Introduction
Tuberculosis (TB) is caused by the etiological agent
Mycobacterium (M.), which primarily affects the lungs
(1). TB was responsible for ~1.5
million deaths in 2018 worldwide (2). M. tuberculosis parasitizes the
macrophages of the host; it manipulates the host's defenses and
consequently the immune response (3). Furthermore, M. tuberculosis can
evade innate immunity to survive and replicate inside macrophages
(4). Therefore, the development of
therapeutics that prevent immune evasion is crucial.
MicroRNAs (miRNAs/miRs) are a class of non-coding
RNAs (~22 nucleotides long) that serve a role in silencing target
gene expression and are associated with immune signaling pathways
(5,6). For example, miR-325-3p promotes M.
tuberculosis survival by targeting ligand of numb-protein X1
and increasing the phosphorylation of STAT3 (7). Overexpression of miR-26a was reported
to modulate the survival of M. tuberculosis in macrophages
(8). miR-125a inactivates NF-κB
signaling by targeting TNF receptor associated factor 6,
attenuating inflammation and facilitating the survival of M.
tuberculosis (9). Although the
potential role of several miRNAs in M. tuberculosis
infection has been examined, this field requires further
investigation.
Rho-associated coiled-coil-forming protein kinase 1
(ROCK1) is a downstream effector of RhoA; it acts as a ‘molecular
switch’ in the activation of the monocyte pro-inflammatory response
(10). Suppression of ROCK1
expression prevents NF-κB signaling in a variety of inflammatory
diseases, such as cervical cancer, and is a hallmark of obstructive
sleep apnea syndrome (11,12). A previous study has shown that
Toll-like receptor (TLR)4 is involved in the regulation of
pulmonary immune responses and recognition of M.
tuberculosis (13). NF-κB is a
downstream effector of the TLR4 signaling pathway and an important
pro-inflammatory factor (14).
Numerous inflammatory cytokines, including TNF-α, IL-1β and IL-6,
regulate the TLR4/NF-κB pathway (15).
In the present study, upregulated miRNAs in patients
with TB were identified using the Gene Expression Omnibus (GEO)
datasets, GSE34608 and GSE116542. miR-502-3p was selected for
further studies. The aim of the present study was to investigate
the function of miR-502-3p in M. tuberculosis-infected
macrophages.
Materials and methods
Bioinformatics
The GEO (https://www.ncbi.nlm.nih.gov/geo) database [GSE34608
(PMID: 22547807) and GSE116542] was used to analyze miR-502-3p
expression in patients with TB and healthy individuals. In
GSE116542, 11 patients with TB (8 men; 3 women; age range, 17–51
years) and 8 healthy individuals (4 men; 4 women; age range, 29–60
years) were collected for exosomal miRNAs extraction. TargetScan
7.2 (http://www.targetscan.org) was used to
predict the binding sites between miR-502-3p and ROCK1.
Cell culture
The human leukemia monocytic THP-1 and the mouse
macrophage-like RAW 264.7 cell lines were purchased from The Cell
Bank of Type Culture Collection of The Chinese Academy of Sciences.
Cells were maintained in an incubator (37°C; 5% CO2; 70%
relative humidity) in RPMI-1640 medium containing 10% fetal bovine
serum (Hyclone; Cytiva).
Transfection
THP-1 and RAW 264.7 cells were plated on 12-well
plates at a seeding density of 3×105 cells/well and
transfected with miR-502-3p mimic, miR-502-3p inhibitor,
pcDNA3.1-ROCK1 (Shanghai GenePharma Co., Ltd.), or their negative
controls (NCs), mimic NC, inhibitor NC and pcDNA3.1-NC (50 pg/well)
at 37°C for 24 h, using Lipofectamine® 3000 (Invitrogen;
Thermo Fisher Scientific, Inc.). At 24 h following transfection,
the transfected macrophages were used for other experimental
assays. The sequences are listed in Table I.
 | Table I.Primer sequences used in
transfection. |
Table I.
Primer sequences used in
transfection.
| Gene | Primer sequence
(5′→3′) |
|---|
| miR-502-3p
mimic |
AAUGCACCUGGGCAAGGAUUCA |
| Mimic NC |
UCACAACCUCCUAGAAAGAGUAGA |
| miR-502-3p
inhibitor |
UGAAUCCUUGCCCAGGUGCAUU |
| Inhibitor NC |
UCUACUCUUUCUAGGAGGUUGUGA |
Infection
M. tuberculosis strain H37Rv (cat. no. 25618;
American Type Culture Collection) was cultured in Middlebrook 7H9
broth media (Beijing Solarbio Science & Technology Co., Ltd.)
containing 10% oleic acid albumin dextrose catalase enrichment
(OADC; BD Biosciences) at 37°C. Transfected THP-1 cells were
incubated with 100 nM phorbol 12-myristate 13-acetate (PMA;
Sigma-Aldrich; Merck KGaA) at 37°C for 48 h until differentiation
into human macrophages occurred. Following transfection and
PMA-differentiation, THP-1 cells and RAW 264.7 cells
(5.0×105) were infected with the M. tuberculosis
strain H37Rv at a multiplicity of infection (MOI) of 1, 2, 5 and 10
at 37°C for 48 h. In subsequent experiments, cells were infected
with M. tuberculosis H37Rv at 37°C for 3, 6, 12, 24 and 48 h
at an MOI of 10.
Reverse transcription-quantitative PCR
(RT-qPCR)
Total RNA was extracted from the transfected and
infected THP-1 and RAW 264.7 cells using TRIzol®
(Invitrogen; Thermo Fisher Scientific, Inc.) according to the
manufacturer's protocol. Total RNA (1 µg) was reverse transcribed
into cDNA using the M-MLV First Strand Kit (Invitrogen; Thermo
Fisher Scientific, Inc.) according to the manufacturer's protocol.
qPCR was subsequently performed using the SYBR-Green PCR master mix
(Applied Biosystems; Thermo Fisher Scientific, Inc.) in a 7900HT
Fast Real-Time PCR System (Applied Biosystems; Thermo Fisher
Scientific, Inc.). qPCR was conducted under the following
conditions: Initial denaturation at 95°C for 20 sec, followed by 40
cycles of 95°C for 5 sec, 60°C for 30 sec and 72°C for 15 sec. The
primers used for qPCR are provided in Table II. For IL-6, TNF-α and IL-1β,
β-actin was used as the internal reference gene. Bulge-loop™ miRNA
RT-qPCR primer sets specific for miR-502-3p were designed by
Guangzhou RiboBio Co., Ltd. For miR-502-3p, U6 was used as the
internal control. Relative expression levels were analyzed using
the 2−ΔΔCq method (16).
 | Table II.Sequences of primers used for reverse
transcription-quantitative PCR. |
Table II.
Sequences of primers used for reverse
transcription-quantitative PCR.
| Gene | Primer sequence
(5′→3′) |
|---|
| miR-502-3p | F:
ACACTCCAGCTGGGAATGCACCTGGGCAAGG |
|
| R:
CTCAACTGGTGTCGTGGA |
| U6 | F:
CTCGCTTCGGCAGCACA |
|
| R:
AACGCTTCACGAATTTGCGT |
| IL-6 (human) | F:
ACAACCACGGCCTTCCCTACT |
|
| R:
CACGATTTCCCAGAGAACATGTG |
| IL-6 (mouse) | F:
GGGCTGCGATGGAGTCAGAG |
|
| R:
TCCCTCACACAGGGCTCGAC |
| TNF-α (human) | F:
GCCTCTTCTCATTCCTGCTTG |
|
| R:
GGCCATTTGGGAACTTCTCA |
| TNF-α (mouse) | F:
TGACCCCCATTACTCTGACC |
|
| R:
TTCAGCGTCTCGTGTGTTTC |
| IL-1β (human) | F:
GTGGCAATGAGGATGACTTGTTC |
|
| R:
GGTGGTCGGAGATTCGTAGCT |
| IL-1β (mouse) | F:
GAGCAACAAGTGGTGTTCTCC |
|
| R:
AACACGCAGGACAGGTACAG |
| β-actin
(human) | F:
CATGTACGTTGCTATCCAGGC |
|
| R:
CTCCTTAATGTCACGCACGAT |
| β-actin
(mouse) | F:
GTGTGGGCATTTGATGAGCC |
|
| R:
AGGTCACTTACCTGGTGCCT |
Colony-forming unit (CFU) assay
Following transfection, cells (3×105
cells/well) were infected with M. tuberculosis (MOI=10) for
48 h and lysed with 0.5% Triton X-100 at 25°C for 20 min. A 10-fold
serial dilution method was used for quantitative culture of
bacterial colonies. The cell lysate was diluted with Middlebrook
7H9 broth media and added to Middlebrook 7H10 agar plates (Beijing
Solarbio Science & Technology Co., Ltd.) supplemented with 10%
OADC. Plates were incubated at 37°C for 3 weeks before the colonies
were quantified manually. A colony referred to the growth of
bacteria on solid medium that could be identified by the naked
eye.
Dual-luciferase reporter assay
TargetScan was used to predict the mRNAs that may
have a target site of miR-502-3p. Wild-type (WT) and mutant (MUT)
ROCK1 3′-untranslated regions (UTRs) were cloned into the firefly
luciferase reporter plasmid psi-CHECK2 (Qiagen China Co., Ltd.) to
synthesize the ROCK1-WT and ROCK1-Mut reporter plasmids. 293T cells
(American Type Culture Collection) at 5×104 cells/well
in 24-well plates were co-transfected with WT or Mut ROCK1 3′-UTR
reporter plasmids and miR-502-3p mimic or mimic NC using
Lipofectamine® 3000 (Invitrogen; Thermo Fisher
Scientific, Inc.) at 37°C. After 48 h transfection, luciferase
activity was detected using a Dual-Luciferase Reporter Assay System
(Promega Corporation), according to the manufacturer's protocol.
Renilla luciferase activity was used for normalization.
Western blotting
Total protein was extracted from THP-1 and RAW 264.7
cells (5×105 cells/well) using RIPA lysis buffer
(Beyotime Institute of Biotechnology). The protein content was
assessed using the BCA method. Total protein (50 µg protein/lane)
was separated by SDS-PAGE on a 12% gel. The separated proteins were
transferred onto a nitrocellulose membrane (Invitrogen; Thermo
Fisher Scientific, Inc.). The membranes were blocked using 5%
skimmed milk for 2 h at 25°C. The membranes were incubated with
primary antibodies against the following: ROCK1 (1:1,000; cat. no.
4035), TLR4 (1:1,000; cat. no. 14358; Cell Signaling Technology,
Inc.; cat. no. ab13556, Abcam), phosphorylated (p)-p65 (1:1,000;
cat. no. 3033; Cell Signaling Technology, Inc.), p65 (1:1,000; cat.
no. 8242; Cell Signaling Technology, Inc.), p-IκBα (1:1,000; cat.
no. 2859; Cell Signaling Technology, Inc.), IκBα (1,000; cat. no.
4812; Cell Signaling Technology, Inc.) and β-actin (1:2,000; cat.
no. 4970; Cell Signaling Technology, Inc.) overnight at 4°C.
Following three washes of 5 min each with TBS-0.1% Tween-20, the
membranes were incubated with HRP-conjugated anti-rabbit secondary
antibodies (1:3,000; cat. no. A0208; Beyotime Institute of
Biotechnology) at 25°C for 1 h. Protein bands were visualized using
an ECL reagent (Beyotime Institute of Biotechnology) and a gel
imaging system (Tanon Science & Technology Co., Ltd.). Protein
bands were semi-quantified using ImageJ software v1.8.0 (National
Institutes of Health). β-actin was used as the loading control.
Immunocytochemistry
THP-1 and RAW 264.7 cells (5×104) were
fixed in 24-well plates using 4% paraformaldehyde at 25°C for 30
min (Beyotime Institute of Biotechnology), permeabilized using 0.3%
Triton X-100 at 25°C for 20 min and blocked with 5% goat serum
(Gibco; Thermo Fisher Scientific, Inc.) at 25°C for 30 min. Cells
were incubated with primary antibody against p-p65 (1:1,600; cat.
no. 3033; Cell Signaling Technology, Inc.) overnight at 4°C. Cells
were subsequently incubated with Alexa Fluor 594-conjugated goat
anti-rabbit IgG secondary antibody (1:500; cat. no. ab150080;
Abcam) for 1 h in the dark at room temperature. Cell nuclei were
stained using DAPI (Beyotime Institute of Biotechnology) at 25°C
for 30 min. Images were captured using a BX51 fluorescence
microscope (Olympus Corporation).
Statistical analysis
All presented data were obtained from at least three
independent experiments. Data are shown as the mean ± SD.
Statistical comparisons between two groups were determined by
Student's unpaired t-test, whereas comparisons between multiple
groups were determined using one-way ANOVA followed by Tukey's
post-hoc test. P<0.05 was considered to indicate a statistically
significant difference.
Results
M. tuberculosis infection induces
miR-502-3p expression in patients with TB
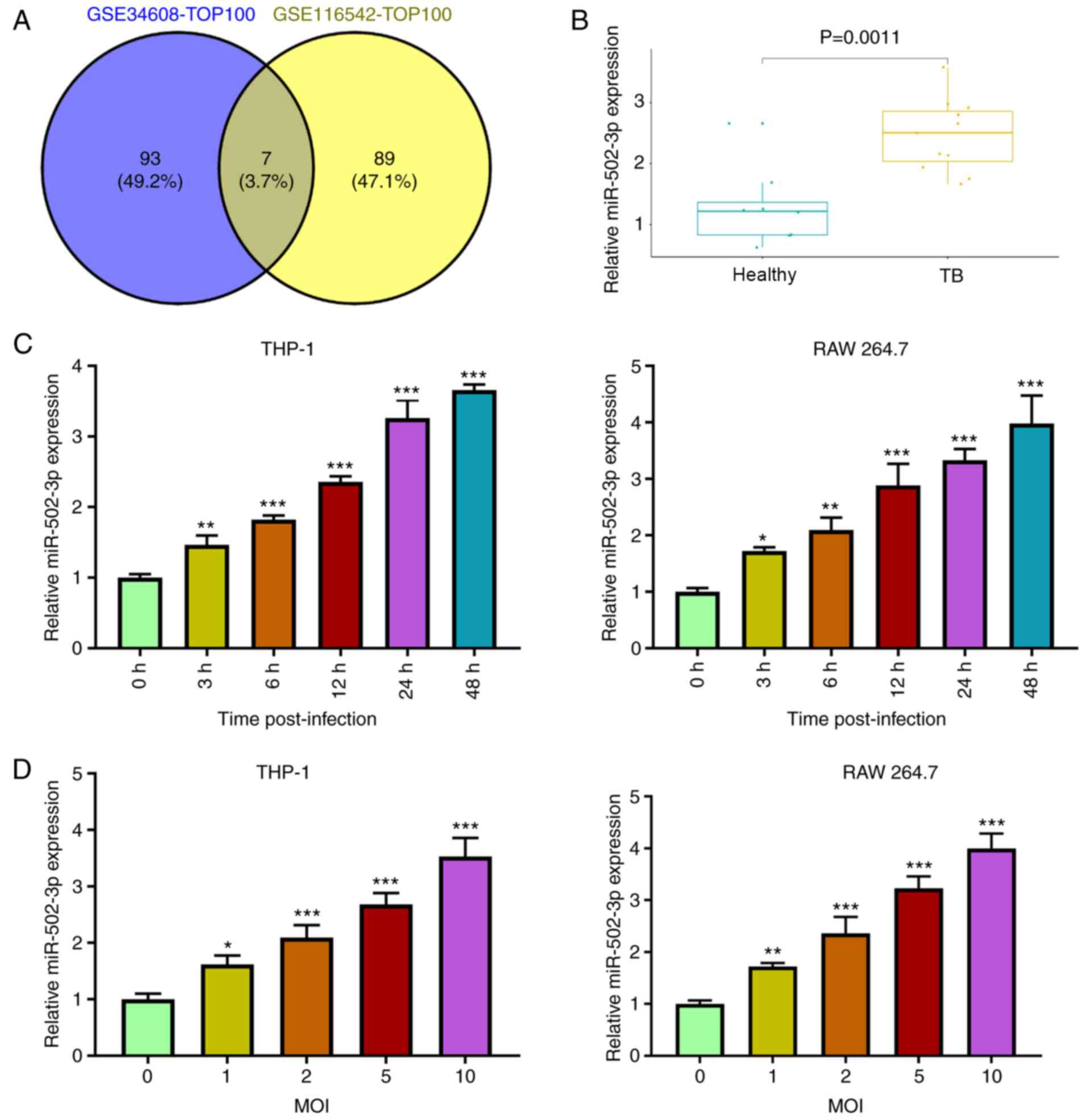
miRNAs with markedly high expression levels in TB
compared with the healthy control group were selected by analyzing
the GSE34608 and GSE116542 datasets from the GEO database. A total
of seven miRNAs were identified as being the same between the
datasets (Fig. 1A). The current
study aimed to investigated ROCK1 in TB. TargetScan predicted the
binding sites between miR-502-3p and ROCK1. Thus, miR-502-3p was
chosen from the seven miRNAs. miR-502-3p expression levels were
significantly elevated in patients with TB compared with healthy
individuals in the GSE116542 database (Fig. 1B). miR-502-3p expression levels in
M. tuberculosis-infected macrophages increased in a time and
dose-dependent manner (Fig. 1C and
D). The expression level of miR-502-3p in macrophages at 48 h
post-infection was >3-fold higher than that in uninfected
control (Fig. 1C). Moreover, there
was a ~3-fold increase in miR-502-3p expression in macrophages
cells at a MOI of 10 for 48 h compared with uninfected cells
(Fig. 1D). Macrophages infected
with M. tuberculosis at an MOI of 10 for 48 h were used for
subsequent experiments.
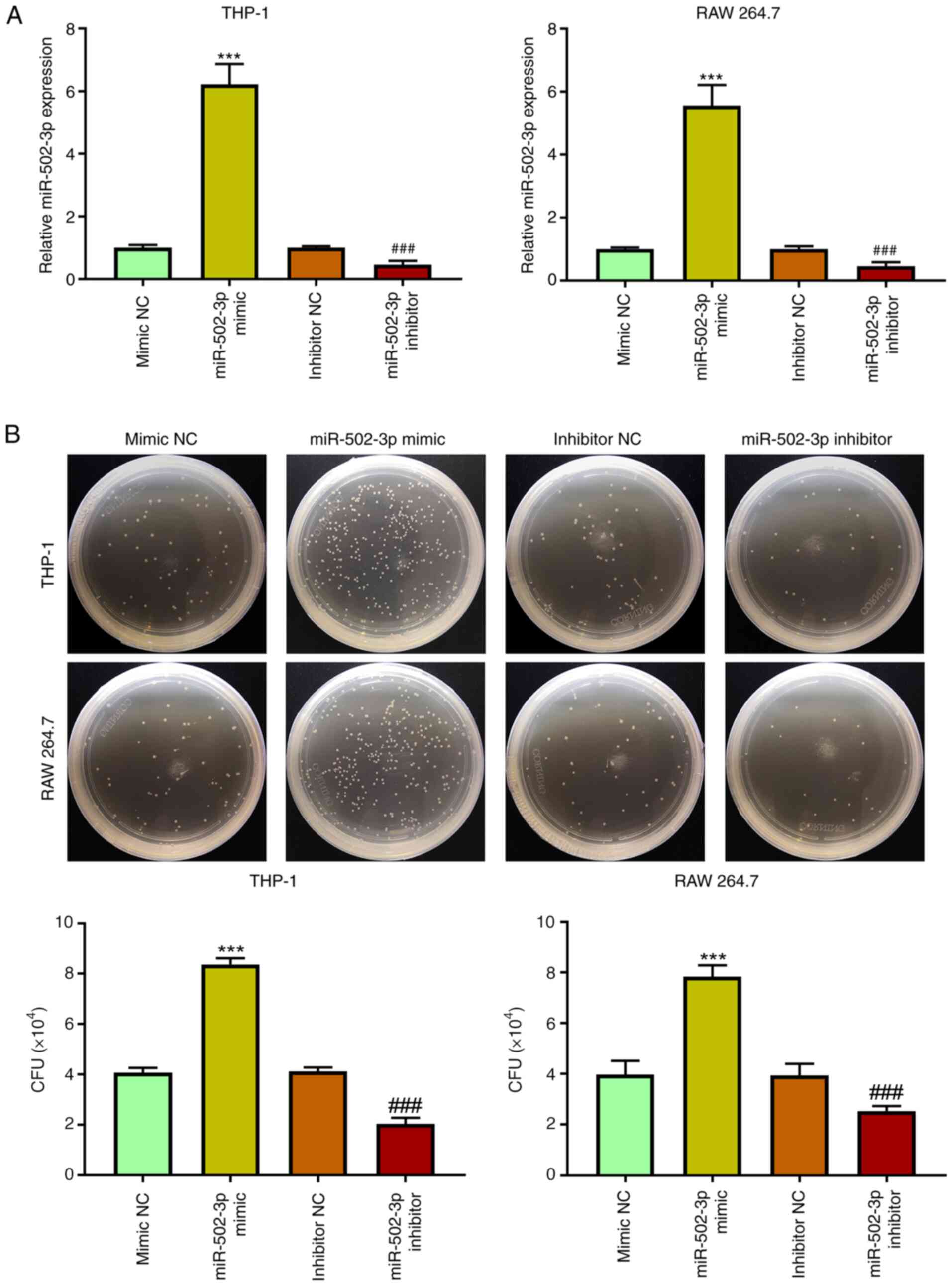
miR-502-3p facilitates M. tuberculosis
survival in macrophages
To further determine the potential role of
miR-502-3p in the cellular immune response during M.
tuberculosis infection, THP-1 and RAW 264.7 cells were
transfected with either miR-502-3p mimic or inhibitor. miR-502-3p
expression levels significantly increased following miR-502-3p
mimic transfection compared with mimic NC, and significantly
decreased following miR-502-3p inhibitor transfection compared with
inhibitor NC (Fig. 2A). The CFU
assay demonstrated that miR-502-3p mimic transfection significantly
increased the survival of M. tuberculosis from infected
macrophages compared with mimic NC, whereas inhibition of
miR-502-3p significantly reduced the intracellular growth of M.
tuberculosis compared with inhibitor NC (Fig. 2B). The high number of M.
tuberculosis colonies means that the phagocytosis of
macrophages to M. tuberculosis is weakened and the survival
of macrophages is reduced.
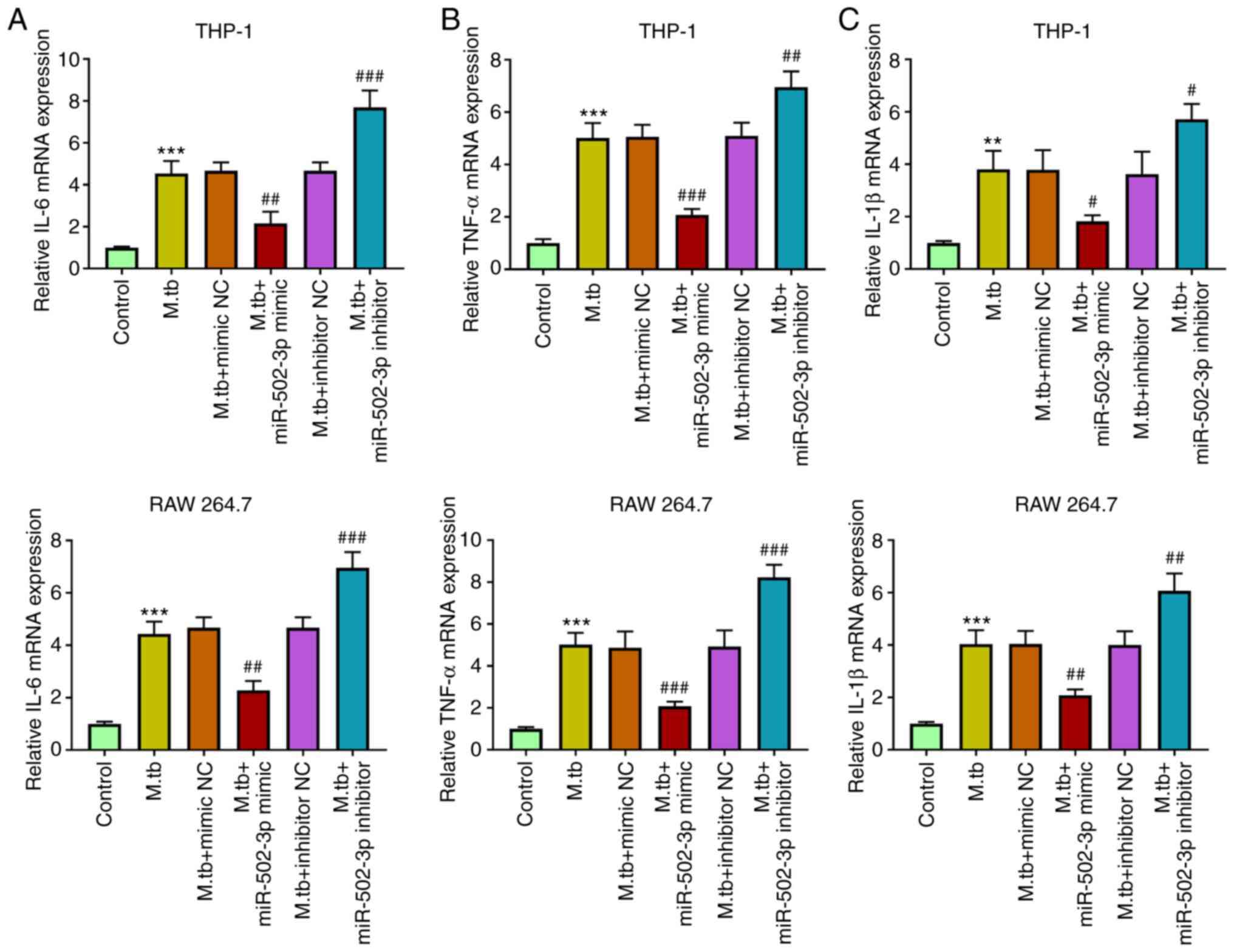
miR-502-3p suppresses cytokine
production in M. tuberculosis-infected macrophages
M. tuberculosis infection promotes
macrophages to produce cytokines (17). The effects of miR-502-3p on
cytokines in M. tuberculosis-infected THP-1 and RAW 264.7
cells were investigated. The mRNA expression levels of IL-6, TNF-α
and IL-1β were significantly induced in M.
tuberculosis-infected macrophages. The mRNA expression levels
of IL-6, TNF-α and IL-1β, were significantly reduced in M.
tuberculosis-infected macrophages transfected with miR-502-3p
mimic, compared with the M. tuberculosis-infected only
group. Moreover, downregulation of miR-502-3p significantly
increased the expression of IL-6, TNF-α and IL-1β, in M.
tuberculosis-infected macrophages compared with the M.
tuberculosis-infected only group (Fig. 3).
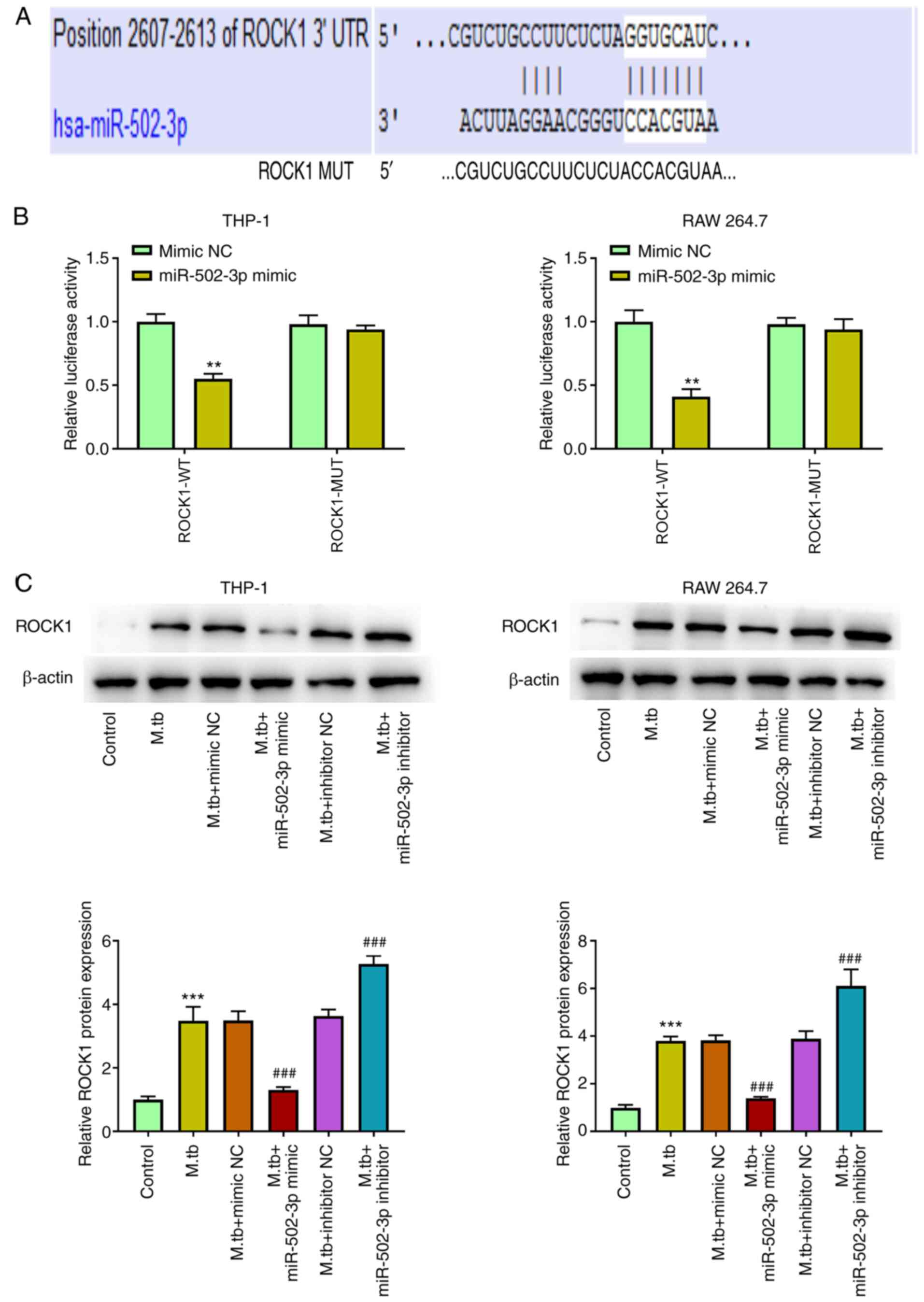
miR-502-3p directly targets ROCK1
Using TargetScan, ROCK1 was predicted to have a
potential miR-502-3p target site in its 3′UTR (Fig. 4A). miR-502-3p mimic transfection
significantly suppressed luciferase activity in 293T cells
containing the ROCK1-WT vector compared with mimic NC, whereas no
change in luciferase activity was observed in cells transfected
with the ROCK1-Mut vector (Fig.
4B). Western blotting demonstrated that the protein expression
levels of ROCK1 were significantly increased in M.
tuberculosis-infected macrophages compared with the control.
Compared with the M. tuberculosis-infected only group,
overexpression of miR-502-3p significantly decreased ROCK1 protein
expression levels, whereas knockdown of miR-502-3p significantly
increased ROCK1 protein expression levels (Fig. 4C).
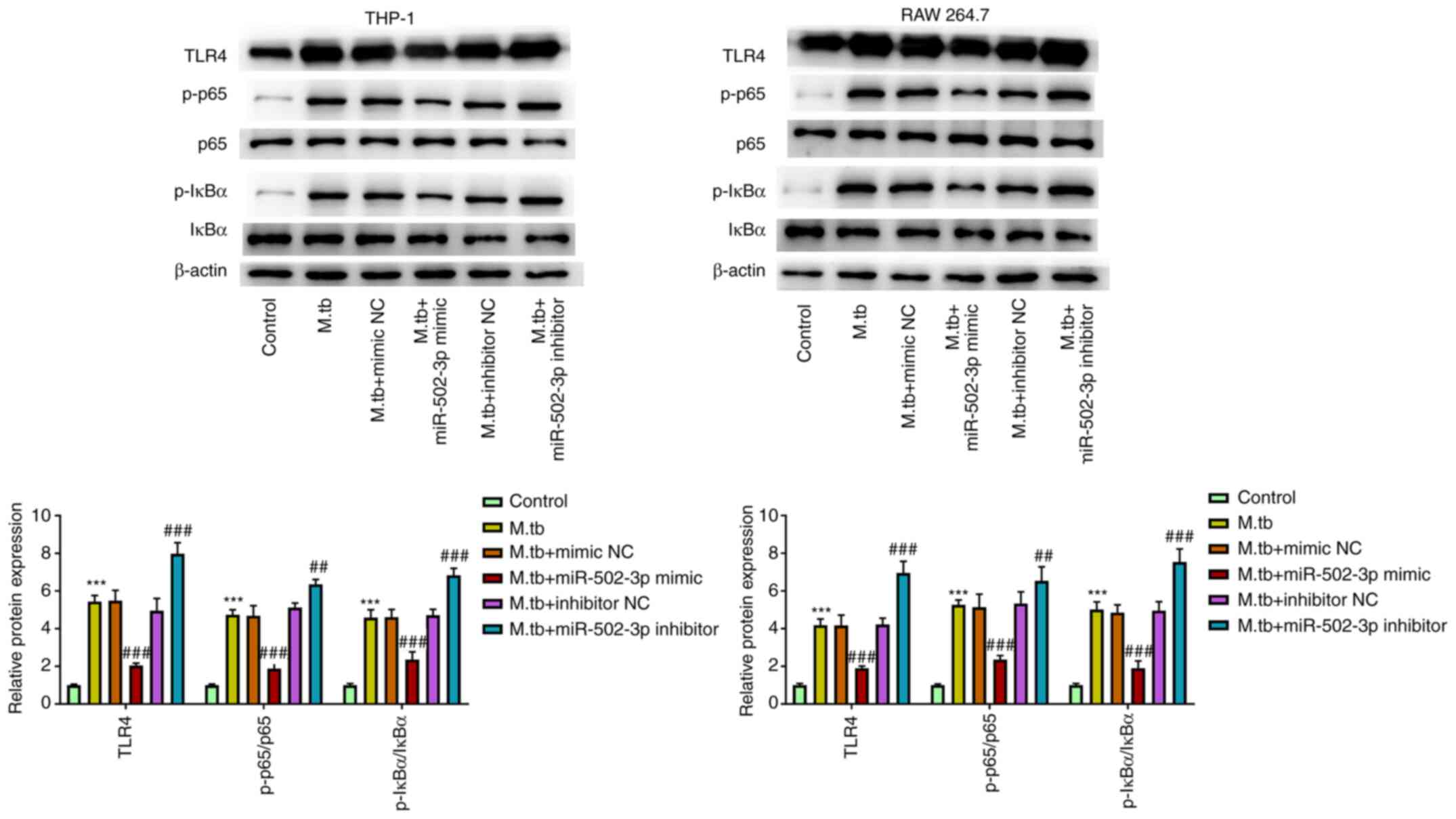
miR-502-3p regulates the TLR4/NF-κB
signaling pathway in M. tuberculosis-infected macrophages
A previous study has shown that TLR-4/miR-125a/NF-κB
signaling modulates the immune response to M. tuberculosis
infection (9). To determine the
mechanism by which miR-502-3p promoted M. tuberculosis
survival in macrophages, the TLR4/NF-κB signaling pathway was
examined. TLR4, p65 phosphorylation and IκBα phosphorylation were
significantly increased in M. tuberculosis-infected THP-1
and RAW 264.7 cells. However, TLR4, p65 phosphorylation and IκBα
phosphorylation were significantly reduced in M.
tuberculosis-infected THP-1 and RAW 264.7 cells transfected
with miR-502-3p mimic compared with the M.
tuberculosis-infected only group. Moreover, the downregulation
of miR-502-3p expression significantly increased TLR4 levels and
the phosphorylation of p65 and IκBα compared with the M.
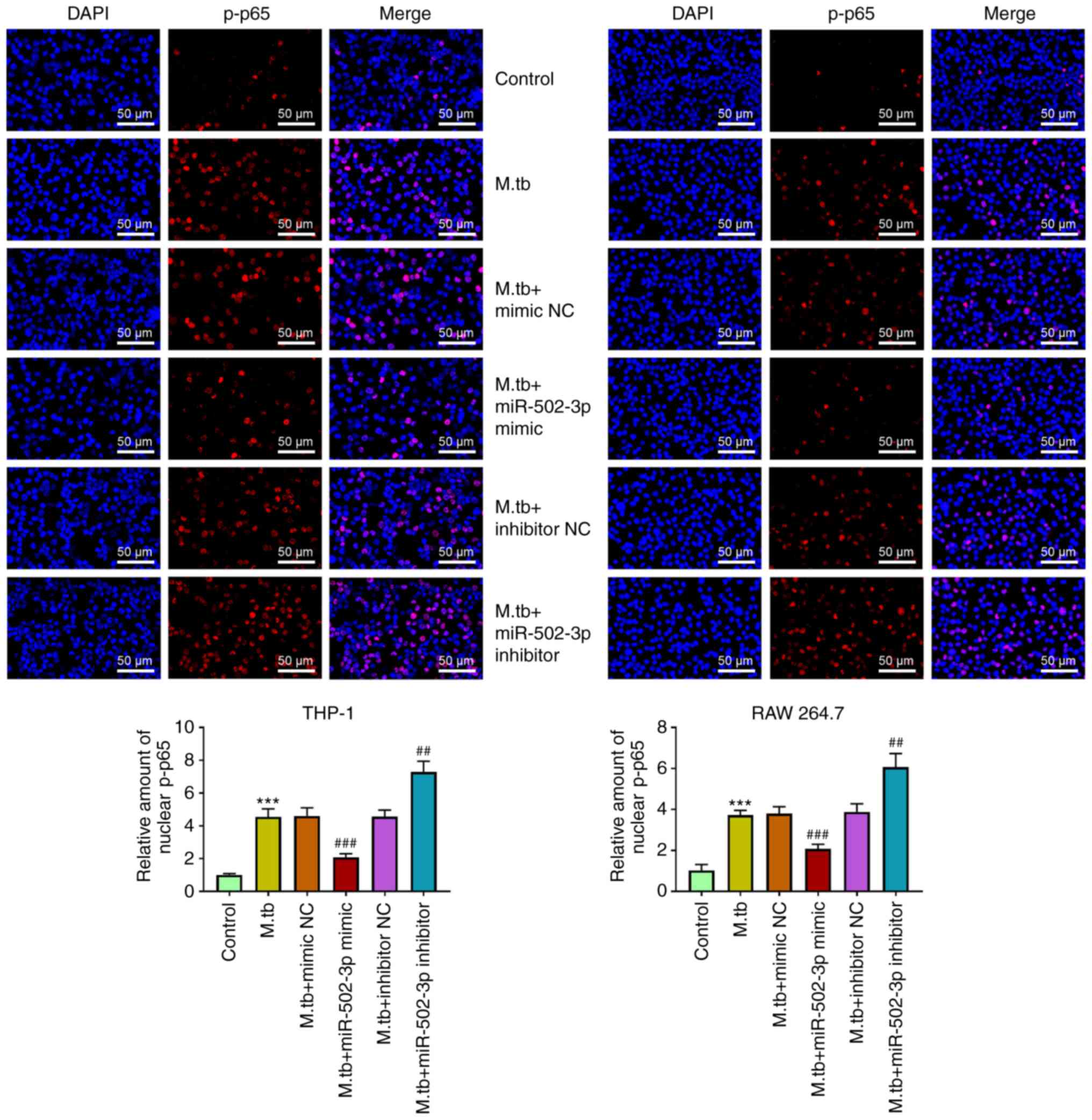
tuberculosis-infected only group (Fig. 5). Furthermore, compared with M.
tuberculosis-infected only group, the expression level of p-p65
in nucleus was significantly reduced by miR-502-3p overexpression
and significantly induced by miR-502-3p inhibition (Fig. 6).
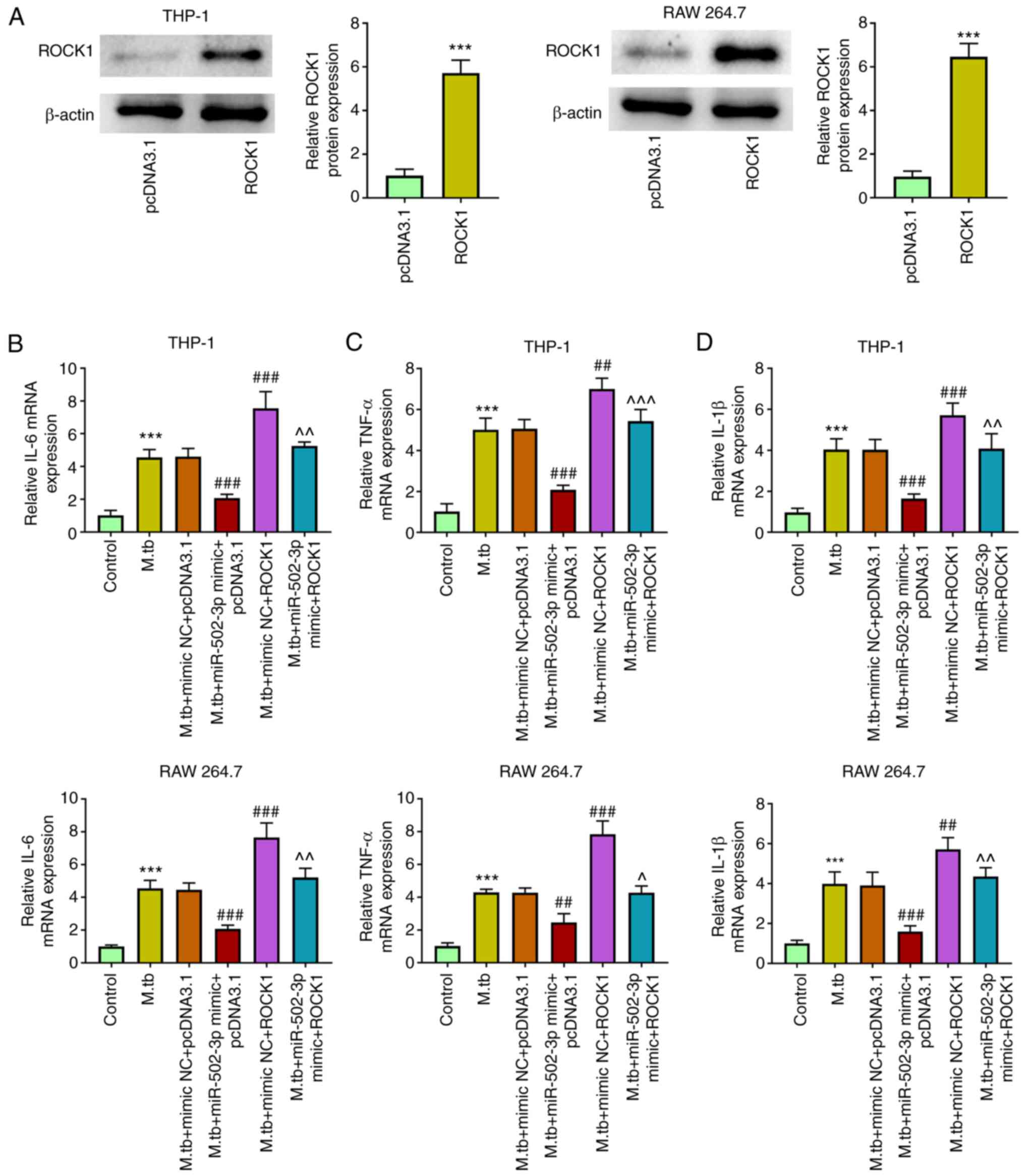
ROCK1 overexpression reverses the
miR-502-3p inhibitory effect on cytokine production in M.
tuberculosis-infected macrophages
The effect of ROCK1 overexpression on cytokine
production in miR-502-3p-overexpressing macrophages was
investigated. ROCK1 overexpression was successfully achieved by
transfecting THP-1 and RAW 264.7 cells with pcDNA3.1-ROCK1
(Fig. 7A). IL-6, TNF-α and IL-1β
mRNA expression levels were increased in M.
tuberculosis-infected THP-1 and RAW 264.7 cells. The miR-502-3p
mimic increased and overexpression of ROCK1 decreased IL-6, TNF-α
and IL-1β mRNA expression in M. tuberculosis-infected THP-1
and RAW 264.7 cells. Overexpression of ROCK1 partially markedly
reversed the inhibitory effects of miR-502-3p mimic on IL-6, TNF-α
and IL-1β levels in M. tuberculosis-infected THP-1 and RAW
264.7 cells compared with the M. tuberculosis + miR-502-3p
mimic group (Fig. 7B-D).
Discussion
miRNAs have been reported to serve important roles
in the host response to intracellular M. tuberculosis
(18–20). For example, increased miR-20a-3p
expression was shown to regulate the host immune response to
promote the growth of M. tuberculosis in human macrophages
(21). In the present study,
miR-502-3p expression was significantly upregulated in M.
tuberculosis-infected THP-1 and RAW 264.7 cells. Furthermore,
miR-502-3p overexpression significantly reduced the mRNA expression
levels of inflammatory cytokines and the activation of NF-κB
signaling through the targeting of ROCK1. miR-502-3p also serves a
role in carcinomatosis, is associated with inflammation and its
expression is significantly decreased following the buildup phase
of venom immunotherapy (22–24).
However, the role of miR-502-3p in the modulation of immune
responses has not been extensively investigated.
The production of inflammatory cytokines by
macrophages is regarded as a bactericidal pathway for M.
tuberculosis infection (25).
However, M. tuberculosis is able to resist these
antimicrobial activities by establishing a niche for survival in
macrophages (26). Previous studies
have determined that miRNAs inhibit the expression of
pro-inflammatory cytokines to generate niches favorable for
mycobacterial survival (9,27,28).
In the present study, it was considered that the detection of mRNA
changes was more direct and convenient to reflect the level of
inflammatory factors. The current study demonstrated that
miR-502-3p overexpression significantly reduced pro-inflammatory
cytokine mRNA expression levels and promoted mycobacterial
intracellular survival.
Furthermore, previous studies have demonstrated that
the association between miRNAs and mRNAs serves an important role
during M. tuberculosis infection. For example, miR-21-5p
enhances mycobacterial survival and weakens inflammatory responses
by targeting Bcl-2 and TLR4 (29).
miR-144 regulates inflammatory cytokine secretion and ERK signaling
by targeting tumor progression locus 2 in M.
tuberculosis-infected macrophages (30). In the present study, miR-502-3p
overexpression promoted mycobacterial survival in macrophages by
directly targeting ROCK1 to evade the immune response via the
ROCK1/TLR4/NF-κB pathway, suggesting that miR-502-3p may have a
similar function as miR-21-5 during M. tuberculosis
infection. Inhibition of ROCK1 alleviates
lipopolysaccharide-induced inflammatory cytokine production and
suppresses TLR4/NF-κB and ERK signaling pathways in corneal
epithelial cells (31).
Furthermore, it has been demonstrated that an M.
tuberculosis nucleoside diphosphate kinase stimulates GTPase
activity of RhoA, RacI and cell division control protein 42
(32). Therefore, the RhoA/ROCK
signaling pathway may be important for M.
tuberculosis-induced inflammatory responses. The regulatory
function of miRNAs through TLRs and the NF-κB signaling pathway in
M. tuberculosis infection has also recently been studied.
For example, miR-148a inhibits TLR4-mediated NF-κB activation in
response to mycobacterial infection (33). The present study demonstrated that
miR-502-3p regulated the host immune response to M.
tuberculosis by directly targeting ROCK1 to possibly attenuate
TLR4-mediated NF-κB signaling pathway activation and consequent
immune responses.
In conclusion, miR-502-3p promoted M.
tuberculosis survival in macrophages and reduced
pro-inflammatory cytokine release in M.
tuberculosis-infected macrophages by targeting the
ROCK1/TLR4/NF-κB pathway. The expression levels of inflammatory
cytokines were determined only via RT-qPCR, and the levels of these
inflammatory factors were not assessed using ELISA. These results
have provided the foundations for the future development of TB
therapeutics.
Acknowledgements
Not applicable.
Funding
No funding was received.
Availability of data and materials
The datasets used and/or analyzed during the current
study are available from the corresponding author on reasonable
request.
Authors' contributions
FL, YZ and ZD designed the study. YL, HY and PW
performed the research and analyzed the data. FL and ZD confirmed
the authenticity of all the raw data. YZ and PW wrote the paper and
FL and ZD reviewed the article. All authors approved the final
version of the manuscript.
Ethics approval and consent to
participate
Not applicable.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Alzahabi KH, Usmani O, Georgiou TK, Ryan
MP, Robertson BD, Tetley TD and Porter AE: Approaches to treating
tuberculosis by encapsulating metal ions and anti-mycobacterial
drugs utilizing nano- and microparticle technologies. Emerg Top
Life Sci. 4:581–600. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Harding E: WHO global progress report on
tuberculosis elimination. Lancet Respir Med. 8:30418–7. 2020.
View Article : Google Scholar
|
|
3
|
Koch A and Mizrahi V: Mycobacterium
tuberculosis. Trends Microbiol. 26:555–556. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Leopold Wager CM, Arnett E and Schlesinger
LS: Mycobacterium tuberculosis and macrophage nuclear receptors:
What we do and don't know. Tuberculosis (Edinb). Apr 25–2019.(Epub
ahead of print). doi: 10.1016/j.tube.2019.04.016. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Correia de Sousa M, Gjorgjieva M, Dolicka
D, Sobolewski C and Foti M: Deciphering miRNAs' Action through
miRNA Editing. Int J Mol Sci. 20:62492019. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Zhang X, Zhu M and Hu X: Integrated miRNA
and mRNA expression profiling to identify mRNA targets of
dysregulated miRNAs in pulmonary tuberculosis. Epigenomics.
10:1051–1069. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Fu B, Xue W, Zhang H, Zhang R, Feldman K,
Zhao Q, Zhang S, Shi L, Pavani KC, Nian W, et al: MicroRNA-325-3p
facilitates immune escape of Mycobacterium tuberculosis through
targeting LNX1 via NEK6 accumulation to promote anti-apoptotic
STAT3 signaling. mBio. 11:00557–20. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Sahu SK, Kumar M, Chakraborty S, Banerjee
SK, Kumar R, Gupta P, Jana K, Gupta UD, Ghosh Z, Kundu M and Basu
J: MicroRNA 26a (miR-26a)/KLF4 and CREB-C/EBPβ regulate innate
immune signaling, the polarization of macrophages and the
trafficking of Mycobacterium tuberculosis to lysosomes during
infection. PLoS Pathog. 13:e10064102017. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Niu W, Sun B, Li M, Cui J, Huang J and
Zhang L: TLR-4/microRNA-125a/NF-κB signaling modulates the immune
response to Mycobacterium tuberculosis infection. Cell Cycle.
17:1931–1945. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Chen LY, Zuraw BL, Liu FT, Huang S and Pan
ZK: IL-1 receptor-associated kinase and low molecular weight GTPase
RhoA signal molecules are required for bacterial
lipopolysaccharide-induced cytokine gene transcription. J Immunol.
169:3934–3939. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Shimizu S, Tahara M, Ogata S, Hashimoto K,
Morishige K, Tasaka K and Murata Y: Involvement of nuclear
factor-kB activation through RhoA/Rho-kinase pathway in LPS-induced
IL-8 production in human cervical stromal cells. Mol Hum Reprod.
13:181–187. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Wang ZH, Zhu D, Xie S, Deng Y, Pan Y, Ren
J and Liu HG: Inhibition of Rho-kinase attenuates left ventricular
remodeling caused by chronic intermittent hypoxia in rats via
suppressing myocardial inflammation and apoptosis. J Cardiovasc
Pharmacol. 70:102–109. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Juarez E, Nuñez C, Sada E, Ellner JJ,
Schwander SK and Torres M: Differential expression of Toll-like
receptors on human alveolar macrophages and autologous peripheral
monocytes. Respir Res. 11:1465–9921. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Guo H, Zhang Y, Cheng BC, Fu X, Zhu P,
Chen J, Chan Y, Yin C, Wang Y, Hossen M, et al: An ethanolic
extract of the aerial part of Siegesbeckia orientalis L. inhibits
the production of inflammatory mediators regulated by AP-1, NF-κB
and IRF3 in LPS-stimulated RAW 264.7 cells. Biosci Trends.
12:330–337. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Sierra-Mondragon E, Molina-Jijon E,
Namorado-Tonix C, Rodríguez-Muñoz R, Pedraza-Chaverri J and Reyes
JL: All-trans retinoic acid ameliorates inflammatory response
mediated by TLR4/NF-κB during initiation of diabetic nephropathy. J
Nutr Biochem. 60:47–60. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(-Delta Delta C(T)) method. Methods. 25:402–408. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Korbel DS, Schneider BE and Schaible UE:
Innate immunity in tuberculosis: Myths and truth. Microbes Infect.
10:995–1004. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Sharma N, Verma R, Kumawat KL, Basu A and
Singh SK: miR-146a suppresses cellular immune response during
Japanese encephalitis virus JaOArS982 strain infection in human
microglial cells. J Neuroinflammation. 12:302015. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Pachathundikandi SK and Backert S:
Helicobacter pylori controls NLRP3 expression by regulating
hsa-miR-223-3p and IL-10 in cultured and primary human immune
cells. Innate Immun. 24:11–23. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Yu T, Ju Z, Luo M, Hu R, Teng Y, Xie L,
Zhong C, Chen L, Hou W, Xiong Y and Feng Y: Elevated expression of
miR-146a correlates with high levels of immune cell exhaustion
markers and suppresses cellular immune function in chronic
HIV-1-infected patients. Sci Rep. 9:188292019. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Cui J, Li Z, Cui K, Gao Y, Zhang B, Niu J
and Wang Y: MicroRNA-20a-3p regulates the host immune response to
facilitate the Mycobacterium tuberculosis infection by targeting
IKKβ/NF-κB pathway. Int Immunopharmacol. 91:1072862021. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Jin H, Yu M, Lin Y, Hou B, Wu Z, Li Z and
Sun J: MiR-502-3P suppresses cell proliferation, migration, and
invasion in hepatocellular carcinoma by targeting SET. Onco Targets
Ther. 9:3281–3289. 2016.PubMed/NCBI
|
|
23
|
Zhang J, Hou L, Liang R, Chen X, Zhang R,
Chen W and Zhu J: CircDLST promotes the tumorigenesis and
metastasis of gastric cancer by sponging miR-502-5p and activating
the NRAS/MEK1/ERK1/2 signaling. Mol Cancer. 18:802019. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Specjalski K, Maciejewska A, Pawłowski R,
Chełmińska M and Jassem E: Changes in the expression of microRNA in
the buildup phase of wasp venom immunotherapy: A pilot study. Int
Arch Allergy Immunol. 170:97–100. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Lv J, He X, Wang H, Wang Z, Kelly GT, Wang
X, Chen Y, Wang T and Qian Z: TLR4-NOX2 axis regulates the
phagocytosis and killing of Mycobacterium tuberculosis by
macrophages. BMC Pulm Med. 17:1942017. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Shi L, Jiang Q, Bushkin Y, Subbian S and
Tyagi S: Biphasic dynamics of macrophage immunometabolism during
Mycobacterium tuberculosis infection. mBio. 10:e02550–18. 2019.
View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Li X, Huang S, Yu T, Liang G, Liu H, Pu D
and Peng N: MIR-140 modulates the inflammatory responses of
Mycobacterium tuberculosis-infected macrophages by targeting TRAF6.
J Cell Mol Med. 23:5642–5653. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Gu X, Gao Y, Mu DG and Fu EQ: MiR-23a-5p
modulates mycobacterial survival and autophagy during Mycobacterium
tuberculosis infection through TLR2/MyD88/NF-κB pathway by
targeting TLR2. Exp Cell Res. 354:71–77. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Zhao Z, Hao J, Li X, Chen Y and Qi X:
MiR-21-5p regulates mycobacterial survival and inflammatory
responses by targeting Bcl-2 and TLR4 in Mycobacterium
tuberculosis-infected macrophages. FEBS Lett. 593:1326–1335. 2019.
View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Liu HY: Down-regulation of miR-144 after
Mycobacterium tuberculosis infection promotes inflammatory factor
secretion from macrophages through the Tpl2/ERK pathway. Cell Mol
Biol. 62:87–93. 2016.PubMed/NCBI
|
|
31
|
Gong J, Guan L, Tian P, Li C and Zhang Y:
Rho kinase type 1 (ROCK1) promotes lipopolysaccharide-induced
inflammation in corneal epithelial cells by activating toll-like
receptor 4 (TLR4)-mediated signaling. Med Sci Monit. 24:3514–3523.
2018. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Chopra P, Koduri H, Singh R, Koul A,
Ghildiyal M, Sharma K, Tyagi AK and Singh Y: Nucleoside diphosphate
kinase of Mycobacterium tuberculosis acts as GTPase-activating
protein for Rho-GTPases. FEBS Lett. 571:212–216. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Wu H, Bao Y, Wang L, Li X and Sun J:
Mycobacterium marinum down-regulates miR-148a in macrophages in an
EsxA-dependent manner. Int Immunopharmacol. 73:41–48. 2019.
View Article : Google Scholar : PubMed/NCBI
|