Introduction
Pancreatic cancer (PC) is a lethal disease with
increasing incidence that causes an estimate number of 227,000
deaths worldwide each year (1). It
is listed as the 14th most common cancer and the seventh major
cause of cancer death in the world (2). The incidence of PC in the developed
world is increasing and its 5-year survival rate is as low as 2% in
some nations despite improved surgical techniques, chemotherapy
regimens and the introduction of neoadjuvant chemoradiotherapy
(3,4). Surgical resection is the mains
curative treatment for PC in its early stages and most operations
have a low mortality rate (5).
However, due to the fact that PC can still metastasize following
surgery, metastasis becomes the leading cause of death in patients
with PC (6). It is hypothesized
that epithelial to mesenchymal transition (EMT) contributes to the
early-stage spread of cancer cells and is essential for PC invasion
and metastasis (7,8). Hence, identifying novel biomarkers for
PC diagnosis and prognosis is of urgent necessity.
Epithelial sodium channel (ENaC) is an
amido-sensitive epithelial sodium channel localized to the
epithelial apical portion of the distal kidney, distal colon, lung
and exocrine gland ducts (9). The
channel is composed of a heteromeric complex of three homologous
subunits α, β and γ, encoded by the sodium channel epithelial 1α
subunit (SCNN1A), SCNN1B and SCNN1G genes respectively (10). There has been evidence that ENaC
channels have key roles in cancer cell biology, such as
proliferation, migration, invasion and apoptosis and serve a role
in tumor development and progression (11,12).
Among them, SCNN1A has been relatively well studied in cancer.
SCNN1A has been shown to induce osteosarcoma tumor growth in
vitro and in vivo (13).
In addition, it has been demonstrated that SCNN1A may serve a key
role in ovarian cancer cell growth, invasion and migration
(14). Nevertheless, the role and
biological significance of SCNN1A in PC remains to be
elucidated.
Homeobox (HOX) genes are a superfamily of genes
involved in embryonic development and cell differentiation
(15). Homeobox D9 (HOXD9) is
associated with the initiation and evolution of numerous malignant
tumors, such as ovarian, glioma and cervical cancers (16–18).
HOXD9 can promote the growth, invasion and metastasis of gastric
cancer cells by transcriptional activation of RUN and FYVE domain
containing 2 (19). Similarity,
another study noted that HOXD9 regulates cervical cancer metastasis
through transcriptional activation of its downstream gene HMCN1
(20). HOXD9 is a transcription
factor for SCNN1A as predicted by the PROMO website. However, the
role of HOXD9 in PC has not been reported. Therefore, it was
hypothesized whether SCNN1A could produce an effect in PC by
regulating HOXD9.
The present study aimed to investigate whether the
transcription factor HOXD9 can activate SCNN1A transcription and
thus affect the proliferation, invasion and migration of PC cells
through the regulation of EMT.
Materials and methods
Online database analysis
Prior to the in vitro experiments, the Gene
Expression Profiling Interactive Analysis (GEPIA2, http://gepia.cancer-pku.cn) (21), The Cancer Genome Atlas (TCGA,
http://tcga-data-nci-nih-gov.elib.tcd.ie/tcga/)
(22) and Genotype-Tissue
Expression (GTEx, http://gtexportal.org) databases (23) were used to compare the expression
levels of SCNN1A and HOXD9 in patients with PC.
The GEPIA, TCGA and GTEx databases and Kaplan-Meier
plotter website (http://kmplot.com/analysis/) (24) were next used to investigate the
association between SCNN1A expression and the prognosis of PC.
JASPAR website (http://jaspar.binf.ku.dk/) was consulted to predict
the binding sites of HOXD9 and SCNN1A promoter sequences. PROMO is
freely available for download at http://acgt.cs.tau.ac.il/promo/. The Human Protein
Atlas database (www.proteinatlas.org/pathology) was used to check the
expression of SCNN1A in PC patient tissues via immunohistochemistry
(IHC).
Cell lines and culture conditions
The human PC cell lines (BxPC-3, SW1990, PANC-1,
CFPAC-1), together with the human non-cancerous pancreatic ductal
epithelium cell line HPDE6c7, were purchased from the Type Culture
Collection of the Chinese Academy of Science (Shanghai, China). The
cells were grown in DMEM (HyClone) supplemented with 10% fetal
bovine serum (FBS; Gibco; Thermo Fisher Scientific, Inc.), 100 U/ml
penicillin and 100 mg/ml streptomycin. All cells were then
incubated in a humidified 5% CO2 atmosphere at 37°C and
the medium was replaced every 3 days.
Cell transfection
SW1990 cells (at 1×104 cells/well) were
transferred to a 6-well plate and subsequently transfected with
small interfering (sh)RNAs [shRNA-negative control (NC),
shRNA-SCNN1A and shRNA-HOXD9] and empty pcDNA3.1 vector and
HOXD9-overexpressing vector (Oe-HOXD9), which were purchased from
Shanghai GenePharma Co., Ltd. The SCNN1A target sequence was
5′-CCAGAACAAATCGGACTGCTT−3′. The HOXD9 target sequence was
5′-GGACTCGCTTATAGGCCAT-3′. When the cells reached 60–70%
confluence, they were transfected with 500 ng transfectants using
2.5 µl Lipofectamine® 3000 (Invitrogen; Thermo Fisher
Scientific, Inc.) according to the manufacturer's protocol.
Following incubation at 37°C for 6 h, the serum-free DMEM was
replaced with fresh DMEM containing 10% FBS, and the cells were
incubated for 24 h.
RNA extraction and reverse
transcription-quantitative (RT-q) PCR
Total RNA was extracted from cells using
TRIzol® (Thermo Fisher Scientific, Inc.) following the
manufacturer's instructions. RNA was reverse transcribed into cDNA
using the PrimeScript RT kit (Qiagen GmbH). RT-qPCR was performed
with SYBR Premix Ex Taq (Qiagen GmbH) to quantify the RNA
expression. The following thermocycling conditions were used for
the qPCR: Initial denaturation at 95°C for 6 min; followed by 40
cycles of initiation at 94°C for 30 sec, annellation at 60°C for 30
sec and elongation at 73°C for 90 sec. The 2−ΔΔCq
(25) method was employed to assay
relative expression levels. The primers for SCNN1A were: Forward
5′-AGGACTCTAGCCCTCCACAG-3′, reverse 5′-GTGGATGGTGGTGTTGTTGC-3′. The
primers for HOXD9 were: Forward 5′-ACGTCCCCAAGCCAGGCTC-3′, reverse
5′-CTATGTAGCTCAGGAATAA-3′. The primers for GAPDH were: Forward
5′-CACCATTGGCAATGAGCGGTTC-3′, reverse
5′-AGGTCTTTGCGGATGTCCACGT-3′.
Western blotting
Total protein was extracted from SW1990 cells with
RIPA lysis buffer supplemented with the proteinase inhibitors PMSF
and PI (Beyotime Institute of Biotechnology), the concentration was
determined using a BCA assay protein assay kit (Beyotime Institute
of Biotechnology). Equal amounts (20 µg per lane) of the isolated
protein was separated by 10% SDS-PAGE and transferred to PVDF
membranes (EMD Millipore). The membranes were blocked in 5%
fat-free milk, and then were probed with the respective primary
antibodies: SCNN1A (Abcam; 1:1,000; cat. no. ab272878), E-cadherin
(Santa Cruz Biotechnology, Inc.; 1:1,000; cat. no. sc-8426),
N-cadherin (Santa Cruz Biotechnology, Inc.; 1:1,000; cat. no.
sc-59987), Vimentin (Santa Cruz Biotechnology, Inc.; 1:1,000; cat.
no. sc-6260), Snail (Abcam; 1:1,000; cat. no. ab229701), HOXD9
(Santa Cruz Biotechnology, Inc.; 1:1,000; cat. no. sc-137134) and
GAPDH (Santa Cruz Biotechnology, Inc.; 1:3,000; cat. no. sc-47724)
at 4°C overnight. After washing three times with TBS-0.2% Tween-20,
membranes were probed with a goat anti-rabbit HRP-conjugated
secondary antibody (Cell Signaling Technology, Inc.; 1:2,000; cat.
no. 7074S) or horse anti-mouse HRP-conjugated secondary antibody
(Cell Signaling Technology, Inc.; 1:2,000; cat. no. 7076S;) at room
temperature for 1 h. Proteins were visualized with an ECL detection
system (Applygen Technologies, Inc.) and subsequently
semi-quantified using ImageJ software (version 1.52r, National
Institutes of Health). The protein expression of the bands was
normalized against the gray value of GAPDH.
Cell proliferation assays
To measure the proliferation rate of SW1990 cells,
Cell Counting Kit (CCK)-8 was used and a cell counting assay was
conducted. Briefly, cells were seeded into 6-well plates and
transfected with small RNA oligonucleotides or plasmids. At 24 h
post-transfection, cells were reseeded. After 0, 24, 48 and 72 h of
incubation, the optical density of each well was measured using a
CCK-8 kit (Beyotime Institute of Biotechnology; cat. no. C0038)
according to the manufacturers' instructions and the number of
cells was counted after incubation 1 h using a Celigo image
cytometer (Nexcelom Bioscience LLC) at 450 nm. For cell colony
assay, cells were seeded into 6-well plates at 500 cells per well.
After 2 weeks, the cells were fixed with 75% ethanol at 37°C for 30
min and then stained with 0.1% crystalline violet at 37°C for 15
min. Finally, the cells were imaged and counted.
Wound-healing assay
The cells were cultured in 6-well plates and allowed
to form cell monolayers overnight to full confluence. Then, a 20-µl
sterile plastic tip was used to scratch a wound line across the
surface of plates. The suspended cells were removed with PBS, while
the rest were incubated with serum-free medium. Images were
captured at 0 and 24 h after scratching using a 600 Autobiochemical
Analyzer (Olympus Corporation), and ImageJ software (version 1.52r,
National Institutes of Health) was used to calculate the average
distance between cells. The width of wound healing was quantified
and compared with baseline values.
Transwell invasion assay
Cells were injected (5×105 cells/well)
into the upper chambers of Transwell chambers (8 µm pore size;
Corning, Inc.) containing serum-free DMEM, while the lower chambers
were supplemented with DMEM containing 10% FBS. The upper surface
of the bottom membrane of Transwell chambers was covered with a
dilution of Matrigel (BD Biosciences) at a ratio of 1:8 overnight
at 37°C with 5% CO2. The chambers were incubated at 37°C
for 30 min. Next, cells remaining in the upper chamber were gently
removed with a cotton swab, while invading cells were fixed in 4%
paraformaldehyde for 15 min at room temperature and stained with
0.1% crystal violet for 20 min at 37°C. Invading SW1990 cells were
counted using an inverted light microscope (Eclipse Ti2; Nikon
Corporation) for cell counts and images were captured, and five
areas were randomly selected for counting.
Luciferase assays
Luciferase activity in SW1990 cells was detected by
interfering with HOXD9 and two SCNN1A promoter deletion mutants,
respectively. The canonical HOXD9 binding motif and PVT1 promoter
were constructed into the pGL3 plasmid (Promega Corporation),
respectively. Cells were transfected with the pGL3-based reporter
construct, and pRL-SV40 was the internal control plasmid. OPTI-MEM
(49 µl) was pipetted onto 24-well plates to dilute 1 µl
Lipofectamine® 2000 reagent (Invitrogen; Thermo Fisher
Scientific, Inc.), and the final volume was 50 µl. After 36 h of
transfection, the cells were lysed and assayed via a
dual-luciferase reporter assay kit (Promega Corporation) and
normalized to that of Renilla luciferase activity.
Chromatin immunoprecipitation (ChIP)
assay
The ChIP assay was performed with a commercial kit
(Beyotime Institute of Biotechnology; cat. no. P2078) according to
the manufacturer's instructions. Briefly, cells were cross-linked
using 1% formaldehyde (Sigma-Aldrich; Merck KGaA) in PBS at 25°C
for 10 min. Termination of cross-linking was achieved by the
addition of glycine (Beijing Solarbio Technology Co., Ltd.) to a
final concentration of 125 µM. Then, 1×106 cells were
collected via centrifugation at 300 × g for 3 min at 25°C and
washed twice with pre-chilled PBS. The cells (1×106)
were treated with 2 µg anti-HOXD9 (Santa Cruz Biotechnology, Inc.;
cat. no. sc-137134), anti-SCNN1A (Abcam; cat. no. ab272878) or
anti-mouse IgG (Beyotime Institute of Biotechnology; cat. no.
A7028) antibodies, which were immunoprecipitated against
cross-linked 100 µg DNA (using a spectrophotometer at 260
nm)/protein and incubated at 4°C for 2 h. A ChIP DNA purification
kit (Beyotime Institute of Biotechnology; cat. no. D0033) was used
to purify the immunoprecipitated DNA and it was amplified via qPCR
as described above. RT-qPCR was applied to the analysis of the
enriched DNA.
Statistical analysis
The association between the pathological stage and
SCNN1A expression as well as the relative levels of SCNN1A and
HOXD9 in the tissues of patients with PC was analyzed by one-way
ANOVA. The Kaplan-Meier test was performed to identify overall
survival and disease-free survival-related clinical characteristics
of patients with PC. Data are shown as mean ± standard deviation.
All experiments were performed at least in triplicate. Unpaired
Student's t-test was used to analyze differences between two
groups. The differences among the multiple groups were analyzed
using one-way ANOVA followed by a Tukey's post hoc test. P<0.05
was considered to indicate a statistically significant
difference.
Results
Highly expressed SCNN1A is found in PC
tissues and is associated with poor prognosis
The GEPIA, TCGA and GTEx databases were used to
analyze the expression level of SCNN1A in patients with PC. The
results showed a high expression of SCNN1A in the tissues of
patients with PC (Fig. 1A). This
result was confirmed via IHC, which demonstrated more
SCNN1A-positive cells in tumor sections of patients with PC
compared with the normal tissues from the Human Protein Atlas
database (Fig. 1B). The expression
of SCNN1A was found to be significantly associated with the
pathological stage of tumors in patients with PC (Fig. 1C). Similarly, the combined results
of GEPIA, TCGA and GTEx databases revealed that patients with PC
with high SCNN1A expression had a low overall survival rate and
disease-free survival rate compared with those with low SCNN1A
expression (Fig. 1D). These results
implied that SCNN1A served a crucial role in PC progression.
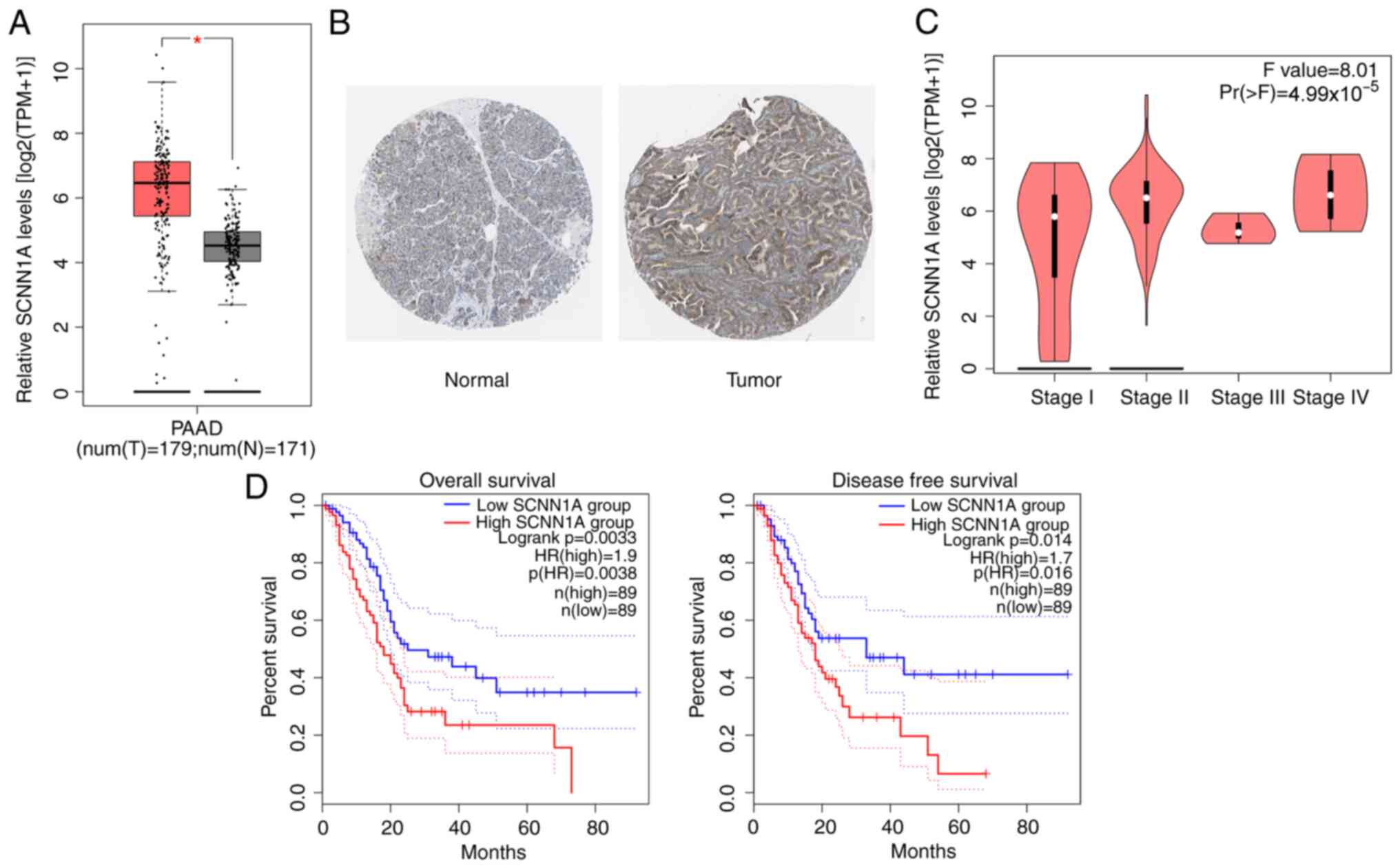 | Figure 1.SCNN1A expression is upregulated in
PC tissues and associated with prognosis. (A) SCNN1A expression in
different disease state (tumor or normal) based on Gene Expression
Profiling Interactive Analysis, The Cancer Genome Atlas and
Genotype-Tissue Expression databases. Left, num(T)=1,798; Right,
num(N)=171. (B) SCNN1A expression in PC tumor sections by
immunohistochemical staining from the Human Protein Atlas database
(magnification, ×20). (C) Differential expression of SCNN1A in
different pathological stage. (D) Kaplan-Meier plotter database was
used for overall survival and disease-free survival analysis.
Dashed lines represents 95% confidence intervals. *P<0.5 vs. N.
SCNN1A, sodium channel epithelial 1α subunit; PC, pancreatic
cancer; T, tumor tissues; N, normal tissues; PAAD, pancreatic
adenocarcinoma. |
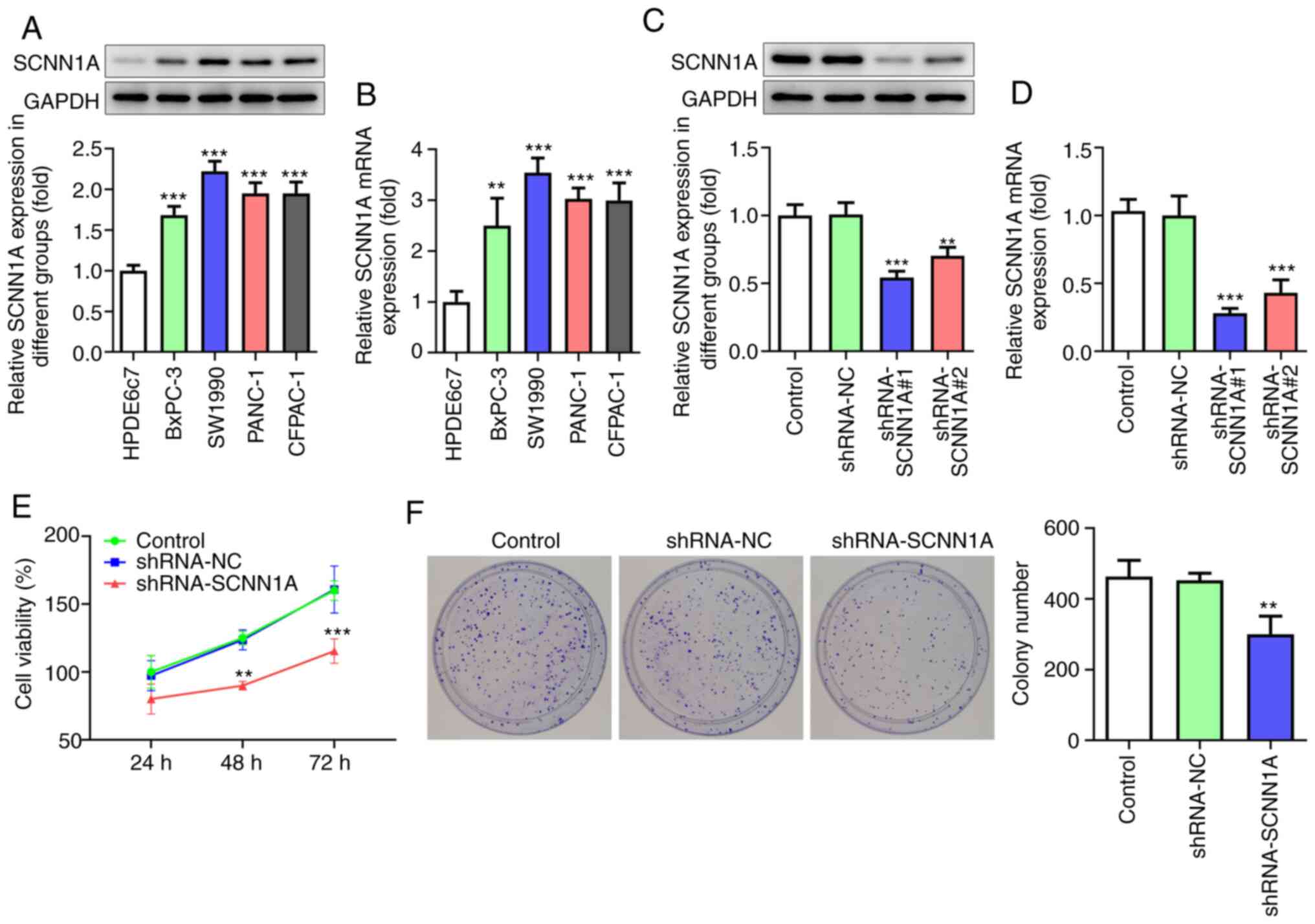
SCNN1A promotes PC cell proliferation,
invasion, migration and EMT
To investigate the role of SCNN1A, qPCR and western
blotting were applied to test the mRNA and protein level of SCNN1A
in PC cell lines (BxPC-3, SW1990, PANC-1 and CFPAC-1) and human
non-cancerous pancreatic ductal epithelium cell line HPDE6c7. The
expression level of SCNN1A was found to be significantly higher in
all cancer cell lines compared with that in HPDE6c7 cells (Fig. 2A and B). As the expression level of
SCNN1A in SW1990 was the highest among all four cancer cell lines,
SW1990 cells were selected for the following experiments. SCNN1A
shRNA was transfected into SW1990 cells to inhibit SCNN1A
expression. It was detected by qPCR and western blotting that
shRNA-SCNN1A#1 had improved efficacy compared with shRNA-SCNN1A#2
(Fig. 2C and D) when it came to
SCNN1A knockdown. Thus, shRNA-SCNN1A#1 was chosen for the following
experiments.
To investigate the effect of SCNN1A on PC cell
proliferation, CCK8 and colony formation assays were performed.
According to the results obtained from the CCK8 assay, the
shRNA-SCNN1A group showed markedly lower optical density value at
48 and 72 h compared with the Control and shRNA-NC group (Fig. 2E). Fig.
2F shows that following knocking down SCNN1A, the number of
colonies was reduced relative to that in the Control and shRNA-NC
groups. These results prove that SCNN1A suppression inhibits SW1990
cell proliferation.
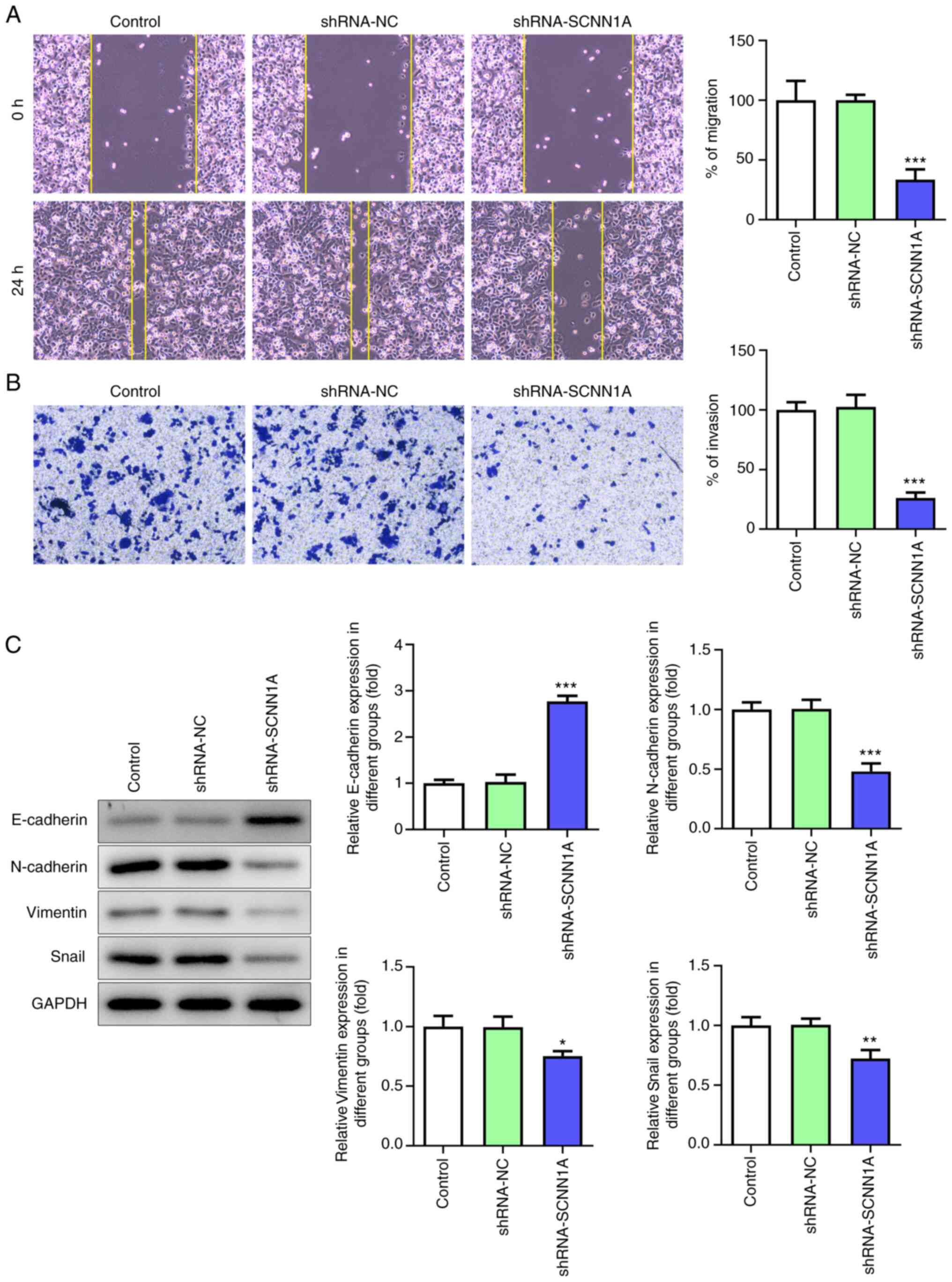
Wound healing assay and Transwell assay were used to
determine the function of SCNN1A in the PC cell migration. After
SCNN1A knockdown, SW1990 cell migrated much slower than the Control
and shRNA-NC group (Fig. 3A). In
the Transwell assay, the number of migrant cells in the SCNN1A
silencing group was markedly fewer than the control group (Fig. 3B). These results indicated that
SCNN1A promotes SW1990 cell migration.
To further explore whether SCNN1A regulates EMT in
PC cells, the expression levels of EMT-related cell markers were
tested via western blotting. As shown in Fig. 3C, knockdown of SCNN1A notably
decreased the expression levels of N-cadherin, Vimentin and Snail
but increased the expression level of E-cadherin. This suggested
that SCNN1A triggers EMT. Overall, these results demonstrated that
SCNN1A promotes PC cell proliferation, invasion, migration and
EMT.
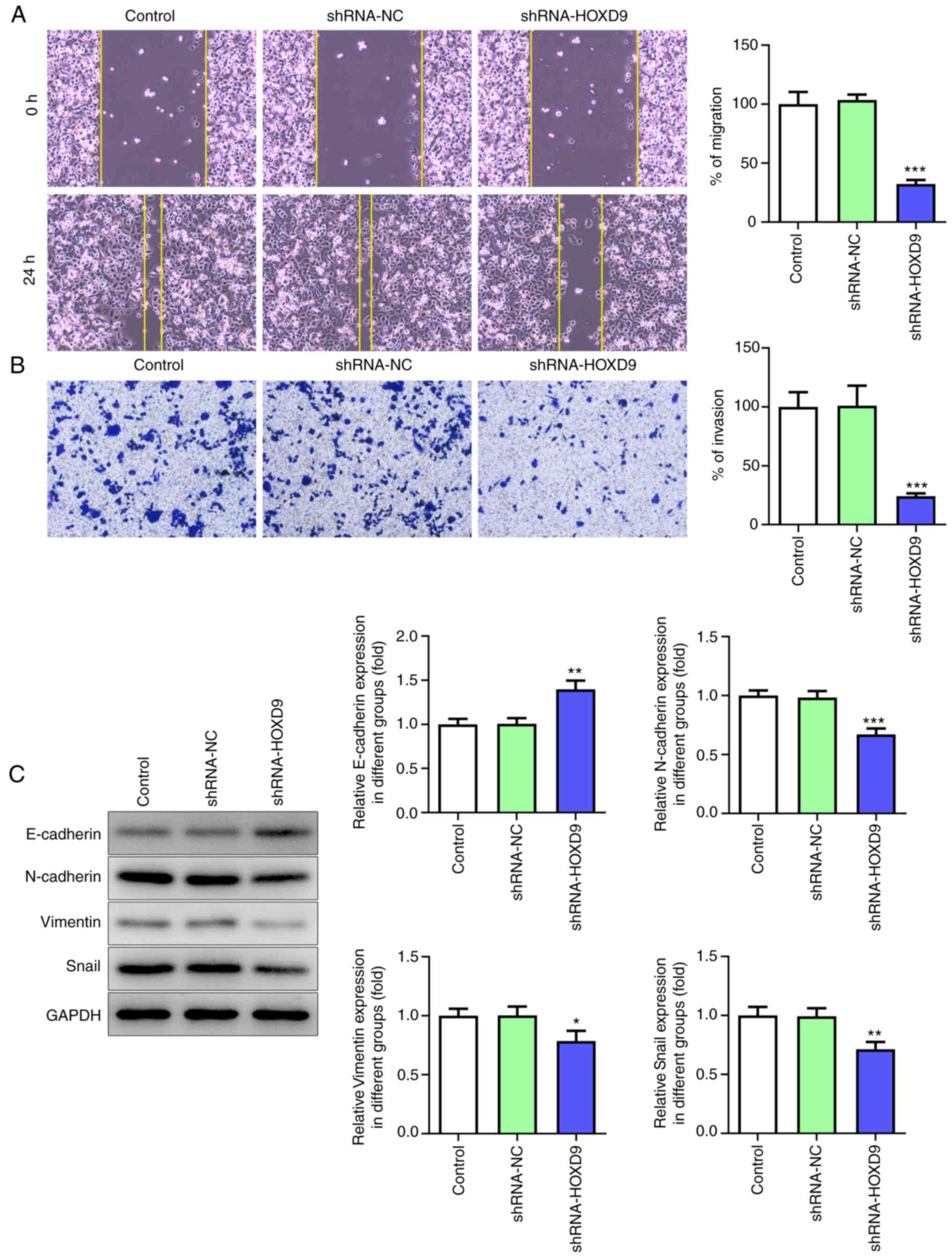
HOXD9 promotes PC cell proliferation,
invasion, migration and EMT
HOXD9 was predicted to be highly expressed in PC
tissues by the GEPIA, TCGA and GTEx databases (Fig. 4A). Furthermore, this prediction was
confirmed via qPCR and western blotting in PC cell lines (Fig. 4B and C). To investigate the effect
of HOXD9 on PC, HOXD9 shRNA and overexpression plasmids Oe-HOXD9
were transfected into SW1990 cells. Fig. 4D and E shows that shRNA-HOXD9#2 was
superior to shRNA-HOXD9#1 at reducing the expression level of HOXD9
and that Oe-HOXD9 up-regulated the expression of HOXD9 effectively.
Therefore, shRNA-HOXD9#2 and Oe-HOXD9 were selected for the
subsequent experiments. The CCK8 results revealed that
downregulated HOXD9 reduced cell viability ratio at 48 and 72 h
significantly (Fig. 4F). Similarly,
the colony formation assay showed that HOXD9 knockdown suppressed
PC cell proliferation (Fig. 4G).
Additionally, HOXD9 silencing was validated to suppress PC cell
migration and invasion (Fig. 5A and
B). The protein expression of E-cadherin, N-cadherin, Vimentin
and Snail was detected to explore the role of HOXD9 in EMT
(Fig. 5C), which supported the
hypothesis that HOXD9 promoted EMT in PC cells.
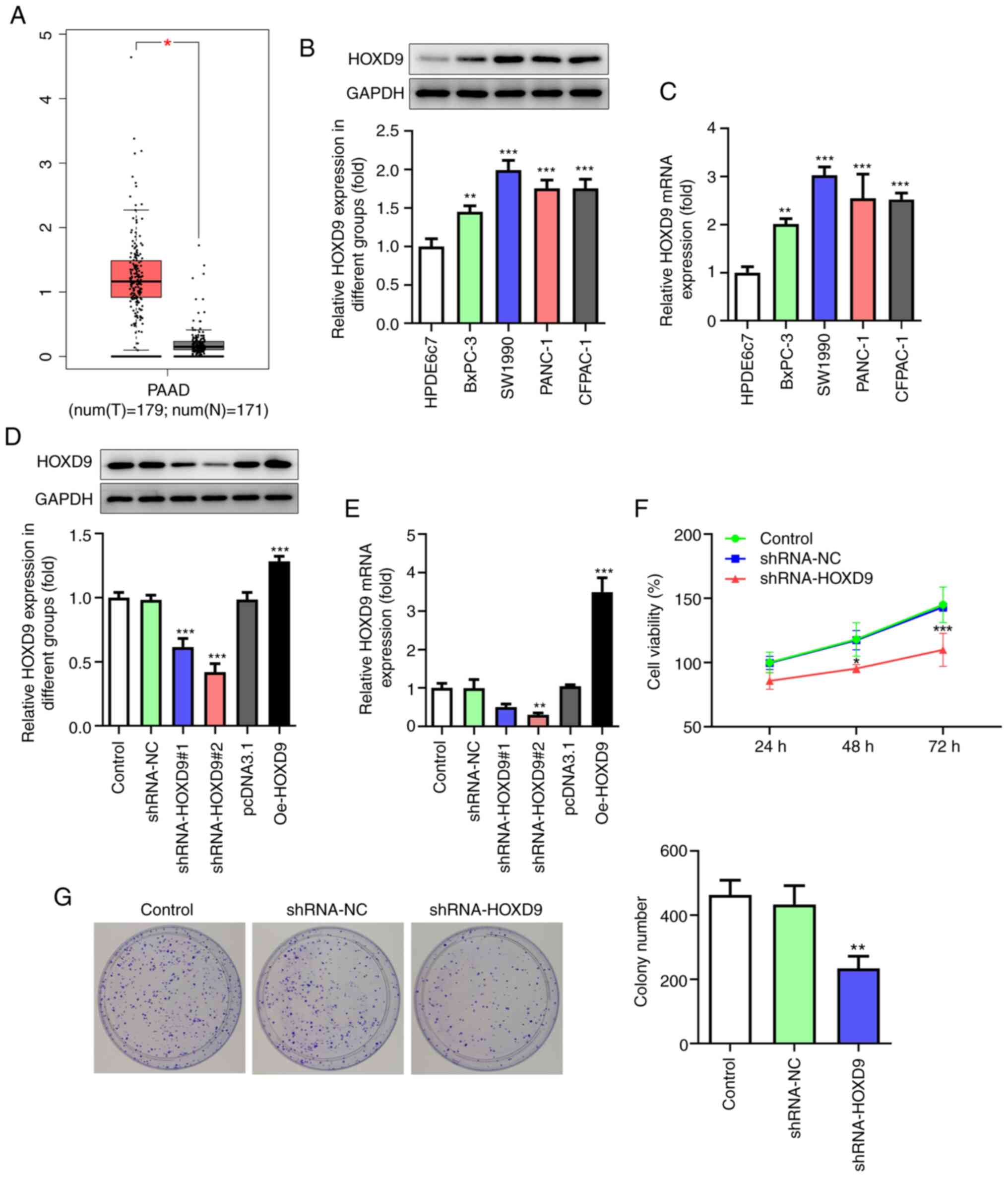 | Figure 4.HOXD9 is highly expressed in PC
tissues and cell lines and promotes PC cell proliferation. (A)
HOXD9 expression in PC tissues based on Gene Expression Profiling
Interactive Analysis, The Cancer Genome Atlas and Genotype-Tissue
Expression databases. (B and C) HOXD9 expression in PC cell lines.
(D and E) HOXD9 expression following transfection with shRNA and
overexpression plasmids. (F) CCK8 assay. (G) Colony formation assay
(magnification, ×100). *P<0.05, **P<0.01, ***P<0.001 vs.
the control. HOXD9, homeobox D9; PC, pancreatic cancer; shRNA,
short hairpin RNA; PAAD, pancreatic adenocarcinoma; T, tumor
tissues; N, normal tissues. |
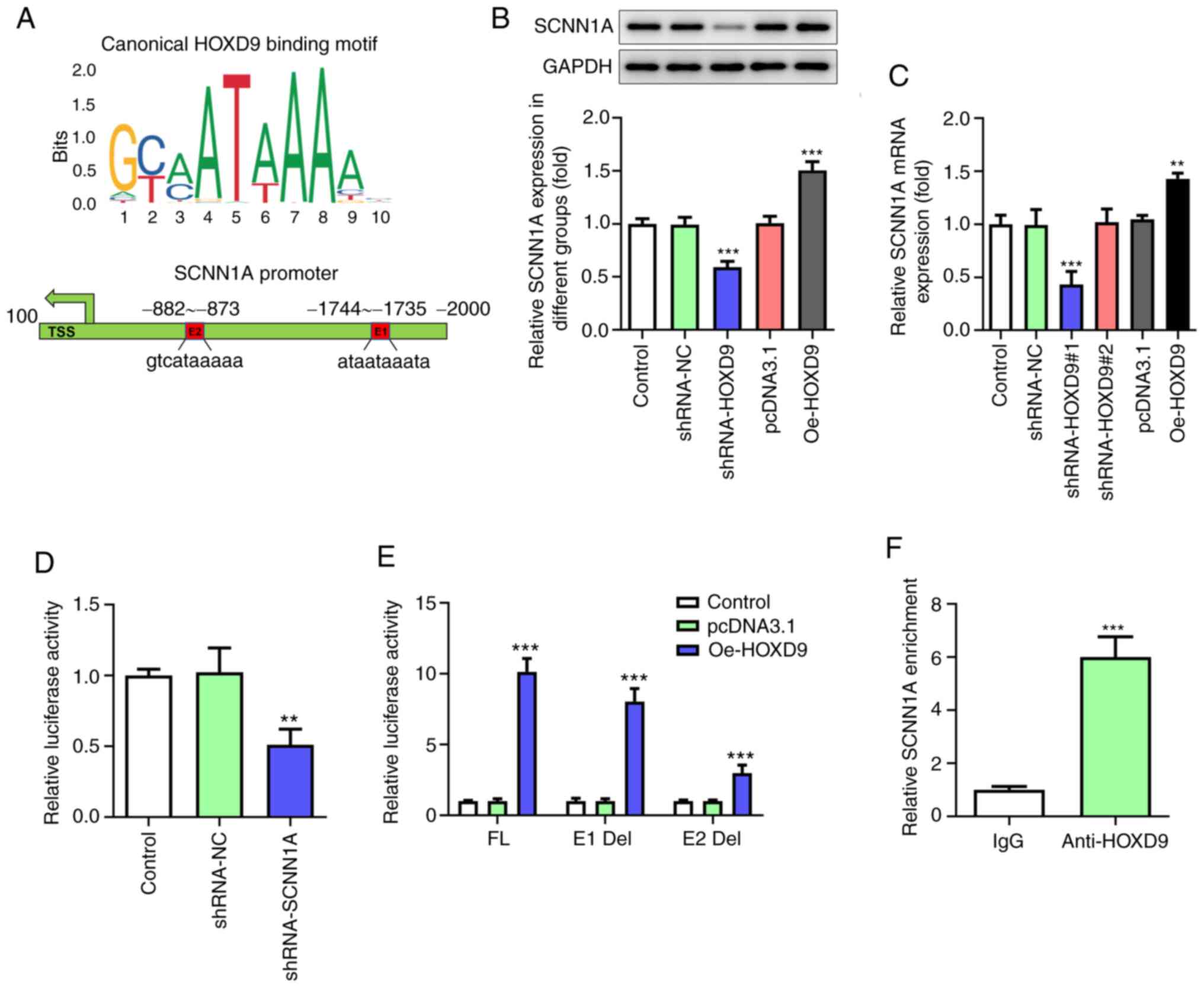
HOXD9 activates SCNN1A transcription
to form a feedback regulatory loop
The JASPAR database was consulted to predict the
binding of HOXD9 to SCNN1A promotor and two putative HOXD9-binding
elements within the SCNN1A promoter region were discovered through
bioinformatics analysis (Fig. 6A).
Given the vital oncogenic roles of SCNN1A, whether HOXD9 could
regulate SCNN1A expression in turn was further investigated. qPCR
and western blotting assays proved that HOXD9 silencing decreased
the expression of SCNN1A, whereas its overexpression had an
opposite effect in SW1990 cells (Fig.
6B and C). This indicated that HOXD9 was positively associated
with SCNN1A transcription. Luciferase assay confirmed that the
SCNN1A promoter was significantly transactivated by HOXD9 (Fig. 6D). Next, to clarify which element
was necessary for HOXD9-mediated SCNN1A expression, the two
predicted HOXD9-binding sites were individually deleted, named E1
Del and E2 Del. Of note, both E1 and E2 absence was found to
notably downregulate the transcriptional activity of SCNN1A, with
E2 having a more pronounced effect (Fig. 6E). To validate this result, ChIP
assay was performed with HOXD9 antibody, followed by PCR detection
with a specific primer for the E2 element. As shown in Fig. 6F, HOXD9 could associate with SCNN1A
promoter, as SCNN1A was enriched within the E2 region (−882-873
bp). These findings suggest that there is a regulatory feedback
loop between HOXD9 and SCNN1A, which may continuously activate
their oncogenic functions.
Discussion
PC is currently one of the most lethal malignancies,
with a low 5-year survival rate (26). Therefore, it is important to study
the causative factors of its rapid proliferation and aggressive
metastasis for early diagnosis and good prognosis of PC and for the
development of effective therapeutic agents. The focus of recent
research into PC has been on searching for powerful prognostic
markers, such as PRMT5, STC2 and Asporin, which are expected to be
valuable to cancer classification aimed at adopting appropriate
therapies and to an accurate forecast of the survival outcomes
(27–29). The present study confirmed the
oncogenic effect of SCNN1A and HOXD9 and validated their prognostic
value in predicting PC. Through in vitro assays, it was
observed that HOXD9 and SCNN1A were highly expressed in PC tissues
and cells. Knockdown of HOXD9 and SCNN1A could inhibit cell
proliferation, migration and EMT. Furthermore, the expression of
HOXD9 and SCNN1A was verified to be positively associated in PC
cells. The combination of high levels of HOXD9 and SCNN1A signified
a poorer prognosis.
SCNN1A has been reported to be involved in a variety
of diseases, such as ovarian cancer, Lam's disease and
pseudoaldosteronism type 1 (30,31).
Its implication in PC is not yet confirmed. SCNN1A has been
validated as exhibiting extremely high sensitivity and specificity
in melanoma detection (32). A
study showed that miR-95 regulates the cell cycle and apoptosis of
osteosarcoma cells by targeting SCNN1A (13). In the present study, the expression
of SCNN1A was first predicted by the databases and was found to be
significantly high in tissue samples from patients with PC and
negatively associated with the prognosis. The role of SCNN1A in PC
was examined through an in vitro model. Experiments have
proved that SCNN1A silence has an inhibitory effect on ovarian
cancer cell growth, migration and invasion (14). The results of the present study
yielded the same conclusion that silencing SCNN1A can inhibit PC
cell proliferation, migration and EMT. Metastasis is the main
feature of cancer and the leading cause of death in ~90% of cancer
patients (33). Upregulation of
EMT-related transcription factors can suppress tumor cell apoptosis
and promote cell lineage tumor invasiveness, leading to
angiogenesis, development of chemotherapy resistance and poor
prognosis for clinical tumor patients. Thus, SCNN1A may promote PC
cell proliferation and migration and may serve as a predictive
marker by regulating EMT.
HOXD9 is a transcription factor of SCNN1A according
to database prediction. HOXD9 also serves a central role in the
development of cancer by regulating the expression of multiple
genes and promoting cell proliferation, migration and invasion. It
has proven to promote the growth, invasion and metastasis of
gastric cancer cells (19). Ectopic
expression of HOXD9 has been shown to promote the invasive
metastasis of colorectal tumor cells (34). The role of HOXD9 in PC was also
explored in the present study. The results indicated that HOXD9 was
overexpressed in PC and that HOXD9 silence inhibited PC cell
proliferation, migration and EMT.
HOXD9 can interact with the promoter region of Zinc
Finger E-Box Binding Homeobox 1 (ZEB1) and promotes ZEB1 expression
(35). HOXD9 promotes the
expression of HPC16 E6 and E7 through direct binding to the P97
promoter, thereby enhancing the proliferation, migration and
metastasis of cervical cancer cells (18). In the present study, the interaction
between HOXD9 and SCNN1A was revealed. Activated HOXD9 stimulated
SCNN1A expression by directly occupying the −882–873 bp motif of
the SCNN1A promoter region. These pieces of evidence supported that
SCNN1A can form a positive feedback loop with its binding partner
HOXD9, thus continuously enhancing its oncogenic effect and
aggravating cancer progression.
The present study also has some limitations. For
example, 3-D growth assay would be superior in detecting EMT, which
we failed to demonstrate well due to the limited conditions.
In conclusion, the present study elucidated that
HOXD9 induced SCNN1A upregulation and promoted PC cell
proliferation and migration by regulating EMT, potentially serving
as a promising prognostic marker. These results propose a possible
role of SCNN1A as a prognostic factor and a candidate therapeutic
target for PC. This may provide a research base for early detection
and clinical management of PC. However, the present in vitro
study does venture conclusions without in vivo data.
Therefore, the results will be further validated with animal models
as well as clinical samples in future experimental studies.
Acknowledgements
Not applicable.
Funding
This work was supported by the Science and
Technology Research Project of Jiangxi Education Department (grant
no. 191405).
Availability of data and materials
The datasets used and/or analyzed during the current
study are available from the corresponding author on reasonable
request.
Authors' contributions
JHC and XTJ designed the present study. XGH, JNN and
XHZ performed the experiments, analyzed the data and prepared the
figures, and drafted the initial manuscript. JHC reviewed and
revised the manuscript. All authors read and approved the final
manuscript. JHC and JNN have read and confirm the authenticity of
the raw data.
Ethics approval and consent to
participate
Not applicable.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Vincent A, Herman J, Schulick R, Hruban RH
and Goggins M: Pancreatic cancer. Lancet. 378:607–620. 2011.
View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Siegel RL, Miller KD and Jemal A: Cancer
statistics, 2017. CA Cancer J Clin. 67:7–30. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
McGuigan A, Kelly P, Turkington RC, Jones
C, Coleman HG and McCain RS: Pancreatic cancer: A review of
clinical diagnosis, epidemiology, treatment and outcomes. World J
Gastroenterol. 24:4846–4861. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Neoptolemos JP, Kleeff J, Michl P,
Costello E, Greenhalf W and Palmer DH: Therapeutic developments in
pancreatic cancer: Current and future perspectives. Nat Rev
Gastroenterol Hepatol. 15:333–348. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Zeng S, Pöttler M, Lan B, Grutzmann R,
Pilarsky C and Yang H: Chemoresistance in pancreatic cancer. Int J
Mol Sci. 20:45042019. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Collisson EA, Bailey P, Chang DK and
Biankin AV: Molecular subtypes of pancreatic cancer. Nat Rev
Gastroenterol Hepatol. 16:207–220. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Rhim AD, Mirek ET, Aiello NM, Maitra A,
Bailey JM, McAllister F, Reichert M, Beatty GL, Rustgi AK,
Vonderheide RH, et al: EMT and dissemination precede pancreatic
tumor formation. Cell. 148:349–361. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Reichert M, Bakir B, Moreira L, Pitarresi
JR, Feldmann K, Simon L, Suzuki K, Maddipati R, Rhim AD, Schlitter
AM, et al: Regulation of epithelial plasticity determines
metastatic organotropism in pancreatic cancer. Dev Cell.
45:696–711.e8. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Canessa CM, Schild L, Buell G, Thorens B,
Gautschi I, Horisberger JD and Rossier BC: Amiloride-sensitive
epithelial Na+ channel is made of three homologous subunits.
Nature. 367:463–467. 1994. View
Article : Google Scholar : PubMed/NCBI
|
|
10
|
Jasti J, Furukawa H, Gonzales EB and
Gouaux E: Structure of acid-sensing ion channel 1 at 1.9 A
resolution and low pH. Nature. 449:316–323. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Liu C, Zhu LL, Xu SG, Ji HL and Li XM:
ENaC/DEG in tumor development and progression. J Cancer.
7:1888–1891. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Xu S, Liu C, Ma Y, Ji HL and Li X:
Potential roles of amiloride-sensitive sodium channels in cancer
development. Biomed Res Int. 2016:21902162016. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Geng Y, Zhao S, Jia Y, Xia G, Li H, Fang
Z, Zhang Q and Tian R: miR95 promotes osteosarcoma growth by
targeting SCNN1A. Oncol Rep. 43:1429–1436. 2020.PubMed/NCBI
|
|
14
|
Wu L, Ling ZH, Wang H, Wang XY and Gui J:
Upregulation of SCNN1A promotes cell proliferation, migration, and
predicts poor prognosis in ovarian cancer through regulating
epithelial-mesenchymal transformation. Cancer Biother Radiopharm.
34:642–649. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Jerkovic I, Ibrahim DM, Andrey G, Haas S,
Hansen P, Janetzki C, González Navarrete I, Robinson PN, Hecht J
and Mundlos S: Genome-wide binding of posterior HOXA/D
transcription factors reveals subgrouping and association with
CTCF. PLoS Genet. 13:e10065672017. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Pernía O, Sastre-Perona A,
Rodriguez-Antolin C, García-Guede A, Palomares-Bralo M, Rosas R,
Sanchez-Cabrero D, Cruz P, Rodriguez C, Diestro M, et al: A novel
role for the tumor suppressor gene ITF2 in tumorigenesis and
chemotherapy response. Cancers (Basel). 12:7862020. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Li ZG, Xiang WC, Shui SF, Han XW, Guo D
and Yan L: 11 Long noncoding RNA UCA1 functions as miR-135a sponge
to promote the epithelial to mesenchymal transition in glioma. J
Cell Biochem. 121:2447–2457. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Hirao N, Iwata T, Tanaka K, Nishio H,
Nakamura M, Morisada T, Morii K, Maruyama N, Katoh Y, Yaguchi T, et
al: Transcription factor homeobox D9 is involved in the malignant
phenotype of cervical cancer through direct binding to the human
papillomavirus oncogene promoter. Gynecol Oncol. 155:340–348. 2019.
View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Zhu H, Dai W, Li J, Xiang L, Wu X, Tang W,
Chen Y, Yang Q, Liu M, Xiao Y, et al: HOXD9 promotes the growth,
invasion and metastasis of gastric cancer cells by transcriptional
activation of RUFY3. J Exp Clin Cancer Res. 38:4122019. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Wen D, Wang L, Tan S, Tang R, Xie W, Liu
S, Tang C and He Y: HOXD9 aggravates the development of cervical
cancer by transcriptionally activating HMCN1. Panminerva Med. May
14–2020.doi: 10.23736/S0031-0808.20.03911-7 (Epub ahead of
print).
|
|
21
|
Liu X, Bing Z, Wu J, Zhang J, Zhou W, Ni
M, Meng Z, Liu S, Tian J, Zhang X, et al: Integrative gene
expression profiling analysis to investigate potential prognostic
biomarkers for colorectal cancer. Med Sci Monit.
26:e9189062020.PubMed/NCBI
|
|
22
|
Tomczak K, Czerwińska P and Wiznerowicz M:
The cancer genome atlas (TCGA): An immeasurable source of
knowledge. Contemp Oncol (Pozn). 19:A68–A77. 2015.PubMed/NCBI
|
|
23
|
GTEx Consortium: The genotype-tissue
expression (GTEx) project. Nat Genet. 45:580–585. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Lacny S, Wilson T, Clement F, Roberts DJ,
Faris P, Ghali WA and Marshall DA: Kaplan-Meier survival analysis
overestimates cumulative incidence of health-related events in
competing risk settings: A meta-analysis. J Clin Epidemiol.
93:25–35. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(-Delta Delta C(T)) method. Methods. 25:402–408. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Xu L, Faruqu FN, Lim YM, Lim KY, Liam-Or
R, Walters AA, Lavender P, Fear D, Wells CM, Tzu-Wen Wang J and
Al-Jamal KT: Exosome-mediated RNAi of PAK4 prolongs survival of
pancreatic cancer mouse model after loco-regional treatment.
Biomaterials. 264:1203692021. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Lin C, Sun L, Huang S, Weng X and Wu Z:
STC2 Is a potential prognostic biomarker for pancreatic cancer and
promotes migration and invasion by inducing epithelial-mesenchymal
transition. Biomed Res Int. 2019:80424892019. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Ge L, Wang H, Xu X, Zhou Z, He J, Peng W,
Du F, Zhang Y, Gong A and Xu M: PRMT5 promotes
epithelial-mesenchymal transition via EGFR-β-catenin axis in
pancreatic cancer cells. J Cell Mol Med. 24:1969–1979. 2020.
View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Wang L, Wu H, Wang L, Zhang H, Lu J, Liang
Z and Liu T: Asporin promotes pancreatic cancer cell invasion and
migration by regulating the epithelial-to-mesenchymal transition
(EMT) through both autocrine and paracrine mechanisms. Cancer Lett.
398:24–36. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Crosby JR, Zhao C, Jiang C, Bai D, Katz M,
Greenlee S, Kawabe H, McCaleb M, Rotin D, Guo S and Monia BP:
Inhaled ENaC antisense oligonucleotide ameliorates cystic
fibrosis-like lung disease in mice. J Cyst Fibros. 16:671–680.
2017. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Dirlewanger M, Huser D, Zennaro MC,
Girardin E, Schild L and Schwitzgebel VM: A homozygous missense
mutation in SCNN1A is responsible for a transient neonatal form of
pseudohypoaldosteronism type 1. Am J Physiol Endocrinol Metab.
301:E467–E473. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
D'Arcangelo D, Scatozza F, Giampietri C,
Marchetti P, Facchiano F and Facchiano A: Ion channel expression in
human melanoma samples: In silico identification and experimental
validation of molecular targets. Cancers (Basel). 11:4462019.
View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Pastushenko I and Blanpain C: EMT
transition states during tumor progression and metastasis. Trends
Cell Biol. 29:212–226. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Liu M, Xiao Y, Tang W, Li J, Hong L, Dai
W, Zhang W, Peng Y, Wu X, Wang J, et al: HOXD9 promote
epithelial-mesenchymal transition and metastasis in colorectal
carcinoma. Cancer Med. 9:3932–3943. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Lv X, Li L, Lv L, Qu X, Jin S, Li K, Deng
X, Cheng L, He H and Dong L: HOXD9 promotes epithelial-mesenchymal
transition and cancer metastasis by ZEB1 regulation in
hepatocellular carcinoma. J Exp Clin Cancer Res. 34:1332015.
View Article : Google Scholar : PubMed/NCBI
|