Introduction
Cervical cancer is a common malignancy of the female
reproductive system, demonstrating the second highest morbidity
rate among female malignancies worldwide (1,2).
Chemotherapy, radiotherapy and surgical treatment are the standard
methods for treating cervical cancer; however, its 5-year survival
rate remains low, with lymph node and distant metastasis being the
contributing factors leading to treatment failure (3,4).
Therefore, to provide potential targets for subsequent
individualized treatment, it is important to study the molecular
mechanisms of the occurrence and development of cervical
cancer.
MicroRNAs (miRNA/miR) are a class of non-coding RNAs
that exist in numerous biological genomes, which have the function
of sequence-specific regulation over gene expression (5,6). The
regulatory role of miRNAs mainly occurs at the post-transcriptional
level, as miRNAs inhibit protein translation and/or promote the
degradation of mRNA (7,8). miRNAs serve important roles not only
in embryonic formation and the growth and development of organisms,
but also in cell proliferation, differentiation, apoptosis and the
occurrence of several human diseases (9,10).
Previous studies have demonstrated that abnormal miRNA expression
critically affected tumor growth, invasion and apoptosis (11,12).
In addition, miR-802 was also revealed to be involved in
tumorigenesis and development, whereby it served distinct roles in
different types of tumor (13,14).
However, studies into the role of miR-802 in cervical cancer are
limited. In one study, Zhang et al (15) found that miR-802 inhibited cell
proliferation and induced apoptosis in human cervical cancer by
targeting serine/arginine-rich splicing factor 9. Thus, the present
study further investigated the effect of miR-802 on the biological
characteristics of cervical cancer cells.
Studies have reported that one miRNA can
simultaneously regulate hundreds of target genes, including
transcription factors, cytokines and receptors (16–18).
The regulatory networks between miRNAs and their target genes have
been revealed to be involved in the occurrence and development of
tumors (19); miRNAs affect cell
proliferation, apoptosis, invasion, angiogenesis, metastasis and
other biological behaviors by regulating the mRNA translational
levels of target genes, which subsequently leads to the occurrence
and development of tumors (20).
In the current study, TargetScan 7.2 software was used to predict
the target genes of miR-802; the results revealed that miR-802 has
a complementary binding site in the 3′-untranslated region (UTR) of
the basic transcription factor 3 (BTF3) gene. Therefore, the
relationship between miR-802 and BTF3 was further investigated to
determine the role of miR-802 in the occurrence and development of
cervical cancer.
Materials and methods
Patient studies
Cervical cancer tissue and adjacent tissue samples
were obtained from 40 female patients (age, 43.57±2.79) with
cervical cancer who attended Jingmen First People's Hospital
between January 2018 and May 2019. The tissue samples were
maintained in liquid nitrogen and stored at −80°C. Exclusion
criteria: i) other gynaecological diseases; ii) severe internal and
external diseases; iii) history of chemoradiotherapy; and iv)
history of pelvic organ surgery. All research subjects provided
written informed consent and the study was approved by the Ethics
Committees of Jingmen No. 1 People's Hospital.
Cell culture
The human endometrial epithelial cell (Ect1/E6E7)
line were purchased from American Type Culture Collection. The
cervical cancer cell lines (HeLa, C-33 A, SiHa and ME-180) were
purchased from the American Type Culture Collection. All the cells
were cultured in DMEM (cat. no. D0819; Sigma-Aldrich; Merck KGaA),
supplemented with 10% FBS (Gibco; Thermo Fisher Scientific, Inc.)
at 37°C in a 5% CO2 atmosphere.
Cell transfection
Among the cervical cancer cell lines, as miR-802 was
expressed at the lowest levels in SiHa cells, SiHa cells were used
in the following experiments. For transfection, 2 ml cell solution,
containing 1×106 cells/well and culture medium, was
plated into 6-well plates and incubated at 37°C in a 5%
CO2 atmosphere until the cell confluence reached 40–60%.
To prepare A transdye, 20 pmol miR-802 mimic (forward,
5′-UCAGUAACAAAGAUUCAUCCUUGU-3′ and reverse,
5′-ACAAGGAUGAAUCUUUGUUACUGA-3′; Shanghai GenePharma Co., Ltd.), the
negative control for the mimic (NC-mimic; a non-targeting sequence;
forward, 5′-UUUUACUACACAAAAGUACUG-3′ and reverse,
5′-CAGUACUUUUGUGUAGUACAAA-3′; Shanghai GenePharma Co., Ltd.), BTF3
overexpression plasmid (pWZL-BTF3; Shanghai GenePharma Co., Ltd.)
or the empty plasmid (pWZL-Blast; pWZL-NC; Shanghai GenePharma Co.,
Ltd.) were dissolved in 50 µl DMEM (Hyclone; GE Healthcare Life
Sciences) and fully mixed. To prepare B transdye, 1 µl
Lipofectamine® 2000 reagent (Invitrogen; Thermo Fisher
Scientific, Inc.) was dissolved in 50 µl DMEM, incubated for 15 min
at room temperature and then mixed with A transdye. This solution
(50 µl) was subsequently added into each well of the plate and
incubated at 37°C with 5% CO2. Following 24 h of
transfection, the medium was changed and the cells were collected
after culture for 72 h.
Experimental grouping
To investigate the effects of miR-802 on the
viability, migration and invasion of cervical cancer cells, the
cells were divided into three groups: Control (untreated cells),
NC-mimic (cells transfected with the NC-mimic) and miR-802 mimic
(cells transfected with the miR-802 mimic) group. To determine the
effects of BTF3 on the viability, migration and invasion of
cervical cancer cells, the cells were divided into five groups:
Control (untreated cells), pWZL-NC + NC-mimic (cells co-transfected
with empty plasmid and NC-mimic), pWZL-BTF3 + NC-mimic (cells
co-transfected with BTF3 overexpression plasmid and NC-mimic),
pWZL-BTF3 + miR-802 mimic (cells co-transfected with BTF3
overexpression plasmid and miR-802 mimic) and miR-802 mimic (cells
transfected with miR-802 mimic) group.
Dual-luciferase activity assay
The target-binding region of miR-802 and BTF3 was
predicted using TargetScan (version 7.2; www.targetscan.org/vert_72/). For the dual-luciferase
reporter assays, the 3′-UTR of BTF3 containing miR-802 binding
sites was inserted into a pmirGLO dual-luciferase vector (Promega
Corporation) to generate wild-type (WT) BTF3-3′-UTR. The mutant
(mut) 3′ UTR of BTF3 was synthesized using a Site-Directed
Mutagenesis kit (Thermo Fisher Scientific, Inc.) and inserted into
the pmirGLO dual-luciferase vector to generate BTF3-3′-UTR-mut. The
pmirGLO vector containing WT or mut BTF3 3′-UTR was co-transfected
with the miR-802 mimic or NC-mimic into SiHa cells using
Lipofectamine® 2000 reagent (Invitrogen; Thermo Fisher
Scientific, Inc.). Following incubation for 48 h, the relative
luciferase activity in the cells was measured using a
Dual-Luciferase Reporter Assay kit (Promega Corporation) according
to the manufacturer's protocol. Firefly luciferase activities were
normalized to Renilla luciferase activities.
Cell migration assay
Following 24 h of transfection, SiHa cells were
resuspended in DMEM without FBS until the cell density reached
1×106 cells/ml. Then, 100 µl cell suspension was plated
into the upper chambers of the Transwell plate (Corning, Inc.) and
cultured at 37°C for 6 h. DMEM with 10% FBS was plated into the
lower chamber. Following the incubation, the non-migratory cells
were removed with a cotton swab and the migratory cells were washed
three times with PBS, and fixed with 4% formaldehyde (cat. no.
P0099; Beyotime Institute of Biotechnology) for 20 min at room
temperature and with 1% Triton X-100 (cat. no. P0096; Beyotime
Institute of Biotechnology) for 5 min at room temperature. After
washing the cells three times with PBS, the cells were stained with
hematoxylin (cat. no. C0107; Beyotime Institute of Biotechnology)
for 20 min at room temperature and then washed under running water.
Stained cells were counted in five randomly selected fields using a
light microscope (magnification, ×200; Olympus Corporation); the
mean number of cells per field of view was calculated.
Cell invasion assay
Following 24 h of transfection, SiHa cells were
resuspended in DMEM without FBS until the cell density reached
1×106 cells/ml. The upper chambers of the Transwell
plates (pore size, 8.0 µm; Corning, Inc.) were precoated with 50 µl
Matrigel (BD Biosciences) for 2 h at 4°C. Then, the cell suspension
was subsequently plated into the upper chamber of the Transwell
plate. DMEM, supplemented with 10% FBS was plated in the lower
chambers. Following incubation for 24 h at 37°C, the invasive cells
in the lower chamber were fixed in 5% gluteraldehyde for 30 min at
room temperature and stained with 0.5% crystal violet for 20 min at
room temperature, while the non-invasive cells remaining in the
upper chamber were removed using a cotton swab. Stained cells were
observed in three randomly selected fields of view using a light
microscope (magnification, ×200).
Cell viability
SiHa cells (2×104 cells/well) were plated
into 96-well plates in DMEM containing 10% FBS and cultured for 24
h at room temperature. Subsequently, 10 µl Cell Counting Kit-8
(cat. no. 96992; Sigma-Aldrich; Merck KGaA) solution was added into
each well, according to the manufacturer's protocol, and incubated
for 4 h at 37°C. The absorbance at 450 nm was used a Multiskan GO
microplate reader (Shanghai Bajiu Industrial Co., Ltd.). The
experiment was conducted in triplicate and the average absorbance
value was calculated.
Western blot analysis
SiHa cells were washed twice with cold PBS and total
protein was extracted from the cells using RIPA lysis buffer (Cell
Signaling Technology, Inc.). Total protein was quantified using the
bicinchoninic acid protein assay kit (Pierce; Thermo Fisher
Scientific, Inc.). The extracted protein was denatured for 5 min at
100°C, and 50 µg/µl protein/lane was separated via 15% SDS-PAGE.
The separated proteins were subsequently transferred onto PVDF
membranes and blocked with 5% milk at room temperature for 1 h. The
membranes were incubated with the following primary antibodies at
4°C overnight: Anti-N-cadherin (1:1,000; cat. no. ab18203; Abcam),
anti-BTF3 (1:1,000; cat. no. ab203517; Abcam), anti-matrix
metallopeptidase (MMP)9 (1:1,000; cat. no. ab73734; Abcam),
anti-MMP2 (1:1,000; cat. no. ab37150; Abcam), anti-E-cadherin
(1:1,000; cat. no. ab40772; Abcam) and anti-GAPDH (1:2,000; cat.
no. ab8245; Abcam). Following the primary antibody incubation, the
membranes were incubated with goat anti-mouse or goat anti-rabbit
IgG (H+L) horseradish peroxidase-conjugated secondary antibodies
(cat. nos. SA00001-1 and SA00001-2; 1:2000; ProteinTech Group,
Inc.) for 2 h at room temperature and then washed with PBS three
times. Protein bands were visualized using an ECL western blotting
kit (cat. no. 93-K820-500; Hangzhou Multi Sciences (Lianke) Biotech
Co., Ltd.) and the expression levels were semi-quantified using
ImageJ v4.7 software (National Institutes of Health).
Reverse transcription-quantitative PCR
(RT-qPCR)
Total RNA was extracted from cells using
TRIzol® reagent (Invitrogen; Thermo Fisher Scientific,
Inc.) and the concentration of RNA was measured using a NanoDrop™
spectrophotometer (NanoDrop Technologies; Thermo Fisher Scientific,
Inc.), which was then diluted to 500 ng/µl. Total RNA was reverse
transcribed into cDNA using a SuperScript™ II First-Strand cDNA
synthesis kit (Invitrogen; Thermo Fisher Scientific, Inc.)
according to the manufacturer's protocol. qPCR was subsequently
performed using a QuantiFast SYBR Green PCR kit (cat. no. 204057;
Qiagen, Inc.) according to the manufacturer's protocol. The
following thermocycling conditions were used for the qPCR: Initial
denaturation at 94°C for 2 min; 35 cycles of 94°C for 30 sec, 63°C
for 30 sec, 72°C for 1 min; and a final extension at 72°C for 7
min, prior to being maintained at 4°C. The following primer
sequences used are listed in Table
I. Expression levels were quantified using the
2−ΔΔCq method (21) and
normalized to the loading controls, GAPDH or U6, for mRNA or
miR-802 expression, respectively.
 | Table I.Primers used for the reverse
transcription-quantitative PCR. |
Table I.
Primers used for the reverse
transcription-quantitative PCR.
| Gene | Primer sequence
(5′→3′) | Species |
|---|
| N-cadherin | F:
ATGAAGAAGGTGGAGGAGA | Human |
|
| R:
AGATCGGACCGGATACT |
|
| E-cadherin | F:
TGGAGGAATTCTTGCTTTGC | Human |
|
| R:
CGTACATGTCAGCCAGCTTC |
|
| Basic transcription
factor 3 | F:
AGCTTGGTGCGGATAGTCTGA | Human |
|
| R:
GTGCTTTTCCATCCACAGATTG |
|
| MMP2 | F:
TCTCCTGACATTGACCTTGGC | Human |
|
| R:
CAAGGTGCTGGCTGAGTAGATC |
|
| MMP9 | F:
TTGACAGCGACAAGAAGTGG | Human |
|
| R:
GCCATTCACGTCGTCCTTAT |
|
| GAPDH | F:
GCTGGCGCTGAGTACGTCGTGGAGT | Human |
|
| R:
CACAGTCTTCTGGGTGGCAGTGATGG |
|
| microRNA-802 | F:
CGTTGTGTAGCTTATCAGACTG | Human |
|
| R:
AATGGTTGTTCTCCACACTCTC |
|
| U6 | F:
TGACTTCCAAGTACCATCGCCA | Human |
|
| R:
TTGTAGAGGTAGGTGTGCAGCAT |
|
Statistical analysis
Statistical analysis was performed using GraphPad
Prism v6.01 software (GraphPad Software, Inc.) and data are
presented as the mean ± SD of ≥3 independent experiments.
Statistical differences between two groups were determined using a
paired Student's t-test, whereas statistical differences among
multiple groups were analyzed using a one-way ANOVA, followed by a
Bonferroni's correction post hoc test. P<0.05 was considered to
indicate a statistically significant difference.
Results
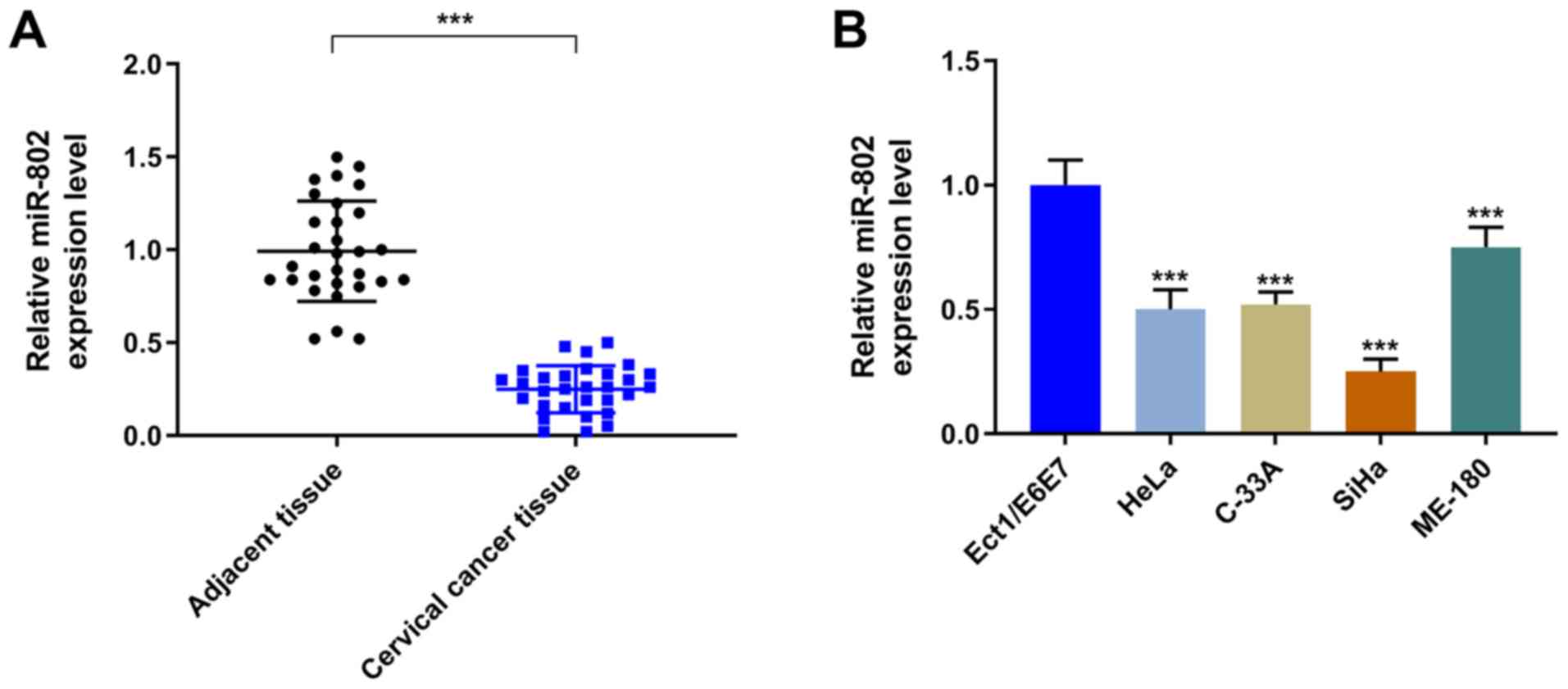
miR-802 expression levels in cervical
cancer tissues and cell lines
The expression levels of miR-802 were significantly
decreased in the cervical cancer tissues compared with the adjacent
tissue (P<0.001; Fig. 1A).
Similarly, the expression levels of miR-802 were observed to be
significantly decreased in the cervical cancer cell lines (HeLa,
C-33 A, SiHa and ME-180) compared with the EEC cell line
(P<0.001; Fig. 1B).
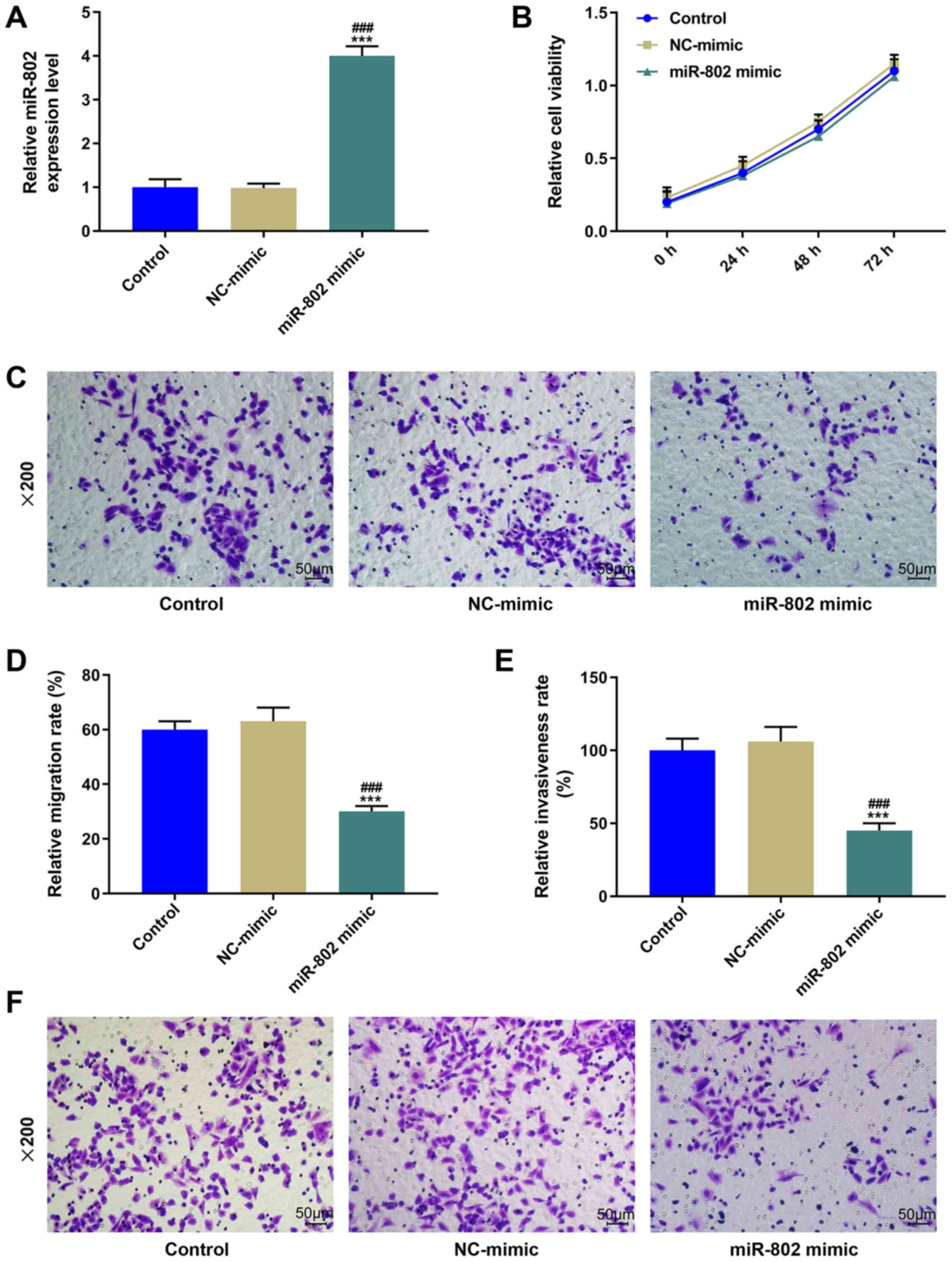
Effect of the overexpression of
miR-802 on cervical cancer cells
The expression levels of miR-802 were significantly
increased following the transfection of the miR-802 mimic into SiHa
cells compared with the control and NC-mimic groups (P<0.001;
Fig. 2A). However, following the
overexpression of miR-802 in the cells, no significant difference
was observed in the cell viability compared with the control group
(P>0.05; Fig. 2B). Moreover, a
Transwell assay was performed to determine the migratory ability of
SiHa cells; the results revealed that the overexpression of miR-802
significantly decreased the migratory rate compared with the
control and NC-mimic groups (P<0.001; Fig. 2C and D). Similarly, the cell
invasive ability was significantly inhibited in the miR-802
mimic-transfected cells compared with the control and NC-mimic
groups (P<0.001; Fig. 2E and
F).
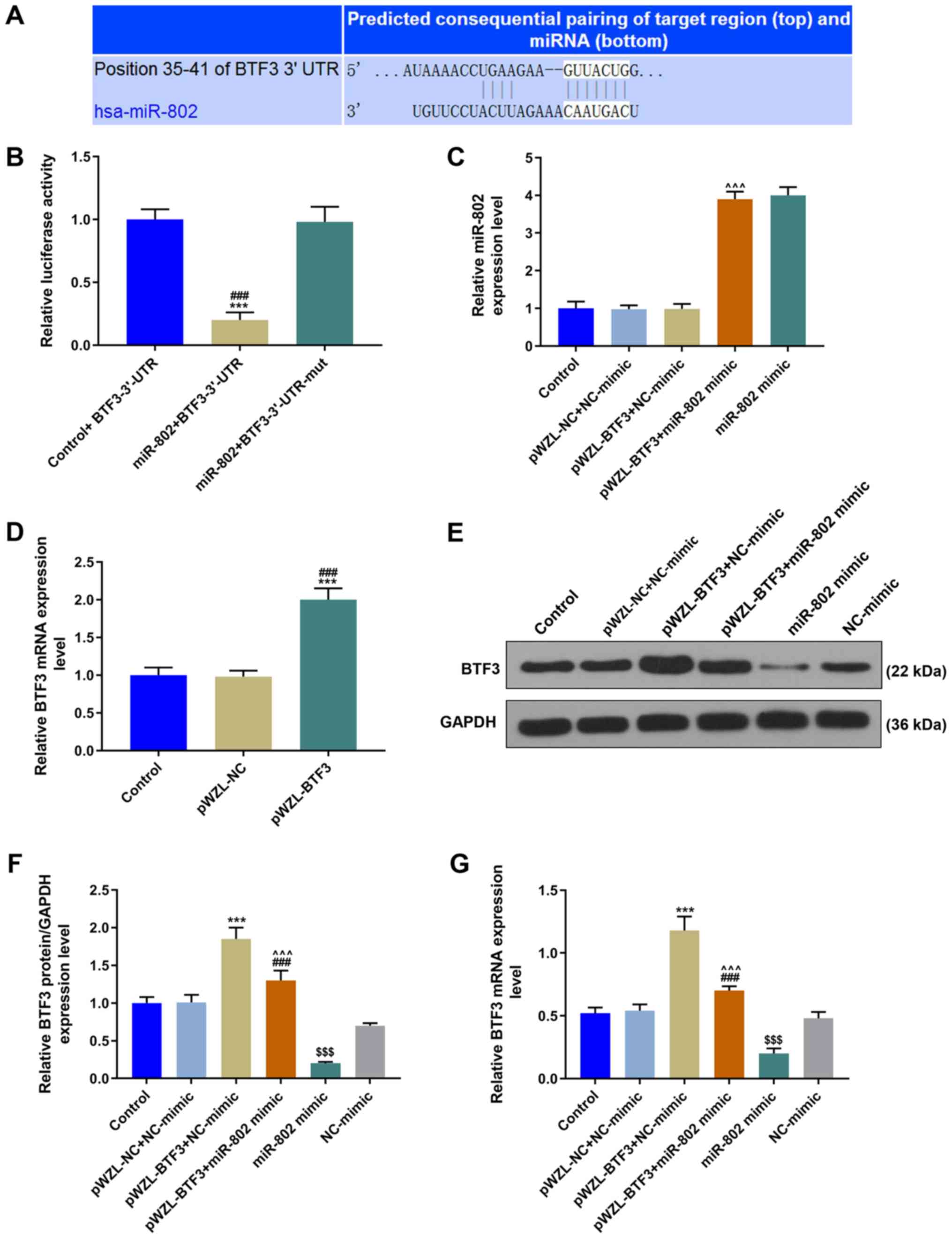
BTF3 is a direct target gene of
miR-802
The possible target genes of miR-802 were predicted
using TargetScan software and it was discovered that miR-802 bound
to the 3′-UTR of BTF3 (Fig. 3A).
Subsequently, two pmirGLO dual-luciferase reporter vectors, namely,
BTF3-3′-UTR and BTF3-3′-UTR-mut, were constructed. BTF3-3′-UTR or
BTF3-3′-UTR-mut were co-transfected with the miR-802 mimic into the
cells. The miR-802 + BTF-3′-UTR group displayed significantly
decreased relative luciferase activity compared with the other two
groups: therefore, the results indicated that BTF3 was a direct
target gene of miR-802 (P<0.001; Fig. 3B). The expression levels of BTF3 in
the pWZL-BTF3 group were significantly increased compared with the
pWZL-NC and control groups, suggesting that the BTF3 overexpression
transfections were successful (P<0.001; Fig. 3D). Furthermore, the expression
levels of miR-802 were significantly increased in the pWZL-BTF3 +
miR-802 mimic group compared with the pWZL-BTF3 + NC-mimic group
(P<0.001; Fig. 3C). The highest
expression levels of BTF3 were observed in the pWZL-BTF3 + NC-mimic
group, whilst the expression levels of BTF3 in the pWZL-BTF3 +
miR-802 mimic group were significantly decreased compared with the
pWZL-BTF3 + NC-mimic group, but significantly increased compared
with the miR-802 mimic group (P<0.001; Fig. 3E-G).
miR-802 suppresses cell migration,
invasion and epithelial-mesenchymal transition (EMT) in cervical
cancer cells through regulating BTF3 expression
The pWZL-BTF3 + NC-mimic group significantly
increased cell viability compared with the pWZL-NC + NC-mimic
group; however, co-transfection with the miR-802 mimic (pWZL-BTF3 +
miR-802 mimic group) significantly reversed the effect of BTF3 on
cell viability (P<0.001; Fig.
4A). Moreover, the overexpression of BTF3 significantly
increased the cell migratory rate compared with the pWZL-NC +
NC-mimic group, whereas this effect was significantly inhibited
following co-transfection with the miR-802 mimic (P<0.001;
Fig. 4B and C). Similarly,
co-transfection with the miR-802 mimic significantly suppressed the
function of BTF3 alone in promoting the cell invasive ability
(P<0.001; Fig. 4D and E). In
addition, the regulatory effect of miR-802 on the EMT process of
cervical cancer cells was investigated. The results demonstrated
that the pWZL-BTF3 + NC-mimic group displayed significantly
increased mRNA and protein expression levels of MMP2, MMP9 and
N-cadherin compared with the pWZL-NC + NC-mimic group, whist
inhibiting the expression levels of E-cadherin expression; however,
the co-transfection of cells with pWZL-BTF and miR-802 mimic
significantly reversed the effects of BTF3 on these genes
(P<0.001; Fig. 4F-H).
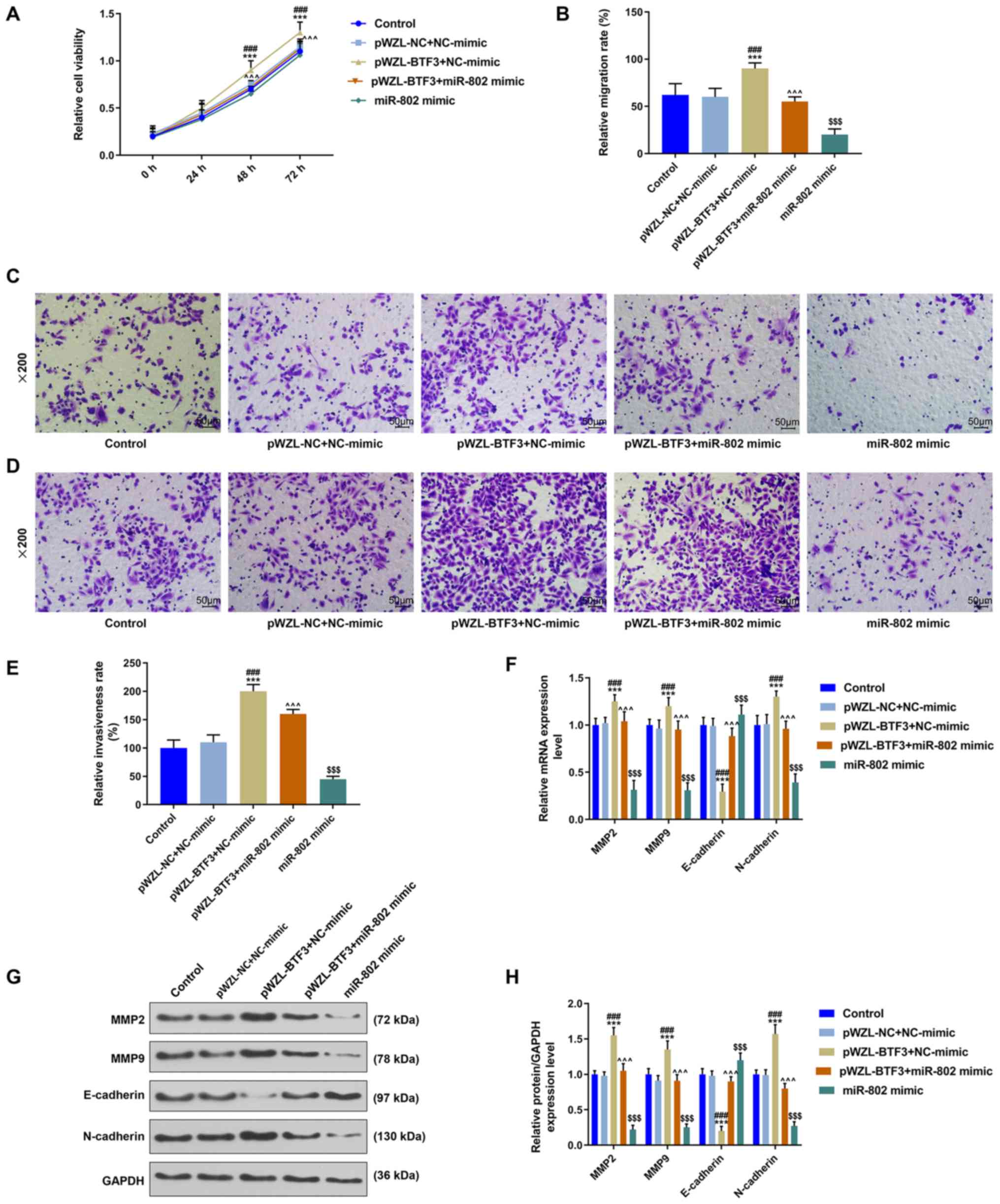 | Figure 4.miR-802 suppresses cell migration,
invasion and epithelial-mesenchymal transition in cervical cancer
cells through regulating BTF3 expression. (A) Cell Counting Kit-8
assay was performed to detect the viability of SiHa cells. n=3;
***P<0.001 vs. control; ###P<0.001 vs. pWZL-NC +
NC-mimic; ^^^P<0.001 vs. pWZL-BTF3 + NC-mimic. (B and
C) Transwell assay was performed to determine the migratory ability
of SiHa cells. Scale bar, 50 µm; magnification, ×200; n=3;
***P<0.001 vs. control; ###P<0.001 vs. pWZL-NC +
NC-mimic; ^^^P<0.001 vs. pWZL-BTF3 + NC-mimic;
$$$P<0.001 vs. pWZL-BTF3 + miR-802 mimic. (D and E)
Invasive ability of SiHa cells was analyzed using a Transwell
assay. Scale bar, 50 µm; magnification, ×200; n=3; ***P<0.001
vs. control; ###P<0.001 vs. pWZL-NC + NC-mimic;
^^^P<0.001 vs. pWZL-BTF3 + NC-mimic;
$$$P<0.001 vs. pWZL-BTF3 + miR-802 mimic. (F)
Expression levels of MMP9, MMP2, E-cadherin and N-cadherin were
detected using reverse transcription-quantitative PCR. N=3;
***P<0.001 vs. control; ###P<0.001 vs. pWZL-NC +
NC-mimic; ^^^P<0.001, vs. pWZL-BTF3 + NC-mimic;
$$$P<0.001 vs. pWZL-BTF3 + miR-802 mimic. (G and H)
Western blotting was used to analyze the expression levels of MMP9,
MMP2, E-cadherin and N-cadherin. N=3; ***P<0.001 vs. control;
###P<0.001 vs. pWZL-NC + NC-mimic;
^^^P<0.001 vs. pWZL-BTF3 + NC-mimic;
$$$P<0.001 vs. pWZL-BTF3 + miR-802 mimic. GAPDH
served as the internal reference control. miR, microRNA; NC,
negative control; BTF3, basic transcription factor 3; MMP, matrix
metallopeptidase. |
Discussion
Due to cervical cytology screening being widely
implemented worldwide, the incidence and mortality rates of
cervical cancer are decreasing (22). In addition, the development and
clinical application of prophylactic tumor vaccines against
cervical cancer are hypothesized to be able to effectively control
cervical cancer in the next 20 to 30 years (23). However, at present, the therapeutic
strategies used to treat patients with cervical cancer are
ineffective, especially for patients presenting with advanced
metastasis (24). Therefore, it is
urgent and of great significance to determine the molecular
mechanisms behind the occurrence, development and metastasis of
cervical cancer.
miR-802, which is a newly discovered endogenous
single-stranded non-coding small RNA molecule, has been found to be
able to regulate the occurrence and development of gastric cancer,
breast cancer and tongue squamous cell carcinoma (14,25,26).
In fact, a previous study observed that miR-802 expression was
increased in osteosarcoma cells compared with the adjacent normal
tissues (27). Currently, the
effects of miR-802 and cervical cancer have not been reported in
detail; thus, the current study aimed to determine the mechanisms
of action of miR-802 in cervical cancer tumors. Consistent with the
expression levels in gastric cancer, breast cancer and tongue
squamous cell carcinoma, it was revealed that miR-802 expression
levels were decreased in cervical cancer tissues and cell lines
compared with the control group.
Moreover, the effects of miR-802 on the biological
activity of cervical cancer cells were investigated; it was
discovered that the overexpression of miR-802 had no significant
effect on the cell viability of cervical cancer cells, but it
inhibited their invasive and migratory abilities, indicating that
miR-802 may exert certain inhibitory effects over the regulation of
cervical cancer cells. Furthermore, miR-802 was reported to promote
osteosarcoma cell proliferation by targeting p27 (27). A previous study also found that
miR-802 promoted the proliferation of lung cancer cells through
negatively regulating the tumor suppressor menin (28), and inhibited the invasion and
migration of gastric cancer cells, whilst promoting cell apoptosis
by targeting RAB23 (14).
TargetScan 7.2 software was used to predict that the
signal transductor and transcriptional activator BTF3 was a
potential target gene of miR-802; this was further validated in the
present study using a dual-luciferase reporter assay, whereby
miR-802 inhibited the activity of the BTF3 3′-UTR reporter gene.
Several studies have previously reported that BTF3 regulated tumor
cell migration and invasion (29–31);
for example, Zhang et al (32) reported that the genetic knockdown
of BTF3 impaired the regulation of proliferation, the cell cycle
and apoptosis in hypopharyngeal squamous cell carcinoma via the
serine-protein kinase ATM signaling pathway (32). BTF3 also promoted prostate cancer
progression through modeling stem-like traits (33). These studies indicated that BTF3
may have a role in promoting cancer. Thus, the roles of miR-802 and
BTF3 in cervical cancer cells was further investigated and it was
revealed that overexpressing miR-802 significantly reversed the
function of BTF3 in promoting the cell viability, migration and
invasion.
In addition, EMT refers to the process via which
epithelial cells acquire invasive mesenchymal phenotypes and it is
considered as an important mechanism in the initial stages of tumor
cell metastasis (34). EMT is
closely related to the occurrence and development of cervical
cancer tumors; therefore, it may be an important target for the
treatment of cervical cancer (35). Studies have increasingly
demonstrated that miRNAs are important regulatory factors related
to EMT; for instance, in cervical cancer, miR-145 and miR-1
inhibited the invasion and migration of cancer cells through
regulating EMT (36,37). Zhang et al (38), also reported that knocking down
BTF3 expression increased E-cadherin expression, but decreased
N-cadherin and ZEB2 expression, which had an overall effect of
downregulating EMT processes in gastric cancer (38). In the current study, the
overexpression of BTF3 increased the expression levels of
N-cadherin and inhibited those of E-cadherin. Notably, the protein
expression levels of E-cadherin were increased in the cells which
overexpressed miR-802, suggesting that miR-802 may suppress EMT in
cervical cancer through targeting BTF3. In addition, MMP2 and MMP9
are considered as proteins related to tumor metastasis, and it was
previously reported that decreasing their expression levels
inhibited the migration and progression of cancer cells (39). In the present study, the
overexpression of miR-802 markedly suppressed the expression levels
of MMP2 and MMP9 in cells by regulating BTF3 expression.
However, this study also has several limitations;
for example, the results obtained would be more convincing if
multiple cervical cancer cell lines were used in the study to
investigate the role of miR-802 in cervical cancer. In addition,
the present study did not include in vivo experiments, which
could verify miR-802 targeting of BTF3.
In conclusion, the findings of the current study
suggested that the overexpression of miR-802 may suppress cervical
cancer progression by decreasing BTF3 expression levels, indicating
that miR-802 may be a potential therapeutic target for the
treatment and prognosis of patients with cervical cancer.
Acknowledgements
Not applicable.
Funding
The present study was supported by the Jingmen
General Science and Technology Project of Hubei Province (grant no.
2019YFYB022).
Availability of data and materials
The datasets used/or analyzed during the study are
available from the corresponding author on reasonable request.
Authors' contributions
XW and LL conceived and designed the study, drafted
the manuscript and critically revised it for important intellectual
content. HZ acquired the data, performed the data analysis and
interpreted the results. All authors read and approved the final
manuscript and agreed to be accountable for all aspects of the work
in ensuring that questions related to the accuracy or integrity of
the work are appropriately investigated and resolved.
Ethics approval and consent to
participate
All research subjects provided written informed
consent and the study was approved by the Ethics Committees of
Jingmen First People's Hospital. All procedures performed in the
studies involving human participants were in accordance with the
ethical standards of the institutional and/or national research
committee, and with the 1964 Declaration of Helsinki and its later
amendments or comparable ethical standards.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Fang J, Zhang H and Jin S: Epigenetics and
cervical cancer: From pathogenesis to therapy. Tumour Biol.
35:5083–5093. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Dehn D, Torkko KC and Shroyer KR: Human
papillomavirus testing and molecular markers of cervical dysplasia
and carcinoma. Cancer. 111:1–14. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Sun XL, Wang HB, Wang ZQ, Cao TT, Yang X,
Han JS, Wu YF, Reilly KH and Wang JL: Effect of transcutaneous
electrical stimulation treatment on lower urinary tract symptoms
after class III radical hysterectomy in cervical cancer patients:
Study protocol for a multicentre, randomized controlled trial. BMC
Cancer. 17:4162017. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Kazumoto T, Kato S, Yokota H, Hasumi Y,
Kino N, Horie K, Yoshida D, Mizukami T and Saito Y: Is a low dose
of concomitant chemotherapy with extended-field radiotherapy
acceptable as an efficient treatment for cervical cancer patients
with metastases to the para-aortic lymph nodes? Int J Gynecol
Cancer. 21:1465–1471. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Xue M, Zhuo Y and Shan B: MicroRNAs, long
noncoding RNAs, and their functions in human disease. Methods Mol
Biol. 1617:1–25. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Mannucci C, Casciaro M, Minciullo PL,
Calapai G, Navarra M and Gangemi S: Involvement of microRNAs in
skin disorders: A literature review. Allergy Asthma Proc. 38:9–15.
2017. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Song Z and Li G: Role of specific
microRNAs in regulation of vascular smooth muscle cell
differentiation and the response to injury. J Cardiovasc Transl
Res. 3:246–250. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Singh RP, Massachi I, Manickavel S, Singh
S, Rao NP, Hasan S, Mc Curdy DK, Sharma S, Wong D, Hahn BH, et al:
The role of miRNA in inflammation and autoimmunity. Autoimmun Rev.
12:1160–1165. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Xu YF, Mao YP, Li YQ, Ren XY, He QM, Tang
XR, Sun Y, Liu N and Ma J: MicroRNA-93 promotes cell growth and
invasion in nasopharyngeal carcinoma by targeting disabled
homolog-2. Cancer Lett. 363:146–155. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Paul P, Chakraborty A, Sarkar D, Langthasa
M, Rahman M, Bari M, Singha RS, Malakar AK and Chakraborty S:
Interplay between miRNAs and human diseases. J Cell Physiol.
233:2007–2018. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Chen YL, Xu QP, Guo F and Guan WH:
MicroRNA-302d downregulates TGFBR2 expression and promotes
hepatocellular carcinoma growth and invasion. Exp Ther Med.
13:681–687. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Wang F, Liu J, Zou Y, Jiao Y, Huang Y, Fan
L, Li X, Yu H, He C, Wei W, et al: MicroRNA-143-3p, up-regulated in
H. pylori-positive gastric cancer, suppresses tumor growth,
migration and invasion by directly targeting AKT2. Oncotarget.
8:28711–28724. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Li N and Qin ZB: Inflammation-induced
miR-802 promotes cell proliferation in cholesteatoma. Biotechnol
Lett. 36:1753–1759. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Zhang XY, Mu JH, Liu LY and Zhang HZ:
Upregulation of miR-802 suppresses gastric cancer oncogenicity via
targeting RAB23 expression. Eur Rev Med Pharmacol Sci.
21:4071–4078. 2017.PubMed/NCBI
|
|
15
|
Zhang Q, Lv R, Guo W and Li X:
microRNA-802 inhibits cell proliferation and induces apoptosis in
human cervical cancer by targeting serine/arginine-rich splicing
factor 9. J Cell Biochem. 120:10370–10379. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Delfino KR and Rodriguez-Zas SL:
Transcription factor-microRNA-target gene networks associated with
ovarian cancer survival and recurrence. PLoS One. 8:e586082013.
View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Zhou W, Wang Y, Wu R, He Y, Su Q and Shi
G: MicroRNA-488 and −920 regulate the production of proinflammatory
cytokines in acute gouty arthritis. Arthritis Res Ther. 19:2032017.
View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Hu J, Ni S, Cao Y, Zhang T, Wu T, Yin X,
Lang Y and Lu H: The angiogenic effect of microRNA-21 targeting
TIMP3 through the regulation of MMP2 and MMP9. PLoS One.
11:e01495372016. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Liu GW, Qin ZM and Shen QH: An ensemble
method integrated with miRNA expression data for predicting miRNA
targets in stomach adenocarcinoma. Cancer Biomark. 20:617–625.
2017. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Li Y, Cai B, Shen L, Dong Y, Lu Q, Sun S,
Liu S, Ma S, Ma PX and Chen J: MiRNA-29b suppresses tumor growth
through simultaneously inhibiting angiogenesis and tumorigenesis by
targeting Akt3. Cancer Lett. 397:111–119. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(-Delta Delta C(T)) Method. Methods. 25:402–408. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Cakmak B and Köseoğlu DR: Comparison of
cervical cytological screening results between postmenopausal and
elderly women. Turk Patoloji Derg. 30:38–42. 2014.PubMed/NCBI
|
|
23
|
Cordeiro MN, De Lima RCP, Paolini F, Melo
ARDS, Campos APF, Venuti A and De Freitas AC: Current research into
novel therapeutic vaccines against cervical cancer. Expert Rev
Anticancer Ther. 18:365–376. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Favero G, Pierobon J, Genta ML, Araujo MP,
Miglino G, Del Carmen Pilar Diz M, de Andrade Carvalho H, Fukushima
JT, Baracat EC and Carvalho JP: Laparoscopic extrafascial
hysterectomy (completion surgery) after primary chemoradiation in
patients with locally advanced cervical cancer: technical aspects
and operative outcomes. Int J Gynecol Cancer. 24:608–614. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Yuan F and Wang W: MicroRNA-802 suppresses
breast cancer proliferation through downregulation of FoxM1. Mol
Med Rep. 12:4647–4651. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Wu X, Gong Z, Sun L, Ma L and Wang Q:
MicroRNA-802 plays a tumour suppressive role in tongue squamous
cell carcinoma through directly targeting MAP2K4. Cell Prolif.
50:502017. View Article : Google Scholar
|
|
27
|
Cao ZQ, Shen Z and Huang WY: MicroRNA-802
promotes osteosarcoma cell proliferation by targeting p27. Asian
Pac J Cancer Prev. 14:7081–7084. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Wang LQ, Chen G, Liu XY, Liu FY, Jiang SY
and Wang Z: microRNA-802 promotes lung carcinoma proliferation by
targeting the tumor suppressor menin. Mol Med Rep. 10:1537–1542.
2014. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Symes AJ, Eilertsen M, Millar M, Nariculam
J, Freeman A, Notara M, Feneley MR, Patel HR, Masters JR and Ahmed
A: Quantitative analysis of BTF3, HINT1, NDRG1 and ODC1 protein
over-expression in human prostate cancer tissue. PLoS One.
8:e842952013. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Wang CJ, Frånbergh-Karlson H, Wang DW,
Arbman G, Zhang H and Sun XF: Clinicopathological significance of
BTF3 expression in colorectal cancer. Tumour Biol. 34:2141–2146.
2013. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Ding J, Wang X, Zhang Y, Sang X, Yi J, Liu
C, Liu Z, Wang M, Zhang N, Xue Y, et al: Inhibition of BTF3
sensitizes luminal breast cancer cells to PI3Kα inhibition through
the transcriptional regulation of ERα. Cancer Lett 440–441. 54–63.
2019. View Article : Google Scholar
|
|
32
|
Zhang Y, Gross N, Li Z, Yin G, Zhong Q,
Liu C and Huang Z: Upregulation of BTF3 affects the proliferation,
apoptosis, and cell cycle regulation in hypopharyngeal squamous
cell carcinoma. Biomed Pharmacother. 118:1092112019. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Hu J, Sun F, Chen W, Zhang J, Zhang T, Qi
M, Feng T, Liu H, Li X, Xing Y, et al: BTF3 sustains cancer
stem-like phenotype of prostate cancer via stabilization of BMI1. J
Exp Clin Cancer Res. 38:2272019. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Hanahan D and Weinberg RA: Hallmarks of
cancer: The next generation. Cell. 144:646–674. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Qureshi R, Arora H and Rizvi MA: EMT in
cervical cancer: Its role in tumour progression and response to
therapy. Cancer Lett 356 (2 Pt B). 321–331. 2015. View Article : Google Scholar
|
|
36
|
Sathyanarayanan A, Chandrasekaran KS and
Karunagaran D: microRNA-145 modulates epithelial-mesenchymal
transition and suppresses proliferation, migration and invasion by
targeting SIP1 in human cervical cancer cells. Cell Oncol (Dordr).
40:119–131. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Cheng Y, Yang M and Peng J: Correlation
the between the regulation of miRNA-1 in c-Met-induced EMT and
cervical cancer progression. Oncol Lett. 17:3341–3349.
2019.PubMed/NCBI
|
|
38
|
Zhang DZ, Chen BH, Zhang LF, Cheng MK,
Fang XJ and Wu XJ: Basic transcription factor 3 is required for
proliferation and epithelial-mesenchymal transition via regulation
of FOXM1 and JAK2/STAT3 signaling in gastric cancer. Oncol Res.
25:1453–1462. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Liu H, Zeng Z, Wang S, Li T, Mastriani E,
Li QH, Bao HX, Zhou YJ, Wang X, Liu Y, et al: Main components of
pomegranate, ellagic acid and luteolin, inhibit metastasis of
ovarian cancer by down-regulating MMP2 and MMP9. Cancer Biol Ther.
18:990–999. 2017. View Article : Google Scholar : PubMed/NCBI
|