Introduction
Esophageal squamous cell carcinoma (ESCC) is defined
as the major histological type of esophageal cancer (1,2) and as
the seventh leading cause of tumor-associated death globally
(3). ESCC is one of the most
malignant and aggressive tumors worldwide, with ~410,000 deaths
occurring annually (4). According
to previous findings, the 3-year overall survival rate in certain
areas of China was only ~30% after radiotherapy alone, whereas the
recurrence rate was as high as 60–80% due to the existence of
certain risk factors (5,6). Despite progress in diagnostic
technologies and therapeutics, including chemoradiotherapy and
surgery, most patients are diagnosed at the advanced stage and the
prognosis of patients with ESCC remains unsatisfactory owing to
recurrence and metastasis (7–9), with
an overall 5-year survival rate <30% (10). Previous studies have reported that
several important parameters, such as life style as well as
environmental and genetic factors, contribute to ESCC tumorigenesis
(11,12); however, the underlying mechanisms
triggering ESCC progression have not been fully defined.
Long non-coding RNAs (lncRNAs) are a class of
non-protein coding transcripts >200 nucleotides in length
(13). lncRNAs serve a crucial role
in a variety of processes in diseases, and their alterations are
recognized as drivers or suppressors for cancer progression,
including ESCC (14,15). Rapidly accumulating evidence has
suggested that lncRNA BRAF-activated non-coding RNA (BANCR) has
crucial roles in the progression of various malignancies, including
colorectal cancer (16), papillary
thyroid carcinoma (17) and
endometrial cancer (18), via
promoting proliferation, migration and invasion of cancer cells.
Moreover, a previous study has suggested that BANCR is highly
expressed in tumor tissues compared with in non-cancerous tissues
of patients with ESCC and its expression level was positively
associated with tumor differentiation, metastasis and tumor stage
(1). Moreover, increased BANCR
expression was observed in the plasma of patients with ESCC, which
was then decreased following tumor resection (5). Additionally, BANCR has been revealed
to exert regulative effects on the migration and invasion of ESCC
cells via the Wnt/β-catenin signaling pathway (19). BANCR expression was also negatively
associated with the survival rate of patients with ESCC, suggesting
BANCR as a novel tumor biomarker for the early detection of ESCC
progression (5). It has been
reported that BANCR mediates ESCC progression by regulating
insulin-like growth factor 1 receptor expression via
microRNA-338-3p (20). However, the
underlying mechanism of BANCR in ESCC pathogenesis has not been
fully understood. Thus, the aim of the present study was to further
investigate the molecular mechanism of BANCR in ESCC
progression.
The aim of the present study was to investigate the
role of BANCR in the pathogenesis of ESCC, and study its potential
molecular mechanism. Thus, ESCC cells were transfected with
overexpression plasmid and short hairpin RNAs (shRNAs) to regulate
BANCR expression in order to determine whether BANCR could regulate
the proliferative, migratory and invasive capabilities of ESCC
cells.
Materials and methods
Cell culture and transfection
ESCC cell lines, including Eca-109, KYSE30, KYSE150
and TE-1, together with the normal esophageal Het-1A cell line,
were obtained from The Cell Bank of Type Culture Collection of The
Chinese Academy of Sciences. The cell lines were cultured in
RPMI-1640 medium with 10% FBS (Gibco; Thermo Fisher Scientific,
Inc., USA) at 37°C with 5% CO2. The MEK inhibitor, U0126
(Selleck Chemicals), was diluted using DMSO to a working
concentration of 20 µM. Eca-109 cells were treated with U0126 for
24 h at 37°C.
Eca-109 cells at the logarithmic growth phase were
transfected with 100 nM BANCR short hairpin (sh)RNAs or plasmids
using Lipofectamine® 2000 (Invitrogen; Thermo Fisher
Scientific, Inc.) at 37°C for 6 h and incubated for another 24 h
before subsequent experimentation, according to the manufacturer's
instructions. The shRNAs were obtained from Shanghai GenePharma
Co., Ltd. The shRNA sequences were as follows: sh-BANCR-1 forward,
5′-GGACUCCAUGGCAAACGUUTT-3′ and reverse,
5′-AACGUUUGCCAUGGAGUCCTT-3′; sh-BANCR-2 forward,
5′-GGAGUGGCGACUAUAGCAATT-3′ and reverse,
5′-UUGCUAUAGUCGCCACUCCTT-3′; and shRNA-NC (scrambled) forward,
5′-UUCUCCGAACGUGUCACGUTT-3′ and reverse,
5′-ACGUGACACGUUCGGAGAATT-3′. The full-length sequences of BANCR
(NR_047671) were inserted into pcDNA3.1 vectors (Invitrogen; Thermo
Fisher Scientific, Inc.) to construct BANCR overexpression plasmid.
The Eca-109 cells transfected with BANCR overexpression plasmid
were classified into the BANCR group, and empty pcDNA3.1 vectors
were used as the control group.
Reverse transcription-quantitative PCR
(RT-qPCR)
After transfection, total RNA was isolated from
Eca-109 cells using TRIzol® reagent (Invitrogen; Thermo
Fisher Scientific, Inc.) and then reverse transcribed using the
PrimeScript RT kit (Takara Bio, Inc.) to synthesize the cDNA. The
prepared reaction mixture was incubated at 37°C for 60 min after
brief centrifugation (14,000 × g, 10 min), followed by incubation
at 85°C for 5 min for RT. Relative expression levels were
determined using quantitative primers and SYBR® Premix
Ex Taq™ reagent (Takara Bio, Inc.). The thermocycling conditions
were as follows: Pre-denaturation at 95°C for 5 min, followed by 40
cycles at 95°C for 15 sec, 60°C for 30 sec and 72°C for 30 sec. The
results were calculated using the 2−ΔΔCq method
(21). The primers used for qPCR
were: BANCR forward, 5′-ACAGGACTCCATGGCAAACG-3′ and reverse,
5′-ATGAAGAAAGCCTGGTGCAGT-3′; and GAPDH (internal reference)
forward, 5′-GGTCTCCTCTGACTTCAACA-3′ and reverse,
5′-AGCCAAATTCGTTGTCATAC-3′.
Cell viability assay
The survival rate of Eca-109 cells in each group was
determined using the Cell Counting Kit-8 (CCK-8; Dojindo Molecular
Technologies, Inc.) assay. Briefly, Eca-109 cells (2×103
cells/well) were seeded into 96-well plates and transfected with
BANCR overexpression plasmids or sh-BANCR, as aforementioned. After
incubation for 24, 48 or 72 h, the CCK-8 reagent (10 µl/well) was
added to Eca-109 cells for another 2 h. Absorbance at 450 nm was
measured using an enzyme-linked immunosorbent assay plate reader
(Bio-Rad Laboratories, Inc.).
Colony formation assay
After transfection, Eca-109 cells
(1×103/well) were seeded into 6-well plates. The cells
were cultured for 14 days at 37°C without disturbance during the
period of incubation to form the cell clusters. Subsequently, cell
colonies were fixed in 4% paraformaldehyde and stained with 0.1%
crystal violet solution for 30 min at room temperature before being
imaged, and colonies with diameters >0.5 mm were imaged and
counted using a digital camera (Nikon Corporation).
Wound healing assay
Eca-109 cells (2×105 cells/well) were
seeded in 6-well plates and transfected until cell confluency
reached ~80-90% on the following day. A straight linear wound was
gently induced using 200-µl sterile pipette tips, and the cells
were cultured in RPMI-1640 culture medium with 1% FBS at 37°C for
24 h. Subsequently, the wound was observed and photographed under a
fluorescence microscope (Olympus IX53; Olympus Corporation) at five
random fields (magnification, ×200). The average distance between
cells was calculated relative to the control by using ImageJ
software (version 1.48; National Institutes of Health) to determine
the wound closure rate.
Transwell chamber assay
After transfection, Eca-109 cells (1×105
cells/well) were resuspended in FBS-free RPMI-1640 medium and
seeded into the upper chambers of Transwell plates precoated with
Matrigel at 37°C for 4 h. The lower chamber was filled with 500 µl
culture RPMI-1640 medium containing 10% FBS, and then maintained at
37°C in 5% CO2 for 24 h. After incubation, the membrane
was fixed with 4% paraformaldehyde for 30 min at 37°C. The
remaining cells on the upper surface of the filter membrane were
removed gently, while cells at the lower surface of the membrane
were stained with 0.1% crystal violet for 30 min at room
temperature. The cells were captured in five random fields with a
fluorescence microscope (Olympus IX53; Olympus Corporation) at ×200
magnification.
Western blotting
Proteins were extracted from transfected Eca-109
cells using RIPA lysis buffer (Takara Bio, Inc.) containing
protease inhibitors according to the manufacturer's instructions,
and then quantified using a BCA assay kit. Total proteins (20
µg/lane) were loaded and separated via SDS-PAGE on 10% gel, and
then separated proteins were transferred onto PVDF membranes. After
membranes were blocked with 5% skimmed milk for 2 h at room
temperature, primary antibodies against the following proteins were
used overnight at 4°C: Matrix metalloproteinase 2 (MMP2; 1:1,000;
cat. no. ab181286; Abcam), MMP9 (1:1,000; cat. no. ab76003; Abcam),
phosphorylated (p)-c-Raf (1:1,000; cat. no. 9423; Cell Signaling
Technology, Inc.), c-Raf (1:1,000; cat. no. 9422; Cell Signaling
Technology, Inc.), p-MEK1/2 (1:1,000; cat. no. 9121; Cell Signaling
Technology, Inc.), MEK1/2 (1:1,000; cat. no. 9122; Cell Signaling
Technology, Inc.), p-ERK1/2 (1:500; cat. no. 9101; Cell Signaling
Technology, Inc.), ERK1/2 (1:1,000; cat. no. 9102; Cell Signaling
Technology, Inc.) and GAPDH (1:1,000; cat. no. 5174; Cell Signaling
Technology, Inc.). After washing with PBS, the membranes were
incubated with HRP-conjugated secondary antibodies (1:5,000; cat.
no. 7074; Cell Signaling Technology, Inc.) at room temperature for
2 h, and then visualized using ECL chemiluminescence (Pierce;
Thermo Fisher Scientific, Inc.). Finally, the gray values of bands
were detected using ImageJ software (version 1.48; National
Institutes of Health) and normalized to GAPDH.
Statistical analysis
Statistical analyses were performed using SPSS 19.0
(IBM Corp.), and data are presented as the mean ± SEM from three
independent experiments. Comparisons of multiple groups were
analyzed with one-way ANOVA followed by Tukey's post-hoc test.
P<0.05 was considered to indicate a statistically significant
difference.
Results
LncRNA BANCR is highly expressed in
ESCC cells compared with in normal esophageal cells
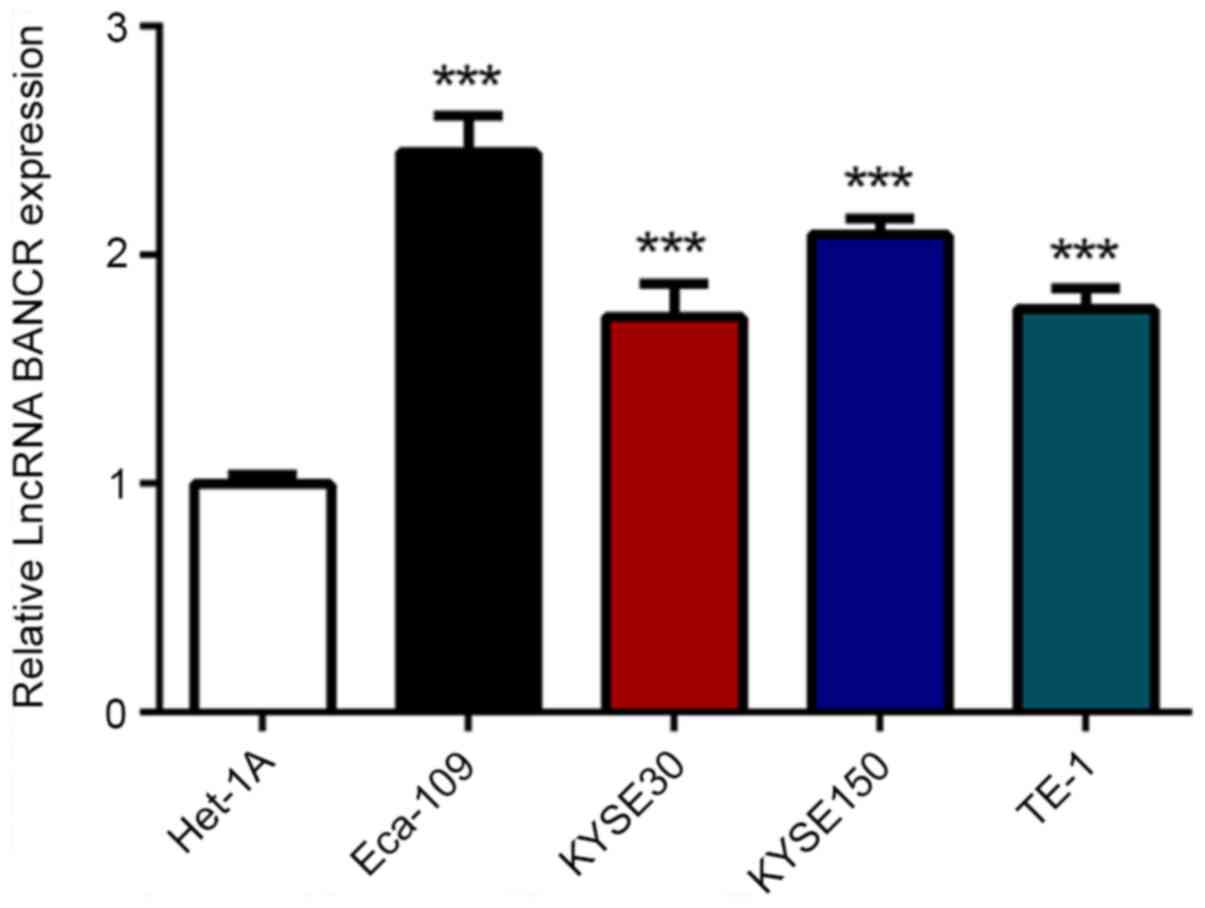
To assess BANCR expression in ESCC cells, RT-qPCR
was applied in the four ESCC cell lines, Eca-109, KYSE30, KYSE150
and TE-1, and one normal esophageal cell line, Het-1A. As shown in
Fig. 1, BANCR expression was
significantly increased in ESCC cells compared with in Het-1A
cells. Moreover, the Eca-109 cell line exhibited the highest level
of BANCR expression and was therefore used for subsequent
experiments.
lncRNA BANCR overexpression promotes
the proliferation, migration and invasion of ESCC cells, and BANCR
silencing induces the opposite effects
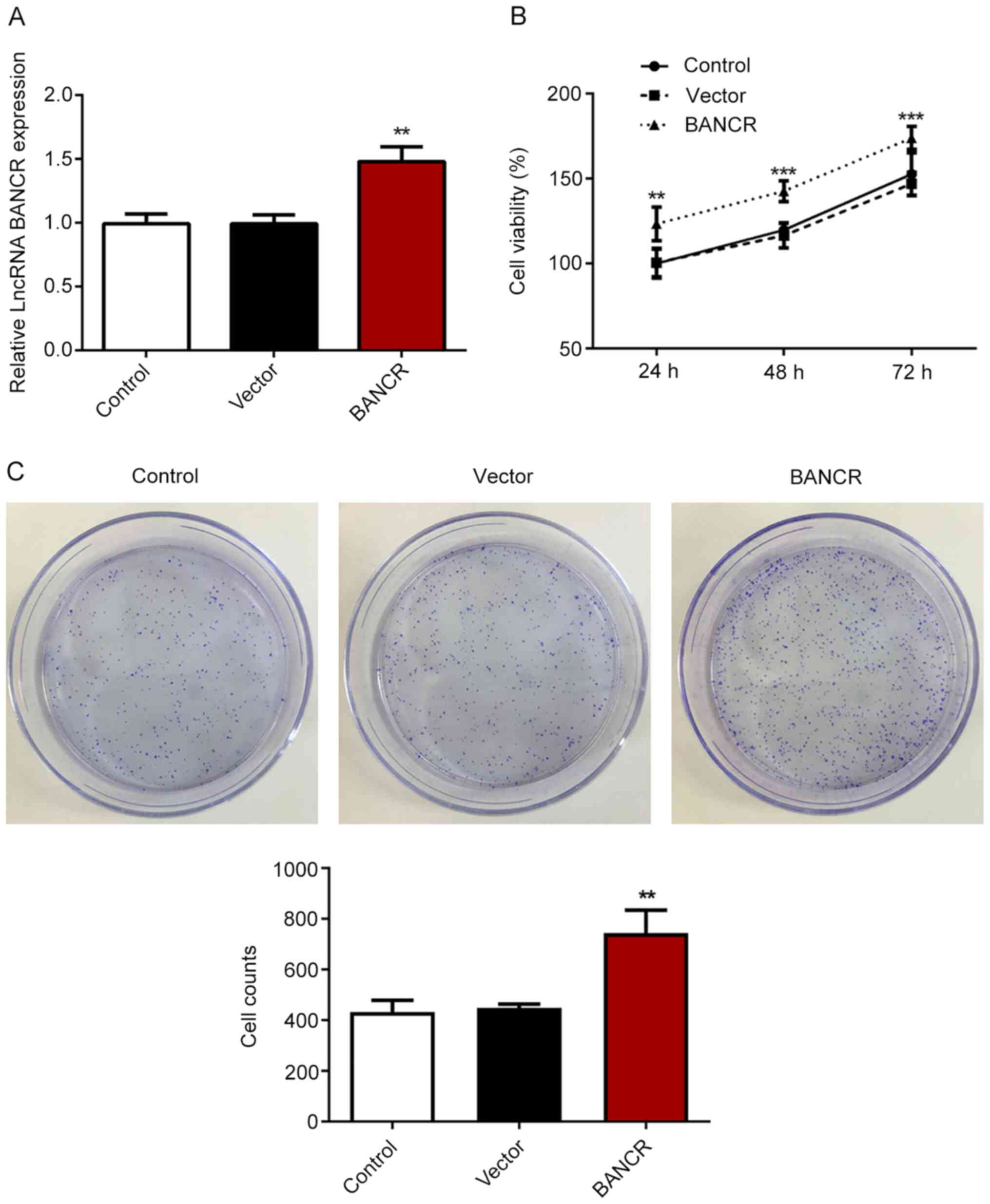
To evaluate the efficiency of BANCR plasmids in
Eca-109 cells, RT-qPCR was performed. The results revealed that
cells transfected with BANCR had a significantly higher BANCR
expression compared with control or vector cells (Fig. 2A). Subsequently, CCK-8 and colony
formation assays were performed to determine the viability and
proliferative ability, respectively. The findings indicated that
BANCR overexpression significantly enhanced the viability and
proliferative ability of Eca-109 cells (Fig. 2B and C), suggesting the promoting
effect of BANCR overexpression on the proliferation of ESCC cells.
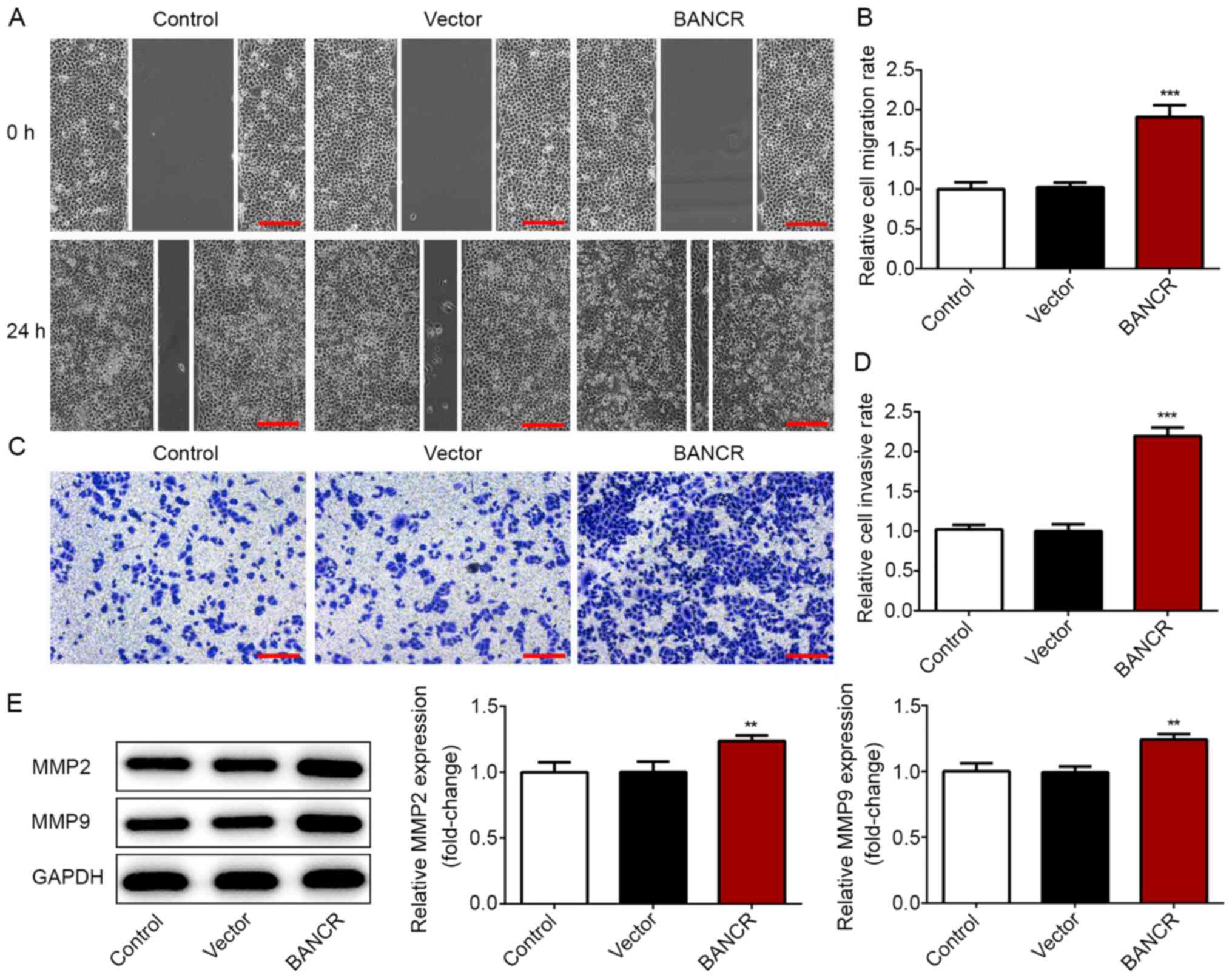
Additionally, wound healing and Transwell chamber assays were
performed to detect the migratory and invasive abilities,
respectively. The data revealed that the migratory and invasive
capacities were significantly elevated in Eca-109 cells from the
BANCR group compared with in those of the control and vector groups
(Fig. 3A-D). Furthermore, western
blot analysis was performed to determine the expression levels of
MMP2 and MMP9, two markers associated with tumor invasion and
metastasis. As shown in Fig. 3E,
BANCR overexpression led to a significant upregulation of MMP2 and
MMP9 expression in Eca-109 cells. These data suggested that BANCR
overexpression enhanced the proliferation, migration and invasion
of ESCC cells.
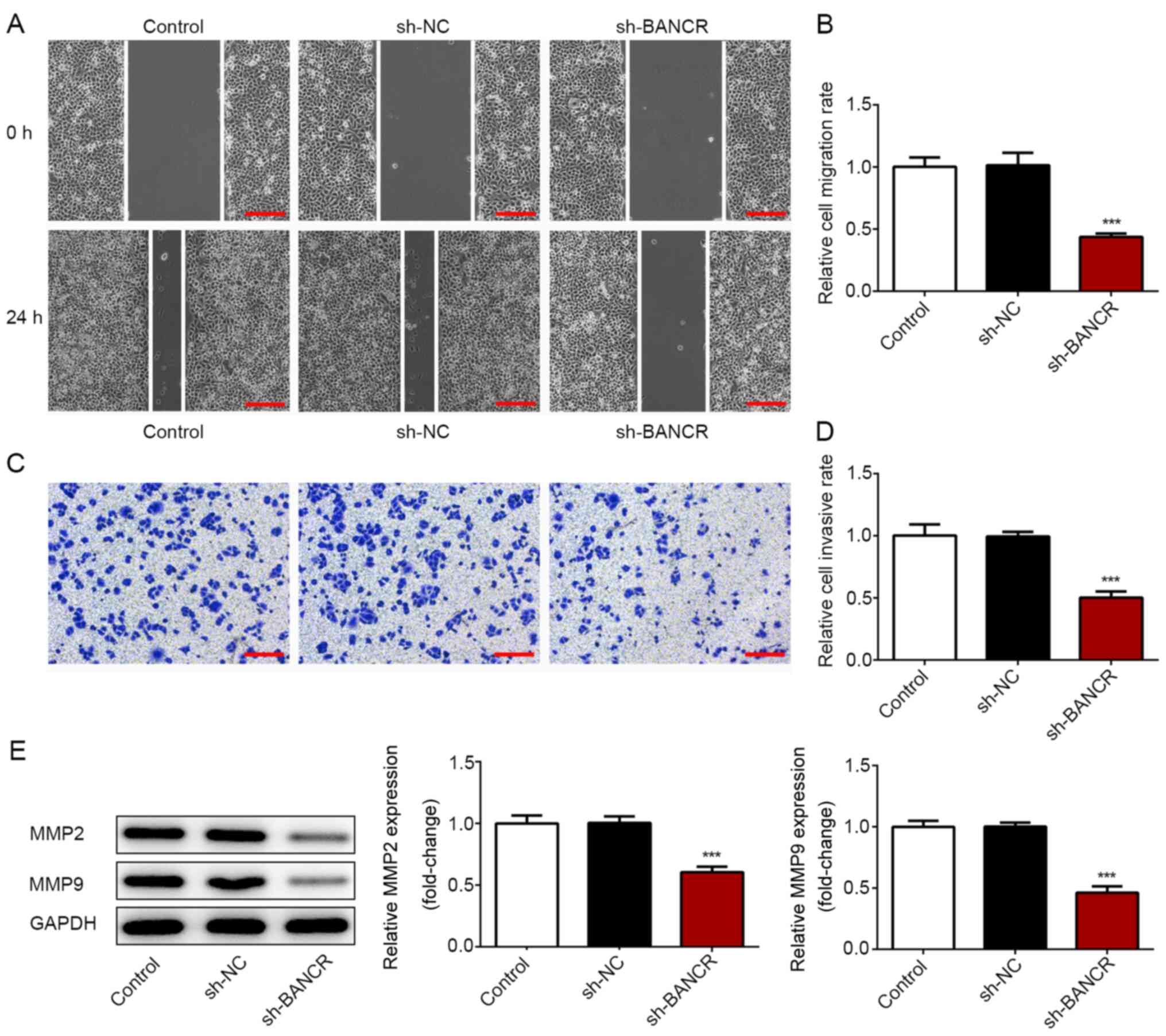
Role of BANCR in the proliferation and
migration of ESCC cells
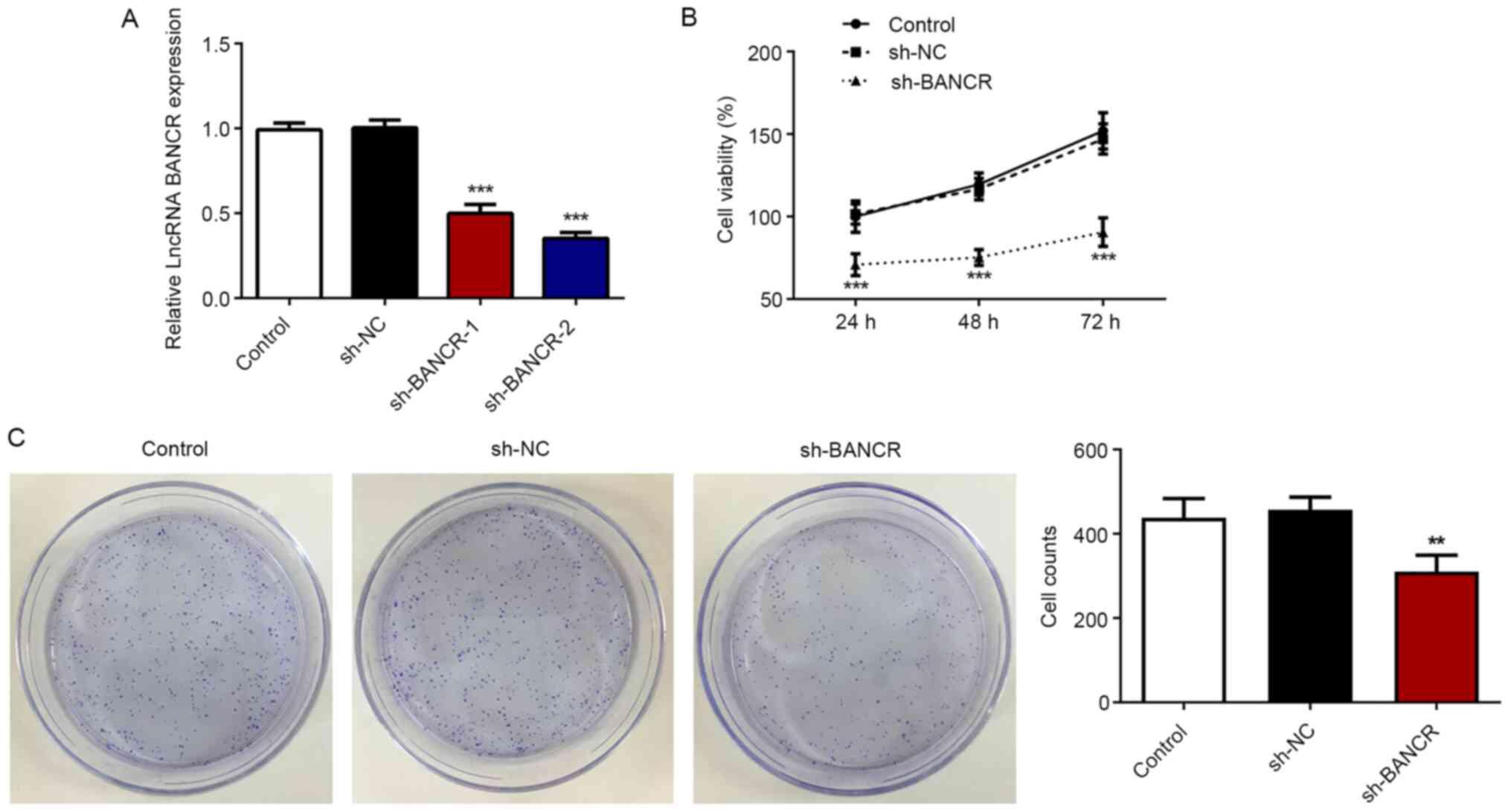
To confirm the role of BANCR in the proliferation
and migration of ESCC cells, the behaviors of Eca-109 cells were
analyzed under the condition of BANCR silencing. sh-BANCR was
established to downregulate BANCR expression. RT-qPCR demonstrated
that a significantly lower BANCR expression was observed in cells
from the sh-BANCR-2 and shRNA-BANCR-1 groups, with a lower BANCR
expression in the sh-BANCR-2 group (Fig. 4A). Therefore, shRNA-BANCR-2 was
employed for further study. In contrast to BANCR overexpression,
BANCR silencing significantly suppressed the viability and
proliferative ability of Eca-109 cells (Fig. 4B and C), as well as the migratory
and invasive abilities (Fig. 5A-D).
Additionally, BANCR-knockdown resulted in a significant decrease in
MMP2 and MMP9 expression (Fig. 5E).
These data indicated that BANCR silencing inhibited the
proliferation, migration and invasion of ESCC cells.
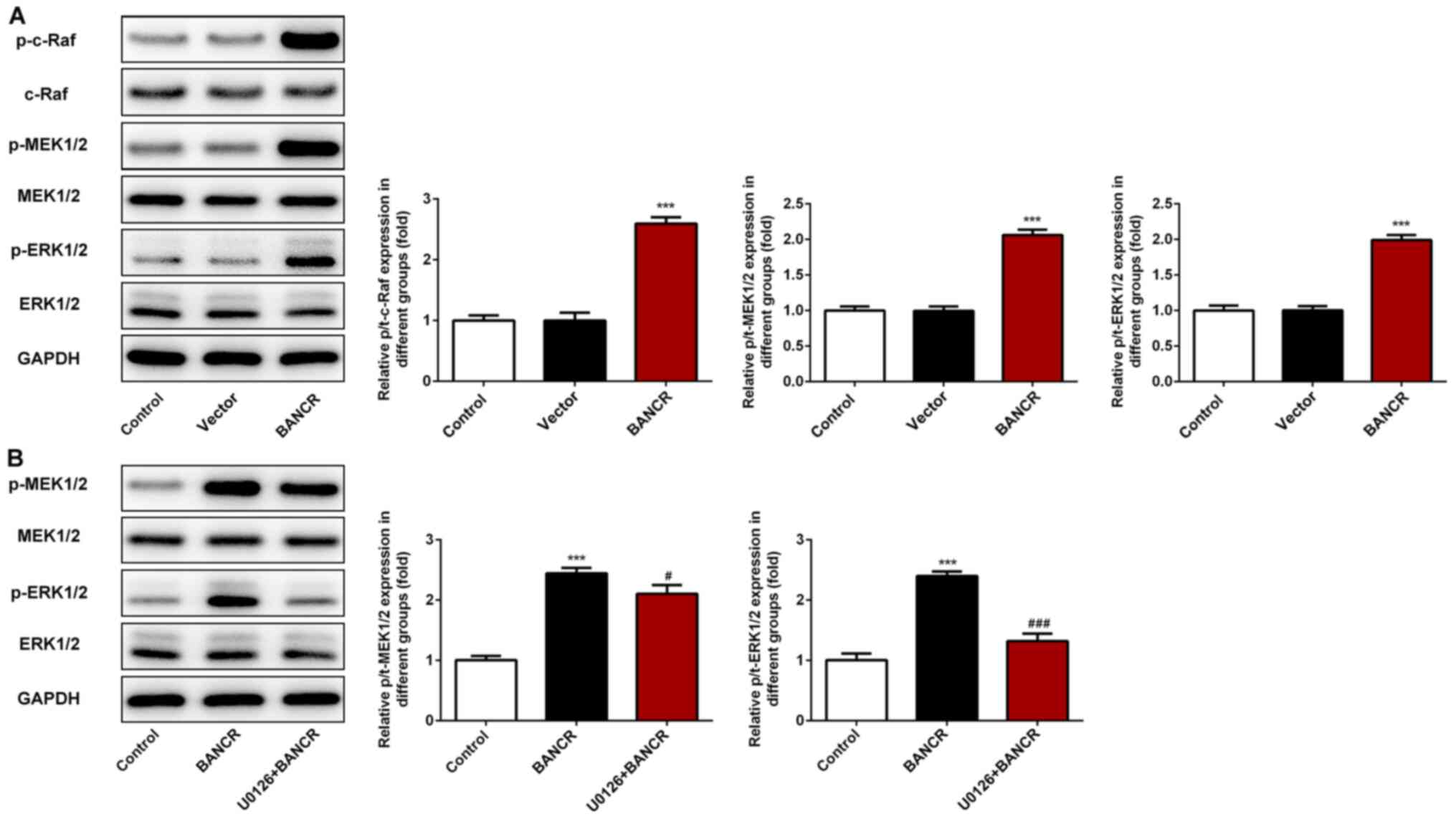
BANCR overexpression induces
activation of the Raf/MEK/ERK signaling pathway in ESCC cells
To investigate the underlying mechanism of BANCR in
ESCC progression, the phosphorylation levels of c-Raf, MEK1/2 and
ERK1/2 were detected using western blot analysis. The results
revealed that BANCR overexpression significantly increased the
levels of p-c-Raf, p-MEK1/2 and p-ERK1/2 (Fig. 6A), suggesting that the Raf/MEK/ERK
signaling pathway may be involved in the role of BANCR in the
proliferation, migration and invasion of ESCC cells.
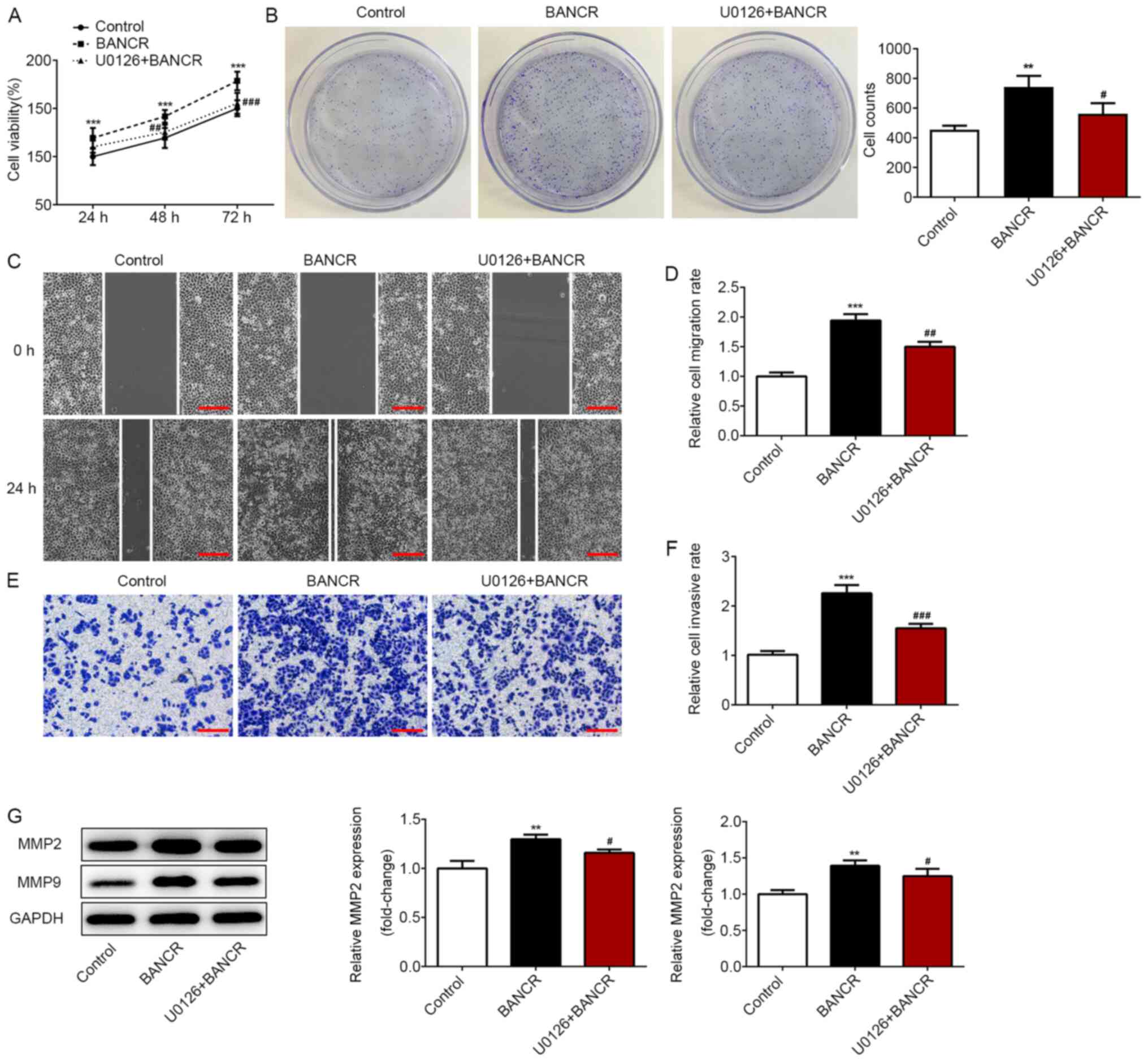
BANCR activates the proliferation,
migration and invasion of ESCC cells via the Raf/MEK/ERK signaling
pathway
To further investigate the mechanism of
BANCR/Raf/MEK/ERK signaling in ESCC progression, the specific MEK
inhibitor (U0126) was employed. Western blot analysis revealed that
U0126 treatment significantly attenuated the increased levels of
p-MEK1/2 and p-ERK1/2 induced by BANCR overexpression (Fig. 6B). In addition, U0126 treatment
significantly suppressed the viability and proliferative ability of
Eca-109 cells transfected with BANCR (Fig. 7A and B). Furthermore, migration and
invasion were significantly inhibited in the U0126+BANCR group
compared with in the BANCR group (Fig.
7C-F), as well as MMP-2 and MMP-9 expression (Fig. 7G). These data revealed that U0126
treatment reversed the effects of BANCR overexpression on
proliferation, migration and invasion of ESCC cells.
Discussion
At present, a variety of lncRNAs have been
identified as master regulators of gene expression, and their
alterations drive or impede cancer progression (22). Tumorigenesis regulated by lncRNAs is
a complicated and multi-stage process, involving complex cell
signaling pathways. Rapidly accumulating evidence has suggested
that lncRNAs serve an important role in oncogenesis, representing a
strategy of lncRNAs as biomarkers for the diagnosis, prognosis and
therapy of various types of cancer, including head and neck cancer
and thyroid cancer (23,24). Recently, increasing evidence has
shown that lncRNA BANCR may be an oncogene or a tumor suppressor in
various human malignancies, including colorectal (16), pancreatic (25), non-small cell lung (26) and ovarian cancer (27). Notably, BANCR expression is
significantly increased in tumor tissues of patients with ESCC
compared with in non-cancerous tissues, and its level is positively
associated with tumor differentiation, metastasis and tumor stage
(1). However, the molecular
mechanism of BANCR in the pathogenesis of ESCC remains unexploited.
Thus, it is of great importance to investigate the underlying
mechanism of BANCR in ESCC progression.
Previously, BANCR expression has been observed to be
upregulated in colorectal cancer tissues, and BANCR downregulation
decreases cell proliferation and invasion, but activates apoptosis
(16). Moreover, BANCR expression
has been reported to be upregulated in retinoblastoma tissues and
cell lines, and positively associated with tumor size and optic
nerve invasion (28). Additionally,
BANCR-knockdown significantly suppresses the proliferation,
migration and invasion of retinoblastoma cells (28). In the present study, it was observed
that BANCR expression was higher in ESCC cell lines than in normal
esophageal cells. In addition, BANCR overexpression enhanced the
proliferation, migration and invasion of ESCC cells, while BANCR
silencing exhibited opposite effects, which was consistent with a
previous study (28). The current
findings indicated that BANC acted as an oncogene in ESCC
progression.
Increasing evidence has revealed that the
Raf/MEK/ERK signaling pathway is closely associated with
tumorigenesis in multiple types of malignant tumor, including colon
and lung cancer (29–31). A previous study has revealed that
diallyl disulfide treatment elevates the apoptotic rate by
downregulating the RAF/MEK/ERK signaling pathway in esophageal
carcinoma cells (32).
Additionally, BANCR promotes the migration and invasion of cancer
cells and induces epithelial-mesenchymal transition in thyroid
cancer through the Raf/MEK/ERK signaling pathway (17). In the present study, BANCR
overexpression induced activation of the Raf/MEK/ERK signaling
pathway in ESCC cells, suggesting the participation of the
Raf/MEK/ERK signaling pathway under BANCR in ESCC progression. In
the current study, U0126, a specific MEK inhibitor, decreased MEK
and ERK expression, and blocked the promotive effects of BANCR
overexpression on the proliferation, migration and invasion of ESCC
cells. Overall, the present results strongly suggested that lncRNA
BANCR accelerated proliferation, migration and invasion of ESCC
cells via the Raf/MEK/ERK signaling pathway.
In conclusion, the current findings provided
evidence to demonstrate that BANCR expression was upregulated in
ESCC cell lines. In addition, lncRNA BANCR overexpression promoted
the proliferation, migration and invasion of ESCC cells, which was
significantly reversed following BANCR downregulation. Thus, lncRNA
BANCR may be a promising target for inhibiting ESCC cell
migration.
Acknowledgements
Not applicable.
Funding
No funding was received.
Availability of data and materials
The datasets used and/or analyzed during the current
study are available from the corresponding author on reasonable
request.
Authors' contributions
XY and GY designed the experiment and drafted the
manuscript. XY, MH and GY performed the experiments and analyzed
the data. GY reviewed the manuscript. XY and GY confirm the
authenticity of all the raw data. All authors read and approved the
final manuscript.
Ethics approval and consent to
participate
Not applicable.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Sadeghpour S and Ghorbian S: Evaluation of
the potential clinical prognostic value of lncRNA-BANCR gene in
esophageal squamous cell carcinoma. Mol Biol Rep. 46:991–995. 2019.
View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Li X, Xiao X, Chang R and Zhang C:
Comprehensive bioinformatics analysis identifies lncRNA HCG22 as a
migration inhibitor in esophageal squamous cell carcinoma. J Cell
Biochem. 121:468–481. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Bray F, Ferlay J, Soerjomataram I, Siegel
RL, Torre LA and Jemal A: Global cancer statistics 2018: GLOBOCAN
estimates of incidence and mortality worldwide for 36 cancers in
185 countries. CA Cancer J Clin. 68:394–424. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Shang QX, Yang YS, Yuan Y, Gu YM, Zhang
HL, Ji AF and Chen LQ: Clinical and prognostic effects of adjuvant
therapy on less advanced esophageal squamous cell carcinoma
patients. Ann Palliat Med. 9:681–699. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Liu Z, Yang T, Xu Z and Cao X:
Upregulation of the long non-coding RNA BANCR correlates with tumor
progression and poor prognosis in esophageal squamous cell
carcinoma. Biomed Pharmacother. 82:406–412. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Zhu Y, Yuan T, Zhang Y, Shi J, Bai L, Duan
X, Tong R and Zhong L: AR-42: A pan-HDAC inhibitor with antitumor
and antiangiogenic activities in esophageal squamous cell
carcinoma. Drug Des Devel Ther. 13:4321–4330. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Shapiro J, van Lanschot JJB, Hulshof M,
van Hagen P, van Berge Henegouwen MI, Wijnhoven BPL, van Laarhoven
HWM, Nieuwenhuijzen GAP, Hospers GAP, Bonenkamp JJ, et al:
Neoadjuvant chemoradiotherapy plus surgery versus surgery alone for
oesophageal or junctional cancer (CROSS): Long-term results of a
randomised controlled trial. Lancet Oncol. 16:1090–1098. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Tachimori Y, Ozawa S, Numasaki H, Ishihara
R, Matsubara H, Muro K, Oyama T, Toh Y, Udagawa H and Uno T;
Registration Committee for Esophageal Cancer of the Japan
Esophageal Society, : Comprehensive registry of esophageal cancer
in Japan, 2011. Esophagus. 15:127–152. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
van Hagen P, Hulshof MC, van Lanschot JJ,
Steyerberg EW, van Berge Henegouwen MI, Wijnhoven BP, Richel DJ,
Nieuwenhuijzen GA, Hospers GA, Bonenkamp JJ, et al: Preoperative
chemoradiotherapy for esophageal or junctional cancer. N Engl J
Med. 366:2074–2084. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
He LR, Liu MZ, Li BK, Jia WH, Zhang Y,
Liao YJ, Chen YC, Zhang LJ, Guan XY, Zeng YX, et al: High
expression of EZH2 is associated with tumor aggressiveness and poor
prognosis in patients with esophageal squamous cell carcinoma
treated with definitive chemoradiotherapy. Int J Cancer.
127:138–147. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Luo J, Wang W, Tang Y, Zhou D, Gao Y,
Zhang Q, Zhou X, Zhu H, Xing L and Yu J: mRNA and methylation
profiling of radioresistant esophageal cancer cells: The
involvement of Sall2 in acquired aggressive phenotypes. J Cancer.
8:646–656. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Ghorbian S and Ardekani AM: Non-invasive
detection of esophageal cancer using genetic changes in circulating
cell-free DNA. Avicenna J Med Biotechnol. 4:3–13. 2012.PubMed/NCBI
|
|
13
|
Liu J, Zhao SY, Jiang Q, Qu Y, Huang X, Du
J, Sun W and Ye Q: Long noncoding RNA MYLK-AS1 promotes growth and
invasion of hepatocellular carcinoma through the EGFR/HER2-ERK1/2
signaling pathway. Int J Biol Sci. 16:1989–2000. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Liu JQ, Deng M, Xue NN, Li TX, Guo YX, Gao
L, Zhao D and Fan RT: lncRNA KLF3-AS1 suppresses cell migration and
invasion in ESCC by impairing miR-185-5p-targeted KLF3 inhibition.
Mol Ther Nucleic Acids. 20:231–241. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Wu D, He X, Wang W, Hu X, Wang K and Wang
M: Long noncoding RNA SNHG12 induces proliferation, migration,
epithelial-mesenchymal transition and stemness of esophageal
squamous cell carcinoma cells via post-transcriptional regulation
of BMI1 and CTNNB1. Mol Oncol. 14:2332–2351. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Ma S, Yang D, Liu Y, Wang Y, Lin T, Li Y,
Yang S, Zhang W and Zhang R: LncRNA BANCR promotes tumorigenesis
and enhances adriamycin resistance in colorectal cancer. Aging
(Albany NY). 10:2062–2078. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Wang Y, Gu J, Lin X, Yan W, Yang W and Wu
G: lncRNA BANCR promotes EMT in PTC via the Raf/MEK/ERK signaling
pathway. Oncol Lett. 15:5865–5870. 2018.PubMed/NCBI
|
|
18
|
Wang D, Wang D, Wang N, Long Z and Ren X:
Long non-coding RNA BANCR promotes endometrial cancer cell
proliferation and invasion by regulating MMP2 and MMP1 via ERK/MAPK
signaling pathway. Cell Physiol Biochem. 40:644–656. 2016.
View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Chen Q, Zheng Y, Wu B, Chen X, Sun F, Ge P
and Wang P: BANCR regulates the cell invasion and migration in
esophageal squamous cell carcinoma through Wnt/beta-catenin
signaling pathway. Onco Targets Ther. 12:9319–9327. 2019.
View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Song W, Wang K, Yang X, Dai W and Fan Z:
Long non-coding RNA BANCR mediates esophageal squamous cell
carcinoma progression by regulating the IGF1R/Raf/MEK/ERK pathway
via miR-338-3p. Int J Mol Med. 46:1377–1388. 2020.PubMed/NCBI
|
|
21
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(-Delta Delta C(T)) method. Methods. 25:402–408. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Peng WX, Koirala P and Mo YY:
LncRNA-mediated regulation of cell signaling in cancer. Oncogene.
36:5661–5667. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Cossu AM, Mosca L, Zappavigna S, Misso G,
Bocchetti M, De Micco F, Quagliuolo L, Porcelli M, Caraglia M and
Boccellino M: Long non-coding RNAs as important biomarkers in
laryngeal cancer and other head and neck tumours. Int J Mol Sci.
20:34442019. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Mahmoudian-Sani MR, Jalali A, Jamshidi M,
Moridi H, Alghasi A, Shojaeian A and Mobini GR: Long non-coding
RNAs in thyroid cancer: Implications for pathogenesis, diagnosis,
and therapy. Oncol Res Treat. 42:136–142. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Wu X, Xia T, Cao M, Zhang P, Shi G, Chen
L, Zhang J, Yin J, Wu P, Cai B, et al: LncRNA BANCR promotes
pancreatic cancer tumorigenesis via modulating
MiR-195-5p/Wnt/β-catenin signaling pathway. Technol Cancer Res
Treat. 18:15330338198879622019. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Yang L and Liu G: lncRNA BANCR suppresses
cell viability and invasion and promotes apoptosis in
non-small-cell lung cancer cells in vitro and in vivo. Cancer Manag
Res. 11:3565–3574. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Xu X, Pan L, Zhuo M, Yang X, Zhang W, Sun
D, Zeng N and Zhang D: Increased expression of LncRNA BANCR and its
prognostic significance in human epithelial ovarian cancer. Eur J
Gynaecol Oncol. 38:449–452. 2017.PubMed/NCBI
|
|
28
|
Su S, Gao J, Wang T, Wang J, Li H and Wang
Z: Long non-coding RNA BANCR regulates growth and metastasis and is
associated with poor prognosis in retinoblastoma. Tumour Biol.
36:7205–7211. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Roberts PJ and Der CJ: Targeting the
Raf-MEK-ERK mitogen-activated protein kinase cascade for the
treatment of cancer. Oncogene. 26:3291–3310. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Wang Z, Ma L, Su M, Zhou Y, Mao K, Li C,
Peng G, Zhou C, Shen B and Dou J: Baicalin induces cellular
senescence in human colon cancer cells via upregulation of DEPP and
the activation of Ras/Raf/MEK/ERK signaling. Cell Death Dis.
9:2172018. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Wu GJ, Pen J, Huang Y, An S, Liu Y, Yang
Y, Hao Q, Guo XX and Xu TR: KAP1 inhibits the Raf-MEK-ERK pathway
to promote tumorigenesis in A549 lung cancer cells. Mol Carcinog.
57:1396–1407. 2018. View
Article : Google Scholar : PubMed/NCBI
|
|
32
|
Yin X, Zhang J, Li X, Liu D, Feng C, Liang
R, Zhuang K, Cai C, Xue X, Jing F, et al: DADS suppresses human
esophageal xenograft tumors through RAF/MEK/ERK and
mitochondria-dependent pathways. Int J Mol Sci. 15:12422–12441.
2014. View Article : Google Scholar : PubMed/NCBI
|