Introduction
Cord blood, also known as placental blood or
umbilical cord blood (UCB), is the blood in the umbilical cord and
blood vessels near the fetal side of the placenta, which may be
collected when the fetus is born. In 1989, experiments by Broxmeyer
et al (1) indicated that
UCB is a potential source of hematopoietic stem/progenitor cells.
UCB stem cells (UCBSCs) are a type of primitive undifferentiated
cells that have the same multidirectional differentiation potential
as bone marrow stem cells. UCBSCs are able to self-renew and
proliferate. They may differentiate into various cell or tissue
types under the influence or induction of specific factors
(2–5). UCBSCs have a wide range of sources
and their application is not limited by ethics concerns and/or
guidelines (6). Therefore, they
are considered an important source of stem cells for
transplantation and have huge potential to be widely used in
clinical tissue engineering and other stem cell therapies (7,8).
The detailed investigation and understanding of the
functions of UCBSCs have laid a foundation for their successful
clinical application. However, the establishment of UCB cells and
their differentiation in vivo remain incomplete (9,10).
Although tracing technology is usually used to understand cell
function, it exhibits several limitations. The traditional tracer
technique cannot differentiate the following generations of cells
from the primary cells at a large scale and the lineage
relationship between the cells is not clear (11,12). Recently, DNA barcode technology
used for cell development tracing has achieved lineage tracing at
single-cell and single-nucleic acid mutation resolution (13). DNA barcoding combined with
sequencing technology clearly demonstrated the relationship among
splinter cells by labeling cells with DNA barcodes, tracing their
developmental history and stacking them to form a lineage
development tree in order to identify their origin, development and
differentiation. This is an effective strategy for tracking large
numbers of cells both spatially and temporally (14). This may aid the understanding of
the self-renewal mechanism of UCBSCs and lays a foundation for
their clinical application.
In the present study, the differentiation
characteristics of UCBSCs were reviewed, including the research
progress of the latest methods of DNA barcode technology. This
information aims to increase the current understanding of the
biological roles and clinical applications of these cells.
Differentiation of UCBSCs
Stem cells possess the potential for self-renewal
and multi-differentiation, which may be used to replace damaged
cells and exert significant therapeutic potential in regenerative
medicine. Several types of stem cells have been detected in UCB,
including the following: Umbilical cord hematopoietic stem cells
(HSCs), endothelial progenitor cells, mesenchymal stem cells
(MSCs), unrestricted somatic stem cells and multipotent progenitor
cells. HSCs, which have a relatively high content in UCB, may be
divided into two cell types, namely CD34+ and
CD34−, among which CD34+ cells account for
>95% of the population. MSCs are mainly derived from UCB and the
bone marrow and their cell phenotypes include CD133, CD34 and CD45.
Although MSCs are rarely found in cord blood, their differentiation
ability is potent. Studies have indicated that UCBSCs may be
induced to differentiate into nerve cells, chondrocytes,
hepatocyte-like cells, fat cells, osteoblasts and islet-like cells
under appropriate microenvironmental conditions. In 2003, Mitchell
et al (15) induced the
differentiation of UCBSCs using β-mercaptoethanol, antioxidants and
dimethylsulfoxide. It was indicated that 80% of the cells exhibited
a neuron-like appearance. Furthermore, a unique Nissl body
structure of neuron cells was noted following 12 h of incubation.
Fu et al (16) cultured
UCBSCs together with the primary cortex of mice for 4 days. In
total,~50% of the cells developed into neural cells, ~33% of the
cells differentiated into astrocytes and ~10% into
oligodendrocytes. This finding indicated the presence of neural
stem cells in UCBSCs, which were able to differentiate into neural
cells. A similar study demonstrated that UCBSCs that were injected
into a rat model at the site of spinal cord injury caused
significant functional improvement following six weeks. These stem
cells were able to differentiate into nerve cells following
transplantation (17). Wang et
al (18) studied the
possibility of the differentiation of UCBSCs into cardiomyocytes
following their treatment with 5-azacytidine or their culture in
cardiomyocyte-conditioned medium. The data indicated that both
conditions resulted in the induction of the expression of the
cardiomyocyte markers N-cadherin and cardiac troponin I.
Furthermore, the multi-lineage potential of UCBSCs was also
validated, indicating that they may be induced into the
chondrogenic, osteogenic and adipogenic lineages in vitro.
To date, multiple studies suggested that UCBSCs differentiate into
hepatocyte-like cells following their transplantation into
liver-damaged mice (19,20). Growth factors are usually added
during these experiments. Kakinuma et al (19) indicated that mice with partial
hepatectomy that received UCBSC transplantation exhibited
transplanted cells in the liver; these cells were differentiated
into hepatocytes and secreted albumin for a year. Similar results
were obtained in animal models of chemically-induced liver disease,
in which infused UCBSCs differentiated into mature hepatocytes
(20). The results of this study
indicated that UCBSCs had the tendency to differentiate into
hepatocyte-like cells under certain conditions.
In addition to the aforementioned findings, Mayani
et al (21) demonstrated
that UCBSCs were able to differentiate into nerve cells,
chondrocytes and liver-like cells. To date, the environmental
conditions that are required for the process of differentiation,
the accuracy of induced differentiation, the assessment of the
relationship between differentiated and normal cells and the
mechanism of differentiation have remained to be determined. In
addition, the majority of experiments that assessed the induction
of differentiation were performed in vitro, which cannot
completely simulate the in vivo conditions. The
differentiation of UCBSCs is still unknown and uncontrollable. The
potential to track the differentiation of stem cells in space and
time by using a more powerful tracer tool will enhance the current
understanding of the biological characteristics of UCBSCs.
In order to investigate the differentiation of
UCBSCs, their in vitro culture is required. Studies have
indicated that UCB contains HSCs and several types of MSC. At
present, the isolation and culture of UCBSCs are mainly focused on
HSCs. The culture methods of UCBSCs mainly include the feeder layer
and the feeder-free culture systems. Shetty et al (22) suggested that the feeder layer
maintains the undifferentiated state of the cells and promotes
self-renewal of UCBSCs. Mouse embryonic fibroblasts inactivated by
γ-rays or mitomycin C are commonly used as feeder layers for UCBSC
culture. Han et al (23)
used DMEM with other substances added to the FBS supplement. In a
subsequent study, Hutton et al (24) replaced the FBS with a serum
substitute and added basic fibroblast growth factor. Demerdash
et al (25) indicated that
the number of expansion times of the feeder culture system were
limited and that the growth ability of UCBSCs was reduced following
differentiation for a certain number of generations. Furthermore,
the authors of that study demonstrated that feeder layer cells
weakened the effect of exogenous factors on UCBSCs and the
animal/human-derived feeder layer was unable to achieve its
clinical effect due to contamination with heterogenic/allogeneic
antigens (23,26–29).
The development of optimized feeder-free culture
systems has been the focus of UCBSC culture. The regeneration of
UCBSCs requires the activation of various signaling pathways, such
as fibroblast growth factor 2, ERK and PI3K/AKT, insulin growth
factor/insulin, 1-phosphate-sphingosine/platelet-derived growth
factor and TGF-β/activin/nodulation protein A/SMAD2/3, by receptor
tyrosine kinases. Zhang et al (30) indicated that the Wnt/β-catenin and
TNF receptor superfamily of proteins were associated with the
survival rate of UCBSCs, while inhibition of bone morphogenetic
protein markers may prevent their independent differentiation.
Several studies have produced feeder-free culture media, such as
TeSRl that contains certain recombinant growth factors, which aim
to enhance the ability of UCBSCs to regenerate (31,32). Zhou et al (33) demonstrated that the combination of
activin inhibitors and MEK/ERK inhibitors increased the induction
rate of UCBSCs. The current focus is to identify a feeder-free
culture system that is able to maintain the normal karyotype of the
cells (34). Naka et al
(35) demonstrated that retaining
the culture system in a state of low oxygen pressure reduces the
differentiation of UCBSCs and the development of chromosomal
abnormalities (36). Lee
(37) highlighted that certain
small molecular weight compounds are able to regulate the
biological function of stem cells by maintaining stem cell
self-renewal, inhibiting differentiation and the activation of the
withering pathway. For instance, stem regenin 1, an antagonist of
the aryl hydrocarbon receptor (38) and the pyrimidodiole derivative
UM1717 (39), may be used to
safely and effectively expand UCBSCs in vitro. Although
these results are preliminary, it is suggested that similar
strategies may be employed in UCBSC culture to reduce the
production and proliferation of abnormal cells.
DNA barcoding
The current understanding of the biological function
of UCBSCs has significantly improved. However, the mechanism of
self-renewal of UCB-derived HSCs remains to be fully elucidated.
Basic and clinical research has focused on investigating the
trajectory inference of UCBSCs, which usually requires labeling
technology. Gene transfection and fluorescent dyes are classical
cell tracing techniques (11,12), which may be easily detected in
viable cells using microscopy imaging technology. The
double-labeled experiments may be performed using a variety of
fluorescent reagents. However, at present, the half-life of the
green fluorescent protein (GFP) is not sufficiently clear to
accurately assess the time period required in the experiments;
furthermore, physical factors, such as high temperature, and
chemical factors, such as strong acid and strong alkali conditions,
may cause structural damage and decomposition. The mechanism of
action of fluorescent dyes is focused on achieving labeling through
the cell membrane. The majority of the fluorescent agents are
degraded following cell proliferation and division, which may lead
to the inability of fluorescent dye labeling to maintain a high
labeling rate for a long time. Seghatoleslam et al (40) used bromodeoxyuracil nucleoside
(BrdU) for UCBSC labeling (Table
I). BrdU is a pyrimidine analog, which competes with endogenous
thymidine during DNA replication. Prior to or during UCBSC
transplantation, the cells proliferate in the presence of BrdU.
Subsequently, labeled UCBSCs continue to proliferate without BrdU
and are finally stained by immunohistochemical methods. However,
certain defects are present in the nucleic acid labeling method,
such as the instability of BrdU during labeling. When the labeling
time increases, the unlabeled surrounding cells are also labeled
due to apoptosis and phagocytosis, which affects the signal
strength of the transplanted UCBSCs.
 | Table I.Labeling and tracing technology of
umbilical cord blood stem cells. |
Table I.
Labeling and tracing technology of
umbilical cord blood stem cells.
| Labeling
method | Labeling agent | Labeling
content | Advantage | Disadvantages | Readout | (Refs.) |
|---|
| Gene transfection
labeling | Protein | DNA | Stable | The half-life is
unclear | Microscopy | (11) |
| Fluorescent dye
labeling | CM-Dil | Cell membrane
lipids | Accurate | Easily
degradable | Microscopy | (12) |
| Nucleic acid
labeling | Pyrimidine
analog | DNA | High
sensitivity | Signal easily
lost | IHC | (40) |
| DNA barcode | DNA, Cre/Cas9 cell
lineage | DNA/RNA | Accurate tracking
of optimization | Sequencing
technology requires | scRNA-seq;
Illumina | (42–45) |
Although gene transfection labeling and fluorescent
dye labeling are able to mark primary cells during the process of
cell division or differentiation, the offspring cells cannot be
distinguished from the primary cells and the lineage relationship
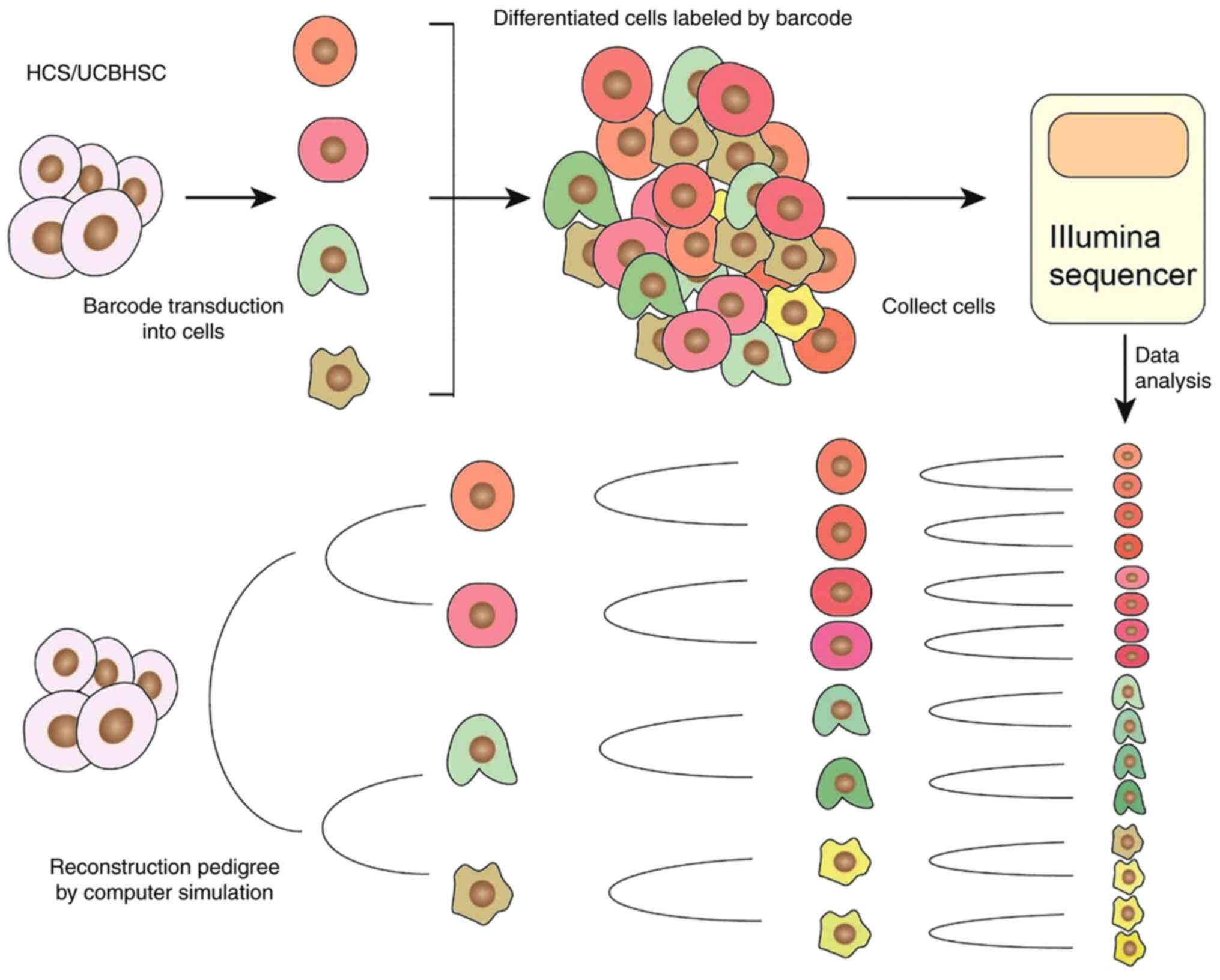
between cells is not clear. DNA barcoding is an emerging technology
that uses unique nucleic acid sequences to label individual cells.
The genetic barcodes are inserted into the genomic DNA of the cells
at a specific time-point and the cells divide to produce progeny
cells, forming cell clones whose progeny inherit the barcodes. The
progeny of the labeled cells is identified by reading the tags
(41). This process is termed
genetic barcoding. This provides insight into cell behavior
regarding space and time. Barcodes are created by using a special
nucleic acid sequence as a permanent or dynamic marker for a single
cell. The number of barcode sequences that may be used is
theoretically infinite and a large population of cells may be
efficiently labeled and tracked at the single-cell level (Table I) (42–48).
Development of the DNA barcode
Initially, the DNA barcode technology was based on
the identification of unique retroviral integration sites and
barcode identities, such as those identified using southern blot or
PCR assays (49,50). As the development of sequencing
technology has promoted the progress of barcode technology,
numerous approaches have created and deployed more complex DNA
barcodes for lineage tracing (Table
II). The basic concept of these methods is lineage tracing by
DNA barcodes and the assessment of the changes induced in their
targets, including whole-genome or mitochondrial-genome sequencing
data (51). The methods based on
targeted barcode approaches are generally divided into the three
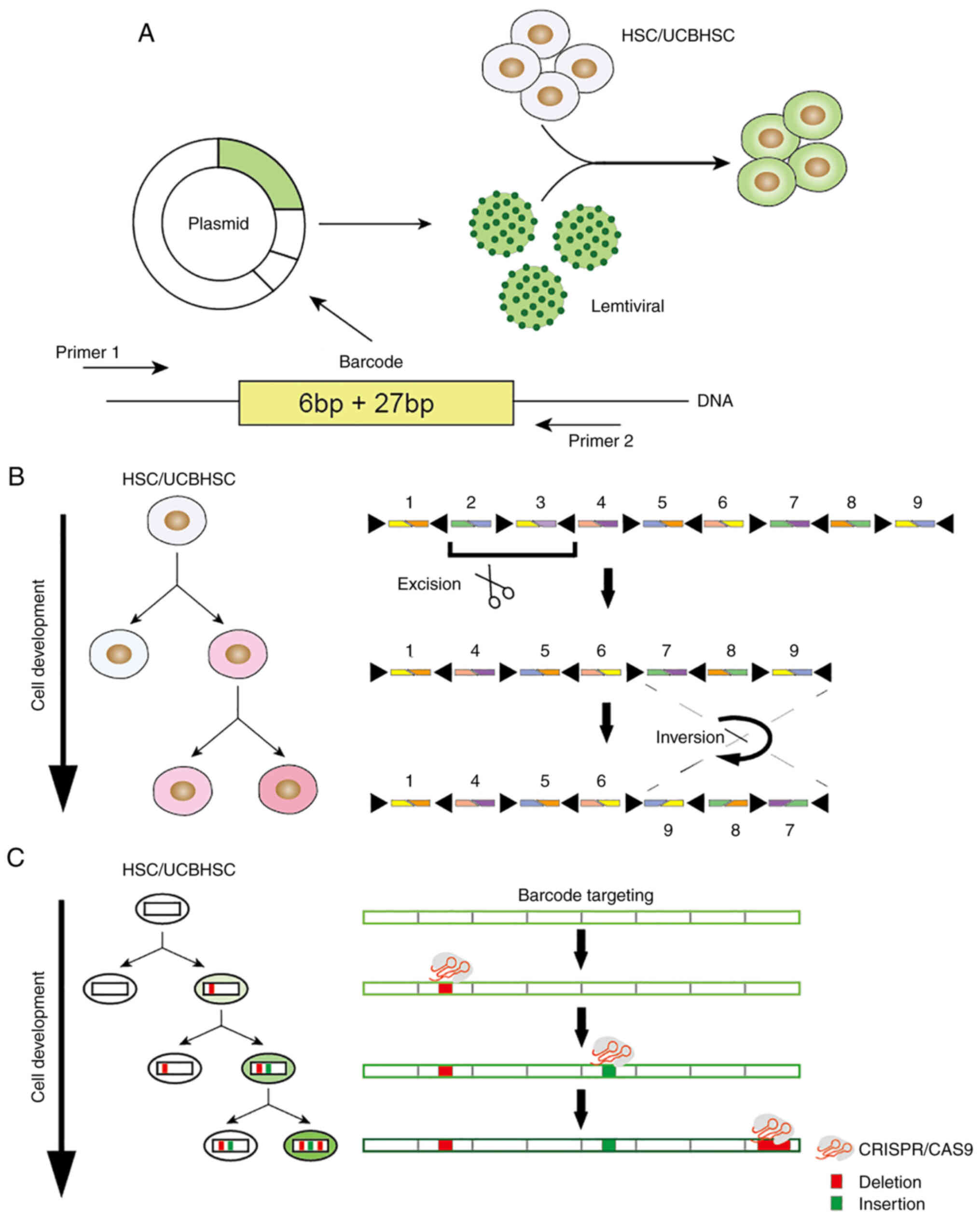
following categories (Fig. 1):
Transposon/retrovirus, Cre-recombinase-based and clustered
regularly interspaced short palindromic repeats
(CRISPR)/CRISPR-associated protein 9 (Cas9)-methods, which mediate
in vivo integration of transgenic DNA targets, in
vivo recombination of transgenic DNA cassettes and in
vivo editing of transgenic DNA targets. In all these methods,
the DNA barcode alters the genome of a single cell, whose progeny
inherits the barcode and may be considered a cloning unit. DNA
barcodes may subsequently be recorded and measured in high
throughput so that thousands of different cloning units may be
tracked in parallel.
 | Table II.Overview of barcoding techniques. |
Table II.
Overview of barcoding techniques.
| Technology | DNA editing
system | Barcode length
(bp) | Diversity | Species | In vivo | Readout | (Refs.) |
|---|
| TracerSeq | Tol2 | 20 | NR | Zebrafish | Yes | Illumina | (52) |
| Embedded viral | Retrovirus | 33 | NR | Mouse | Yes | Illumina | (54) |
| CellTag | Retrovirus | 8 | NR | Human | No | scRNA-seq;
Illumina | (53) |
| Polylox | Cre-loxP | 1,942 | 849 | Mouse | Yes | Pacbio | (55,56) |
| GESTALT | Cas9 | 266 | 4195 | Zebrafish | Yes | Illumina | (58) |
| mSCRIBe | Cas9 | 70 | 1890 | Zebrafish | Yes | scRNA-seq;
Illumina | (60) |
| Homing
barcodes | Cas9 | 240 | NR | Mouse | Yes | Illumina | (61,62) |
| MEMOIR | Cas9 | 256 | −256 | Mouse | Yes | FISH | (59) |
| CARLIN | Cas9 | 276 | 4,400 | Mouse | Yes | scRNA-seq;
Illumina | (64) |
Transposon/retrovirus-based DNA
barcode
Certain studies have used a predefined library of
barcodes that are easy to interpret and track by sequencing. For
instance, TracerSeq is a clonal barcode method demonstrated in
zebrafish (52). TracerSeq
utilizes ongoing transposase activity to continuously integrate a
predefined library of barcodes into embryos as an injection plasmid
library. Progressive integration of plasmids provides clonal and
subclone heritable tags for the genome. CellTag is an integration
method of 8-nt barcodes based on lentiviral delivery (53). In this design, the location of the
cell tag is the 3′untranslated region of the GFP gene, followed by
the simian virus 40 polyadenylation signaling sequence. By allowing
>1 barcode to mark each cell, the diversity of the combination
is expanded. Bramlett et al (54) developed a detailed protocol for a
viral barcoding program (Fig.
1A). The barcode was 33-bp in length and included a 6-bp
library ID and a random 27-bp barcode. In theory, 427-bp barcodes
may be generated. New users may use this protocol to create custom
barcode libraries in their laboratories and easily set barcodes at
low cost (54). However, these
methods limit the possibility of using additional experimental
methods and reduce the diversity of the barcodes. These strategies
are also limited to short-term culture and tolerant cell separation
systems.
Cre recombinase-based DNA barcode
The Polylox system (Fig. 1B) integrated a 2.1-kb synthetic
gene from Arabidopsis into the Rosa26 site of the mouse and
inserted 10 loxP sites into this gene, which resulted in the
induction of the transient expression of the Cre recombinase
enzyme. Following exposure to Cre recombinase, these loxP sites
were randomly excised or flipped from the DNA sequence, which
subsequently generated a barcode in the HSCs, and was finally
combined with sequencing technology in order to evaluate their
differentiation fate in vivo. However, certain shortcomings
may exist in the barcodes, which are based on the Cre-loxP system.
Cre is inherently more prone to excision than inversion. Therefore,
the size of the target array will be reduced over time. An
additional disadvantage is that barcode target arrays are long and
repetitive due to the low diversity of recombinase recognition
sites and the nature of their minimum spacing requirements. To
achieve high barcode diversity, the specific target array must
contain more fragments, which requires the barcode to be read by a
low-flux long read sequence. This drawback may be mitigated by
future improvements in length sequencing technology (55,56).
CRISPR-based DNA barcode
The development of CRISPR/Cas9 technology has
enabled the use of this novel technology instead of DNA recombinase
in order to assess DNA barcoding (Table II). In the barcode scheme, which
is based on CRISPR editing, CRISPR/Cas9-induced double-strand break
of genomic DNA is usually repaired during cell division based on
the non-homologous end joining (NHEJ) (57) mechanism common in mismatch repair.
Therefore, short random insertions and deletions are gradually
introduced into the barcode region of the DNA to mark cells and
construct lineage relationships.
The genome editing of synthetic target arrays for
lineage tracing (GESTALT) system (Fig. 1C and Table II) was the first system
introduced to confirm this principle (58). The GESTALT system utilizes the
CRISPR/Cas9 genome labeling technology to accumulate the combined
sequence diversity and form a compact, multi-target and
information-intensive barcode, which combines and accumulates the
mutations generated by the dense target site array, records the
cell lineage information on a large scale and is able to query the
lineage information from at least hundreds of thousands of cells on
a large scale. Only one barcode sequence is read in each cell and
the generated barcode is used for lineage tracking of the
zebrafish.
An additional similar method is memory by engineered
mutagenesis with optical in situ readout (MEMOIR) system
(Table II), a method of inducing
mutation memory and optical in situ reading by designing a set of
barcode recording elements, which is based on the fact that the
target-induced mutation of CRISPR/Cas9 irreversibly changes the
state of a given barcode recording element and subsequently reads
its sequence in a single cell by multiple single molecule RNA
fluorescence hybridization. This analyzes the final state of a log
in a single cell (59).
Therefore, lineage information may be reconstructed from the cell
community. In addition, the combination of the endogenous gene
expression analysis with lineage reconstruction of the same cell
enables the deduction of the dynamic changing rate of embryonic
stem cells between two gene expression states. Finally, computer
simulation reconstruction is used to indicate the way by which a
parallel MEMOIR system, which runs in the same cell, is able to
record and read the history of dynamic cell events.
The methods used to further increase the diversity
of barcodes have been redeveloped and are included in the mammalian
synthetic cell recorder integrating biological events (mSCRIBE)
(60) and Homing CRISPR barcodes
(Table II) (61,62). Both methods share the same
principle and they are designed to target the spacer sequence of
their own genome by mutated guide RNA (gRNA), which is termed
self-targeting guide (stg) RNA in mSCRIBE and homing gRNA (hgRNA)
in the homing barcode. Despite its different names, its working
principles are interlinked. Initially, stgRNA/hgRNA cleavage is
performed, while the mutated genomic DNA produces barcode diversity
and subsequently, the mutated site produces new stgRNA/hgRNA, which
is targeted to the mutated genomic site. New barcodes are
continuously generated until the spacer is truncated to <16-18
nt or the protospacer-adjacent motif sequence is lost. The
generation of new barcodes would not be terminated and may even be
continuously updated by specific mouse models. Another innovation
that increases the diversity of the Cas9-edited barcodes is the use
of terminal deoxynucleotidyl transferase (TdT) as an additional
transgenic component of the barcodes. In the presence of
double-strand breaks, TdT catalyzes the random binding of
nucleotides at DNA cleavage sites, which increases the
insertion-based editing frequency (63).
More recently, Bowling et al (64) established a mouse cell line for
CRISPR array repair lineage tracing (CARLIN) and its corresponding
analytical tools (Table II).
CARLIN is able to generate transcriptional recognition barcodes (up
to 44,000 barcodes), which are compatible with sequencing barcode
coding and fully genetically defined in an induced manner at any
time-point in mouse development or adulthood. For the CARLIN
system, 10 single guide RNAs were designed based on GESTALT
tracking technology to ensure efficient cleavage of target sites in
the presence of Cas9 (58). The
gRNAs were designed in a quadratic iterative manner, with one
expressing the gRNA driven by the U6 promoter and the second
carrying the tetO operon upstream of each gRNA. Constitutive
expression of molecular tracer arrays is also based on constitutive
CAG promoter-driven fluorescent proteins. All these elements are
integrated in mouse embryonic stem cells, which are mediated by
recombinase and inserted together into the widely used Col1a1 site.
Finally, doxycycline induction was used to control the expression
of the Cas9 gene and promote the fragmentation of double-stranded
DNA in the target array. These breaks were repaired, resulting in
the expression and stable inheritance of a variety of altered DNA
sequences in the CARLIN allele. This system may produce gene
deletions ranging from 1 to 252 bp, as well as gene insertions up
to 51 bp in length. Based on CRISPR-Cas9 gene editing technology,
CARLIN may perform lineage tracing in the phylogenetic process and
heterogeneity analysis on cell populations, as well as control the
extent of gene editing when doxycycline is added. It is a model
that allows simultaneous cell lineage tracing and single-cell level
transcriptome analysis in vivo, providing an important tool
platform for research on multiple cell lineages.
The CRISPR/Cas9 method is promising for achieving
high diversity, labeling of various tissues and organs of organisms
and generation of in vivo barcodes over time. However, the
diversity generated in practice is considerably lower than that in
theory due to the repair mode of the NHEJ, which selects the
generation of deletions rather than insertions, leading to the
gradual shortening of the CRISPR barcodes over time. However, the
development of this methodology has enabled the use of multiple
barcodes of a single cell compared with one single-cell barcode,
which greatly increases the diversity produced. With the rapid
development of CRISPR barcoding technology, these limitations may
be reduced.
Delivery and reading of DNA
barcodes
The manual assignment of a single barcode
(one-to-one) to the labeled cells is the most ideal delivery
method, which may guarantee that they are completely and not
repeatedly labeled. This delivery approach is not acceptable in
terms of cost and time. The assignment of barcodes to individual
cells is difficult and may only be used under limited conditions.
At present, the most effective delivery methods still rely on in
vitro production of barcode vectors, such as the use of
retroviral transfection (41), as
well as plasmid injection and electroporation (44). It should be noted that the nature
of the cells may be altered during barcode transduction. Although
several studies have indicated that lentiviral integration does not
result in significant changes in transduced cells (65–69), it is still possible to randomly
insert specific lentiviral vectors into certain genomic regions and
alter cell behavior. Therefore, experimental replicates and
controls must be used rigorously during the experiments to rule out
this rare possibility.
To date, the majority of research studies that have
investigated reading of the DNA barcodes depend on the extraction
of nucleic acids and subsequently, the barcodes are detected or
quantified in vitro. The method of barcode reading has also
undergone tremendous changes in recent decades. It was initiated
with PCR amplification and sizing, continued with Sanger sequencing
(46,70) and eventually developed to
high-throughput sequencing. The reduction of the sequencing cost is
an important factor that promotes the development of barcode
reading technology. The development of single-cell RNA sequencing
provided significant progress in barcode detection. It is able to
read the cell development information carried by the barcode and at
the same time obtain the transcription status and cell type of the
cell (Fig. 2). However, during
the sequencing of a single cell, its spatial position is lost
(71).
Applications of DNA barcoding. DNA barcode
technology is a powerful pedigree tracing tool used for UCBSCs and
other stem cells. Cheung et al (72) exposed CD34+ UCBSCs to a
barcode lentiviral library and subsequently inoculated two
non-obese diabetic (Nod)/severe combined immunodeficient (Scid)/IL2
receptor (IL2R)y−/− mice with 1×105 barcoded
cells, respectively. This method was initially combined with
sequencing analysis and it confirmed that CD34+ UCBSCs
were able to differentiate into human CD3+ T cells.
Belderbos et al (73)
transplanted lentiviral barcode CD34+ cells from 20 UCB
donors into Nod/Scid/IL2Ry−/− mice. Following 10 weeks,
human B, T and myeloid cells were detected. This indicated that
UCB-derived CD34+ cells were able to be traced for
multiple generations by the DNA barcode in mouse
xenotransplantation. This study further indicated donor-to-donor
heterogeneity in the clonal dynamics of transplanted human UCBSCs
in mouse xenografts. Several research groups have tracked the
activity of various HSCs following single recipient
transplantation, suggesting that HSCs only have a small role in
daily hematopoiesis under stable conditions (42,47,74). Due to the significant reduction in
the sequencing cost and the development of synthetic biology, which
includes the improvement of DNA barcode technology, it is expected
that additional novel barcode strategies will be widely used in
research of UCBSCs. The differentiation plasticity of UCBSCs
remains controversial. Research on UCB cell biology is highly
significant to further the understanding of its cellular and
molecular mechanisms, as well as the difficulties encountered in
the clinical applications of UCBSCs. It is considered that DNA
barcode tracing cannot only be applied to UCBSC research, but also
extended to several fields, such as neuroanatomy, cell activity
recording and cancer research (75–77). The barcode method allows the
reconstruction of stem cell lineages during development based on
barcode similarities and tracing of cell relationships in a single
experiment (78). In fate-mapping
studies, barcodes may be used to count the number of stem cell
divisions in heterogeneous cell populations (79). In addition, the barcode may also
aid the identification of the cellular origin of occurrence,
recurrence and metastasis of cancer stem cells and the
heterogeneous responses of cancer stem cells to treatment (80).
An important challenge remains to be addressed prior
to reconstructing the pedigree tree with the barcodes. A
sufficiently high level of barcode diversity is required so that
every cell may be uniquely labeled at the end of the experiment.
Insufficient diversity will either terminate pedigree tracking
prior to the end-point or seriously hinder tree reconstruction,
since the cells in the distant pedigrees will share the same
barcode. As a result, the complexity of the DNA sequence increases
exponentially with the length and diversity of the barcodes.
However, the current in situ reading of the barcode or cell
transcription is considerably slow, inefficient or biased (49–51,81,82). Additional technical development is
required. DNA barcode technology exhibits high potential as a
genealogy tracking tool and the rapid development of this field is
expected to occur in the next years.
Summary
Currently, research on UCBSC has achieved periodical
success and additional experiments are required to verify its wide
clinical applications. The combination of cell barcode and
single-cell sequencing technologies may continuously record the
development history of cells, which is a milestone in lineage
tracking technology. The mechanism of UCBSC development following
transplantation may be further elucidated and the time required for
these methods to be used in large-scale clinical applications is
reduced. However, the current cell barcode technology is not able
to generate sufficient diversity to perform specific labels on each
cell. In addition, the efficiency of reading the barcode is not
sufficiently high based on the current sequencing technology, the
reading process exhibits certain bias and potential off-target
effects are present. Additional technical development is required
to develop a more simple and feasible lineage tracing technique
with high sensitivity, strong specificity, low external
interference, no apparent toxicity and low false-positive rate. It
is expected that future tracer techniques will be able to fully
describe the development of UCBSCs and their differentiation and
tissue formation ability during tissue homeostasis or disease
development.
Acknowledgements
Not applicable.
Funding
This work was supported by the National Natural
Science Foundation of China (grant nos. 81872412, 81602303 and
31700736), the Natural Science Foundation of Hubei Province (grant
nos. 2017CFB786 and 2019CFB591), Hubei Province Scientific and
Technological Research Project (grant no. D20201306).
Availability of data and materials
Data sharing is not applicable to this article, as
no datasets were generated or analyzed during the current
study.
Authors' contributions
M-YW, X-CP and H-WX conceived and designed the
present review. The manuscript was drafted by M-YW. Manuscript
revisions and modifications were carried out by YZ, G-SL, QH, W-QC,
Z-WH, YW, ZM, X-WW, YX, S-XF and X-CP. Final changes were made by
M-YW and H-WX. All authors read and approved the final manuscript.
Data authentication is not applicable.
Ethics approval and consent to
participate
Not applicable.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
Glossary
Abbreviations
Abbreviations:
|
UCBSC
|
umbilical cord blood stem cells
|
|
HSCs
|
hematopoietic stem cells
|
|
MSCs
|
mesenchymal stem cells
|
|
GFP
|
green fluorescent protein
|
|
Cre
|
cyclization recombination
|
|
GESTALT
|
genome editing of synthetic target
arrays for lineage tracing
|
|
CARLIN
|
CRISPR array repair lineage
tracing
|
|
mSCRIBe
|
mammalian synthetic cell recorder
integrating biological events
|
|
MEMOIR
|
memory by engineered mutagenesis with
optical in situ readout
|
References
|
1
|
Broxmeyer HE, Douglas GW, Hangoc G, Cooper
S, Bard J, English D, Arny M, Thomas L and Boyse EA: Human
umbilical cord blood as a potential source of transplantable
hematopoietic stem/progenitor cells. Proc Natl Acad Sci USA.
86:3828–3832. 1989. View Article : Google Scholar
|
|
2
|
Buzańska L, Jurga M and Domańska-Janik K:
Neuronal differentiation of human umbilical cord blood neural
stem-like cell line. Neurodegener Dis. 3:19–26. 2006. View Article : Google Scholar
|
|
3
|
Zhang J, Huang X, Guo B, Cooper S,
Capitano ML, Johnson TC, Siegel DR and Broxmeyer HE: Effects of
eupalinilide E and UM171, alone and in combination on cytokine
stimulated ex-vivo expansion of human cord blood hematopoietic stem
cells. Blood Cells Mol Dis. 84:1024572020. View Article : Google Scholar
|
|
4
|
Sunitha MM, Srikanth L, Kumar PS,
Chandrasekhar C and Sarma P: Down-regulation of PAX2 promotes in
vitro differentiation of podocytes from human CD34+
cells. Cell Tissue Res. 370:477–488. 2017. View Article : Google Scholar
|
|
5
|
Alatyyat SM, Alasmari HM, Aleid OA,
Abdel-Maksoud MS and Elsherbiny N: Umbilical cord stem cells:
Background, processing and applications. Tissue Cell.
65:1013512020. View Article : Google Scholar
|
|
6
|
Francese R and Fiorina P: Immunological
and regenerative properties of cord blood stem cells. Clin Immunol.
136:309–322. 2010. View Article : Google Scholar
|
|
7
|
Fatrai S, Schepers H, Tadema H, Vellenga
E, Daenen SM and Schuringa JJ: Mucin1 expression is enriched in the
human stem cell fraction of cord blood and is upregulated in
majority of the AML cases. Exp Hematol. 36:1254–1265. 2008.
View Article : Google Scholar
|
|
8
|
Castillo-Melendez M, Yawno T, Jenkin G and
Miller SL: Stem cell therapy to protect and repair the developing
brain: A review of mechanisms of action of cord blood and amnion
epithelial derived cells. Front Neurosci. 7:1942013. View Article : Google Scholar
|
|
9
|
Cairo MS and Wagner JE: Placental and/or
umbilical cord blood: An alternative source of hematopoietic stem
cells for transplantation. Blood. 90:4665–4678. 1997. View Article : Google Scholar
|
|
10
|
Mayani H and Lansdorp PM: Biology of human
umbilical cord blood-derived hematopoietic stem/progenitor cells.
Stem Cells. 16:153–165. 1998. View Article : Google Scholar
|
|
11
|
Liu G, Ye X, Zhu Y, Li Y, Sun J, Cui L and
Cao Y: Osteogenic differentiation of GFP-labeled human umbilical
cord blood derived mesenchymal stem cells after cryopreservation.
Cryobiology. 63:125–128. 2011. View Article : Google Scholar
|
|
12
|
Zheng JH, Zhang JK, Kong DS, Song YB, Zhao
SD, Qi WB, Li YN, Zhang ML and Huang XH: Quantification of the
CM-Dil-labeled human umbilical cord mesenchymal stem cells migrated
to the dual injured uterus in SD rat. Stem Cell Res Ther.
11:2802020. View Article : Google Scholar
|
|
13
|
Kebschull JM and Zador AM: Cellular
barcoding: Lineage tracing, screening and beyond. Nat Methods.
15:871–879. 2018. View Article : Google Scholar
|
|
14
|
Wagner DE and Klein AM: Lineage tracing
meets single-cell omics: Opportunities and challenges. Nat Rev
Genet. 21:410–427. 2020. View Article : Google Scholar
|
|
15
|
Mitchell KE, Weiss ML, Mitchell BM, Martin
P, Davis D, Morales L, Helwig B, Beerenstrauch M, Abou-Easa K,
Hildreth T, et al: Matrix cells from Wharton's jelly form neurons
and glia. Stem Cells. 21:50–60. 2003. View Article : Google Scholar
|
|
16
|
Fu YS, Shih YT, Cheng YC and Min MY:
Transformation of human umbilical mesenchymal cells into neurons in
vitro. J Biomed Sci. 11:652–660. 2004. View Article : Google Scholar
|
|
17
|
Rodrigues LP, Iglesias D, Nicola FC,
Steffens D, Valentim L, Witczak A, Zanatta G, Achaval M, Pranke P
and Netto CA: Transplantation of mononuclear cells from human
umbilical cord blood promotes functional recovery after traumatic
spinal cord injury in Wistar rats. Braz J Med Biol Res. 45:49–57.
2012. View Article : Google Scholar
|
|
18
|
Wang HS, Hung SC, Peng ST, Huang CC, Wei
HM, Guo YJ, Fu YS, Lai MC and Chen CC: Mesenchymal stem cells in
the Wharton's jelly of the human umbilical cord. Stem Cells.
22:1330–1337. 2004. View Article : Google Scholar
|
|
19
|
Kakinuma S, Tanaka Y, Chinzei R, Watanabe
M, Shimizu-Saito K, Hara Y, Teramoto K, Arii S, Sato C, Takase K,
et al: Human umbilical cord blood as a source of transplantable
hepatic progenitor cells. Stem Cells. 21:217–227. 2003. View Article : Google Scholar
|
|
20
|
Tang XP, Zhang M, Yang X, Chen LM and Zeng
Y: Differentiation of human umbilical cord blood stem cells into
hepatocytes in vivo and in vitro. World J Gastroenterol.
12:4014–4019. 2006. View Article : Google Scholar
|
|
21
|
Mayani H, Wagner JE and Broxmeyer HE: Cord
blood research, banking, and transplantation: Achievements,
challenges, and perspectives. Bone Marrow Transplant. 55:48–61.
2020. View Article : Google Scholar
|
|
22
|
Shetty P, Cooper K and Viswanathan C:
Comparison of proliferative and multilineage differentiation
potentials of cord matrix, cord blood, and bone marrow mesenchymal
stem cells. Asian J Transfus Sci. 4:14–24. 2010. View Article : Google Scholar
|
|
23
|
Han JY, Goh RY, Seo SY, Hwang TH, Kwon HC,
Kim SH, Kim JS, Kim HJ and Lee YH: Cotransplantation of cord blood
hematopoietic stem cells and culture-expanded and
GM-CSF-/SCF-transfected mesenchymal stem cells in SCID mice. J
Korean Med Sci. 22:242–247. 2007. View Article : Google Scholar
|
|
24
|
Hutton JF, D'Andrea RJ and Lewis ID:
Potential for clinical ex vivo expansion of cord blood haemopoietic
stem cells using non-haemopoietic factor supplements. Curr Stem
Cell Res Ther. 2:229–237. 2007. View Article : Google Scholar
|
|
25
|
Demerdash Z, El-Baz HG, Maher K, Hassan S,
Salah F, Hassan M, Elzallat M, El-Shafei M and Taha T: Effect of
repeated passaging and cell density on proliferation and
differentiation potential of cord blood unrestricted somatic stem
cells. New Horiz Transl Med. 2:672015.
|
|
26
|
Esmaeili M, Niazi V, Pourfathollah AA,
Hosseini MKM, Nakhlestani M, Golzadeh K, Taheri M, Ghafouri-Fard S
and Atarodi K: The impact of parathyroid hormone treated
mesenchymal stem cells on ex-vivo expansion of cord blood
hematopoietic stem cells. Gene Rep. 17:1004902019. View Article : Google Scholar
|
|
27
|
Mokhtari S, Baptista PM, Vyas DA, Freeman
CJ, Moran E, Brovold M, Llamazares GA, Lamar Z, Porada CD, Soker S
and Almeida-Porada G: Evaluating interaction of cord blood
hematopoietic stem/progenitor cells with functionally integrated
three-dimensional microenvironments. Stem Cells Transl Med.
7:271–282. 2018. View Article : Google Scholar
|
|
28
|
Chaurasia P, Gajzer DC, Schaniel C,
D'Souza S and Hoffman R: Epigenetic reprogramming induces the
expansion of cord blood stem cells. J Clin Invest. 124:2378–2395.
2014. View Article : Google Scholar
|
|
29
|
Li Q, Zhao D, Chen Q, Luo M, Huang J, Yang
C, Wang F, Li W and Liu T: Wharton's jelly mesenchymal stem
cell-based or umbilical vein endothelial cell-based serum-free
coculture with cytokines supports the ex vivo expansion/maintenance
of cord blood hematopoietic stem/progenitor cells. Stem Cell Res
Ther. 10:3762019. View Article : Google Scholar
|
|
30
|
Zhang B, Wu X, Zhang X, Sun Y, Yan Y, Shi
H, Zhu Y, Wu L, Pan Z, Zhu W, et al: Human umbilical cord
mesenchymal stem cell exosomes enhance angiogenesis through the
Wnt4/β-catenin pathway. Stem Cells Transl Med. 4:513–522. 2015.
View Article : Google Scholar
|
|
31
|
Rim YA, Nam Y and Ju JH: Application of
cord blood and cord blood-derived induced pluripotent stem cells
for cartilage regeneration. Cell Transplant. 28:529–537. 2019.
View Article : Google Scholar
|
|
32
|
Zheng YL, Sun YP, Zhang H, Liu WJ, Jiang
R, Li WY, Zheng YH and Zhang ZG: Mesenchymal stem cells obtained
from synovial fluid mesenchymal stem cell-derived induced
pluripotent stem cells on a matrigel coating exhibited enhanced
proliferation and differentiation potential. PLoS One.
10:e01442262015. View Article : Google Scholar
|
|
33
|
Zhou RQ, Wu JH, Gong YP, Guo Y and Xing
HY: Transcription factor SCL/TAL1 mediates the phosphorylation of
MEK/ERK pathway in umbilical cord blood CD34+ stem cells
during hematopoietic differentiation. Blood Cells Mol Dis.
53:39–46. 2014. View Article : Google Scholar
|
|
34
|
Ajami M, Soleimani M, Abroun S and Atashi
A: Comparison of cord blood CD34 + stem cell expansion in coculture
with mesenchymal stem cells overexpressing SDF-1 and
soluble/membrane isoforms of SCF. J Cell Biochem. 120:15297–15309.
2019. View Article : Google Scholar
|
|
35
|
Naka K, Muraguchi T, Hoshii T and Hirao A:
Regulation of reactive oxygen species and genomic stability in
hematopoietic stem cells. Antioxid Redox Signal. 10:1883–1894.
2008. View Article : Google Scholar
|
|
36
|
Bonifazi F, Dan E, Labopin M, Sessa M,
Guadagnuolo V, Ferioli M, Rizzi S, De Carolis S, Sinigaglia B,
Motta MR, et al: Intrabone transplant provides full stemness of
cord blood stem cells with fast hematopoietic recovery and low GVHD
rate: Results from a prospective study. Bone Marrow Transplant.
54:717–725. 2019. View Article : Google Scholar
|
|
37
|
Lee YH: Clinical utilization of cord blood
over human health: Experience of stem cell transplantation and cell
therapy using cord blood in Korea. Korean J Pediatr. 57:110–116.
2014. View Article : Google Scholar
|
|
38
|
Li X, Ma X, Chen Y, Peng D, Wang H, Chen
S, Xiao Y, Li L, Zhou H, Cheng F, et al: Coinhibition of activated
p38 MAPKα and mTORC1 potentiates stemness maintenance of HSCs from
SR1-expanded human cord blood CD34+ cells via inhibition
of senescence. Stem Cells Transl Med. 9:1604–1616. 2020. View Article : Google Scholar
|
|
39
|
Fares I, Chagraoui J, Gareau Y, Gingras S,
Ruel R, Mayotte N, Csaszar E, Knapp DJ, Miller P, Ngom M, et al:
Cord blood expansion. Pyrimidoindole derivatives are agonists of
human hematopoietic stem cell self-renewal. Science. 345:1509–1512.
2014. View Article : Google Scholar
|
|
40
|
Seghatoleslam M, Jalali M, Alamdari DH,
Nikravesh MR, Hosseini SM and Fazel AR: Effect of incubation time
on the in vitro labeling of umbilical cord blood hematopoietic stem
cells with bromodeoxyuridine (BrdU). Clin Biochem. 44
(Suppl):S1532011. View Article : Google Scholar
|
|
41
|
Walsh C and Cepko CL: Widespread
dispersion of neuronal clones across functional regions of the
cerebral cortex. Science. 255:434–440. 1992. View Article : Google Scholar
|
|
42
|
Gerrits A, Dykstra B, Kalmykowa OJ, Klauke
K, Verovskaya E, Broekhuis MJ, de Haan G and Bystrykh LV: Cellular
barcoding tool for clonal analysis in the hematopoietic system.
Blood. 115:2610–2618. 2010. View Article : Google Scholar
|
|
43
|
Zorita E, Cuscó P and Filion GJ: Starcode:
Sequence clustering based on all-pairs search. Bioinformatics.
31:1913–1919. 2015. View Article : Google Scholar
|
|
44
|
Schepers K, Swart E, van Heijst JW,
Gerlach C, Castrucci M, Sie D, Heimerikx M, Velds A, Kerkhoven RM,
Arens R and Schumacher TN: Dissecting T cell lineage relationships
by cellular barcoding. J Exp Med. 205:2309–2318. 2008. View Article : Google Scholar
|
|
45
|
Kristiansen TA, Jaensson Gyllenbäck E,
Zriwil A, Björklund T, Daniel JA, Sitnicka E, Soneji S, Bryder D
and Yuan J: Cellular barcoding links B-1a B cell potential to a
fetal hematopoietic stem cell state at the single-cell level.
Immunity. 45:346–357. 2016. View Article : Google Scholar
|
|
46
|
Lu R, Neff NF, Quake SR and Weissman IL:
Tracking single hematopoietic stem cells in vivo using
high-throughput sequencing in conjunction with viral genetic
barcoding. Nat Biotechnol. 29:928–933. 2011. View Article : Google Scholar
|
|
47
|
Naik SH, Perié L, Swart E, Gerlach C, van
Rooij N, de Boer RJ and Schumacher TN: Diverse and heritable
lineage imprinting of early haematopoietic progenitors. Nature.
496:229–232. 2013. View Article : Google Scholar
|
|
48
|
Verovskaya E, Broekhuis MJ, Zwart E,
Ritsema M, van Os R, de Haan G and Bystrykh LV: Heterogeneity of
young and aged murine hematopoietic stem cells revealed by
quantitative clonal analysis using cellular barcoding. Blood.
122:523–532. 2013. View Article : Google Scholar
|
|
49
|
Keller G, Paige C, Gilboa E and Wagner EF:
Expression of a foreign gene in myeloid and lymphoid cells derived
from multipotent haematopoietic precursors. Nature. 318:149–154.
1985. View Article : Google Scholar
|
|
50
|
Lemischka IR, Raulet DH and Mulligan RC:
Developmental potential and dynamic behavior of hematopoietic stem
cells. Cell. 45:917–927. 1986. View Article : Google Scholar
|
|
51
|
Ludwig LS, Lareau CA, Ulirsch JC,
Christian E, Muus C, Li LH, Pelka K, Ge W, Oren Y, Brack A, et al:
Lineage tracing in humans enabled by mitochondrial mutations and
single-cell genomics. Cell. 176:1325–1339.e22. 2019. View Article : Google Scholar
|
|
52
|
Wagner DE, Weinreb C, Collins ZM, Briggs
JA, Megason SG and Klein AM: Single-cell mapping of gene expression
landscapes and lineage in the zebrafish embryo. Science.
360:981–987. 2018. View Article : Google Scholar
|
|
53
|
Guo C, Kong W, Kamimoto K, Rivera-Gonzalez
GC, Yang X, Kirita Y and Morris SA: CellTag Indexing: Genetic
barcode-based sample multiplexing for single-cell genomics. Genome
Biol. 20:902019. View Article : Google Scholar
|
|
54
|
Bramlett C, Jiang D, Nogalska A, Eerdeng
J, Contreras J and Lu R: Clonal tracking using embedded viral
barcoding and high-throughput sequencing. Nat Protoc. 15:1436–1458.
2020. View Article : Google Scholar
|
|
55
|
Pei W, Feyerabend TB, Rössler J, Wang X,
Postrach D, Busch K, Rode I, Klapproth K, Dietlein N, Quedenau C,
et al: Polylox barcoding reveals haematopoietic stem cell fates
realized in vivo. Nature. 548:456–460. 2017. View Article : Google Scholar
|
|
56
|
Pei W, Wang X, Rössler J, Feyerabend TB,
Hofer T and Rodewald HR: Using Cre-recombinase-driven Polylox
barcoding for in vivo fate mapping in mice. Nat Protoc.
14:1820–1840. 2019. View Article : Google Scholar
|
|
57
|
Cong L, Ran FA, Cox D, Lin S, Barretto R,
Habib N, Hsu PD, Wu X, Jiang W, Marraffini LA and Zhang F:
Multiplex genome engineering using CRISPR/Cas systems. Science.
339:819–823. 2013. View Article : Google Scholar
|
|
58
|
McKenna A, Findlay GM, Gagnon JA, Horwitz
MS, Schier AF and Shendure J: Whole-organism lineage tracing by
combinatorial and cumulative genome editing. Science.
353:aaf79072016. View Article : Google Scholar
|
|
59
|
Frieda KL, Linton JM, Hormoz S, Choi J,
Chow KK, Singer ZS, Budde MW, Elowitz MB and Cai L: Synthetic
recording and in situ readout of lineage information in single
cells. Nature. 541:107–111. 2017. View Article : Google Scholar
|
|
60
|
Perli SD, Cui CH and Lu TK: Continuous
genetic recording with self-targeting CRISPR-Cas in human cells.
Science. 353:aag05112016. View Article : Google Scholar
|
|
61
|
Kalhor R, Mali P and Church GM: Rapidly
evolving homing CRISPR barcodes. Nat Methods. 14:195–200. 2017.
View Article : Google Scholar
|
|
62
|
Kalhor R, Kalhor K, Mejia L, Leeper K,
Graveline A, Mali P and Church GM: Developmental barcoding of whole
mouse via homing CRISPR. Science. 361:eaat98042018. View Article : Google Scholar
|
|
63
|
Loveless TB, Grotts JH, Schechter MW,
Forouzmand E, Carlson CK, Agahi BS, Liang G, Ficht M, Liu B, Xie X
and Liu CC: DNA writing at a single genomic site enables lineage
tracing and analog recording in mammalian cells. bioRxiv.
6391202019.
|
|
64
|
Bowling S, Sritharan D, Osorio FG, Nguyen
M, Cheung P, Rodriguez-Fraticelli A, Patel S, Yuan WC, Fujiwara Y,
Li BE, et al: An engineered CRISPR-Cas9 mouse line for simultaneous
readout of lineage histories and gene expression profiles in single
cells. Cell. 181:1410–1422.e27. 2020. View Article : Google Scholar
|
|
65
|
Nguyen LV, Cox CL, Eirew P, Knapp DJ,
Pellacani D, Kannan N, Carles A, Moksa M, Balani S, Shah S, et al:
DNA barcoding reveals diverse growth kinetics of human breast
tumour subclones in serially passaged xenografts. Nat Commun.
5:58712014. View Article : Google Scholar
|
|
66
|
Naik SH, Schumacher TN and Perie L:
Cellular barcoding: A technical appraisal. Exp Hematol. 42:598–608.
2014. View Article : Google Scholar
|
|
67
|
Nguyen LV, Pellacani D, Lefort S, Kannan
N, Osako T, Makarem M, Cox CL, Kennedy W, Beer P, Carles A, et al:
Barcoding reveals complex clonal dynamics of de novo transformed
human mammary cells. Nature. 528:267–271. 2015. View Article : Google Scholar
|
|
68
|
McKenzie JL, Gan OI, Doedens M, Wang JC
and Dick JE: Individual stem cells with highly variable
proliferation and self-renewal properties comprise the human
hematopoietic stem cell compartment. Nat Immunol. 7:1225–1233.
2006. View
Article : Google Scholar
|
|
69
|
Gonzalez-Murillo A, Lozano ML, Montini E,
Bueren JA and Guenechea G: Unaltered repopulation properties of
mouse hematopoietic stem cells transduced with lentiviral vectors.
Blood. 112:3138–3147. 2008. View Article : Google Scholar
|
|
70
|
Golden JA, Fields-Berry SC and Cepko CL:
Construction and characterization of a highly complex retroviral
library for lineage analysis. Proc Natl Acad Sci USA. 92:5704–5708.
1995. View Article : Google Scholar
|
|
71
|
Adamson B, Norman TM, Jost M, Cho MY,
Nuñez JK, Chen Y, Villalta JE, Gilbert LA, Horlbeck MA, Hein MY, et
al: A multiplexed single-cell CRISPR screening platform enables
systematic dissection of the unfolded protein response. Cell.
167:1867–1882.e21. 2016. View Article : Google Scholar
|
|
72
|
Cheung AM, Nguyen LV, Carles A, Beer P,
Miller PH, Knapp DJ, Dhillon K, Hirst M and Eaves CJ: Analysis of
the clonal growth and differentiation dynamics of primitive
barcoded human cord blood cells in NSG mice. Blood. 122:3129–3137.
2013. View Article : Google Scholar
|
|
73
|
Belderbos ME, Jacobs S, Koster TK, Ausema
A, Weersing E, Zwart E, de Haan G and Bystrykh LV: Donor-to-donor
heterogeneity in the clonal dynamics of transplanted human cord
blood stem cells in murine xenografts. Biol Blood Marrow
Transplant. 26:16–25. 2020. View Article : Google Scholar
|
|
74
|
Sun J, Ramos A, Chapman B, Johnnidis JB,
Le L, Ho YJ, Klein A, Hofmann O and Camargo FD: Clonal dynamics of
native haematopoiesis. Nature. 514:322–327. 2014. View Article : Google Scholar
|
|
75
|
Cai WQ, Zeng LS, Wang LF, Wang YY, Cheng
JT, Zhang Y, Han ZW, Zhou Y, Huang SL, Wang XW, et al: The latest
battles between EGFR monoclonal antibodies and resistant tumor
cells. Front Oncol. 10:12492020. View Article : Google Scholar
|
|
76
|
Han ZW, Lyv ZW, Cui B, Wang YY, Cheng JT,
Zhang Y, Cai WQ, Zhou Y, Ma ZW, Wang XW, et al: Correction to: The
old CEACAMs find their new role in tumor immunotherapy. Invest New
Drugs. 38:1899–1900. 2020. View Article : Google Scholar
|
|
77
|
Wang YY, Lyu YN, Xin HY, Cheng JT, Liu XQ,
Wang XW, Peng XC, Xiang Y, Xin VW, Lu CB, et al: Identification of
putative UL54 (ICP27) transcription regulatory sequences binding to
Oct-1, v-Myb, Pax-6 and hairy in herpes simplex viruses. J Cancer.
10:430–440. 2019. View Article : Google Scholar
|
|
78
|
Jensen P and Dymecki SM: Essentials of
recombinase-based genetic fate mapping in mice. Methods Mol Biol.
1092:437–454. 2014. View Article : Google Scholar
|
|
79
|
Herring CA, Chen B, McKinley ET and Lau
KS: Single-cell computational strategies for lineage reconstruction
in tissue systems. Cell Mol Gastroenterol Hepatol. 5:539–548. 2018.
View Article : Google Scholar
|
|
80
|
Liu XQ, Xin HY, Lyu YN, Ma ZW, Peng XC,
Xiang Y, Wang YY, Wu ZJ, Cheng JT, Ji JF, et al: Oncolytic herpes
simplex virus tumor targeting and neutralization escape by
engineering viral envelope glycoproteins. Drug Deliv. 25:1950–1962.
2018. View Article : Google Scholar
|
|
81
|
Woodworth MB, Girskis KM and Walsh CA:
Building a lineage from single cells: Genetic techniques for cell
lineage tracking. Nat Rev Genet. 18:230–244. 2017. View Article : Google Scholar
|
|
82
|
Xu J, Nuno K, Litzenburger UM, Qi Y,
Corces MR, Majeti R and Chang HY: Single-cell lineage tracing by
endogenous mutations enriched in transposase accessible
mitochondrial DNA. Elife. 8:e451052019. View Article : Google Scholar
|