Introduction
Over recent years, numerous studies have suggested
that vitamin D (VD) is not only associated with bone growth and
development (1,2) but that it is also closely associated
with the cognitive functioning in psychotic disorders (3), autoimmune diseases (4), cardiovascular diseases (5) and metabolic diseases (6,7).
According to the data on standard VD requirements, VD insufficiency
or deficiency is globally widespread, and is present in 50–80% of
the total global population (8).
Previous studies have reported that the VD receptor (VDR) is
present in the human placenta (9),
uterine decidua, pituitary (10)
and ovarian granulosa cells (11,12).
VD can promote the production of estradiol, estrone, progesterone
and insulin-like growth factor binding protein 1 in the ovary, and
these hormones and growth factors have positive roles in the
development of follicles and embryos (12). Moreover, VD deficiency can reduce
endometrial receptivity, which can, in turn, affect reproductive
functions (13). Jiang et al
(14) have revealed that
1,25(OH)2D3 can upregulate the mRNA and protein expression levels
of homeobox gene A10 (Hoxa10) by binding with endometrial stromal
cell receptors. Hoxa10 is essential for female fertility and embryo
implantation, and lack of VD could reduce the expression level of
Hoxa10, thus affecting female fertility. Furthermore, Wei et
al (15) have reported that VDR
is a key regulator of inflammatory responses and β-cell
survival.
Several studies have reported that microRNAs
(miRNAs/miRs) are abundant in the female reproductive system, among
which miR-26a and miR-125b are most abundantly expressed in the
ovaries (16). miR-378d is a member
of the miR-378 family, which has eight members (miR-378
a/b/c/d/e/f/h/i). As these share similar seed sequences, such as a
sequence of 6–8 nucleotides that are critical for mRNA target
recognition, they are considered to have similar activity and
targets (17). Research has mainly
focused on tumor, heart and liver diseases, where miR-378a is the
most widely studied member, and miR-378d the much less studied
factor (18–21). Ishida et al (22) reported that miR-378 was closely
associated with adiponectin expression. Gungormez et al
(23) revealed that the expression
levels of miRNAs from the miR-378 family are decreased in the tumor
tissues of patients with stage II colon cancer compared with the
normal tissues, which could be used as a biomarker for early colon
cancer. Moreover, miR-378d-2 has been reported to be closely
associated with tumor generation in obese patients with breast
cancer (24).
The present study aimed to examine miR-378d,
Glut4 and aromatase (Cyp19a) expression levels in
mouse ovarian tissues. GLUT4 is a member of the facilitative GLUT
family, characterized by preferential expression in muscle and
adipose tissue, and is responsible for insulin-stimulated glucose
uptake (25). The CYP19 gene, which
is widely known to encode aromatase, is the key enzyme for estrogen
production (26).
The effect of VD on the reproductive system in
female mice is achieved by the differential expression of miRNAs
(27). In the present study, a
mouse model of VD deficiency was established to investigate the
molecular mechanisms underlying the effects of miRNAs on ovarian
granulosa cells in female mice with VD deficiency.
Materials and methods
Animals
Female C57BL/6 J mice (age, 3 weeks; weight, 12–18
g) were obtained from the Carvens Animal Laboratory Technology
Company [http://www.cavens.com.cn/index.php; animal certificate
no. SCXK(SU)2016-0010]. All procedures related to animal use were
approved by the Animal Care and Use Committee of Nanjing Medical
University. This study was carried out in strict accordance with
the recommendations in the Guide for the Care and Use of Laboratory
Animals of the National Institutes of Health (28). Mice were housed in a specific
pathogen-free environment under standard housing conditions with
controlled temperature (22±1°C) and humidity (52±5%). Mice were
exposed to a 12-h light/dark cycle and had ad libitum access
to the assigned food and water. Mice were weighed weekly and
monitored for changes in health status.
After acclimatization for 1 week, the mice were
randomized to VD-deficient (25 IU VD3/kg; cat. no. Dyets D 119289)
or control diet (5,000 IU VD3/kg; cat. no. Dyets D 119290) groups
for 8 weeks. All diets were procured from Dyets, Inc.
In the present study a total of 16 mice were used,
of which 5 were the control group and 6 were VD deficient group,
and the remaining 5 were used to extract mouse ovarian granulosa
cells. After 8 weeks, mice from each group were abdominally
anesthetized with 4% chloral hydrate (370–400 mg/kg) and then bled
from the retro-orbital venous plexus, after which they were
sacrificed by cervical dislocation; efforts were made to minimize
animal suffering (28). The blood
(20–50 µl per mouse) was centrifuged at 1,000 × g for 10 min at
room temperature, and the serum was collected for 25-(OH) D3
analysis. Serum 25-(OH) D3 levels were measured using an ELISA kit
(cat. no. AC-57SF1; Immunodiagnostic Systems, Ltd.) to confirm VD
deficiency. Ovarian tissues were collected for RNA sequencing,
reverse transcription-quantitative (RT-q) PCR and western blot
analysis.
In order to confirm whether miR-378d
post-transcriptionally regulates Rsbn1 in ovaries, normal
mouse granulosa cells were isolated from ovarian follicles and
cultured. The granulosa cells were cultured in DMEM/F-12 (1:1; cat.
no. 11330–057) supplemented with 10% fetal bovine serum (cat. no.
10270) and 100 U/ml penicillin plus 100 µg/ml streptomycin (cat.
no. 15140; all Thermo Fisher Scientific, Inc.) at 37°C with 5%
CO2. The miR-378d mimic (5′-ACUGGCCUUGGAGUCAGAAGGU-3′),
inhibitor (5′-ACCUUCUGACUCCAAGGCCAGU-3′) and their control
sequences were synthesized by Shanghai GenePharma Co., Ltd. Once
cells reached 80% confluency, they were digested by 0.25% trypsin
(Thermo Fisher Scientific, Inc.) at room temperature for 2 min,
counted by Countess 3 Automated Cell Counters (Thermo Fisher
Scientific, Inc.) and seeded into 6-well plates in growth medium
without antibiotics at a density of 2.0×105 cells/well.
After culturing at 37°C for 18–24 h when 70% confluency was
reached, cells were transfected with 50 nM miR-378d mimic or
miR-378d inhibitor using Lipofectamine® RNAi MAX reagent
(Invitrogen; Thermo Fisher Scientific, Inc.), according to the
manufacturer's protocol. At 48 h post transfection, cells were
harvested for RNA isolation or protein extraction. Transfection
efficiency was measured by RT-qPCR for miR-378d. All transfection
experiments were performed in duplicate wells and repeated three
times before protein and mRNA expression levels of Rsbn1, Glut4 and
Cyp19a were assessed.
RNA sequencing
The total RNA of each sample was extracted using
TRIzol® reagent (Invitrogen; Thermo Fisher Scientific,
Inc.) and an miRNeasy Mini kit (Qiagen, Inc.), and was quantified
using an Agilent 2100 Bioanalyzer (Agilent Technologies, Inc.),
NanoDrop system (Thermo Fisher Scientific Inc.) and 1% agarose gel.
A total of 2 µg total RNA with RNA integrity number >7.5 was
used for subsequent library preparation. Next-generation sequencing
library preparations were constructed according to the
manufacturer's protocol (NEBNext® Multiplex Small RNA
Library Prep Set for Illumina®; Illumina, Inc.). The 3′
SR Adaptor for Illumina was ligated to the small RNA using
3′Ligation Enzyme. To prevent adaptor-dimer, the excess of 3′ SR
Adaptor was hybrid with SR RT Primer for Illumina. The 5′ SR
Adaptor for Illumina was ligated to the small RNA using 5′Ligation
Enzyme, and the first-strand cDNA was synthesized using ProtoScript
II Reverse Transcriptase (New England Biolabs). Each sample was
then amplified by PCR for 12 cycles using P5
(GTTCAGAGTTCTACAGTCCGACGATCNNNN) and P7 (NNNNTGGAATTCTCGGGTGCCAAGG;
both NextFlex) primers, with both primers carrying sequences that
can anneal with the flow cell to perform bridge PCR, and the P7
primer carrying a six-base index allowed for multiplexing. PCR was
performed under the following conditions: Initial denaturation at
94°C for 30 sec, followed by 12 cycles of 94°C for 15 sec, 62°C for
30 sec and 70°C for 15 sec and final extension at 70°C for 5
min.
RT was performed according to the instructions of
the Vazyme RT kit (cat. no. R223-01; Vazyme Biotech Co., Ltd.). The
reactants (volume, 10 µl) were added into 1.5 ml centrifuge tube in
the following order: 5X HiScript II Select qRT SuperMix (2 µl),
Oligo(dT) (1 µl), Total RNA (1 µl) and RNase Free H2O
(add water to 10 µl). PCR reaction condition were as follows:
Initial denaturation at 95°C for 30 sec, followed by 40 cycles at
95°C for 10 sec, 60°C for 20 sec and 72°C for 1 min. The RT primer
sequence was
5′-GTCGTATCCAGTGCAGGGTCCGAGGTATTCGCACTGGATACGACACCTTC-3′. Further
details of the primers are provided in Table I.
 | Table I.List of forward and reverse
primers. |
Table I.
List of forward and reverse
primers.
| Gene | Forward primer | Reverse primer |
|---|
| GAPDH |
5′-AACGACCCCTTCATTGAC-3′ |
5′-TCCACGACATACTCAGCAC-3′ |
| Rsbn1 |
5′-AGGTCTCAGAGCGATGACGG-3′ |
5′-GGGTTCACTTGTCCGAGGTAG-3′ |
| Glut4 |
5′-TGGCCTTCTTTGAGATTGGC-3′ |
5′-AACCCATGCCGACAATGAAG-3′ |
| Cyp19a |
5′-AACAACAACCCGAGCCTTTG-3′ |
5′-AATGCTGCTTGATGGACTCC-3′ |
| U6 |
5′-CTCGCTTCGGCAGCACA-3′ |
5′-AACGCTTCACGAATTTGCGT-3′ |
The PCR products of ~140 bp were validated using an
Agilent 2100 Bioanalyzer (Agilent Technologies, Inc.), and
quantified using a Qubit 2.0 Fluorometer (Invitrogen; Thermo Fisher
Scientific, Inc.). Then, libraries with different indexes were
multiplexed and loaded on an Illumina HiSeq instrument according to
the manufacturer's instructions (Illumina, Inc.). Sequencing was
performed using a 1×50 single-end (SE)/2×150 paired-end
configuration; image analysis and base calling were conducted using
the HiSeq Control Software v2.2.58 + OLB + GAPipeline-1.6
(Illumina, Inc.) on the HiSeq instrument. The sequences were
processed and analyzed by Genewiz, Inc. Libraries were sequenced,
and ovaries tissue miRNA RNA sequences were generated with the
Illumina HiSeq X sequencing platform by Cofactor Genomics, Inc.
Gene Ontology (GO)-TermFinder (https://metacpan.org/pod/GO::TermFinder)
was used to identify GO terms that annotate a list of enriched
genes with a significant P<0.05. Kyoto Encyclopedia of Genes and
Genomes (KEGG) is a collection of databases dealing with genomes,
biological pathways, diseases, drugs and chemical substances
(https://www.genome.jp/kegg). In-house
scripts were used to enrich significant differentially expressed
genes in KEGG pathways.
Prediction of target genes and
luciferase reporter assays
Using miRNA target gene prediction programs
TargetScan 7.2 (targetscan.org/vert_72/) and miRDB (mirdb.org/cgi-bin/search.cgi), the putative
miR-378d binding sites at the 3′untranslated region (UTR) of
potential miR-378d target genes in mouse were analyzed. Based on
the seed region sequences of the candidate target genes provided by
the National Center for Biotechnology Information, primers
targeting the gene segments containing the seed sequences were
designed with SpeI-HindIII restriction enzyme sites
incorporated. In addition, mutagenic primers were designed to
specifically mutate 3–5 bases in the seed region of the candidate
target genes. The primer sequences are listed in Table I. After primer annealing, candidate
target sequences containing wild-type (WT) and mutant (MUT) seed
regions were synthesized. The WT and MUT sequences were cloned into
pMIR-Report Luciferase [General Biosystems (Anhui) Co. Ltd.] to
construct WT and MUT reporter plasmids, respectively.
The WT and MUT sequences of the 3′UTR of round
spermatid basic protein 1 (RSBN1) were developed and named
RSBN1−3′UTR WT and RSBN1−3′UTR MUT. Next, 100 ng
RSBN1−3′UTR WT or RSBN1−3′UTR MUT and 0.25 µl
miR-378d mimic and inhibitor (20 µM), or suitable negative controls
(NC) were mixed with LipoPlus (B. Braun Medical Indonesia)
following the manufacturer's instruction. Following the plasmid
mixture into the transfected media, the plasmid was co-transfected
into the C57B/L6 mouse granulosa cell line. The cells were cultured
at 37°C with 5% CO2 for 48 h. When the cells reached
70–80% confluence, they were harvested for further analysis. After
the cells were harvested, Renilla luciferase and firefly
luciferase activities were measured using a luciferase assay kit
(Thermo Fisher Scientific, Inc.) and a chemiluminescence reader
(FL600 Fluorescence Microplate Reader; BioTek Instruments,
Inc).
In order to evaluate the ability of miR-378d binding
to the 3′UTR of RSBN1 in mouse, a luciferase reporter assay was
conducted in mouse granulosa cells. Female C57BL/6 mice (age, 21
days; weight, 12–18 g) were injected intraperitoneally (i.p.) with
5 IU pregnant mare serum gonadotropin (PMSG; Prospec-Tany
TechnoGene, Ltd.) to initiate follicular development. Mice were
then euthanized by cervical dislocation 48 h after PMSG injection,
then the ovaries were removed. Granulosa cells were obtained from
the ovarian tissue via follicular puncture and placed in PBS. The
cell suspension was filtered through a 70-µm mesh sieve three times
to remove oocytes and centrifuged at 1,000 × g for 5 min at room
temperature. The pellet was re-suspended in serum-free DMEM F12
insulin-transferrin-sodium selenite (Sigma-Aldrich; Merck
KGaA).
PMSG injection to initiate follicular
development
Next, the miR-378d binding capability to the 3′UTR
of RSBN1 was examined in mice, and a luciferase reporter assay was
conducted in mouse granulosa cells. Female C57BL/6 mice (age, 21
days; weight, 12–18 g) were injected i.p. with 5 IU PMSG
(Prospec-Tany, TechnoGene, Ltd.) to initiate follicular
development. Next, the mice were sacrificed by cervical dislocation
48 h later after PMSG injection, after which the ovaries were
removed, using a 1 ml syringe with a needle. The PMSG injection was
performed by the method as described previously (29).
Rescue experiments
To construct Rsbn1 overexpression plasmid,
RSBN1 forward and reverse (RSBN1 forward,
5′-TAAGCTTGGatgttcatctctggacga-3′ and reverse,
5′-TGGATCCGAGtaaaggacaaatcagact-3′) primers were used to amplify
the RSBN1 sequence. The amplified product was digested by
HindIII and NotI enzymes and connected with pcDNA3.1
(+) digested by the same enzymes to form a recombinant plasmid. The
recombinant plasmid was transformed into E. coli to obtain
stable Rsbn1 overexpression plasmid strain.
For the rescue experiments, mouse granulosa cells
were first transfected using Lipofectamine™ 2000 transfection
reagent (Invitrogen; Thermo Fisher Scientific, Inc.) according to
the manufacturer's instructions with Rsbn1 small interfering
(si)RNA(5′-GCAUACAUGGACGAACUCUTT-3′, Shanghai GenePharma Co.,
Ltd.), negative control (NC; 5′-GTCCAGAACGAATTTATAAGT-3′, Shanghai
GenePharma Co., Ltd.), miR-378d mimic, miR-378d inhibitor (50
pmol/ml)or Rsbn1 OE (1.67 µg/ml) at 37°C for 4 h. The
following groups were established in the C57B/L6 mouse granulosa
cells: NC + control vector, miR378d mimic + control vector, NC +
Rsbn1 OE, miR378d mimic + Rsbn1 OE, inhibitor NC +
siRNA NC, miR-378d inhibitor + siRNA NC, inhibitor NC +
Rsbn1 KD, miR-378d inhibitor + Rsbn1 KD. After
another 48 h, cells were harvested for qPCR and western blot
analysis.
RT-qPCR
Total RNA was isolated from ovary tissue or cultured
cells and extracted using TRIzol® reagent (Invitrogen;
Thermo Fisher Scientific, Inc.) according to the manufacturer's
instructions. The concentration of total RNA was measured by
Ultraviolet Spectrophotometer at 260 nm (Thermo Fisher Scientific,
Inc.). cDNA was synthesized from 1 µg total RNA using an All-in-one
TM miRNA qPCR Detection kit (GeneCopoeia, Inc.) or from 500 ng
total RNA using a HiScript II RT SuperMix according to the
manufacturer's instructions (Vazyme Biotech Co., Ltd.). qPCR
analysis was performed using AceQ qPCR SYBR Green master mix
(Vazyme Biotech Co., Ltd.) on an ABI ViiA 7 Real-Time PCR system
(ABI, USA). The qPCR was performed under the following conditions:
Initial denaturation at 95°C for 30 sec, followed by 40 cycles of
95°C for 10 sec, 60°C for 20 sec and 72°C for 1 min. U6 snRNA was
used as an internal control for miR-378d and GAPDH was used as an
internal control for other genes. Experimental data were analyzed
using a relative quantification method (2−∆∆Cq)
(30). All experiments were
repeated three times. The RT-qPCR primers are described in detail
in Table II.
 | Table II.Primers targeting the seed region of
the candidate target genes. |
Table II.
Primers targeting the seed region of
the candidate target genes.
| Gene | Sequence |
|---|
| 3′UTR sequence of
Rsbn1 |
5′-ACTAGTAGCAGATGCCATCCTGTCATCTAAGCTGGTCATTACTAATACACAAGGAGACTGTCTCCTGACAGCCAGCACTGTGCAATCACTCAGGAACCAGCGGATCTGCAAAGACCAAGCTT-3′ |
| Mutation sequence
of Rsbn1 |
5′-ACTAGTAGCAGATGCCATCCTGTCATCTAAGCTGGTCATTACTAATACACAAGGAGAGTCTGTGCTGACACCGACCACTGTGCAATCACTCAGGAACCAGCGGATCTGCAAAGACCAAGCTT-3′ |
Western blotting
The mouse ovary tissue and granulosa cells were
rinsed with PBS, treated with 0.25% trypsin (Gibco; Thermo Fisher
Scientific, Inc.) for 2 min at 37°C, harvested and transferred to
centrifuge tubes. Cellular proteins were extracted using the RIPA
lysis buffer (cat. no. R0020; Beijing Solarbio Science &
Technology Co., Ltd.) and the protein concentration was measured
using a BCA kit (cat. no. P0012S; Beyotime Institute of
Biotechnology), after which 50 µg protein samples were separated
via SDS-PAGE on a 12% gel and transferred to a PVDF membrane (cat.
no. 3010040001; Sigma-Aldrich; Merck KGaA). The membrane was
blocked for 2 h with blocking buffer (cat. no. P0023B; Beyotime
Institute of Biotechnology) at room temperature and then incubated
with the following primary antibodies (all 1:1,000) diluted with
primary antibody dilution buffer (cat. no. P0023A; Beyotime
Institute of Biotechnology): Anti-Rsbn1 rabbit polyclonal
IgG (cat. no. NBP1-57724; Novus Biologicals, LLC), Ras-related
nuclear-binding protein 10 (RANBP10) rabbit polyclonal IgG (cat.
no. ab172730; Abcam) or GAPDH mouse monoclonal IgG1 (cat. no.
G8795; Sigma-Aldrich; Merck KGaA) for 12 h at 4°C on a shaker. The
membrane was then incubated with corresponding secondary antibodies
(goat anti-rabbit IgG-HRP, cat. no. AP307P, Sigma-Aldrich; Merck
KGaA; or goat anti-mouse IgG-HRP, cat. no. ab97023, Abcam; 1:5,000)
at room temperature for 1 h. The protein was detected by
chemiluminescence using Pierce ECL Western Blotting Substrate
(Thermo Fisher Scientific, Inc.). The optical density of protein
fragments was quantified by ImageJ (version 1.25p; National
Institutes of Health).
Statistical analysis
GraphPad Prism 5.01 (GraphPad Software, Inc.) and
ImageJ (National Institutes of Health) were used for raw data
analysis. Data are presented as the means ± SD (n=3). The
differences between two groups were analyzed using unpaired
Student's t-test. For multiple groups, one-way ANOVA followed by
Bonferroni test was used. P<0.05 was considered to indicate a
statistically significant difference.
Results
Mouse model of VD deficiency
C57BL/6J mice that were nutritionally deficient in
VD were established as previously described (27). After 8 weeks on a diet, VD-mice
(n=6) had 25-(OH) D3 levels of 5.99±2.30 ng/ml compared with
control mice (n=5) that had 25-(OH) D3 levels of 46.70±10.17 ng/ml
(P<0.0001; Table SI). No
significant changes were observed in serum calcium and phosphorus
concentrations at 8 weeks on VD-deficient or control diet (data not
shown). The average weight was similar in mice on VD- and control
chow over the 8 weeks (Table
SI).
Functional enrichment analysis and transcriptional
signature analysis of ovary tissues from VD- and control mice. RNA
sequencing was conducted using total RNA isolated from ovary
tissues of control and VD-mice at the age of 11 weeks. A total of
672 differentially expressed miRNAs were identified, while
transcripts per million (TPM) values of >4 in both libraries
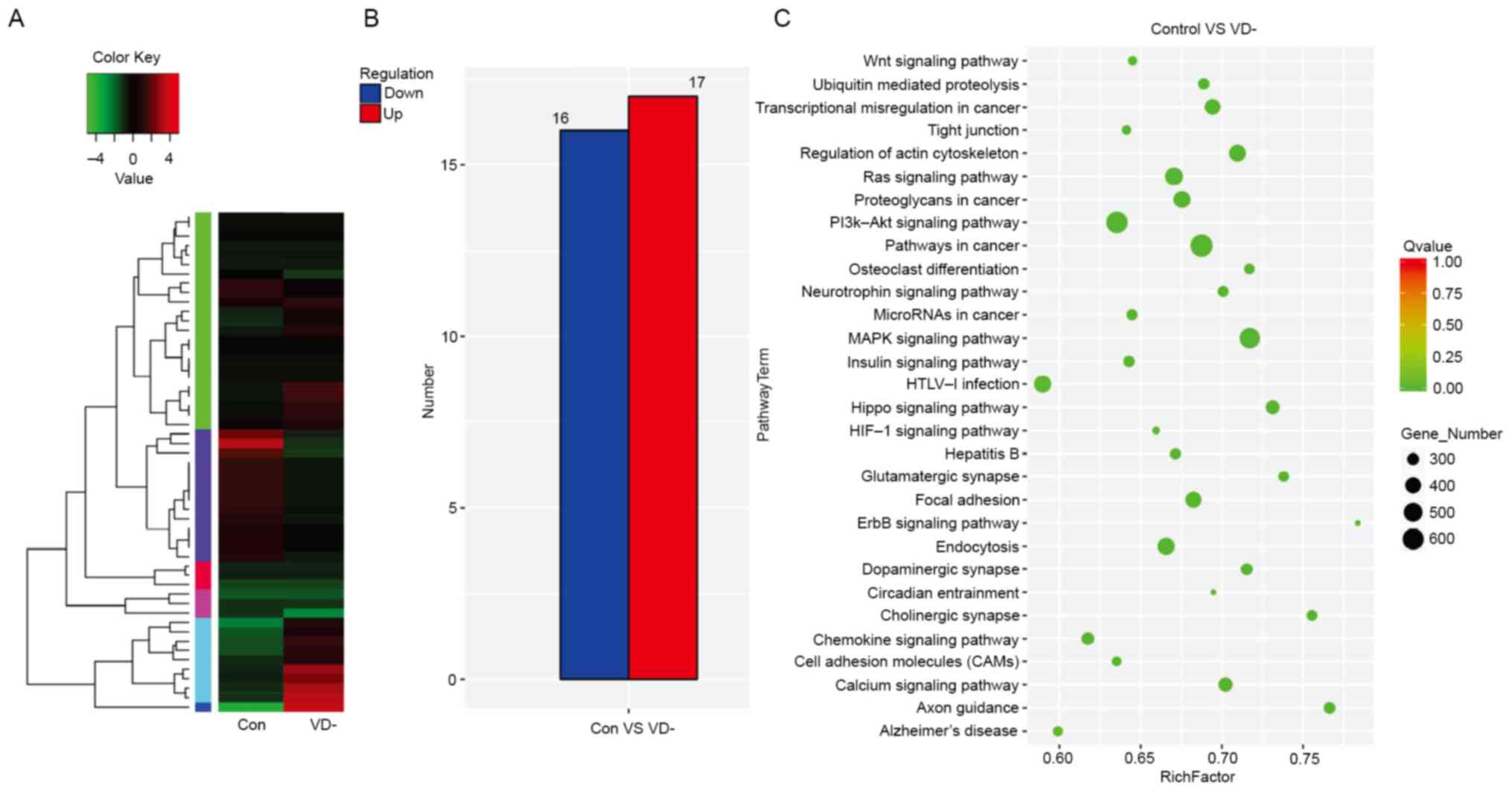
were retained for further analysis (Fig. 1A). Among them, 33 miRNAs were
reported having log2-fold change and false discovery rate ≤0.05) in
upregulation or downregulation, regardless of miRNA abundance
(Table SII). Furthermore, 17
miRNAs were significantly upregulated, while 16 miRNAs were
downregulated in the ovarian tissue of VD-mice compared with the
control (Fig. 1B).
KEGG enrichment analysis was used to analyze the
significant biological processes. Most of the target genes were
associated with ‘cell binding’ and ‘catalytic activity’ and were
involved in multiple functions such as ‘biological regulation’ and
‘molecular process of organelles’. Numerous essential biological
processes were changed, such as ‘ubiquitin-mediated proteolysis’,
‘cell cycle’, ‘Wnt signaling pathway’, ‘calcium signaling pathway’,
‘MAPK signaling pathway’ and ‘hypoxia inducible factor 1 (HIF-1)
signaling pathway’, amongst others (Fig. 1C).
Expression of miR-378d in mouse
ovarian tissue as determined via RT-qPCR
RNA-seq has demonstrated miR-378d was significantly
upregulated in the ovaries of VD mouse models (data not shown).
Importantly, a recent study reported that hsa-miR-378d may be a
novel and potential biomarker for improving the diagnosis of
colorectal cancer (31).
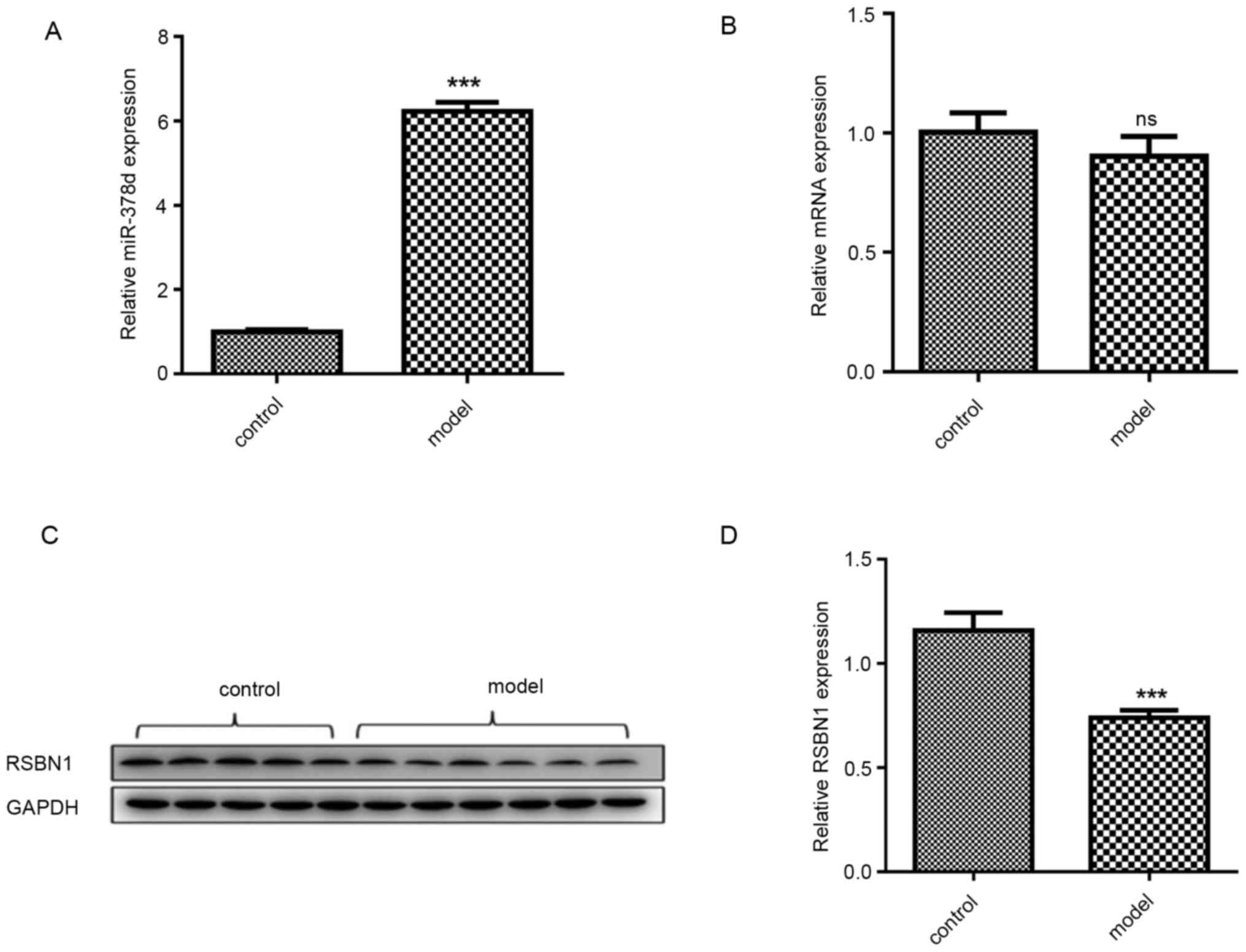
miR-378d expression in VD-mouse ovarian tissue was
detected via qPCR and compared with the control group. The miR-378d
expression was significantly higher compared with that in the
control group (P<0.001; Fig.
2A). Bioinformatic analysis using the two online prediction
algorithms that were previously mentioned was first conducted to
predict the genes targeted by miR-378d. As a result, a total of 11
potential target genes were identified, including methyl-CpG
binding protein 2, neurofibromin 1, BICD cargo adaptor 2, ETS
proto-oncogene 1, growth arrest specific 7, GTP binding protein 2,
LIM domain containing 1, polycomb group ring finger 3, RSBN1, SIM
bHLH transcription factor 1 and tetraspanin 9. The bioinformatic
analysis demonstrated that there is a high complementary sequence
between Rsbn110 3′UTR and miR-378d. Rsbn1 protein
expression level was significantly downregulated in the
VD-deficiency group compared with the control group (P<0.001;
Fig. 2C and D), while Rsbn1
mRNA expression level was not significantly altered (Fig. 2B).
miR-378d regulates RSBN1 in mouse
granulosa cells
RT-qPCR results identified that the miR-378d
expression levels were significantly upregulated in the miR-378d
mimics group compared with the NC group (P<0.001). Moreover, the
miR-378d expression level was significantly downregulated in the
miR-378d inhibitor group compared with the inhibitor NC group
(P<0.001). These results indicated the successful overexpression
or knockdown of miR-378d in mouse granulosa cells (Fig. 3A).
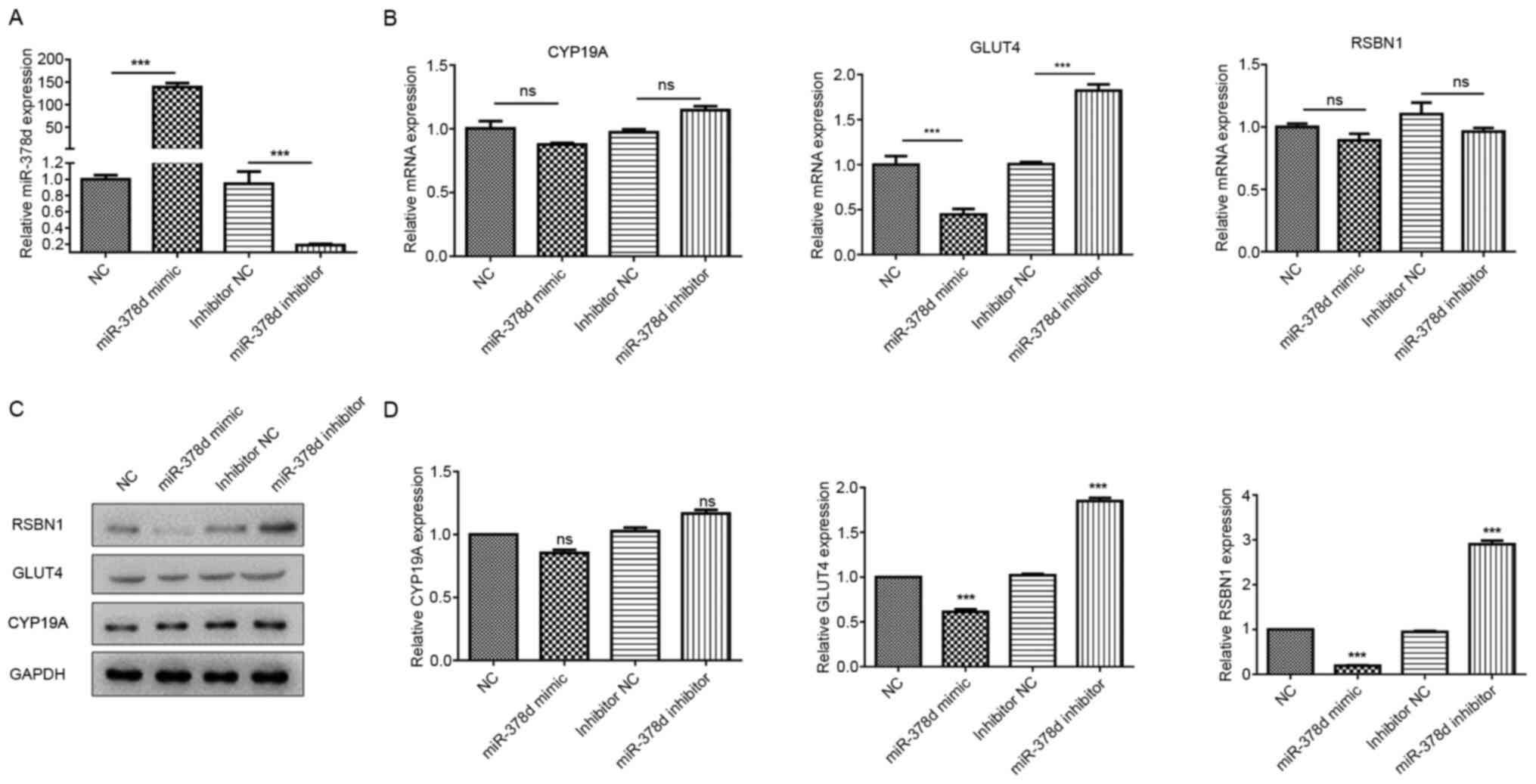 | Figure 3.miR-378d post-transcriptionally
regulates Rsbn1 in ovaries. (A) Reverse transcription-quantitative
PCR results showed that mir-378d was significantly upregulated in
the mimic group and significantly downregulated in the inhibitor
group. (B) Glut4 mRNA expression was decreased significantly
in the mimic group and increased significantly in the inhibitor
group, while Rsbn1 and Cyp19a mRNA expression levels
showed no significant differences. (C) Western blot analyses of
Rsbn1, Glut4 and Cyp19a expression levels in mouse
granulosa cells transfected with miR-378d mimics or inhibitors. (D)
Expression levels of Rsbn1 and Glut4 were decreased
after mir-378d overexpression, and the expression levels of
Rsbn1 and Glut4 were increased after mir-378d
inhibition, while Cyp19a showed no significant change.
***P<0.001 vs. respective NC. ns, non-significant; VD, vitamin
D; miR, microRNA; Rsbn1, round spermatid basic protein 1;
NC, negative control; Glut4, glucose transporter 4;
Cyp19a, aromatase. |
Glut4 mRNA expression was significantly
decreased in the miR-378d mimic group and significantly increased
in the inhibitor group, while Rsbn1 and Cyp19a mRNA
expression levels showed no significant differences (Fig. 3B). The western blotting results
demonstrated that the expression levels of Rsbn1 and
Glut4 were decreased after miR-378d overexpression, but were
increased after miR-378d inhibition, while Cyp19a showed no
significant change (Fig. 3C and D).
These results further verified that miR-378d directly regulates
RSBN1 in mouse granulosa cells.
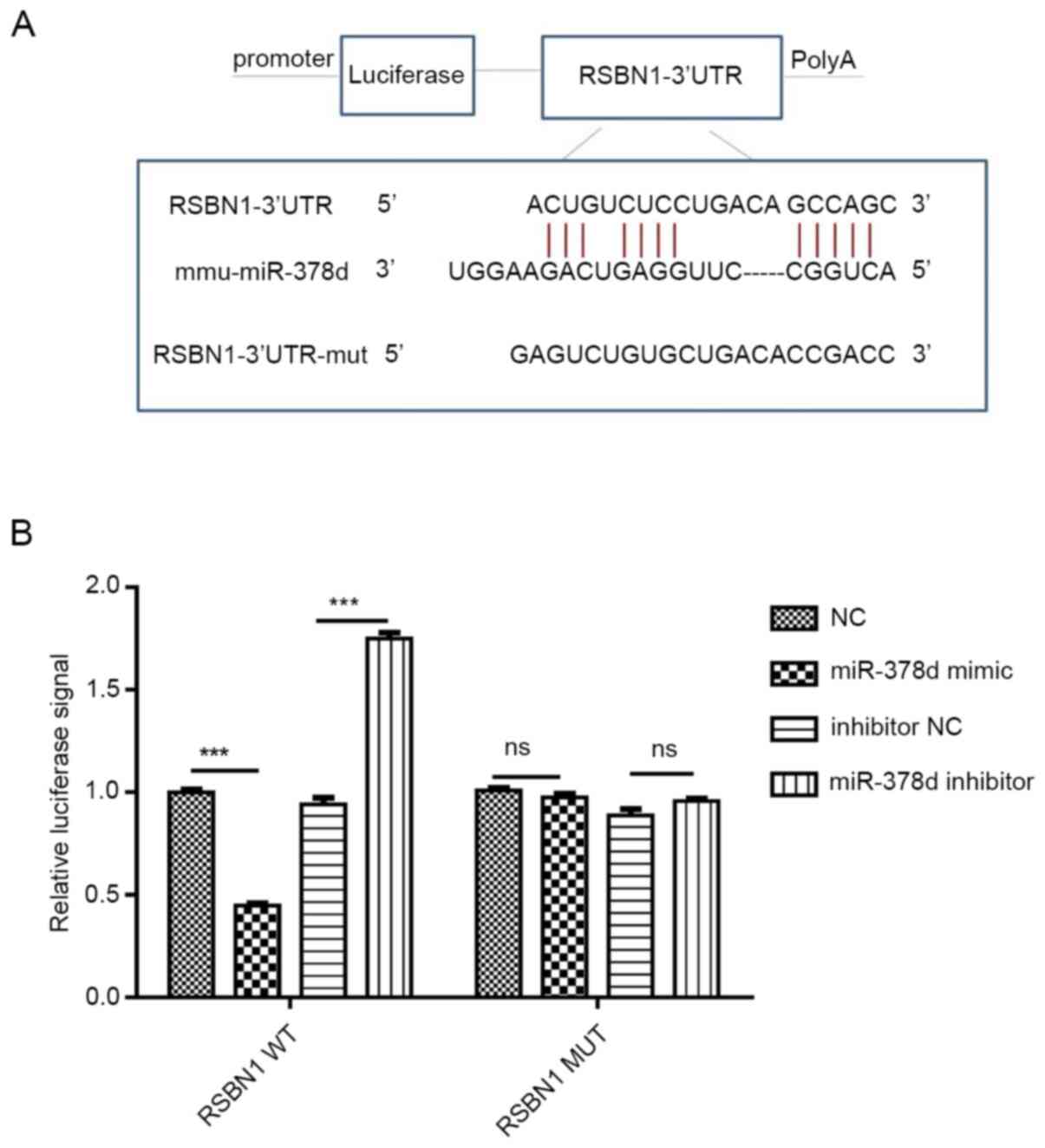
miR-378d targets 3′UTR of RSBN1
mRNA
WT and MUT plasmids of Rsbn1 3′UTR were
constructed according to their binding sites. After co-transfection
with miR-378d mimic or inhibitor, the results of luciferase
reporter gene assays revealed the following: WT Rsbn1 3′UTR
activity was decreased by mir-378d mimic (P<0.001) and increased
by miR-378d inhibitor, while miR-378d mimic and inhibitor had no
significant effect on MUT 3′UTR. These results indicate that
miR-378d can interact with Rsbn1 mRNA and bind to
Rsbn1 mRNA (Fig. 4A and B),
thus having a targeting role.
It was then investigated using rescue experiments
whether the Rsbn1 was a real functional target of miR-378d
in mouse granulosa cells. Western blot analysis demonstrated that
overexpression of miR-378d could downregulate the expression levels
of Rsbn1 and Glut4. After overexpression of
Rsbn1, the protein expression levels of Rsbn1 and
Glut4 were significantly upregulated. Moreover, after
co-transfection with Rsbn1 OE and miR378d mimic,
Rsbn1 and Glut4 expression levels were not
significantly change (Fig. 5A and
B). It was found that the downregulation of Rsbn1 after
the overexpression of miR-378d was reversed by the overexpression
of Rsbn1, while Cyp19a showed no significant change.
Inhibition of miR-378d could upregulate the expression levels of
Rsbn1 and Glut4 (Fig.
5C). After knocking down Rsbn1, the protein expression
levels of Rsbn1 and Glut4 were significantly
downregulated. Furthermore, after co-transfection with Rsbn1
knockdown and miR378d inhibitor, the protein expression levels of
Rsbn1 and Glut4 did not significantly change in
comparison with NC, which may be because of the upregulating
ability of miR-378d on Rsbn1 (Fig. 5C). It was identified that
Glut4 was decreased by the knockdown of Rsbn1, while
Cyp19a expression did not significantly change (Fig. 5C). In addition, RT-qPCR results
demonstrated that, compared with the control group, the mRNA
expression of Glut4 was significantly decreased after
overexpression of miR-378d, while the mRNA expression levels of
Rsbn1 and Cyp19a did not significantly change
(Fig. 5D). After the overexpression
of Rsbn1, the mRNA expression levels of Rsbn1 and
Glut4 were significantly increased, but Cyp19a mRNA
expression did not change (Fig.
5D).
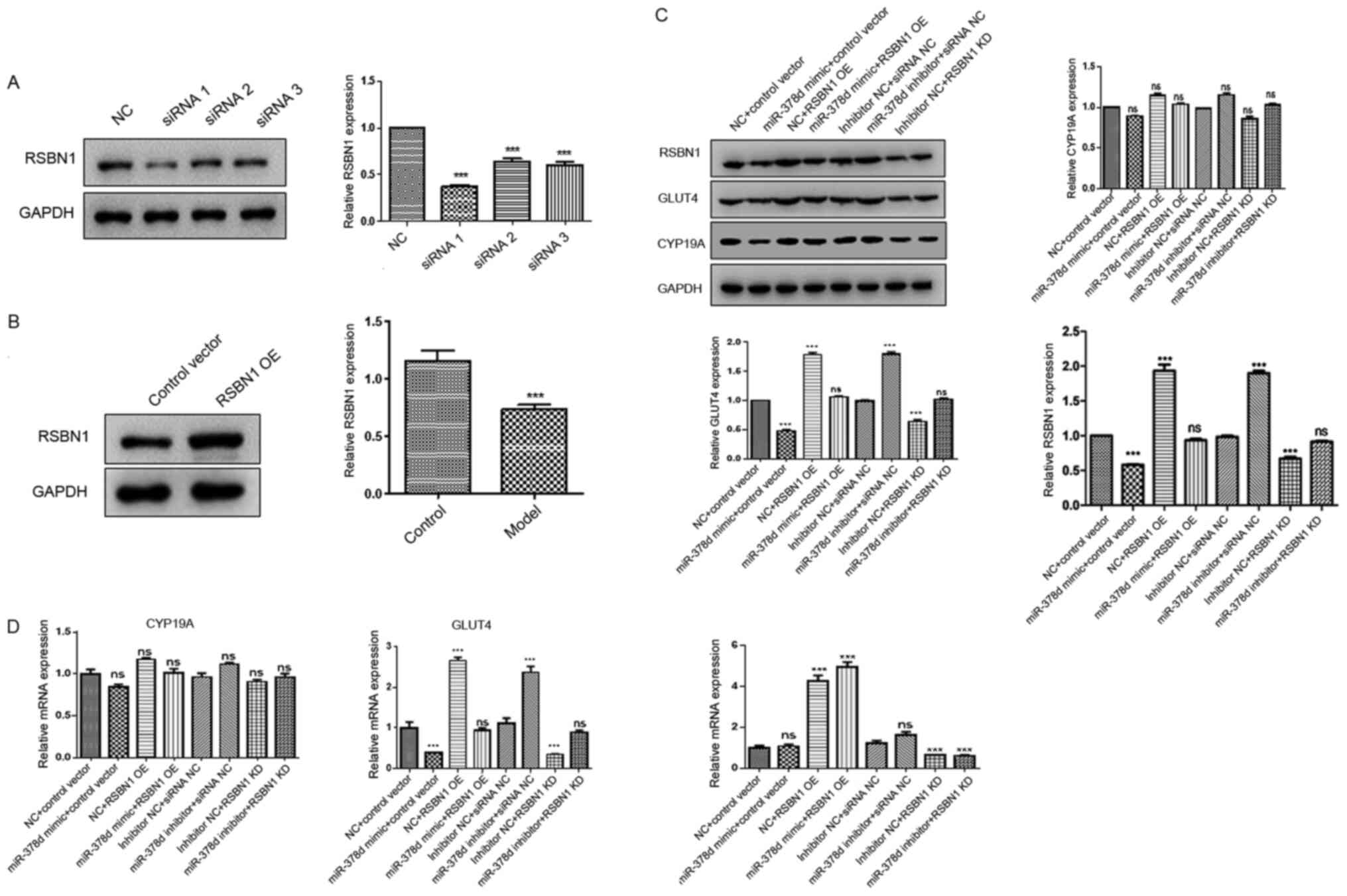 | Figure 5.Rsbn1 is a functional target of
miR-378d in mouse granulosa cells. (A) Western blotting results
showed that siRNA 1 interference was the most efficient. (B)
Western blotting results demonstrated that Rsbn1 expression was
upregulated in the overexpression group. (C) Expression levels of
Rsbn1, Glut4 and Cyp19a were detected via western
blotting after overexpression or knockdown of miR-378d. (D) mRNA
expression levels of Rsbn1, Glut4 and Cyp19a were
detected via reverse transcription-quantitative PCR after miR-378d
interference. ***P<0.001. miR, microRNA; Rsbn1, round
spermatid basic protein 1; NC, negative control; Glut4,
glucose transporter 4; Cyp19a, aromatase; OE,
overexpression; siRNA, small interfering RNA; ns,
non-significant. |
Discussion
Accumulating evidence has suggested that VD is
directly or indirectly associated with fertility disorders in women
(32). Moreover, VD deficiency is
associated with a decrease in calcium and phosphorus. Sun et
al (33) reported that when
calcium and phosphorus are supplemented to the mice with VDR or 1-a
hydroxylase gene knockout; their reproductive dysfunctions could be
corrected, suggesting that the effects of VD on the fertility in
female mice is achieved by regulating the levels of calcium and
phosphorus. From a genetic point of view, mutations in VD-related
genes in diabetic patients may be associated with glucose tolerance
and inflammatory responses (34).
Aghajafari et al (35)
conducted a systematic review and meta-analysis, which revealed
that VD deficiency in pregnant women could increase the risk of
gestational diabetes, preeclampsia and premature birth.
Furthermore, VD supplementation can antagonize the negative effects
of preeclampsia on the number and function of fetal endothelial
cell colony-forming cells (36).
Another study that performed in vitro fertilization and
embryo transfer (IVF-ET) for 132 infertile women reported that the
25(OH)D levels in the serum and follicular fluid of non-pregnant
women were significantly lower compared with those of pregnant
women. At the same time, implantation rates were also relatively
low in patients with low VD levels (37). A different study that determined the
VD levels in the serum and follicular fluid of 173 women after
IVF-ET showed that there were no significant differences in the
gonadotropin dose and endometrial thickness between the two groups
of women with adequate or insufficient 25(OH)D3 levels (13). Moreover, Polyzos et al
(38) found that VD was associated
with pregnancy rates during assisted reproductive technology
treatment. During the treatment with ovulation drugs, both the
number of formed mature oocytes and pregnancy rates were much
higher in the patients with high VD levels compared with those
found in patients with low VD levels.
The application of VD-deficient feed combined with a
UV-free environment is one of the most commonly used methods to
establish animal models with VD deficiency, which is also in line
with the current characteristics of reduced VD levels caused by
unbalanced diet structure and reduced daylight exposure (39). The mouse model of VD deficiency was
established as previously described (28), and transcriptome sequencing analysis
revealed that miR-378d was abnormally overexpressed in the ovarian
tissues of VD-deficient mice, which was further confirmed using
RT-qPCR of the extracted ovarian tissues from the mice. At present,
there are few studies on the mechanisms of miR-378d, and these
mainly focus on the analysis of expression differences (40,41).
Kurowska et al (42) have
reported that the downregulation of miR-378d may be used as a
molecular marker of arthritis progression. Moreover, spectrum
analysis of miRNA differences using a microarray platform revealed
that the expression of miR-378d was significantly higher in colitis
mucosa compared with in uninflamed mucosa (43).
The essential aspect of studying the biological
functions and mechanisms of miRNAs is the accurate identification
of the target genes of the miRNAs (44). In order to further investigate the
molecular mechanisms of miR-378d in the ovaries of the VD-deficient
mice, bioinformatics software was used in the current to perform
prediction analysis, which revealed that Rsbn1 may be the
target gene of miR-378d.
Rsbn1 is a potential target gene of HIF-1α,
which may be involved in a variety of pathophysiological processes.
Ohyama et al (45) reported
that the expression of Rsbn1 in mature testis is a sign of
spermatogenesis since its expression increases during the
development of round spermatids in male mice. Rsbn1 is also
associated with the expression of anti-Mullerian hormone (AMH), as
well as is closely associated with the production of steroid
hormones (45,46). Herein, it was found that, compared
with the control group, the protein expression levels of
Rsbn1 in the ovaries of VD-deficient mice were significantly
decreased, while there were no significant changes in the mRNA
expression levels. Furthermore, the double reporter gene
experiments confirmed that miR-378d could bind with Rsbn1
mRNA to exert its targeting role. Functional rescue experiments
demonstrated that overexpression of miR-378d could downregulate the
expression levels of Rsbn1 and Glut4 proteins.
Moreover, knocking down Rsbn1 could downregulate the protein
expression levels of Rsbn1 and Glut4, while
simultaneously inhibiting miR-378d and knocking down Rsbn1
had no significant effect on the protein expression levels of
Rsbn1 and Glut4. These results suggested that in the
absence of VD, miR-378d was abnormally increased, and the
expression of Rsbn1 protein was downregulated, thus
affecting the expression of downstream molecule Glut4, which
may lead to insulin resistance. Regardless of overexpression or
knockdown of miR-378d, Cyp19a showed no obvious changes,
therefore suggesting that miR-378d may not be significantly
associated with the production of steroid hormones. This finding is
not in line with a previous study reporting that miR-378d could
regulate the production of estradiol in pig ovaries by targeting
the aromatase (47). At the same
time, it suggests that there is no clear correlation between VD and
AMH, which is consistent with the conclusions of Lata et al
(48) and Pearce et al
(49).
In summary, the present study demonstrated that
miR-378d was abnormally upregulated in the ovaries of VD deficient
mice, which could negatively regulate Rsbn1, eventually
affecting the expression of its downstream molecule Glut4.
Furthermore, this study revealed a potential molecular marker for
the mechanistic studies of VD deficiency, as well as provided novel
insights for the diagnosis and treatment of insulin resistance.
Supplementary Material
Supporting Data
Acknowledgements
Not applicable.
Funding
The present study was supported by the Changezhou
Sci & Tech Program (grant no. CJ20200103), Changezhou Sci &
Tech Program (grant no. CJ20180028), the Young Scientists
Foundation of Changzhou No. 2 People's Hospital (grant no.
2019K009) and the Guiding Science and Technology Project of
Changzhou Municipal Health Commission (grant no. WZ201815).
Availability of data and materials
The datasets used and/or analyzed during the present
study are available from the corresponding author on reasonable
request.
Authors' contributions
HS and FX conducted the studies, participated in
collecting data and drafted the manuscript. HS, YShang and XC are
responsible for confirming the authenticity of the raw data. YShi,
HS and FX performed the statistical analysis and participated in
its design. HS, YShang and XC participated in acquisition, analysis
and interpretation of data, and draft the manuscript. All authors
read and approved the final manuscript.
Ethics approval and consent to
participate
All procedures related to animal use were approved
by the Animal Care and Use Committee of Nanjing Medical University.
This study was carried out in strict accordance with the
recommendations in the Guide for the Care and Use of Laboratory
Animals of the National Institutes of Health.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests
References
|
1
|
Holick MF: Vitamin D deficiency. N Engl J
Med. 357:266–281. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Holick MF, Binkley NC, Bischoff-Ferrari
HA, Gordon CM, Hanley DA, Heaney RP, Murad MH and Weaver CM:
Guidelines for preventing and treating vitamin D deficiency and
insufficiency revisited. J Clin Endocrinol Metab. 97:1153–1158.
2012. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Nerhus M, Berg AO, Simonsen C, Haram M,
Haatveit B, Dahl SR, Gurholt TP, Bjella TD, Ueland T, Andreassen
OA, et al: Vitamin D Deficiency Associated With Cognitive
Functioning in Psychotic Disorders. J Clin Psychiatry.
78:e750–e757. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Ponsonby AL, McMichael A and van der Mei
I: Ultraviolet radiation and autoimmune disease: Insights from
epidemiological research. Toxicology. 181-182:71–78. 2002.
View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Ford JA, MacLennan GS, Avenell A, Bolland
M, Grey A and Witham M; RECORD Trial Group, : Cardiovascular
disease and vitamin D supplementation: Trial analysis, systematic
review, and meta-analysis. Am J Clin Nutr. 100:746–755. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Gagnon C, Lu ZX, Magliano DJ, Dunstan DW,
Shaw JE, Zimmet PZ, Sikaris K, Ebeling PR and Daly RM: Low serum
25-hydroxyvitamin D is associated with increased risk of the
development of the metabolic syndrome at five years: results from a
national, population-based prospective study (The Australian
Diabetes, Obesity and Lifestyle Study: AusDiab). J Clin Endocrinol
Metab. 97:1953–1961. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Muscogiuri G, Mitri J, Mathieu C,
Badenhoop K, Tamer G, Orio F, Mezza T, Vieth R, Colao A and Pittas
A: Mechanisms in endocrinology: Vitamin D as a potential
contributor in endocrine health and disease. Eur J Endocrinol.
171:R101–R110. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
van Schoor N and Lips P: Global Overview
of Vitamin D Status. Endocrinol Metab Clin North Am. 46:845–870.
2017. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Tanamura A, Nomura S, Kurauchi O, Furui T,
Mizutani S and Tomoda Y: Purification and characterization of
1,25(OH)2D3 receptor from human placenta. J Obstet Gynaecol (Tokyo
1995). 21:631–639. 1995. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Pérez-Fernandez R, Alonso M, Segura C,
Muñoz I, García-Caballero T and Diguez C: Vitamin D receptor gene
expression in human pituitary gland. Life Sci. 60:35–42. 1997.
View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Thill M, Becker S, Fischer D, Cordes T,
Hornemann A, Diedrich K, Salehin D and Friedrich M: Expression of
prostaglandin metabolising enzymes COX-2 and 15-PGDH and VDR in
human granulosa cells. Anticancer Res. 29:3611–3618.
2009.PubMed/NCBI
|
|
12
|
Parikh G, Varadinova M, Suwandhi P, Araki
T, Rosenwaks Z, Poretsky L and Seto-Young D: Vitamin D regulates
steroidogenesis and insulin-like growth factor binding protein-1
(IGFBP-1) production in human ovarian cells. Horm Metab Res.
42:754–757. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Zhao J, Huang X, Xu B, Yan Y, Zhang Q and
Li Y: Whether vitamin D was associated with clinical outcome after
IVF/ICSI: A systematic review and meta-analysis. Reprod Biol
Endocrinol. 16:132018. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Jiang R, Ding L, Zhou J, Huang C, Zhang Q,
Jiang Y, Liu J, Yan Q, Zhen X, Sun J, et al: Enhanced HOXA10
sumoylation inhibits embryo implantation in women with recurrent
implantation failure. Cell Death Discov. 3:170572017. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Wei Z, Yoshihara E, He N, Hah N, Fan W,
Pinto AFM, Huddy T, Wang Y, Ross B, Estepa G, et al: Vitamin D
Switches BAF Complexes to Protect β Cells. Cell. 173:1135–1149.e15.
2018. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Liang Y, Ridzon D, Wong L and Chen C:
Characterization of microRNA expression profiles in normal human
tissues. BMC Genomics. 8:1662007. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Jiang XP, Ai WB, Wan LY, Zhang YQ and Wu
JF: The roles of microRNA families in hepatic fibrosis. Cell
Biosci. 7:342017. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Weng WH, Leung WH, Pang YJ and Hsu HH:
Lauric acid can improve the sensitization of Cetuximab in KRAS/BRAF
mutated colorectal cancer cells by retrievable microRNA-378
expression. Oncol Rep. 35:107–116. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Ji KX, Cui F, Qu D, Sun RY, Sun P, Chen
FY, Wang SL and Sun HS: miR-378 promotes the cell proliferation of
non-small cell lung cancer by inhibiting FOXG1. Eur Rev Med
Pharmacol Sci. 22:1011–1019. 2018.PubMed/NCBI
|
|
20
|
Ganesan J, Ramanujam D, Sassi Y, Ahles A,
Jentzsch C, Werfel S, Leierseder S, Loyer X, Giacca M, Zentilin L,
et al: miR-378 controls cardiac hypertrophy by combined repression
of mitogen-activated protein kinase pathway factors. Circulation.
127:2097–2106. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Hyun J, Wang S, Kim J, Rao KM, Park SY,
Chung I, Ha CS, Kim SW, Yun YH and Jung Y: MicroRNA-378 limits
activation of hepatic stellate cells and liver fibrosis by
suppressing Gli3 expression. Nat Commun. 7:109932016. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Ishida M, Shimabukuro M, Yagi S, Nishimoto
S, Kozuka C, Fukuda D, Soeki T, Masuzaki H, Tsutsui M and Sata M:
MicroRNA-378 regulates adiponectin expression in adipose tissue: A
new plausible mechanism. PLoS One. 9:e1115372014. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Gungormez C, Gumushan Aktas H, Dilsiz N
and Borazan E: Novel miRNAs as potential biomarkers in stage II
colon cancer: Microarray analysis. Mol Biol Rep. 46:4175–4183.
2019. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Meerson A, Eliraz Y, Yehuda H, Knight B,
Crundwell M, Ferguson D, Lee BP and Harries LW: Obesity impacts the
regulation of miR-10b and its targets in primary breast tumors. BMC
Cancer. 19:862019. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Dupont J and Scaramuzzi RJ: Insulin
signalling and glucose transport in the ovary and ovarian function
during the ovarian cycle. Biochem J. 473:1483–1501. 2016.
View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Laganà AS, Vitale SG, Ban Frangež H,
Vrtačnik-Bokal E and D'Anna R: Vitamin D in human reproduction: The
more, the better? An evidence-based critical appraisal. Eur Rev Med
Pharmacol Sci. 21:4243–4251. 2017.PubMed/NCBI
|
|
27
|
Zarnani AH, Shahbazi M, Salek-Moghaddam A,
Zareie M, Tavakoli M, Ghasemi J, Rezania S, Moravej A, Torkabadi E,
Rabbani H, et al: Vitamin D3 receptor is expressed in the
endometrium of cycling mice throughout the estrous cycle. Fertil
Steril. 93:2738–2743. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Li P, Xu X, Cao E, Yu B, Li W, Fan M,
Huang M, Shi L, Zeng R, Su X, et al: Vitamin D deficiency causes
defective resistance to Aspergillus fumigatus in mice via
aggravated and sustained inflammation. PLoS One. 9:e998052014.
View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Lamas S, Carvalheira J, Gartner F and
Amorim I: C57BL/6J mouse superovulation: Schedule and age
optimization to increase oocyte yield and reduce animal use.
Zygote. Jan 15–2021.(Epub ahead of print). doi:
10.1017/S0967199420000714. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(-Delta Delta C(T)) Method. Methods. 25:402–408. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Mao Z, Zhao H, Qin Y, Wei J, Sun J, Zhang
W and Kang Y: Post-Transcriptional Dysregulation of microRNA and
Alternative Polyadenylation in Colorectal Cancer. Front Genet.
11:642020. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Lerchbaum E and Obermayer-Pietsch B:
Vitamin D and fertility: A systematic review. Eur J Endocrinol.
166:765–778. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Sun W, Xie H, Ji J, Zhou X, Goltzman D and
Miao D: Defective female reproductive function in
1,25(OH)2D-deficient mice results from indirect effect mediated by
extracellular calcium and/or phosphorus. Am J Physiol Endocrinol
Metab. 299:E928–E935. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Takiishi T, Gysemans C, Bouillon R and
Mathieu C: Vitamin D and diabetes. Rheum Dis Clin North Am.
38:179–206. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Aghajafari F, Nagulesapillai T, Ronksley
PE, Tough SC, O'Beirne M and Rabi DM: Association between maternal
serum 25-hydroxyvitamin D level and pregnancy and neonatal
outcomes: Systematic review and meta-analysis of observational
studies. BMJ. 346 (mar26 4):f11692013. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
von Versen-Höynck F, Brodowski L, Dechend
R, Myerski AC and Hubel CA: Vitamin D antagonizes negative effects
of preeclampsia on fetal endothelial colony forming cell number and
function. PLoS One. 9:e989902014. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Fung JL, Hartman TJ, Schleicher RL and
Goldman MB: Association of vitamin D intake and serum levels with
fertility: Results from the Lifestyle and Fertility Study. Fertil
Steril. 108:302–311. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Polyzos NP, Anckaert E, Guzman L,
Schiettecatte J, Van Landuyt L, Camus M, Smitz J and Tournaye H:
Vitamin D deficiency and pregnancy rates in women undergoing single
embryo, blastocyst stage, transfer (SET) for IVF/ICSI. Hum Reprod.
29:2032–2040. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Duchow EG, Cooke NE, Seeman J, Plum LA and
DeLuca HF: Vitamin D binding protein is required to utilize
skin-generated vitamin D. Proc Natl Acad Sci USA. 116:24527–24532.
2019. View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Liu C, Lai Y, Ying S, Zhan J and Shen Y:
Plasma exosome-derived microRNAs expression profiling and
bioinformatics analysis under cross-talk between increased
low-density lipoprotein cholesterol level and ATP-sensitive
potassium channels variant rs1799858. J Transl Med. 18:4592020.
View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Zhang X, Mens MMJ, Abozaid YJ, Bos D,
Darwish Murad S, de Knegt RJ, Ikram MA, Pan Q and Ghanbari M:
Circulatory microRNAs as potential biomarkers for fatty liver
disease: The Rotterdam study. Aliment Pharmacol Ther. 53:432–442.
2021.PubMed/NCBI
|
|
42
|
Kurowska W, Kuca-Warnawin E, Radzikowska
A, Jakubaszek M, Maślińska M, Kwiatkowska B and Maśliński W:
Monocyte-related biomarkers of rheumatoid arthritis development in
undifferentiated arthritis patients - a pilot study. Reumatologia.
56:10–16. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
43
|
Valmiki S, Ahuja V and Paul J: MicroRNA
exhibit altered expression in the inflamed colonic mucosa of
ulcerative colitis patients. World J Gastroenterol. 23:5324–5332.
2017. View Article : Google Scholar : PubMed/NCBI
|
|
44
|
Bolha L, Pižem J, Frank-Bertoncelj M,
Hočevar A, Tomšič M and Jurčić V: Identification of microRNAs and
their target gene networks implicated in arterial wall remodelling
in giant cell arteritis. Rheumatology (Oxford). 59:3540–3552. 2020.
View Article : Google Scholar : PubMed/NCBI
|
|
45
|
Ohyama K, Ohta M, Hosaka YZ, Tanabe Y,
Ohyama T and Yamano Y: Expression of anti-Müllerian hormone and its
type II receptor in germ cells of maturing rat testis. Endocr J.
62:997–1006. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
46
|
Abu-Jamous B, Buffa FM, Harris AL and
Nandi AK: In vitro downregulated hypoxia transcriptome is
associated with poor prognosis in breast cancer. Mol Cancer.
16:1052017. View Article : Google Scholar : PubMed/NCBI
|
|
47
|
Xu S, Linher-Melville K, Yang BB, Wu D and
Li J: Micro-RNA378 (miR-378) regulates ovarian estradiol production
by targeting aromatase. Endocrinology. 152:3941–3951. 2011.
View Article : Google Scholar : PubMed/NCBI
|
|
48
|
Lata I, Tiwari S, Gupta A and Yadav S and
Yadav S: To Study the Vitamin D Levels in Infertile Females and
Correlation of Vitamin D Deficiency with AMH Levels in Comparison
to Fertile Females. J Hum Reprod Sci. 10:86–90. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
49
|
Pearce K, Gleeson K and Tremellen K: Serum
anti-Mullerian hormone production is not correlated with seasonal
fluctuations of vitamin D status in ovulatory or PCOS women. Hum
Reprod. 30:2171–2177. 2015. View Article : Google Scholar : PubMed/NCBI
|