Introduction
Stroke is the second leading cause of death and
disability worldwide (1). Ischemic
stroke, which accounts for 80% of all strokes, usually causes
severe neuronal damage and secondary neuronal death (2). Cerebral ischemia and reperfusion
injury are complex pathological processes that involve
inflammation, apoptosis and neuronal stress that rely on
progressive changes in protein expression (3,4).
Currently, there are very few effective drugs that can treat
cerebral ischemia and reperfusion injury in the clinic (5). Medicinal plants, such as the popular
traditional Chinese herb Panax notoginseng, also known as
Sanqi, are often used to treat cerebrovascular and cardiovascular
disorders in Traditional Chinese medicine (6). Panax notoginseng saponins
(PNS) can be extracted from Sanqi, and its use can result in an
effective neuroprotective effect and promote stroke recovery
(7,8). Compounds in the PNS mainly include
ginsenoside Rb1, ginsenoside Rd, ginsenoside Rg1,notoginsenoside R1
and ginsenoside Re (9–11). A previous study found that
ginsenoside Rb1 had anti-apoptotic, anti-oxidative and
anti-inflammatory effects that ameliorated brain water content,
promoted bioactivities associated with neurogenesis potential and
had a neuroprotective effect (12). Ginsenoside Rd is known to attenuate
ischemic stroke-induced damage through an anti-oxidative stress and
anti-inflammatory response (13).
Ginsenoside Rg1 treatment attenuated infarct volume and
neurological function score, which demonstrated that it is a
potential neuroprotective drug that could be used to treat ischemic
stroke (14). A previous study
found that PNS treatment attenuated oxygen-glucose
deprivation/reoxygenation-induced injury in human SH-SY5Y cells
(15). PNS treatment has also been
demonstrated to inhibit the secretion of inflammatory factors,
regulate Nogo-A, NgR and p75NGF expression, and to regulate the
RhoA/ROCK2 pathway in permanent middle cerebral artery occlusion
(MCAO) model rats to prevent neurological damage (8,16,17).
However, the molecular mechanisms underlying how PNS protects
against ischemic injury remain unclear.
The purpose of the present study was to determine
the gene expression profile in PNS-treated MCAO rats after ischemic
stroke using mRNA deep sequencing, which may provide insight into
the molecular mechanisms involved in PNS treatment of ischemic
stroke.
Materials and methods
Treatments
PNS was purchased from Guangxi Wuzhou Pharmaceutical
Group Co., Ltd., (China Food and Drug Administration approval no.
Z20025652), which was mostly made up of ginsenoside Rb1 (29.7%),
ginsenoside Rd (7.3%), ginsenoside Rg1 (28.0%), notoginsenoside R1
(6.9%) and ginsenoside Re (3.8%).
Establishing MCAO model and treatment
conditions
The present study obtained approval from the Ethics
Committee of Youjiang Medical College for Nationalities
Institutional Review Board. A total of 60 male Sprague-Dawley (SD)
rats of a specific pathogen-free grade (age, 7-weeks; weight,
200±20 g) were purchased from Changsha Tianqin Biotechnology Co.,
Ltd. [License Number: SCXK(Xiang)2014-0011]. SD rats were housed
under 50–60% humidity, 12/12 h light/darkness, and 20±2°C room
temperature with free access to drinking water and food. SD rats
were randomly divided into three groups: Control group (rats
without any treatment, n=10), Sham operation group (sham, n=10) and
MCAO group (model, n=40). SD rats were fasted overnight, but
allowed ad libitum access to water in the Youjiang Medical
College [License Number: SYXK(Gui)2017-0004]. Rats were
anesthetized for surgery by inhalation of isoflurane (4% isoflurane
for induction and 2% for maintenance). After 3 min, SD rats were
checked to ensure they were fully anesthetized, this was performed
using the following methods: i) induction box was shaken gently, if
the rat's body fell over to the side-lying position and it did not
try to restore its prone position, it indicated that the animal was
fully anesthetized; and ii) the toes of the rat were lightly
clamped by tweezers, if the rat did not react after pinching, the
rat was judged to be in a deep anesthesia. Then, MCAO rat models
were established as previously described by Longa et al
(18). Briefly, the common carotid
artery (CCA), internal carotid artery (ICA) and external carotid
artery (ECA) were carefully exposed after anesthesia. Then, the CCA
and ECA were tied with a loose surgical knot. The ICA was tied with
a loose knot with a silk suture. A small incision was made in the
ECA and a silicon-coated monofilament was introduced through the
CCA into the ICA 20 mm beyond the carotid bifurcation to occlude
the MCA for 1 h. Following surgery, the rats were housed
individually and closely monitored for changes in behavior and
vital signs. Neurological scores were assessed when rats awoke
according to neurological deficit scores, where a score of 3–5
indicates that the model was successfully established (18). Then, MCAO rats were randomly
divided into four groups: i) MCAO group; ii) 50 mg/kg PNS treatment
group; iii) 100 mg/kg PNS treatment group; and iv) 150 mg/kg PNS
treatment group. PNS treatment groups were given PNS by
intragastric treatment once every 2 days. The rats in the control
and sham groups were treated with the same volume of normal saline.
Rats in the sham groups underwent an operation to expose the CCA,
ICA and ECA after anesthesia but the ligation was not performed.
Rats in the control group did not undergo surgery.
In order to minimize pain, the following
characteristics were considered humane endpoints to the study that
required immediate intervention: Infection at the surgical site,
wound dehiscence, rapid weight loss (>20% body weight loss),
becoming cachectic, self-harming, biting or aggressiveness, and
difficulty eating, drinking or moving around freely. No rat was
euthanized throughout the experiment. After treatment for 7 days,
the rats were euthanized with pentobarbital (intraperitoneal
injection, 120 mg/kg). Cervical dislocation was used to confirm
death.
Evaluation of neurological deficit and
2,3,5-triphenyltetrazolium chloride (TTC) staining
After treatment, the neurological deficit was
evaluated by two researchers who were unaware of the groupings
using a neurological deficit score as previously described
(18,19). Then, the extent and localization of
the cerebral lesion were determined by TTC staining. Briefly, the
brain was dissected in the coronal plane at the bifurcation of MCA
and anterior cerebral artery, and the cerebral lesion was
visualized by TTC staining. Briefly, 2-mm thick brain tissue
sections were stained with 2% TTC solution at 37°C for 30 min,
followed by fixation with 4% paraformaldehyde. TTC-stained sections
were photographed with a D5600 camera (Nikon). The digital images
were analyzed using Image-Pro Plus 6.0 software (Media Cybernetics,
Inc.).
mRNA sequencing and differential gene
analysis
Total RNA from sham, MCAO and 100 mg/kg PNS
treatment groups was extracted using TRIzol®
(Invitrogen; Thermo Fisher Scientific, Inc.). RNA concentration was
measured using a NanoDrop™ 2000 spectrophotometer (Thermo Fisher
Scientific, Inc.) at 260/280 nm and a Bioanalyzer 4200 (Agilent
Technologies, Inc.). The libraries were prepared using a VAHTS
mRNA-seq v2 Library Prep Kit for Illumina® (cat. no.
NR601-01; Vazyme Biotech Co., Ltd.). The reverse transcription was
conducted at 25°C for 10 min, 42°C for 15 min, then 70°C for 15
min. Adapters were ligated at the 3′ and 5′ ends, and templates
were amplified using PCR. The PCR products were further purified
using VAHTS™ RNA Clean Beads (cat. no. N412-01; Vazyme Biotech Co.,
Ltd.) according to the manufacturer's protocol and used for
sequencing. mRNAs were analyzed by deep sequencing using a HiSeq X
Ten Reagent kit v2.5 (cat. no. FC-501-2521; Illumina, Inc.) on an
Illumina HiSeq X sequencing platform (pair-ended; 150-bp reads;
Illumina, Inc.).
The sequencing data was filtered using SOAPnuke
(version 1.5.2), and clean reads were obtained after removing: i)
reads containing sequencing adapters; ii) reads with low-quality
base ratio >20% (base quality ≤5); and iii) reads with unknown
base (N base) ratio >5%. Differentially expressed genes were
selected as having >two-fold differences between their
geometrical mean expression in the compared groups, and a
statistically significant P-value (<0.05) by analysis of DEseq2.
Gene Ontology (GO) analysis of differentially expressed genes was
performed with an R package with cluster profiler using P<0.05
to define statistically enriched GO categories (20–22).
Pathway analysis was used to determine the significant pathway of
the differential genes according to Kyoto Encyclopedia of Genes and
Genomes (KEGG) database (http://www.genome.jp/kegg/) (23–25).
Protein-protein interactions (PPI) were analyzed with the Search
Tool for the Retrieval of Interacting Genes/Proteins (STRING;
http://string-db.org) (26). Hub genes were analyzed by CytoHubba
app in Cytoscape (27).
Reverse transcription-quantitative PCR
(RT-qPCR)
cDNA synthesis was carried out using PrimeScript™ RT
reagent kit (Takara Biotechnology Co., Ltd.). The conditions of
reverse transcription were as follows: 30°C for 10 min and 42°C for
60 min, followed by 85°C for 10 min. RT-qPCR was performed using
the SYBR® Premix ExTaq™ II kit (Takara Biotechnology
Co., Ltd.) on a 7500 Real-Time PCR System (Applied Biosystems;
Thermo Fisher Scientific, Inc.). The RT-qPCR thermocycling
conditions consisted of an initial denaturation at 95°C for 10 min,
followed by 40 cycles at 95°C for 15 sec and 65°C for 32 sec. The
relative mRNA expression levels were calculated using the
2−ΔΔCq method (28).
Glyceraldehyde 3-phosphate dehydrogenase (GAPDH) was used as the
internal reference gene. The sequences of primers are shown in
Table I.
 | Table I.Primer sequences for reverse
transcription-quantitative PCR. |
Table I.
Primer sequences for reverse
transcription-quantitative PCR.
| Gene | Forward
(5′→3′) | Reverse
(5′→3′) |
|---|
| UBE2V2 |
GCGGTGCGTCTGGTAGAA |
CGTGCATCCACCATTCCACT |
| SUMO1 |
GGTGAATCCACCGACACCAT |
CTCACTGCTGTCCTGTCCAA |
| SSB |
GCTGGGTACCTTTGGAAACA |
TATCATCAAGGGTGGCATCA |
| FAU |
GCGGGTAAGTAGCCAACATG |
AGCCTGCCAGAAGCACGAC |
| CEP290 |
TCCAGTGGGCTACAGAGCAA |
CTTTGTCAGGCTGTGGACCT |
| POLR2K |
CCGGTAGTGTCTCTTGCTTC |
CGAGCATCAAAAACCACCAATCT |
| CUL4B |
AGTGACTCCGGCAAAAAGAGG |
GGCGCTCTTGATTGGAGGTT |
| MATR3 |
CCAAGAGGAAATCTGGGTGCT |
ATGTGAACAACTCGGCTGGT |
| KDR |
TCACGGTTGGGCTACTGC |
AGACCTTCTGCCATCACG |
| PRPF40A |
ACACTGGAGAGGAACCACCT |
CATTGGAGGGTACCCGCTTT |
Statistical analysis
Experimental data are presented as the mean ±
standard deviation. All the statistical analyses were performed
using SPSS 19.0 statistical software (IBM Corp.) with one-way ANOVA
followed by a Tukey's post hoc test. P<0.05 was considered to
indicate a statistically significant difference.
Results
Aberrant mRNA expression profiles in
MCAO rats treated with PNS
Initially, no neurological deficit and infarction
were observed in the Control and Sham groups. Compared with the
Model group, it was found that PNS treatment attenuated the
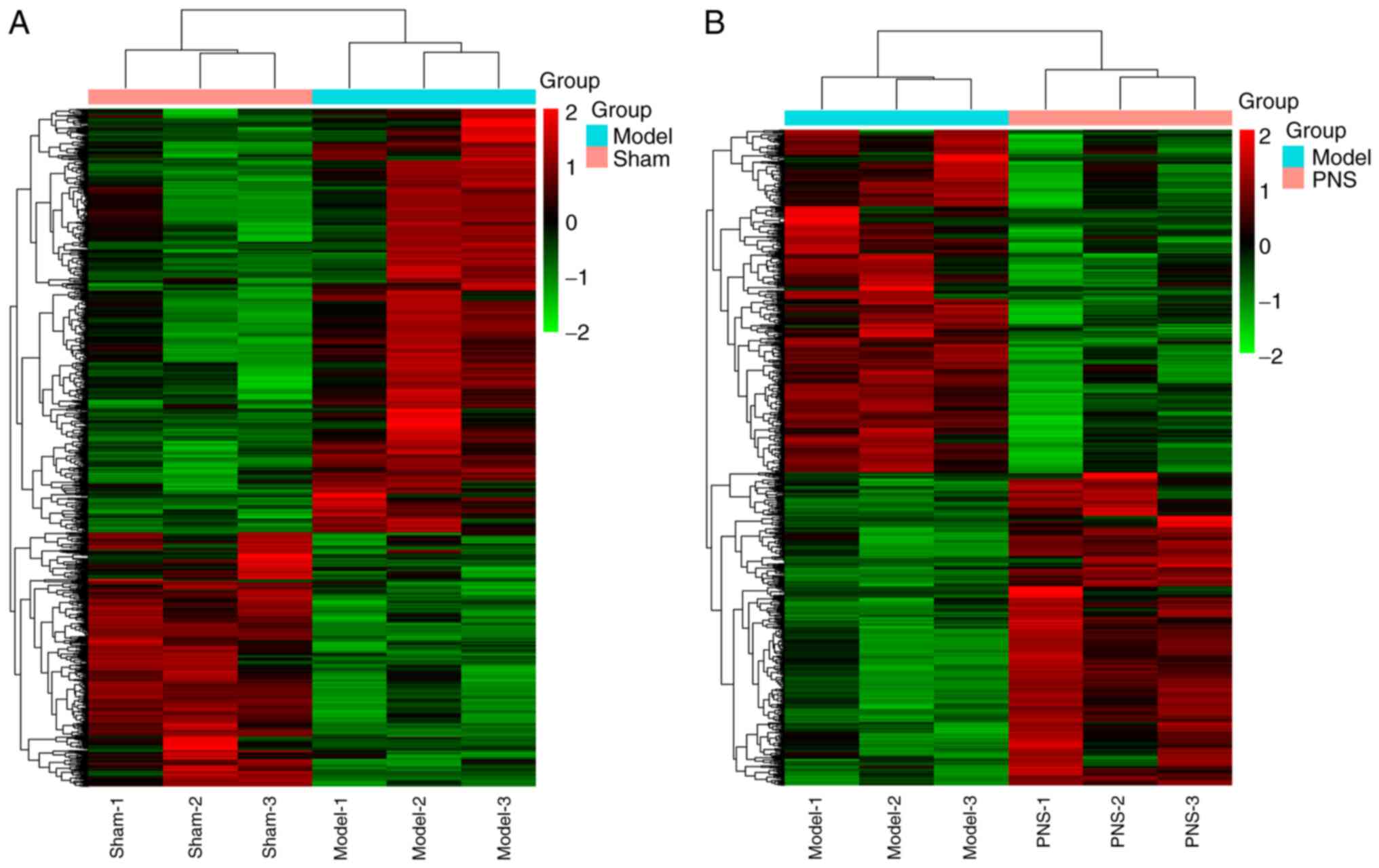
neurological deficit and infarction area (Fig. S1). Additionally, aberrant mRNA
expression profiles were analyzed between the Sham and Model
groups, and between the Model and PNS-treated groups. According to
the cut-off criteria 1,104 genes of interest were selected, where
690 genes were upregulated and 414 were downregulated in the Model
group compared with the Sham group. Additionally, 817 genes of
interest were selected, where 390 genes were upregulated and 427
were downregulated in the PNS-treated group compared with the Model
group. Aberrant mRNA expression profiles between the Sham and Model
groups, and between the Model and PNS-treated groups were
identified using hierarchical cluster analysis of the data
(Fig. 1). The 10 most upregulated
and downregulated genes between the Sham and Model groups, and
between the Model and PNS-treated groups are shown in Table II.
 | Table II.Top 10 upregulated and downregulated
aberrant mRNAs between the Sham group and Model group and between
the Model group and PNS group. |
Table II.
Top 10 upregulated and downregulated
aberrant mRNAs between the Sham group and Model group and between
the Model group and PNS group.
| A, Differentially
expressed mRNAs between Sham and Model groups |
|---|
|
|---|
| Gene |
log2Fold |
|---|
| Shox2 | 9.66 |
| LOC108348145 | 9.67 |
| Gns | 12.41 |
| Selenbp1 | 21.56 |
| Ppcdc | 21.71 |
| LOC100911572 | 21.74 |
| LOC100912537 | 22.64 |
| LOC103690164 | 22.72 |
| Gatd1 | 23.39 |
| LOC103692166 | 24.71 |
| Calcoco1 | −24.61 |
| LOC108348078 | −24.52 |
| Wbp11 | −23.43 |
| Rufy2 | −23.32 |
| Prmt6 | −22.25 |
| LOC100912585 | −21.77 |
| Map2k3 | −21.53 |
| AABR07035479.1 | −21.47 |
| Pidd1 | −21.38 |
| AABR07002870.2 | −21.20 |
|
| B,
Differentially expressed mRNAs between Model and PNS
groups |
|
| Gene |
log2Fold |
|
| Calcoco1 | 24.66 |
| Rufy2 | 23.58 |
| Wbp11 | 23.39 |
| AC109542.1 | 21.64 |
| AABR07035479.1 | 20.63 |
| Cttn | 12.68 |
| Rsl1d1l1 | 11.91 |
| LOC103690108 | 10.90 |
| Knop1 | 9.32 |
| Map2k3 | 9.10 |
| LOC100911572 | −21.78 |
| Isl1 | −21.95 |
| LOC100910990 | −22.56 |
| LOC100912537 | −22.67 |
| LOC103690164 | −22.78 |
| Gatd1 | −23.43 |
| LOC100911881 | −23.91 |
| Cpne1 | −24.71 |
| LOC103692166 | −25.05 |
| LOC108348101 | −25.78 |
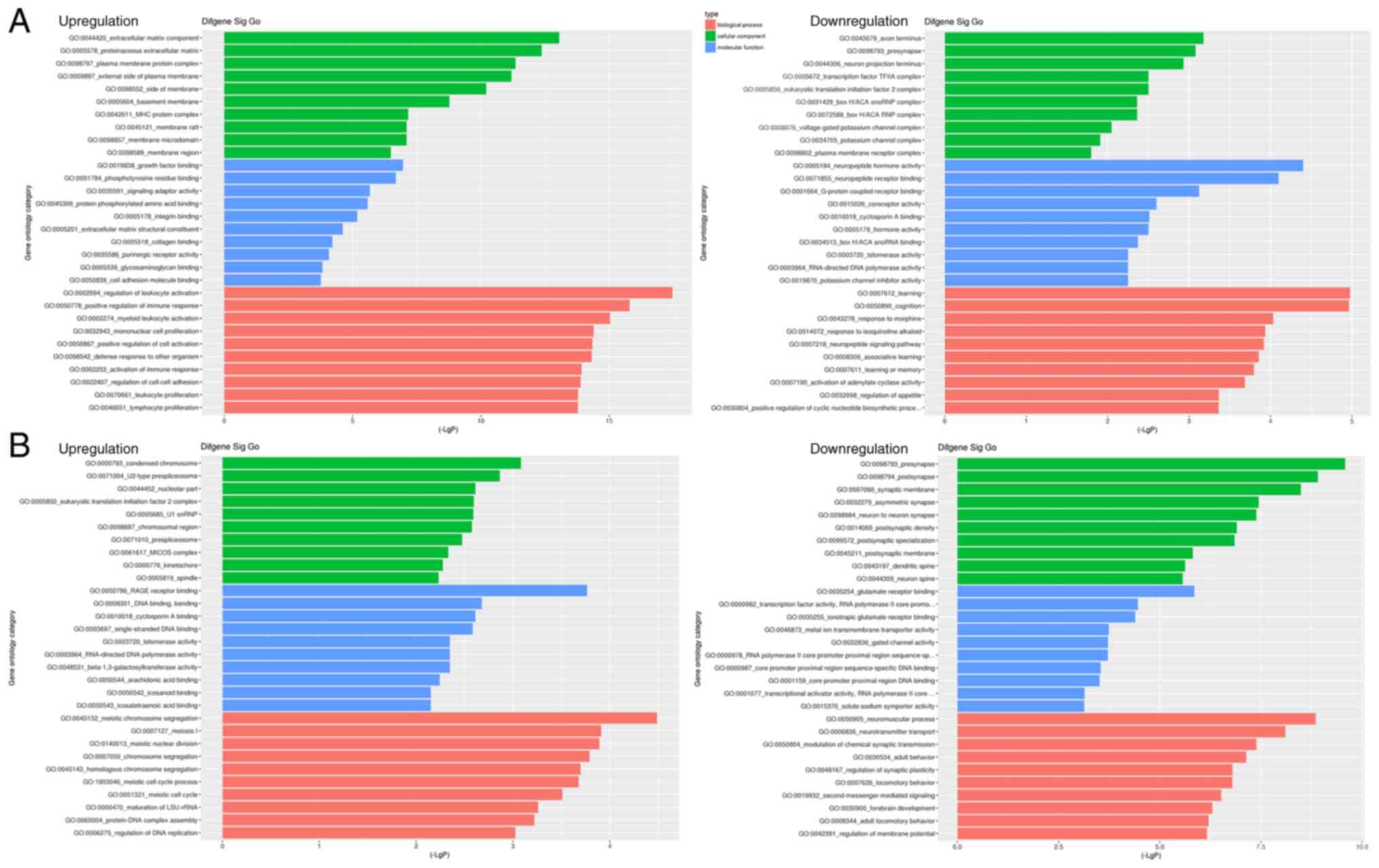
GO analysis and KEGG pathway
analysis
GO functional enrichment analysis revealed that when
comparing the Sham and Model groups, the upregulated genes involved
3,232 ‘biological processes’, 477 ‘cellular components’ and 830
‘molecular functions’. The downregulated genes involved 3,240
‘biological processes’, 350 ‘cellular components’ and 561
‘molecular functions’. When comparing the Model and PNS-treated
groups, GO functional enrichment analysis revealed that the
upregulated genes involved 2,610 ‘biological processes’, 352
‘cellular components’ and 445 ‘molecular functions’. The
downregulated genes involved 3,963 ‘biological processes’, 384
‘cellular components’ and 723 ‘molecular functions’. The top 10
significant ‘biological process’, ‘cellular component’ and
‘molecular functions’ identified by GO enrichment analysis for
differentially expressed genes between the Sham and Model groups,
and between the Model and PNS-treated groups are shown in Fig. 2. GO analysis suggested that genes
upregulated following PNS treatment were primarily related to
‘condensed chromosome’, ‘RAGE receptor binding’ and ‘meiotic
chromosome segregation’. Downregulated genes after PNS treatment
were mainly involved in ‘presynapse’, ‘glutamate receptor binding’,
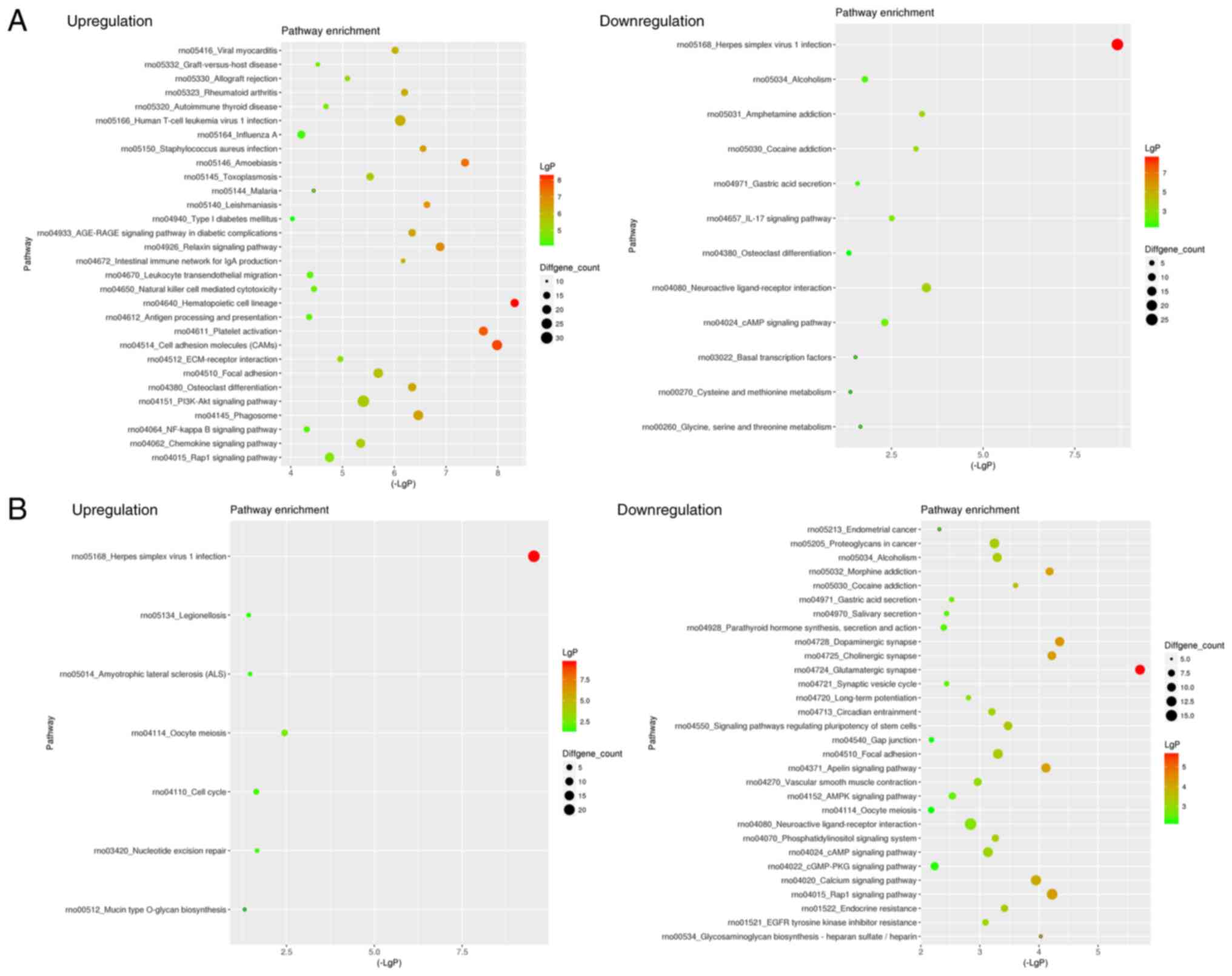
‘neuromuscular process’. KEGG analysis was performed to enrich the
signaling pathways of differentially expressed genes between the
Sham group and Model group or between the Model group and
PNS-treated groups. KEGG analysis revealed that the upregulated
genes between the Sham and Model groups were involved in 258
signaling pathways, whereas the downregulated genes between the
Sham and Model groups were involved in 164 signaling pathways.
Additionally, KEGG analysis revealed that the upregulated genes
between the Model and PNS-treated groups were involved in 258
signaling pathways, whereas the downregulated genes between the
Model and PNS-treated groups were involved in 226 signaling
pathways. The top enrichment results are shown in Fig. 3. KEGG analysis indicated that
upregulated genes after PNS treatment were primarily involved in
the ‘Herpes simplex virus 1 infection’, ‘Oocyte meiosis’ and ‘Cell
cycle’, whereas downregulated genes after PNS treatment were
related to ‘Glutamatergic synapses’, ‘Rap1 signaling pathway’ and
‘Calcium signaling pathway’.
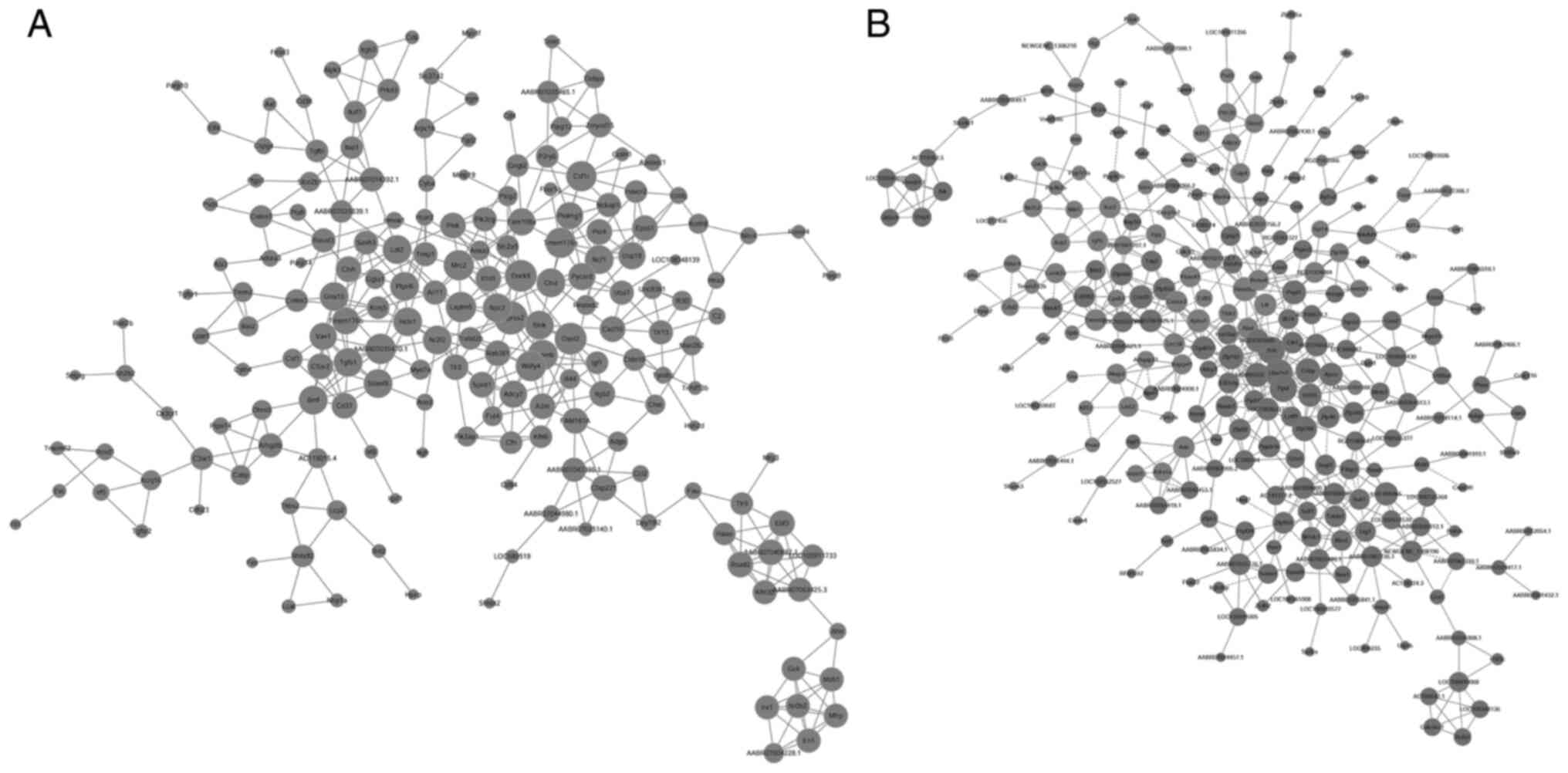
PPI network analysis is important for understanding
the interactions that occur between the proteins. The results of
the PPI network analysis are shown in Fig. 4. Based on PPI network analysis,
transmembrane protease, serine 2 (Tmprss2), dedicator of
cytokinesis 8 (Dock8), 2′-5′ oligoadenylate synthase
(Oasl2), mannose receptor, C type 2 (Mrc2),
glycoprotein (transmembrane) nmb (Gpnmb), lysosomal protein
transmembrane 5 (Laptm5), colony stimulating factor 1
receptor (Csf1r), transmembrane protein 176A
(Tmem176a), AABR07035470.1 and WDFY family member 4
(Wdfy4) were the top 10 differentially expressed genes
between the Sham and Model groups. Furthermore, the top 10
differentially expressed genes between the Model group and
PNS-treated groups were ubiquitin conjugating enzyme E2 variant 2
(UBE2V2), small RNA binding exonuclease protection factor La
(SSB), peptidylprolyl isomerase D (Ppid), CCHC-type
zinc finger, nucleic acid binding protein (Cnbp), Zinc
finger protein 788 (Zfp788), zinc finger protein 182
(Zfp182), AABR07035107.1, polyribonucleotide
nucleotidyltransferase 1 (Pnpt1), required for meiotic
nuclear division 5 homolog A (Rmnd5a) and C1GALT1-specific
chaperone 1 (C1galt1c1).
Validating mRNA expression by
RT-qPCR
To analyze the aberrant expression of mRNAs that
have important roles in ischemic stroke and the underlying
mechanisms of PNS treatment, the overlapping genes of interest
identified between Sham and Model groups, and between the Model and
PNS-treated groups were analyzed using Venn diagrams, which
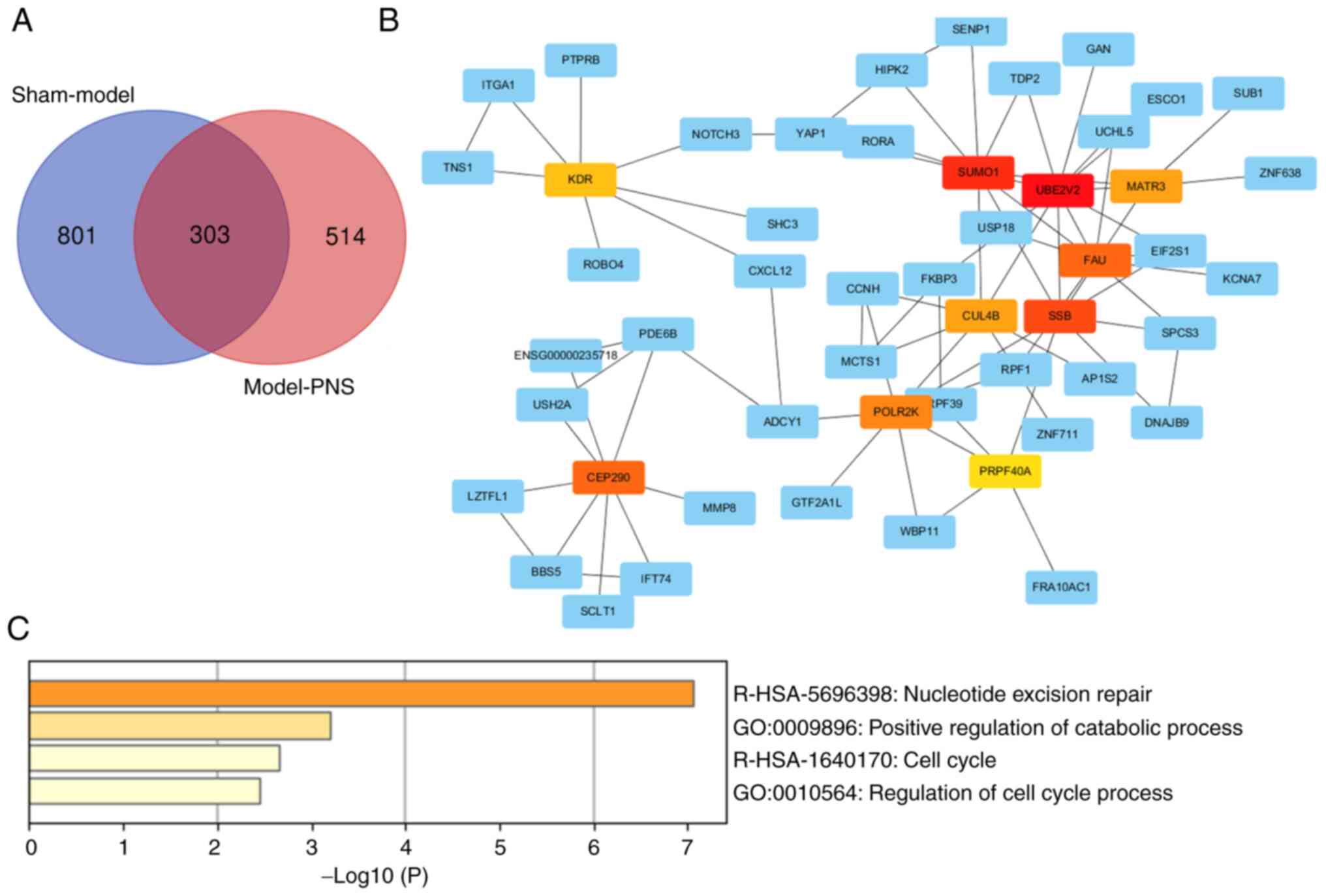
demonstrated 303 overlapping genes (Fig. 5A). These 303 mRNAs were analyzed
using STRING, and the hub genes were analyzed using CytoHubba to
identify maximum neighborhood component (MNC) centrality in the
Cytoscape software. Based on MNC centrality, the top 10
differentially expressed genes were identified as UBE2V2,
small ubiquitin-related modifier 1 (SUMO1), SSB,
Finkel-Biskis-Reilly murine sarcoma virus ubiquitously expressed
(FAU), centrosomal protein 290 kDa (CEP290),
DNA-directed RNA polymerase II subunit K (POLR2K), cullin-4B
(CUL4B), matrin-3 (MATR3), vascular endothelial
growth factor receptor 2 (KDR) and pre-mRNA-processing
factor 40 homolog A (PRPF40A) (Fig. 5B). Additionally, the present study
also found that these top 10 genes were involved ‘nucleotide
excision repair’, ‘positive regulation of catabolic process’, ‘cell
cycle’ and ‘regulation of cell cycle process’ (Fig. 5C). The changes in expression in the
top 10 genes are shown in Table
III. The results showed that the expression levels of
UBE2V2, SUMO1, SSB, CEP290, POLR2K, CUL4B, PRPF40A and
MATR3 were significantly reduced in the Model group compared
with the Sham group, but was upregulated in the PNS-treated group.
Expression of FAU and KDR was significantly increased
in the Model group compared with the Sham group, but decreased in
the PNS-treated group. Finally, the expression levels of UBE2V2,
SUMO1, SSB, FAU, CEP290, POLR2K, CUL4B, MATR3, PRPF40A and
KDR were analyzed using RT-qPCR in the Control, Sham, Model,
50 mg/kg PNS, 100 mg/kg PNS and 150 mg/kg PNS treatment groups
(Fig. 6). The expression levels of
UBE2V2, SUMO1, SSB, CEP290, POLR2K, CUL4B, PRPF40A and
MATR3 were significantly reduced, whereas FAU and
KDR expression was significantly increased in the Model
group compared with the Sham group. PNS treatment appeared to
reverse the trends in gene expression regulation; the effects of
100 and 150 mg/kg PNS treatment groups were more significant than
that of the 50 mg/kg PNS treatment group.
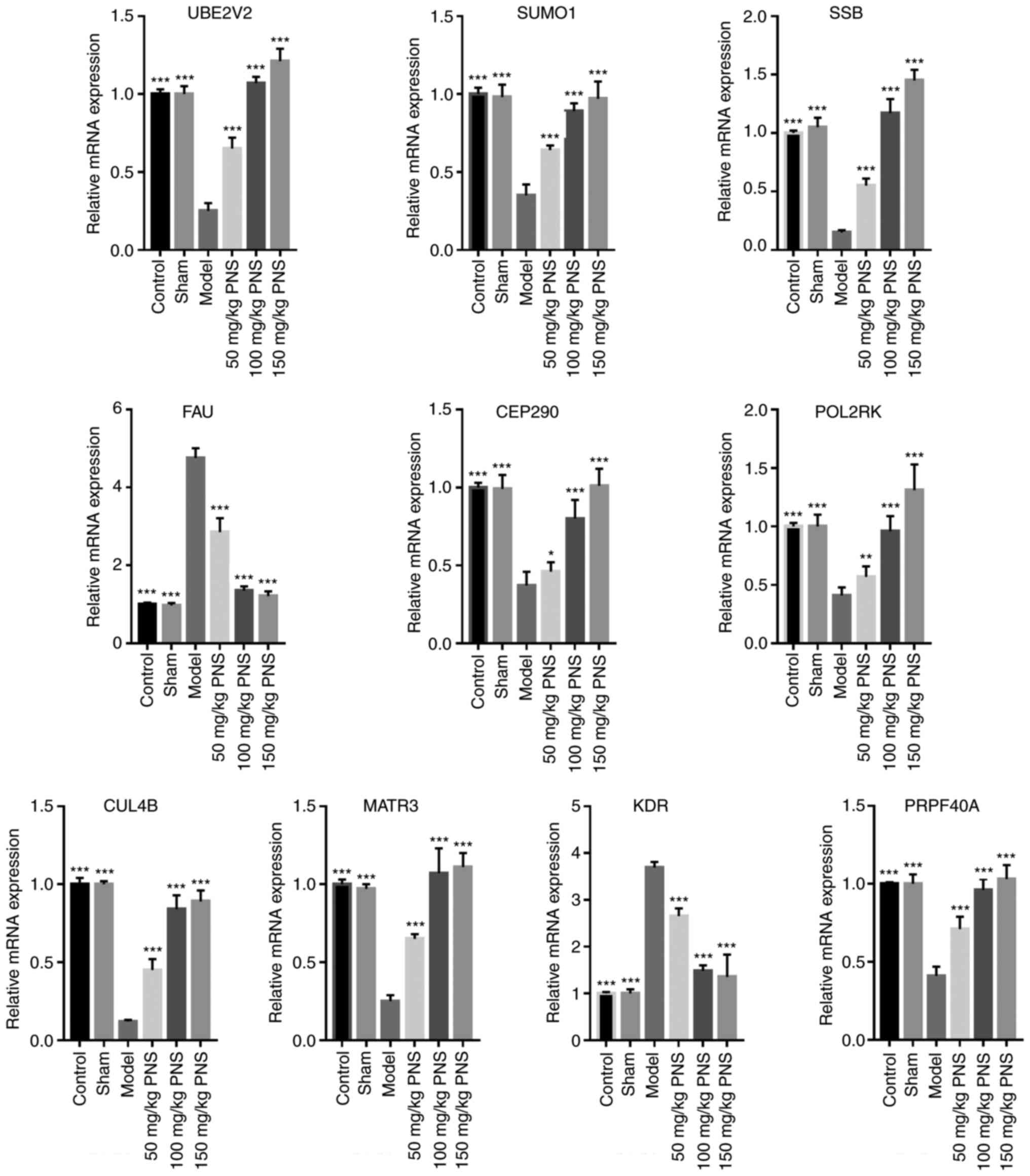 | Figure 6.Expression of top 10 hub genes were
validated by reverse transcription-quantitative PCR. n=10, samples
include the rats used for mRNA deep sequencing. *P<0.05,
**P<0.01 and ***P<0.001 vs. Model group. PNS, Panax
notoginseng saponin; UBE2V2, ubiquitin conjugating enzyme E2
variant 2; SUMO1, small ubiquitin-related modifier 1; SSB, small
RNA binding exonuclease protection factor La; FAU,
Finkel-Biskis-Reilly murine sarcoma virus ubiquitously expressed;
CEP290, centrosomal protein 290 kDa; POLR2K, DNA-directed RNA
polymerase II subunit K; CUL4B, cullin-4B; MATR3, matrin-3; KDR,
vascular endothelial growth factor receptor 2; PRPF40A,
pre-mRNA-processing factor 40 homolog A. |
 | Table III.Expression change of the top 10
differentially expressed mRNAs based on MNC centrality. |
Table III.
Expression change of the top 10
differentially expressed mRNAs based on MNC centrality.
| Gene | log2Fold
(Model/Sham) | log2Fold
(PNS/Model) |
|---|
| UBE2V2 | −1.74 | 2.02 |
| SUMO1 | −1.14 | 1.19 |
| SSB | −1.81 | 1.92 |
| FAU | 1.18 | −1.18 |
| CEP290 | −1.10 | 1.13 |
| POLR2K | −1.09 | 1.08 |
| CUL4B | −1.25 | 1.03 |
| MATR3 | −1.37 | 1.09 |
| KDR | 1.20 | −1.52 |
| PRPF40A | −1.03 | 1.00 |
Discussion
Various studies have demonstrated that PNS treatment
can prevent neurological damage in MCAO model rats to promote
recovery after a stroke (8,16,17).
The present study also found that PNS treatment attenuated the
neurological deficit and infarction area. However, the exact
molecular mechanisms are unclear and require further study. In the
present study, mRNA sequencing was employed to investigate
differential gene expression between the Model and Sham groups, and
between the Model and PNS-treated groups. When comparing the Model
and Sham groups, 1,104 genes of interest were identified, including
690 upregulated and 414 downregulated genes. When the Model group
was compared with the PNS-treated groups, 817 genes of interest
were identified, which included 390 upregulated and 427
downregulated genes. These genes may be important in the underlying
mechanisms of PNS in the treatment of ischemic stroke.
The genes identified via GO and KEGG analyses in the
present study may indicate novel molecules or pathways underlying
PNS treatment of ischemic stroke. Glutamatergic synapses and
calcium signaling are known to play important roles in cerebral
ischemia, contributing to neuronal death after a stroke (29,30).
Under ischemic stroke conditions, neuronal calcium signaling
participates in endothelial restoration to stabilize intravascular
thrombi (31). Rap1 is a member of
the Ras family of small GTPases that activates the ERK pathway
(32). Rap1 is also known to
regulate cell-cell adhesion (33),
promote angiogenesis and help maintain endothelial barrier
functions in the event of a stroke (34,35).
The apelinergic system, which consists of apelin and apelin
receptor, is temporally dysregulated during an ischemic stroke
(36). Moreover, apelin signaling
is known to reduce neuronal apoptosis and facilitate angiogenesis
in post-stroke recovery (36).
cAMP and phosphatidylinositol signaling pathways can attenuate
cerebral ischemic injury by inhibiting neuronal apoptosis and
stimulating angiogenesis (37–39).
These results indicate that PNS attenuates ischemic stroke injury
through a variety of signaling pathways. Wang et al
(40) investigated the underlying
mechanisms of Xuesaitong (XST; a Chinese medicine extracted from
Panax notoginseng roots) in preventing stroke using
microarray on a MCAO rat animal model, and found that the top
differentially expressed pathways of XST against stroke included
‘focal adhesion’ and ‘ECM-receptor interaction’, which were also
found in the present study.
Additionally, the present study found that 303
aberrantly expressed genes overlapped when comparing the Model and
Sham groups, and the Model and PNS-treated groups. The top 10
differentially expressed genes among the 303 genes included
UBE2V2, SUMO1, SSB, FAU, CEP290, POLR2K, CUL4B, MATR3,
PRPF40A and KDR, which were identified using STRING and
Cytoscape analysis. Additionally, the top 10 differentially
expressed genes were involved in ‘nucleotide excision repair’,
‘positive regulation of catabolic process’, ‘cell cycle’ and
‘regulation of cell cycle process’. Expression of UBE2V2, SUMO1,
SSB, CEP290, POLR2K, CUL4B, PRPF40A and MATR3 was
significantly reduced in the Model group compared with the Sham
group, whereas expression of FAU and KDR was
significantly increased according to mRNA sequencing and RT-qPCR
results. PNS treatment reversed these patterns of gene expression.
UBE2V2 can reduce DNA damage responses, leading to worse
neurological outcomes after stroke (41,42).
SUMO1 is known to have a neuroprotective role in
ischemia/reperfusion injury by inhibiting neuron apoptosis
(43). FAU is involved in
apoptosis and metastasis in tumor progression and may affect the
cell apoptosis in the nervous system following stroke (44). CEP290 knockout mice exhibit early
vision loss, nephropathy and die from hydrocephalus (45). Hydrocephalus also occurs in the
brain of patients with ischemic stroke, suggesting that CEP290 may
be a target for the treatment of ischemic stroke. CUL4B can
regulate Wnt/β-catenin signaling, autophagy and Janus Kinase (JNK)
signaling (46,47). Wnt/β-catenin signaling, autophagy
and JNK signaling have important roles in oxidative stress, the
breakdown of the blood-brain barrier and mitochondrial dysfunction
in ischemic stroke (48–50). KDR is a surface marker of
endothelial progenitor cells, and its expression is significantly
reduced in patients with ischemic stroke (51,52).
These results showed that these genes of interest, including
UBE2V2, SUMO1, FAU, CEP290 and CUL4B, are critical
during PNS treatment of ischemic stroke. Additionally, SSB,
POLR2K, MATR3, PRPF40A and KDR are novel genes found in
the present study that may have important effects in the
development of ischemic stroke and in the PNS treatment of ischemic
stroke. Wang et al (40)
investigated the mechanism by which XST prevents strokes and found
that fatty acid-binding protein 4 adipocyte (FABP4),
urokinase-type plasminogen activator (PLAU), heme oxygenase
1 (HMOX1), leukotriene C4 synthase (LTC4S),
insulin-like growth factor I (Igf1) and secreted
phosphoprotein 1 (SPP1) expression were increased by MCAO
induction, and decreased by XST treatment (40). It was also found that FABP4,
PLAU, HMOX1 and SPP1 expression were increased by MCAO
and Igf1 expression was inhibited by PNS treatment. The
results indicated that there are some differences in the targets of
PNS and XST in the treatment of stroke. The possible reasons are:
i) composition of the two drugs may differ; ii) doses of the drugs
used in the two studies are different; and iii) sequencing sample
is too small, which may result in false negative results.
The present study has some limitations. Because the
sequencing samples are small, a number of differentially expressed
genes may not be identified effectively, alternatively, some genes
may appear that are false positives. Additionally, the lack of a
positive control to evaluate the therapeutic effect of PNS was also
a limitation of the present study. Furthermore, the top 10
differentially expressed genes was only verified at the mRNA
expression level, and western blotting and immunohistochemistry
were not performed to investigate the protein expression of the
target genes. Finally, at present there are no data to prove that
these genes are altered in human patients who suffer from a
stroke.
In conclusion, the present study found that 303
genes of interest overlapped between comparisons of the Model group
and the Sham group, and between the Model group and the PNS-treated
group. The 10 most notable hub genes among the 303 genes included
UBE2V2, SUMO1, SSB, FAU, CEP290, POLR2K, CUL4B, MATR3
and KDR. These genes are potentially important during PNS
treatment against ischemic stroke. The present findings provided
novel insight into the pathogenesis of ischemic stroke.
Supplementary Material
Supporting Data
Acknowledgements
Not applicable.
Funding
The present study was supported by the National
Natural Science Foundation of China (grant no. 81460614).
Availability of data and materials
The datasets used and/or analyzed during the current
study are available from the corresponding author on reasonable
request.
Authors' contributions
JL, PL, LS and LM conceived and designed the present
study, and developed the methodology. JL, PL, QH, CJ, JH, XT, XL
and YL performed the experiments and collected the data. JL, XH,
WH, PL and LM analyzed and interpreted the data. JL and PL drafted
the manuscript. All authors read and approved the final
manuscript.
Ethics approval and consent to
participate
The present study was approved by the Ethics
Committee of Youjiang Medical College for National Institutional
Review Board.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Fuentes B and Tejedor ED: Stroke: The
worldwide burden of stroke-a blurred photograph. Nat Rev Neurol.
10:127–128. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Moskowitz MA, Lo EH and Iadecola C: The
science of stroke: mechanisms in search of treatments. Neuron.
67:181–198. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Duan J, Cui J, Yang Z, Guo C, Cao J, Xi M,
Weng Y, Yin Y, Wang Y, Wei G, et al: Neuroprotective effect of
Apelin 13 on ischemic stroke by activating AMPK/GSK-3β/Nrf2
signaling. J Neuroinflammation. 16:242019. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Bi BL, Wang HJ, Bian H and Tian ZT:
Identification of therapeutic targets of ischemic stroke with DNA
microarray. Eur Rev Med Pharmacol Sci. 19:4012–4019.
2015.PubMed/NCBI
|
|
5
|
Wang J, Cao B, Han D, Sun M and Feng J:
Long Non-coding RNA H19 induces cerebral ischemia reperfusion
injury via activation of autophagy. Aging Dis. 8:71–84. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Liu N, Shan D, Li Y, Chen H, Gao Y and
Huang Y: Panax notoginseng saponins attenuate phenotype switching
of vascular smooth muscle cells induced by notch3 silencing. Evid
Based Complement Alternat Med. 2015:1621452015. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Li F, Zhao H, Han Z, Wang R, Tao Z, Fan Z,
Zhang S, Li G, Chen Z and Luo Y: Xuesaitong may protect against
ischemic stroke by modulating microglial phenotypes and inhibiting
neuronal cell apoptosis via the STAT3 Signaling pathway. CNS Neurol
Disord Drug Targets. 18:115–123. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Liu L, Zhu L, Zou Y, Liu W, Zhang X, Wei
X, Hu B and Chen J: Panax notoginseng saponins promotes stroke
recovery by influencing expression of Nogo-A, NgR and p75NGF, in
vitro and in vivo. Biol Pharm Bull. 37:560–568. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Yang PF, Song XY and Chen NH: Advances in
pharmacological studies of Panax notoginseng saponins on brain
ischemia-reperfusion injury. Yao Xue Xue Bao. 51:1039–1046.
2016.(In Chinese).
|
|
10
|
Xie W, Meng X, Zhai Y, Zhou P, Ye T, Wang
Z, Sun G and Sun X: Panax notoginseng saponins: A review of its
mechanisms of antidepressant or anxiolytic effects and network
analysis on phytochemistry and pharmacology. Molecules. 23:9402018.
View Article : Google Scholar
|
|
11
|
Xu C, Wang W, Wang B, et al: Analytical
methods and biological activities of Panax notoginseng
saponins: Recent trends. J Ethnopharmacol. 236:443–465. 2019.
View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Shi YH, Li Y, Wang Y, Xu Z, Fu H and Zheng
GQ: Ginsenoside-Rb1 for ischemic stroke: A systematic review and
meta-analysis of preclinical evidence and possible mechanisms.
Front Pharmacol. 11:2852020. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Nabavi SF, Sureda A, Habtemariam S and
Nabavi SM: Ginsenoside Rd and ischemic stroke; a short review of
literatures. J Ginseng Res. 39:299–303. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Xie CL, Wang WW, Xue XD, Zhang SF, Gan J
and Liu ZG: A systematic review and meta-analysis of
Ginsenoside-Rg1 (G-Rg1) in experimental ischemic stroke. Sci Rep.
5:77902015. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Meng L, Lin J, Huang Q, Liang P, Huang J,
Jian C, Lin C and Li X: Panax notoginseng saponins attenuate
oxygen-glucose deprivation/reoxygenation-induced injury in human
SH-SY5Y Cells by regulating the expression of inflammatory factors
through miR-155. Biol Pharm Bull. 42:462–467. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Shi X, Yu W, Yang T, Liu W, Zhao Y, Sun Y,
Chai L, Gao Y, Dong B and Zhu L: Panax notoginseng saponins provide
neuroprotection by regulating NgR1/RhoA/ROCK2 pathway expression,
in vitro and in vivo. J Ethnopharmacol. 190:301–312. 2016.
View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Shi X, Yu W, Liu L, Liu W, Zhang X, Yang
T, Chai L, Lou L, Gao Y and Zhu L: Panax notoginseng saponins
administration modulates pro-/anti-inflammatory factor expression
and improves neurologic outcome following permanent MCAO in rats.
Metab Brain Dis. 32:221–233. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Longa EZ, Weinstein PR, Carlson S and
Cummins R: Reversible middle cerebral artery occlusion without
craniectomy in rats. Stroke. 20:84–91. 1989. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Bederson JB, Pitts LH, Tsuji M, Nishimura
MC, Davis RL and Bartkowski H: Rat middle cerebral artery
occlusion: Evaluation of the model and development of a neurologic
examination. Stroke. 17:472–476. 1986. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Ashburner M, Ball CA, Blake JA, Botstein
D, Butler H, Cherry JM, Davis AP, Dolinski K, Dwight SS, Eppig JT,
et al: Gene ontology: Tool for the unification of biology. The Gene
Ontology Consortium. Nat Genet. 25:25–29. 2000. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
The Gene Ontology Resource: 20 years and
still GOing strong. Nucleic Acids Res. 47:D330–D338. 2019.
View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Yu G, Wang LG, Han Y and He QY:
ClusterProfiler: An R package for comparing biological themes among
gene clusters. Omics. 16:284–287. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Kanehisa M and Goto S: KEGG: kyoto
encyclopedia of genes and genomes. Nucleic Acids Res. 28:27–30.
2000. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Kanehisa M, Sato Y, Furumichi M, Morishima
K and Tanabe M: New approach for understanding genome variations in
KEGG. Nucleic Acids Res. 47:D590–D595. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Kanehisa M: Toward understanding the
origin and evolution of cellular organisms. Protein Sci.
28:1947–1951. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Szklarczyk D, Gable AL, Lyon D, Junge A,
Wyder S, Huerta-Cepas J, Simonovic M, Doncheva NT, Morris JH, Bork
P, et al: STRING v11: Protein-protein association networks with
increased coverage, supporting functional discovery in genome-wide
experimental datasets. Nucleic Acids Res. 47:D607–D613. 2019.
View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Shannon P, Markiel A, Ozier O, Baliga NS,
Wang JT, Ramage D, Amin N, Schwikowski B and Ideker T: Cytoscape: A
software environment for integrated models of biomolecular
interaction networks. Genome Res. 13:2498–2504. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(-Delta Delta C(T)) method. Methods. 25:402–408. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Huang L, Wang C, Zhao S, Ge R, Guan S and
Wang JH: PKC and CaMK-II inhibitions coordinately rescue
ischemia-induced GABAergic neuron dysfunction. Oncotarget.
8:39309–39322. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Chen BH, Park JH, Lee YL, Kang IJ, Kim DW,
Hwang IK, Lee CH, Yan BC, Kim YM, Lee TK, et al: Melatonin improves
vascular cognitive impairment induced by ischemic stroke by
remyelination via activation of ERK1/2 signaling and restoration of
glutamatergic synapses in the gerbil hippocampus. Biomed
Pharmacother. 108:687–697. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Secondo A, Bagetta G and Amantea D: On the
role of store-operated calcium entry in acute and chronic
neurodegenerative diseases. Front Mol Neurosci. 11:872018.
View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Hattori M and Minato N: Rap1 GTPase:
Functions, regulation, and malignancy. J Biochem. 134:479–484.
2003. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Kooistra MR, Dube N and Bos JL: Rap1: A
key regulator in cell-cell junction formation. J Cell Sci.
120:17–22. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Chrzanowska-Wodnicka M: Rap1 in
endothelial biology. Curr Opin Hematol. 24:248–255. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Chrzanowska-Wodnicka M: Distinct functions
for Rap1 signaling in vascular morphogenesis and dysfunction. Exp
Cell Res. 319:2350–2359. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Wu Y, Wang X, Zhou X, Cheng B, Li G and
Bai B: Temporal expression of apelin/apelin receptor in ischemic
stroke and its therapeutic potential. Front Mol Neurosci. 10:12017.
View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Hu S, Cao Q, Xu P, Ji W, Wang G and Zhang
Y: Rolipram stimulates angiogenesis and attenuates neuronal
apoptosis through the cAMP/cAMP-responsive element binding protein
pathway following ischemic stroke in rats. Exp Ther Med.
11:1005–1010. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Bai H, Zhao L, Liu H, Guo H, Guo W, Zheng
L, Liu X, Wu X, Luo J, Li X, et al: Adiponectin confers
neuroprotection against cerebral ischemia-reperfusion injury
through activating the cAMP/PKA-CREB-BDNF signaling. Brain Res
Bull. 143:145–154. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Kim YS, Yoo A, Son JW, Kim HY, Lee YJ,
Hwang S, Lee KY, Lee YJ, Ayata C, Kim HH and Koh SH: Early
activation of phosphatidylinositol 3-kinase after ischemic stroke
reduces infarct volume and improves long-term behavior. Mol
Neurobiol. 54:5375–5384. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Wang L, Yu Y, Yang J, Zhao X and Li Z:
Dissecting Xuesaitong's mechanisms on preventing stroke based on
the microarray and connectivity map. Mol Biosyst. 11:3033–3039.
2015. View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Zhao Y, Long MJC, Wang Y, Zhang S and Aye
Y: Ube2V2 Is a rosetta stone bridging redox and ubiquitin codes,
coordinating DNA damage responses. ACS Cent Sci. 4:246–259. 2018.
View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Li P, Stetler RA, Leak RK, Shi Y, Li Y, Yu
W, Bennett MVL and Chen J: Oxidative stress and DNA damage after
cerebral ischemia: Potential therapeutic targets to repair the
genome and improve stroke recovery. Neuropharmacology. 134:208–217.
2018. View Article : Google Scholar : PubMed/NCBI
|
|
43
|
Zhang H, Wang Y, Zhu A, Huang D, Deng S,
Cheng J, Zhu MX and Li Y: SUMO-specific protease 1 protects neurons
from apoptotic death during transient brain ischemia/reperfusion.
Cell Death Dis. 7:e24842016. View Article : Google Scholar : PubMed/NCBI
|
|
44
|
Perina D, Korolija M, Hadzija MP, Grbeša
I, Belužić R, Imešek M, Morrow C, Marjanović MP, Bakran-Petricioli
T, Mikoč A and Ćetković H: Functional and structural
characterization of FAU gene/protein from marine sponge suberites
domuncula. Mar Drugs. 13:4179–4196. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
45
|
Rachel RA, Yamamoto EA, Dewanjee MK,
May-Simera HL, Sergeev YV, Hackett AN, Pohida K, Munasinghe J,
Gotoh N, Wickstead B, et al: CEP290 alleles in mice disrupt
tissue-specific cilia biogenesis and recapitulate features of
syndromic ciliopathies. Hum Mol Genet. 24:3775–3791. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
46
|
He YM, Xiao YS, Wei L, Zhang JQ and Peng
CH: CUL4B promotes metastasis and proliferation in pancreatic
cancer cells by inducing epithelial-mesenchymal transition via the
Wnt/β-catenin signaling pathway. J Cell Biochem. 119:5308–5323.
2018. View Article : Google Scholar : PubMed/NCBI
|
|
47
|
Li Y, Zhou X, Zhang Y, Yang J, Xu Y, Zhao
Y and Wang X: CUL4B regulates autophagy via JNK signaling in
diffuse large B-cell lymphoma. Cell Cycle. 18:379–394. 2019.
View Article : Google Scholar : PubMed/NCBI
|
|
48
|
Jean LeBlanc N, Menet R, Picard K, Parent
G, Tremblay ME and ElAli A: Canonical Wnt pathway maintains
blood-brain barrier integrity upon ischemic stroke and its
activation ameliorates tissue plasminogen activator therapy. Mol
Neurobiol. 56:6521–6538. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
49
|
Wang P, Shao BZ, Deng Z, Chen S, Yue Z and
Miao CY: Autophagy in ischemic stroke. Prog Neurobiol.
163-164:98–117. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
50
|
Zheng J, Dai Q, Han K, Hong W, Jia D, Mo
Y, Lv Y, Tang H, Fu H and Geng W: JNK-IN-8, a c-Jun N-terminal
kinase inhibitor, improves functional recovery through suppressing
neuroinflammation in ischemic stroke. J Cell Physiol.
235:2792–2799. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
51
|
Deng Y, Wang J, He G, Qu F and Zheng M:
Mobilization of endothelial progenitor cell in patients with acute
ischemic stroke. Neurol Sci. 39:437–443. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
52
|
Marti-Fabregas J, Delgado-Mederos R,
Crespo J, Peña E, Marín R, Jiménez-Xarrié E, Fernández-Arcos A,
Pérez-Pérez J, Martínez-Domeño A, Camps-Renom P, et al: Circulating
endothelial progenitor cells and the risk of vascular events after
ischemic stroke. PLoS One. 10:e01248952015. View Article : Google Scholar : PubMed/NCBI
|