Introduction
Plant-based herbal medicines have been extensively
applied traditionally for centuries. Recently, attention towards
these types of medicine has increased worldwide owing to their
nutraceutical value. Eurycoma (E.) longifolia Jack is
one of the most popular traditional plants found in the tropical
rainforests of Southeast Asia, especially in Malaysia and
Indonesia. This herb belongs to the Simaroubaceae family that can
grow slender up to 15 m high, able to grow in different types of
soil (1–4). The plant is known locally as ‘Tongkat
Ali’, ‘Pasak Bumi’, ‘Cay ba binh’ and ‘Ian-don’ in Malaysia,
Indonesia, Vietnam and Thailand, respectively (5–7).
E. longifolia is a promising natural source of biologically
active compounds, including quassinoids, mainly those in the 13β
family, 18-dihydroeurycomanol, eurycomanol-2-O-β-D-glucopyranoside,
eurycomanol and eurycomanone (6,8,9).
Some of the constituents of E. longifolia have been
previously found to exert antiamoebic (10), in vitro anticancer or
antiproliferative effects against cancer cells (11) and plasmodial activities (12). A previous study revealed that
semi-purified eurycomanone conferred cytotoxic activity towards
MCF-7 breast cancer cells, demonstrating the potential anticancer
property of this plant (9,13). In addition, E. longifolia
root extract has also demonstrated significant cytotoxicity against
human lung cancer (A-549) and human breast cancer (MCF-7) cell
lines (14) in other previous
studies.
A number of studies have previously demonstrated the
efficacy of E. longifolia as an effective booster of
testosterone and aphrodisiac supplement to improve strength and
power during sexual activity (15,16).
Additionally, E. longifolia has been found to either prevent
and/or alleviate erectile dysfunction in men (17). The roots and leaves of this plant
have long been used to treat a number of diseases (6,7). In
addition to fever, intestinal parasites, mouth ulcers and headache
(6,18), the roots of E. longifolia
have been applied as a traditional ‘anti-aging’ remedy to help
older individuals adapt to the reduced energy, mood and libido
associated with aging (3,19). The roots of this plant have also
been documented to enhance blood flow that function as an herbal
supplement for women following childbirth (6,20).
By contrast, the leaves of the plant have been used to prevent gum
diseases, treat ulcers and sexually-transmitted infections such as
syphilis and gonorrhoea (3).
Leydig cells are the primary sites of
steroidogenesis in the testis, which serve as the main source of
testosterone in this organ (21).
Testosterone production in Leydig cells is stimulated by
luteinizing hormone, which is secreted by the anterior pituitary
gland through the hypothalamic pituitary-gonadal axis (22). Testosterone is the primary male sex
hormone, which is crucial for the differentiation of the male
urogenital system, spermatogenesis and accumulation of bone and
muscle mass in men (23).
The spermatogenic effect of E. longifolia is
attributed to the presence of quassinoids in the extracts (8,24).
Quassinoids are biologically active compounds that contribute to
E. longifolia as a testosterone production booster, such
that extracts enriched with E. longifolia quassinoids have
been found to exert more potent effects. The effects of E.
longifolia extracts on steroidogenesis have been previously
investigated in animal models (3,25–28).
However, despite preliminary findings demonstrating the effect of
quassinoid-rich E. longifolia on rat interstitial cells
(8), the in vitro effects
of E. longifolia extracts in Leydig cells, for example,
remain poorly understood. Therefore, the present study aimed to
investigate the impact of a standardized E. longifolia root
extract (F2) on testosterone production, growth and cell cycle
progression in mouse Leydig cells. It is hoped that data from the
present study will lead to an improved understanding on the
mechanism of action mediated by the E. longifolia extract so
it can be further utilized as a natural product for treating human
diseases.
Materials and methods
E. longifolia standardised extract and
chemical preparation
A standardised root methanolic extract of E.
longifolia, F2, and the pure compound, eurycomanone (EN;
>96% purity), were received from Professor Chan Kit Lam at the
School of Pharmaceutical Science, Universiti Sains Malaysia
(Penang, Malaysia). Both the extract and the pure compound were
dissolved in double-sterile water (ddH2O). The aromatase
inhibitor, formestane (FM; >98% purity), which was purchased
from MedChem Express, was dissolved in DMSO.
TM3 cell culture
TM3 (CRL-1714™) Leydig cells, purchased from the
American Type Culture Collection, were cultured in a 1:1 mixture of
Gibco Ham's F12 medium (Invitrogen; Thermo Fisher Scientific, Inc.)
and Dulbecco's modified eagle's medium (DMEM; Invitrogen; Thermo
Fisher Scientific, Inc.) supplemented with 5% (v/v) horse serum
(Invitrogen; Thermo Fisher Scientific, Inc.) and 2.5% (v/v) foetal
bovine serum (Invitrogen; Thermo Fisher Scientific, Inc.).
Penicillin (100 U/ml) and streptomycin (100 µg/ml) were added to
the growth medium to prevent contamination. The culture was
incubated in a humidified atmosphere and supplied with 5%
CO2 at 37°C. The Leydig cells were sub-cultured every
2–3 days depending on the confluence of the cell culture. The use
of a mouse Leydig cell line may be a limitation of the present
study. Human Leydig cells would have been a better cell line for
proper verification of the hypothesis.
Measurement of testosterone levels in
F2-, EN- and FM-treated Leydig cells using ELISA
TM3 cells were seeded onto 24-well plates at a
density of 2.0×104 cells/well. After being cultured at
37°C in an incubator overnight, the cells were treated with 0.1,
1.0 and 10 mg/ml F2 or 0.1, 1.0 and 10 µM EN or FM. Each treatment
was carried out in triplicate for four treatment time-points: 24,
48, 72 and 96 h in ≥2 independent experiments, following which the
culture supernatant was collected at each treatment time-point. The
collected culture supernatants were used for the measurement of
testosterone level using a mouse testosterone (T) enzyme-linked
immunosorbent assay (ELISA) kit (cat. no. CSB-E05101m; Cusabio
Biotech Co., Ltd.), according to the manufacturer's instructions.
Briefly, the wells of the microtitre plates were pre-coated with a
goat anti-rabbit antibody specific for testosterone. The reactions
were then incubated with a secondary horseradish
peroxidase-conjugated antibody from the aforementioned kit for the
target hormone signal development. A substrate solution was also
added to enhance the color development to indicate the presence of
the target hormone. The intensity of the signal for each plate was
measured at 450 nm by using a Multiskan™ spectrum
spectrophotometric plate reader (Thermo Fisher Scientific, Inc.). A
standard curve was plotted to estimate the level of testosterone (%
relative to control) in each well.
Viability analysis of F2-, EN- and
FM-treated Leydig cells using MTT assay
Firstly, 2.5×103 TM3 cells per well were
seeded onto 96-well plates. The cells were cultured overnight in
growth medium at 37°C and treated with 0.1, 1.0, and 10 mg/ml F2
and 0.1, 1.0 and 10 µM EN or FM in triplicate for four treatment
time-points: 24, 48, 72 and 96 h for ≥2 independent experiments at
37°C. Cell viability was measured by using a standard
3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide (MTT)
assay. At each time-point, 24 µl MTT reagent (2.5 mg/ml) was added
to each well before the reactions were incubated for 4 h at room
temperature. Subsequently, 100 µl DMSO was added to each well for
color development. The color or optical density (OD) was then read
at 570 nm by using a Multiskan™ spectrum spectrophotometric plate
reader (Thermo Fisher Scientific, Inc.). The cell viability in each
well was calculated using the following formula: [(OD of treated
sample-OD of blank)/(OD of control-OD of blank)] ×100%. The level
of cell viability was indicated as % relative to control.
Cell cycle progression analysis of
F2-treated Leydig cells using flow cytometry
A density of 6.0×104 TM3 cells/well was
seeded onto 6-well plates and incubated at 37°C with 5%
CO2 overnight. On the next day, the cells were treated
with F2 at concentrations of 0.1, 1.0 and 10 mg/ml in triplicate at
37°C for four treatment time-points: 24, 48, 72 and 96 h in ≥2
independent experiments. The culture supernatants and the cells
were then harvested from each well and transferred to new 15 ml
Falcon tubes at each time-point. The tubes were centrifuged at
1,000 × g for 5 min. The supernatant was discarded, and the cell
pellet was washed with PBS and centrifuged again at 1,000 × g for 5
min. After discarding the supernatant, 500 µl PBS was added to the
cell pellets. All these procedures were carried out at room
temperature. The number of cells in the cell suspension was then
counted and adjusted to 1×106 cells, which was suspended
in 500 µl using 70% cold absolute ethanol. The suspension was mixed
gently and stored at 4°C to facilitate ethanol fixation. Before
cell cycle analysis, the ethanol-fixed cells were centrifuged at
1,000 × g for 10 min and washed three times by resuspending the
cells in PBS at room temperature. The suspension was then
centrifuged again at 1,000 × g at room temperature for 10 min and
the supernatant was discarded. The cells were stained with 500 µl
FxCycle™ propidium iodide (PI)/RNase staining solution (Thermo
Fisher Scientific, Inc.) at room temperature in the dark for 30
min. The stained samples were then transferred to new sterile flow
tubes and maintained on ice until the samples were analysed by flow
cytometry using a BD FACSCanto™ II flow cytometer (BD Biosciences).
The % of the cell population at each cell cycle phase was then
calculated.
mRNA gene expression profile analysis
of F2-treated Leydig cells
TM3 cells were seeded onto 24-well plates at a
density of 2.0×104 cells/well and incubated at 37°C with
5% CO2 overnight. The next day, cells were treated with
500 µl 0.1 mg/ml F2 in triplicate for 72 and 96 h. A total of three
wells of treatment solution and treated cells were then harvested
and pooled as one sample for RNA extraction. The total RNA
extraction was performed by using the RNeasy Mini Kit (QIAGEN
Inc.), according to the manufacturer's protocol. The concentration
and purity of the extracted total RNA were determined by using a
NanoPhotometer® (Implen GmbH), whereas the integrity of
the RNA was assessed by 1% (w/v) agarose gel electrophoresis
(Bio-Rad Laboratories Inc.). The integrity of the total RNA was
confirmed further using a 2100 Bioanalyzer (Agilent Technologies
Inc.) coupled with the RNA 6000 Pico LabChip® Kit (cat.
no. 5067-1513; Agilent Technologies Inc.). Good quality RNA
(showing 2 ribosomal peaks and RNA Integrity Number ≥8) was used
for steroidogenesis gene expression profiling using Affymetrix
GeneChip™ microarray technology (Thermo Fisher Scientific, Inc.).
The copy RNA (cRNA) targets for the Affymetrix platform were
synthesized, purified, labelled and hybridised, according to the
manufacturer's instructions. Briefly, ~100 ng extracted total RNA
was converted to cDNA and was then amplified to cRNA following the
procedures of Affymetrix GeneChip™ Whole Transcript (WT) PLUS
Reagent Kit (cat. no. 902280; Thermo Fisher Scientific, Inc.). The
cRNA was then purified and quantified before being synthesised into
single-stranded (ss)-cDNA using the same kit, according to the
manufacturer's protocols. The ss-cDNA was then fragmented and
labelled with biotin before being hybridised onto the array strip
for GeneChip™ WT Expression Array (Affymetrix; Thermo Fisher
Scientific, Inc.). The gene expression profiles were scanned using
the Affymetrix GeneArray scanner 3000 7G system (Affymetrix; Thermo
Fisher Scientific, Inc.) at 488 nm and 3-µm resolution. The images
of the patterns were analysed using the Affymetrix Microarray Suite
software v.5.0 (MAS5). The raw data .txt file obtained from the
GeneArray scanner was then loaded onto the GeneSpring Gx software
v.12.0 (Agilent Technologies Inc.) for further data processing,
including normalization and statistical analysis. The expression
ratios for >22,000 genes in non-treated and F2-treated Leydig
cells at the respective time-points were computed and filtered by
comparative analysis of the MAS5 values. The log2 values
were used to identify differentially expressed genes (DEGs) in
F2-treated compared with non-treated Leydig cells. Genes with a
fold-change ≥2 were identified as statistically significant
DEGs.
Identified gene expression validation
in F2-treated Leydig cells using reverse transcription-quantitative
(RT-q)PCR
Leydig cells were seeded onto 24-well plates at a
density of 2.0×104 cells/well and incubated at 37°C with
5% CO2 overnight. The next day, the cells were treated
with 500 µl either 0.1 or 1.0 mg/ml F2 in triplicate for 72 and 96
h. A total of three wells of treatment solution and treated cells
were then harvested and pooled as one sample for RNA extraction
using the RNeasy Mini Kit (QIAGEN Inc.), according to
manufacturer's protocols. The concentration of the extracted total
RNA was determined by using a NanoPhotometer and the integrity of
extracted RNA was assessed by using 1% (w/v) agarose gel
electrophoresis as aforementioned. Good quality total RNA was
reverse-transcribed to cDNA at 60°C using the RevertAid First
Strand cDNA Synthesis Kit (Thermo Fisher Scientific, Inc.) and the
cDNA was stored at −20°C until it was used to analyse the gene
expression by qPCR. All primers used were designed using the Primer
Express™ software v.3.0.1 (Thermo Fisher Scientific, Inc.) and are
listed in Table I. qPCR was
performed by using the Applied Biosystems Power SYBR®
Green PCR Master Mix and the 7500 Fast Sequence Detection System
(both Thermo Fisher Scientific, Inc.). Each reaction mixture
contained 12.5 µl of 2X SYBR® Green PCR Master Mix, 20
pmol each of forward and reverse primer and 5 µl cDNA (25 ng). A
sufficient volume of deionised water was added to the mixture to
bring the final reaction volume to 25 µl. The following
thermocycler program was used for the amplification: 50°C for 3 min
and 95°C for 5 min for initiation, followed by 35–40 cycles of 95°C
for 10 sec and 60°C for 30 sec. Finally, a melting curve for each
amplicon was generated with 1°C temperature increments from
72–95°C. All reactions were performed in triplicate and included
three non-template reactions as negative controls. The experiment
was repeated ≥2 times, whereby the mRNA expression levels of the
target genes were normalised to that of β-actin in the test sample.
The mRNA expression level (relative to control) of the target gene
was then calculated using the 2−ΔΔCq method (29).
 | Table I.List of primers used for quantitative
PCR analysis. |
Table I.
List of primers used for quantitative
PCR analysis.
| Gene name | Primer
sequence |
|---|
| CDKL3 | Forward:
5′-TGTCCTCCTGTCTGGTGCATT-3′ |
|
| Reverse:
5′-CTACCCGAGAAGACCCTTTTCAG-3′ |
| Cyclin
G1 | Forward:
5′-AAGGTACAGGCGAAGCATCTTG-3′ |
|
| Reverse:
5′-CCTTTCCTCTTCAGTCGCTTTC-3′ |
| CDKN1A | Forward:
5′-GACCACTGGACCTAGCAATTCAC-3′ |
|
| Reverse:
5′-GTAGGAGTCACCGTCCTGTTTACC-3′ |
| Cyclin
L1 | Forward:
5′-AATAGGCGAAGTCGATCTGGAA-3′ |
|
| Reverse:
5′-TATGATGTCTTCGAGGGCTTTCA-3′ |
| CDKL2 | Forward:
5′-CAAATGCCAACTGTCTCCATGA-3′ |
|
| Reverse:
5′-GTTGACATTGCTGCAATCTCTGA-3′ |
| HMOX1 | Forward:
5′-GGTGTCCAGAGAAGGCTTTAAGC-3′ |
|
| Reverse:
5′-TGCGCTCTATCTCCTCTTCCA-3′ |
| Cyclin
G2 | Forward:
5′-GGAGTGTTTTGATCCCCAACA-3′ |
|
| Reverse:
5′-GAACAGCTAAGGATGACCTCGAA-3′ |
| Cyclin
B2 | Forward:
5′-GTGAAGTCCTGGAAGTCATGCA-3′ |
|
| Reverse:
5′-CGATGAACTTGGTACGGTTGTC-3′ |
| CDKN2C | Forward:
5′-AACCATCCCAGTCCTTCTGTCA-3′ |
|
| Reverse:
5′-CCCCTTTCCTTTGCTCCTAATC-3′ |
| β-actin | Forward:
5′-GCTCTGGCTCCTAGCACCAT-3′ |
|
| Reverse:
5′-GCTGATCCACATCTGCTGGAA-3′ |
Functional enrichment analysis
To further study the interaction of identified DEGs
at a functional level, Gene Ontology (GO) functional enrichment
analysis was performed using GeneMANIA Cytoscape program v.3.6.0
(https://genemania.org/). GeneMANIA discovered
functionally similar genes using a wealth of genomics and
proteomics data from a query gene. The algorithm of the modal was
then weighted each functional genomic dataset, according to its
predictive value for the query, whereby a fold change discovery
≤0.05 was set as the cut-off criterion for the enrichment analysis
(30). A diagram of network weight
for CDKL2 was plotted by -log10 (DWeight).
Statistical analysis
All data were presented as the means ± standard
deviation from experimental triplicates, and all statistical
analyses were performed using GraphPad Prism 7.05 (GraphPad
Software, Inc.). The significant levels of cell viability, hormone
and target gene expression were analysed by using one-way ANOVA
with the Tukey's post hoc test. P<0.005 was considered to
indicate a statistically significant difference.
Results
Comparison of testosterone production
in F2-, EN- and FM-treated Leydig cells
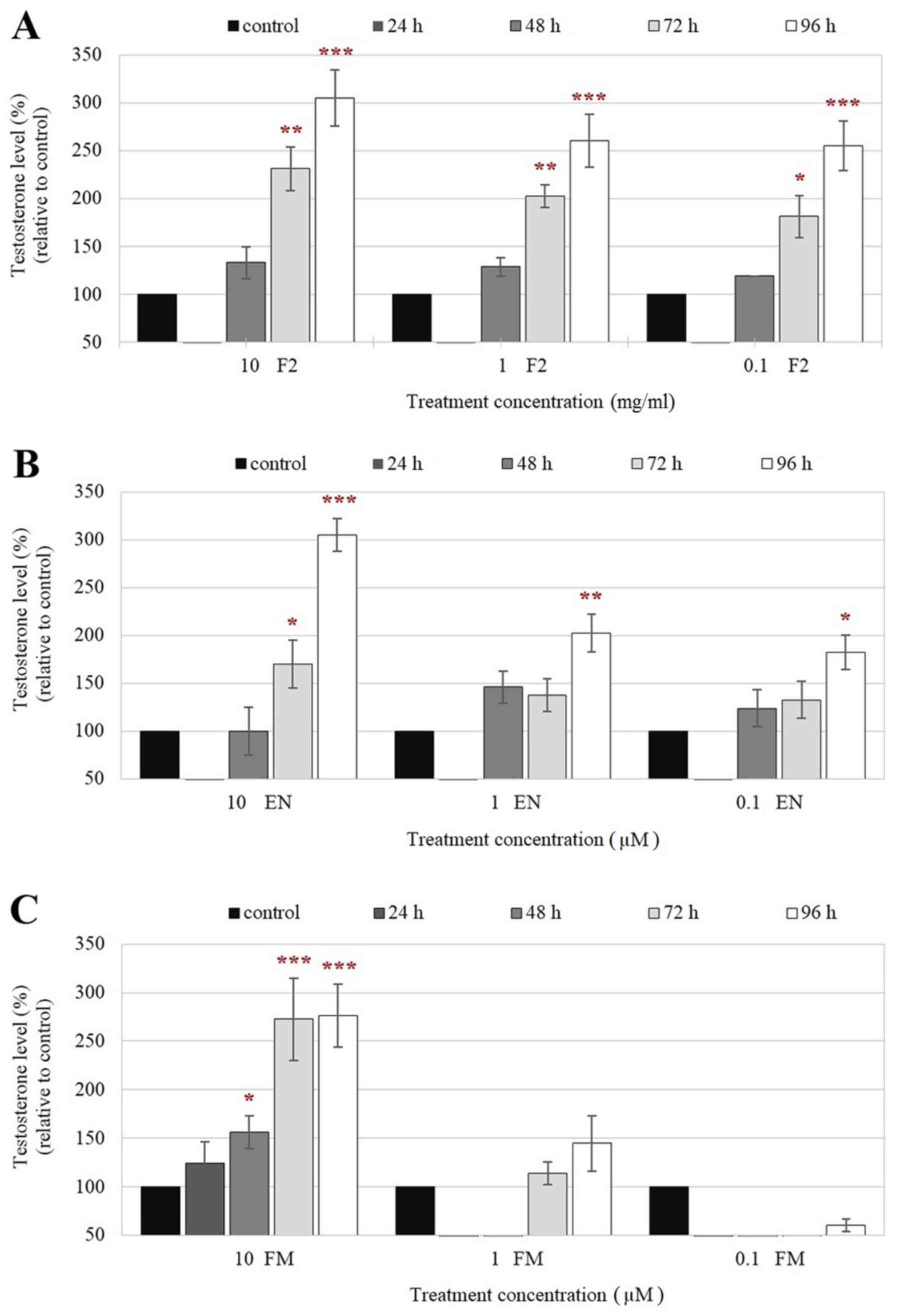
Testosterone levels in culture supernatants of F2-
and EN-treated TM3 cells, as measured by ELISA, were first
inhibited 24 h after treatment. Following that, the levels were
induced with increasing treatment time-points compared with those
of control (Fig. 1A and B),
indicating the ability of F2 to stimulate testosterone production
by these cells. The experiments further validated the highest
activity of F2 on hormone production by TM3 cells 96 h after
treatment. FM, a known aromatase inhibitor, also demonstrated a
similar effect and induced high levels of testosterone production
in TM3 cells but only ≥72 h after treatment with the highest
concentration of FM (10 µM) tested (Fig. 1C). Therefore, these results suggest
that F2 and EN may exert similar effects as aromatase inhibitors in
regulating testosterone synthesis and production. In addition, it
should also be noted that high levels of testosterone production
(~250% increase) were also observed in F2-treated TM3 cells even at
lower concentrations (0.1 and 1.0 mg/ml; Fig. 1A). Therefore, F2 may be more potent
at stimulating testosterone production compared with EN and FM in
Leydig cells.
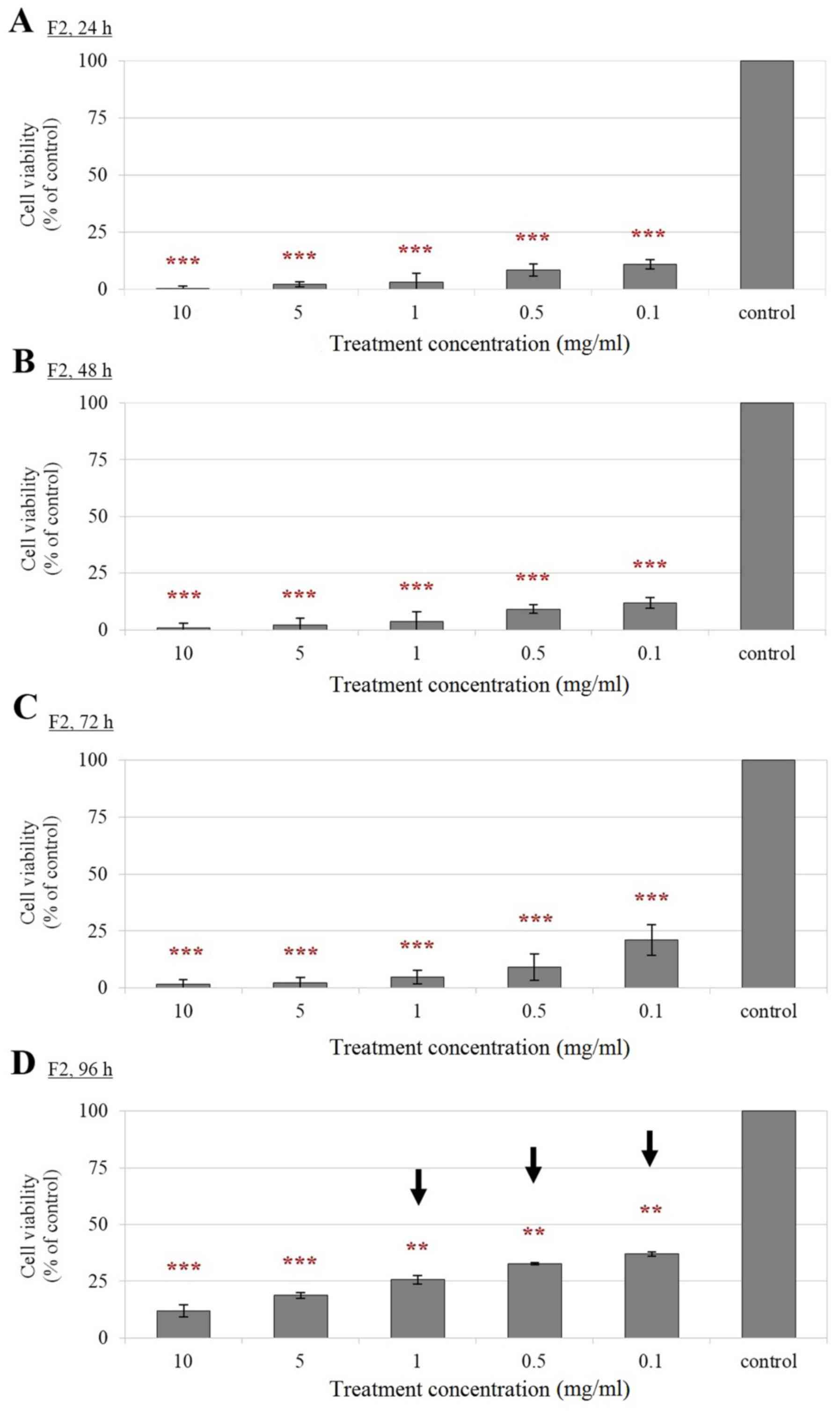
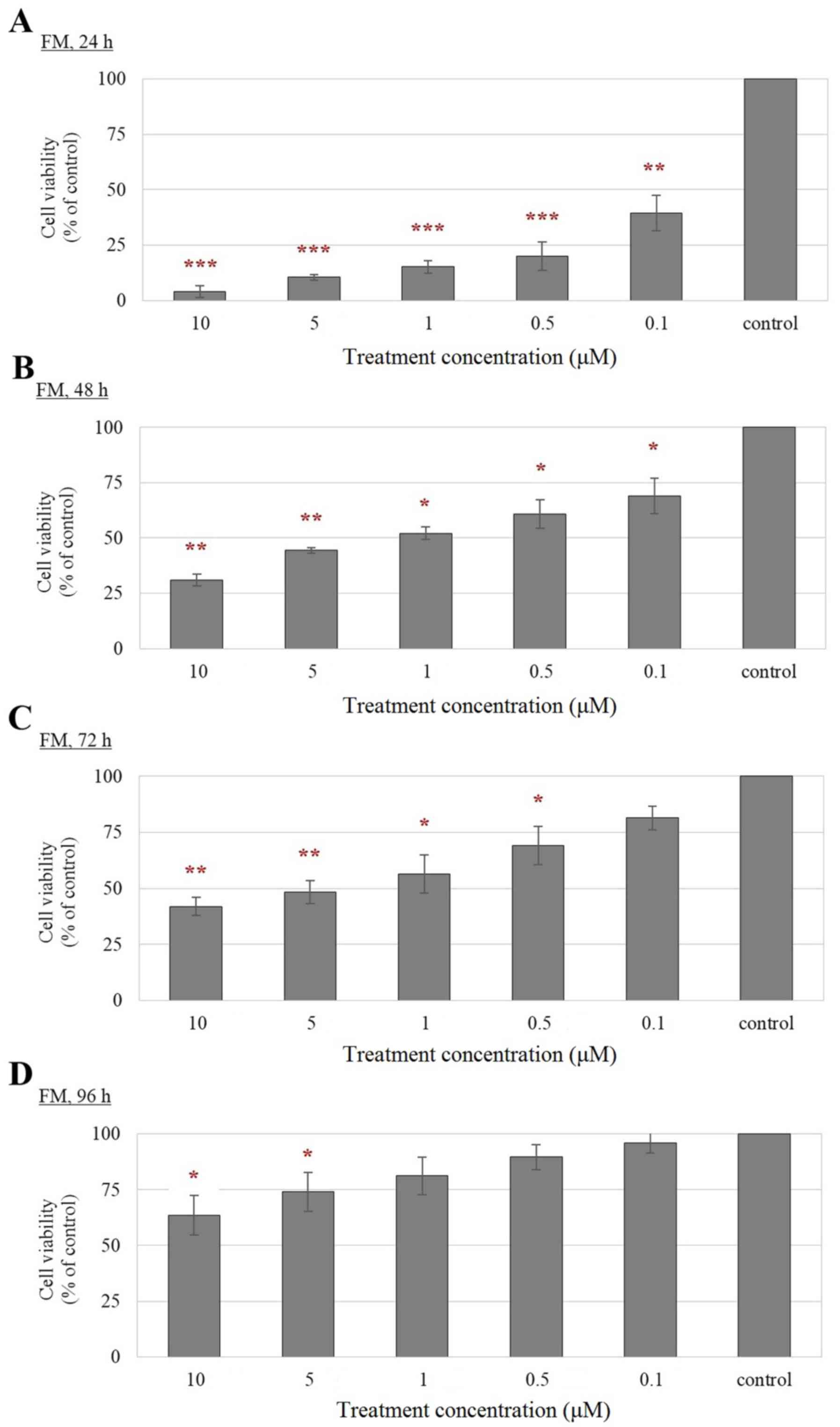
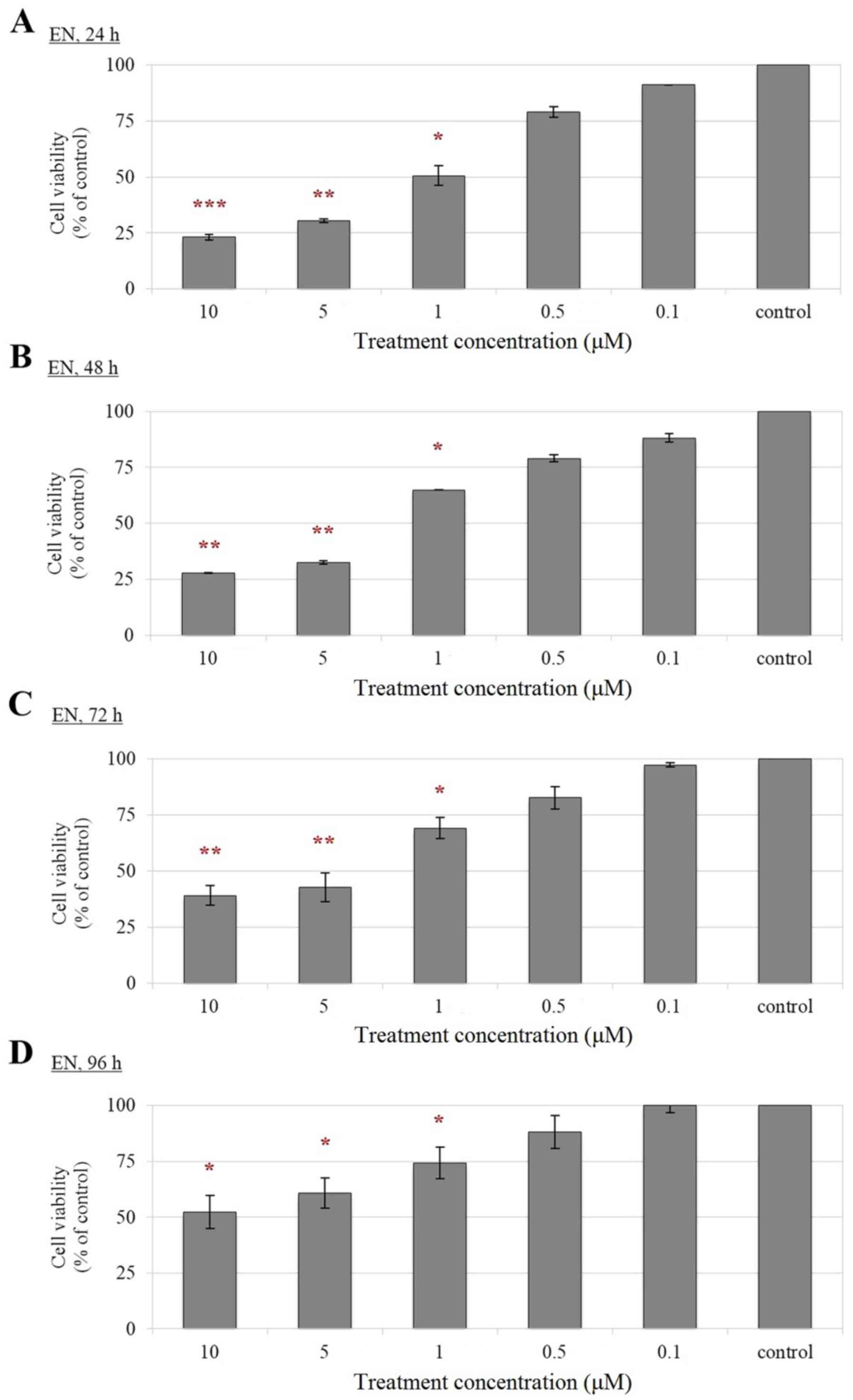
Reduced cell viability of F2-, EN- and
FM-treated Leydig cells compared with control cells
According to the results from MTT assay (Figs. 2–4), a significant reduction in the
viability of F2-, EN- and FM-treated TM3 cells was observed
compared with that of their corresponding controls in a
dose-dependent manner. Treatment of Leydig cells with F2
demonstrated a significant reduction in cell viability at all
concentrations and treatment time-points tested (Fig. 2). The viability of TM3 cells
treated with lower concentrations of F2, including 0.1, 0.5 and 1.0
mg/ml, were able to revive 96 h following treatment, although the
viability of the treated cells remained low at this treatment
time-point (Fig. 2D). Notably, a
less potent inhibitory effect on cell viability was observed for
EN, whereby cell treated with EN were viable throughout all
experimental time-points tested (Fig.
3). A gradual increment of cell viability inhibition on TM3 was
only observed when higher concentrations of EN, including 1, 5 and
10 µM were used (Fig. 3). In the
FM-treated cells, a stronger growth inhibitory effect was observed
at earlier treatment time-points compared with that observed in EN
treated cells (Fig. 4). Notably,
the FM-treated cells remained viable at 96 h, although a
dose-dependent inhibition of cell viability was also observed at
higher concentrations (5 and 10 µM) of FM treatment (Fig. 4D). The revival of the treated cells
could be attributed to the reduction in the drug effect with
prolonged treatment duration. Therefore, F2, EN and FM are suitable
for long term treatments of Leydig cells at low concentrations.
S-phase cell cycle arrest in
F2-treated Leydig cells
Cell cycle analysis of TM3 cells by flow cytometry
(Fig. 5A) demonstrated that the
percentage of cells at the G0/G1 phase was
significantly reduced after treatment with 0.1, 1.0 and 10 mg/ml
compared with that in the untreated cells (control) 24 h after
treatment (Fig. 5B). The reduced
percentage of F2-treated TM3 cells at the
G0/G1 phase was accompanied by an
accumulation in the percentage of cells at the S phase (Fig. 5B). This phenomenon suggests that
the growth of F2-treated cells was affected by cell cycle arrest in
the S phase. This observation persisted 48 h following treatment,
particularly in the cells treated with higher concentrations of F2
(1.0 and 10 mg/ml; Fig. 5C).
However, the percentage of cells in the G0/G1
phase appeared to be revived 48 h after treatment with 0.1 mg/ml F2
(Fig. 5C). The opposite trend was
observed in the percentage of cells in the S phase (Fig. 5C). Cells treated with higher
concentrations of F2 were only revived 72 h following treatment
(Fig. 5D); the number of cells in
Go/G1 cell cycle phase 72 h following treatment was higher compared
with the number of cells in the same phase 48 h following treatment
(Fig. 5C). Following 96 h of F2
treatment, reductions in the proportion of TM3 cells in the
G0/G1 phases treated with 0.1 mg/ml F2
observed at 24, 48 and 72 h time-points were reversed (Fig. 5E). Likewise, increments in the
proportion of TM3 cells at the S phase treated with F2 observed at
24, 48 and 72 h were also reversed after 96 h, particularly in
cells treated with 0.1 mg/ml F2 (Fig.
5E). The percentages of treated cells at the G2
phase post F2 treatment demonstrated no difference compared with
those of control following F2 treatment for 24–96 h (Fig. 5).
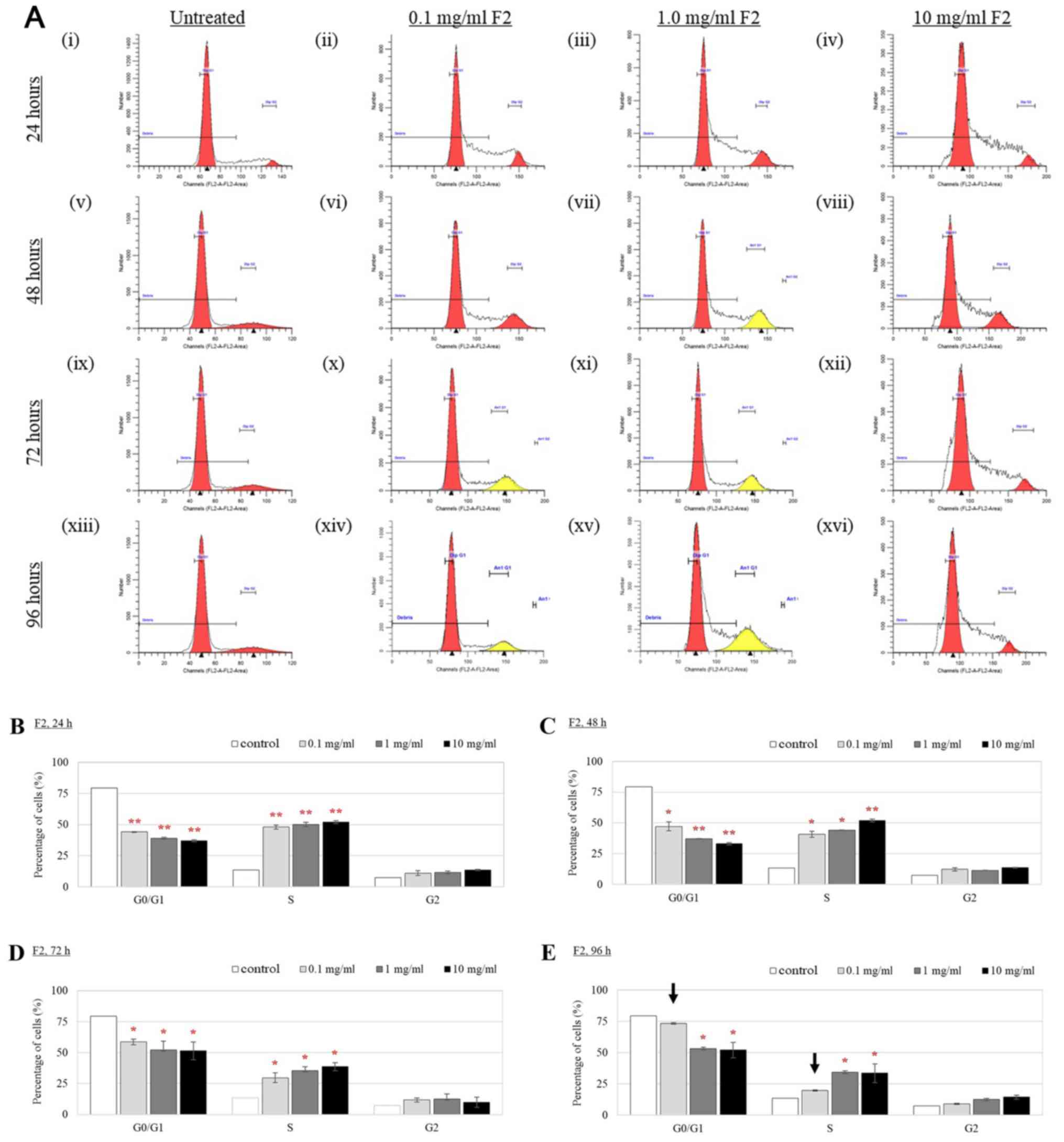 | Figure 5.Cell cycle distribution of Leydig
cells after F2 treatment. (A) Histograms of cell cycle
distribution. (i) Untreated Leydig cells (control), Leydig cells
treated with (ii) 0.1 mg/ml F2, (iii) 1.0 mg/ml F2, and (iv) 10
mg/ml F2 for 24 h; (v) Untreated Leydig cells (control), Leydig
cells treated with (vi) 0.1 mg/ml F2, (vii) 1.0 mg/ml F2, and
(viii) 10 mg/ml F2 for 48 h; (ix) Untreated Leydig cells (control),
Leydig cells treated with (x) 0.1 mg/ml F2, (xi) 1.0 mg/ml F2, and
(xii) 10 mg/ml F2 for 72 h; (xiii) Untreated Leydig cells
(control), Leydig cells treated with (xiv) 0.1 mg/ml F2, (xv) 1.0
mg/ml F2, and (xvi) 10 mg/ml F2 for 96 h. Quantification of cell
cycle distribution (B) 24, (C) 48, (D) 72 and (E) 96 h after F2
treatment. Data are expressed as means ± SD (n=3). *P<0.05 and
**P<0.01 indicate a significant change of cell cycle phase in
treated cells vs. control at specific time-points. Arrows (→)
indicate the cells were significantly revived. F2, Eurycoma
longifolia extract. |
Functional level of identified DEGs in
F2-treated Leydig cells
Microarray analysis revealed 237 and 324 genes to be
up- and downregulated, respectively, in Leydig cells 72 h following
F2 treatment (Fig. 6A). However,
the number of genes that were found to be up- and downregulated in
Leydig cells was reduced to 192 and 212, respectively, 96 h
following F2 treatment (Fig. 6A).
These DEGs could indicate the mechanism on the revival of
F2-treated Leydig cells and should be studied in depth to improve
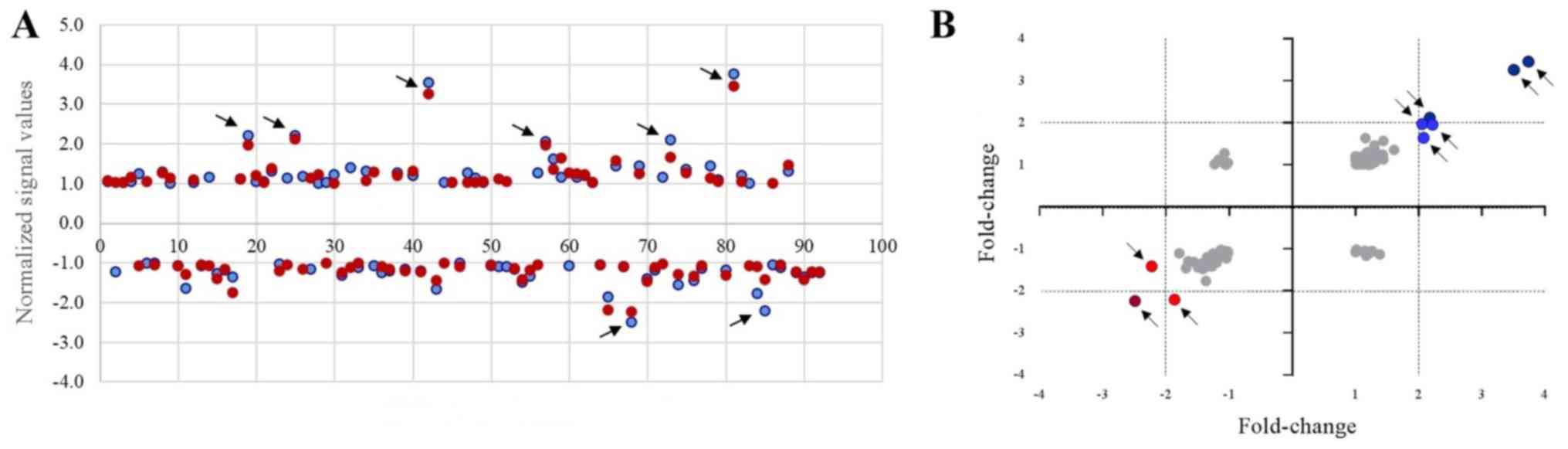
the understanding and safety of F2 consumption. A scatter plot was
generated to assess the variations in DEGs in the cyclin family,
where its related members among non-treated (control) and
F2-treated Leydig cells 72 and 96 h following treatment with F2
were compared (Fig. 6B). The DEGs
in F2-treated Leydig cells were clearly observed in the plots,
whereby the top 9 cyclin-related genes that demonstrated >2-fold
up- and downregulated expression in F2-treated Leydig cells for 72
and 96 h were selected. Gene ontology of the DEGs revealed the role
of the selected genes encoding the cyclin family of proteins and
cyclin-dependent kinases (Table
II). The selected genes, including cyclin-dependent
kinase-like-3, cyclin G1, cyclin-dependent kinase inhibitor 1A
(CDKN1A), cyclin L1, cyclin-dependent kinase-like 2 (CDKL2), heme
oxygenase 1, cyclin G2, cyclin B2 and cyclin-dependent kinase
inhibitor 2C were then measured using RT-qPCR. The analysis
revealed consistent expression of all 9 genes in Leydig cells
treated with 0.1 mg/ml F2 at all treatment time-points (Fig. 7). However, when 1.0 mg/ml F2 was
used for treatment, a significant reduction of CDKL2 expression was
observed in Leydig cells treated for 72 and 96 h (Fig. 7E). The CDKL2 expression was reduced
to 0.708 (P<0.05) and 0.743 (P<0.05) fold-change in Leydig
cells treated with 1.0 mg/ml F2 for 72 and 96 h, respectively,
compared to the CDKL2 expression in 1.0 mg/ml F2-treated Leydig
cells for 24 h (1.000 fold-change). Comparison of the CDKL2
expression in the Leydig cells treated with 1.0 mg/ml F2 (0.708
fold change; P<0.05) than that with 0.1 mg/ml F2 (0.991
fold-change) for 72 h also showed significant reduction of the
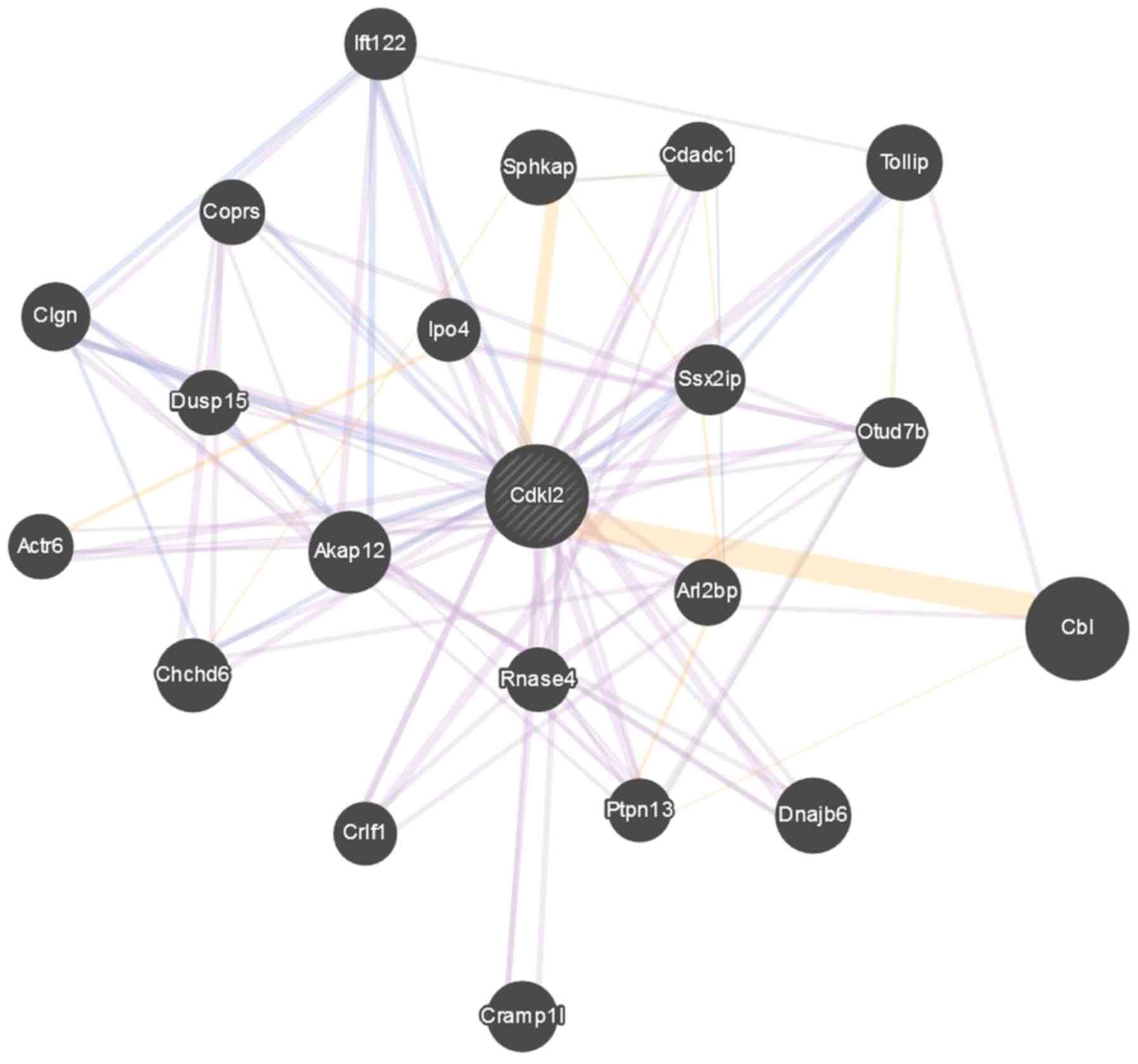
CDKL2 expression. Gene-gene interaction studies demonstrated that
CDKL2 closely interacted with Casitas B-lineage lymphoma (CBL) and
sphingosine kinase 1-interactor-A-kinase adaptor protein
domain-containing (Sphkap), which may also serve important roles in
the mechanism and action of cell revival in F2-treated Leydig cells
(Fig. 8).
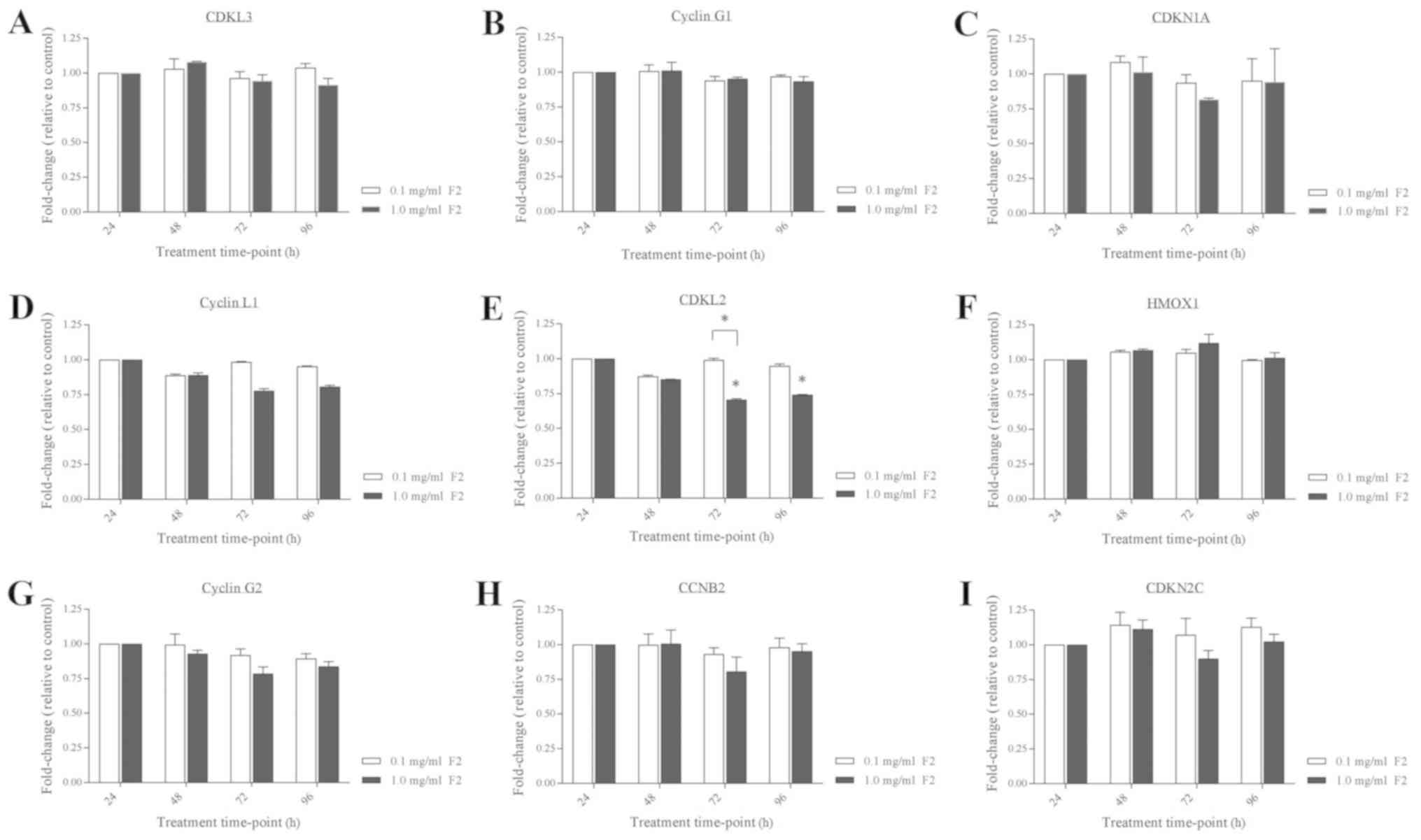 | Figure 7.Gene expression validation in
F2-treated Leydig cells. (A) CDKL3, (B) Cyclin G1, (C) CDKN1A, (D)
Cyclin L1, (E) CDKL2, (F) HMOX1, (G) Cyclin G2, (H) CCNB2 and (I)
CDNK2C mRNA expression in TM3 cells were measured after F2
treatment. Data are expressed as means ± SD (n=3). *P<0.05
indicates a significant fold-change of Leydig cells treated with
1.0 mg/ml F2 after specific time-point vs. the same treated cells
after 24 h or vs. the cells treated with 0.1 mg/ml F2 after the
same time-point. CDKL3, cyclin-dependent kinase-like-3; CDKN1A,
cyclin-dependent kinase inhibitor 1A; CDKL2, cyclin-dependent
kinase-like 2; HMOX1, heme oxygenase 1; CCNB2, cyclin B2; CDKN2C,
cyclin-dependent kinase inhibitor 2C. |
 | Table II.Top nine selected genes showing
>2-fold up- and downregulation by microarray analysis. The
Leydig cells were treated with 0.1 mg/ml F2 for 72 and 96 h. |
Table II.
Top nine selected genes showing
>2-fold up- and downregulation by microarray analysis. The
Leydig cells were treated with 0.1 mg/ml F2 for 72 and 96 h.
| No. | Gene name | 72 h (fold) | 96 h (fold) | Gene function |
|---|
| 1 | Cyclin-dependent
kinase like-3 | 2.204 | 1.959 | A member of the
cyclin family that encodes a CMGC-type serine/threonine protein
kinase for an unknown function. |
| 2 | Cyclin G1 | 2.189 | 2.114 | A transcriptional
target of p53 and is induced by DNA damage in a p53 dependent
manner. |
| 3 | Cyclin-dependent
kinase inhibitor 1A | 3.531 | 3.249 | Encodes a potent
cyclin-dependent kinase inhibitor that binds to and inhibits the
activity of cyclin-cyclin-dependent kinase 2 or -cyclin-dependent
kinase4 complexes. A regulator of cell cycle progression at
G1. |
| 4 | Cyclin L1 | 2.056 | 1.959 | A regulatory
subunit of CDK11 and involved in splicing process for mRNA
production. |
| 5 | Cyclin-dependent
kinase-like 2 | 2.085 | 1.647 | A member of a large
family of CDC2-related serine/threonine protein kinases. |
| 6 | Heme oxygenase
1 | 3.758 | 3.424 | An enzyme that
catalyzes the degradation of heme to produce biliverdin, ferrous
iron and carbon monoxide. |
| 7 | Cyclin G2 | −2.479 | −2.235 | Contains a
C-terminal PEST protein destabilization motif, suggesting that
their expression is tightly regulated through the cell cycle. |
| 8 | Cyclin B2 | −2.219 | −1.424 | A member of the
cyclin family that regulate PLK1 activity at G2/M
Transition. |
| 9 | Cyclin-dependent
kinase inhibitor 2C | −1.866 | −2.189 | A protein coding
gene for the member of the INK4 family that contains 5 ankyrin
repeats. |
Discussion
The importance of exploiting naturally occurring
medicines from herbal plants has been growing around the world. An
increasing number of studies are recognising the use of herbal
medicine for therapy due to fewer side effects,
environmentally-friendly and increased cost-effectiveness (1,31,32).
Previous studies by Low et al (8,26)
revealed that quassinoid (EN) found in E. longifolia
increased testosterone levels in male rats testicular Leydig
cell-rich interstitial cells. However, the scientific evidence for
the mechanism behind this phenomenon remains unknown. The present
study demonstrated that Leydig cells treated with F2, EN and FM
exhibited higher levels of testosterone in the culture
supernatants. F2 acted as a potential testosterone booster because
induction could also be achieved at lower concentrations (0.1
mg/ml) according to results in the in vitro assays. Although
F2 treatment did not affect the accumulated testosterone level,
treatment of Leydig cells with as little as 0.1 mg/ml F2 reduced
the viability of the treated cells. A previous study revealed that
the treatment of a rat cell suspension at doses 0.1–10 µM EN
demonstrated non-cytotoxicity to the cells; >95% cell viability
remained (26). The cell
suspension contained ~40% Leydig cells with a viability of >97%.
However, the effect of F2 on the viability of the cell suspension
was not mentioned in this previous study (26). The present study revealed that high
viability (>95%) of EN-treated cells was only observed when
lower concentrations of EN (<0.5 µM) were used. By contrast, a
50% reduction in cell viability was found when higher
concentrations of EN were used. The current formulation of F2
contains 7.46% EN, which may be a factor to consider in future
formulations of F2, which should contain ≤0.5 µM EN to reduce the
inhibitory effect of F2 on Leydig cells (33). The reduction of cell viability
could be attributed to the biologically active components in F2
(13,21-dihydroeurycomanone, 13α(21)-epoxyeurycomanone or eurycomanol)
that have been previously found to inhibit cell replication,
division and proliferation, in turn terminating the growth of the
treated cells.
Cell cycle is a continuously organised process
during which replication and division of a cell occurs. The primary
principle of cell cycle assays is to quantify the cellular DNA
content at each phase of the cycle (34). PI, which binds to the DNA of the
cell and produces a fluorescent signal, has been used extensively
for cell cycle analyses (34). The
cell cycle consists of two major phases; interphase (90%) and
mitosis (10%) (34). The
interphase can be divided further into another three sub-phases,
which are G0/G1, S and G2, and the
mitotic phase, which is broken down further into mitosis and
cytokinesis (35). The present
study revealed that Leydig cells treated with F2 demonstrated
reductions in cell viability by reducing the cell population in the
G0/G1 phase, which could have been caused by
cell cycle arrest at the S phase. However, cell viability was
revived, especially following 96 h treatment with F2, which may be
due to changes in the number of cells at the
G0/G1 and S phases. Following 96 h treatment
with 0.1 mg/ml F2, the proportions of Leydig cells at the
G0/G1 and S phases were not found to be
significantly different compared with those of the control cells.
The cell revival may also be attributed to the reduced drug effect
and adaptation of cells to F2 treatment. Therefore, further
identification of the potential molecular target(s) that regulate
this mechanism is warranted.
A number of studies have previously demonstrated
cell cycle arrest at the S phase upon treatment of Leydig cells
with different compounds (36–38).
A study recently investigated the effects of calretinin on the
testicular Leydig cell line, MLTC-1, whereby cell cycle analysis
demonstrated a significantly high cell number at the S phase,
suggesting that the reduction in cell viability was due to S-phase
cell cycle arrest (36).
Similarly, a study conducted a cell cycle test by using primary
Leydig cells treated with different concentrations of
dehydroepiandrosterone (DHEA) and discovered that after 48 h 50 µM
DHEA treatment, the cell number at S phase was significantly
increased following a reduction in the cell number at the
G2 phase compared to that in the control group (37). These observations indicate that
cell cycle arrest in S phase inhibited the growth of Leydig cells.
To the best of our knowledge, only two articles have reported the
effect of E. longifolia extracts on the cell cycle
progression. Tong et al (38) previously reported that a
standardised E. longifolia extract, SQ40, could inhibit
LNCaP cell development by suppressing cell growth via
G0/G1 phase arrest accompanied by
downregulation of cyclin dependant kinase (CDK)2, CDK4, Cyclin D1
and Cyclin D3 expression and the upregulation in the protein level
of p21CDKN1A. By contrast, Al-Salahi et al (39) demonstrated that TAF273 from the
E. longifolia root methanolic extract exerted a potent
cytotoxic effect on K562 cells via cell cycle arrest at the
G1 and S phases. In the present study, the ability of F2
to regain the cell population at the G0/G1
phase could be attributed to the revival of the F2-treated Leydig
cells following 96 h of treatment. Therefore, it is reasonable to
suggest that the G0/G1 and S phases are the
critical factors to explore in improving the viability of
F2-treated Leydig cells.
At the molecular level, CDKL2 expression was found
to be significantly affected in Leydig cells treated with 1.0 mg/ml
F2 for 72 and 96 h compared with that of respective controls,
implying the essential role of CDKL2 in regulating the viability of
F2-treated Leydig cells. However, the detection of CDKL2 expression
does not clearly define the mechanisms that were being studied. The
CDKL2 expression profiles in Leydig cells treated with 0.1 and 1.0
mg/ml F2 for 72 and 96 h did not clearly reveal the mechanisms of
S-phase cell cycle arrest and reduced cell viability in the cells.
Leydig cells treated with ≤1.0 mg/ml F2 for 72 h were not revived
compared with those treated with the same concentrations of F2 for
96 h, which started to revive. In addition, the populations of
Leydig cells treated with 0.1 mg/ml F2 for 96 h in
G0/G1 and S phases of the cell cycle were not
significantly different compared with those of the respective
controls, indicative of cell revival. Compared with the populations
of Leydig cells treated with 1.0 mg/ml F2 for 96 h at the
G0/G1 and S phases of the cell cycle, there
was a significant difference compared with those of the respective
controls. CDKL2 is a cdc2-related serine/threonine protein kinase
that is postnatally expressed in various brain regions, including
the cerebral cortex, entorhinal cortex, hippocampus, amygdala and
dorsal thalamus (40). The
upregulation of CDKL2 in these regions suggests that it has a role
in cognition and emotion. Although studies have elucidated the
physiological role of CDKL2 in mice models lacking Cdkl2,
detailed studies on Cdkl2 remain rare. CDKL2 may also be
involved in gastric cancer progression, where a previous study
exploring the clinical effect of CDKL2 on gastric cancer indicated
that the loss of CDKL2 was positively associated with several
clinical and pathological characteristics (40–42).
Patients with low CDKL2 expression levels exhibited significantly
worse disease-free and overall survival rates compared with those
with high CDKL2 expression levels (41). Cellular studies revealed that CDKL2
overexpression impaired the proliferation and invasion of gastric
cancer cells (41); therefore, the
loss of CDKL2 may serve as a biomarker for predicting the outcome
in patients with gastric cancer and is a potential therapeutic
target for gastric cancer treatment. To the best of our knowledge,
only one study has previously demonstrated that the expression of
CDKL2 was upregulated in human breast cancer tissues and cells
compared with that in normal breast tissues and cells (43).
CDKL2 is similar to another member of the
cyclin-dependent kinase family, CDKL1, and is presumably derived
from an early vertebrate duplication (41). Reanalysis of genome-wide
association studies suggested that CDKL2 may contribute to breast
cancer, where human mammary gland epithelial cells expressing CDKL2
demonstrated increased epithelial-mesenchymal transition and stem
cell properties, which were obtained from the activation of a
positive feedback loop comprising of Zinc finger E-box-binding
homeobox 1, E-cadherin and β-catenin (43). In addition, CDKL2 promoted
xenograft proliferation and metastasis in vivo (41,43).
In particular, CDKL2 expression was found to be upregulated in
mesenchymal breast cancer cells compared with that in epithelial
cells, where its upregulation is negatively associated with
disease-free survival (41,43).
The present study demonstrated that the reduced growth of Leydig
cells following treatment with high concentrations of F2 may be due
to the downregulation of CDKL2 expression. Therefore, further
investigation on the regulation of this specific molecular target
and its associated targets, such as CBL and Sphkap, which may act
in concert with CDKL2 in F2-treated Leydig cells, is warranted.
In conclusion, the present study demonstrated the
elevation of testosterone production in Leydig cells treated with
the standardised E. longifolia extract, F2. However, this
phenomenon was accompanied by the reduced viability of treated
cells. Also, the present study revealed CDKL2 as an important
molecular target for the regulation of these F2-dependent
mechanisms. However, further studies are warranted to improve
understanding of the growth inhibitory effect caused by F2
treatment in Leydig cells.
Acknowledgements
Not applicable.
Funding
Universiti Sains Malaysia Fellowship Scheme
supported Nor Amira Khurshid Ahmed (grant no. P-NFM0006/16R). The
present study was supported by the Ministry of Agriculture and
Agro-based Industry, Malaysia, under the National Key Economic
Areas (NKEA) Research Grant Scheme (grant no.
304/PFARMASI/650733/k123).
Availability of data and materials
The datasets used and/or analysed during the current
study are available from the corresponding author on reasonable
request.
Authors' contributions
HS and CL contributed to the conception of the
present study. LY, KY and CL made substantial contributions to the
design of the present study. NA and LK performed the experiments
under technical support provided by GP, HS and CL. NA, GP and KY
interpreted the results, drafted and revised the manuscript. All
authors read and approved the final manuscript.
Ethics approval and consent to
participate
Not applicable.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
Glossary
Abbreviations
Abbreviations:
|
EN
|
eurycomanone
|
|
FM
|
formestane
|
|
MTT
|
3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide
|
|
OD
|
optical density
|
|
PI
|
propidium iodide
|
|
WT
|
whole transcript
|
|
ss
|
single-stranded
|
|
DEGs
|
differentially expressed genes
|
|
CDKL2
|
cyclin-dependent kinase-like 2
|
|
CBL
|
Casitas B-lineage lymphoma
|
|
Sphkap
|
SPHK1 interactor-AKAP
domain-containing
|
|
DHEA
|
dehydroepiandrosterone
|
References
|
1
|
Ekor M: The growing use of herbal
medicines: Issues relating to adverse reactions and challenges in
monitoring safety. Front Pharmacol. 4:1772014. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Nasri H, Baradaran A, Shirzad H and
Rafieian-Kopaei M: New concepts in nutraceuticals as alternative
for pharmaceuticals. Int J Prev Med. 5:1487–1499. 2014.PubMed/NCBI
|
|
3
|
Rehman SU, Choe K and Yoo HH: Review on a
traditional herbal medicine, Eurycoma longifolia Jack
(Tongkat Ali): Its traditional uses, chemistry, evidence-based
pharmacology and toxicology. Molecules. 21:3312016. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Yuan H, Ma Q, Ye L and Piao G: The
traditional medicine and modern medicine from natural products.
Molecules. 21:5592016. View Article : Google Scholar
|
|
5
|
Chan KL, Choo CY, Morita H and Itokawa H:
High performance liquid chromatography in phytochemical analysis of
Eurycoma longifolia. Planta Med. 64:741–745. 1998.
View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Chan KL, Low BS, Teh CH and Das PK: The
effect of Eurycoma longifolia on sperm quality of male rats.
Nat Prod Commun. 4:1331–1336. 2009.PubMed/NCBI
|
|
7
|
Meng D and Li X, Han L, Zhang L, An W and
Li X: Four new quassinoids from the roots Eurycoma
longifolia jack. Fitoterapia. 92:105–110. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Low BS, Das PK and Chan KL: Standardized
quassinoid-rich Eurycoma longifolia extract improved
spermatogenesis and fertility in male rats via the
hypothalamic-pituitary-gonadal axis. J Ethnopharmacol. 145:706–714.
2013. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Mohamed AN, Vejayan J and Yusoff MM:
Review on Eurycoma longifolia pharmacological and
phytochemical properties. J Appl Sci. 15:831–844. 2015. View Article : Google Scholar
|
|
10
|
Farouk AE and Benafri A: Antibacterial
activity of Eurycoma longifolia Jack. A Malaysian medicinal
plant. Saudi Med J. 28:1422–1424. 2007.PubMed/NCBI
|
|
11
|
Nurhanan MY, Azimahtol Hawariah LP, Mohd
Ilham A and Mohd Shukri MA: Cytotoxic effects of the root extracts
of Eurycoma longifolia Jack. Phytother Res. 19:994–996.
2005. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Wernsdorfer WH, Ismail S, Chan KL,
Congpuong K and Wernsdorfer G: Activity of Eurycoma
longifolia root extract against plasmodium falciparum in vitro.
Wien Klin Wochenschr. 121 (Suppl 3):S23–S26. 2009. View Article : Google Scholar
|
|
13
|
Chuen CS and Azimahtol HLP: Eurycomanone
exerts antiproliferative activity via apoptosis upon MCF-7 cells.
Proceedings of the 4th Annual Seminar of National Science
Fellowship. BIO04:2004.
|
|
14
|
Kuo PC, Damu AG, Lee KH and Wu TS:
Cytotoxic and antimalarial constituents from the roots of
Eurycoma longifolia. Bioorg Med Chem. 12:537–544. 2004.
View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Noor MM, Mohd Nor AHS and Hassan LC: The
effect of Eurycoma longifolia Jack (Tongkat Ali) on sexual
behaviour and sperm quality in rats. Malaysia J Pharm Sci. 2:53–60.
2004.
|
|
16
|
Park S, Nhiem NX, Kiem PV, Minh CV, Tai
BH, Kim N, Yoo HH, Song JH, Ko HJ and Kim SH: Five new quassinoids
and cytotoxic constituents from the roots of Eurycoma
longifolia. Bioorg Med Chem Lett. 24:3835–3840. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Ang HH, Ikeda S and Gan EK: Evaluation of
the potency activity of aphrodisiac in Eurycoma longifolia
jack. Phytother Res. 15:435–436. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Shuid AN, El-arabi E, Effendy NM, Razak
HS, Muhammad N, Mohamed N and Soelaiman IN: Eurycoma
longifolia upregulates osteoprotegerin gene expression in
androgen-deficient osteoporosis rat model. BMC Complement Altern
Med. 12:1522012. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Zhari I, Norhayati I and Jaafar L:
Malaysian herbal monograph. Malaysian Monograph Committee. 1:67–70.
1999.
|
|
20
|
Bhat R and Karim AA: Tongkat ali
(Eurycoma longifolia Jack): A review on its ethnobotany and
pharmacological importance. Fitoterapia. 81:669–679. 2010.
View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Foley GL: Overview of male reproductive
pathology. Toxicol Pathol. 29:49–63. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Erasmus N, Solomon MC, Fortuin KA and
Henkel RR: Effect of Eurycoma longifolia Jack (Tongkat Ali)
extract on human spermatozoa in vitro. Andrologia. 44:308–314.
2012. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Isidori AM, Buvat J, Corona G, Goldstein
I, Jannini EA, Lenzi A, Porst H, Salonia A, Traish AM and Maggi M:
A critical analysis of the role of testosterone in erectile
function: From pathophysiology to treatment-a systematic review.
Eur Urol. 65:99–112. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Talbott SM, Talbott JA, George A and Pugh
M: Effect of Tongkat Ali on stress hormones and psychological mood
state in moderately stressed subjects. J Int Soc Sports Nutr.
10:282013. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Miyake K, Tezuka Y, Awale S, Li F and
Kadota S: Quassinoids from Eurycoma longifolia. J Nat Prod.
72:2135–2140. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Low BS, Choi SB, Abdul Wahab H, Das PK and
Chan KL: Eurycomanone, the major quassinoid in Eurycoma
longifolia root extract increases spermatogenesis by inhibiting
the activity of phosphodiesterase and aromatase in steroidogenesis.
J Ethnopharmacol. 149:201–207. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Solomon MC, Erasmus N and Henkel RR: In
vivo effects of Eurycoma longifolia Jack (Tongkat Ali)
extract on reproductive functions in the rat. Andrologia.
46:339–348. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Jayusman PA, Mohamed IN, Alias E, Mohamed
N and Shuid AN: The effects of quassinoid-rich Eurycoma
longifolia extract on bone turnover and histomorphometry
indices in the androgen-deficient osteoporosis rat model.
Nutrients. 10:7992018. View Article : Google Scholar
|
|
29
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(-Delta Delta C(T)) method. Methods. 25:402–408. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
30
|
The Gene Ontology Consortium, . The gene
ontology resource: 20 years and still GOing strong. Nucleic Acids
Res. 47:D330–D338. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Fokunang CN, Ndikum V, Tabi OY, Jiofack
RB, Ngameni B, Guedje NM, Tembe-Fokunang EA, Tomkins P, Barkwan S,
Kechia F, et al: Traditional medicine: Past, present and future
research and development prospects and integration in the national
health system of cameroon. Afr J Tradit Complement Altern Med.
8:284–295. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Wachtel-Galor S and Benzie IFF: Herbal
medicine: An introduction to its history, usage, regulation,
current trends, and research needs. Herbal Medicine: Biomolecular
and Clinical Aspects. 2nd edition. CRC Press /Taylor & Francis;
Boca Raton, FL: 2011
|
|
33
|
Thu HE, Hussain Z, Mohamed IN and Shuid
AN: Eurycoma longifolia, a promising suppressor of
RANKL-induced differentiation and activation of osteoclasts: An in
vitro mechanistic evaluation. J Ayurveda Integr Med. 10:102–110.
2019. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Elledge SJ: Cell cycle checkpoints:
Preventing an identity crisis. Science. 274:1664–1672. 1996.
View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Campbell JB, Reece JB, Urry LA, Cain ML,
Wasserman SA, Minorsky PV and Jackson RB: Biology. 8th edition.
Pearson Eucation Inc.; San Francisco, CA: 2008
|
|
36
|
Xu W, Zhu Q, Zhang B, Liu S, Dai X, Gao C,
Gao L and Cui Y: Protective effect of calretinin on testicular
leydig cells via the inhibition of apoptosis. Aging (Albany NY).
9:1269–1279. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Liu L, Wang D, Li L, Ding X and Ma H:
Dehydroepiandrosterone inhibits cell proliferation and improves
viability by regulating S phase and mitochondrial permeability in
primary rat leydig cells. Mol Med Rep. 14:705–714. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Tong KL, Chan KL, AbuBakar S, Low BS, Ma
HQ and Wong PF: The in vitro and in vivo anti-cancer activities of
a standardized quassinoids composition from Eurycoma
longifolia on LNCaP human prostate cancer cells. PLoS One.
10:e01217522015. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Al-Salahi OSA, Ji D, Majid AMSA, Kit-Lam
C, Abdullah WZ, Zaki A, Din SKKJ, Yusoff NM and Majid ASA:
Anti-tumor activity of Eurycoma longifolia root extracts
against K-562 cell line: In vitro and in vivo study. PLoS One.
9:e838182014. View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Gomi H, Sassa T, Thompson RF and Itohara
S: Involvement of cyclin-dependent kinase-like 2 in cognitive
function required for contextual and spatial learning in mice.
Front Behav Neurosci. 4:172010.PubMed/NCBI
|
|
41
|
Fang CL, Uen YH, Chen HK, Hseu YC, Lin CC,
Hung ST, Sun DP and Lin KY: Loss of cyclin-dependent kinase-like 2
predicts poor prognosis in gastric cancer, and its overexpression
suppresses cells growth and invasion. Cancer Med. 7:2993–3002.
2018. View Article : Google Scholar
|
|
42
|
Zhang J, Su G, Lin Y, Meng W, Lai JKL,
Qiao L, Li X and Xie X: Targeting cyclin-dependent kinases in
gastrointestinal cancer therapy. Discov Med. 27:27–36.
2019.PubMed/NCBI
|
|
43
|
Li L, Liu C, Amato RJ, Chang JT, Du G and
Li W: CDKL2 promotes epithelial-mesenchymal transition and breast
cancer progression. Oncotarget. 5:10840–10853. 2014. View Article : Google Scholar : PubMed/NCBI
|