Introduction
For worldwide, ovarian cancer (OVC) is the one of
top 10 most common type of women cancer, while as the same common
as in China (1). Early diagnosis
of OVC is difficult to achieve, as the main symptoms are not
OVC-specific (2). Therefore, most
patients with OVC are diagnosed with metastatic cancer, with a
5-year survival rate of 29% (3,4).
OVC can metastasize through two different mechanisms (5–7):
i) The primary tumor can expand and directly infiltrate adjacent
organs, including the bladder and colon; or ii) cancer cells can
become isolated from the primary tumor and spread to the peritoneal
cavity, an event usually associated with the formation of ascites.
OVC is easy to metastasize, and patients with OVC often present
with advanced pelvic disease (8),
including expansion of the cancer into the uterus, fallopian tubes
and ovaries (9). In vivo
animal models are essential tools for cancer research (10); they have enabled the
identification of carcinogens, the development of cancer therapies
(11) and high-throughput drug
screening (12), and improved our
understanding of the molecular mechanisms of tumor growth and
metastasis (13).
OVC models must be established to enhance our
current understanding of the biological characteristics of OVC and
to develop novel and improved therapeutics (14). Mice are often used as in
vivo models of human disease owing to high similarities of gene
homologous. Previous experiments on Microtus fortis found
that in some established strains the natural incidence of OVC in
adult female voles is markedly higher than that exhibited in other
animals in the Department of Laboratory Animal Science, Xiangya
Medical College, Central South University (15). Through clinical and pathological
observations, it is demonstrated that M. fortis OVC shares a
number of similarities with human OVC (16,17). To elucidate the underlying
mechanisms of human OVC pathogenesis, the pathological
characteristics and histological classification of spontaneous
epithelial OVC in M. fortis can be used.
Unlike mouse models, less available biological
information is the limitation of M. fortis as a novel
experimental animal model. It is therefore challenging to study the
underlying mechanisms of primary OVC in M. fortis using
techniques such as flow cytometry, western blotting and microarrays
(18). De novo RNA-seq
technology is useful to analyze gene expression and identify novel
genes (19). Therefore, the
present study aimed to sequence and compare the M. fortis
transcriptomes in OVC and healthy ovarian (OV) tissues and between
fallopian tube cancer (FTC) and healthy fallopian tube (FT)
tissues, using Illumina sequencing platform. The results
demonstrated the suitability of short-read sequencing for de novo
transcriptome assembly and for the annotation of genes expressed in
a eukaryote with no previous genome information. The present study
also identified differentially expressed genes (DEGs) between FTC
and FT and between OVC and OV tissue samples. Gene Ontology (GO)
and Kyoto Encyclopedia of Genes and Genomes (KEGG) enrichment
analyses were performed to determine the functional relationships
of the identified DEGs. The results demonstrated that cysteine-rich
angiogenic inducer 61 (CYR61), Ras-related protein Rap-1b (RAP1B),
Ras homolog family member A (RHOA), RAC1 and BTG antiproliferation
factor (BTG) 3 may serve an important role in the pathogenesis of
primary OVC in M. fortis.
Materials and methods
Animals and sample preparation
M. fortis, originating from the Dongting Lake
area (Yueyang, China) were bred in the laboratory. All animals were
raised in cages maintained in a 12-h light/dark cycle at a
temperature of 22±1.5°C and 50±5% relative humidity. Animals were
fed with standard formula feed for M. fortis (sterilized by
γ-radiation from a Cobalt-60 source) and sterile water. The housing
environment was controlled by an automatic heating and ventilation
device, and the bedding materials were sterilized in advance.
During the pedigree breeding process, M. fortis were found
to have OVC or FTC with an unstable ratio (~4%) as the number of
animals with OVC or FTC changed in each generation. In the present
study, a total of 110 M. fortis females were produced from
the breeding process and four were obtained with cancer. The
average age of the animals discovered with cancer was 1 year while
the average lifetime is 3-4 years. The animals with cancer were
observed to have ascites and sluggish movements (maximum weight
gain 100 g; Fig. S1). M.
fortis were euthanized by 4% isoflurane overdose, following an
induction with 4% isoflurane, using an anesthesia machine (ABM
model; Shanghai Yuyan Instruments Co., Ltd.); animals underwent
prolonged exposure to 4% isoflurane to ensure death (>3 min).
Death was verified when there was no breathing for >2 min and no
blink reflex (20,21). All animal experiments were
performed according to the Laboratory Animal Guidelines for
Euthanasia (T/CALAS 31-2017; Chinese Association for Laboratory
Animal Sciences; http://www.calas.org.cn/index.php?m=content&c=index&a=show&catid=19&id=1007)
within the recommendations of the Laboratory of Animal Welfare
& Ethics Committee of China. The protocol was approved by the
Laboratory Animal Welfare and Ethics Committee of Central South
University (Changsha, China; approval no. 2018sydw0236). Cancerous
and healthy tissues were collected from four animals with OVC and
two healthy animals. Samples were stored in an ultra-low
temperature freezer (−80°C).
Pathological observations
For individuals with ascites, cancer was diagnosed
using histopathologic hematoxylin and eosin (H&E) staining
(Fig. S1). The fresh tissue was
fixed with a FAA fixative solution (catalogue no. G1103-500ML;
ServiceBio, Inc.) for >24 h and then dehydrated by gradient
alcohol in the dehydrator (Histocentre 3; Thermo Fisher Scientific,
Inc.) and soaked in paraffin wax. The paraffin wax-soaked tissues
were cooled on a −20°C freezing table and then sliced to of 3 µm
using a rotary microtome (Finesse E+; Thermo Fisher Scientific,
Inc.). The slices were floated in 40°C warm water to flatten the
tissue and then baked in an oven at 60°C 1 h. Slices were dried and
then stored at room temperature. The sections were incubated with
xylene for 40 min, absolute ethanol for 10 min and 75% ethanol for
5 min, then washed with distilled water, stained with hematoxylin
for 5 min and then differentiated with 1% hydrochloric acid and
alcohol for 10 sec (all at room temperature). The slices were
washed with distilled water, then returned to blue for 20 min at
room temperature with 0.6% ammonia water and rinsed with running
distilled water. Slices were separately dehydrated 5 min at room
temperature with 85 and 95% alcohol in succession and then stained
in eosin solution for 5 min. The slices were successively put into
anhydrous ethanol 15 min and xylene 15 min at room temperature
until they were transparent and were then sealed with neutral gum.
All the above incubation, dewaxing and dyeing processes were
performed at room temperature. The morphological changes in the
tissues were observed under an optical light microscope for 22
fields (BX43; Olympus Corp.).
cDNA library construction and Illumina
sequencing
Total RNA of 12 M. fortis ovary (two healthy
and four cancerous) and fallopian tube (two healthy and four
cancerous) tissue samples was extracted using the RNeasy mini kit
(Qiagen GmbH). RNA degradation and contamination were monitored on
1% agarose gels. RNA purity was checked using a
NanoPhotometer® spectrophotometer (Implen, Inc.). RNA
concentration was measured using a Qubit® RNA Assay kit
and a Qubit® 2.0 Fluorometer (Thermo Fisher Scientific,
Inc.). RNA integrity was assessed using the RNA Nano 6000 Assay kit
and the Agilent Bioanalyzer 2100 system (Agilent Technologies,
Inc.). A total amount of 1.5 µg RNA per sample was used as input
material. Sequencing libraries were generated using the
NEBNext® Ultra™ RNA Library Prep kit for
Illumina® (catalogue no. E7530L; New England BioLabs,
Inc.) according to the manufacturer's protocol. Briefly, mRNA was
purified from total RNA using poly-T oligo-attached magnetic beads.
mRNA was fragmented when mixed with fragmentation buffer. cDNA was
synthesized using the mRNA fragments as templates. To select cDNA
fragments of the right length, the library fragments were purified
using the AMPure XP system (Beckman Coulter, Inc.). Subsequently, 3
µl USER enzyme (New England BioLabs, Inc.) was incubated with the
size-selected, adaptor-ligated cDNA at 37°C for 15 min followed by
5 min at 95°C prior to PCR. PCR was performed using Phusion
High-Fidelity DNA polymerase (catalogue no. M0530L; NEB, Inc.),
universal PCR primers and PCR index (Table SI), following initial
denaturation (94°C, 1 min); 12 of cycles of denaturation (94°C, 30
sec), annealing (58°C, 30 sec) and elongation (72°C, 30 sec), and
final extension (72°C, 2 min). Finally, products were purified
(AMPure XP system) and library quality was assessed using the
Agilent Bioanalyzer 2100 system. From these libraries (3–7 ng/µl,
measured by Qubit), 150 bp paired-end and strand-specific sequence
reads were produced using the Illumina HiSeq X Ten (Illumina, Inc.)
at E-GENE Co., Ltd.
Quality control and de novo assembly
for transcriptome analysis
The raw reads were initially processed using
Illumina sequencing, aforementioned. Quality control of raw reads
was performed using FastQC (version 0.11.5; www.bioinformatics.babraham.ac.uk/projects/fastqc),
and clean reads were obtained following the removal of low-quality
reads with Trimmomatic (version 0.36; http://github.com/usadellab/Trimmomatic) software
(22). Subsequently, de novo
transcriptome assembly was performed using the short reads assembly
program, Trinity (version 2.3.2, http://github.com/trinityrnaseq/trinityrnaseq/wiki)
(23). The final sequences
obtained after assembly were termed unigenes. Clean reads were
mapped to the de novo assembly transcriptome reference sequences of
M. fortis using Bowtie2 (version 2.4.1, http://bowtie-bio.sourceforge.net/bowtie2/index.shtml).
The expression levels of unigenes were calculated using the RSEM
algorithm. The hierarchical cluster analysis was performed via
function hclust in vegan R package (version 2.51. http://www.R-project.org/) (24).
Functional annotation
TransDecoder (version 2.0.1;
github.com/TransDecoder/TransDecoder/wiki) was used to identify
open reading frames (ORFs) and to predict potential coding
sequences in the assembled unigenes. After ORFs were extracted from
the assembly, the Trinotate (version 2.0.2
github.com/Trinotate/Trinotate) pipeline was used to annotate the
M. fortis transcript ORF dataset. Both nucleotide
transcripts and protein sequences were used to search against the
UniProt Knowledgebase (KB)/Swiss-Prot databases (www.uniprot.org), using basic local alignment search
tool BLASTx and BLASTp [version 2.2.28+; cut-off level (E-value),
1×10−5], respectively. The UniProtKB/Swiss-Prot database
is a collection of functional information on proteins, with
accurate, consistent and rich annotation. Protein domains were
annotated using the Pfam domain database with HMMER (version 3.1,
http://www.hmmer.org/) (25). Potential signal peptides were
predicted using SignalP (version 4.1) (26). All M. fortis transcriptome
annotations were loaded into the SQLite database (https://www.sqlite.org/index.html) and the
annotated results were exported to an excel file.
Abundance estimating and screening of
DEGs
The expression of each unigene was quantified by
RNA-seq quantification analysis using RNA-Seq by
Expectation-Maximization (RSEM) software (version 1.2.31,
http://github.com/deweylab/RSEM).
Unigenes that were expressed at low levels (FPKM in all samples
<1.0) were removed from the database. DEGs between OVC and OV
and between FTC and FT tissues were determined using the DESeq2 R
package (27) (q-value <0.001;
log2 fold-change >1). GO terms (geneontology.org) were
distributed into ‘Biological Processes’ (BP), ‘Cellular Components’
(CC) and ‘Molecular Functions’ (MF). KEGG (www.kegg.jp) is the main public database related to
pathways, usually used for enrichment analysis of DEGs.
Subsequently, GO term and KEGG pathway enrichment analyses for the
DEGs were executed using GOstats (version 2.50.0, http://bioconductor.riken.jp/packages/release/bioc/html/GOstats.html)
and GSEABase (version 1.46.0, http://bioconductor.riken.jp/packages/3.0/bioc/html/GSEABase.html)
packages with P<0.01 as a threshold, according to guidance from
Trinity.
Results
Illumina sequencing and de novo
transcriptome assembly
To obtain an overview of the M. fortis gene
expression profile, cDNA from four OVC, two OV, four FTC and two FT
tissues was generated and sequenced using the Illumina sequencing
platform. After removing the adaptor sequences and low-quality
reads, 331,138,454 clean paired-end reads were obtained from the 12
sequenced samples (average size, 7.86±1.06 Gb). The average GC
content and average Q30 values were 48.67 and 96.72%, respectively
(Table SII). The clean reads
were assembled using Trinity software. A total of 521,853 contigs
and 339,830 unigenes (average length, 874.6 bp; N50, 1,444 bp) were
obtained (Table SIII). The read
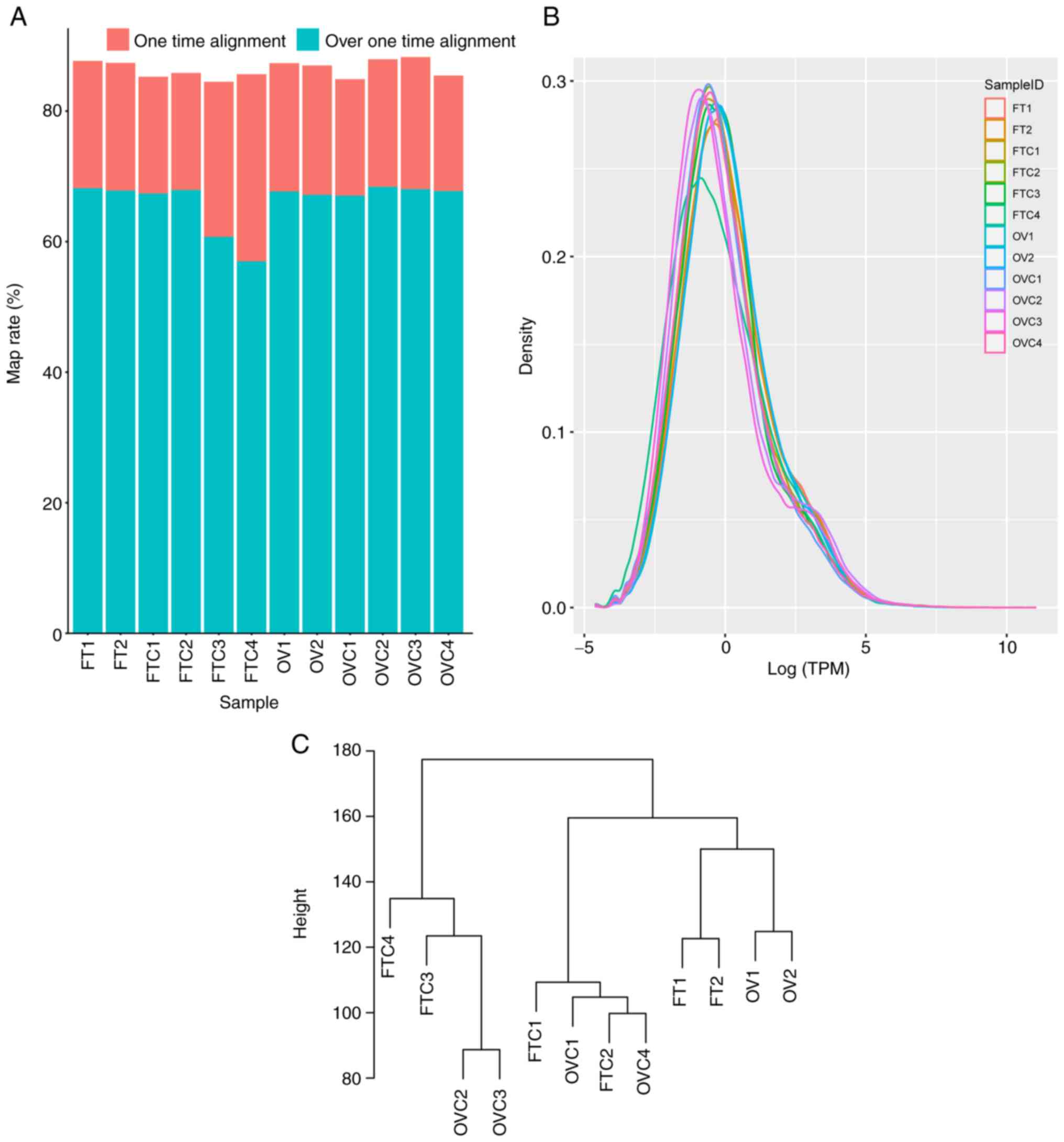
mapping ratio of all samples was >80% (Fig. 1A). Density analysis of all unigene
expression levels in almost all samples displayed a similar
expression pattern except FTC4, while the lower expression level
genes of FTC4 were more compared with the other samples (Fig. 1B). After removing low expression
(FPKM <1.0 in all samples) unigenes, 104,443 unigenes remained.
Cluster analysis indicated that there were not only differences in
expression between healthy and cancerous tissues, but also between
different organs (Fig. 1C).
Functional annotation and
classification of assembled transcripts
To annotate the unigenes, sequences of unigenes were
searched using BLASTx against the Swiss-Prot protein and Pfam
databases with a cut-off E-value of 1×10−5. A total of
44,511 (42.62%) genes and 13,949 (13.35%) unigenes were identified
as significant hits using the respective databases (Table SIV). These annotated results
demonstrated that the annotated sequences percentages of species,
Mus musculus (52.25%), Homo sapiens (13.84%) and
Rattus norvegicus (13.48%) were highly homologous with M.
fortis. In the GO database 40,725 unigenes were annotated to
15659 GO number. Among these annotated unigenes, 11,277 (27.69%)
were categorized as cytoplasm belonged to CC terms, 9,017 (22.14%)
as nucleus belonged to CC, and 8,935 (21.94%) as metal ion binding
belonged to MF (Table SV).
In the present study, 21,206 unigenes were mapped to
479 KEGG pathways (Table SVI),
including ‘chromosome and associated proteins’ (2,148 unigenes to
734 koIDs; 10.13% of sequences), ‘membrane trafficking’ (2101
unigenes to 547 koIDs; 10.40% of sequences), ‘ubiquitin system’
(1,830 unigenes to 548 koIDs; 8.63% of sequences), ‘exosome’ (1,725
unigenes to 508 koIDs; 8.13% of sequences), ‘transcription factors’
(1,222 unigenes to 461 koIDs; 5.78% of sequences), ‘transporters’
(1107 unigenes to 496 koIDs; 5.22% of sequences), ‘protein kinases’
(1042 unigenes to 367 koIDs; 4.91% of sequences) ‘peptidases’ (983
unigenes to 397 koIDs; 4.64% of sequences), ‘Mitochondrial
biogenesis’ (982 unigenes to 230 koIDs; 4.63% of sequences) and
‘pathways in cancer’ (922 unigenes to 346 koIDs; 4.35% of
sequences). Out of the 479 KEGG pathways, 16 were identified as
being involved in cancer, including, ‘pathways in cancer’ (922
unigenes to 346 koIDs; 4.35%), ‘proteoglycans in cancer’ (426
unigenes to 143 koIDs; 2.01%), ‘microRNAs in cancer’ (297 unigenes
to 115 koIDs; 1.40%), ‘gastric cancer’ (247 unigenes to 90 koIDs;
1.16%) and ‘transcriptional misregulation in cancer’ (246 unigenes
to 125 koIDs; 1.16%). The catalog of the identified unigenes
provided a broad gene transcription profile of M. fortis and
a foundation for the screening of DEGS to reveal the underlying
mechanisms of primary OVC in M. fortis.
DEGs involved in M. fortis primary
OVC
Identification and characterization of DEGs derived
from comparing FTC with FT and OVC with OV group were crucial for
revealing the underlying mechanisms of primary OVC. To investigate
the DEGs, RNA-seq technology was used. Clean reads from the four
M. fortis tissue groups were mapped to the de novo assembly
transcriptome reference sequences of M. fortis using Bowtie2
(version 2.4.1, http://bowtie-bio.sourceforge.net/bowtie2/index.shtml)
(28). The expression levels of
unigenes were calculated using the RSEM algorithm (29). In the present study,
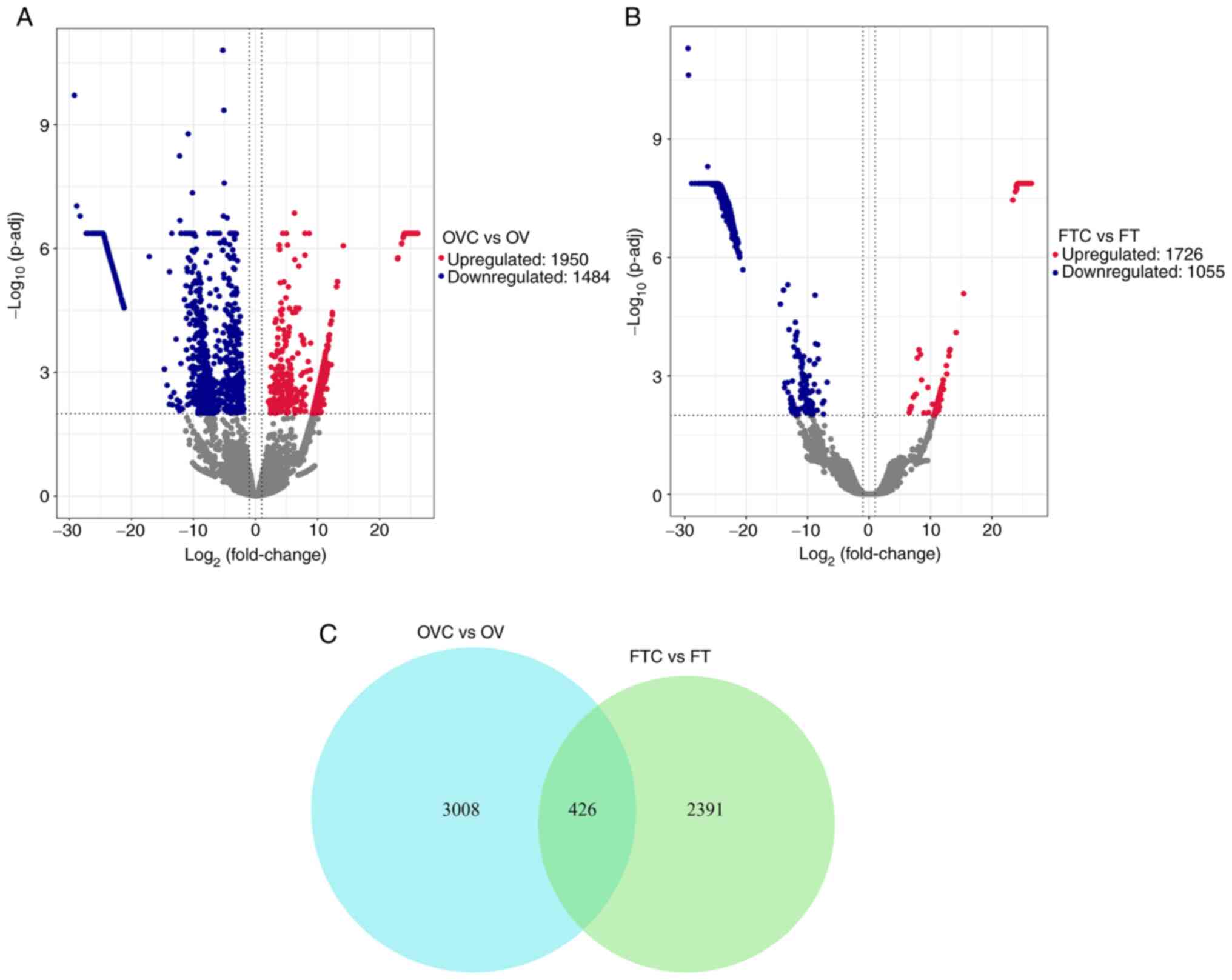
bioinformatics analysis identified 3,434 DEGs between the OVC and
the OV groups (Table SVII),
including 1,950 significantly upregulated and 1,484 significantly
downregulated genes (Fig. 2A). In
the comparison between the FTC and the FT group, 2,817 DEGs were
identified (Table SVIII),
including 1,762 significantly upregulated and 1,055 significantly
downregulated genes (Fig. 2B).
The number of overlapping DEGs between fallopian tube tissues and
ovarian tissues was 426 (Fig.
2C).
In the OVC vs. OV group, there were seven
upregulated expression genes, containing TNNC1,
RAP1B,RAC1,HA12,BTG3,CYR61 and RHOA, while only one downregulated
gene, P53. Among the seven upregulated expression genes, five
significantly upregulated DEGs were identified that were related to
OVC cell proliferation and migration: CYR61, RAP1B, RHOA, RAC1 and
BTG3. In the FTC vs. FT group, two significantly upregulated DEGs,
troponin C (TNNC1) and hyaluronan dodecasaccharides (HA12) were
identified (Fig. 3).
Overexpression of TNNC1 may be beneficial for cell transfer
(30) and high expression of HA12
may promote endothelial cell morphogenesis (31).
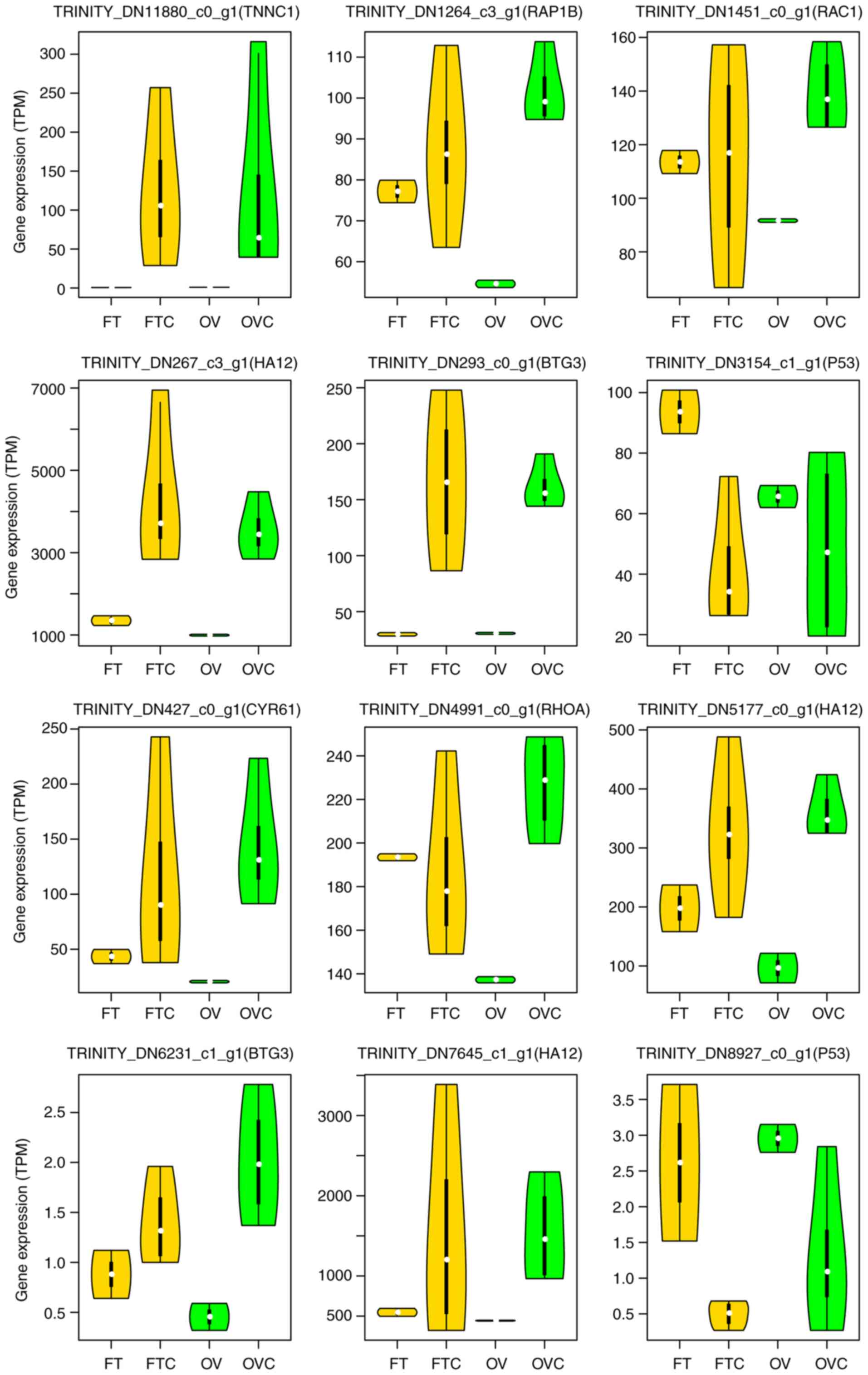 | Figure 3.Violin plots of DEGs related to
cancer development. DEGs analyzed include CYR61, RAP1B, RHOA, RAC1,
BTG3, TNNC1, HA12 and p53 (different transcripts could be annotated
with the same gene symbol). BTG3, BTG antiproliferation factor 3;
CYR61, cysteine-rich angiogenic inducer 61; DEGs, differentially
expressed genes; FT, healthy fallopian tubes; FTC, fallopian tube
cancer; HA12, hyaluronan dodecasaccharides; RAP1B, Ras-related
protein Rap-1b; RHOA, Ras homolog family member A; OV, healthy
ovarian tissue; OVC, ovarian cancer; TNNC1, troponin C. |
KEGG functional enrichment analysis of
DEGs
Investigating the statistically enriched KEGG
pathways related to the DEGs revealed that the most significant and
most frequently enriched terms for upregulated unigenes in the OVC
compared with the OV group were signaling pathway, cell growth and
proliferation-associated terms, including ‘cytoskeleton proteins’,
‘cellular senescence’, ‘cell cycle’, ‘cytokines and growth
factors’, ‘exosome’, ‘cytokine-cytokine receptor interaction’,
‘cell adhesion molecules (CAMs)’, ‘hippo signaling pathway’ and
‘TNF signaling pathway’ (Fig. 4A;
Table SIX). Moreover, the
downregulated DEGs in the OVC compared with the OV group were
enriched in steroid biosynthesis-related terms, including ‘steroid
biosynthesis’, ‘steroid hormone biosynthesis’ and ‘ovarian
steroidogenesis’ (Fig. 4B;
Table SX). In the FTC vs. FT
group comparisons, upregulated DEGs were mainly enriched in the
terms ‘exosome’ and ‘messenger RNA biogenesis’ (Fig. 4C; Table SXI), whereas downregulated DEGs
were mainly enriched in the terms ‘Ras signaling pathway’,
‘cGMP-PKG signaling pathway’, ‘G protein-coupled receptors’,
‘calcium signaling pathway’, ‘cAMP signaling pathway’ and ‘Wnt
signaling pathway’ (Fig. 4D;
Table SXII).
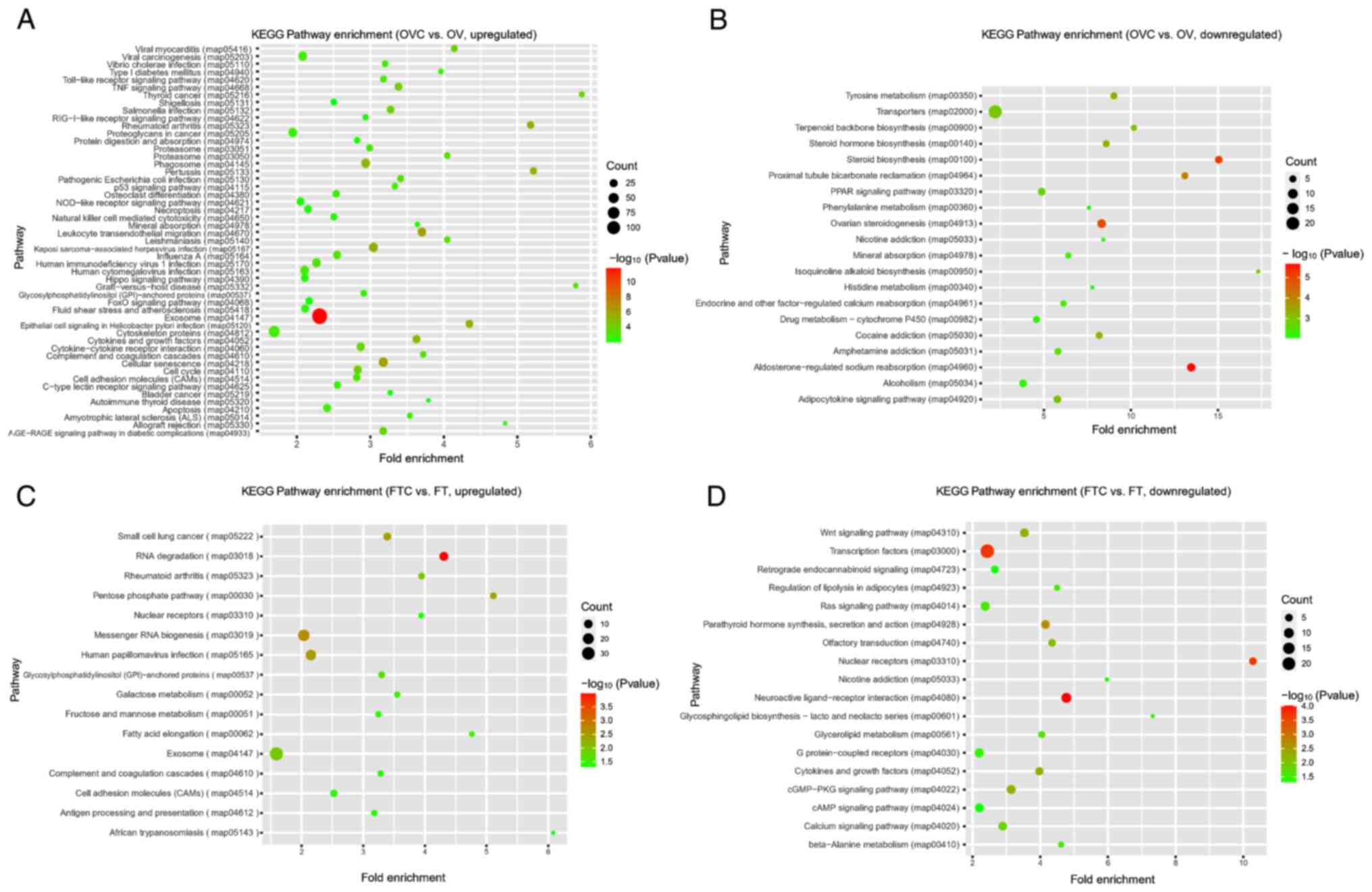 | Figure 4.KEGG enrichment analysis of DEGs. (A)
KEGG enrichment analysis of upregulated DEGs between OVC and OV
groups. (B) KEGG enrichment analysis of downregulated DEGs between
OVC and OV groups. (C) KEGG enrichment analysis of upregulated DEGs
between FTC and FT groups. (D) KEGG enrichment analysis of
downregulated DEGs between FTC and FT groups. AGE, advanced
glycation end products; cGMP, cyclic guanosine monophosphate; DEGs,
differentially expressed genes; FT, healthy fallopian tubes; FTC,
fallopian tube cancer; GTP, guanosine triphosphate; KEGG, Kyoto
Encyclopedia of Genes and Genomes; NOD, nucleotide-binding
oligomerization domain; OV, healthy ovarian tissue; OVC, ovarian
cancer; PKG, protein kinase G; PPAR, peroxisome
proliferator-activated receptor; RAGE, receptor for advanced
glycation end products. |
Discussion
The pathogenesis of OVC is complicated and,
therefore, improved animal models are required to investigate the
underlying mechanisms of OVC. Nowadays, more induced and less
spontaneous models of small animals were used for disease
researches, however spontaneous cancer model similar with human
metabolism can offer more translatable results (32). In the present study, four M.
fortis females with spontaneous OVC were identified, some of
which were hereditary. The cancerous individuals displayed ascites
and histopathological H&E staining demonstrated the presence of
papillary cancer cell structures in the ovarian tissues.
Spontaneous cancer models are valuable and understanding the
pathogenesis of spontaneous OVC in M. fortis at the genetic
level is of great importance to the stabilization of this
spontaneous ovarian cancer animal model.
During the DEG analysis, biomarkers related to the
development of cancer were identified. p53, located in the
chromosome 17 (17p13.1) and known as cancer suppressor gene, can
determine the degree of DNA variation (33). If the variation is small, cell
regeneration may be promoted; if the variation is large, apoptosis
may be induced. Furthermore, the induction of apoptosis is closely
related to cell proliferation and differentiation, with 50% of
human tumors possessing a mutation in p53 (33). However, CYR61 can reduce p53
promoter activity, thereby reducing the expression level of p53
(34). In the present study, the
expression of p53 (TRINITY_DN3154_c1_g1 and TRINITY_DN8927_c0_g1)
in cancerous tissues (both OVC and FTC) was also lower compared
with normal tissues. This suggested that in OVC tissues, CYR61 may
increase cell proliferation by inhibiting the expression of
p53.
Tumor invasion and metastasis, which are both
hallmarks of tumor malignancy (35), frequently coincide with the loss
of E-cadherin-mediated cell-cell adhesion (36). Previous studies have shown that
the overexpression of RHOA (37),
RAP1B (38) and RAC1 (39) is beneficial for tumor invasion. In
the present study, RAP1B, RHOA and RAC1, were markedly upregulated
in the OVC group compared with the OV group, which indicated that
these genes may serve a key role in promoting cell metastasis in
OVC tissues. Previous studies have reported that BTG3 can inhibit
cell proliferation and metastasis, therefore affecting cancer
development (40–42). However, it has been demonstrated
that mRNA expression levels of transducer of ERBB-2,1 (TOB1) and
BTG2 are decreased in most types of cancer compared with healthy
tissues, whereas BTG3 is upregulated (43); in the present study, the gene
expression of BTG3 was upregulated in both OVC and FTC tissues.
Further analysis of CYR61, RAP1B, RHOA, RAC1 and BTG3 may help
reveal the underlying mechanisms of primary OVC.
As M. fortis does not yet have a completely
sequenced genome, the present study employed de novo RNA-seq
technology to identify DEGs. An unclear exon and intron structure
and the poor preservation state of the residual RNA led to a lack
of molecular biology verification experiments for the identified
DEGs. In future work, following the ongoing study of the de novo
assembly of the M. fortis genome, molecular biology
verification experiments will be performed and molecular breeding
technology (using DNA markers that are tightly linked to phenotypic
traits to assist in a selection scheme for a particular breeding
objective) will be employed to build a stable natural OVC incidence
model of M. fortis.
In summary, to the best of our knowledge, the
present study was the first to characterize the M. fortis de
novo transcriptome and to perform RNA-seq analysis to determine the
genetic differences between OVC, OV, FTC and FT animal groups. The
results demonstrated that the enriched biological pathways differed
between cancerous and healthy tissues. Future analysis of these
pathways will help to further reveal the pathogenesis of primary
OVC in M. fortis. The some DEGs in this study also were
reported as marker genes associated with human OVC, such as CYR61
(44), RAP1B (45), RHOA (46), RAC1 (47) and BTG3 (48). Hence, M. fortis could be
used to better understand human OVC.
Supplementary Material
Supporting Data
Supplementary Material
Supporting Data
Supplementary Material
Supporting Data
Supplementary Material
Supporting Data
Supplementary Material
Supporting Data
Supplementary Material
Supporting Data
Supplementary Material
Supporting Data
Supplementary Material
Supporting Data
Supplementary Material
Supporting Data
Supplementary Material
Supporting Data
Supplementary Material
Supporting Data
Supplementary Material
Supporting Data
Supplementary Material
Supporting Data
Acknowledgements
Not applicable.
Funding
The present study was supported by The Natural Science
Foundation of Hunan Province (grant no. 2020JJ4110) and The Science
and Technology Planning Project of Changsha (grant no.
kq1801070).
Availability of data and materials
All raw sequencing data has been deposited in the
National Centre for Biotechnology Information Sequence Read Archive
(BioProject identifier, PRJNA687349; SRA accessions,
SRR13329310-SRR13329323).
Authors' contributions
ZZ and QH designed the experiments. QL, WZ, JW, BL
and SH performed the experiments and sample collection. QH, DZ and
MG conducted the bioinformatics analysis. QH, DZ and BL confirm the
authenticity of all the data. MG drafted the work manuscript. BL,
QH and ZZ revised the manuscript critically for important
intellectual content. All authors read and approved the final
manuscript.
Ethics approval and consent to
participate
This study was performed in strict accordance with
the recommendations of the Laboratory of Animal Welfare &
Ethics Committee of China. The protocol was approved by the
Laboratory Animal Welfare and Ethics Committee of Central South
University (Changsha, China; approval no. 2018sydw0236).
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Chen W, Zheng R, Baade PD, Zhang S, Zeng
H, Bray F, Jemal A, Yu XQ and He J: Cancer statistics in China,
2015. CA Cancer J Clin. 66:115–132. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Ishizuka B: Current Understanding of the
etiology, symptomatology, and treatment options in premature
ovarian insufficiency (POI). Front Endocrinol (Lausanne).
12:6269242021. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Chen SN, Chang R, Lin LT, Chern CU, Tsai
HW, Wen ZH, Li YH, Li CJ and Tsui KH: MicroRNA in ovarian cancer:
Biology, pathogenesis, and therapeutic opportunities. Int J Environ
Res Public Health. 16:15102019. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Stewart C, Ralyea C and Lockwood S:
Ovarian cancer: An integrated review. Semin Oncol Nurs. 35:151–156.
2019. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Lutgendorf SK and Sood AK: Biobehavioral
factors and cancer progression: Physiological pathways and
mechanisms. Psychosom Med. 73:724–730. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Bigorie V, Morice P, Duvillard P, Antoine
M, Cortez A, Flejou JF, Uzan S, Darai E and Barranger E: Ovarian
metastases from breast cancer: Report of 29 cases. Cancer.
116:799–804. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Mikula-Pietrasik J, Uruski P, Tykarski A
and Książek K: The peritoneal ‘soil’ for a cancerous ‘seed’: A
comprehensive review of the pathogenesis of intraperitoneal cancer
metastases. Cell Mol Life Sci. 75:509–525. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Parashar D, Nair B, Geethadevi A, George
J, Nair A, Tsaih SW, Kadamberi IP, Gopinadhan Nair GK, Lu Y,
Ramchandran R, et al: Peritoneal spread of ovarian cancer harbors
therapeutic vulnerabilities regulated by FOXM1 and EGFR/ERBB2
signaling. Cancer Res. 80:5554–5568. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Levy A, Medjhoul A, Caramella C, Zareski
E, Berges O, Chargari C, Boulet B, Bidault F, Dromain C and
Balleyguier C: Interest of diffusion-weighted echo-planar MR
imaging and apparent diffusion coefficient mapping in gynecological
malignancies: A review. J Magn Reson Imaging. 33:1020–1027. 2011.
View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Li Z, Zheng W, Wang H, Cheng Y, Fang Y, Wu
F, Sun G, Sun G, Lv C and Hui B: Application of animal models in
cancer research: Recent progress and future prospects. Cancer Manag
Res. 13:2455–2475. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Overgaard NH, Fan TM, Schachtschneider KM,
Principe DR, Schook LB and Jungersen G: Of Mice, Dogs, Pigs, and
Men: Choosing the appropriate model for immuno-oncology research.
ILAR J. 59:247–262. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Jayson GC, Kohn EC, Kitchener HC and
Ledermann JA: Ovarian cancer. Lancet. 384:1376–1388. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Katt ME, Placone AL, Wong AD, Xu ZS and
Searson PC: In Vitro tumor models: Advantages, disadvantages,
variables, and selecting the right platform. Front Bioeng
Biotechnol. 4:122016. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Kamdem JP, Duarte AE, Ibrahim M, Lukong
KE, Barros LM and Roeder T: Bibliometric analysis of personalized
humanized mouse and Drosophila models for effective
combinational therapy in cancer patients. Biochim Biophys Acta Mol
Basis Dis. 1866:1658802020. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Yu YJ, Su ZJ, Zhou ZJ and Ma YD:
Pathological observation on spontaneous epithelial ovarian cancer
in reed vole. Chin J V Sci. 1070–1073. 2008.(In Chinese).
|
|
16
|
Zhou ZJ, Yu YJ, Su ZJ and Tang LF:
Expression of heat shock proteins HSP27, HSP70 and HSP90 in
spontaneous ovarian cancer of Microtus fortis. Chin J Comp
Med. 4:26–28. 2005.(In Chinese).
|
|
17
|
Yu Y: An investigation of pathological
changes in Captive-bred Microtus fortis. Chin J Zool.
38:71–73. 2003.(In Chinese).
|
|
18
|
Zheng M, Hu Y, Gou R, Nie X, Li X, Liu J
and Lin B: Identification three LncRNA prognostic signature of
ovarian cancer based on genome-wide copy number variation. Biomed
Pharmacother. 124:1098102020. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Hu Y, Xu Y, Lu W, Yuan Z, Quan H, Shen Y
and Cao J: De novo assembly and transcriptome
characterization: Novel insights into the natural resistance
mechanisms of Microtus fortis against Schistosoma japonicum.
BMC Genomics. 15:4172014. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Marquardt N, Feja M, Hünigen H, Plendl J,
Menken L, Fink H and Bert B: Euthanasia of laboratory mice: Are
isoflurane and sevoflurane real alternatives to carbon dioxide?
PLoS One. 13:e02037932018. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Laferriere CA, Leung VS and Pang DS:
Evaluating intrahepatic and intraperitoneal sodium pentobarbital or
ethanol for mouse euthanasia. J Am Assoc Lab Anim Sci. 59:264–268.
2020. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Bolger AM, Lohse M and Usadel B:
Trimmomatic: A flexible trimmer for Illumina sequence data.
Bioinformatics. 30:2114–2120. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Grabherr MG, Haas BJ, Yassour M, Levin JZ,
Thompson DA, Amit I, Adiconis X, Fan L, Raychowdhury R, Zeng Q, et
al: Full-length transcriptome assembly from RNA-Seq data without a
reference genome. Nat Biotechnol. 29:644–652. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
R Core Team: R: A Language and Environment
for Statistical Computing. R Foundation for Statistical Computing,
Vienna, Austria. Computing,. 14:pp12–21. 2009.
|
|
25
|
Eddy SR: Profile hidden markov models.
Bioinformatics. 14:755–763. 1998. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Almagro Armenteros JJ, Tsirigos KD,
Sønderby CK, Petersen TN, Winther O, Brunak S, von Heijne G and
Nielsen H: SignalP 5.0 improves signal peptide predictions using
deep neural networks. Nat Biotechnol. 37:420–423. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Love MI, Huber W and Anders S: Moderated
estimation of fold change and dispersion for RNA-seq data with
DESeq2. Genome Biol. 15:5502014. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Langmead B and Salzberg SL: Fast
gapped-read alignment with Bowtie 2. Nat Methods. 9:357–359. 2012.
View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Li B and Dewey CN: RSEM: Accurate
transcript quantification from RNA-Seq data with or without a
reference genome. BMC Bioinformatics. 12:3232011. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Leung CS, Yeung TL, Yip KP, Pradeep S,
Balasubramanian L, Liu J, Wong KK, Mangala LS, Armaiz-Pena GN,
Lopez-Berestein G, et al: Calcium-dependent FAK/CREB/TNNC1
signalling mediates the effect of stromal MFAP5 on ovarian cancer
metastatic potential. Nat Commun. 5:50922014. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Takahashi Y, Li L, Kamiryo M, Asteriou T,
Moustakas A, Yamashita H and Heldin P: Hyaluronan fragments induce
endothelial cell differentiation in a CD44- and
CXCL1/GRO1-dependent manner. J Biol Chem. 280:24195–24204. 2005.
View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Onaciu A, Munteanu R, Munteanu VC, Gulei
D, Raduly L, Feder RI, Pirlog R, Atanasov AG, Korban SS, Irimie A
and Berindan-Neagoe I: Spontaneous and induced animal models for
cancer research. Diagnostics (Basel). 10:6602020. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Cicalese A, Bonizzi G, Pasi CE, Faretta M,
Ronzoni S, Giulini B, Brisken C, Minucci S, Di Fiore PP and Pelicci
PG: The tumor suppressor p53 regulates polarity of self-renewing
divisions in mammary stem cells. Cell. 138:1083–1095. 2009.
View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Yue XT, Zhao Y, Xu Y, Zheng M, Feng Z and
Hu W: Mutant p53 in Cancer: Accumulation, Gain-of-function, and
therapy. J Mol Biol. 429:1595–1606. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Barkal AA, Brewer RE, Markovic M, Kowarsky
M, Barkal SA, Zaro BW, Krishnan V, Hatakeyama J, Dorigo O, Barkal
LJ and Weissman IL: CD24 signalling through macrophage Siglec-10 is
a target for cancer immunotherapy. Nature. 572:392–396. 2019.
View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Lee KB, Byun HJ, Park SH, Park CY, Lee SH
and Rho SB: CYR61 controls p53 and NF-κB expression through
PI3K/Akt/mTOR pathways in carboplatin-induced ovarian cancer cells.
Cancer Lett. 315:86–95. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
von Karstedt S, Conti A, Nobis M,
Montinaro A, Hartwig T, Lemke J, Legler K, Annewanter F, Campbell
AD, Taraborrelli L, et al: Cancer cell-autonomous TRAIL-R signaling
promotes KRAS-driven cancer progression, invasion, and metastasis.
Cancer Cell. 27:561–573. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Lin KT, Yeh YM, Chuang CM, Yang SY, Chang
JW, Sun SP, Wang YS, Chao KC and Wang LH: Glucocorticoids mediate
induction of microRNA-708 to suppress ovarian cancer metastasis
through targeting Rap1B. Nat Commun. 6:59172015. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Leng R, Liao G, Wang H, Kuang J and Tang
L: Rac1 expression in epithelial ovarian cancer: Effect on cell EMT
and clinical outcome. Med Oncol. 32:3292015. View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Ren XL, Zhu XH, Li XM, Li YL, Wang JM, Wu
PX, Lv ZB, Ma WH, Liao WT, Wang W, et al: Down-regulation of BTG3
promotes cell proliferation, migration and invasion and predicts
survival in gastric cancer. J Cancer Res Clin Oncol. 141:397–405.
2015. View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Mao D, Qiao L, Lu H and Feng Y: B-cell
translocation gene 3 overexpression inhibits proliferation and
invasion of colorectal cancer SW480 cells via Wnt/β-catenin
signaling pathway. Neoplasma. 63:705–716. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Deng B, Zhao Y, Gou W, Chen S, Mao X,
Takano Y and Zheng H: Decreased expression of BTG3 was linked to
carcinogenesis, aggressiveness, and prognosis of ovarian carcinoma.
Tumor Biol. 34:2617–2624. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
43
|
Bai Y, Qiao L, Xie N, Shi Y, Liu N and
Wang J: Expression and prognosis analyses of the Tob/BTG
antiproliferative (APRO) protein family in human cancers. PLoS One.
12:e01849022017. View Article : Google Scholar : PubMed/NCBI
|
|
44
|
Liu T, Yang Y, Xie Z, Luo Q, Yang D, Liu
X, Zhao H, Wei Q, Liu Y, Li L, et al: The RNA binding protein QKI5
suppresses ovarian cancer via downregulating transcriptional
coactivator TAZ. Mol Ther Nucleic Acids. 26:388–400. 2021.
View Article : Google Scholar : PubMed/NCBI
|
|
45
|
Cui G, Wang C, Lin Z, Feng X, Wei M, Miao
Z, Sun Z and Wei F: Prognostic and immunological role of
Ras-related protein Rap1b in pan-cancer. Bioengineered.
12:4828–4840. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
46
|
Wei X, Lou H, Zhou D, Jia Y, Li H, Huang
Q, Ma J, Yang Z, Sun C, Meng Y, et al: TAGLN mediated
stiffness-regulated ovarian cancer progression via RhoA/ROCK
pathway. J Exp Clin Cancer Res. 40:2922021. View Article : Google Scholar : PubMed/NCBI
|
|
47
|
Wang X, Wei Z, Tang Z, Xue C, Yu H, Zhang
D, Li Y, Liu X, Shi Y, Zhang L, et al: IL-37bΔ1-45 suppresses the
migration and invasion of endometrial cancer cells by targeting the
Rac1/NF-κB/MMP2 signal pathway. Lab Invest. 101:760–774. 2021.
View Article : Google Scholar : PubMed/NCBI
|
|
48
|
Yanagida S, Taniue K, Sugimasa H, Nasu E,
Takeda Y, Kobayashi M, Yamamoto T, Okamoto A and Akiyama T: ASBEL,
an ANA/BTG3 antisense transcript required for tumorigenicity of
ovarian carcinoma. Sci Rep. 3:13052013. View Article : Google Scholar : PubMed/NCBI
|