Introduction
Diabetes mellitus (DM) is a chronic disease
worldwide, and is a major risk factor for cardiovascular disease.
The incidence of diabetes is increasing; according to the
International Diabetes Federation Diabetes Atlas (8th edition,
2017), >425 million individuals are living with diabetes, 50% of
whom are undiagnosed (1). Among
the complications of DM, cardiovascular disease has been confirmed
to be the leading cause of death (2). DM can result in both structural and
functional changes in the myocardium, thus leading to diabetic
cardiomyopathy (DCM) (3).
In the past four decades, despite extensive
molecular and cellular based research, which have unraveled the
mechanisms underlying DM and DCM, the pathogenesis of DCM remains
controversial. DCM is a lifelong and progressive disease, the
progress of which can be divided into three stages: Early, advanced
and late stage (4). During the
early stage, the majority of the patients with DCM typically
exhibit changes in the structure of the heart, without any
noticeable symptoms, such as diastolic and contractile dysfunction.
Therefore, these patients remain asymptomatic during the early
stages. As the disease advances, it progresses to irreversible
heart failure, for which there is no proven effective treatment
(5). Hence, unveiling the
underlying pathogenesis and identifying early and specific
diagnostic indicators of DCM is paramount for preventing heart
failure. Recently, numerous imaging techniques and biochemical
markers have been used to address this issue. In type 2 diabetes
mellitus (T2DM) mice, 2D-Echo-Doppler was used to detect early
changes in diastolic function and myocardial hypertrophy (6). Shaver et al (7) identified a panel of biomarkers to
detect modifications in cardiac structure and function; although
none of these were conclusive, and only served as a compensatory
index. Thus, there is an urgent need for accurate early detection
and effective therapeutics for the diagnosis and treatment of DCM.
Therefore, an improved understanding of the pathogenesis of DCM is
required, particularly for early-stage DCM.
Circular RNAs (circRNAs) are a unique type of
endogenous non-coding RNAs, which were initially considered to be
abnormal splicing byproducts with limited functional potential
(8). However, in recent years,
increasing evidence has demonstrated that circRNAs play important
roles in a range of diverse physiological and pathological
processes (9). Although the
functions of the majority of circRNAs remains to be fully
elucidated, the established functions include acting as a sponge to
sequester certain microRNAs (miRNAs), modulating transcription and
interfering with splicing, and even translation into polypeptides
(10–12). Characterized by their stability and
tissue-specificity, circRNAs may not only serve as an ideal
biomarker, but may also assist in elucidating the underlying
mechanisms in various disease, including DM. For example, Zhao
et al (13) detected
circRNAs in the peripheral blood of patients with T2DM and found
that hsa_circ_0054633 may be used as a diagnostic biomarker of
pre-diabetes. Fang et al (14) showed that circANKRD36 was
associated with inflammation in patients with T2DM. Nevertheless,
relatively less is known on the expression profile and potential
role of circRNAs in early-stage DCM. Therefore, in the present
study, secondary sequencing technology was used to examine the
expression of circRNAs in early-stage DCM mice. Key circRNAs were
screened for further analysis in the pathogenesis of early-stage
DCM, as well as their potential as biomarkers, for early diagnosis
and treatment of DCM.
Materials and methods
Animals
A total of 5 male C57BL/BKS-Leprdb (db/db) mice (8
weeks old; weight, 42–47 g) were obtained from Gempharmatech Co.,
Ltd. A total of 5 monogenic LepR mutated mice (db-/db-; 8 weeks
old; weight, 20–22 g) from the same company were used as control
mice. Mice were maintained under controlled conditions at 20–22°C
with 12-h light/dark cycles and 40–60% humidity, with free access
to food and water. Body weight was measured and the fasting blood
glucose levels were detected using the glucose oxidase-peroxidase
method (15). All experiments were
approved by the Institutional Animal Care and Use Committee of
Fudan University (China).
Myocardium sample collection and
RNAseq
Mice were anesthetized by intraperitoneal injection
of 1% pentobarbital sodium solution at 55 mg/kg, and then
sacrificed by cervical dislocation and the myocardium samples were
harvested for RNAseq. A total of 5 µg RNA in each db/db and
control sample was extracted for preparation of RNA samples.
Epicentre Ribo-Zero™ rRNA Removal kit (Epicentre; Illumina, Inc.)
was used to remove ribosomal RNA. To obtain enriched pure circRNA,
linear RNA was digested with 3 U of RNase R (Epicentre; Illumina,
Inc.) per µg of RNA. According to the manufacturer's protocol, the
sequencing libraries were constructed using a NEBNext®
Ultra™ Directional RNA Library Prep kit for Illumina®
(New England Biolabs, Inc.). Briefly, circRNAs were interrupted
randomly by adding a fragment reagent. Subsequently, the first
strand cDNA was synthesized using random hexamer primers and M-MuLV
reverse transcriptase (RNaseH), and subsequently the second strand
of cDNA was synthesized. After the repair of the end of the DNA
strand and adenylation of 3′ends of DNA fragments, AMPure XP beads
were used to select cDNA fragments with the appropriate length.
Subsequently, 3 µl USER Enzyme (New England Biolabs, Inc.) was used
to degrade the second strand containing cDNA. Finally, PCR was
performed to amplify the circRNA library using the same materials
and methods as below, and products were purified using the AMPure
XP system.
Reverse transcription-quantitative
(RT-q)PCR
Total RNAs in heart tissue were extracted using
TRIzol® (Invitrogen; Thermo Fisher Scientific, Inc.)
according to the manufacturer's protocol. Using a PrimeScript
RT-PCR kit (cat. no. RR014; Takara Bio, Inc.) extracted RNA was
reverse transcribed to cDNA following the 20 µl system protocol,
the reaction condition were as follows: 37°C for 5 min, 85°C for 15
sec and 4°C for preservation. The expression levels of circRNAs and
related mRNAs, brain natriuretic peptide (BNP), atrial natriuretic
peptide (ANP) and interleukin (IL)-6 were detected using a SYBR
Green PCR kit (Qiagen, Inc.) in a 7500 Fast Real-Time PCR system
(Applied Biosystems; Thermo Fisher Scientific, Inc.). The
thermocycling conditions were: Initial denaturation at 95°C for 2
min; followed by 40 cycles of 95°C for 5 sec and annealing at 60°C
for 34 sec; then 95°C for 15 sec; 60°C for 1 min and 95°C for 15
sec. The miRNA levels were detected using miRNA First Strand cDNA
Synthesis kit (cat. no. B532451; Sangon Biotech Co., Ltd.),
according to manufacturer's protocols. U6 and β-actin were used as
internal controls. The 2−δδCq method (16) was used for quantitative analysis of
gene expression. The primer sequences are presented in Table I.
 | Table I.Sequences of the primers used in the
present study. |
Table I.
Sequences of the primers used in the
present study.
| Gene | Primer sequence
(5′→3′) |
|---|
| BNP | F:
GAGGTCACTCCTATCCTCTGG |
|
| R:
GCCATTTCCTCCGACTTTTCTC |
| ANP | F:
GTGCGGTGTCCAACACAGAT |
|
| R:
TCCAATCCTGTCAATCCTACCC |
| IL-6 | F:
CCAAGAGGTGAGTGCTTCCC |
|
| R:
CTGTTGTTCAGACTCTCTCCCT |
|
mmu_circ_0001697 | F:
AGATGGCTTCTGAGCTGCTTT |
|
| R:
TAGCTTTCCGCTGGTGGTTG |
|
mmu_circ_0001160 | F:
TGGTGTAATTGCCTCTGCCATC |
|
| R:
CTGCCAATCCGGCCAATATG |
|
novel_circ_0008273 | F:
CCAGAGATCTGGGAGGAGTAGA |
|
| R:
CCTCAGGAACTGAAGGTAAAGT |
|
novel_circ_0009344 | F:
TGATGCTGGCTTTGTTCCCAA |
|
| R:
TTCAAACCCCGACTGGAGCTA |
|
mmu_circ_0001625 | F:
CATCCTGGCATTGGTTTTGCC |
|
| R:
GGGCTCATGATTTTCGTGACTT |
|
mmu_circ_0000431 | F:
ACTCTGAACGGCGAGATCCT |
|
| R:
TGTCATCTCTAACCATCACCAACA |
|
mmu_circ_0000652 | F:
GCAGGAGACAAGGAGCTACA |
|
| R:
AGTTGCTGGTGTAAGAGGCA |
|
mmu_circ_0000058 | F:
GGTAGACCTGACTGATGCCA |
|
| R:
TAGTAAAGTGTTCGCCCTCG |
|
mmu_circ_0001058 | F:
AGGAGCGTCTGAATGAGGACT |
|
| R:
GCGATACTGTGAACACCAGGG |
|
mmu_circ_0000680 | F:
ATTCAAACTGTGCCTTCCCA |
|
| R:
TTCCAGGGAAACAAAGTGACA |
|
novel_circ_0000824 | F:
GAAGTGCCTCTTCAGGGGTG |
|
| R:
AGTCCTTCTCTCTGTGTTGCTC |
|
mmu_circ_0000547 | F:
GGCGACGGCAGATGAAAACA |
|
| R:
GTCAGACAGTGGTCGTGGC |
|
novel_circ_0004285 | F:
GGTTGAAGAATGGAGGAGGGT |
|
| R:
GCAGATACTCGTGAAGGAAGCA |
| IGF-1 | F:
AAATCAGCAGCCTTCCAACTC |
|
| R:
GCACTTCCTCTACTTGTGTTCTT |
| FOXO3A | F:
GGGGAACCTGTCCTATGCC |
|
| R:
TCATTCTGAACGCGCATGAAG |
| CAB39 | F:
TGCTGTTGGACAGACACAACT |
|
| R:
GGAGGTTCATCATTAGCTTGAGG |
| BCL2 | F:
GCTACCGTCGTGACTTCGC |
|
| R:
CCCCACCGAACTCAAAGAAGG |
| SPRY1 | F:
GGTCATAGGTCAGATCGGGTC |
|
| R:
GTCCCGTATTCCACCATGCT |
| miR-195 | F:
TAGCAGCACAGAAATATTGGC |
|
| R: Universal PCR
Primer R |
| miR-21 | F:
TAGCTTATCAGACTGATGTTGA |
|
| R: Universal PCR
Primer R |
| miR-320 | F:
AAAAGCTGGGTTGAGAGGGCGA |
|
| R: Universal PCR
Primer R |
| miR-451 | F:
AAACCGTTACCATTACTGAGTT |
|
| R: Universal PCR
Primer R |
| miR-30d | F:
TGTAAACATCCCCGACTGGAAG |
|
| R: Universal PCR
Primer R |
TUNEL assay
Cardiomyocyte apoptosis was detected using terminal
deoxynucleotidyl transferase dUTP nick-end labeling (TUNEL)
technique. Mice were sacrificed by cervical dislocation following
anesthesia, then heart tissue was isolated and immediately fixed in
4% formalin solution for 24 h at 4°C and then embedded in paraffin.
Next, the specimens were sectioned into 4-µm thick slides and
stained with a DeadEnd™ Fluorometric TUNEL system kit at 37°C for 1
h (cat. no. G3250; Promega Corporation), following the
manufacturer's instructions.
Prediction of circRNA-miRNA
interactions and identification of key circRNAs
The bioinformatics platform miRanda version 3.3a
(microrna.org/) was used to predict putative miRNAs that exhibited
a potential association with one of the 13 differential circRNAs
identified by RT-qPCR. The major parameters were as follows: i)
-sc, 140; ii) -en, −10; iii) -scale, 4; iv) -strict. The
circRNA-miRNA interaction network was constructed using Cytoscape
version 3.01 (17). Subsequently,
Gene Ontology (GO) enrichment analysis (18–20)
was used to identify the processes with the highest degree of
enrichment, in which the mRNA targets of the miRNAs were involved.
Similarly, Kyoto Encyclopedia of Genes and Genomes (KEGG) analysis
was performed to identify the significant pathways associated with
these mRNAs (genome.jp/kegg/) (21). To identify the key circRNAs, known
DCM-related miRNAs were searched by reviewing the relevant
literature in pubmed (pubmed.ncbi.nlm.nih.gov/), and the common
miRNAs between the known DCM-related miRNAs and the predicted
miRNAs were extrapolated to identify key miRNAs. The circRNAs that
were associated with these key miRNAs were used to construct an
integral circRNA-miRNA-mRNA regulatory network using Cytoscape
version 3.01. Finally, these key miRNAs and their target mRNAs were
verified by RT-qPCR, following the aforementioned protocol.
Internal ribosome entry site (IRES)
and open reading frame (ORF) prediction of circRNAs
To extensively explore the protein-coding function
of the 13 differentially expressed circRNAs, IRES finder
(github.com/xiaofengsong/IRESfinder) and circAtlas 2.0
(circatlas.biols.ac.cn) were used to predict the presence of IRES
and ORF elements, which are essential elements of circRNAs with
protein coding capacity. The IRES score and ORF numbers were
plotted using R software version 3.5 (r-project.org/).
Statistical analysis
Data are presented as the mean ± standard deviation
of three independent experiments each performed in triplicate.
Statistical significance was calculated using a Student's t-test in
GraphPad Prism version 5 (GraphPad Software, Inc.). P<0.05 was
considered to indicate a statistically significant difference.
Results
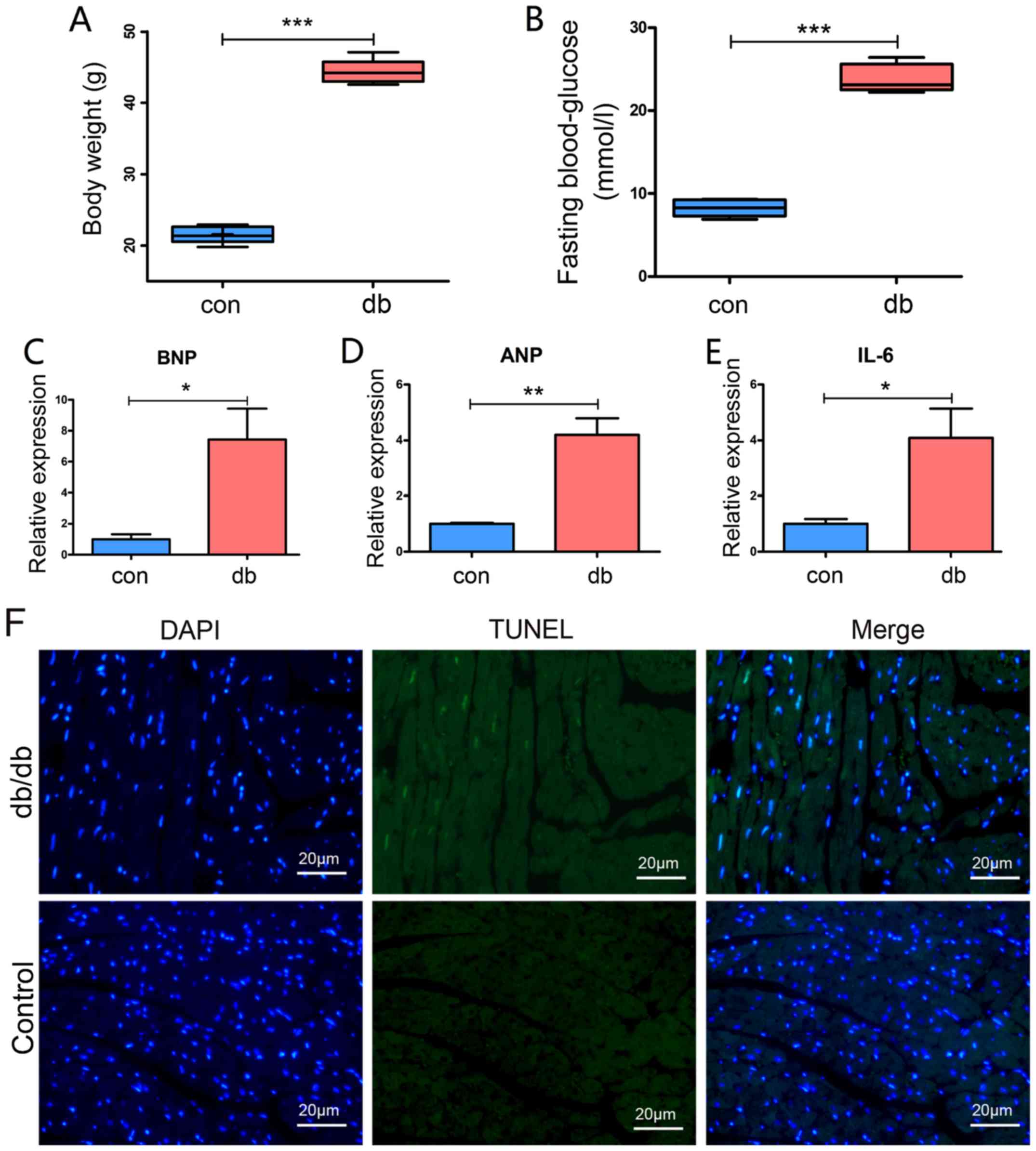
Early changes in the diabetic
myocardium
A series of biochemical biomarkers have been
determined to crudely indicate functional changes in early-stage
DCM (22,23). BNP, ANP and IL-6 expression levels
were assessed to detect early modifications in db/db myocardial
tissue. The body weight of the five db/db mice were measured and
the fasting blood-glucose levels were measured three times on
different days. The average weight of db/db mice was 44.34 g,
whereas the control mice were 22.8 g (Fig. 1A). The fasting blood-glucose level
of db/db mice ranged from 22.2–26.4 mmol/l with a mean of 23.8
mmol/l (Fig. 1B). Furthermore, the
qPCR results exhibited a significant increase in the expression
levels of BNP, ANP and IL-6 (P<0.05). In addition, TUNEL
staining also exhibited an increasing trend of cardiomyocyte
apoptosis in the db/db model (Fig.
1F), suggesting successful establishment of a model of
early-stage DCM.
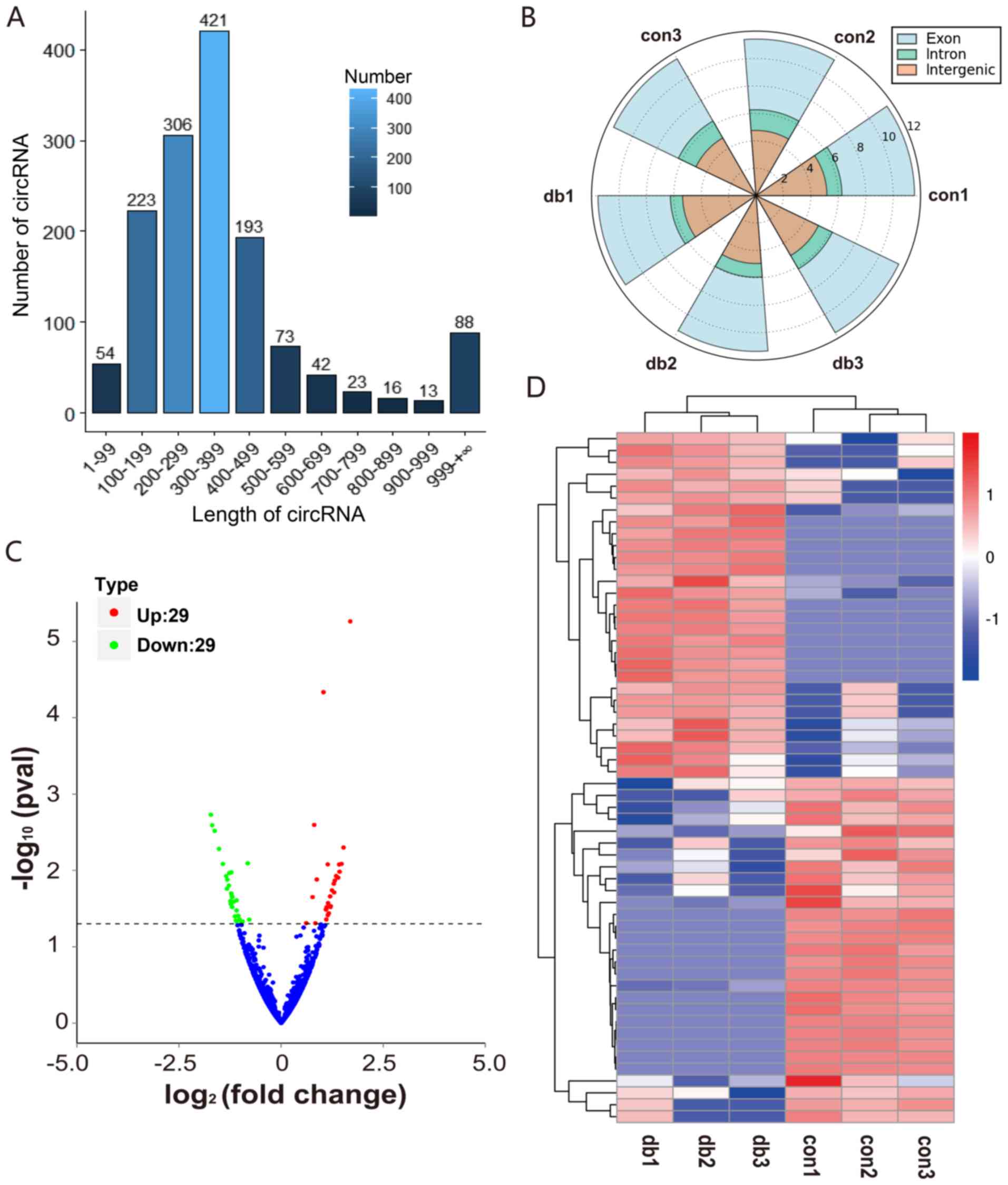
circRNA expression in the myocardial
tissue of db/db mice
To detect circRNA expression in the myocardial
tissues of db/db mice, RNA-seq analysis was performed. Then, a
volcano plot and heatmap were constructed using R software version
3.5 (r-project.org/). The length of the detected circRNAs was
primarily in the 100–500 bp range (Fig. 2A), and a majority of the reads were
covered in the exon of the genome (Fig. 2B). Overall, 58 circRNAs were
determined to be significantly differentially expressed (P<0.05;
Table SI), including 29
upregulated circRNAs and 29 downregulated circRNAs (Fig. 2C and D). Of these, three-quarters
were newly identified circRNAs. In addition, all candidate circRNAs
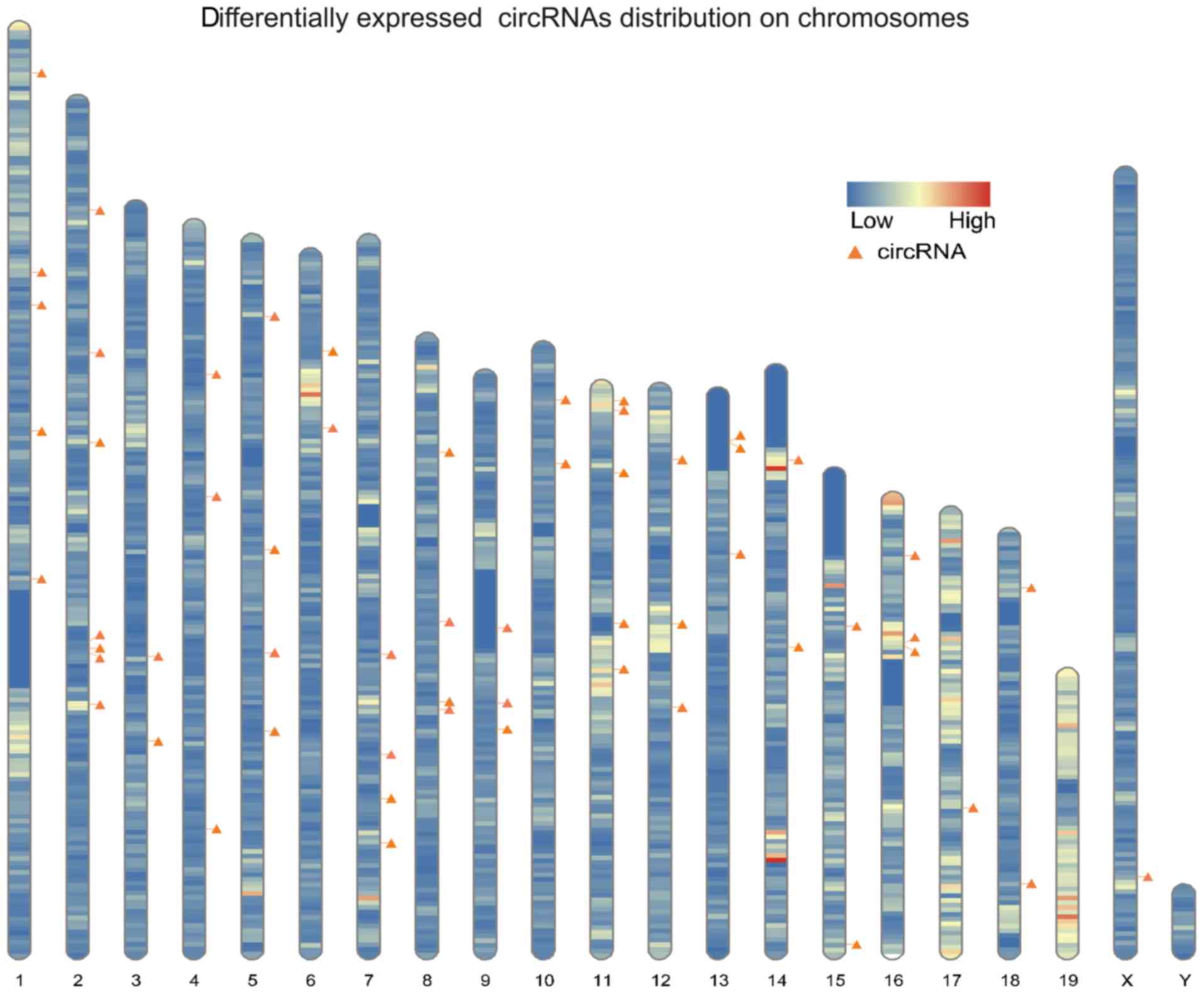
were found to be distributed among all the chromosomes (Fig. 3).
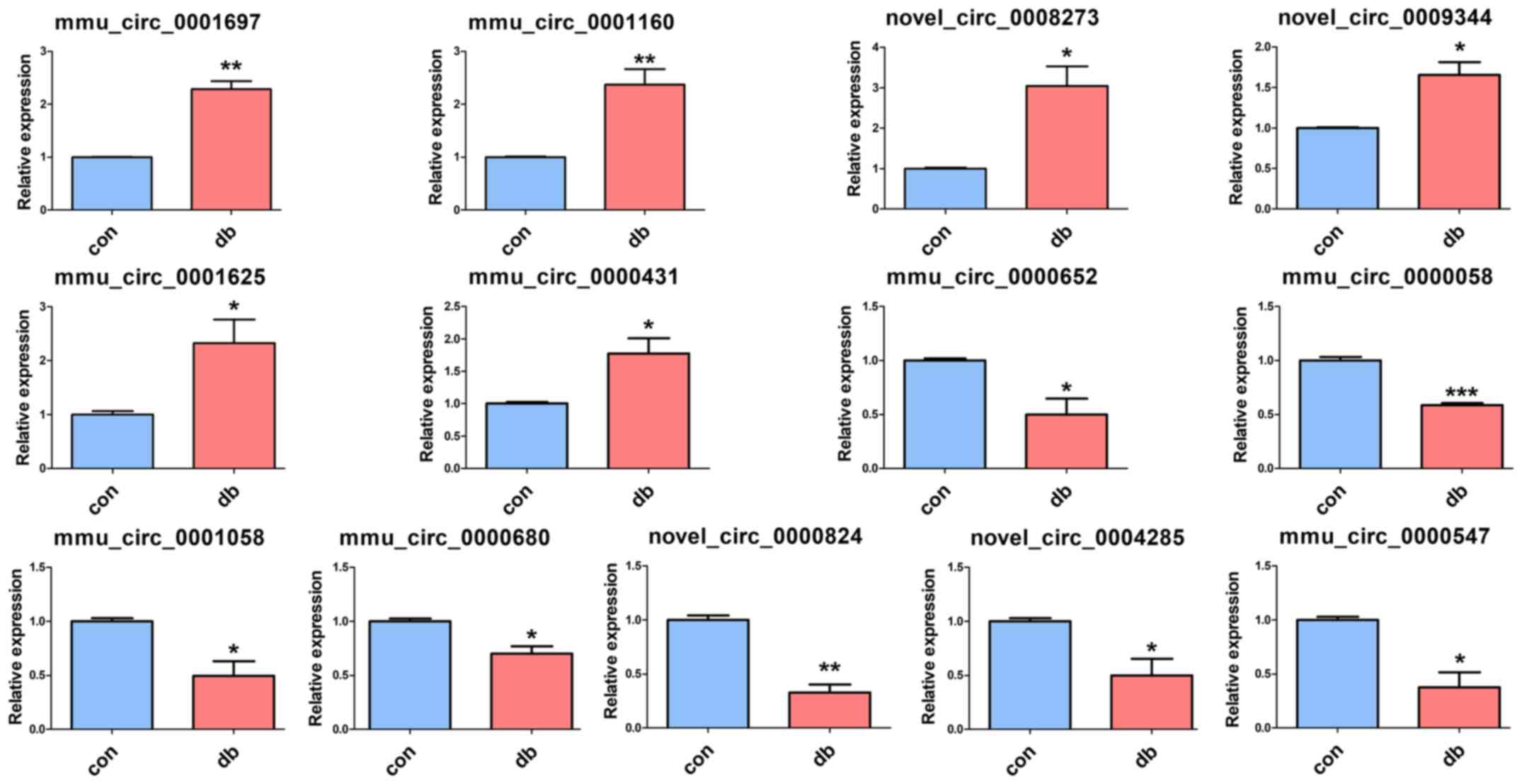
To validate the RNA-seq results, RT-qPCR was
performed to analyze circRNA expression. According to the RT-qPCR
results, six upregulated circRNAs (mmu_circ_0001697,
mmu_circ_0001160, novel_circ_0008273, novel_circ_0009344,
mmu_circ_0001625 and mmu_circ_0000431) and seven downregulated
circRNAs (mmu_circ_0000652, mmu_circ_0000058, mmu_circ_0001058,
mmu_circ_0000680, novel_circ_0000824, mmu_circ_0000547 and
novel_circ_0004285) were verified to be differentially expressed in
the tissue samples (Fig. 4).
Establishment of the circRNA-miRNA
network
Increasing evidence has suggested that circRNAs may
regulate the function of miRNAs by acting as competing endogenous
RNAs (ceRNAs) (24). In an attempt
to identify the functions of these differentially-expressed
circRNAs in DCM, circRNA-miRNA co-expression networks were
constructed based on the miRanda algorithm. The results showed
there were 610 related miRNAs that exhibited close binding
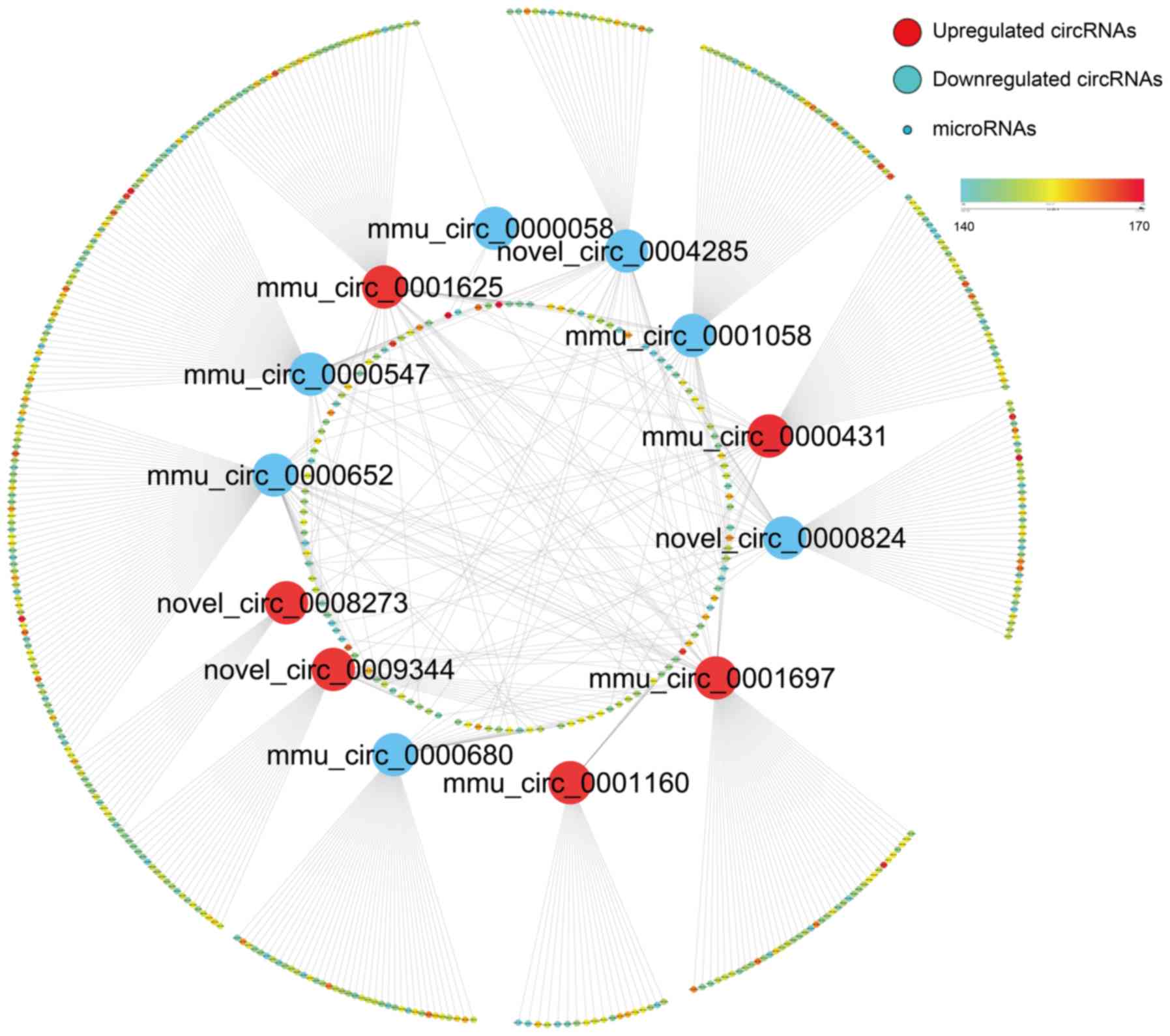
competency to these circRNAs (Fig.
5), the detailed data are listed in Table SII. Total scores of the targeting
relationship for all binding sites were predicted and displayed on
a color scale, where higher values indicated an increased
likelihood of targeting. To further probe the function of these
target genes related to these differentially-expressed circRNAs, GO
enrichment analysis was performed. The results showed that these
target genes participated in various biological processes, such as
‘metabolic process’, ‘binding’ and ‘negative regulation of cellular
process’ (Fig. S1). KEGG analysis
showed these target genes may have effects on several vital
signaling pathways, such as ‘glycerophospholipid metabolism’,
‘glycolysis/gluconeogenesis’, ‘ether lipid metabolism’ and
‘fructose and mannose metabolism’ (Fig. S2). The results suggested that the
detected circRNAs may be related to glucose and lipid metabolism,
which is consistent with the fasting blood-glucose result. However,
whether these circRNAs function in DCM and the underlying
mechanisms need to be further explored.
mmu_circ_0000652 and mmu_circ_0001058
may play an important role in early-stage DCM
To construct a potential ceRNA network in
early-stage DCM, the functions of the predicted miRNAs were
explored. Based on bioinformatics analysis and literature
retrieval, five miRNAs (miR-30d, miR-320, miR-451, miR-195 and
miR-21) were determined to be associated with DCM (Table II). Additionally, their possible
regulatory mechanisms has been confirmed in previous studies
(25–29). The predicted binding sites between
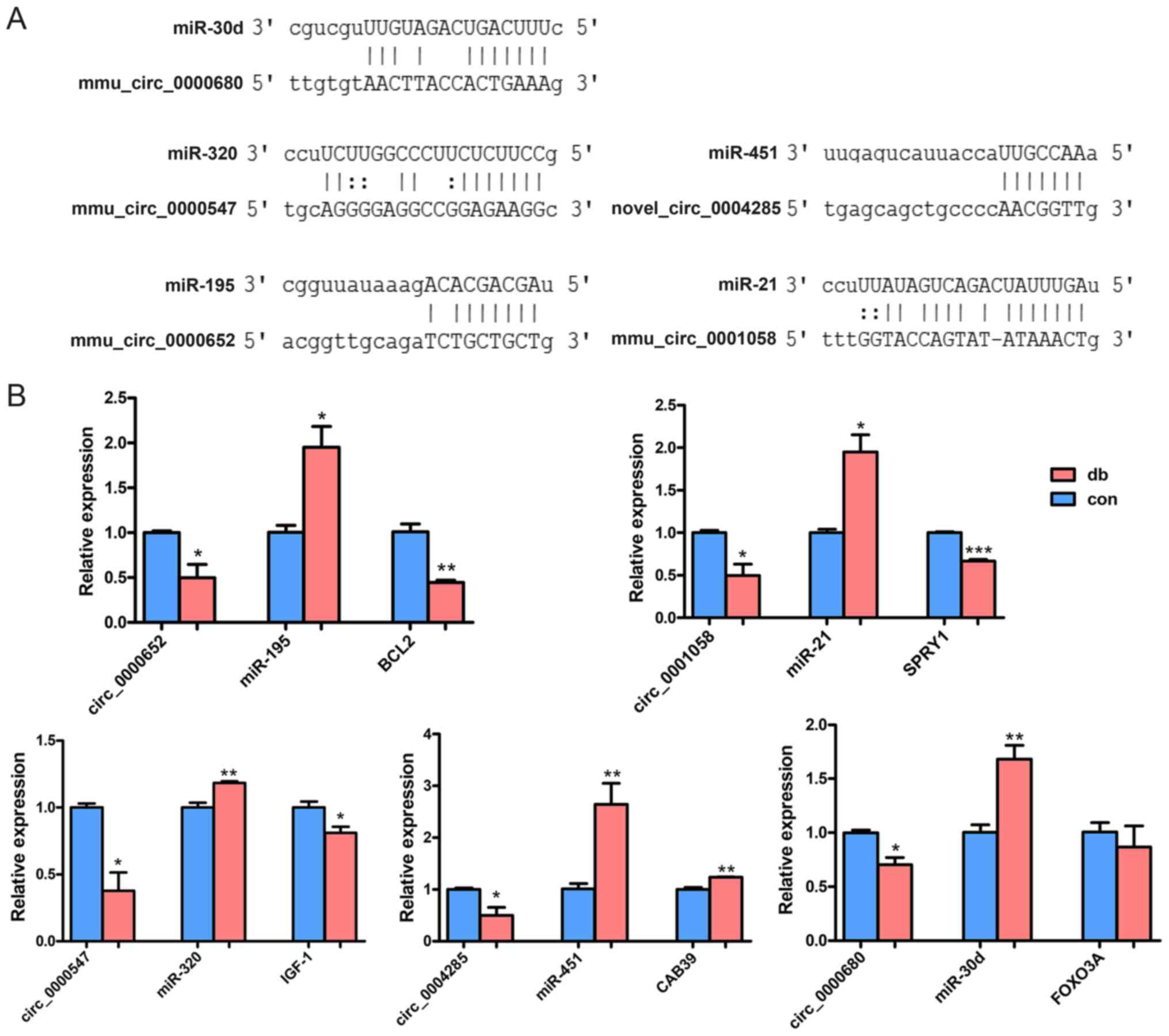
these key circRNAs and DCM-related miRNAs are exhibited in Fig. 6A. To further investigate the
interactions between these RNAs, the five DCM-related miRNAs and
the target mRNAs were also verified by RT-Qpcr (Fig. 6B). The results showed that two
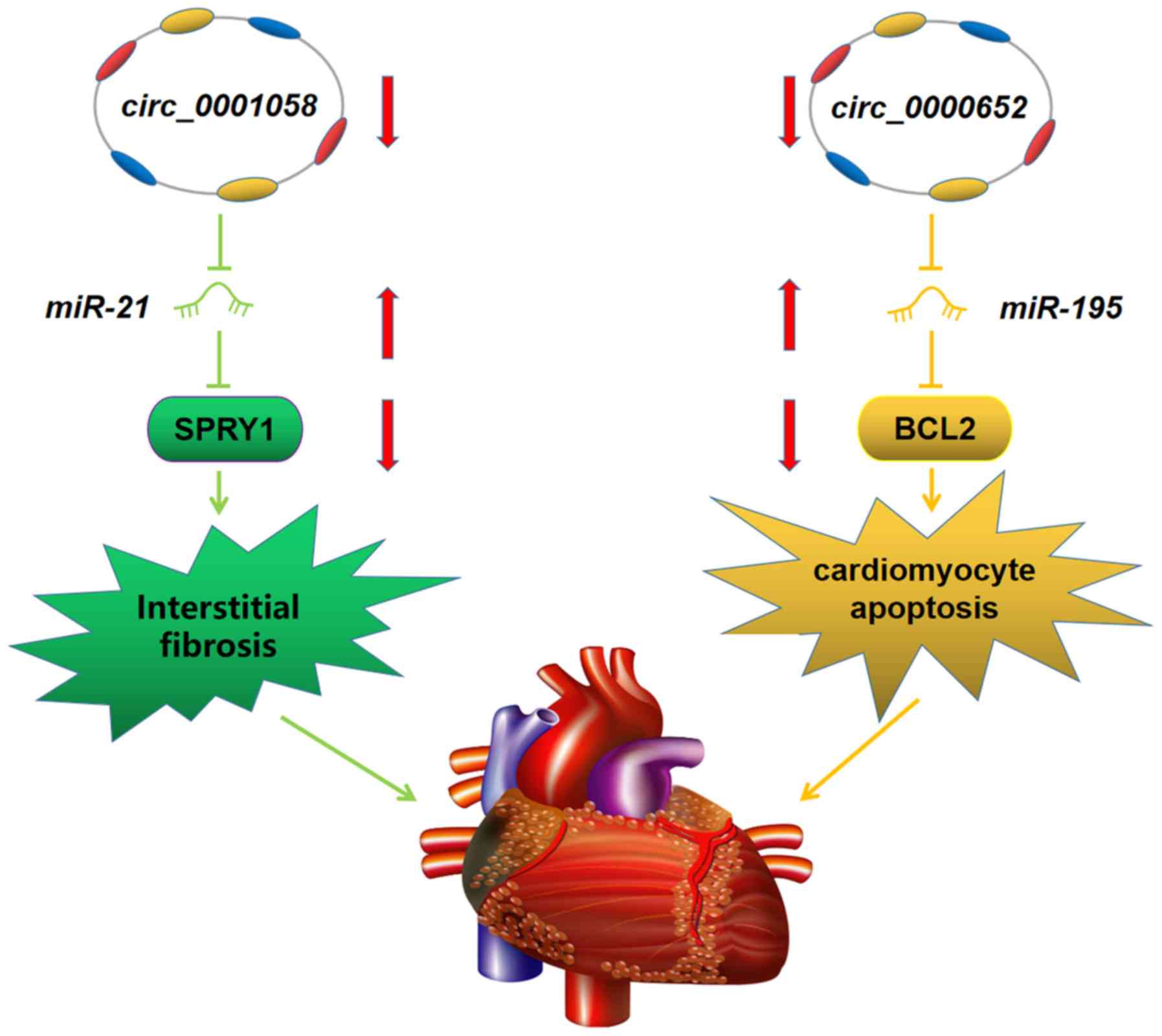
regulatory networks, mmu_circ_0000652/miR-195/BCL2 and
mmu_circ_0001058/miR-21/sprouty homolog 1 (SPRY1), were
representative of a classical ceRNA regulatory mechanism. A
schematic plot of the two potential ceRNA networks is presented in
Fig. 7. As BCL2 serves as a key
regulatory molecule in the process of apoptosis and SPRY1 is
associated with interstitial fibrosis (30,31),
the results suggested that mmu_circ_0000652 and mmu_circ_0001058
may serve an important role in early-stage DCM.
 | Table II.The five potential ceRNA regulatory
networks and functions in DCM. |
Table II.
The five potential ceRNA regulatory
networks and functions in DCM.
| circRNA | miRNA | mRNA | Function | Refs. |
|---|
|
mmu_circ_0000680 | miR-30d | FOXO3A | Cardiomyocyte
pyroptosis | (26) |
|
mmu_circ_0000547 | miR-320 | IGF-1 | Impaired
angiogenesis | (28) |
|
novel_circ_0004285 | miR-451 | CAB39 | Cardiomyocyte
lipotoxicity | (25) |
|
mmu_circ_0000652 | miR-195 | BCL2 | Cardiomyocyte
apoptosis | (27) |
|
mmu_circ_0001058 | miR-21 | SPRY1 | Interstitial
fibrosis | (29) |
Mmu_circ_0001160 exhibits high
protein-coding potential
Recent studies have reported that a fraction of
circRNAs exhibit protein-coding potential based on the presence of
IRES and ORF elements (32–34).
IRES and ORF are both important regulatory elements for circRNA
translation without the need for a 5′ cap structure. To extensively
explore the function of these circRNAs, IRES-finder software and
circAtlas 2.0 were used to predict whether a circRNA sequence
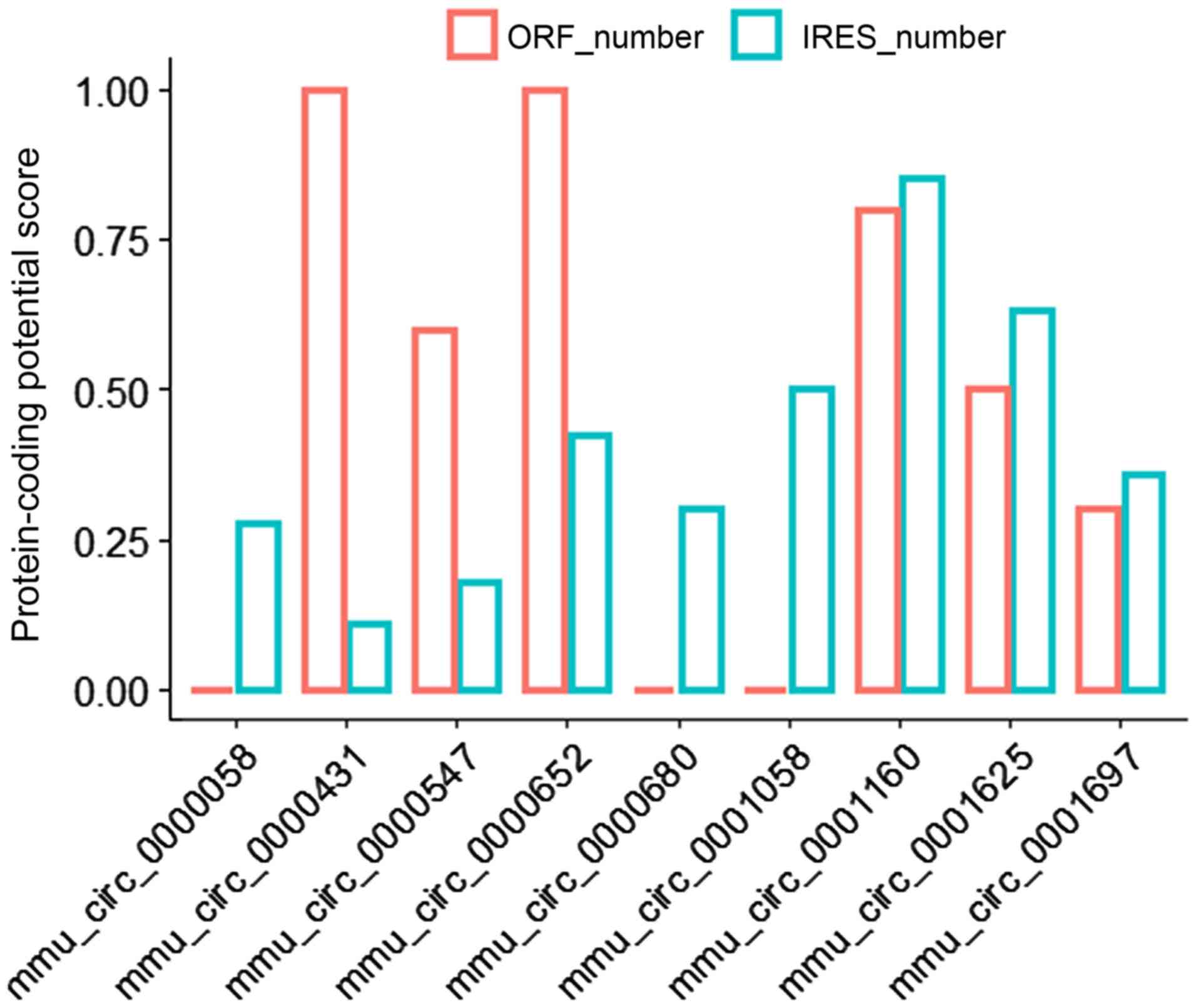
possessed potential IRES and ORF elements. Among the differentially
expressed circRNAs, there were 9 circRNAs that had a potential IRES
element. The closer the score value was to 1, the higher the
credibility of the IRES in the circRNA was (Fig. 8). Based on the results,
mmu_circ_0001160 had a relatively high IRES and ORF score,
suggesting that this circRNA may function in a novel way by
producing a polypeptide, the potential regulatory mechanisms of
which are worthy of future studies.
Discussion
circRNAs can regulate the expression of genes at the
transcriptional or post-transcriptional levels (24,35).
In recent years, expression of circRNAs have been shown to be
dysregulated in several types of diseases, including DM (1,11,36–39).
However, their function and the underlying mechanisms in DCM
remains largely unknown. In the present study, the circRNA
expression profile was analyzed in a mouse model of early-stage DCM
to improve our understanding of the pathogenesis of early-stage DCM
and to find potential circRNA biomarkers. To the best of our
knowledge, the present study was the first to explore the
significance of circRNAs in early-stage DCM. Two promising circRNAs
and their regulatory networks were identified, which may serve an
important role in early-stage DCM.
An increasing number of studies have suggested that
circRNAs serve a significant role in DM (13,14,36,40–42).
For example, Zhao et al (13) found that hsa_circ_0054633 may serve
as a diagnostic biomarker for pre-diabetes. However, relatively
less is known about circRNAs in early-stage DCM. In the present
study, 58 circRNAs were shown to be differentially expressed in a
mouse model of early-stage DCM. RT-qPCR analysis results were
consistent with the results of RNA-seq analysis, confirming the
reliability of the RNA-seq data. KEGG analyses indicated that the
related mRNAs were primarily involved in glucose metabolism and
lipid metabolism pathways, both of which serve a significant role
in triggering further alterations of the myocardial tissue. For
example, lipid metabolism disorders may contribute to cardiomyocyte
lipotoxicity, which is an initial deleterious response in the DM
heart through the formation of fatty acid metabolites, as well as
the release of mitochondrial and cytosolic ROS (43,44).
Although the alterations in the KEGG pathway may be caused by high
levels of blood-glucose in our model, the results still
preliminarily suggested that these circRNAs may have an association
with early stage DCM and the function of these circRNAs needs
further investigation.
Numerous studies have shown that circRNAs may
negatively regulate the inhibitory effects of miRNAs on their
target mRNAs by directly binding to miRNAs, thereby acting as miRNA
sponges (24,35,40,45–47).
In the present study, mmu_circ_0000652 and mmu_circ_0001058 were
found to potentially interact with miR-195 and miR-21, both of
which have been shown to serve roles in the metabolism of DCM.
miR-195 is associated with a number of pathological myocardial
conditions, including hypertrophy, fibrosis, apoptosis, arrhythmia
and heart failure (27). Zheng
et al (48) showed that
silencing miR-195 attenuated myocardial hypertrophy, promoted
coronary circulation and reduced myocardial dysfunction in
streptozotocin-induced diabetic mice. In addition, further
experiments showed that these protective effects were associated
with decreased apoptosis, generated by the induction of the direct
targets of miR-195, BCL2 and sirtuin 1. In the present study,
upregulation of miR-195 and downregulation of BCL2 were observed,
suggesting that downregulation of mmu_circ_0000652 may promote
apoptosis by indirectly inhibiting BCL2 in the db/db model.
Apoptosis is a relatively early event in DCM, when apoptotic
cardiomyocytes are lost, collagen is deposited to replace dead
cardiomyocytes, which contributes to cardiac fibrosis and
dysfunction (49). The function of
miR-21 differs from miR-195. miR-21 regulates the ERK-MAP kinase
signaling pathway by affecting SPRY1, ultimately resulting in
fibrosis, hypertrophy and cardiac dysfunction (29). Similarly, mmu_circ_0001058 serves
an important role in the progress of DCM by interfering with
miR-21. Further studies on mmu_circ_0000652 and mmu_circ_0001058
may highlight the roles of these circRNAs in the underlying
pathogenesis of early-stage DCM, thus potentially improving early
diagnosis and treatment of early-stage DCM and minimizing its
complications.
In addition to acting as an miRNA sponge, studies
have reported that circRNAs exhibit protein/peptide coding
capacity. To exhibit protein-coding potential, an IRES and ORF are
required. The present results showed that mmu_circ_0001160
possessed a relatively high coding possibility amongst the
identified circRNAs. Zn2+ transporter 7 (ZNT7), the
linear parental gene of mmu_circ_0001160, has been shown to be
associated with DM by interfering with insulin secretion (50,51).
Furthermore, Tuncay et al (52) demonstrated that the novel role of
ZNT7 in hyperglycemic cardiomyocytes by affecting
sarco(endo)plasmic reticulum-mitochondria coupling. Based on the
hypothesis that circRNAs may affect its linearly expressed gene, it
was hypothesized that mmu_circ_0001160 may interfere with
early-stage DCM by producing a ZNT7-related protein. However, to
validate whether this circRNA could encode a protein, in-depth
studies are required. As the function of circRNA still remains to
be fully elucidated, further functional experiments on
mmu_circ_0001160 are required to extensively explore the role of
circRNAs in various diseases.
There are several limitations of the present study.
First, since the metabolic disturbances could cause a series of
cardiac structural and functional alterations in the early stage of
DCM (53), some additional
structure and function parameters, such as echocardiographic
examination and extra cardiac function biomarkers, are also
important to confirm the successful establishment of our model
(54), it will be applied in our
future experiments. Second, although mice exhibit a high degree of
homology with humans, the expression profile of its homologous
sequences in humans have yet to be determined. Therefore, whether
these results can be applied to humans requires further
confirmation. In addition, due to the low validation rate of
circRNA RNA-seq data compared with the RT-qPCR experiments, it is
not possible to exclude the possibility of other dysregulated
circRNAs that participate in the development of DCM. Thus,
additional studies on these issues are required. As circRNAs are
stably expressed in the blood, identifying circulating circRNAs in
patients with DCM may provide a promising avenue for research, and
should thus be an integral part of future studies.
In conclusion, the present study provided a
comprehensive analysis of the circRNA expression profile during
early phase DCM, and identified two promising circRNA-miRNA-mRNA
axes. Further functional studies are required to determine the
underlying pathogenesis of early-stage DCM, and the potential role
of these axes in diagnosis and treatment of DCM.
Supplementary Material
Supporting Data
Acknowledgements
Not applicable.
Funding
This work was supported by grants from the Key
Projects of National Natural Science Foundation of China (grant no.
81430047) and National Key Research and Development Program (grant
no. 2018 YFC0807202).
Availability of data and materials
The datasets used and/or analyzed during the present
study are available from the corresponding author on reasonable
request.
Authors' contributions
SD was a major contributor in writing the manuscript
and performing the experiments. JL and YS designed the study and
were in charge of reviewing the manuscript. SD, CT, XY and SC
performed the experiments and analyzed the data. LL, MZ, AX, ZZ and
BC interpreted the data and edited the important intellectual
content of the manuscript. All authors read and approved the final
manuscript.
Ethics approval and consent to
participate
All experiments were approved by the Institutional
Animal Care and Use Committee of Fudan University (approval no.
20190221-124).
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Cho NH, Shaw JE, Karuranga S, Huang Y, da
Rocha Fernandes JD, Ohlrogge AW and Malanda B: IDF diabetes atlas:
Global estimates of diabetes prevalence for 2017 and projections
for 2045. Diabetes Res Clin Pract. 138:271–281. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Marwick TH, Ritchie R, Shaw JE and Kaye D:
Implications of underlying mechanisms for the recognition and
management of diabetic cardiomyopathy. J Am Coll Cardiol.
71:339–351. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Jia G, Hill MA and Sowers JR: Diabetic
cardiomyopathy: An update of mechanisms contributing to this
clinical entity. Circ Res. 122:624–638. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Murtaza G, Virk HUH, Khalid M, Lavie CJ,
Ventura H, Mukherjee D, Ramu V, Bhogal S, Kumar G, Shanmugasundaram
M and Paul TK: Diabetic cardiomyopathy-A comprehensive updated
review. Prog Cardiovasc Dis. 62:315–326. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Lam CS: Diabetic cardiomyopathy: An
expression of stage B heart failure with preserved ejection
fraction. Diab Vasc Dis Res. 12:234–238. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Tadic M, Celic V, Cuspidi C, Ilic S,
Pencic B, Radojkovic J, Ivanovic B, Stanisavljevic D, Kocabay G and
Marjanovic T: Right heart mechanics in untreated normotensive
patients with prediabetes and type 2 diabetes mellitus: A Two- and
three-dimensional echocardiographic study. J Am Soc Echocardiog.
28:317–327. 2015. View Article : Google Scholar
|
|
7
|
Shaver A, Nichols A, Thompson E, Mallick
A, Payne K, Jones C, Manne ND, Sundaram S, Shapiro JI and Sodhi K:
Role of serum biomarkers in early detection of diabetic
cardiomyopathy in the west virginian population. Int J Med Sci.
13:161–168. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Li X, Yang L and Chen LL: The biogenesis,
functions, and challenges of circular RNAs. Mol Cell. 71:428–442.
2018. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Zhang HD, Jiang LH, Sun DW, Hou JC and Ji
ZL: CircRNA: A novel type of biomarker for cancer. Breast Cancer.
25:1–7. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Pamudurti NR, Bartok O, Jens M,
Ashwal-Fluss R, Stottmeister C, Ruhe L, Hanan M, Wyler E,
Perez-Hernandez D, Ramberger E, et al: Translation of CircRNAs. Mol
Cell. 66:9–21.e7. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Meng S, Zhou H, Feng Z, Xu Z, Tang Y, Li P
and Wu M: Functions and properties of a novel potential biomarker
for cancer. Mol Cancer. 16:942017. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Huang C and Shan G: What happens at or
after transcription: Insights into circRNA biogenesis and function.
Transcription. 6:61–64. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Zhao Z, Li X, Jian D, Hao P, Rao L and Li
M: Hsa_circ_0054633 in peripheral blood can be used as a diagnostic
biomarker of pre-diabetes and type 2 diabetes mellitus. Acta
Diabetol. 54:237–245. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Fang Y, Wang X, Li W, Han J, Jin J, Su F,
Zhang J, Huang W, Xiao F, Pan Q and Zou L: Screening of circular
RNAs and validation of circANKRD36 associated with inflammation in
patients with type 2 diabetes mellitus. Int J Mol Med.
42:1865–1874. 2018.PubMed/NCBI
|
|
15
|
Karkalas J: An improved enzymic method for
the determination of native and modified starch. J Sci Food Agr.
36:1019–1027. 1985. View Article : Google Scholar
|
|
16
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(-Delta Delta C(T)) method. Methods. 25:402–408. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Shannon P, Markiel A, Ozier O, Baliga NS,
Wang JT, Ramage D, Amin N, Schwikowski B and Ideker T: Cytoscape: A
software environment for integrated models of biomolecular
interaction networks. Genome Res. 13:2498–2504. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Mi H, Muruganujan A, Ebert D, Huang X and
Thomas PD: PANTHER version 14: More genomes, a new PANTHER GO-slim
and improvements in enrichment analysis tools. Nucleic Acids Res.
47:D419–D426. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
The Gene Ontology Consortium, . The gene
ontology resource: 20 years and still GOing strong. Nucleic Acids
Res. 47:D330–D338. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Ashburner M, Ball CA, Blake JA, Botstein
D, Butler H, Cherry JM, Davis AP, Dolinski K, Dwight SS, Eppig JT,
et al: Gene ontology: Tool for the unification of biology. The Gene
Ontology Consortium. Nat Genet. 25:25–29. 2000. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Kanehisa M, Goto S, Sato Y, Kawashima M,
Furumichi M and Tanabe M: Data, information, knowledge and
principle: Back to metabolism in KEGG. Nucleic Acids Res 42
(Database Issue). D199–D205. 2014. View Article : Google Scholar
|
|
22
|
Jellis CL, Sacre JW, Wright J, Jenkins C,
Haluska B, Jeffriess L, Martin J and Marwick TH: Biomarker and
imaging responses to spironolactone in subclinical diabetic
cardiomyopathy. Eur Heart J Cardiovasc Imaging. 15:776–786. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Nunes S, Soares E, Fernandes J, Viana S,
Carvalho E, Pereira FC and Reis F: Early cardiac changes in a rat
model of prediabetes: Brain natriuretic peptide overexpression
seems to be the best marker. Cardiovasc Diabetol. 12:442013.
View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Cai J, Chen Z, Wang J, Wang J, Chen X,
Liang L, Huang M, Zhang Z and Zuo X: circHECTD1 facilitates
glutaminolysis to promote gastric cancer progression by targeting
miR-1256 and activating β-catenin/c-Myc signaling. Cell Death Dis.
10:5762019. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Kuwabara Y, Horie T, Baba O, Watanabe S,
Nishiga M, Usami S, Izuhara M, Nakao T, Nishino T, Otsu K, et al:
MicroRNA-451 exacerbates lipotoxicity in cardiac myocytes and
high-fat diet-induced cardiac hypertrophy in mice through
suppression of the LKB1/AMPK pathway. Circ Res. 116:279–288. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Li X, Du N, Zhang Q, Li J, Chen X, Liu X,
Hu Y, Qin W, Shen N, Xu C, et al: MicroRNA-30d regulates
cardiomyocyte pyroptosis by directly targeting foxo3a in diabetic
cardiomyopathy. Cell Death Dis. 5:e14792014. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Zhu H, Yang Y, Wang Y, Li J, Schiller PW
and Peng T: MicroRNA-195 promotes palmitate-induced apoptosis in
cardiomyocytes by down-regulating Sirt1. Cardiovasc Res. 92:75–84.
2011. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Wang XH, Qian RZ, Zhang W, Chen SF, Jin HM
and Hu RM: MicroRNA-320 expression in myocardial microvascular
endothelial cells and its relationship with insulin-like growth
factor-1 in type 2 diabetic rats. Clin Exp Pharmacol Physiol.
36:181–188. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Thum T, Gross C, Fiedler J, Fischer T,
Kissler S, Bussen M, Galuppo P, Just S, Rottbauer W, Frantz S, et
al: MicroRNA-21 contributes to myocardial disease by stimulating
MAP kinase signalling in fibroblasts. Nature. 456:980–984. 2008.
View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Liu J, Liu W, Lu Y, Tian H, Duan C, Lu L,
Gao G, Wu X, Wang X and Yang H: Piperlongumine restores the balance
of autophagy and apoptosis by increasing BCL2 phosphorylation in
rotenone-induced Parkinson disease models. Autophagy. 14:845–861.
2018. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Chai C, Song LJ, Han SY, Li XQ and Li M:
MicroRNA-21 promotes glioma cell proliferation and inhibits
senescence and apoptosis by targeting SPRY1 via the PTEN/PI3K/AKT
signaling pathway. Cns Neurosci Ther. 24:369–380. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Liang WC, Wong CW, Liang PP, Shi M, Cao Y,
Rao ST, Tsui SK, Waye MM, Zhang Q, Fu WM and Zhang JF: Translation
of the circular RNA circβ-catenin promotes liver cancer cell growth
through activation of the Wnt pathway. Genome Biol. 20:842019.
View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Yang Y, Gao X, Zhang M, Yan S, Sun C, Xiao
F, Huang N, Yang X, Zhao K, Zhou H, et al: Novel role of FBXW7
circular RNA in repressing glioma tumorigenesis. J Natl Cancer
Inst. 110:304–315. 2018. View Article : Google Scholar
|
|
34
|
Li LJ, Leng RX, Fan YG, Pan HF and Ye DQ:
Translation of noncoding RNAs: Focus on lncRNAs, pri-miRNAs, and
circRNAs. Exp Cell Res. 361:1–8. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Liu H, Liu Y, Bian Z, Zhang J, Zhang R,
Chen X, Huang Y, Wang Y and Zhu J: Circular RNA YAP1 inhibits the
proliferation and invasion of gastric cancer cells by regulating
the miR-367-5p/p27Kip1 axis. Mol Cancer. 17:1512019. View Article : Google Scholar
|
|
36
|
Altesha MA, Ni T, Khan A, Liu K and Zheng
X: Circular RNA in cardiovascular disease. J Cell Physiol.
234:5588–5600. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Lin F, Zhao G, Chen Z, Wang X, Lv F, Zhang
Y, Yang X, Liang W, Cai R, Li J, et al: circRNA-miRNA association
for coronary heart disease. Mol Med Rep. 19:2527–2536.
2019.PubMed/NCBI
|
|
38
|
Patop IL and Kadener S: circRNAs in
cancer. Curr Opin Genet Dev. 48:121–127. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Fischer JW and Leung AK: CircRNAs: A
regulator of cellular stress. Crit Rev Biochem Mol. 52:220–233.
2017. View Article : Google Scholar
|
|
40
|
Yang F, Li A, Qin Y, Che H, Wang Y, Lv J,
Li Y, Li H, Yue E, Ding X, et al: A novel circular RNA mediates
pyroptosis of diabetic cardiomyopathy by functioning as a competing
endogenous RNA. Mol Ther Nucleic Acids. 17:636–643. 2019.
View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Liu C, Ge HM, Liu BH, Dong R, Shan K, Chen
X, Yao MD, Li XM, Yao J, Zhou RM, et al: Targeting
Pericyte-endothelial cell crosstalk by circular RNA-cPWWP2A
inhibition aggravates diabetes-induced microvascular dysfunction.
Proc Natl Acad Sci USA. 116:7455–7464. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Wu H, Wu S, Zhu Y, Ye M, Shen J, Liu Y,
Zhang Y and Bu S: Hsa_circRNA_0054633 is highly expressed in
gestational diabetes mellitus and closely related to glycosylation
index. Clin Epigenetics. 11:222019. View Article : Google Scholar : PubMed/NCBI
|
|
43
|
Fuentes-Antrás J, Picatoste B, Ramírez E,
Egido J, Tuñón J and Lorenzo Ó: Targeting metabolic disturbance in
the diabetic heart. Cardiovasc Diabetol. 14:172015. View Article : Google Scholar : PubMed/NCBI
|
|
44
|
Duncan JG: Mitochondrial dysfunction in
diabetic cardiomyopathy. Biochim Biophys Acta. 1813:1351–1359.
2011. View Article : Google Scholar : PubMed/NCBI
|
|
45
|
Huang P, Qi B, Yao H, Zhang L, Li Y and Li
Q: Circular RNA cSMARCA5 regulates the progression of cervical
cancer by acting as a microRNA-432 sponge. Mol Med Rep.
21:1217–1223. 2020.PubMed/NCBI
|
|
46
|
Salzman J: Circular RNA expression: Its
potential regulation and function. Trends Genet. 32:309–316. 2016.
View Article : Google Scholar : PubMed/NCBI
|
|
47
|
Han D, Li J, Wang H, Su X, Hou J, Gu Y,
Qian C, Lin Y, Liu X, Huang M, et al: Circular RNA circMTO1 acts as
the sponge of microRNA-9 to suppress hepatocellular carcinoma
progression. Hepatology. 66:1151–1164. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
48
|
Zheng D, Ma J, Yu Y, Li M, Ni R, Wang G,
Chen R, Li J, Fan GC, Lacefield JC and Peng T: Silencing of miR-195
reduces diabetic cardiomyopathy in C57BL/6 mice. Diabetologia.
58:1949–1958. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
49
|
Gu J, Wang S, Guo H, Tan Y, Liang Y, Feng
A, Liu Q, Damodaran C, Zhang Z, Keller BB, et al: Inhibition of p53
prevents diabetic cardiomyopathy by preventing early-stage
apoptosis and cell senescence, reduced glycolysis, and impaired
angiogenesis. Cell Death Dis. 9:822018. View Article : Google Scholar : PubMed/NCBI
|
|
50
|
Tuncay E and Turan B: Intracellular
Zn2+ increase in cardiomyocytes induces both electrical
and mechanical dysfunction in heart via endogenous generation of
reactive nitrogen species. Biol Trace Elem Res. 169:294–302. 2016.
View Article : Google Scholar : PubMed/NCBI
|
|
51
|
Huang L, Yan M and Kirschke CP:
Over-expression of ZnT7 increases insulin synthesis and secretion
in pancreatic beta-cells by promoting insulin gene transcription.
Exp Cell Res. 316:2630–2643. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
52
|
Tuncay E, Bitirim CV, Olgar Y, Durak A,
Rutter GA and Turan B: Zn2+-transporters ZIP7 and ZnT7 play
important role in progression of cardiac dysfunction via affecting
Sarco(endo)plasmic reticulum-mitochondria coupling in hyperglycemic
cardiomyocytes. Mitochondrion. 44:41–52. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
53
|
Jia G, Whaley-Connell A and Sowers JR:
Diabetic cardiomyopathy: A hyperglycaemia- and
insulin-resistance-induced heart disease. Diabetologia. 61:21–28.
2018. View Article : Google Scholar : PubMed/NCBI
|
|
54
|
Lorenzo-Almoros A, Tunon J, Orejas M,
Cortes M, Egido J and Lorenzo O: Diagnostic approaches for diabetic
cardiomyopathy. Cardiovasc Diabetol. 16:282017. View Article : Google Scholar : PubMed/NCBI
|