of
gestational diabetes. Diabetes Res Clin Pract. 132:1–9. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Tagoma A, Alnek K, Kirss A, Uibo R and
Haller-Kikkatalo K: MicroRNA profiling of second trimester maternal
plasma shows upregulation of miR-195-5p in patients with
gestational diabetes. Gene. 672:137–142. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Floris I, Descamps B, Vardeu A, Mitić T,
Posadino AM, Shantikumar S, Sala-Newby G, Capobianco G, Mangialardi
G, Howard L, et al: Gestational diabetes mellitus impairs fetal
endothelial cell functions through a mechanism involving
microRNA-101 and histone methyltransferase enhancer of zester
homolog-2. Arterioscler Thromb Vasc Biol. 35:664–674. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Lewis PW, Muller MM, Koletsky MS, Cordero
F, Lin S, Banaszynski LA, Garcia BA, Muir TW, Becher OJ and Allis
CD: Inhibition of PRC2 activity by a gain-of-function H3 mutation
found in pediatric glioblastoma. Science. 340:857–861. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Xu CR, Li LC, Donahue G, Ying L, Zhang YW,
Gadue P and Zaret KS: Dynamics of genomic H3K27me3 domains and role
of EZH2 during pancreatic endocrine specification. EMBO J.
33:2157–2170. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
International Association of Diabetes and
Pregnancy Study Groups Consensus Panel, . Metzger BE, Gabbe SG,
Persson B, Buchanan TA, Catalano PA, Damm P, Dyer AR, Leiva A, Hod
M, et al: International association of diabetes and pregnancy study
groups recommendations on the diagnosis and classification of
hyperglycemia in pregnancy. Diabetes Care. 33:676–682. 2010.
View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Matsumoto T, Takaoka E, Ishida K, Nakayama
N, Noguchi E, Kobayashi T and Kamata K: Abnormalities of
endothelium-dependent responses in mesenteric arteries from Otsuka
long-evans Tokushima fatty (OLETF) rats are improved by chronic
treatment with thromboxane A2 synthase inhibitor. Atherosclerosis.
205:87–95. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(-Delta Delta C(T)) method. Methods. 25:402–408. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Zinman B, Genuth S and Nathan DM: The
diabetes control and complications trial/epidemiology of diabetes
interventions and complications study: 30th anniversary
presentations. Diabetes Care. 37:82014. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Cefalu WT and Ratner RE: The diabetes
control and complications trial/epidemiology of diabetes
interventions and complications study at 30 years: The ‘gift’ that
keeps on giving! Diabetes Care. 37:5–7. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Hales CN and Barker DJ: The thrifty
phenotype hypothesis. Br Med Bull. 60:5–20. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Morwessel NJ: The genetic basis of
diabetes mellitus. AACN Clin Issues. 9:539–554. 1998. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Pishavar E and Behravan J: miR-126 as a
therapeutic agent for diabetes mellitus. Curr Pharm Des.
23:3309–3314. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Assmann TS, Duarte GCK, Brondani LA, de
Freitas PHO, Martins EM, Canani LH and Crispim D: Polymorphisms in
genes encoding miR-155 and miR-146a are associated with protection
to type 1 diabetes mellitus. Acta Diabetol. 54:433–441. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Morais Junior GS, Souza VC, Machado-Silva
W, Henriques AD, Melo Alves A, Barbosa Morais D, Nobrega OT, Brito
CJ and Dos Santos Silva RJ: Acute strength training promotes
responses in whole blood circulating levels of miR-146a among older
adults with type 2 diabetes mellitus. Clin Interv Aging.
12:1443–1450. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Shan K, Liu C, Liu BH, Chen X, Dong R, Liu
X, Zhang YY, Liu B, Zhang SJ, Wang JJ, et al: Circular noncoding
RNA HIPK3 mediates retinal vascular dysfunction in diabetes
mellitus. Circulation. 136:1629–1642. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Banerjee RR: Piecing together the puzzle
of pancreatic islet adaptation in pregnancy. Ann N Y Acad Sci.
1411:120–139. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Lane-Cordova AD, Gunderson EP, Carnethon
MR, Catov JM, Reiner AP, Lewis CE, Dude AM, Greenland P and Jacobs
DR Jr: Pre-pregnancy endothelial dysfunction and birth outcomes:
The coronary artery risk development in young adults (CARDIA)
study. Hypertens Res. 41:282–289. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Magee TR, Ross MG, Wedekind L, Desai M,
Kjos S and Belkacemi L: Gestational diabetes mellitus alters
apoptotic and inflammatory gene expression of trophobasts from
human term placenta. J Diabetes Complications. 28:448–459. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Li J, Ying H, Cai G, Guo Q and Chen L:
Impaired proliferation of pancreatic beta cells, by reduced
placental growth factor in pre-eclampsia, as a cause for
gestational diabetes mellitus. Cell Prolif. 48:166–174. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Tryggestad JB, Vishwanath A, Jiang S,
Mallappa A, Teague AM, Takahashi Y, Thompson DM and Chernausek SD:
Influence of gestational diabetes mellitus on human umbilical vein
endothelial cell miRNA. Clin Sci (Lond). 130:1955–1967. 2016.
View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Jin J, Wang C, Ouyang Y and Zhang D:
Elevated miR-195-5p expression in deep vein thrombosis and
mechanism of action in the regulation of vascular endothelial cell
physiology. Exp Ther Med. 18:4617–4624. 2019.PubMed/NCBI
|
|
30
|
Xu J, Wang Z, Lu W, Jiang H, Lu J, Qiu J
and Ye G: EZH2 promotes gastric cancer cells proliferation by
repressing p21 expression. Pathol Res Pract. 215:1523742019.
View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Li Y, Li D, Zhao M, Huang S, Zhang Q, Lin
H, Wang W, Li K, Li Z, Huang W, et al: Long noncoding RNA SNHG6
regulates p21 expression via activation of the JNK pathway and
regulation of EZH2 in gastric cancer cells. Life Sci. 208:295–304.
2018. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Lian R, Ma H, Wu Z, Zhang G, Jiao L, Miao
W, Jin Q, Li R, Chen P, Shi H and Yu W: EZH2 promotes cell
proliferation by regulating the expression of RUNX3 in laryngeal
carcinoma. Mol Cell Biochem. 439:35–43. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Yan JJ, Chang Y, Zhang YN, Lin JS, He XX
and Huang HJ: miR-195 inhibits cell proliferation via targeting
AEG-1 in hepatocellular carcinoma. Oncol Lett. 13:3118–3126. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Yin Y, Hong S, Yu S, Huang Y, Chen S, Liu
Y, Zhang Q, Li Y and Xiao H: MiR-195 inhibits tumor growth and
metastasis in papillary thyroid carcinoma cell lines by targeting
CCND1 and FGF2. Int J Endocrinol. 2017:61804252017. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Han Li C and Chen Y: Targeting EZH2 for
cancer therapy: Progress and perspective. Curr Protein Pept Sci.
16:559–570. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Li Z, Hou P, Fan D, Dong M, Ma M, Li H,
Yao R, Li Y, Wang G, Geng P, et al: The degradation of EZH2
mediated by lncRNA ANCR attenuated the invasion and metastasis of
breast cancer. Cell Death Differ. 24:59–71. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Pepin ME, Bickerton HH, Bethea M, Hunter
CS, Wende AR and Banerjee RR: Prolactin receptor signaling
regulates a pregnancy-specific transcriptional program in mouse
islets. Endocrinology. 160:1150–1163. 2019. View Article : Google Scholar : PubMed/NCBI
|