Introduction
Bradyarrhythmias are various life-threatening
conditions such as sinus syndrome and atrioventricular blockage and
are caused by cardiac pacing or conduction dysfunction (1). Implanted electronic pacemakers are the
preferred treatment for such diseases (2). However, electronic pacemakers still
produce adverse reactions such as infection, metal allergy,
electrode dislocation, electronic interference and lack of response
to neurohormones. Therefore, biological pacemakers provide new
options for the treatment of arrhythmia. Bone marrow mesenchymal
stem cells (MSCs) are among the main sources of cardiac repair and
regenerative cell therapy because these cells are readily available
and have little inherent immunogenicity to any adverse immune
response (3). In vitro,
5-Azacytidine (5-AZA) was found to induce the differentiation of a
variety of the MSCs into cardiomyocytes (4–7). In
vivo, the transplantation of MSCs has been shown to improve
cardiac function in damaged hearts by myocardial cell regeneration
and paracrine effects that promote angiogenesis, reduce cardiac
fibrosis and prevent host myocardial cell apoptosis (8). However, just a small percentage of the
MSCs express cardiac-specific proteins during differentiation,
which significantly limits their potential clinical application
(3,9). Induction efficiency can be increased
by transfection of key genes and transcription factors or
simulating specific myocyte growth microenvironment (10–12).
However, these methods have a number of disadvantages including
complicated operating procedures, high costs and a low induction
efficiency.
Dimethylation of histone H3 at lysine 9 (H3K9me2)
has, in recent years, been found to be closely related to the
process of cell differentiation, proliferation and reprogramming
(13). Following H3K9me2 methylase,
euchromatic histone-lysine N-methyltransferase 2 (G9a) was
inhibited and H3K9me2 was decreased, the levels of cardiac
transcription factors GATA binding protein 4 (GATA-4), NK2 Homeobox
5 (Nkx2.5) and myocardial protein produced by Wnt11 treatment were
2.6–5.6 times greater compared with those of the untreated group
(14). Zhang et al (15) found that H3K9me2 lysine demethylase
3A (KDM3A) could control pathological cardiac hypertrophy and
fibrosis. In their study, JIB-04 inhibits KDM3A and suppresses the
transcription of fibrotic genes that overlap with genes
downregulated in Kdm3A-KO mice vs. wild-type controls. This
suggests that KDM3A may be involved in cardiac development.
As part of the present study, in a system for the
5-AZA induction of MSCs into cardiomyocytes, the expression of
H3K9me2 was affected by the knockdown of G9a and KDM3A, resulting
in a change in differentiation efficiency during induction. The
role of H3K9me2 in the differentiation of the MSCs into
cardiomyocytes was identified, thus optimizing the induction
efficiency of cardiomyocytes in vitro and laying a
foundation for clinical applications.
Materials and methods
Culture, expansion and
characterization of rat MSCs
A total of 20 healthy male and 20 female Wistar rats
(weight, 150–200 g; age, 6–8 weeks) were obtained from Yangzhou
University and three rats were used in each of three individual
experiments. Rats received standard care under a 12 h dark/light
cycle (25°C with humidity of 60%) and given a free access to food
and water. Before the experiment, adaptive feeding was carried out
for one week. The animal's health and behavior were monitored daily
until mortality. All procedures involving the care and use of
animals conformed to the guidelines of Animal Research: Reporting
of in vivo Experiments (ARRIVE guidelines 2.0; http://arriveguidelines.org/arrive-guidelines) and
were approved by the animal care guidelines of the Laboratory
Animal Management and Experimental Animal Ethics Committee of
Yangzhou University (approval no. 201903478). Bone mesenchymal stem
cells (MSCs) were obtained as described by Huang et al
(16) and Nippert et al
(17). All rats were anesthetized
with 20% sodium urethane (0.2 g/ml; 1.0 g/kg intraperitoneal
injection) and subsequently sacrificed by cervical dislocation.
Animal mortality was verified by ascertaining cardiac and
respiratory arrest. MSCs were extracted from the tibias and femurs.
The cell suspension from three rats was mixed together and the
cells were dispersed by pipetting, following which they were placed
in a centrifuge at 1,100 × g for 4 min at 37°C. The supernatant and
adipose tissue were removed. The cell suspension was transferred to
a 15 ml centrifuge tube containing 5 ml Percoll (1.073 g/ml;
Sigma-Aldrich; Merck KGaA). Cells were dispersed by pipetting again
and centrifuged at 1,500 × g for 30 min at 4°C. The mononuclear
cells in the middle layer were obtained, washed three times with
phosphate buffered saline (PBS) and then cultured in Dulbecco's
Modified Eagle's Medium (DMEM) (Thermo Fisher Scientific, Inc.)
containing 10% fetal bovine serum (FBS; Biowest) at 37°C in a 5%
CO2 incubator. During the expansion and proliferation of
MSCs, the culture medium was replaced every three days and cells
were passaged once they reached 70% confluency. For some
experiments, MSCs were cultured in the presence or absence of 1 µM
BIX01294 (MedChemExpress) for 12 h.
For cell phenotypic characterization, MSCs of the
third passage were incubated overnight at 4°C with mouse anti-CD29
(1:100; cat. no. sc-9970; Santa Cruz Biotechnology, Inc.),
anti-CD44 (1:100; cat. no. sc-7297; Santa Cruz Biotechnology, Inc.)
and anti-CD45 (1:100; cat. no. sc-1178; Santa Cruz Biotechnology,
Inc.) and then with an appropriate secondary antibody conjugated to
PE (1:100; cat. no. sc-516141; Santa Cruz Biotechnology, Inc.) for
1 h at room temperature. The cells were subsequently detected using
a BD FACSCanto™ II flow cytometer (BD Biosciences). The results
were analyzed and processed by FlowJo version 10.0 (FlowJo
LLC).
Construction of G9a and KDM3A
lentivirus interference vectors
According to the CDS-region sequences of G9a and
KDM3A provided by National Center of Biotechnology Information
(https://www.ncbi.nlm.nih.gov), three
interference targets were designed. Meanwhile, an unrelated
sequence was designed as a negative control (Table I). After the company (Genomediotech)
synthesized the upstream and downstream sequences, an oligo double
chain was formed by the annealing process and then three
recombinant interference plasmids were obtained by means of
connection with pGMLV-SC5 RNA interference vectors through
BamHI and EcoRI double-enzyme digestion,
respectively. These were called short hairpin (sh)1-G9a, sh2-G9a,
sh3-G9a, sh1-KDM3A, sh2-KDM3A, sh3-KDM3A and sh-negative control
(NC). The recombinant interfering plasmids were then subjected to
sequencing for further verification. Following successful
sequencing, the vector with the strongest interference activity was
delivered to Shanghai Jiman Co., Ltd. for lentivirus parcel (titer:
2.5×108 TU/ml) after verification of activity.
 | Table I.Primers used for plasmid
construction. |
Table I.
Primers used for plasmid
construction.
| Gene | Primer sequence
(5′-3′) |
|---|
| sh1-G9a | F:
GATCCGCCTGTACTATGACGCGTACTCTCGAGAGTACGCGTCATAGTACAGGCTTTTTT |
|
| R:
AATTAAAAAAGCCTGTACTATGACGCGTACTCTCGAGAGTACGCGTCATAGTACAGGCG |
| sh2-G9a | F:
GATCCGGTTTGCACTGCAGCTCAATCCTCGAGGATTGAGCTGCAGTGCAAACCTTTTTT |
|
| R:
AATTAAAAAAGGTTTGCACTGCAGCTCAATCCTCGAGGATTGAGCTGCAGTGCAAACCG |
| sh3-G9a | F:
GATCCGCGGCTGCTCCAGGAGTTTAACTCGAGTTAAACTCCTGGAGCAGCCGCTTTTTT |
|
| R:
AATTAAAAAAGCGGCTGCTCCAGGAGTTTAACTCGAGTTAAACTCCTGGAGCAGCCGCG |
| sh1-KDM3A | F:
GATCCGCAAGTCTTCTGAGAATAATGCTCGAGCATTATTCTCAGAAGACTTGCTTTTTT |
|
| R:
AATTAAAAAAGCAAGTCTTCTGAGAATAATGCTCGAGCATTATTCTCAGAAGACTTGCG |
| sh2-KDM3A | F:
GATCCGCAGCCAATTCTCCACCTAACCTCGAGGTTAGGTGGAGAATTGGCTGCTTTTTT |
|
| R:
AATTAAAAAAGCAGCCAATTCTCCACCTAACCTCGAGGTTAGGTGGAGAATTGGCTGCG |
| sh3-KDM3A | F:
GATCCGCAGGTGTCACTAGGCTTAATCTCGAGATTAAGCCTAGTGACACCTGCTTTTTT |
|
| R:
AATTAAAAAAGCAGGTGTCACTAGGCTTAATCTCGAGATTAAGCCTAGTGACACCTGCG |
| sh-NC | F:
UUCUCCGAACGUGUCACGUTT |
|
| R:
ACGUGACACGUUCGGAGAATT |
Cell transfection
MSCs were cultured in DMEM supplemented with 10% FBS
and the cells were kept at 37°C and 5% CO2 containing
atmosphere. The cells were plated in 24-well plates, grown to a
confluency of 70–80% and were transfected with 1 µg/plate shRNA-G9a
or shRNA-KDM3A lentivirus, which were diluted in polybrene
(Sigma-Aldrich; Merck KGaA). sh-NC were transfected with plasmids
that were non-targeting and indistinguishable for KDM3A and G9A.
The cells that were not treated were used as controls. Following
treatment, the cells were observed daily through the use of a
fluorescent microscope (magnifications, ×100, 200 and 400; TE2000;
Nikon Corporation). After 72 h, the cells were collected for
further experiments.
Cardiac differentiation induction
The MSCs were divided into four separate groups: i)
MSCs group: MSCs were untreated; ii) control group: MSCs were
transfected with control lentivirus; iii) shKDM3A group: MSCs were
transfected with lentivirus encoding shRNA-KDM3A; and iv) shG9a
group: MSCs were transfected with lentivirus encoding shRNA-G9a.
All groups treated with 10 µM of 5-AZA (Sigma-Aldrich; Merck KGaA).
Following overnight incubation, the medium was aspirated and the
cells were washed with PBS three times for three to five min. New
culture medium was added and was then replaced every three days.
Following 5-AZA treatment, cell morphology was observed daily using
a phase-contrast microscope (DMIL-PH1; Leica Microsystems
GmbH).
Western blotting
Cells from different treatment groups were lysed
with RIPA lysis buffer (Applygen Technologies, Inc.) and samples
containing 25–50 µg of total protein lysates (after Bradford
estimation) were separated by 10% SDS-PAGE and transferred onto a
PVDF membrane (EMD Millipore), which were then semi-dry blocked by
TBS-0.05% Tween 20 containing 5% fetal calf serum (Gibco; Thermo
Fisher Scientific, Inc.) for 1 h at room temperature. The membranes
were incubated overnight at 4°C with primary antibodies targeted
against: H3K9me2 (1:1,000; cat. no. 39239; Active Motif, Inc.), H3
(1:3,000; cat. no. H0164; Sigma-Aldrich; Merck KGaA), cTnT
(1:3,000; cat. no. MABT368; Sigma-Aldrich; Merck KGaA), GATA-4
(1:3,000; cat. no. SAB4501128; Sigma-Aldrich; Merck KGaA), Nkx2.5
(1:5,000; cat. no. 13921-1-AP; ProteinTech Group, Inc.), MEF2c
(1:3,000; cat. no. SAB2103534; Sigma-Aldrich; Merck KGaA) and
β-actin (1:1,000; cat. no. SAB3500350; Sigma-Aldrich; Merck KGaA).
The blots were then washed and incubated for 2 h with an
appropriate secondary antibody (1:3,000, cat. no. A9169; 1:5,000,
cat. no. AP162P; 1:5,000, cat. no. AP160P; all Sigma-Aldrich; Merck
KGaA). The membranes were then washed and enhanced
chemiluminescence (ECL) detection was performed using Pierce ECL
kit (Thermo Fisher Scientific, Inc.). Densiometric analysis was
performed using Image Lab software (version 3.0, Bio-Rad
Laboratories, Inc.).
Total RNA isolation and reverse
transcription-quantitative (RT-q) PCR
A RNeasy Mini kit (Qiagen GmbH) was used to extract
the total RNA from the MSCs of different treatment groups
post-seeding, according to the manufacturer's instructions. cDNA
was amplified by PCR through the use of a Qiagen PCR kit according
to the manufacturer's instructions at 50°C for 30 min, 95°C for 3
min, 95°C for 15 sec and 60°C for 30 sec. GATA-4, Nkx2.5 and MEF2c
expressions were detected by RT-qPCR. The 20 µl PCR amplification
reaction included 2 µl cDNA, 10 µl SYBR Taq, 0.8 µl forward primer,
0.8 µl reverse primer, 0.4 µl RoxII and 6 µl double-distilled
water. Subsequently, PCR reaction was achieved on the basis of the
two-step procedure (95°C for 15 min; 95°C for 10 sec and 60°C for
32 sec) and procedure was repeated 40 times. Primers applied were
shown in Table II. The PCR
instrument for RT-qPCR was ABIPRISM 7500 (Applied Biosystems;
Thermo Fisher Scientific, Inc.). Each experimental condition was
repeated in triplicate. The relative mRNA quantities were
determined using the 2−ΔΔCq method (18).
 | Table II.Primers used for RT-qPCR. |
Table II.
Primers used for RT-qPCR.
| Gene | Primer sequence
(5′-3′) |
|---|
| cTnT | F:
TTCGACCTGCAGGAAAAGTT |
|
| R:
GTGCCTGGCAAGACCTAGAG |
| Nkx 2.5 | F:
ACCGCCCCTACATTTTATCC |
|
| R:
GACAGGTACCGCTGTTGCTT |
| GATA4 | F:
TCTCACTATGGGCACAGCAG |
|
| R:
CCGAGCAGGAATTTGAAGAG |
| MEF2c | F:
CTCCCCTGTGGACAGTCTGA |
|
| R:
CAGAGGGGCTTTCTCTGTCC |
| G9a | F:
GCTACCATGACTGCGTTCTG |
|
| R:
TCCCGGCAGATGATCTTCTC |
| KDM3A | F:
GGCAGTTCAAGCTCTTCTCG |
|
| R:
TGGACAGATGGGCTTCACAT |
| GAPDH | F:
GGAAAGCTGTGGCGTGATGG |
|
| R:
GTAGGCCATGAGGTCCACCA |
Flow cytometry
Subsequent to treatment with 5-AZA, the expression
of cTnT was detected in four groups on different days. Cells were
incubated overnight at 4°C with anti-rat cTnT (1:200; cat. no.
15513-1-AP; ProteinTech Group, Inc.). Then, goat anti-rabbit
fluorescein isothiocyanate (PE)-labeled mouse anti-rabbit IgG
(1:500; cat. no. sc-3753; Santa Cruz Biotechnology, Inc.) was used
as the secondary antibody and incubated with the cells for 1 h at
room temperature. The percentage of fluorescent protein-positive
cells was detected by flow cytometry using a BD FACSCanto™ II flow
cytometer (BD Biosciences). The results were analyzed and processed
by FlowJo version 10.0 (FlowJo LLC).
Statistical analysis
The results were expressed as mean ± standard error.
Data were analyzed by one way ANOVA and Tukey's post hoc test using
SPSS v23 (IBM Corp.). Unpaired t-test was used to evaluate
significance between two groups. P<0.05 was considered to
indicate a statistically significant difference.
Results
Culture and characterizations of
isolated rat MSCs
The obtained cells were cultured for 48 h. Under the
microscope, some adherent cells with fusiform growth could be seen
and a few cells were polygonal. After 7 to 10 days, cell growth
reached 80–90% confluence and was arranged in a spiral pattern
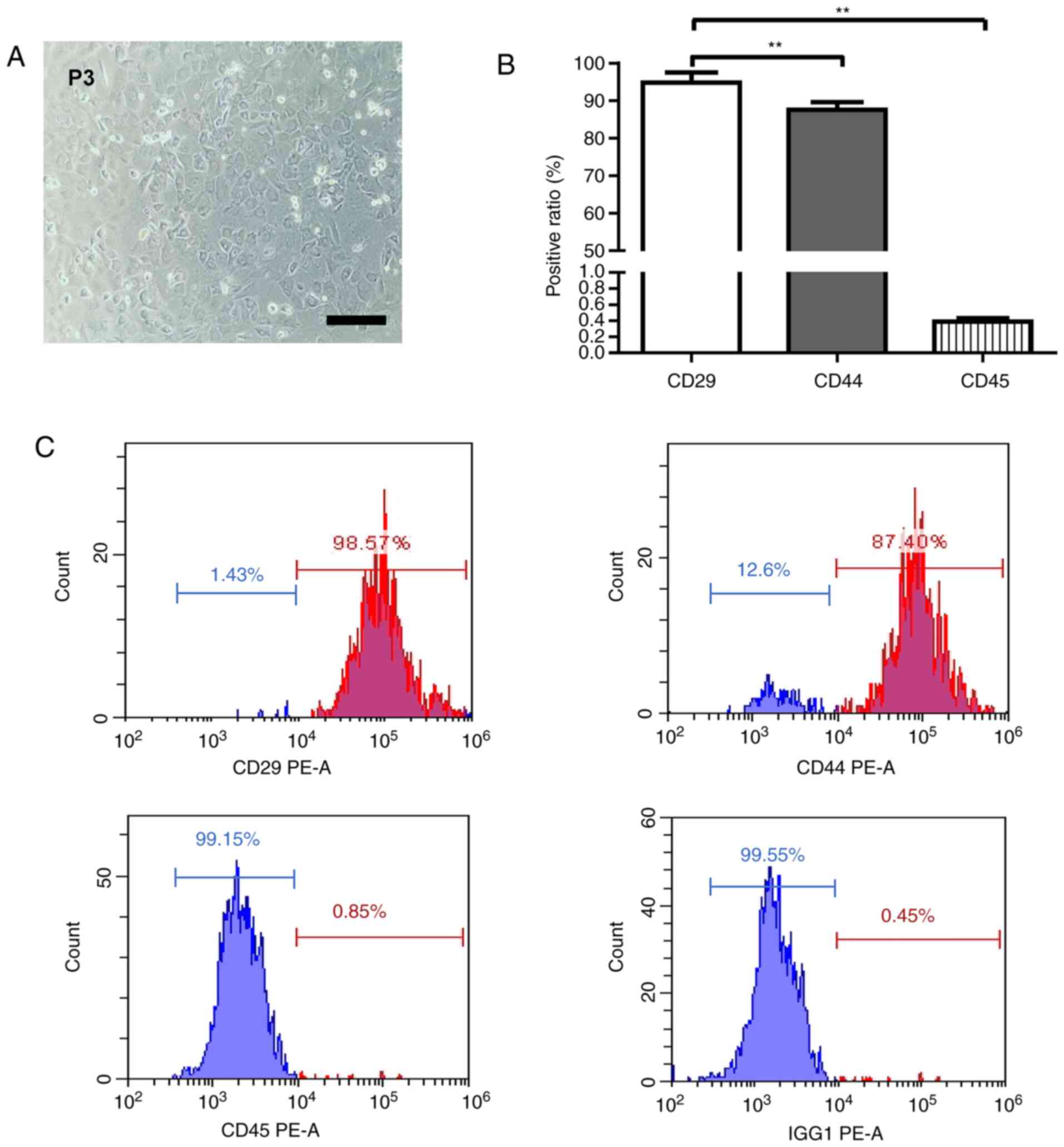
(Fig. 1A). To characterize the
MSCs, they were analyzed with antibodies against CD44, CD29 and
CD45 by means of flow cytometry. As shown in Fig. 1B and C, the majority of cells
expressed CD29 and CD44 at moderate to high levels while these
cells were negative for CD45 (P<0.01). This confirmed that the
MSCs isolated in this study expressed cell markers consistently
with those reported previously (9).
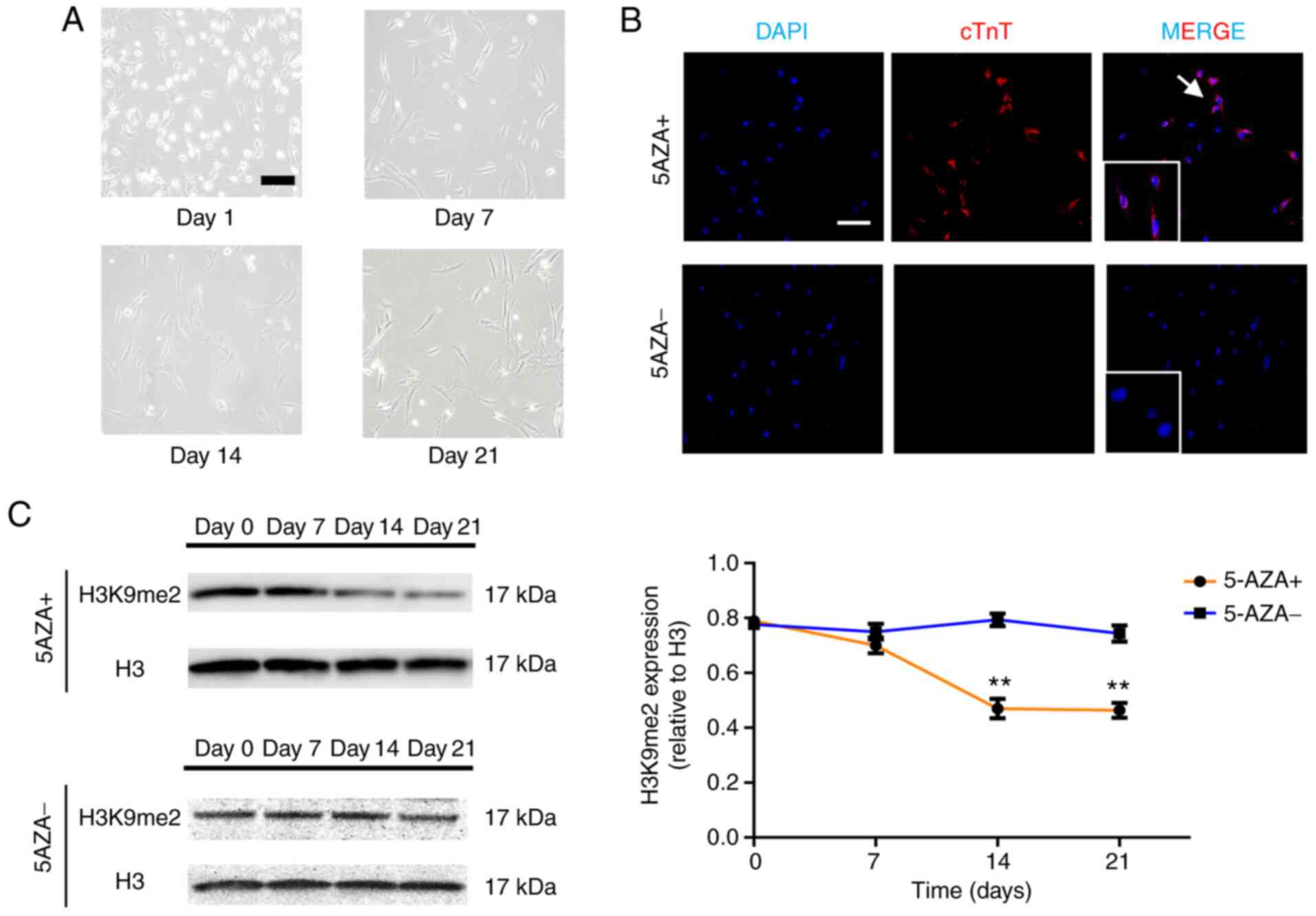
Cardiac differentiation of the MSCs
with 5-AZA induction
In order to construct a system for the
differentiation of MSCs into cardiomyocytes, the cells were treated
with 5-AZA; ≤50% of the MSCs died and detached from the plate. On
the seventh day following 5-AZA treatment, the cell volume
increased significantly and gradually became short columnar with
the same direction of arrangement. With the extension of culture
time, the cell morphology was varied, the cytoplasm was rough and
the myofilament structure appeared after two weeks. After three
weeks it was observed that the cells were long, and spindle shaped
or short and column shaped and were closely connected with adjacent
cells (Fig. 2A). After three weeks
the cell proliferation slowed down, and a portion of the cells
died. No cell pulsation was observed during this process.
Immunohistochemical staining results demonstrated that cTnT
positive cells appeared following 5-AZA treatment (Fig. 2B). These data indicated that the
system for the differentiation of MSCs into cardiomyocytes was
successfully constructed.
To observe the changes of H3K9me2 in the process of
induced differentiation, western blotting was used to detect the
expression of H3K9me2. It was found that H3K9me2 decreased
gradually with the increase in days, but there was no significant
change in untreated group (Fig.
2C). This suggests that H3K9me2 may be involved in the process
of differentiation.
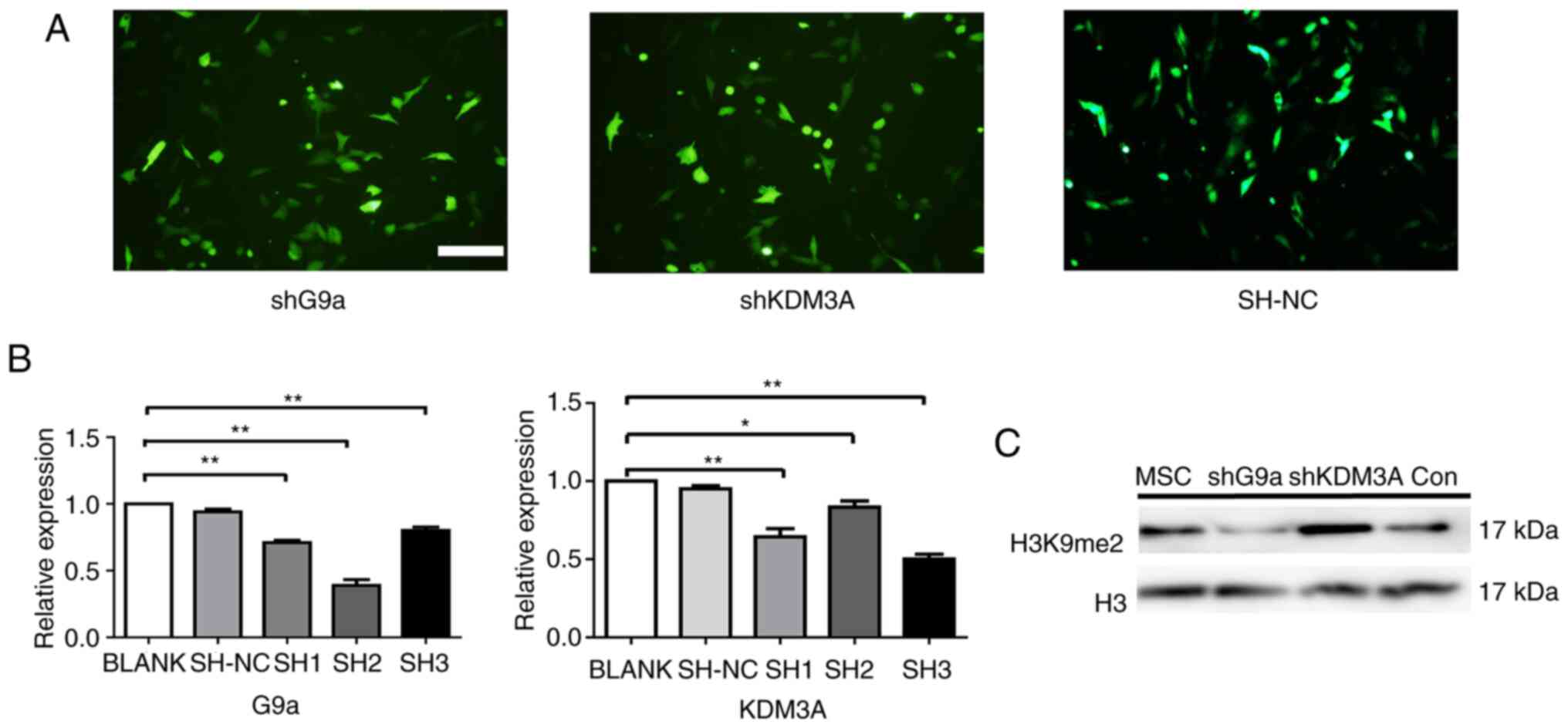
The interference vectors of G9a and
KDM3A genes can affect the expression of H3K9me2
In order to investigate the function of H3K9me2 in
the differentiation of the MSCs into cardiomyocytes, three
interference carriers of G9a and KDM3A, respectively were
constructed (Fig. 3A). The
expressions of G9a and KDM3A were detected 48 h following
transfection of the MSCs with the interference vector thus
constructed. Among them, RT-qPCR demonstrated the strongest
interference activity with respect to sh3-KDM3A. Similarly, sh2-G9a
was screened for the best interference (P<0.05; Fig. 3B). Thus, sh3-KDM3A and sh2-G9a were
selected for further study.
To observe H3K9me2 expression following H3K9me2
methylation modification enzyme knockdown, H3K9me2 expression was
detected at 72 h following lentivirus transfection of rat MSCs.
Western blotting results demonstrated that, compared with the
control group, the H3K9me2 was significantly reduced when G9a was
knocked down but the level of H3K9me2 was significantly increased
following KDM3A knockdown (Fig.
3C). This suggests that the knockdown of H3K9me2 methylation
modifying enzyme could affect the expression of H3K9me2.
The knockdown of G9a enhances cardiac
differentiation of the MSCs treated with 5-AZA
In order to compare the effect of H3K9me2 on the
final induction efficiency, the expression of cardiac protein cTnT
was detected by means of flow cytometry, western blotting and
RT-qPCR. Following 5-AZA treatment, cTnT-positive cells in all
groups increased gradually as more days passed. However, there were
significant differences in the percentages of cTnT-positive cells
between the groups with different treatments as shown in Fig. 4A (P<0.05). In the control group,
less than 20% of the differentiating cells that were cTnT positive
after three weeks of culturation. When KDM3A were knocked down,
slightly more than 10% of cells were positive for cTnT after three
weeks of differentiation. However, almost 30% of the cells stained
positive for cTnT following G9a knockdown (Fig. 4B). Western blotting demonstrated the
same results (Fig. 4C). Similarly,
the mRNA expression of the shG9a group was significantly
upregulated for the specific cardiomyocyte markers on day 21
(P<0.05; Fig. 4D). The results
demonstrated that the reduction of H3K9me2 by interfering with G9a
could increase the efficiency of the MSCs induced differentiation
into cardiomyocytes.
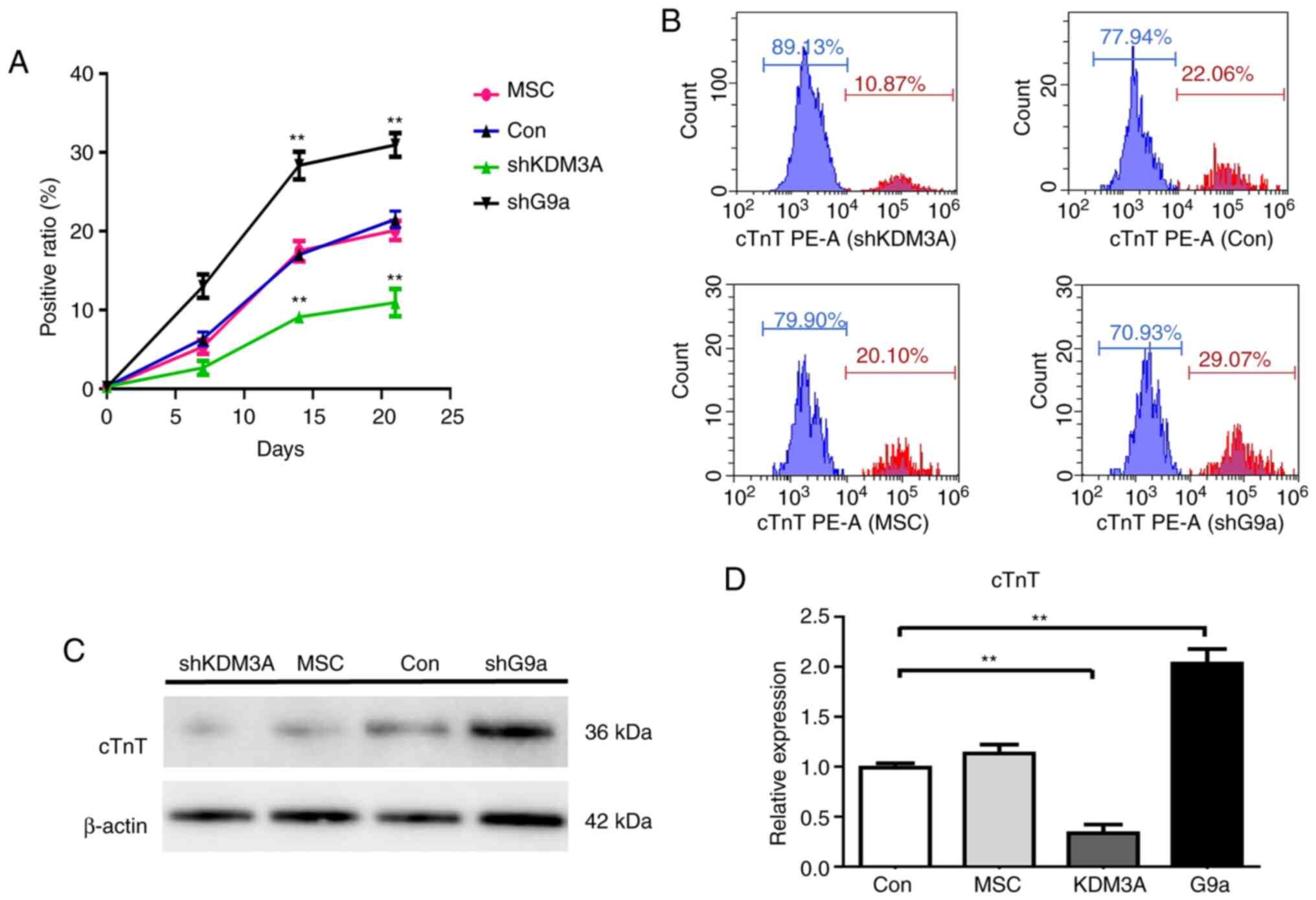 | Figure 4.The levels of cTnT following 5-AZA
treatment. (A) Flow-cytometry analysis demonstrated that
cTnT-positive cells rate gradually increased in all groups
following 5-AZA induction. The results on different days
demonstrated that cTnT-positive cell rate in the G9a group was the
highest and the KDM3A group was the lowest. The mean ± standard
deviation is shown; n=3 independent experiments. P by t-test. (B)
Fluorescence intensity was detected by means of flow cytometry,
which demonstrated that the proportion of cTnT-positive cells rate
was ~30% in G9a knockdown group on day 21 and ~11% in the KDM3A
knockdown group. (C) Cardiomyocyte differentiation of the MSCs with
5-AZA treatment was detected by western blot analysis, which
demonstrated that cTnT protein levels were upregulated following
G9A knockdown on day 21. β-actin was used as an internal control.
(D) Cardiomyocyte differentiation of the MSCs with 5-AZA treatment
was detected by reverse transcription-quantitative PCR, which
demonstrated that G9a knockdown increased the expression of cTnT
gene mRNA by 1.5 times compared with the control group. The
opposite result was found when KDM3A was knocked down (n=3,
**P<0:01 vs. the control group). cTnT, cardiac troponin-T;
5-AZA, 5-Azacytidine; G9a, euchromatic histone-lysine
N-methyltransferase 2; KDM3A, H3K9me2 lysine demethylase 3A; MSCs,
rat mesenchymal stem cells; sh, short hairpin. |
The knockdown of G9a and KDM3A can
affect the expression of myocardial specific transcription
factors
To investigate the effect of H3K9me2 on MSCs-induced
differentiated cardiomyocyte, changes in cell morphology and the
expression of early transcription factors GTA-4, Nkx2.5 and MEF2c
following the knockdown of G9a and KDM3A were observed. There was
no significant difference in cell morphology between the groups
(Fig. 5A). The results of western
blotting demonstrated that the myocardial specific transcription
factors GTA-4, Nkx2.5 and MEF2c were increased in all groups
(Fig. 5B). Compared with the MSCs
and control groups, RT-qPCR demonstrated that, following the
knockdown of G9a, the GATA-4, Nkx2.5 and MEF2c mRNA expression was
higher on days 7, 14 and 21 (P<0.05). Following the knockdown of
KDM3A, the expression levels of GTA-4, Nkx2.5 and MEF2c were
significantly downregulated (P<0.05; Fig. 5C). These results suggest that
inhibition of H3K9me2 by knockdown G9a can increase the expression
of cardiac early transcription factors.
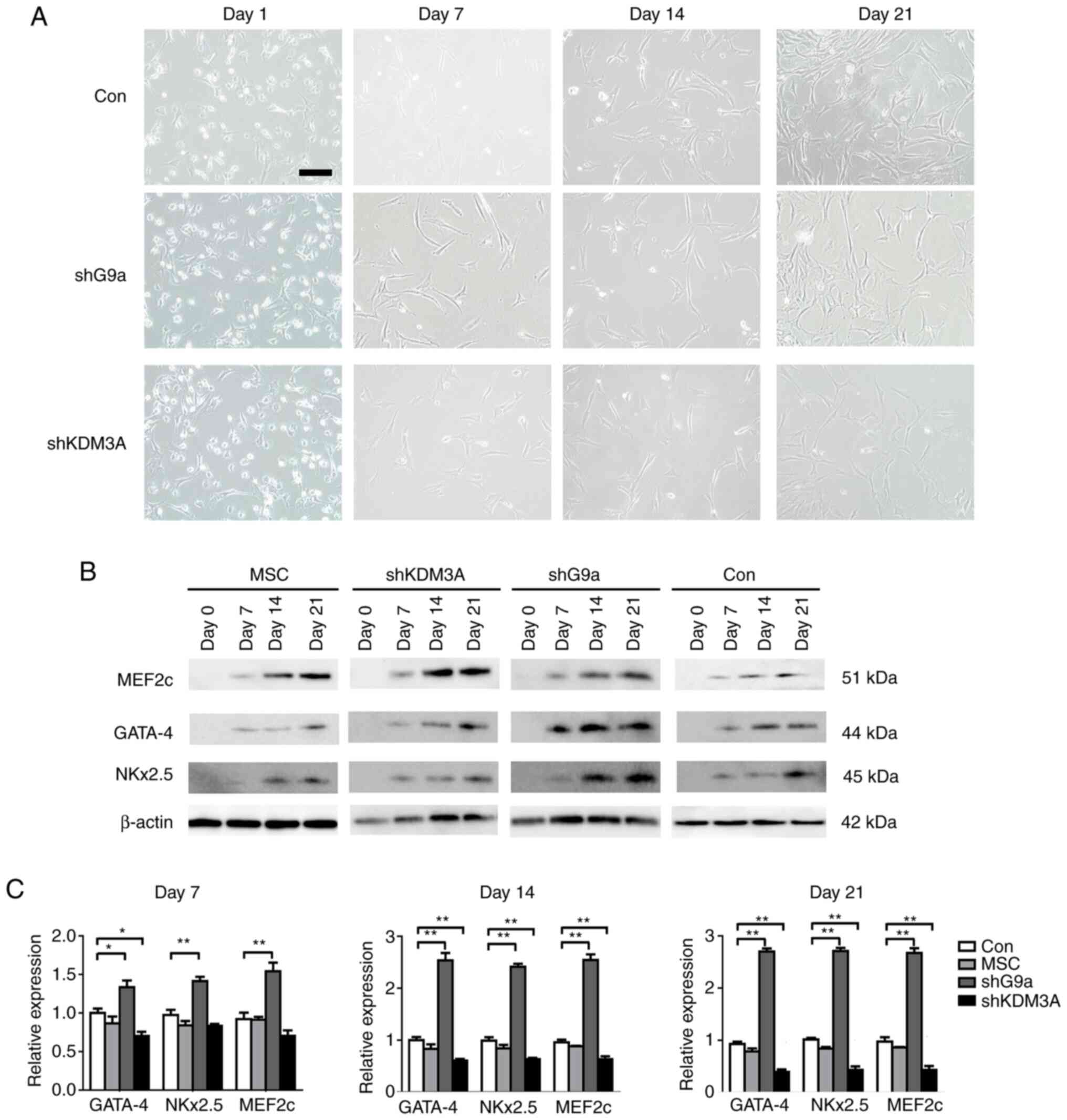 | Figure 5.Morphological and early transcription
factor expression changes in the mesenchymal stem cells treated
with 5-AZA. (A) The cells in all groups partly succumbed following
5-AZA treatment and the surviving cells began to proliferate and
differentiate. After a week the cells were enlarged, partially
fusiform and regularly arranged and after three weeks they formed a
myotubular-like structure by connecting with adjacent cells. Scale
bar=50 µm. (B) Cardiomyocyte differentiation of the MSCs with 5-AZA
treatment was detected by western blotting, which demonstrated the
upregulation of MEF2c, GATA-4 and Nkx2.5 mRNA expression on days 7,
14 and 21 in all groups. β-actin was used as a reference gene. (C)
Cardiomyocyte differentiation of the MSCs with 5-AZA treatment was
detected by reverse transcription-quantitative PCR. When G9a was
knocked down, GATA-4, Nkx2.5 and MEF2c mRNA expression was
upregulated on days 7, 14 and 21 compared with those of the control
group. By contrast, GATA-4, Nkx2.5 and MEF2c mRNA expression were
decreased following KDM3A knockdown. GAPDH was used as a reference
gene. *P<0.05 and **P<0.01 vs. control group. 5-AZA,
5-Azacytidine; MSCs, rat mesenchymal stem cells; MEF2c, myocyte
enhancer factor 2c; GATA-4, GATA binding protein 4; Nkx2.5, NK2
Homeobox 5; sh, short hairpin; MSCs, untreated MSCs; control, MSCs
transfected with control lentivirus; shG9a group, MSCs transfected
with lentivirus encoding shRNA-G9a; KDM3A group, MSCs transfected
with lentivirus encoding shRNA-KDM3A; G9a, euchromatic
histone-lysine N-methyltransferase 2; KDM3A, H3K9me2 lysine
demethylase 3A. |
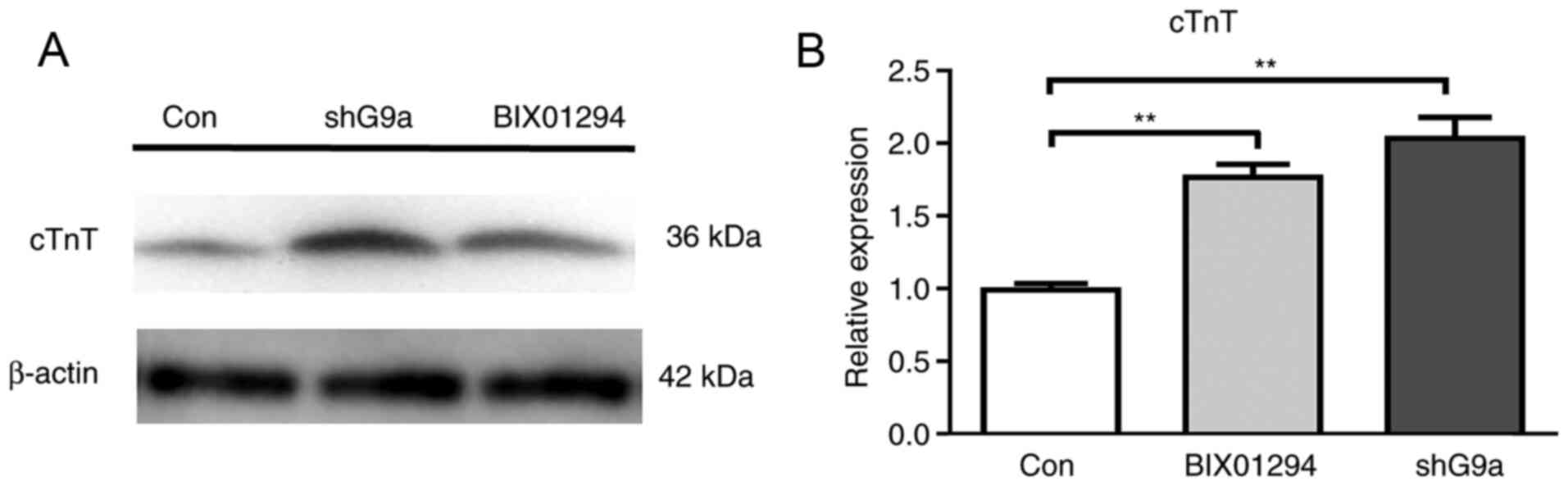
BIX01294 stimulates differentiation of
MSCs
The present study investigated the effect of
BIX01294 on the process of differentiation to demonstrate that
inhibiting G9a can improve the efficiency of the differentiation of
MSCs into cardiomyocytes. MSCs were divided into three groups:
Treated with 1 µM BIX01294 for 12 h, transfected with shRNA-G9a and
control. All groups were induced by 5-AZA. The expression of cTnT
on the 21st day was detected by western blotting and RT-qPCR
(Fig. 6A and B). The results
demonstrated that both BIX01294 group and shG9a group can increase
the expression of cTnT, but there was no significant difference
between two groups. The results further demonstrated that H3K9me2
was involved in the process of cardiomyocyte' differentiation.
Discussion
Compared with implantable electronic pacemakers,
biological pacemakers are more suitable for treating chronic
arrhythmia and pathological sinus syndrome (2). Currently, the efficiency of stem cell
induction into cardiomyocytes is low, which limits its clinical
application. Our previous study treated very small embryonic-like
stem cells with 5-AZA and the positive expression rates of cTnT and
α-actin were 18.41 and 19.43%, respectively on day 14 (19). In the present study, epigenetics was
used to improve the differentiation efficiency of stem cells.
5-AZA, widely used in the cardiac differentiation of
stem cells, can regulate the histone demethylation and DNA
methylation (20). A number of stem
cells have been used to differentiate into cardiomyocytes by 5-AZA
(19–23). The mechanism of 5-AZA may be related
to CpG base-pair demethylation and regulation of early myocardial
transcription factors (24–27). However, the efficiency of 5-AZA in
inducing the differentiation of stem cells into cardiomyocytes
varies between different laboratories. Antonitsis et al
(24) induced MSCs into
cardiomyocytes with an efficiency of 15%. After embryonic stem
cells were treated with 5-AZA, Choi et al (28) found that the proportion of
sarcomeric α-actin positive cells accounted for 6.48% on day 15
measured by means of flow cytometry. In the present study, MSCs
isolated by density centrifugation expressed CD29 and CD44, but not
CD45, which is consistent with previously reported expression
profiles of the MSCs (12). By
using 5-AZA, cTnT-expressing cells were successfully induced, and
it is suggested that the present study successfully constructed a
system of 5-AZA inducing MSCs into cardiomyocytes. In the present
study, cTnT-positive cells were 19% following 5-AZA induction,
which was not different from previous studies. The low efficiency
with respect to the induced cardiomyocyte differentiation of the
MSCs could possibly be due to the reduced response to other factors
produced in the microenvironment that could be involved in the
process of cardiac differentiation following the treatment of 5-AZA
(12). Therefore, an effective way
to improve the induction efficiency is required.
It is becoming clear that histone modifications
regulate transcription and control cell fate during cell
development. During cardiac development, Histone
acetylation/methylation and DNA methylation were both involved in
regulating GATA binding protein 4 (GATA-4) expression (29), while H3K27me1, H3K27me3, H3K36me3
and H3K4me3 are associated with the common histone methylation
modifications of H3K9me3/me2. H3K27me3 and H3K9me3/me2 are
associated with gene silencing and H3K27me1, H3K4me3 and H3K36me3
are associated with gene activation (30). During the maturation of
cardiomyocytes, the expression of H3K9me2 can be regulated to
affect the expression of genes related to cardiac development and
protect the heart from pathological hypertrophy caused by
overexpression of this gene (20).
In the present study, H3K9me2 decreased gradually with the increase
of induction days, suggesting that H3K9me2 may be involved in the
differentiation of the MSCs into cardiomyocytes. In addition, the
changes of H3K9me2 expression were not significant on the 7th day,
but were from the 14th day, which was consistent with the changes
in the expression of cTnT. This may be because the differentiation
of MSCs into cardiomyocytes is a dynamic process. The results of
the present study are consistent with previous studies (9,20).
Unlike G9a, the mechanism by which 5-AZA regulates histone
methylation remains unclear. A previous study also demonstrated
that 5-AZA inhibits H3K9me2 rather than G9a/Glp expression
(31).
It was hypothesized that the differentiation
efficiency could be affected by regulating the expression of
H3K9me2. G9a and KDM3A are important H3K9me2 enzymes for
methylation modification. G9a can modify histones at H3K9 and H3K27
sites in promoter regions of some genes through its histone lysine
methyltransferases activity, thus regulating gene transcriptional
silencing (32). Previous studies
have found that knockdown of G9a or use of G9a inhibitor can
significantly reduce the level of H3K9me2 in vivo (33,34).
In the present study, knockdown of G9a also reduced the expression
of H3K9me2, which was consistent with their results. Qin et
al (35) found that the
knockout of KDM3A significantly increased H3K9me2/me1 levels. The
present study found that knockdown of KDM3A increased the
expression of H3K9me2, which was consistent with previous research
results (15,35). However, the knockdown of G9a and
KDM3A did not cause changes in cell morphology during the induction
of 5-AZA. It was considered that the changes in H3K9me2 expression
caused by knockdown of G9a and KDM3A only caused changes in the
transcription level of genes regulated by chromatin, which did not
involve changes in DNA sequencing.
Additionally, the present study compared the final
cTnT-positive cell rate and the differentiation efficiency increase
following G9a knockdown, which was greater compared with the
previous efficiency following treatment with 5-AZA alone (19,24,28).
The same effect can be achieved with G9a inhibitors BIX012094.
5-AZA can induce the differentiation of bone marrow mesenchymal
stem cells into cardiomyocytes by participating in the gene
expression regulation of the transcription factors GATA-4 and/or
Nkx2.5 in early myocardial development (36,37).
In the present study, following 5-AZA treatment, the early
transcription factors GATA-4, NKx2.5 and MEF2c increased in all
groups of the MSCs, indicating that the treated cells had reached
the myocardial phenotype, consistent with the results of
differentiation induced by different inducers (9,12,16).
The results of the present study found that the expressions of
GATA-4, NKx2.5 and MEF2c were highest following G9a knockdown but
that the expression of transcription factors in early cardiac
development was decreased following KMD3A knockdown, indicating
that H3K9me2 may be involved in the differentiation of MSCs into
cardiomyocytes. The final results of the present study demonstrated
that the reduction H3K9me2 by G9a knockdown can improve the
efficiency of 5-AZA inducing the differentiation of MSCs into
cardiomyocytes. This may be because H3K9me2 recruits inhibitory
factors, such as members of the HP1 family, leading to tighter
binding of DNA and histones, thereby inhibiting transcription of
key genes in cardiac cell development (38). Unfortunately, the present study did
not compare the efficiency of 5-AZA and G9a against H3K9me2
inhibition directly. Therefore, in the next stage of the study,
further research will be conducted on this aspect.
In conclusion, the present study suggested that
H3K9me2 may be involved in the process of differentiation MSCs into
cardiomyocytes. Interference with G9a is an effective way to
increase 5-AZA induction of the MSCs into cardiomyocytes in
vitro.
Acknowledgements
The authors would like to thank Professor Lei Sun of
the Cardiology Department of Northern Jiangsu People's Hospital
(Yangzhou, China) for help with the experimental design.
Funding
The present study was funded by the National Natural
Science Foundation of China (grant no. 81370305).
Availability of data and materials
The datasets used and/or analyzed during the current
study are available from the corresponding author on reasonable
request.
Authors' contributions
XS, HL, PX, ML and XG designed the study, collected
the data, performed the statistical analyses and contributed to the
writing of the manuscript. QZ and BL helped supervise the
experiments. PX and YZ helped with the data collection, study
design and supervised the study. QZ and BL participated in the
study design and helped to critically revise the manuscript. XS and
XG confirm the authenticity of all the raw data. All authors read
and approved the final manuscript.
Ethics approval and consent to
participate
All procedures involving the care and use of animals
conformed to the Animal Research: Reporting of in vivo
Experiments (ARRIVE) guidelines and were approved by the Laboratory
Animal Management and Experimental Animal Ethics Committee of
Yangzhou University (approval no. 201903478).
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Barstow C and McDivitt JD: Cardiovascular
disease update: Bradyarrhythmias. FP Essent. 454:18–23.
2017.PubMed/NCBI
|
|
2
|
Burns CG and Burns CE: Canonical Wnt
signaling sets the pace. Dev Cell. 50:675–676. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Bagno L, Hatzistergos KE, Balkan W and
Hare JM: Mesenchymal stem cell-based therapy for cardiovascular
disease: Progress and challenges. Mol Ther. 26:1610–1623. 2018.
View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Fukuda K: Development of regenerative
cardiomyocytes from mesenchymal stem cells for cardiovascular
tissue engineering. Artif Organs. 25:187–193. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Hou J, Lü AL, Liu BW, Hou J, Xing YJ, Da
J, Hou ZL and Ai SY: Combination of BMP-2 and 5-AZA is advantageous
in rat bone marrow-derived mesenchymal stem cells differentiation
into cardiomyocytes. Cell Biol Int. 37:1291–1299. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Xu W, Zhang X, Qian H, Zhu W, Sun X, Hu J,
Zhou H and Chen Y: Mesenchymal stem cells from adult human bone
marrow differentiate into a cardiomyocyte phenotype in vitro. Exp
Biol Med (Maywood). 229:623–631. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Cheng F, Zou P, Yang H, Yu Z and Zhong Z:
Induced differentiation of human cord blood mesenchymal
stem/progenitor cells into cardiomyocyte-like cell in vitro. J
Huazhong Univ Sci Technolog Med Sci. 23:154–157. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Luo L, Tang J, Nishi K, Yan C, Dinh PU,
Cores J, Kudo T, Zhang J, Li TS and Cheng K: Fabrication of
synthetic mesenchymal stem cells for the treatment of acute
myocardial infarction in mice. Circ Res. 120:1768–1775. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Li J, Zhu K, Wang Y, Zheng J, Guo C, Lai H
and Wang C: Combination of IGF1 gene manipulation and 5-AZA
treatment promotes differentiation of mesenchymal stem cells into
cardiomyocyte-like cells. Mol Med Rep. 11:815–820. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Saito Y, Nakamura K, Yoshida M, Sugiyama
H, Takano M, Nagase S, Morita H, Kusano KF and Ito H:
HCN4-overexpressing mouse embryonic stem cell-derived
cardiomyocytes generate a new rapid rhythm in rats with
bradycardia. Int Heart J. 59:601–606. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Gorabi AM, Hajighasemi S, Tafti HA, Atashi
A, Soleimani M, Aghdami N, Saeid AK, Khori V, Panahi Y and Sahebkar
A: TBX18 transcription factor overexpression in human-induced
pluripotent stem cells increases their differentiation into
pacemaker-like cells. J Cell Physiol. 234:1534–1546. 2019.
View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Yang G, Tian J, Feng C, Zhao LL, Liu Z and
Zhu J: Trichostatin a promotes cardiomyocyte differentiation of rat
mesenchymal stem cells after 5-azacytidine induction or during
coculture with neonatal cardiomyocytes via a mechanism independent
of histone deacetylase inhibition. Cell Transplant. 21:985–996.
2012. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Singh PB, Belyakin SN and Laktionov PP:
Biology and physics of heterochromatin-like domains/complexes.
Cells. 9:18812020. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Yang J, Kaur K, Edwards JG, Eisenberg CA
and Eisenberg LM: Inhibition of histone methyltransferase, histone
deacetylase, and β-catenin synergistically enhance the cardiac
potential of bone marrow cells. Stem Cells Int. 2017:34649532017.
View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Zhang QJ, Tran TAT, Wang M, Ranek MJ,
Kokkonen-Simon KM, Gao J, Luo X, Tan W, Kyrychenko V, Liao L, et
al: Histone lysine dimethyl-demethylase KDM3A controls pathological
cardiac hypertrophy and fibrosis. Nat Commun. 9:52302018.
View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Huang YL, Qiu RF, Mai WY, Kuang J, Cai XY,
Dong YG, Hu YZ, Song YB, Cai AP and Jiang ZG: Effects of
insulin-like growth factor-1 on the properties of mesenchymal stem
cells in vitro. J Zhejiang Univ Sci B. 13:20–28. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Nippert F, Schreckenberg R and Schlüter
KD: Isolation and cultivation of adult rat cardiomyocytes. J Vis
Exp. 128:566342017.PubMed/NCBI
|
|
18
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(-Delta Delta C(T)) method. Methods. 25:402–408. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Sun X, Li H, Zhu Y, Xu P, Zuo Q, Li B and
Gu X: 5-Azacytidine-induced cardiomyocyte differentiation of very
small embryonic-like stem cells. Stem Cells Int. 2020:51623502020.
View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Thienpont B, Aronsen JM, Robinson EL,
Okkenhaug H, Loche E, Ferrini A, Brien P, Alkass K, Tomasso A,
Agrawal A, et al: The H3K9 dimethyltransferases EHMT1/2 protect
against pathological cardiac hypertrophy. J Clin Invest.
127:335–348. 2017. View
Article : Google Scholar : PubMed/NCBI
|
|
21
|
Abou-ElNaga A, El-Chennawi F, Ibrahim
Kamel S and Mutawa G: The potentiality of human umbilical cord
isolated mesenchymal stem/stromal cells for cardiomyocyte
generation. Stem Cells Cloning. 13:91–101. 2020.PubMed/NCBI
|
|
22
|
Soltani L, Rahmani HR, Daliri Joupari M,
Ghaneialvar H, Mahdavi AH and Shamsara M: Ovine fetal mesenchymal
stem cell differentiation to cardiomyocytes, effects of co-culture,
role of small molecules; reversine and 5-azacytidine. Cell Biochem
Funct. 34:250–261. 2016. View
Article : Google Scholar : PubMed/NCBI
|
|
23
|
Jain M, Minocha E, ripathy NK, Singh N,
Chaturvedi CP and Nityanand S: Comparison of the cardiomyogenic
potency of human amniotic fluid and bone marrow mesenchymal stem
cells. Int J Stem Cells. 12:449–456. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Antonitsis P, Ioannidou-Papagiannaki E,
Kaidoglou A and Papakonstantinou C: In vitro cardiomyogenic
differentiation of adult human bone marrow mesenchymal stem cells.
The role of 5-azacytidine. Interact Cardiovasc Thorac Surg.
6:593–597. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Larsen F, Gundersen G, Lopez R and Prydz
H: CpG islands as gene markers in the human genome. Genomics.
13:1095–1107. 1992. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Ruan ZB, Zhu L, Yin YG and Chen GC: The
mechanism underlying the differentiation of human umbilical
cord-derived mesenchymal stem cells into myocardial cells induced
by 5-azacytidine. Indian J Med Sci. 64:402–407. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Enright BP, Kubota C, Yang X and Tian XC:
Epigenetic characteristics and development of embryos cloned from
donor cells treated by trichostatin A or 5-aza-2′-deoxycytidine.
Biol Reprod. 69:896–901. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Choi SC, Yoon J, Shim WJ, Ro YM and Lim
DS: 5-Azacytidine induces cardiac differentiation of P19 embryonic
stem cells. Exp Mol Med. 36:515–523. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Xu H, Yi Q, Yang C, Wang Y, Tian J and Zhu
J: Histone modifications interact with DNA methylation at the GATA4
promoter during differentiation of mesenchymal stem cells into
cardiomyocyte-like cells. Cell Prolif. 49:315–329. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Gilsbach R, Schwaderer M, Preissl S,
Grüning BA, Kranzhöfer D, Schneider P, Nührenberg TG,
Mulero-Navarro S, Weichenhan D, Braun C, et al: Distinct epigenetic
programs regulate cardiac myocyte development and disease in the
human heart in vivo. Nat Commun. 9:3912018. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Wang X, Wu H, Ma J, Xu S, Li S, Bao S and
Wang F: threshold inhibition of methyltransferase G9a/Glp
exacerbates neuropathic hypersensitivity through mediating GRIN2B
methylation. Sci Insigt. 29:33–47. 2019. View Article : Google Scholar
|
|
32
|
Schones DE, Chen X, Trac C, Setten R and
Paddison PJ: G9a/GLP-dependent H3K9me2 patterning alters chromatin
structure at CpG islands in hematopoietic progenitors. Epigenetics
Chromatin. 7:232014. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Liang L, Zhao JY, Kathryn T, Bekker A and
Tao YX: BIX01294, a G9a inhibitor, alleviates nerve injury-induced
pain hypersensitivities during both development and maintenance
periods. Transl Perioper Pain Med. 6:106–114. 2019.PubMed/NCBI
|
|
34
|
Fu L, Yan FX, An XR and Hou J: Effects of
the histone methyltransferase inhibitor UNC0638 on Histone H3K9
dimethylation of cultured ovine somatic cells and development of
resulting early cloned embryos. Reprod Domest Anim. 49:e21–e25.
2014. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Qin L, Xu Y, Yu X, Toneff MJ, Li D, Liao
L, Martinez JD, Li Y and Xu J: The histone demethylase Kdm3a is
required for normal epithelial proliferation, ductal elongation and
tumor growth in the mouse mammary gland. Oncotarget. 8:84761–84775.
2017. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Almalki SG and Agrawal DK: Key
transcription factors in the differentiation of mesenchymal stem
cells. Differentiation. 92:41–51. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Cao F, Niu LL, Meng L, Zhao LX, Zheng M,
Yue W, Bai CX, Jia GL and Pei XT: Cadiomyocyte-like differentiation
of human bone marrow mesenchymal stem cells after exposure to
5-azacytidine in vitro. Shi Yan Sheng Wu Xue Bao. 37:118–124.
2004.PubMed/NCBI
|
|
38
|
Dong W, Oya E, Zahedi Y, Prasad P,
Svensson JP, Lennartsson A, Ekwall K and Durand-Dubief M: Abo1 is
required for the H3K9me2 to H3K9me3 transition in heterochromatin.
Sci Rep. 10:60552020. View Article : Google Scholar : PubMed/NCBI
|