Introduction
Ovarian cancer is one of the most common types of
gynecologic cancer, resulting in the mortality of hundreds of
thousands of women every year worldwide. Ovarian cancer is the
seventh most common cause of cancer-associated mortality in women
(1). Emerging evidence has
supported the incessant ovulation theory as the mechanism
underlying ovarian cancer; this theory suggests that recurrent
ovulation-induced repair of the ovarian surface epithelium raises
the likelihood of DNA damage and carcinogenesis, and thereby
increases the risk of developing ovarian cancer (2,3).
Over the past decade, optimal resection, chemotherapy and
combination therapy have evolved as ovarian cancer treatments to
substantially improve patient outcomes and 5-year survival rates
(4). Nevertheless, the overall
patient outcome is far from ideal; thus, there remains a need for
revolutionary new treatments of ovarian cancer. One strategy for
identifying novel treatment targets and developing treatments
involves elucidating the regulatory molecular mechanisms and key
signaling pathways that underlie tumor growth and metastasis in
ovarian cancer.
MicroRNAs (miRNAs/miRs) are small noncoding
endogenous RNAs that mediate expression of their target gene via
genetic transcription or mRNA cleavage (5). Although the precise regulatory
mechanisms of miRNAs have not been fully elucidated, they are known
to function in various biological processes, including cell
proliferation, differentiation, morphogenesis and apoptosis
(6). Previously, the abnormal
expression of miRNAs has been associated with carcinogenesis and
tumor development in human cancers (7–9).
Thus, miRNAs are speculated to function as oncogenes or tumor
suppressors, depending on their functional targets; consequently,
they have great therapeutic potential as cancer treatment
targets.
Previous studies have indicated that miR-196a plays
a critical role in cell proliferation and tumor progression. For
example, overexpression of miR-196a has been shown to accelerate
cell differentiation of osteoporosis in a mouse model through
regulation of the GNAS-Hedgehog signaling pathway (10). In addition, Ren et al
(11), demonstrated that
inhibition of miR-196a attenuated cell proliferation and
glycolysis, but elicited apoptosis, in hepatocellular carcinoma by
targeting suppressor of cytokine signaling 2. Furthermore, high
levels of miR-196a expression have been found to be related to poor
survival in patients with ovarian cancer; thus, it is a potential
biomarker for the clinical prognosis of ovarian carcinoma (12,13).
A preliminarily evaluation of the regulatory mechanisms of miR-196a
in ovarian cancer suggested that miR-196a may act as an oncogene to
promote migration and invasion through the downstream target gene
homeobox A10 (HOXA10) (14).
Despite these findings, the regulatory mechanisms of miR-196a in
ovarian cancer are yet to be fully elucidated.
DEAD box protein RNA helicase 3 (DDX3) belongs to
the DDX protein family, which is highly conserved in all
eukaryotes. DDX3 has attracted increasing attention in cancer
research due to its critical function in cancer progression. For
example, the downregulation of DDX3 was shown to increase cell
metastasis in colorectal cancer (15), whereas the activation of DDX3
promoted cell proliferation in breast epithelial cells (16); thus, the role of DDX3 in cancer
progression is complex. In preliminary bioinformatics analysis, the
target prediction software DIANA was used to determine the
potential targets of miR-196a: DDX3 was found to be a direct target
of miR-196a. Therefore, the relationship between miR-196a and DDX3
may be important in the regulation of ovarian cancer
development.
In the present study, this relationship was further
investigated. It was revealed that upregulation of miR-196a in
ovarian cancer promoted cell proliferation but inhibited apoptosis.
It was also demonstrated that miR-196a may have an important
regulatory role in ovarian cancer progression by targeting DDX3
through regulation of the PTEN/PI3K/AKT signaling pathway.
Together, these findings suggested that miR-196a and DDX3 might be
potential targets for future treatments of ovarian cancer.
Materials and methods
Clinical samples and cell lines
Ovarian cancer samples and matched normal tissue
samples were collected from 30 patients (median age 50, range 30–69
years) with ovarian cancer who were treated at The Second
Affiliated Hospital of Soochow University (Suzhou, China) between
January 2018 and July 2019. The inclusion criteria was as follows:
Age 18–80; newly diagnosed with epithelial carcinoma of the ovary,
which was histologically or pathologically confirmed; no prednisone
use within 14 days of sampling; no previous chemotherapy or
radiotherapy; no concurrent diseases. The patients had not received
chemoradiotherapy before samples were collected and all ovarian
cancer tissue samples were dissected from resected tumors. Written
informed consent was obtained from all patients, and the present
study was approved by the Institutional Ethics Committee and
Academic Committee of The Second Affiliated Hospital of Soochow
University.
The ovarian cancer cell lines OVCAR3, CAOV-3, A2780
and ES-2, along with the immortalized ovarian epithelial cell line
HS832, were obtained from ATCC. Cells were cultured in DMEM (Gibco;
Thermo Fisher Scientific, Inc.) containing 10% fetal calf serum
(HyClone; GE Healthcare Life Sciences) at 37°C and 5%
CO2. In order to elucidate the regulatory mechanisms of
the interaction between miR-196a and DDX3, OVCAR3 ovarian cancer
cells were treated with LY294002 (10 µM; cat. no. 19-142;
Sigma-Aldrich; Merck KGaA) or DMSO for 6 h, and then the following
methods were used to determine cell proliferation and apoptosis in
OVCAR3 ovarian cancer cells treated with LY294002.
Cell transfection
miR-196a mimic (5-GCTCTGGCT CCGTGTCTTCACTCCC-3),
miR-negative control (NC) mimic (5-UUCUCCGAACGUGUCACGUTT-3),
miR-196a inhibitor (5-GGGAGTGAAGACACGGAGCCAGAGC-3) and miR-NC
inhibitor (5-CAGUACUUUUGUGUA GUACAA-3) were provided by Shanghai
Sangon Biotech Corporation. The miRNAs (50 nM) were mixed with
Lipofectamine® 2000 reagent (GE Healthcare Life
Sciences) and were incubated for 15 min at room temperature.
Subsequently, the sequences were transfected into OVCAR3 and CAOV-3
cells plated at 1×105 cells/well in 6-well plates. Cells
were collected for analysis 48 h after transfection.
Reverse transcription-quantitative PCR
(RT-qPCR)
Total RNA from tissues and cell lines was extracted
with TRIzol® reagent (Invitrogen; Thermo Fisher
Scientific, Inc.), according to the manufacturers protocol, and
RNAs were reverse transcribed into cDNA at 42°C for 1 h, then 90°C
for 5 min, using a TaqMan microRNA RT kit (Invitrogen; Thermo
Fisher Scientific, Inc.). To determine the relative expression
levels of targeted genes, RT-qPCR was performed using a SYBR-Green
PCR kit (Invitrogen; Thermo Fisher Scientific, Inc.) on a real-time
PCR system (Bio-Rad Laboratories, Inc.). The primer sequences were
as follows: miR-196a, forward (F) 5-GCTCTGGCTCCGTGTCTTCACTCCC-3,
reverse (R) 5-TGCCCCAGCACAGCCCCCGTCCCTC-3; DDX3, F
5-CGGCAGCATCAAATGTTTCAG-3, R 5-AAC TGGCAGGTAGAAGGCAACTC-3; GAPDH, F
5-AATGGGCAGCCGTTAGGAAA-3, R 5 TGAAGG GGTCATTGATGGCA-3; and U6, F
5-CTCGCTTCG GCAGCACATATA-3 and R 5 ACGCTTCACGAATTT GAGTGTC-3. The
PCR thermocycling conditions were as follows: Initial denaturation
at 95°C for 2 min, and 40 cycles of 95°C for 10 sec and 60°C for 1
min. miR-196a and DDX3 mRNA levels were quantified using the
2−ΔΔCq method and normalized to the internal reference
gene U6 or GAPDH, respectively (17).
Plasmid and short hairpin RNA (shRNA)
transfection
OVCAR3 and CAOV-3 cells were plated into a 6-well
plate (1×105 cells/well), cultured overnight, and were
then transfected with 50 nM corresponding vectors using
Lipofectamine® 2000 (Invitrogen; Thermo Fisher
Scientific, Inc.), according to the manufacturers instructions. The
shRNA targeting DDX3 (shDDX3), control shRNA (shCtrl), pcDNA-DDX3
and the control plasmid pcDNA3.1 were provided by Novogene Co.,
Ltd. The sequences of the targeted genes were as follows: shDDX3, F
5-AACCCACCACAGCUAGAACTT-3, R 5-AAG UUCUAGCUGUGGUGGGTT-3; shCtrl, F
5 UUCUCC GAACGUGUCACGUTT-3, R 5 ACGUGACACGUU CGGAGAATT-3; and
pcDNA-DDX3, F 5 GAAGCT ACTAGAGGTTTCTAC-3 and R 5 TCTCAACATCAC
TGAAACTTTC-3. Analyses were performed 48 h after transfection and
replicated three times to produce the experimental data.
MTT assays
OVCAR3 and CAOV-3 cells were added to a 96-well dish
at 1×103 cells/well and were then transfected with 50 nM
miR-196 mimic, miR-NC mimic, miR-196 inhibitor, miR-NC inhibitor,
pcDNA-DDX3, pcDNA-3.1, shDDX3 or shCtrl. Subsequently, 10 µl MTT
solution was added to each well at 0, 24, 48, 72 and 96 h. The
supernatant was discarded after 4 h of incubation at 37°C and then
100 µl DMSO was added to each well to dissolve the formazan. The
absorbance at 570 nm was determined using an enzyme-labeling
instrument (EL808; BioTek Instruments, Inc.).
Cell proliferation assay
Cell proliferation assays were performed using a
cell proliferation colorimetric ELISA-BrdU kit (Roche Diagnostics),
according to the manufacturers protocol. Briefly, OVCAR3 and CAOV-3
cells were cultured in a 96-well plate at a density of
1×103 cells/well. Overnight, the cells were transfected
with 50 nM miR-196 mimic, miR-NC mimic, miR-196 inhibitor, miR-NC
inhibitor, pcDNA-DDX3, pcDNA-3.1, shDDX3 or shCtrl at 37°C. After
24 h of incubation, cell proliferation was analyzed.
Apoptosis analysis
Cell apoptosis assays were conducted via flow
cytometry using an Annexin V-FITC Apoptosis Detection kit (Beyotime
Institute of Biotechnology). Briefly, the OVCAR3 and CAOV-3 cells
were seeded into 6-well plates at a density of 1×105
cells/well. After transfection, cells were collected using trypsin
diluted in PBS and the supernatant was removed. Subsequently,
Annexin-V/FITC (5 µl) and propidium iodide (10 µl) were added to
cells, and incubated for 15 min at room temperature in the dark.
The percentage of apoptotic cells was analyzed within 1 h using a
FACSCalibur flow cytometer (BD Biosciences) and BD CellQuestTM
software (v5; BD Biosciences). The Q3 data reflected the apoptotic
rate in this study.
Luciferase reporter assay
Using the online tool DIANA (v8; DIANA FEA BV), DDX3
was considered to be a putative target gene of miR-196a. For
generation of the DDX3 luciferase reporter plasmid, the
3-untranslated region (3-UTR) containing the miR-196a binding site
or mutant site was cloned into pmirGLO luciferase vectors (Promega
Corporation). Subsequently, 1×106 OVCAR3 cells were
co-transfected with 2 ϻg constructed luciferase vectors accompanied
with miR-196a mimic or miR-NC mimic (50 nM) by
Lipofectamine® 2000 (Invitrogen; Thermo Fisher
Scientific, Inc.). Renilla plasmid (hRluc-neo) was used as an
internal control. Cells were collected and lysed after 48 h of
transfection. The fluorescence activity was detected using the
luciferase reporter assay system (Promega Corporation), according
to the manufacturers instructions. The firefly luciferase activity
was normalized to Renilla luciferase activity.
Western blotting
In the present study, western blot analysis was
conducted according to the methods detailed below. Total proteins
were extracted from OVCAR3 cells using a Protein Isolation kit
(Tiangen Biotech Co., Ltd.), and the protein concentrations were
quantified using a Pierce bicinchoninic acid assay (Pierce; Thermo
Fisher Scientific, Inc.). Equal amounts of protein samples (40 µg)
were separated by SDS-PAGE on a 10% gel before being transferred to
a PVDF membrane (EMD Millipore) and blocked with 5% skimmed milk at
25°C for 1 h. After blocking, the proteins were incubated with the
following primary antibodies overnight at 4°C, and then probed with
the secondary antibody at 37°C for 1 h. The antibodies against DDX3
(1:1,000; cat. no. PA5-17165; Invitrogen; Thermo Fisher Scientific,
Inc.), PTEN (1:1,000; cat. no. 9188), phosphorylated (p)-PI3K (1:
1,000; cat. no.17366), total (t)-PI3K (t-PI3K) (1:1,000; cat. no.
4255), p-AKT (1:1,000; cat. no. 4060) and t-AKT (1:1,000; cat. no.
4685) (all obtained from Cell Signaling Technology, Inc.) were used
as primary antibodies, whereas peroxidase-conjugated anti-IgG
(1:5,000; cat. no. 7074; Cell Signaling Technology, Inc.) was used
as a secondary antibody. Monoclonal α-tubulin (1:1,000; cat. no.
2125; Cell Signaling Technology, Inc.) and GAPDH antibodies
(1:1,000; cat. no. 5174; Cell Signaling Technology, Inc.) were used
as controls. Bands were detected using an enhanced chemiluminescent
kit (Pierce; Thermo Fisher Scientific, Inc.), and data were
semi-quantified using ImageJ software v1.41 (National Institutes of
Health).
Statistical analysis
GraphPad Prism 5 software v8.0 (GraphPad Software,
Inc.) was used for all statistical analyses. All experiments were
carried out in triplicate, and data are presented as the mean ± SD.
A paired Students t-test was performed to distinguish differences
between two groups and one-way ANOVA followed by Tukeys post hoc
test were performed for multiple comparisons. P<0.05 was
considered to indicate a statistically significant difference.
Results
miR-196a is upregulated in ovarian
cancer cells and tumor tissues
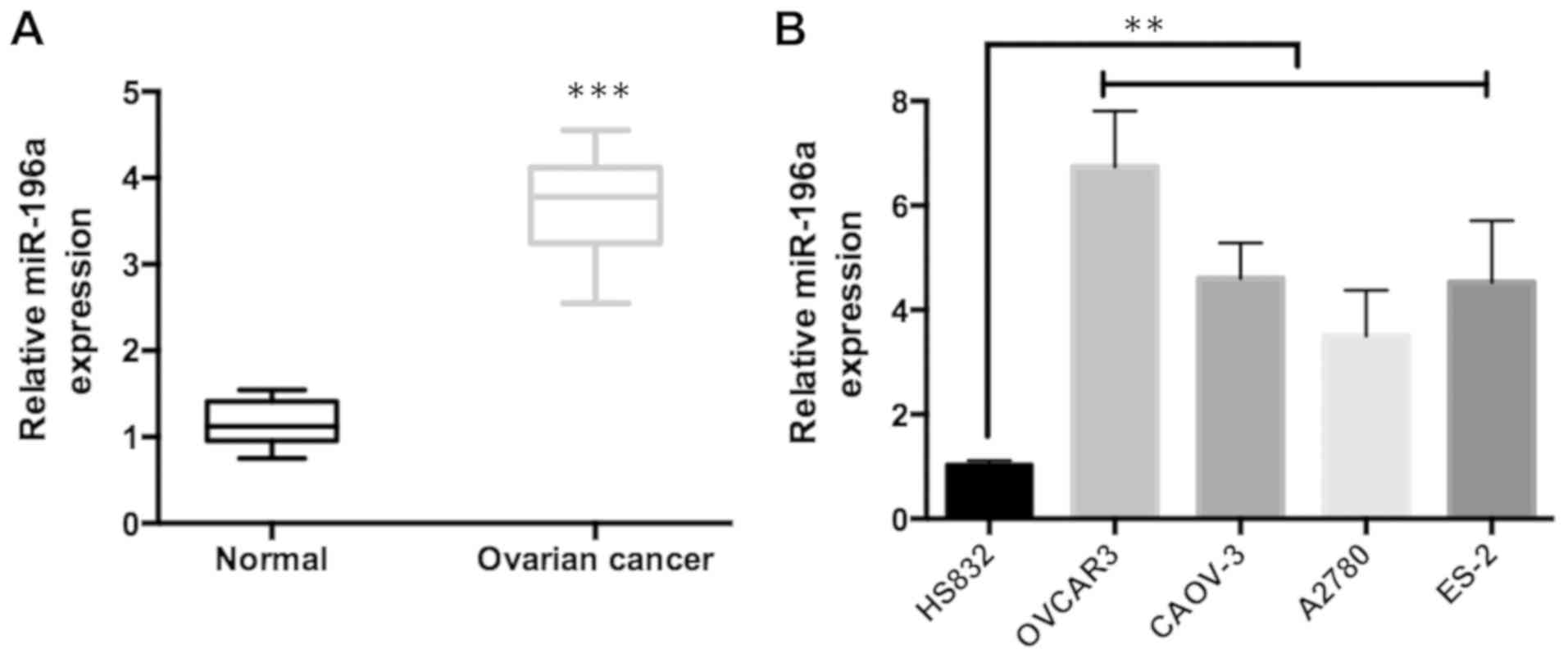
To investigate the relationship between miR-196a and
ovarian cancer cells, the expression levels of miR-196a in 30 pairs
of epithelial ovarian cancer tissues and corresponding non-tumor
tissues were measured using RT-qPCR. It was observed that the
expression of miR-196a was significantly higher in ovarian cancer
tissues relative to non-tumor tissues (Fig. 1A). Furthermore, the expression of
miR-196a was examined in ovarian cancer cell lines (OVCAR3, CAOV-3,
A2780 and ES-2) and the immortalized ovarian epithelial cell line
HS832. The results were similar to those from the clinical data:
The expression of miR-196a was significantly upregulated in ovarian
cancer cells (Fig. 1B). Taken
together, these findings indicated that miR-196a expression was
upregulated in ovarian cancer.
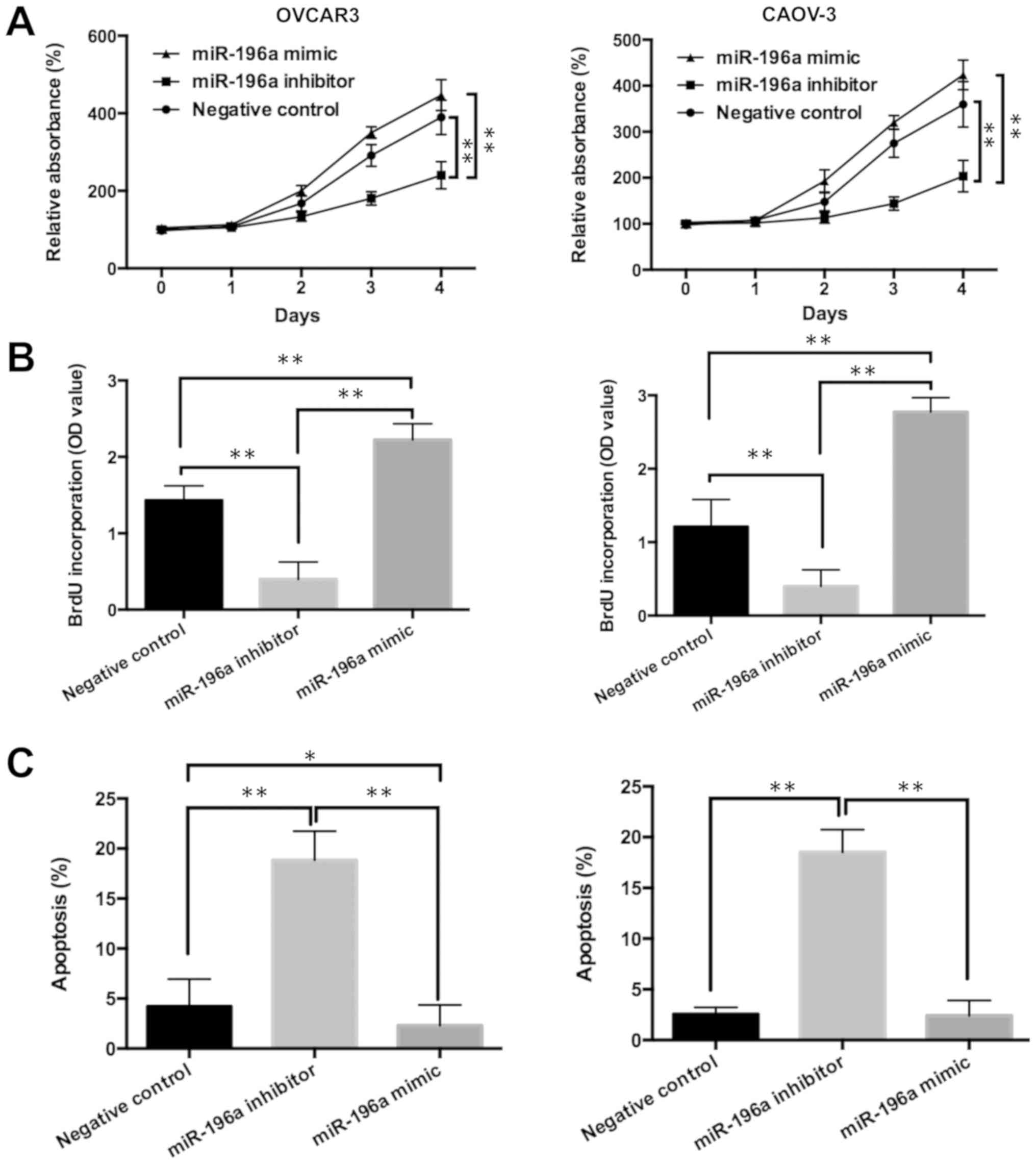
miR-196a promotes cell proliferation
and attenuates apoptosis of ovarian cancer cells
To investigate the function of miR-196a in ovarian
cancer progression, miR-196a mimic and miR-196a inhibitor were
transfected into the ovarian cancer cell lines OVCAR3 and CAOV-3 to
induce overexpression or silencing of miR-196a, respectively. OVAR3
and CAOV-3 were chosen for these subsequent experiments, as
miR-196a expression was higher in these cell lines than in the
other cell lines tested, and both cell lines are the two most
commonly used in human ovarian cancer research (18,19).
The miR-196a mimic significantly increased miR-196a expression
levels compared with miR-NC mimic both in OVCAR3 and CAOV-3 cell
lines, whereas the miR-196a inhibitor significantly decreased
miR-196a expression compared with the miR-NC inhibitor (Fig. S1). As miR-NC mimic and miR-NC
inhibitor transfection had no effect on the expression of miR-196a,
the control data of the mimic and inhibitor were combined as a NC
for comparative analysis. It was also shown that, compared with the
NC, miR-196a mimic promoted cell viability and proliferation
(Fig. 2A and B). In contrast,
miR-196a inhibitor increased the apoptosis of ovarian cancer cells
(Figs. 2C and S2). Collectively, these data suggested
that miR-196a may promote cell proliferation and attenuate
apoptosis in ovarian cancer.
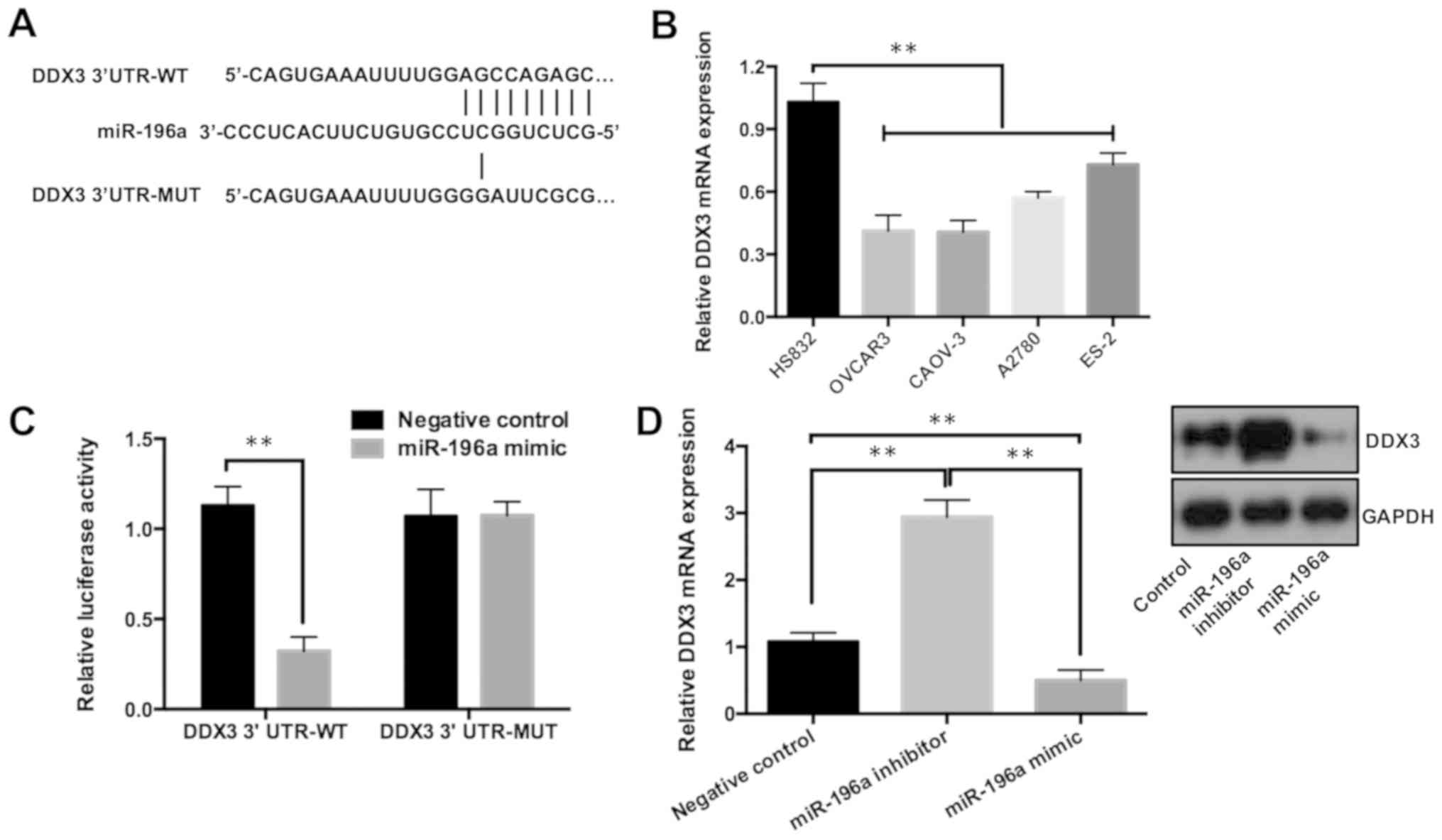
DDX3 is a direct target of
miR-196a
Previous studies have shown that DDX3 mediated miRNA
expression, and therefore regulated cell proliferation and
metastasis in various cancers (15,20).
Using the DIANA tool to uncover potential targets of miR-196a, DDX3
was found to be predicted as a direct target (Fig. 3A). Furthermore, RT-qPCR analysis
showed that the expression of DDX3 was downregulated in various
ovarian cancer cell lines (Fig.
3B). Luciferase reporter assays were performed to verify the
direct interaction between miR-196a and DDX3. It was demonstrated
that miR-196a mimic significantly reduced the luciferase activity
of the reporter that contained the wild-type DDX3 3-UTR; in
contrast, the luciferase activity of the DDX3 3-UTR containing a
mutation of the miR-196a binding site showed no change in activity
(Fig. 3C). In addition, DDX3
expression in cells transfected with miR-196a mimic or inhibitor
were examined by RT-qPCR and western blotting. It was shown that
DDX3 mRNA expression was significantly reduced by the miR-196a
mimic but increased by the miR-196a inhibitor (Fig. 3D). Similar to the RT-qPCR data, the
protein expression levels of DDX3 were markedly increased when the
miR-196a inhibitor was transfected, whereas they were decreased
when miR-196a was overexpressed (Fig.
3D).
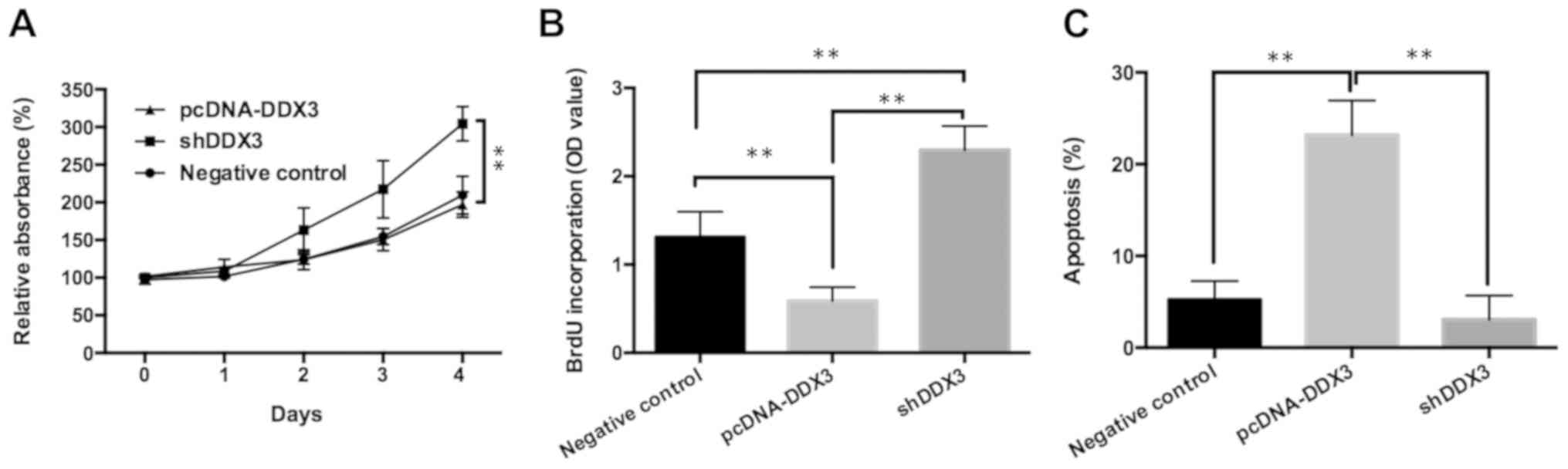
Inhibition of DDX3 promotes cell
proliferation and decreases apoptosis of ovarian cancer cells
To confirm that miR-196a regulated ovarian cancer
cell proliferation and apoptosis by targeting DDX3, OVCAR3 cells
were transfected with pcDNA-DDX3, shDDX3, or control pcDNA3.1
vector and shCtrl into and analyzed using MTT, cell proliferation
and flow cytometry assays. pcDNA-DDX3 was transfected into OVCAR3
cells to significantly upregulate DDX3 expression compared with the
control, whereas shDDX3 was transfected into cells to markedly
downregulate DDX3 expression (Fig.
S3). As pcDNA3.1 and shCtrl transfection had no effect on the
expression of DDX3, the control data of the pcDNA vector and shCtrl
were combined as a NC for comparative analysis. Increased cell
viability and proliferation, as well as reduced apoptosis, were
observed in the shDDX3 group relative to the pcDNA-DDX3 or control
groups (Figs. 4A-C and S4). In summary, these results indicated
that miR-196a may promote cell proliferation and attenuate
apoptosis in ovarian cancer through the targeting of DDX3.
DDX3 reverses the effects of miR-196a
on ovarian cancer cells via the PTEN/PI3K/AKT signaling
pathway
To investigate the molecular mechanism underlying
miR-196a targeting of DDX3 in ovarian cancer, miR-196a mimic,
miR-196a inhibitor, pcDNA-DDX3, shDDX3 and a corresponding control
were transfected into OVCAR3 ovarian cancer cells to assess the
effects on the PTEN/PI3K/AKT pathway. As shown in Fig. 5A, the miR-196a mimic decreased
protein expression of PTEN, while also inducing PI3K and AKT
phosphorylation. Previous studies have reported that inhibition of
PTEN as a tumor suppressor may activate the expression of PI3K and
AKT, thereby increasing survival of various tumors (21,22).
Notably, in the present study, downregulation of DDX3 attenuated
the expression of PTEN and therefore increased the expression of
p-PI3K and p-AKT (Fig. 5B); thus,
DDX3 may play a role in regulating ovarian cancer cell
proliferation. Furthermore, the effect of PTEN/PI3K/AKT pathway
inhibition on cell proliferation and apoptosis was explored. It was
revealed that the miR-196a mimic or shDDX3-mediated upregulation of
cell proliferation and miR-196a inhibitor or pcDNA-DDX3-mediated
inhibition of apoptosis were blocked by pretreatment with LY294002,
which is an AKT inhibitor (Figs.
5C-F and S5). Taken together,
these results suggested that miR-196a may promote ovarian cancer
cell proliferation and attenuate apoptosis by targeting DDX3
through PTEN/PI3K/AKT signaling pathway regulation.
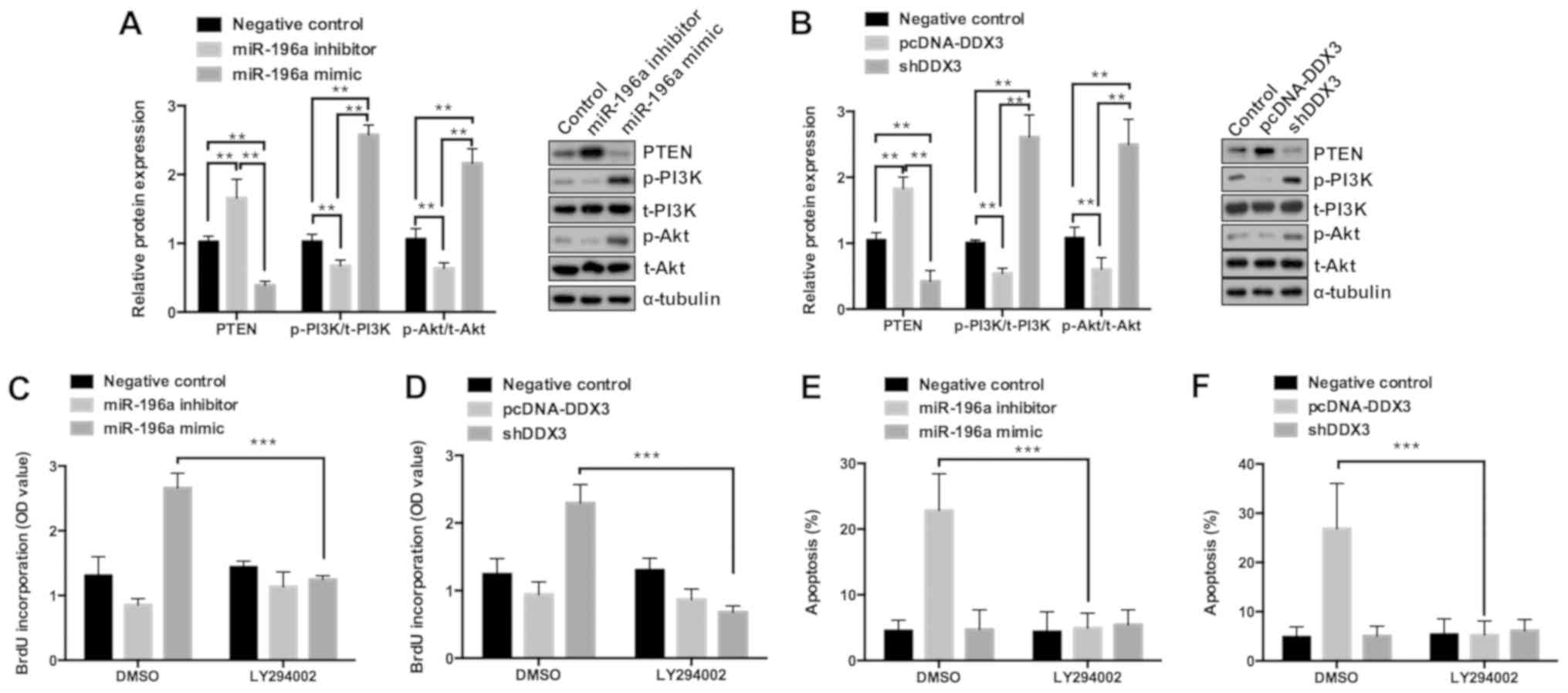 | Figure 5.miR-196a promotes cell proliferation
by targeting DDX3 via regulating the PTEN/PI3K/AKT signaling
pathway. (A and B) Western blot analysis was performed to determine
the expression levels of PTEN, p-PI3K, t-PI3K, p-Akt and t-Akt in
OVCAR3 cells transfected with miR-196a mimic, miR-196a inhibitor,
pcDNA-DDX3, shDDX3 and corresponding control. OVCAR3 cells were
pretreated with DMSO or LY294002 for 6 h and then transfected with
miR-196a mimic, miR-196a inhibitor, pcDNA-DDX3, shDDX3 and
corresponding control for 24 h. (C and D) Results of the ELISA-BrdU
assays performed to analyze cell proliferation and (E and F)
results of the flow cytometry used to evaluate the percentage of
apoptotic cells. **P<0.01 and ***P<0.001. miR, microRNA; p,
phosphorylated; t, total; sh, short hairpin; DDX3, DEAD box RNA
helicase 3; OD, optical density. |
Discussion
miR-196a is highly expressed in ovarian cancer and
is associated with various other types of cancer; therefore, it
could represent an important biomarker for cancer diagnosis and
prognosis (12,13). In epithelial ovarian cancer
progression, Yang et al (14) found that miR-196a promoted ovarian
cancer cell migration and invasion through the downstream target
gene HOXA10; however, these authors did not report the regulatory
mechanisms of miR-196a and HOXA10. Notably, there are few studies
on the precise regulatory mechanisms of miR-196a in ovarian cancer
progression. Therefore, in the present study, the relationship
between miR-196a and ovarian cancer was investigated in detail. It
was found that the expression of miR-196a was increased in ovarian
cancer tissues and cell lines, and that miR-196a promoted ovarian
cell proliferation and suppressed cell apoptosis. In addition,
transfection of miR-196a mimics reduced DDX3 expression levels in
cells and miR-196a bound to the 3-UTR of DDX3. Thus, DDX3 was
demonstrated to be a direct target of miR-196a, and importantly,
attenuated the effect of miR-196a on ovarian cancer. After further
mechanistic studies, it was revealed that miR-196a promoted cell
viability and proliferation but attenuated cell apoptosis by
inhibiting DDX3 via PTEN/PI3K/AKT pathway regulation. To the best
of our knowledge, the present study is the first to demonstrate an
association between miR-196a and DDX3 in ovarian cancer, and has
provided a novel insight into the molecular mechanisms of ovarian
cancer progression.
As previously reported, the DDX3 gene is similar to
a ‘double-edged sword’: Depending on the type of cancer, it either
contributes to cancer progression or functions as a tumor
suppressor (23). For example,
DDX3 exhibited oncogenic functions in breast cancer, in which
overexpression of the gene promoted cell growth and proliferation
in breast epithelial cancer cells (16,24).
However, in colorectal cancer, the knockdown of DDX3 promoted colon
cancer progression via the Snail/E-cadherin pathway (15). Although the mechanism of this dual
role of DDX3 in various types of cancer has not been fully
elucidated, the results of the present study suggested that the
regulatory function of miRNAs may have an important role in these
processes. In previous studies, several DDX genes, including DDX1
and DDX10, have been reported to play tumor-suppressing roles in
ovarian cancer progression (25,26).
In addition, the present study demonstrated a downregulation of
DDX3 in ovarian cancer cell lines. Furthermore, overexpression of
DDX3 decreased cell proliferation and induced apoptosis, which
suggested that, similar to other DDX genes, DDX3 may act as an
ovarian cancer tumor suppressor.
The present study indicated that miR-196a targeted
DDX3 to promote cell proliferation through the PTEN/PI3K/AKT
signaling pathway. PTEN, a well-established tumor suppressor, is
known to contribute to cancer development and progression (27). In addition, the PTEN/PI3K/AKT
signaling pathway has a confirmed link to the carcinogenesis of
various tumors, including hepatocellular carcinoma, gastric cancer
and pancreatic cancer (28,29).
PTEN has been reported to attenuate cancer cell viability and
growth by suppressing the PI3K/AKT signaling pathway, while
downregulation of PTEN expression may result in the activation of
AKT, and the promotion of cell proliferation and angiogenesis
(30). Collectively, the results
of the present study supported the hypothesis that overexpression
of miR-196a promoted ovarian cell growth and proliferation via
downregulation of PTEN expression. These findings suggested that
miR-196a may be a negative regulator of the PTEN/PI3K/AKT signaling
pathway. Moreover, it was suggested that overexpression of DDX3
reversed miR-196a-mediated tumor progression by activating PTEN
expression and suppressing AKT activity, thereby blocking the
miR-196a-mediated effects of the PTEN/PI3K/AKT signaling
pathway.
In summary, the present study provides a new insight
into the regulatory mechanisms that underlie the effects of
miR-196a on ovarian cancer progression. To the best of our
knowledge, the present study is the first to report an association
between miR-196a and DDX3, and has demonstrated that miR-196a may
promote cell proliferation and inhibit apoptosis in ovarian cancer
through attenuation of DDX3 expression via PTEN/PI3K/AKT signaling
pathway regulation. It is therefore suggested that miR-196a and
DDX3 are potential targets for the development of future ovarian
cancer treatments.
Supplementary Material
Supporting Data
Acknowledgements
Not applicable.
Funding
No funding was received.
Availability of data and materials
The datasets used and/or analyzed during the current
study are available from the corresponding author on reasonable
request.
Authors contributions
JN and WZ conceived and designed the experiments;
JN, LC, LL, MW and QR performed the experiments and analyzed the
data; JN, LC and WZ wrote the manuscript. All authors read and
approved the final manuscript.
Ethics approval and consent to
participate
Written informed consent was obtained from all
patients, and the present study was approved by the Institutional
Ethics Committee and Academic Committee of The Second Affiliated
Hospital of Soochow University.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Webb PM and Jordan SJ: Epidemiology of
epithelial ovarian cancer. Best Pract Res Clin Obstet Gynaecol.
41:3–14. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Fathalla MF: Incessant ovulation--a factor
in ovarian neoplasia? Lancet. 2:1631971. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Cramer DW and Welch WR: Determinants of
ovarian cancer risk. II. Inferences regarding pathogenesis. J Natl
Cancer Inst. 71:717–721. 1983.PubMed/NCBI
|
|
4
|
Orr B and Edwards RP: Diagnosis and
treatment of ovarian cancer. Hematol Oncol Clin North Am.
32:943–964. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Lu TX, Rothenberg ME and Micro RNA:
MicroRNA. J Allergy Clin Immunol. 141:1202–1207. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Rupaimoole R and Slack FJ: MicroRNA
therapeutics: Towards a new era for the management of cancer and
other diseases. Nat Rev Drug Discov. 16:203–222. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Thorsen SB, Obad S, Jensen NF, Stenvang J
and Kauppinen S: The therapeutic potential of microRNAs in cancer.
Cancer J. 18:275–284. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Nana-Sinkam SP and Croce CM: Clinical
applications for microRNAs in cancer. Clin Pharmacol Ther.
93:98–104. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Tutar L, Tutar E, Özgür A and Tutar Y:
Therapeutic Targeting of microRNAs in Cancer: Future Perspectives.
Drug Dev Res. 76:382–388. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Zhong LN, Zhang YZ, Li H, Fu HL, Lv CX and
Jia XJ: Overexpressed miR-196a accelerates osteogenic
differentiation in osteoporotic mice via GNAS-dependent Hedgehog
signaling pathway. J Cell Biochem. 120:19422–19431. 2019.
View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Ren W, Wu S, Wu Y, Liu T, Zhao X and Li Y:
MicroRNA-196a/-196b regulate the progression of hepatocellular
carcinoma through modulating the JAK/STAT pathway via targeting
SOCS2. Cell Death Dis. 10:3332019. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Fan Y, Fan J, Huang L, Ye M, Huang Z, Wang
Y, Li Q and Huang J: Increased expression of microRNA-196a predicts
poor prognosis in human ovarian carcinoma. Int J Clin Exp Pathol.
8:4132–4137. 2015.PubMed/NCBI
|
|
13
|
Lukács J, Soltész B, Penyige A, Nagy B and
Póka R: Identification of miR-146a and miR-196a-2 single nucleotide
polymorphisms at patients with high-grade serous ovarian cancer. J
Biotechnol. 297:54–57. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Yang B, Li SZ, Ma L, Liu HL, Liu J and
Shao JJ: Expression and mechanism of action of miR-196a in
epithelial ovarian cancer. Asian Pac J Trop Med. 9:1105–1110. 2016.
View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Su CY, Lin TC, Lin YF, Chen MH, Lee CH,
Wang HY, Lee YC, Liu YP, Chen CL and Hsiao M: DDX3 as a strongest
prognosis marker and its downregulation promotes metastasis in
colorectal cancer. Oncotarget. 6:18602–18612. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Botlagunta M, Vesuna F, Mironchik Y, Raman
A, Lisok A, Winnard P Jr, Mukadam S, Van Diest P, Chen JH,
Farabaugh P, et al: Oncogenic role of DDX3 in breast cancer
biogenesis. Oncogene. 27:3912–3922. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(-Delta Delta C(T)) Method. Methods. 25:402–408. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Blomquist CH, Leung BS, Zhang R, Zhu Y and
Chang PM: Properties and regulation of 17 β-hydroxysteroid
oxidoreductase of OVCAR-3, CAOV-3, and A431 cells: Effects of
epidermal growth factor, estradiol, and progesterone. J Cell
Biochem. 59:409–417. 1995. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Cheung LWT, Leung PCK and Wong AST:
Gonadotropin-releasing hormone promotes ovarian cancer cell
invasiveness through c-Jun NH2-terminal kinase-mediated activation
of matrix metalloproteinase (MMP)-2 and MMP-9. Cancer Res.
66:10902–10910. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
You S, Wang F, Hu Q, Li P, Zhang C, Yu Y,
Zhang Y, Li Q, Bao Q, Liu P, et al: Abnormal expression of YEATS4
associates with poor prognosis and promotes cell proliferation of
hepatic carcinoma cell by regulation the TCEA1/DDX3 axis. Am J
Cancer Res. 8:2076–2087. 2018.PubMed/NCBI
|
|
21
|
Kim MJ, Lee SJ, Ryu JH, Kim SH, Kwon IC
and Roberts TM: Combination of KRAS gene silencing and PI3K
inhibition for ovarian cancer treatment. J Control Release.
318:98–108. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Patch AM, Christie EL, Etemadmoghadam D,
Garsed DW, George J, Fereday S, Nones K, Cowin P, Alsop K, Bailey
PJ, et al Australian Ovarian Cancer Study Group, : Whole-genome
characterization of chemoresistant ovarian cancer. Nature.
521:489–494. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
He Y, Zhang D, Yang Y, Wang X, Zhao X,
Zhang P, Zhu H, Xu N and Liang S: A double-edged function of DDX3,
as an oncogene or tumor suppressor, in cancer progression (Review).
Oncol Rep. 39:883–892. 2018.(Review). PubMed/NCBI
|
|
24
|
Heerma van Voss MR, Schrijver WA, Ter
Hoeve ND, Hoefnagel LD, Manson QF, van der Wall E, Raman V and van
Diest PJ; Dutch Distant Breast Cancer Metastases Consortium, : The
prognostic effect of DDX3 upregulation in distant breast cancer
metastases. Clin Exp Metastasis. 34:85–92. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Gai M, Bo Q and Qi L: Epigenetic
down-regulated DDX10 promotes cell proliferation through Akt/NF-κB
pathway in ovarian cancer. Biochem Biophys Res Commun.
469:1000–1005. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Han C, Liu Y, Wan G, Choi HJ, Zhao L, Ivan
C, He X, Sood AK, Zhang X and Lu X: The RNA-binding protein DDX1
promotes primary microRNA maturation and inhibits ovarian tumor
progression. Cell Rep. 8:1447–1460. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Carnero A and Paramio JM: The
PTEN/PI3K/AKT Pathway in vivo, Cancer Mouse Models. Front Oncol.
4:2522014. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Wise HM, Hermida MA and Leslie NR:
Prostate cancer, PI3K, PTEN and prognosis. Clin Sci (Lond).
131:197–210. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Nakahata S, Ichikawa T, Maneesaay P, Saito
Y, Nagai K, Tamura T, Manachai N, Yamakawa N, Hamasaki M,
Kitabayashi I, et al: Loss of NDRG2 expression activates PI3K-AKT
signalling via PTEN phosphorylation in ATLL and other cancers. Nat
Commun. 5:33932014. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Xia M, Tong JH, Ji NN, Duan ML, Tan YH and
Xu JG: Tramadol regulates proliferation, migration and invasion via
PTEN/PI3K/AKT signaling in lung adenocarcinoma cells. Eur Rev Med
Pharmacol Sci. 20:2573–2580. 2016.PubMed/NCBI
|