Introduction
Renal cell carcinoma (RCC) is the ninth most common
malignant tumor, accounting for 2–3% of all adult malignancies
(1). In the USA, RCC is the sixth
leading cause of cancer-related deaths in men, while the eighth
leading cause in women (2).
Currently, the worldwide incidence rate of malignancy increases by
~2% every year (3). In 2012,
~84,400 new RCC cases were diagnosed, of which 34,700 resulted in
kidney cancer-related deaths in the European Union (3). Surgery is still the optimal treatment
for primary RCC, while for advanced metastatic RCC several targeted
therapies have been approved by the Food and Drug Administration
(FDA), including sunitinib (4).
Although these FDA-approved therapies have extended the survival
time of patients with advanced RCC, the response rate of targeted
therapy is weak and the 5-year survival rate is <10% (5). Therefore, studies have focused on
novel, efficacious strategies for the treatment of metastatic
RCC.
The spalt-like gene family consists of four members,
including spalt like transcription factor (SALL) 1, SALL2, SALL3
and SALL4 (6). SALL4 was cloned
based on the DNA sequence homology to the homeotic gene in
Drosophila, spalt (7,8).
SALL4 is enriched in embryonic cells and plays a major role in
self-renewal capability, while its expression is silenced in mature
adults (9,10). However, SALL4 can also be
re-expressed in various cancer types (11–16);
it was first recognized as an oncogene in leukemia (17). Previous studies have shown that
SALL4 is overexpressed in various tumors, and may play a role in
tumorigenesis and tumor progression (18). Furthermore, SALL4 may have a
function in different subclasses of hepatocellular carcinoma (HCC),
and is a key factor in maintaining the properties of cancer stem
cells (19,20). Therefore, targeting the
SALL4 gene as a potential therapeutic strategy has been
demonstrated in various cancer types. In acute myeloid leukemia and
HCC, a peptide that can compete with SALL4 to interact with the
HDAC complex has been used to treat patients (21,22).
However, the expression level and function of SALL4
in different subtypes of RCC are not fully understood. The present
study aimed to investigate the expression level and function of
SALL4 using the data from The Cancer Gene Atlas (TCGA) to
understand the molecular mechanisms underlying SALL4 expression.
The present study also assessed the function of SALL4 in different
types of RCC to ascertain whether it has vital clinical
implications. If SALL4 promotes RCC malignancy, then therapeutic
strategies targeting SALL4 using PTEN (23) or entinostat (24) may have clinical therapeutic
efficacy.
Materials and methods
TCGA
The RNA-seq data from the cohorts of 604 clear cell
RCC (ccRCC), 320 papillary RCC (pRCC) and 89 chromophobe RCC
(chRCC) cases were extracted from TCGA database (https://xena.ucsc.edu/). In addition, the clinical
outcomes including the pathological Tumor-Node-Metastasis (TNM)
stage (25), T and M stages and
the overall survival (OS) were assessed using the Xena platform
(http://xena.ucsc.edu/). These three cohorts
included ~20,500 gene data points. In addition, clinical
information, including the time to last follow-up, survival state
and sex of each patient from the TCGA database was extracted. In
addition to SALL4 gene data, copy number data and the clinical
relevance were retrieved from TCGA. The SALL4 expression level
between tumor and normal tissues in different tumors was analyzed
using FireBrowse software (Broad Institute GDAC Firebrowse version
1.1.35; http://firebrowse.org/).
Association between SALL4 gene and
survival
The three types of patients with RCC were divided
into two groups based on the level of SALL4 mRNA expression (high
expression SALL4 or low expression SALL4 group) or copy number
(SALL4 high copy number or SALL4 low copy number group).
Kaplan-Meier analysis was performed using GraphPad Prism 7
(GraphPad Software, Inc.) or Xena (http://xena.ucsc.edu/).
SALL4-associated gene expression and
enriched pathway analysis
In total, the gene expression of 10 patients from
TCGA databse with high expression of SALL4 and 10 with low
expression from the TCGA database were analyzed. These data were
obtained using the WebMeV cloud platform for analyzing and
visualizing cancer genomic data (http://mev.tm4.org/#/datasets/tcga) using the voom
function.
SALL4 expression and its function in
RCC cells and samples
Between September 2018 and June 2019, 10 patients
with RCC and 10 healthy control patients were enrolled in the
present study at the Department of Shanghai Tenth People's
Hospital, Tongji University. Preoperative clinical data for each
patient, including complete blood count, were entered into a
computerized database. Then, two different types of tissues from
each patient with RCC, including RCC tumor tissue and another
tumor-free sample was taken at >2 cm from the tumor edge
following surgical resection. These specimens were preserved in 10%
formaldehyde solution at 62°C for 1 h and embedded in paraffin. The
detail information of the patients is documented in Table I. The current study was performed
according to the protocol approved by the Ethics Committee of
Shanghai Tenth People's Hospital, Tongji University School of
Medicine. Written informed consent for participation was obtained
from each patient.
 | Table I.Patient data of the RCC cases (N=10)
and the healthy controls (N=10). |
Table I.
Patient data of the RCC cases (N=10)
and the healthy controls (N=10).
|
|
|
|
| TNM stage | Pathology |
|---|
|
|
|
|
|
|
|
|---|
| Group | Sex | Patients | Mean age,
years | T1a | T1b | T2 | ccRCC | pRCC | chRCC |
|---|
| RCC | Total | 10 | 65.1 | 7 | 2 | 1 | 8 | 2 | 0 |
|
| Male | 7 | 68.6 | 6 | 1 |
| 6 | 1 | 0 |
|
| Female | 3 | 57.0 | 1 | 1 | 1 | 2 | 1 | 0 |
| Control | Total | 10 | 63.3 |
|
|
|
|
|
|
|
| Male | 6 | 64.8 |
|
|
|
|
|
|
|
| Female | 4 | 61.0 |
|
|
|
|
|
|
Cell culture
OSRC-2, HK2, ACHN, 293T and SW839 cell lines were
purchased from the American Type Culture Collection. All cell lines
were cultured in DMEM (Invitrogen; Thermo Fisher Scientific, Inc.)
supplemented with 10% FBS (Thermo Fisher Scientific, Inc.),
penicillin (25 U/ml), streptomycin (25 g/ml) and 1% L-glutamine.
All cell lines were cultured in a 5% CO2 humidified
incubator at 37°C.
Lentivirus packaging
The pLKO-sh-SALL4, the psAX2 packaging plasmid and
pMD2G envelope plasmid from George Whipple lab of University of
Rochester were transfected into 293T cells using the standard
calcium chloride transfection method (26) for 48 h to get the lentivirus soup.
The lentivirus soup was collected and concentrated by density
gradient centrifugation at 4,000 × g for 20 min at 4°C, and then
frozen at −80°C. The cells were transfected using
Lipofectamine® 2000 (Invitrogen; Thermo Fisher
Scientific, Inc.) The standard transfection method of the SW839 and
OSRC-2 cells with sh-SALL4 was as follows: 4 µl aqueous solution
containing 4 µg of DNA was mixed with 10 µl 2.5 M CaCl2
solution. The dispersion was incubated for 5 min. The volume of the
dispersion was adjusted to 100 µl with water and 100 µl HEPES
buffered saline solution (280 mM NaCl, 10 mM KCl, 1.5 mM
Na2HPO4, 12 mM dextrose, 50 mM HEPES, pH
7.05±0.01) was added. Cell culture medium (RPMI 1640 with 15% fetal
calf serum; Thermo Fisher Scientific, Inc.) was added up to 1 ml.
The culture medium was removed was removed from the cells, and the
transfection mixture was added. After 7-h incubation at 37°C, the
transfection mixture was replaced by fresh cell culture medium. The
sequences of sh-SALL4 were as follows: Forward,
5′-CGCGTCCAGAGAATCCCTGTGACTTTACGGACCCGGTCGACGTCCGTAAAGTCACAGGGATTCTCTGGCATTTTTG-3′
and reverse,
5′-CGCAAAAACCAGAGAATCCCTGTGACTTTACGGACGTCGACCGGGTCCGTAAAGTCACAGGGATTCTCTGGCATATCTA-3′.
Immunohistochemistry (IHC)
Human RCC sections (thickness, 5 µm) were
deparaffinized in a xylene solution (100%) and rehydrated using
gradient ethanol concentrations (70% for 5 min, 80% for 5 min, 90%
for 5 min and 100% for 5 min). Endogenous peroxidase activity was
blocked with 3% hydrogen peroxide for 10 min at 37°C. Heat-induced
antigen retrieval was performed for all sections with 0.01 M sodium
citrate (pH 6.0) at 98°C for 30 min. Then, IHC staining was
performed with specific primary antibodies against SALL4 at 37°C
for 120 min (cat. no. ab57577; dilution, 1:100; Abcam). The
sections were subsequently incubated with a horseradish
peroxidase-conjugated IgG H&L secondary antibody (cat. no.
ab205719; dilution, 1:1,000; Abcam) at 37°C for 60 min. Staining
was performed using diaminobenzidine for 5 min at room temperature
followed by counterstaining with hematoxylin for 1 min at room
temperature. The sections were dehydrated and fixed using a graded
ethanol series (70% for 5 min, 80% for 5 min, 90% for 5 min and
100% for 5 min), treated with xylene for 10 min at room temperature
and mounted with Permount™ mounting medium (Thermo Fisher
Scientific, Inc.) The slides were observed using a light microscope
under five random high-power fields (magnification, ×400).
Reverse transcription-quantitative PCR
(RT-qPCR)
For RNA extraction, total RNAs were isolated using
TRIzol® reagent (Invitrogen; Thermo Fisher Scientific,
Inc.). Then, 1 µg of total RNA was subjected to RT using an RT-PCR
kit (Takara Bio, Inc.) according to the manufacturer's
instructions. qPCR was subsequently performed in triplicate for
each sample using a SYBR® ExScript Real-time PCR kit
(Takara Biotechnology Co., Ltd.). A 20 µl reaction mixture was
used, containing 2 µl template DNA, 1 µl primers, 10 µl SYBR premix
and 7 µl ddH2O. The primer sequences were as follows: β-actin
forward, 5′-TGAAGGTGACAGCAGTCGGTT-3′ and reverse,
5′-AGAAGTGGGGTGGCTTTTAGGA-3′; SALL4 forward,
5′-TTGCGACCACCCAAGTAT-3′ and reverse, 5′-AACCCACAGAACCAACCAC-3′.
PCR was performed using a 7900HT Fast Real-Time PCR machine
(Applied Biosystems; Thermo Fisher Scientific, Inc.) under the
following conditions: 95°C for 30 sec, 40 cycles of 95°C for 5 sec
and 60°C for 30 sec. PCR results were quantified using the
−2ΔΔCq method (27).
ELISA
SALL4 in the serum was measured using a Human
Sal-like protein 4(SALL4) ELISA kit (cat. no. EL020676HU; Miltenyi
Biotec GmbH) according to the manufacturer's instructions. The
optical density (OD) at 450 nm was determined. A standard curve was
established using OD450 as the y-axis and the concentration of a
standard substance as the x-axis; from this standard curve the
level of protein was determined. Data are presented as the
concentration of SALL4 (ng/ml) in samples.
Cell invasion assay
The invasive capability of OSRC-2 cells was
determined by the Transwell assay (8-µm pore size; Corning, Inc.).
OSRC-2 cells were harvested and seeded at 5×104
cells/well with serum-free DMEM into the upper chambers pre-coated
with Matrigel (37°C for 60 min), and the lower chambers contained
DMEM with 10% FBS. Then, the Transwell assay was incubated for 24 h
at 37°C. Following incubation, the invasive cells attached to the
lower surface of the membrane were fixed by 4% paraformaldehyde for
10 min at room temperature and stained with 1% toluidine blue for
10 min at room temperature. The number of cells penetrating across
membrane was counted under a light microscope in ten random visual
fields (magnification, ×100).
MTT assay
Cell viability was assessed using MTT assay. After
24 h transfection with plasmids, SW839 or OSRC-2 cells were seeded
at 1,000 per well in a 96-well plate. The cell proliferation assay
was performed on days 1, 2, 3 and 4. MTT reagent (5 mg/ml) was
added to each well and the plate was incubated for 2 h at 37°C. The
formazan crystals formed were solubilized in 100 µl DMSO for 10
min. Before the endpoint of incubation, the absorbance was measured
at 450 nm. Each sample was assayed in triplicate.
Statistical analysis
Statistical analyses were performed using SPSS 20
statistical software (SPSS, Inc.) and GraphPad Prism 7 (GraphPad
Software, Inc.). Differences in mean values between two groups were
analyzed by a two-tailed Student's t-test and the mean values of
>2 groups were compared with one-way ANOVA with the Bonferroni
post hoc test for multiple comparisons. Kaplan-Meier curves were
calculated to determine if SALL4 expression was related to patient
survival. Pearson's correlation was used to assess correlations
between SALL4 gene expression and SALL4 copy number. P<0.05 was
considered to indicate a statistically significant difference.
Results
SALL4 expression is related to the
survival of patients with RCC
The mRNA expression level of SALL4 and
clinical information of 604 cases of ccRCC were obtained from TCGA
using UCSC Xena (http://xena.ucsc.edu/). The patients with ccRCC were
divided into SALL4-high or SALL4-low groups according to the median
SALL4 mRNA expression level. The Kaplan-Meier curve was used to
analyze if the expression level of SALL4 was related to the
survival of patients with RCC. A significant difference
(P<0.0001) was found between the two groups, where patients with
lower SALL4 expression level had longer survival time compared with
patients with higher SALL4 expression levels (Fig. 1A).
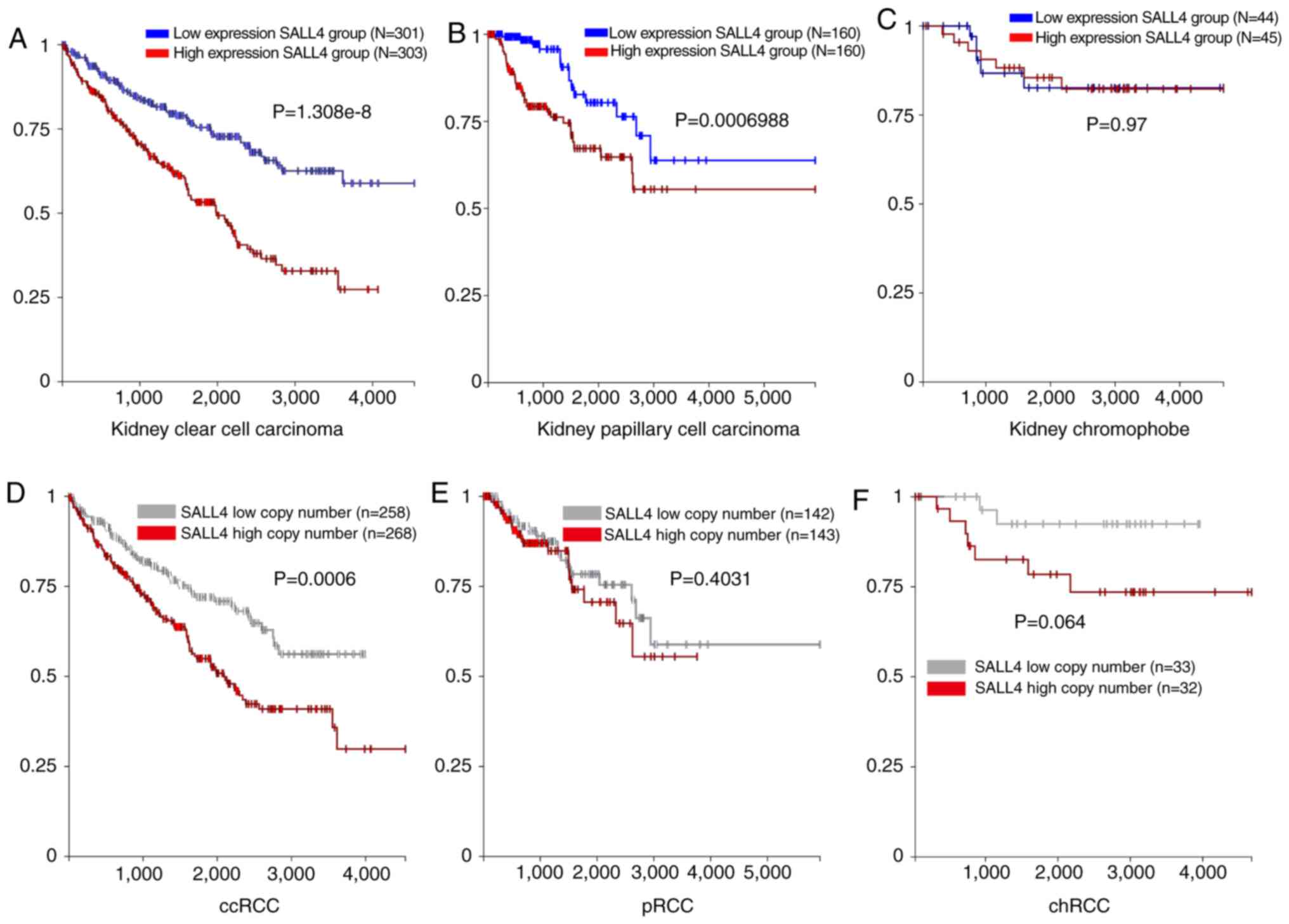 | Figure 1.mRNA and copy number of SALL4, and
SALL4 association with survival in ccRCC, pRCC and chRCC.
Kaplan-Meier analysis of SALL4 mRNA expression level and OS in (A)
ccRCC, (B) pRCC and (C) chRCC. Kaplan-Meier analysis of SALL4 copy
number and OS in (D) ccRCC, (E) pRCC and (F) chRCC. OS, overall
survival; SALL4, spalt like transcription factor 4; RCC, renal cell
carcinoma; ccRCC, clear cell RCC; pRCC, papillary RCC; chRCC,
chromophobe RCC. |
Similar analyses were carried out in patients with
pRCC (n=320) and chRCC (n=89). A significant difference was found
between SALL4 high and low expression groups in the pRCC cases
(P=0.0006; Fig. 1B). However, no
significant difference was detected in chRCC in both groups
(P=0.97; Fig. 1C). Therefore, the
present results suggested that patients with pRCC and low
SALL4 mRNA expression have a longer survival time compared
with patients with high SALL4 expression.
In addition, the prognostic relevance of sex was
analyzed in RCC, as RCC is reported to have a sex bias with a male
to female ratio of 2.3:1 (28).
However, in the present study no significant difference was found
in ccRCC, pRCC and chRCC in both SALL4 groups (data not shown).
Therefore, while the incidence of RCC is higher in males compared
with females, the outcome of RCC is the same in both sex.
SALL4 expression level in different
stage and grades of RCC
The present study analyzed the expression level of
SALL4 in different pathological TNM stages in the three RCCs
groups. It was demonstrated that the expression level of SALL4 was
higher in M1 compared with M0 (P=0.0092). Moreover, as the
pathology T and stage increased, the expression level of SALL4 was
also significantly increased in ccRCC (Fig. 2A). Similar results were identified
in patients with chRCC (Fig. 2C).
However, the expression level of SALL4 showed no difference between
different pathology T, M and stage in pRCC (Fig. 2B). Collectively, the present
results suggested that SALL4 promotes the progression of ccRCC and
chRCC, but not pRCC.
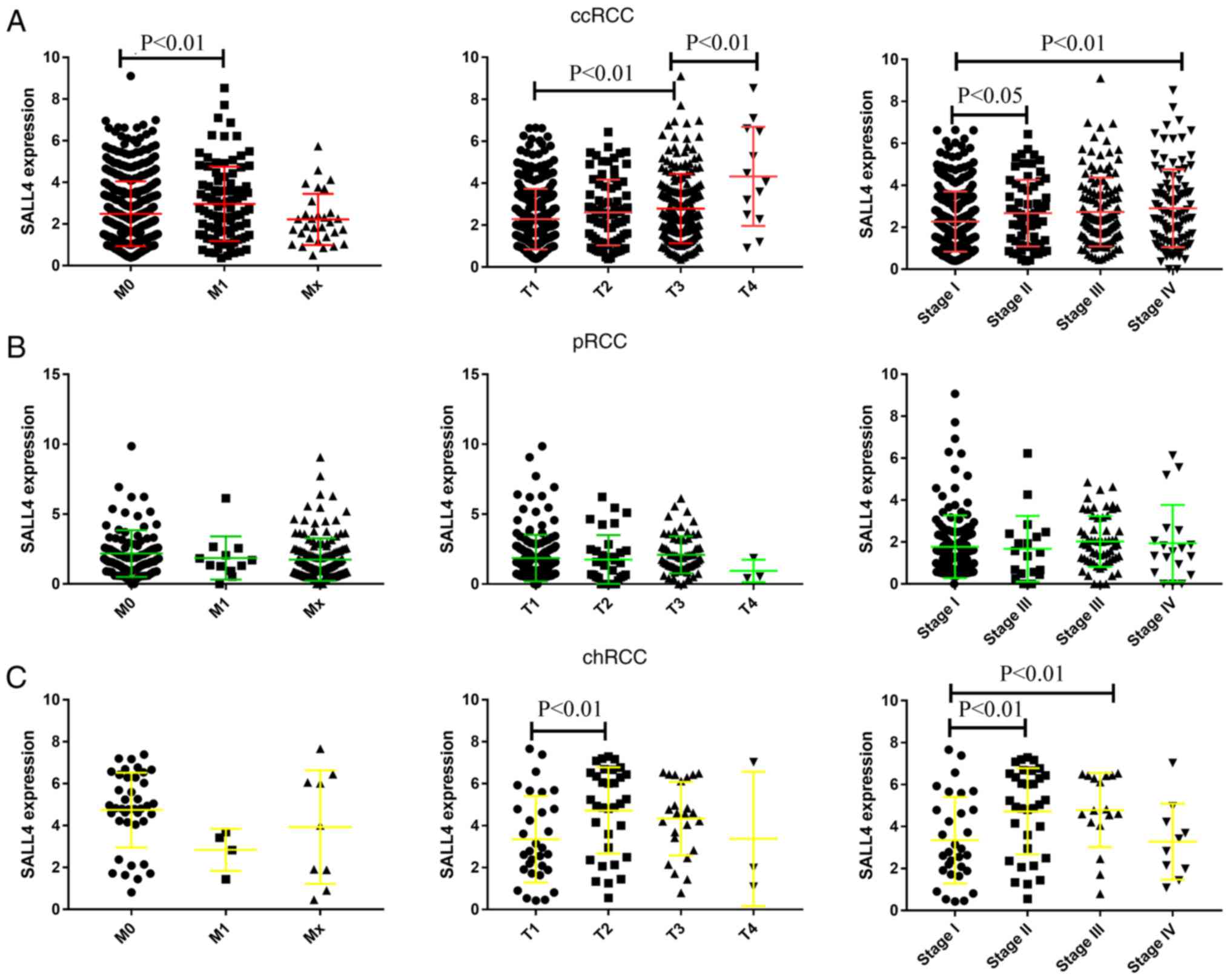 | Figure 2.mRNA expression level of SALL4 and
its association with pathology M, T and stage in ccRCC, pRCC and
chRCC. (A) mRNA expression of SALL4 and its association with
pathology M, T and stage in ccRCC. (B) mRNA expression of SALL4 and
its association with pathology M, T and stage in pRCC. (C) mRNA
expression of SALL4 and its association with pathology M, T and
stage in chRCC. Data are presented as the mean ± SD. SALL4, spalt
like transcription factor 4; RCC, renal cell carcinoma; ccRCC,
clear cell RCC; pRCC, papillary RCC; chRCC, chromophobe RCC. |
SALL4 expression level between tumor
and normal tissues in different tumors
To investigate the expression level of SALL4 in
tumor and normal tissues, SALL4 expression level was assessed in
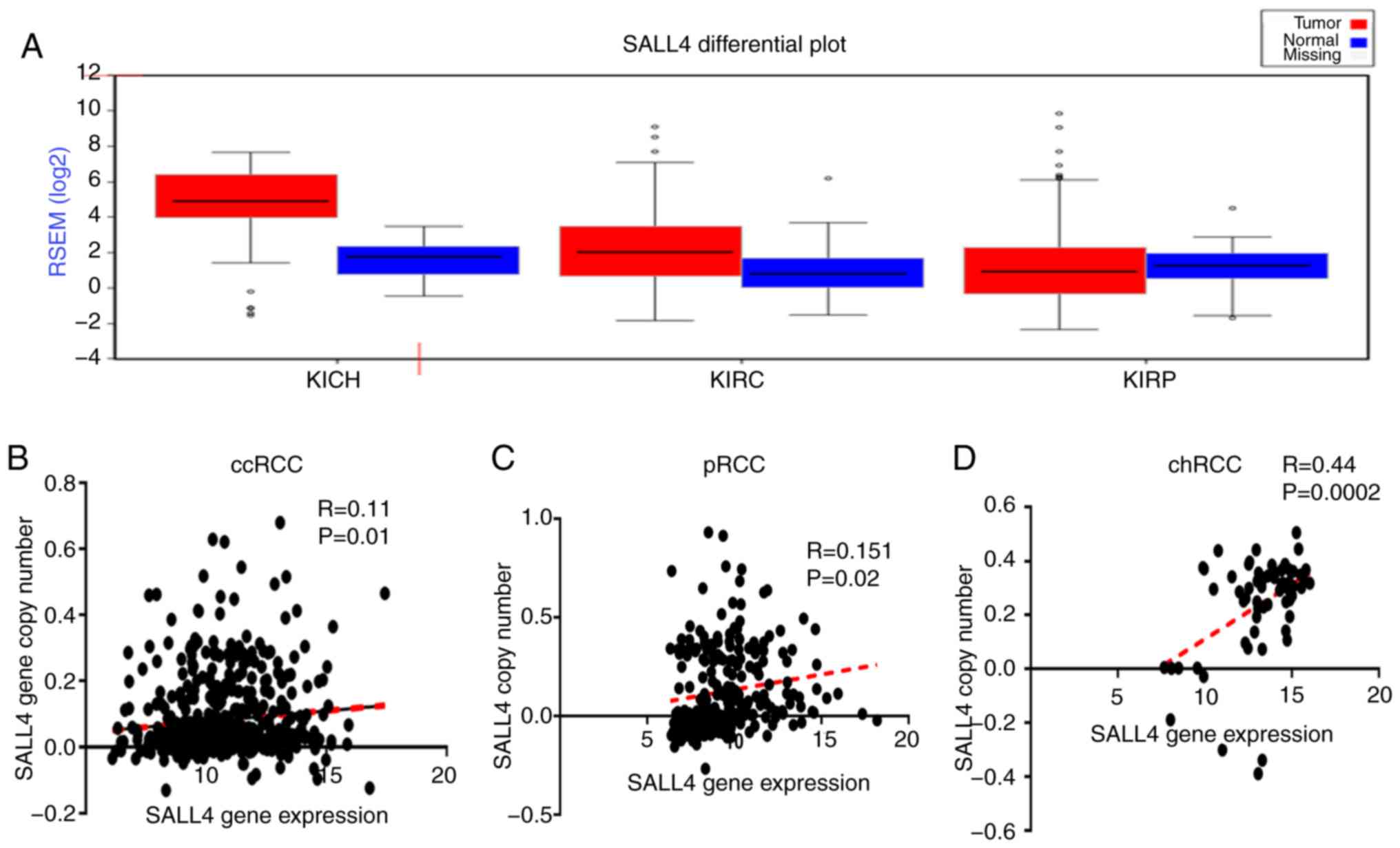
different cancer types using FireBrowse software. It was found that
the expression level of SALL4 was higher in almost all cancer
tissues compared with the normal tissues (Fig. S1A). For the three types of RCCs
[KICH (chRCC), KIRC (ccRCC) and KIRP (pRCC)], it was demonstrated
that the expression level of SALL4 was higher in the tumor tissue
compared with the normal in ccRCC and chRCC. However, no difference
was detected in pRCC (Fig. 3A).
Therefore, the present results indicated that SALL4 may promote the
progression of ccRCC and chRCC, but not pRCC.
Copy number of SALL4 in RCC
An increase in the copy number of SALL4 may be the
putative mechanism underlying the high expression level of SALL4 in
RCC. Thus, the present study analyzed the copy number of SALL4. The
expression levels of the molecule and the clinical information of
the 526 cases of ccRCC were obtained from TCGA using UCSC Xena
(http://xena.ucsc.edu/). The patients with ccRCC
were divided into SALL4-high or SALL4-low groups according to the
median SALL4 copy number expression. The Kaplan-Meier curve was
used to analyze the SALL4 copy number expression related to the
survival of patients with RCC. A significant difference was found
between the two groups (P=0.0006), similar to the results found for
the mRNA expression level of SALL4 (Fig. 1D). However, no difference was
observed in pRCC and chRCC (Fig. 1E
and F). Furthermore, the correlation between SALL4 gene
expression level and SALL4 copy number in ccRCC, pRCC and chRCC was
found to have weak positive correlation between SALL4 gene
expression and SALL4 copy number in ccRCC (R=0.11; P=0.01) and pRCC
(R=0.151; P=0.02), and a moderate correlation in chRCC (R=0.44;
P=0.0002; Fig. 3B-D).
Collectively, the present results suggested that the
increase in SALL4 copy number may be one of the mechanisms for
elevated SALL4 expression level in pRCC. However, the mechanism of
high expression of SALL4 in ccRCC requires further
investigation.
Enriched pathways and related genes
correlates with SALL4 expression in RCC
Additionally, the present study examined the
potential pathways, and identified the genes correlated to SALL4
using WebMeV (http://mev.tm4.org/#/datasets/tcga). In total, 10
patients with high expression levels of SALL4 and 10 with low
expression levels were analyzed. It was found that 19 pathways were
significantly associated with SALL4 expression level in ccRCC
(Fig. S1B). The top three
pathways include translation, eukaryotic translation initiation and
cap-dependent translation initiation, suggesting that SALL4 may be
involved in the translation pathway.
Moreover, genes with expression levels that
correlated with SALL4 expression levels in ccRCC were analyzed
using WebMeV (http://mev.tm4.org/#/datasets/tcga). The expression
levels of ~2,674 genes were correlated with the expression level of
SALL4 (data not shown). The top 20 genes whose expression levels
were related to SALL4 are listed in Table II. Moreover, the present study did
not identify any cancer stem cell genes such as NANOG, SOX2 and
OCT4.
 | Table II.List of top 20 genes that are related
to SALL4 gene expression in ccRCC. |
Table II.
List of top 20 genes that are related
to SALL4 gene expression in ccRCC.
| Gene | logFC | P-value | adj. P-value |
|---|
| AGR3 | −3.0309 | 0.0010 | 0.0561 |
| RAB25 | −2.7967 | 0.0180 | 0.1876 |
| CHGB | −2.6242 | 0.0053 | 0.1090 |
| SCNN1B | −2.6181 | 0.0333 | 0.2504 |
| APCDD1L | −2.4312 | 0.0122 | 0.1555 |
| MUC15 | −2.336 | 0.0488 | 0.3009 |
| HBG1 | −2.3191 | 0.0065 | 0.1181 |
| GPC5 | −2.2762 | 0.0090 | 0.1372 |
| CALML3 | −2.1989 | 0.0223 | 0.2081 |
| LY6H | −2.0310 | 0.0078 | 0.1277 |
| DCT | 1.9643 | 0.0000 | 0.0076 |
| SLC17A2 | 1.9728 | 0.0049 | 0.1054 |
|
HIST1H2AJ | 2.0104 | 0.0033 | 0.0888 |
| MSLNL | 2.0332 | 0.0009 | 0.0526 |
|
B4GALNT4 | 2.0702 | 0.0028 | 0.0816 |
| ADCY2 | 2.1198 | 0.0001 | 0.0019 |
| MOGAT1 | 2.1340 | 0.0018 | 0.0680 |
| ADAM18 | 2.3234 | 0.0138 | 0.1663 |
| TRIM72 | 2.3842 | 0.0071 | 0.1218 |
| MSLN | 2.5377 | 0.0003 | 0.0333 |
High expression levels of SALL4 in RCC
samples and serum
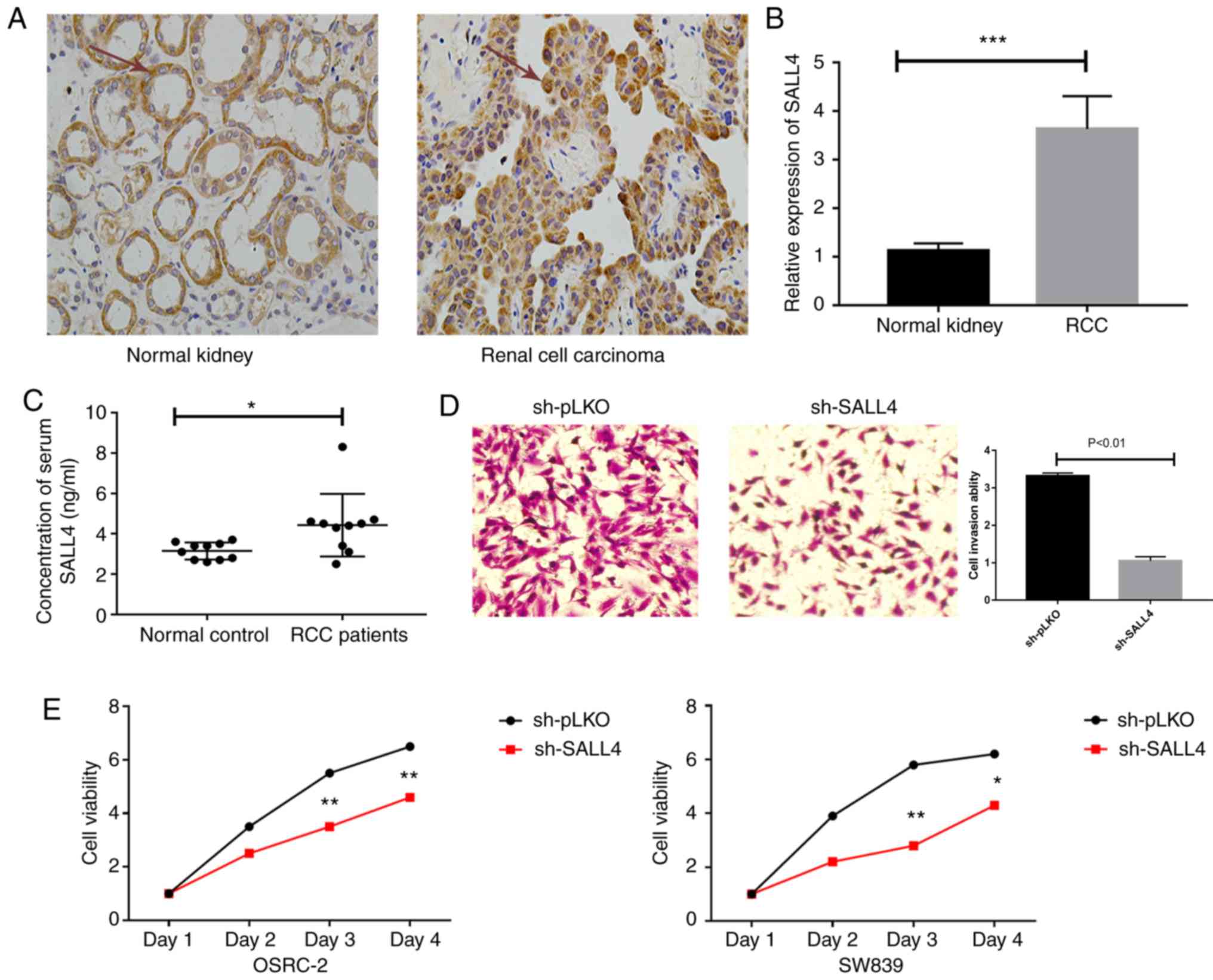
To further examine the function of SALL4 in patients
with RCC, the present study measured the protein expression levels
of SALL4 in RCC tumors and healthy specimens using RT-qPCR and IHC.
It was found that SALL4 protein expression level (yellow color) was
higher in tumor samples compared with healthy specimens (Fig. 4A). Furthermore, the mRNA expression
level of SALL4 was significantly higher in tumors compared with
healthy specimens (Fig. 4B). In
addition, expression level of SALL4 in the serum of 10 patients
with RCC and 10 controls was measured. It was demonstrated that
significantly higher serum SALL4 levels were presented in the
patients with RCC compared with the controls. Furthermore, the mean
serum SALL4 level was 4.03±0.61 ng/ml in the ccRCC group compared
with 3.45±0.38 ng/ml in the control group (P<0.05; Fig. 4C). The present study also measured
the mRNA expression level in human renal tubular epithelial cells
and the three RCC cell lines. It was found that SALL4 was
significantly highly expressed in the three RCC cell lines
(Fig. S2A).
SALL4 promotes cell viability and
invasion in RCC cells
The present study performed lentivirus packaging of
pLKO and sh-SALL4, which was then transduced into OSRC-2 and SW839
cells. RT-qPCR was used to examine the efficiency of transfection
(Fig. S2B).
An invasion assay was performed in OSRC-2 cells, and
the results indicated that knockdown of SALL4 decreased cell
invasion compared with pLKO (3.32±0.08 vs. 1.05±0.11; P<0.01;
Fig. 4D).
In addition, an MTT assay was used to assess the
viability of OSRC-2 and SW839 cells. It was demonstrated that SALL4
knockdown decreased cell viability compared with the pLKO group
(P=0.003 in OSRC-2 and P=0.001 in SW839 at day 3; Fig. 4E).
Discussion
In 2018, ~403,000 new cases of kidney cancer were
diagnosed worldwide, with a higher than 43% mortality rate in
patients (29). A high incidence
of small renal cancer is reported due to improved diagnosis, and
~1/3 of the patients with RCC develop metastatic lesions during the
development of the disease (30).
Moreover, nephrectomy (radial or partial) is the primary treatment
for localized RCC; however, >40% of patients with localized RCC
exhibit a relapse or metastasis after surgery (31). Currently, the therapeutic targeting
of the vascular endothelial growth factor using sunitinib,
sorafenib, pazopanib, axitinib, tivozanib and cabozantinib, or of
mTOR using everolimus, temsirolimus, and bevacizumab combined with
interferon-α have been applied clinically to prolong survival in
metastatic RCC (32). However,
some patients are naturally resistant to these methods and most
develop resistance (33), thus the
treatment of RCC can be difficult. Therefore, further
investigations on the molecular mechanisms underlying the
metastasis or progression of ccRCC, and into new novel targets are
urgently required. Currently, there is no reliable biomarker for
RCC, unlike prostate-specific antigen for prostate cancer (34). Thus, identifying novel and reliable
prognostic biomarkers for patients with RCC is important to predict
patient outcomes and facilitate effective clinical management.
SALL4, a member of the spalt-like gene family, is a
critical stem cell factor. SALL4 is a zinc finger transcription
factor that is enriched in the embryonic cell. Moreover, SALL4
plays a major role in the self-renewal capability, while its
expression is silenced in the mature adult (35). A previous study showed that SALL4
is crucial for maintaining the stemness properties of embryonic
stem cells (36). Another previous
study demonstrated that SALL4 controls the stemness properties of
embryonic stem cells at both the transcriptional and epigenetic
levels via direct or indirect interaction with Nanog and OCT4
(20). However, it has also been
showed that SALL4 is re-expressed in various cancer types (11–16)
and was first recognized as an oncogene in leukemia (17,37).
Yakaboski et al (38) found
that in SALL4-positive HCC, the expression of certain
progenitor-like genes is high. Other studies have also confirmed a
critical role of SALL4 in cell survival and tumorigenicity by
knocking down SALL4 (39). Zhang
et al (20) demonstrated
that the overexpression of SALL4 in gastric cancer cells promotes
cell stemness by increasing the expression levels of other cancer
stem cell markers such as CD133, SOX2, Bmi-1 and Lin28.
Furthermore, SALL4 has been identified as a core factor in the
SALL4/Nanog/Oct4 network (9).
Moreover, as a transcription factor, SALL4 can activate Oct4 and
interact with Nanog (9,40). Therefore, as a cancer stem cell
marker, SALL4 plays a major role in cancer formation. However, the
present study searched the top 20 genes related to SALL4, and
identified genes that may be attributed to the varied SALL4 pathway
in different cancer types.
The present study investigated the clinical value of
SALL4 in RCC and demonstrated that the expression level of SALL4
was associated with survival, stage and pathology T, thus
indicating that SALL4 is a poor prognostic factor for a poor
outcome in RCC. In addition to being a biomarker for RCC diagnosis,
SALL4 may also be a potential therapeutic target. A previous study
found that the inhibition of SALL4 expression by siRNA reduces cell
survival, and impairs the migration and invasion of indistinct
cancer cells in vitro (20). Moreover, targeting SALL4 using PTEN
(23) or entinostat (24) may have therapeutic efficacy in both
acute myeloid leukemia and lung cancer. Thus, it can be
hypothesized that targeting SALL4 using miRNA, PTEN or entinostat
may have therapeutic efficacy in RCC.
The mechanism underlying the high expression level
of SALL4 and its downstream genes in RCC is not fully understood.
The present results suggested that the copy number of SALL4 was
increased in ccRCC. Additionally, the copy number of SALL4 was
positively associated with the survival curve. Moreover, a positive
correlation was established between SALL4 mRNA and copy number in
ccRCC, pRCC and chRCC, indicating that an increased copy number may
be the mechanism underlying the high expression of SALL4 in RCC.
The enriched pathways analysis results identified several genes and
pathways that may be associated with SALL4; however, the cancer
stem cell marker gene and related pathway were not deduced as SALL4
is involved in multiple pathways promoting cancer progression.
Nevertheless, the downstream genes and the mechanism via which
SALL4 promotes RCC progression requires further investigation. The
present study used specimens and serum from patients with RCC to
identify the high expression level of SALL4. Furthermore, using the
MTT and invasion assays, it was found that SALL4 promotes cell
viability and invasion in RCC cells. To the best of our knowledge,
this is the first study to investigate the expression levels of
SALL4 in the serum and tumor specimens in RCC patients. Moreover,
targeting this newly identified SALL4 signaling pathway may
facilitate the development of novel therapies to treatment RCC and
improved survival rates. However, there were limitations to the
present study. Firstly, only three most common types of RCC were
analyzed due to TCGA data limitation. Secondly, the downstream
pathways or factors of SALL4 were not identified. In addition, the
patients with RCC only had 1 year follow-up data, thus the survival
curve is not valuable. Furthermore, the cause of increased SALL4
expression levels in patients with RCC is still unknown.
In conclusion, the present results suggested that
SALL4 may be a sensitive and specific cancer biomarker in ccRCC and
pRCC. Thus, targeting SALL4 may improve RCC therapy and prolong the
survival of patients with ccRCC and pRCC.
Supplementary Material
Supporting Data
Acknowledgements
The results of this study are partially based on the
data generated by the TCGA Research Network (https://www.cancer.gov/tcga).
Funding
This study was funded by the National Natural
Science Foundation of China (grant no. 81802518).
Availability of data and materials
The datasets used and/or analyzed during the current
study are available from the corresponding author on reasonable
request.
Authors' contributions
JC, CG, and PW were involved in creating/designing
the study, data collection and data analysis. JC and CG wrote the
manuscript. JZ, GW and XY were involved in project development and
data collection. All authors have read and approved the final
version of the manuscript.
Ethics approval and consent to
participate
The present study analyzed the raw data that is
published in The Cancer Genome Atlas. The current study was
performed according to the protocol approved by the Ethics
Committee of Shanghai Tenth People's Hospital, Tongji University
School of Medicine. Written informed consent for participation was
obtained from each patient.
Patient consent for publication
Not applicable.
Competing interest
The authors declare that they have no competing
interests.
References
|
1
|
Rini BI, Campbell SC and Escudier B: Renal
cell carcinoma. Lancet. 373:1119–1132. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
King SC, Pollack LA, Li J, King JB and
Master VA: Continued increase in incidence of renal cell carcinoma,
especially in young patients and high grade disease: United States
2001 to 2010. J Urol. 191:1665–1670. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Ljungberg B, Bensalah K, Canfield S,
Dabestani S, Hofmann F, Hora M, Kuczyk MA, Lam T, Marconi L,
Merseburger AS, et al: EAU guidelines on renal cell carcinoma: 2014
update. Eur Urol. 67:913–924. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Linehan WM and Ricketts CJ: Decade in
review-kidney cancer: Discoveries, therapies and opportunities. Nat
Rev Urol. 11:614–616. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Choueiri TK and Motzer RJ: Systemic
therapy for metastatic renal-cell carcinoma. N Engl J Med.
376:354–366. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Al-Baradie R, Yamada K, St Hilaire C, Chan
WM, Andrews C, McIntosh N, Nakano M, Martonyi EJ, Raymond WR,
Okumura S, et al: Duane radial ray syndrome (Okihiro syndrome) maps
to 20q13 and results from mutations in SALL4, a new member of the
SAL family. Am J Hum Genet. 71:1195–1199. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Kohlhase J, Schuh R, Dowe G, Kühnlein RP,
Jäckle H, Schroeder B, Schulz-Schaeffer W, Kretzschmar HA, Köhler
A, Müller U, et al: Isolation, characterization, and organ-specific
expression of two novel human zinc finger genes related to the
Drosophila gene spalt. Genomics. 38:291–298. 1996.
View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Tatetsu H, Kong NR, Chong G, Amabile G,
Tenen DG and Chai L: SALL4, the missing link between stem cells,
development and cancer. Gene. 584:111–119. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Yang J, Gao C, Chai L and Ma Y: A novel
SALL4/OCT4 transcriptional feedback network for pluripotency of
embryonic stem cells. PLoS One. 5:e107662010. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Rao S, Zhen S, Roumiantsev S, McDonald LT,
Yuan GC and Orkin SH: Differential roles of Sall4 isoforms in
embryonic stem cell pluripotency. Mol Cell Biol. 30:5364–5380.
2010. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Cao D, Li J, Guo CC, Allan RW and Humphrey
PA: SALL4 is a novel diagnostic marker for testicular germ cell
tumors. Am J Surg Pathol. 33:1065–1077. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Zhang L, Yan Y, Jiang Y, Cui Y, Zou Y,
Qian J, Luo C, Lu Y and Wu X: The expression of SALL4 in patients
with gliomas: High level of SALL4 expression is correlated with
poor outcome. J Neurooncol. 121:261–268. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Cao D, Guo S, Allan RW, Molberg KH and
Peng Y: SALL4 is a novel sensitive and specific marker of ovarian
primitive germ cell tumors and is particularly useful in
distinguishing yolk sac tumor from clear cell carcinoma. Am J Surg
Pathol. 33:894–904. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Mei K, Liu A, Allan RW, Wang P, Lane Z,
Abel TW, Wei L, Cheng H, Guo S, Peng Y, et al: Diagnostic utility
of SALL4 in primary germ cell tumors of the central nervous system:
A study of 77 cases. Mod Pathol. 22:1628–1636. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Li A, Jiao Y, Yong KJ, Wang F, Gao C, Yan
B, Srivastava S, Lim GS, Tang P, Yang H, et al: SALL4 is a new
target in endometrial cancer. Oncogene. 34:63–72. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Ushiku T, Shinozaki A, Shibahara J,
Iwasaki Y, Tateishi Y, Funata N and Fukayama M: SALL4 represents
fetal gut differentiation of gastric cancer, and is diagnostically
useful in distinguishing hepatoid gastric carcinoma from
hepatocellular carcinoma. Am J Surg Pathol. 34:533–540. 2010.
View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Ma Y, Cui W, Yang J, Qu J, Di C, Amin HM,
Lai R, Ritz J, Krause DS and Chai L: SALL4, a novel oncogene, is
constitutively expressed in human acute myeloid leukemia (AML) and
induces AML in transgenic mice. Blood. 108:2726–2735. 2006.
View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Wu M, Yang F, Ren Z, Jiang Y, Ma Y, Chen Y
and Dai W: Identification of the nuclear localization signal of
SALL4B, a stem cell transcription factor. Cell Cycle. 13:1456–1462.
2014. View
Article : Google Scholar : PubMed/NCBI
|
|
19
|
Jeong HW, Cui W, Yang Y, Lu J, He J, Li A,
Song D, Guo Y, Liu BH and Chai L: SALL4, a stem cell factor,
affects the side population by regulation of the ATP-binding
cassette drug transport genes. PLoS One. 6:e183722011. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Zhang X, Yuan X, Zhu W, Qian H and Xu W:
SALL4: An emerging cancer biomarker and target. Cancer Lett.
357:55–62. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Yong KJ, Chai L and Tenen DG: Oncofetal
gene SALL4 in aggressive hepatocellular carcinoma. N Engl J Med.
369:1171–1172. 2013.PubMed/NCBI
|
|
22
|
Zhang L, Xu Z, Xu X, Zhang B, Wu H, Wang
M, Zhang X, Yang T, Cai J, Yan Y, et al: SALL4, a novel marker for
human gastric carcinogenesis and metastasis. Oncogene.
33:5491–5500. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Gao C, Dimitrov T, Yong KJ, Tatetsu H,
Jeong HW, Luo HR, Bradner JE, Tenen DG and Chai L: Targeting
transcription factor SALL4 in acute myeloid leukemia by
interrupting its interaction with an epigenetic complex. Blood.
121:1413–1421. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Yong KJ, Li A, Ou WB, Hong CK, Zhao W,
Wang F, Tatetsu H, Yan B, Qi L, Fletcher JA, et al: Targeting SALL4
by entinostat in lung cancer. Oncotarget. 7:75425–75440. 2016.
View Article : Google Scholar : PubMed/NCBI
|
|
25
|
May M, Surcel C, Capitanio U, Dell'Oglio
P, Klatte T, Shariat S, Ecke T, Wolff I, Vergho D, Wagener N, et
al: Prognostic and discriminative power of the 7th TNM
classification for patients with surgically treated papillary renal
cell carcinoma: Results of a multi-institutional validation study
(CORONA subtype project). Scand J Urol. 51:269–276. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Zhai W, Sun Y, Guo C, Hu G, Wang M, Zheng
J, Lin W, Huang Q, Li G, Zheng J and Chang C: LncRNA-SARCC
suppresses renal cell carcinoma (RCC) progression via altering the
androgen receptor(AR)/miRNA-143-3p signals. Cell Death Differ.
24:1502–1517. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(-Delta Delta C(T)) method. Methods. 25:402–408. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
28
|
He D, Li L, Zhu G, Liang L, Guan Z, Chang
L, Chen Y, Yeh S and Chang C: ASC-J9 suppresses renal cell
carcinoma progression by targeting an androgen receptor-dependent
HIF2α/VEGF signaling pathway. Cancer Res. 74:4420–4430. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Bray F, Ferlay J, Soerjomataram I, Siegel
RL, Torre LA and Jemal A: Global cancer statistics 2018: GLOBOCAN
estimates of incidence and mortality worldwide for 36 cancers in
185 countries. CA Cancer J Clin. 68:394–424. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Petejova N and Martinek A: Renal cell
carcinoma: Review of etiology, pathophysiology and risk factors.
Biomed Pap Med Fac Univ Palacky Olomouc Czech Repub. 160:183–194.
2016. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Ravaud A, Motzer RJ, Pandha HS, George DJ,
Pantuck AJ, Patel A, Chang YH, Escudier B, Donskov F, Magheli A, et
al: Adjuvant Sunitinib in High-Risk Renal-Cell Carcinoma after
Nephrectomy. N Engl J Med. 375:2246–2254. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Ljungberg B, Albiges L, Abu-Ghanem Y,
Bensalah K, Dabestani S, Fernández-Pello S, Giles RH, Hofmann F,
Hora M, Kuczyk MA, et al: European association of urology
guidelines on renal cell carcinoma: The 2019 update. Eur Urol.
75:799–810. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Rini BI and Atkins MB: Resistance to
targeted therapy in renal-cell carcinoma. Lancet Oncol.
10:992–1000. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Pezaro C, Woo HH and Davis ID: Prostate
cancer: Measuring PSA. Intern Med J. 44:433–440. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Yang J: SALL4 as a transcriptional and
epigenetic regulator in normal and leukemic hematopoiesis. Biomark
Res. 6:12018. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Yang L, Liu L, Gao H, Pinnamaneni JP,
Sanagasetti D, Singh VP, Wang K, Mathison M, Zhang Q, Chen F, et
al: The stem cell factor SALL4 is an essential transcriptional
regulator in mixed lineage leukemia-rearranged leukemogenesis. J
Hematol Oncol. 10:1592017. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Farawela HM, Zawam HM, Al-Wakeel HA,
El-Nagdy MH, El-Refaey FA and Abdel-Rahman HA: Expression pattern
and prognostic implication of SALL4 gene in myeloid leukemias: A
case-control study. Scand J Clin Lab Invest. 79:65–70. 2019.
View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Yakaboski E, Jares A and Ma Y: Stem cell
gene SALL4 in aggressive hepatocellular carcinoma: A cancer stem
cell-specific target? Hepatology. 60:419–421. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
He J, Zhou M, Chen X, Yue D, Yang L, Qin
G, Zhang Z, Gao Q, Wang D, Zhang C, et al: Inhibition of SALL4
reduces tumorigenicity involving epithelial-mesenchymal transition
via Wnt/β-catenin pathway in esophageal squamous cell carcinoma. J
Exp Clin Cancer Res. 35:982016. View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Wang J, Rao S, Chu J, Shen X, Levasseur
DN, Theunissen TW and Orkin SH: A protein interaction network for
pluripotency of embryonic stem cells. Nature. 444:364–368. 2006.
View Article : Google Scholar : PubMed/NCBI
|