Introduction
Colorectal cancer (CRC), a frequent malignant tumour
of the digestive tract, ranks third in terms of morbidity and is
the second most common cause of cancer-related deaths worldwide
(1). Annually, ~1.3 million novel
CRC cases occur globally, which cause >0.6 million deaths
(2). Curative resection along with
chemotherapy can be applied in a small proportion of patients,
whereas a great proportion of patients with CRC are diagnosed at
late stage or suffer from post-surgical diseases such as recurrence
or distant metastasis (3). Although
marked improvements have been identified in CRC therapy, the
therapeutic efficacy for patients at advanced stages has not
distinctly improved (4). Notably,
the major causes for worse clinical outcomes are delayed diagnosis,
tumour relapse and distant metastasis (5). Therefore, elucidating the mechanisms
underlying CRC pathogenesis is urgently required in order to find
novel putative targets for diagnosis and treatment.
Long noncoding RNAs (lncRNAs) are a type of RNA
transcript with >200 nucleotides that have lost protein-coding
capacity (6). Although lncRNAs were
previously regarded as useless, they have been revealed to produce
considerable effects in numerous biological behaviours, including
cell differentiation, the cell cycle, cytogenetic processes and
epigenetic modification (7).
Notably, lncRNAs have been described to perform important
regulatory actions in promoting and maintaining tumour initiation
and progression (8). To date, a
growing number of lncRNAs have been confirmed to be dysregulated in
CRC and closely associated with malignant characteristics (9). The differentially expressed lncRNAs
play carcinogenic or anti-oncogenic actions and consequently
participate in the control of CRC progression (10).
MicroRNAs (miRNAs) are a family of
non-protein-coding RNAs with ~17–23 nucleotides (11). They have been reported to exhibit
crucial functions in posttranscriptional regulation of genes via
binding to their 3′untranslated regions (3′UTRs), triggering
transcriptional silencing or mRNA reduction (12). The contribution of miRNAs in CRC has
received widespread attention in the past decade, and extensive
miRNAs have been confirmed to be implicated in CRC progression by
exerting tumour-promoting or tumour-inhibiting roles (13–15).
Specifically, the competitive endogenous RNA (ceRNA) theory has
been recognized and has indicated that lncRNAs execute a
‘sponge-like’ activity on certain miRNAs, thereby abolishing or
weakening the modulatory activities of miRNAs on their targets
(16). Subsequently, comprehending
the dysregulated lncRNAs in CRC and clarifying their working
mechanisms may be instrumental in the development of attractive
targets for cancer diagnosis and therapy.
Long intergenic nonprotein coding RNA 649
(LINC00649) has been identified as a functional regulator in acute
myeloid leukaemia (17). In
addition, the lncRNA has been validated as a hub lncRNA that may be
strongly correlated with colorectal carcinogenesis and progression
(18). However, the detailed roles
of LINC00649 in CRC have not been fully confirmed. Therefore, the
specific roles of LINC00649 in CRC and the related mechanisms were
explored.
Materials and methods
Patient tissues
The present research was authorized (approval no.
EC-FPHCQLND-20150815) by the Ethics Committee of The First People's
Hospital of Chongqing Liangjiang (Chongqing, China) and was
implemented in strict accordance with the Declaration of Helsinki.
Written informed consent was provided by all participants. CRC
tissues were acquired from 52 patients with CRC (30 male patients
and 22 female patients; age range, 41–72 years) at The First
People's Hospital of Chongqing Liangjiang (Chongqing, China)
between June 2015 and January 2016. Adjacent normal tissues were
acquired 3 cm away from tumour tissues. All patients included in
the present study underwent a form of anticancer treatment before
the operation. Additionally, patients who were diagnosed with other
types of human cancer were excluded. All patient tissues were
preserved in liquid nitrogen.
Cell culture
A normal human colon epithelium cell line FHC was
acquired from the American Type Culture Collection (ATCC), and
cultured in DMEM:F12 medium (cat. no. 11320033; Gibco; Thermo
Fisher Scientific, Inc.) with 25 mM HEPES, 10 ng/ml cholera toxin,
0.005 mg/ml insulin, 0.005 mg/ml transferrin, 100 ng/ml
hydrocortisone, 20 ng/ml human recombinant epidermal growth factor
and 10% foetal bovine serum (FBS; Gibco; Thermo Fisher Scientific
Inc.). HCT 116 (ATCC), a CRC cell line, was cultured in McCoy's 5a
medium (cat. no. 16600108; Gibco; Thermo Fisher Scientific, Inc.)
and 10% FBS. Another two CRC cell lines, SW480 and SW620 (both from
the National Collection of Authenticated Cell Cultures; Shanghai,
China), were maintained in 10% FBS-supplemented L-15 medium (cat.
no. 11415114; Gibco; Thermo Fisher Scientific Inc.). All these
cells were cultured at 37°C and 5% CO2.
Transfection
Specific small interfering RNAs (siRNAs) against
LINC00649 (si-LINC00649), non-targeting negative control siRNA
(si-NC), miR-432-5p mimic, NC miRNA mimic (miR-NC), miR-432-5p
inhibitor (anti-miR-432-5p) and miRNA inhibitor control
(anti-miR-NC) were all constructed by Shanghai GenePharma Co., Ltd.
The sequences are presented in Table
I. The pcDNA3.1-hepatoma-derived growth factor (pc-HDGF)
plasmid was obtained from Shanghai GenePharma Co., Ltd. First,
cells were inoculated into 6-well plates and cultured to ~60-80%
density. Next, the cells were transfected with siRNAs (100 pmol),
mimic (100 pmol), miRNA inhibitor (100 pmol) or plasmid (4 µg) by
applying Lipofectamine® 2000 (Invitrogen; Thermo Fisher
Scientific, Inc.) according to the manufacturer's instructions.
After incubation for 6 h with transfection reagent at 37°C, the
medium was replaced with fresh medium. Reverse
transcription-quantitative (RT-q)PCR, flow cytometry and Transwell
migration and invasion assays were carried out at 48 h
post-transfection. After incubation for 24 h, Cell Counting Kit-8
(CCK-8) assay was conducted.
 | Table I.Sequences of siRNAs, miRNA
oligonucleotides and shRNAs. |
Table I.
Sequences of siRNAs, miRNA
oligonucleotides and shRNAs.
| Type | Sequences
(5′→3′) |
|---|
| si-LINC00649#1 |
TAGATGTTATTGACAAAATAAGC |
| si-LINC00649#2 |
AAGTTAAAGCTTCCAAAAATAGA |
| si-NC |
CACGATAAGACAATGTATTT |
| miR-432-5p |
UCUUGGAGUAGGUCAUUGGGUGG |
| mimic |
|
| miR-NC |
CGAUCGCAUCAGCAUCGAUUGC |
| miR-432-5p |
CCACCCAAUHACCUACUCCAAGA |
| inhibitor |
|
| anti-miR-NC |
UGAGCUGCAUAGAGUAGUGAUUA |
| sh-LINC00649 |
CCGGTAGATGTTATTGACAAAATAA |
|
|
GCCTCGAGGCTTATTTTGTCAATAA |
|
| CATCTATTTTTG |
| sh-NC |
CCGGCACGATAAGACAATGTATTTC |
|
|
TCGAGAAATACATTGTCTTATCGTG |
|
| TTTTTG |
RT-qPCR
Small RNA was extracted from cultured cells or
tissues utilizing RNAiso for Small RNA and reverse-transcribed
utilizing a Mir-X miRNA First-Strand Synthesis Kit (both from
Takara Biotechnology Co., Ltd.) according to the manufacturer's
instructions. For quantification of miR-432-5p, qPCR was executed
with Mir-X miRNA RT-qPCR TB Green® Kit (Takara
Biotechnology Co., Ltd.) according to the manufacturer's
instructions, and the data were normalized to U6 small nuclear RNA.
The thermocycling conditions were as follows: 95°C for 10 sec; 40
cycles at 95°C for 5 sec and 60°C for 20 sec; and 95°C for 60 sec,
55°C for 30 sec and 95°C for 30 sec.
Tissues or cells were immersed in 1 ml TRIzol
(Invitrogen; Thermo Fisher Scientific, Inc.), after which they were
incubated at room temperature for 5 min. Prior to being centrifuged
at 1,000 × g at 4°C for 15 min, 200 µl chloroform was supplemented
into the Eppendorf tube and rigorous shaking was executed for 15
sec. The supernatant fluid was transferred to a novel Eppendorf,
followed by the addition of 500 µl isopropanol. After
centrifugation at 1,000 × g at 4°C for 10 min, the supernatant
fluid was discarded, and the precipitant was washed with 1 ml 75%
alcohol and centrifuged at 1,000 × g at 4°C for 5 min. The RNA was
collected and dissolved in RNA-free water. Then, the isolated RNA
was reverse transcribed into cDNA by PrimeScript™ RT reagent Kit
(Takara Biotechnology Co., Ltd.) according to the manufacturer's
instructions. To assess LINC00649 and HDGF expression, TB
Green® Premix Ex Taq™ (Takara Biotechnology Co., Ltd.)
was applied for PCR amplification. The thermocycling conditions
were as follows: Initial denaturation at 95°C for 30 sec, followed
by 40 cycles at 95°C for 3 sec, 60°C for 30 sec and 72°C for 30
sec. GAPDH served as the internal reference for LINC00649 and HDGF.
The 2−ΔΔCq method (19)
was used to analyse the expression of genes.
The primers were designed as follows: LINC00649
forward, 5′-CTTGCAGTTTGATCTCAGACTGC-3′ and reverse,
5′-ACCACGAGACTATATCCCACACC-3′; HDGF forward,
5′-GTGACGGTGATAAGAAGGGGAAT-3′ and reverse,
5′-TTCAACGCTCCTTTCTCGTTCT-3′; GAPDH forward,
5′-AGTCAACGGATTTGGTCGTATTG-3′ and reverse,
5′-AAACCATGTAGTTGAGGTCAATGAA-3′; U6 forward,
5′-CTCGCTTCGGCAGCACA-3′ and reverse, 5′-AACGCTTCACGAATTTGCGT-3′;
miR-497-5p forward, 5′-TCGGCAGGCAGCAGCACACUG-3′; miR-432-5p
forward, 5′-TCGGCAGGUCUUGGAGUAGG-3′; miR-28-3p forward,
5′-TCGGCAGGAGGUCCUCGAG-3′; miR-195-5p forward,
5′-TCGGCAGGUAGCAGCACAG-3′; miR-15b-5p forward,
5′-TCGGCAGGUAGCAGCACAUCA-3′. The universal reverse sequence for
miRNA was 5′-CACTCAACTGGTGTCGTGGA-3′ (reverse).
CCK-8 assay
Each well of 96-well plates was supplemented with
100 µl cell suspension containing 2×103 transfected
cells. Subsequently, 10 µl CCK-8 reagent (Nanjing KeyGen Biotech
Co., Ltd.) was added, and the culture plates were incubated at 37°C
for 2 h. The absorbance was measured with a microplate reader at a
wavelength of 450 nm. The assay was performed for three consecutive
days, and the obtained results were used for growth curve
construction.
Flow cytometric analysis for
apoptosis
Two days following cell injection, 0.25% trypsin was
employed for cell harvest (2×106). Phosphate-buffered
saline (PBS) was pre-cooled at 4°C and used to wash the collected
cells. In accordance with the instructions of the Annexin V-FITC
Apoptosis Detection Kit (Beyotime Institute of Biotechnology), the
obtained cells were resuspended in 195 µl Annexin V-FITC, followed
by mixing with 5 µl Annexin V-FITC and 10 µl propidium iodide.
Subsequently, the cells were cultured at 25°C for 15 min and
continually maintained in darkness. The early + late apoptotic
cells were detected by a flow cytometer (FACScan™; BD Biosciences)
and analysed with CellQuest software v.2.9 (BD Biosciences).
Transwell cellular migration and
invasion assays
The migratory and invasive capacities of CRC cells
were evaluated employing a Transwell chamber (8-µm pore size; BD
Biosciences). For the invasion assay, Matrigel (BD Biosciences) was
used to precoat the lower surface of the upper chamber at 37°C for
2 h, whereas a migration assay was implemented without this step.
For both assays, 5×104 transfected cells resuspended in
FBS-free culture medium (McCoy's 5a medium for HCT116, L-15 medium
for SW480) were seeded into the upper chambers. In the lower
chambers, 600 µl complete medium mixed with 20% FBS was plated and
functioned as the chemoattractant. Cells were cultured at 37°C for
one day, after which the non-migrated or non-invasive cells were
removed from the upper face of the membrane. The migrated and
invasive cells were fixed with ethanol at room temperature for 15
min and staining with 0.1% crystal violet at room temperature for
30 min. After washing with PBS, the number of stained cells was
observed under a light microscope (magnification, ×200; Olympus
Corporation).
Tumour xenograft in nude mice
Short hairpin RNA (shRNA) for LINC00649
(sh-LINC00649) and NC shRNA (sh-NC) were synthesized by Shanghai
GenePharma Co., Ltd. A 2nd lentiviral system was used in the
production of lentiviruses. The sequences are presented in Table I. The shRNAs were cloned into a
lentivirus vector prior to being transduced into 293T cells
(National Collection of Authenticated Cell Cultures) together with
psPAX2 and pMD2.G. A total of 30 µg plasmids were used in
lentivirus packaging, and the ratio of lentiviral plasmid: psPAX2:
pMD2.G was 2:1:1. After 6 h of cultivation at 37°C, the medium was
replaced with fresh 10% FBS-supplemented DMEM with 1% glutamax, 1%
non-essential amino acids and 1% sodium pyruvate solution (all
purchased from Gibco; Thermo Fisher Scientific, Inc.). Following 2
days of incubation, the lentiviruses carrying either sh-LINC00649
or sh-NC were harvested and utilized to infect HCT 116 cells with
the multiplicity of infection (MOI) 5. The stable cell line was
screened out by treatment with 2 µg/ml puromycin, and maintained
with 1 µg/ml puromycin.
The animal study was performed with the approval
(approval no. IACUC-FPHCQLND-20190601) of the Institutional Animal
Care and Use Committee of The First People's Hospital of Chongqing
Liangjiang (Chongqing, China). A total of 6 BALB/c nude mice (male;
age, 4–6 weeks; mean weight, 20.12 g) were supplied by Vital River
Laboratory Animal Technology Co., Ltd. The mice were housed under
specific pathogen-free conditions at 25°C and 50% humidity, with a
10/14 h light/dark cycle and ad libitum access to food and
water. A total of 2×106 HCT 116 cells stably
overexpressing sh-LINC00649 or sh-NC were resuspended in 100 µl PBS
and subcutaneously inoculated into the flank of mice. The width and
length of the subcutaneous tumours were recorded weekly. The tumour
volume was calculated with the equation: Volume=0.50 × length ×
width2. On the 28th day, the mice were euthanized by
means of cervical dislocation, and tumour xenografts were resected
for tumour weighing.
Bioinformatic prediction and
luciferase reporter assay
starBase 3.0 (http://starbase.sysu.edu.cn/) was employed to identify
potential target miRNA of LINC00649. The putative targets of
miR-432-5p were searched by applying TargetScan (version 7.2;
http://www.targetscan.org/) and starBase
3.0. The Cancer Genome Atlas (TCGA; http://portal.gdc.cancer.gov/) database (20) was employed to analyse LINC00649
expression in CRC (TCGA-COAD and TCGA-READ Projects.).
Fragments of LINC00649 and HDGF carrying the
wild-type (wt) miR-432-5p binding sequences were synthesized by
Shanghai GenePharma Co., Ltd. After inserting into the pmirGLO
reporter vector (Promega Corporation), the recombinant reporter
plasmids were named LINC00649-wt and HDGF-wt. Similarly, the
LINC00649-mutant (mut) and HDGF-mut were constructed in a similar
manner. For the reporter assay, the reporter plasmids alongside
miR-432-5p mimic or miR-NC were transfected into CRC cells using
Lipofectamine 2000 (Invitrogen; Thermo Fisher Scientific, Inc.),
followed by 48 h of culture at 37°C in an atmosphere containing 5%
CO2. Transfected cells were immersed in 100 µl Passive
Lysis Buffer provided from the Dual-Luciferase Reporter Assay kit
(Promega Corporation) according to the manufacturer's instructions.
The luminometer tubes were loaded with 50 µl LAR II that was
supplemented with 10 µl cell lysis buffer. After the measurement of
firefly luciferase activity, 50 µl L Stop & GLoR Reagent
(Promega Corporation) was added, followed by the detection of
Renilla luciferase activity. Renilla luciferase
activity was used as the reference.
Subcellular fractionation
analysis
The Cytoplasmic & Nuclear RNA Purification Kit
(Norgen Biotek Corp.) was applied for subcellular fractionation
according to the manufacturer's instructions. After total RNA
extraction from nuclear and cytoplasmic fractions, RT-qPCR was
carried out to measure the relative amount of LINC00649 in both
fractions. GAPDH and U6 functioned as normalizing cytoplasmic and
nuclear controls, respectively.
RNA immunoprecipitation (RIP)
The binding of LINC00649 and miR-432-5p to the
Argonaute-2 (Ago2) protein was explored with a RIP assay, which was
carried out employing a Magna RIP RNA-Binding Protein
Immunoprecipitation Kit (cat. no. 17–700; MilliporeSigma) according
to the manufacturer's instructions. CRC cells were immersed in RIP
lysis buffer (MilliporeSigma) to obtain whole cell extracts. A 10%
cell extract was used as the input, and the other parts were
treated with RIP buffer containing magnetic beads that were
conjugated with human anti-Ago2 (1:500; cat. no. ab32381; Abcam) or
anti-IgG antibodies (1:5,000; included in the kit mentioned above)
at 4°C overnight. The input served as the positive control, while
IgG was used as the negative control. The magnetic beads were
harvested and probed with Proteinase K (Beyotime Institute of
Biotechnology) at 55°C for 30 min with vibration to remove the
protein. The immunoprecipitated RNA was examined with RT-qPCR to
determine LINC00649 and miR-432-5p enrichment, and the RT-qPCR was
performed according to the manufacturer's (Takara Biotechnology
Co., Ltd.) instructions as outlined above.
Western blotting
Transfected cells were washed three times with
precooled PBS, and RIPA buffer (Nanjing KeyGen Biotech Co., Ltd.)
was added to isolate total protein. Next, a BCA Protein Assay Kit
was adopted for protein quantification. Proteins (30 µg/lane) were
loaded and electrophoresed on a 10% SDS-PAGE, followed by
transferring to PVDF membranes and blocking the membranes at room
temperature for 2 h with 5% non-fat milk powder. Subsequent to
overnight incubation at 4°C with primary antibodies specifically
targeting HDGF (cat. no. ab128921; 1:1,000) or GAPDH (cat. no.
ab128915; 1:1,000; both from Abcam), the membranes were probed with
a horseradish peroxidase (HRP)-conjugated secondary antibody (cat.
no. ab205718; 1:5,000; Abcam) at room temperature for 2 h. The
protein signals were detected with an ECL detection kit (Nanjing
KeyGen Biotech Co., Ltd.) and images were captured with ChemiDoc™
XRS+imaging system (Bio-Rad Laboratories, Inc.). Densitometry was
performed using Quantity One software (version 4.6.6; Bio Rad
Laboratories, Inc.).
Statistical analysis
The mean ± standard deviation (SD) values of three
independent replicates were employed to present the experimental
data. The Kaplan-Meier method estimated the survival curves, which
were then analysed with log-rank testing. Both paired and unpaired
Student's t-tests were applied to detect comparisons between two
groups. The differences between multiple groups were analysed by
one-way analysis of variance (ANOVA), and Tukey's post hoc test was
implemented following ANOVA. Pearson's correlation analysis was
conducted to compare the correlation between the expression of
LINC00649, miR-432-5p and HDGF. P<0.05 was considered to
indicate a statistically significant difference.
Results
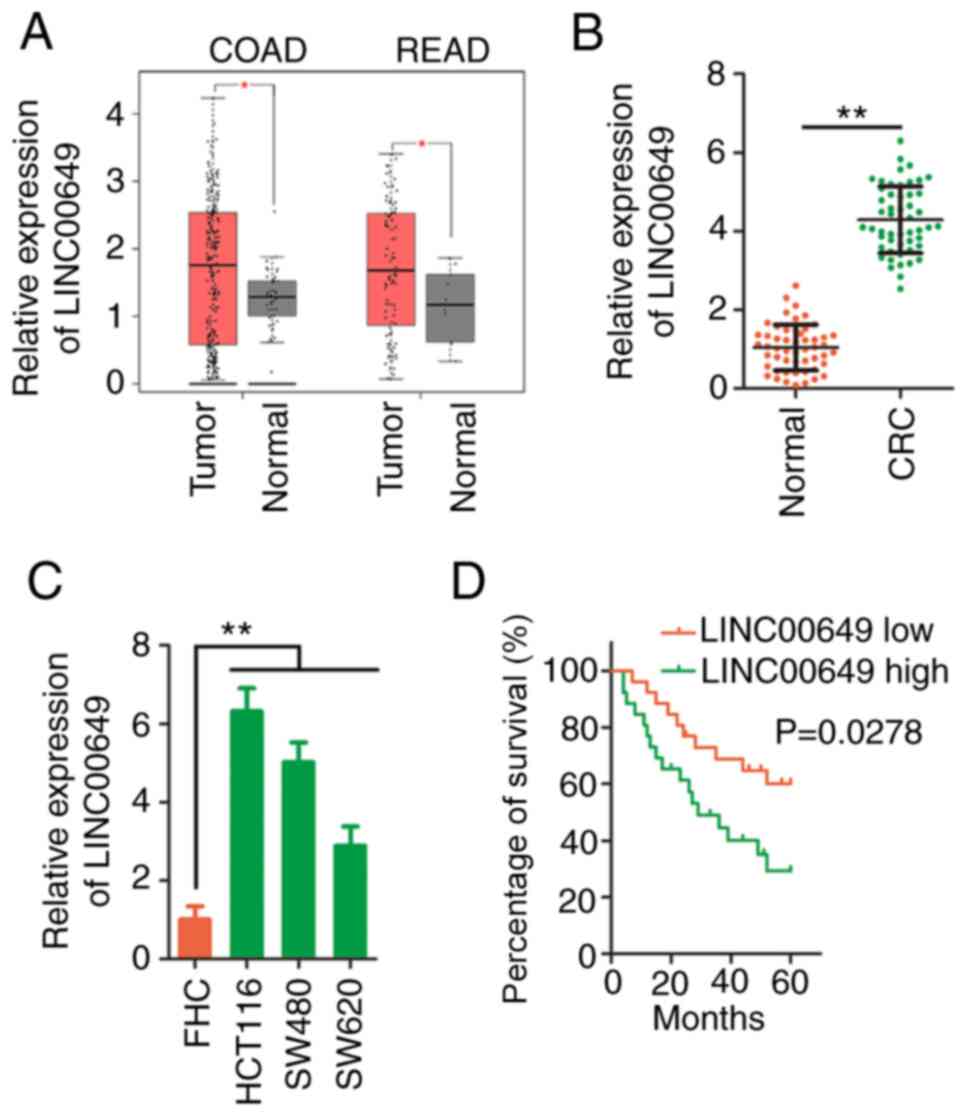
LINC00649 is overexpressed in CRC
First, LINC00649 expression in CRC was analysed
using a dataset from TCGA. As compared with normal tissues,
LINC00649 was upregulated in colon adenocarcinoma (COAD) and rectum
adenocarcinoma (READ) (Fig. 1A).
Additionally, LINC00649 was detected in the cohort of the present
study. LINC00649 was highly expressed in CRC tissues compared with
adjacent normal tissues (Fig. 1B).
Moreover, the three assessed CRC cell lines overexpressed LINC00649
compared with FHC (Fig. 1C). Next,
utilizing the median value of LINC00649 expression in CRC tissues
as the cut-off point, all subjects were subdivided into
LINC00649-low or LINC00649-high expression groups. Survival
analysis was performed, and the results revealed that a high
expression level of LINC00649 was associated with shorter overall
survival (Fig. 1D). Therefore,
overexpressed LINC00649 was negatively associated with the
prognosis of patients with CRC.
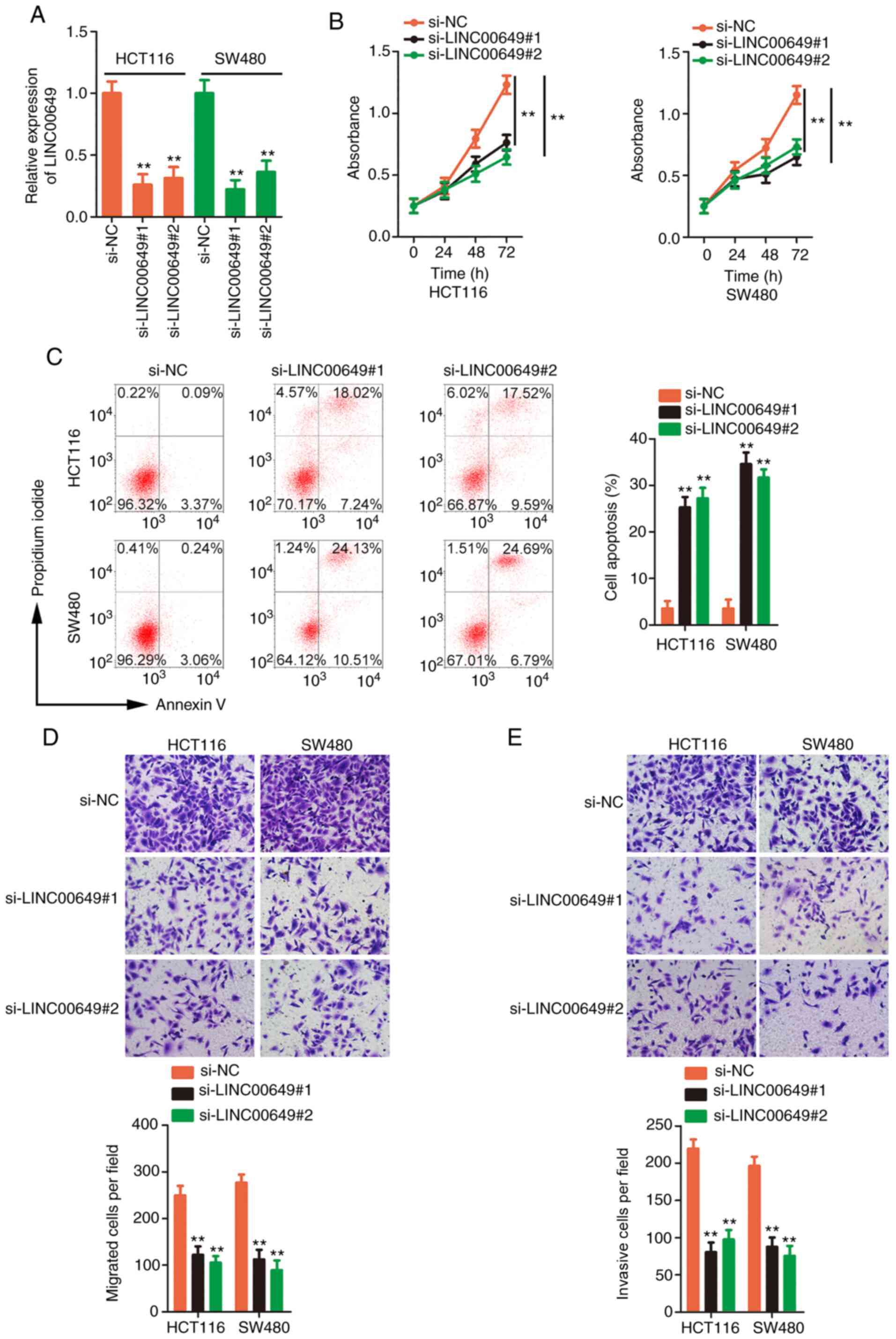
Silencing of LINC00649 impairs the
malignant behaviours of CRC cells
The HCT 116 and SW480 cell lines expressed the
highest expression of LINC00649 and accordingly, they were applied
in subsequent research. To understand the detailed functions of
LINC00649 in CRC progression, two siRNAs targeting LINC00649 were
used to efficiently knock down LINC00649 expression (Fig. 2A). Then, CCK-8 assays confirmed that
proliferative activity was hindered in CRC cells after the
silencing of LINC00649 (Fig. 2B).
Flow cytometric analysis also revealed that transfection with
si-LINC00649 led to the promotion of CRC cell apoptosis (Fig. 2C). Furthermore, loss of LINC00649
expression suppressed the migratory (Fig. 2D) and invasive (Fig. 2E) properties of CRC cells. Briefly,
these results confirmed the oncogenic actions of LINC00649 in CRC
cells.
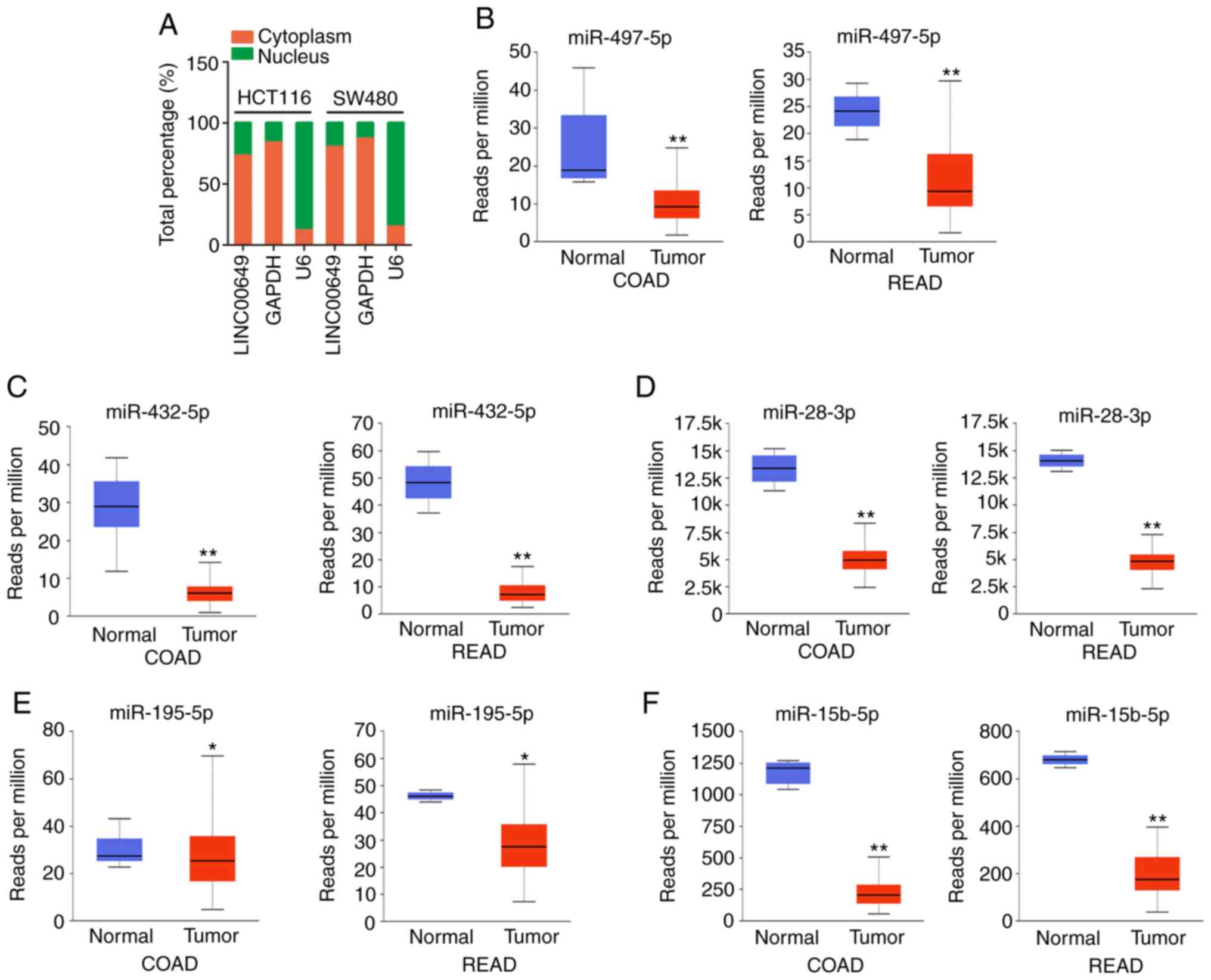
LINC00649 acts as a ceRNA by sponging
miR-432-5p in CRC cells
To elucidate the molecular events of LINC00649,
subcellular fractionation analysis was implemented to reveal the
localization of LINC00649 in CRC cells. LINC00649 was principally
distributed in CRC cell cytoplasm (Fig.
3A). Thus, LINC00649 may function in a ceRNA manner. starBase
3.0 was applied to predict the target of LINC00649. In total, 12
miRNAs, including miR-28-3p, miR-16-5p, miR-15a-5p, miR-15b-5p,
miR-195-5p, miR-424-5p, miR-497-5p, miR-432-5p, miR-1323,
miR-4766-5p, miR-6838-5p and miR-548o-5p, contained complementary
sequences to LINC00649. Utilizing TCGA database, miR-497-5p,
miR-432-5p, miR-28-3p, miR-195-5p and miR-15b-5p were revealed to
be downregulated in CRC (Fig.
3B-F); hence, the five miRNAs were selected for subsequent
experimental confirmation.
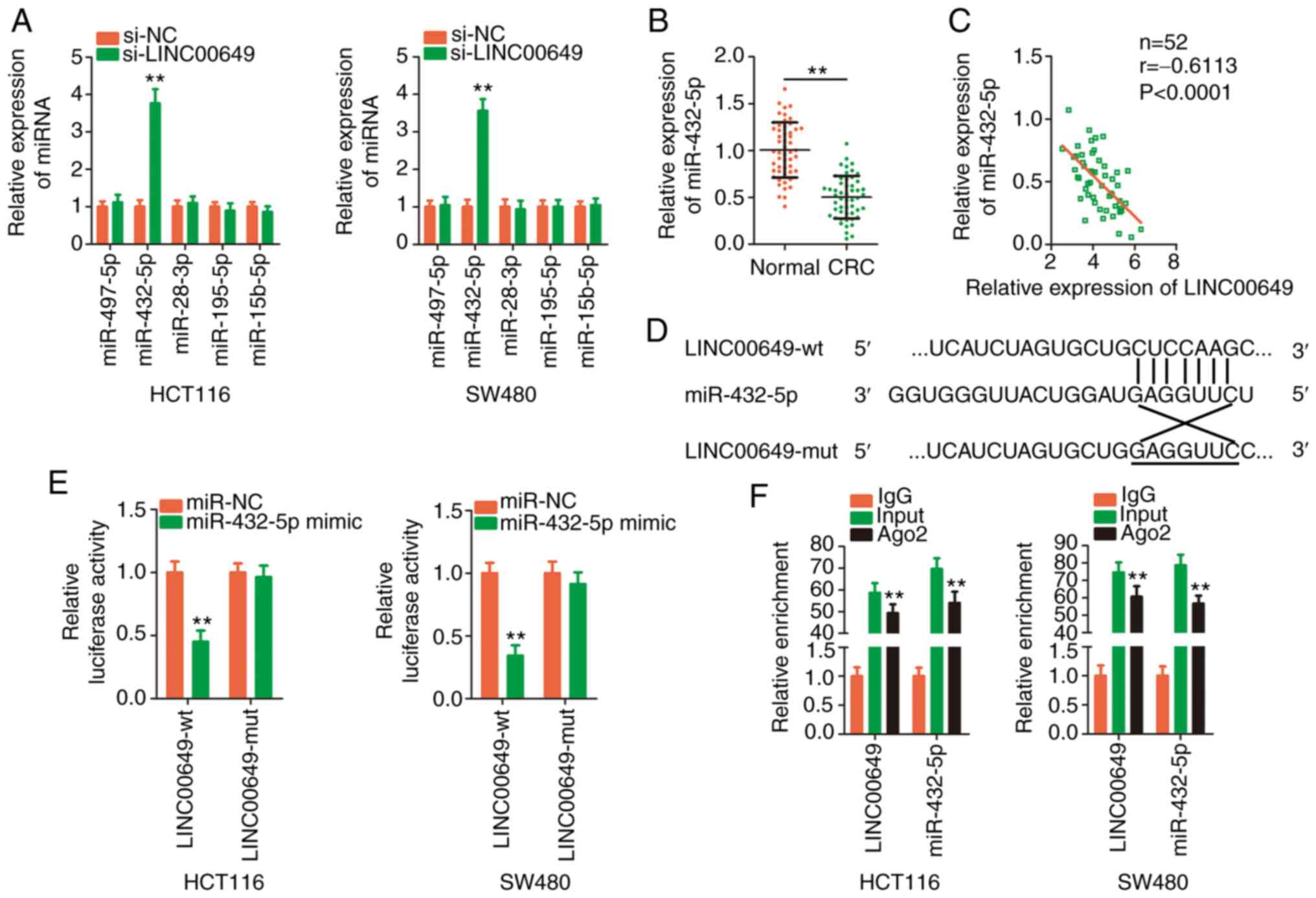
Following LINC00649 knockdown in CRC cells, their
expression was detected via RT-qPCR. The expression level of
miR-432-5p was significantly increased by LINC00649 deficiency,
whereas the expression of other candidates was unaltered in CRC
cells upon silencing of LINC00649 (Fig.
4A). Furthermore, miR-432-5p exhibited relatively lower
expression in CRC tissues (Fig. 4B)
and presented a negative expression correlation with LINC00649
(Fig. 4C). The direct binding
between LINC00649 and miR-432-5p (Fig.
4D) was then confirmed by employing a luciferase reporter
assay. Overexpressed miR-432-5p decreased the luciferase activity
of LINC00649-wt, whereas the suppression was counteracted when the
binding site was mutated (Fig. 4E).
As evidenced by RIP, LINC00649 and miR-432-5p were both enriched in
Ago2-containing beads in comparison with IgG-containing beads
(Fig. 4F). Briefly, LINC00649
sponged miR-432-5p in CRC cells.
miR-432-5p directly targets and
decreases HDGF in CRC cells
Transfection with miR-432-5p mimic caused a
significant upregulation of miR-432-5p (Fig. 5A). Exogenous miR-432-5p expression
decreased CRC cell proliferation (Fig.
5B) and facilitated cell apoptosis (Fig. 5C). Cell migration and invasion
(Fig. 5D and E) were restricted in
CRC cells after miR-432-5p mimic transfection. As predicted by
bioinformatics analysis, a potential binding site was revealed
between miR-432-5p and the HDGF 3′UTR (Fig. 5F). Next, to confirm the prediction,
a luciferase reporter assay was performed, and the results revealed
that the luciferase activity of HDGF-wt, but not HDGF-mut, was
attenuated by miR-432-5p (Fig. 5G).
The regulatory activities of miR-432-5p overexpression on HDGF
expression were then determined. HDGF expression was clearly
downregulated in CRC cells when miR-432-5p was overexpressed
(Fig. 5H and I). Collectively,
these results confirmed HDGF as a downstream target of
miR-432-5p.
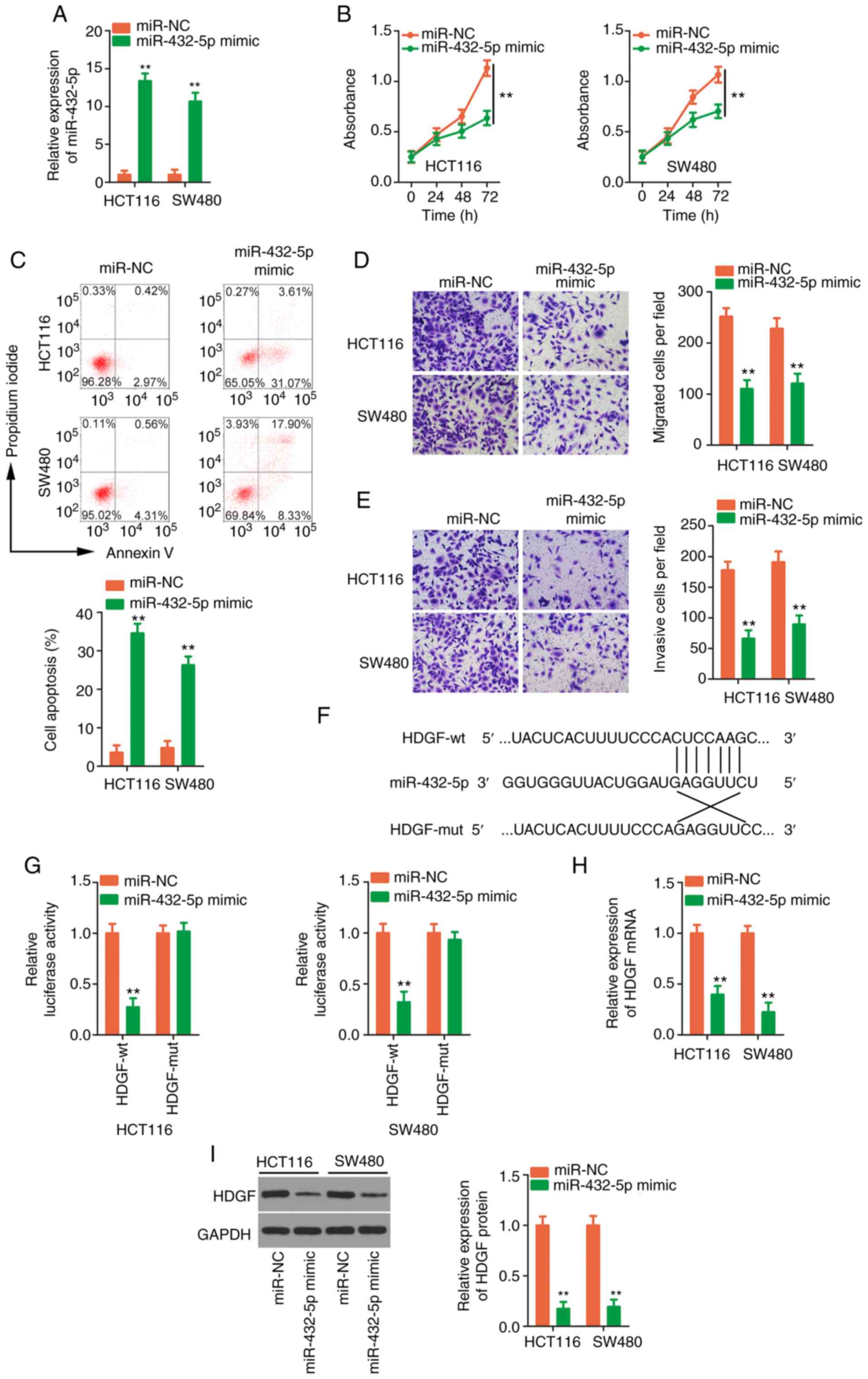 | Figure 5.Confirmation of HDGF as a target of
miR-432-5p. (A) RT-qPCR was employed to gauge the transfection
efficiency of miR-432-5p mimic. (B and C) The proliferation and
apoptosis of CRC cells after miR-432-5p overexpression were
examined by Cell Counting Kit-8 assay and flow cytometric analysis,
respectively. (D and E) Cell motility was analysed by Transwell
cellular migration and invasion assays in CRC cells upon miR-432-5p
overexpression (magnification, ×200). (F) The target sequences
between miR-432-5p and the HDGF 3′UTR were predicted by
bioinformatics analysis. (G) Luciferase activity was quantified in
CRC cells after being transfected with miR-432-5p mimic or miR-NC
and HDGF-wt or HDGF-mut. (H and I) The measurement of HDGF in
miR-432-5p overexpressed-CRC cells was performed by applying
RT-qPCR and western blotting. **P<0.01 vs. miR-NC. HDGF,
hepatoma-derived growth factor; miR, microRNA; RT-qPCR, reverse
transcription-quantitative PCR; CRC, colorectal cancer; UTR,
untranslated region; NC, negative control; wt, wild-type; mut,
mutant. |
si-LINC00649-induced effects on CRC
cells are dependent on the miR-432-5p/HDGF axis
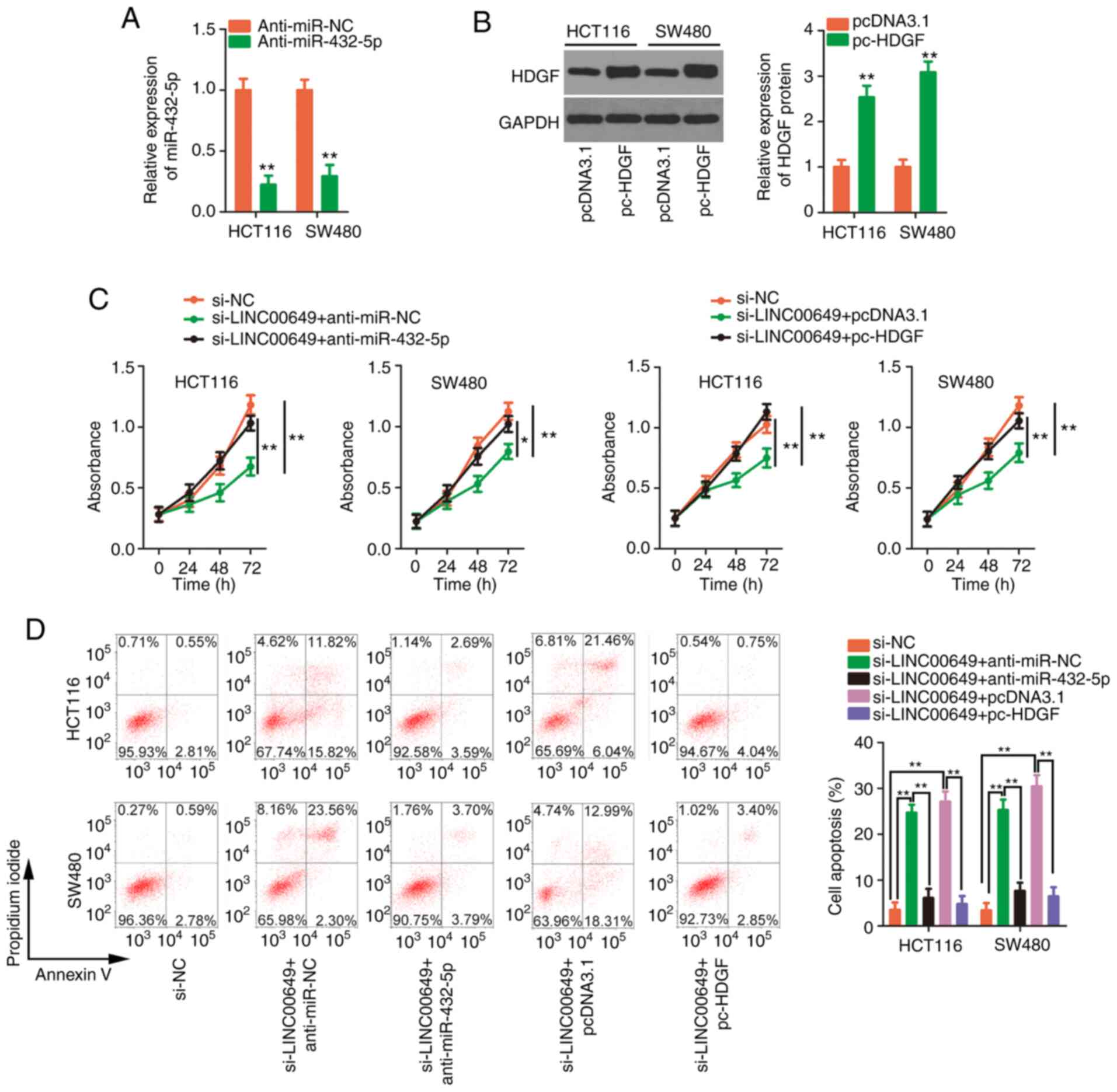
To assess the association between LINC00649,
miR-432-5p and HDGF, Pearson's correlation analysis was applied,
and the results revealed that highly expressed HDGF (Fig. 6A) in CRC tissues was inversely
correlated with miR-432-5p (Fig.
6B). The positive expression correlation between LINC00649 and
HDGF in CRC tissues was significant (Fig. 6C). Next, silencing of LINC00649
significantly decreased HDGF expression (Fig. 6D and E), which could be restored in
CRC cells after anti-miR-432-5p co-transfection (Fig. 6F and G). Additionally, inhibiting
miR-432-5p reversed the stimulatory effect of si-LINC00649 on
miR-432-5p expression in CRC cells (Fig. 6H). RIP assays revealed that,
compared with the IgG group, the three RNAs, including LINC00649,
miR-432-5p and HDGF (Fig. 6I), were
all enriched in Ago2-containing beads, implying that they coexisted
in the RNA induction-silencing complex. The aforementioned results
confirmed that LINC00649 could sponge miR-432-5p in CRC cells and
subsequently increase HDGF expression.
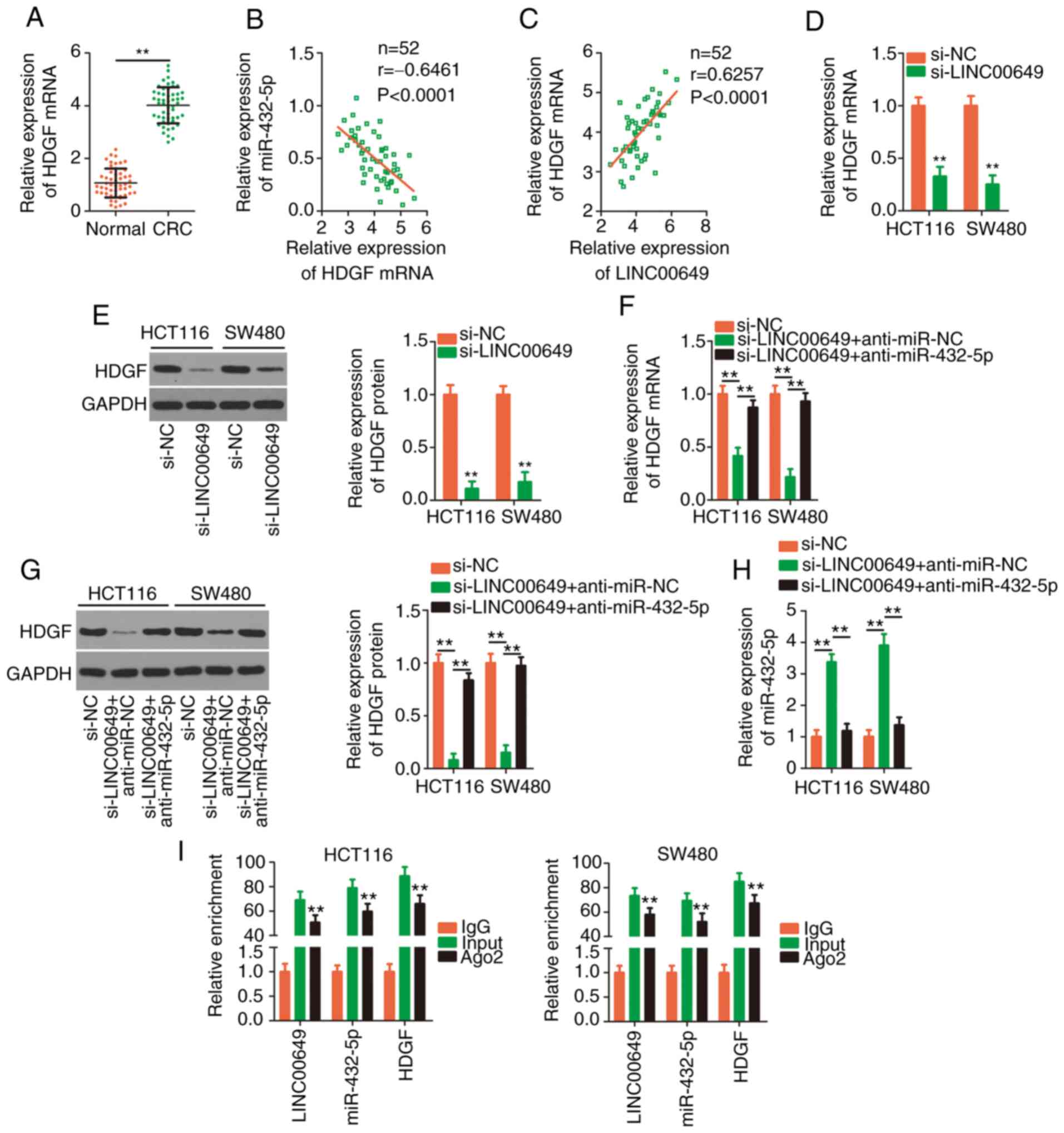 | Figure 6.LINC00649 upregulates HDGF expression
in CRC cells by adjusting miR-432-5p. (A) Detection of HDGF
expression in CRC tissues was performed via RT-qPCR. **P<0.01.
(B) Pearson's correlation analysis verified the negative expression
correlation between HDGF and miR-432-5p. (C) The positive
expression correlation between LINC00649 and HDGF. (D and E)
RT-qPCR and western blotting detected HDGF expression in CRC cells
when LINC00649 was silenced. **P<0.01 vs. si-NC. (F and G)
si-LINC00649 alongside anti-miR-432-5p or anti-miR-NC was injected
into CRC cells, followed by the detection of HDGF expression. (H)
The change in miR-432-5p expression in the aforementioned cells was
analysed by RT-qPCR. **P<0.01. (I) RIP assays confirmed the
target binding among LINC00649, miR-432-5p and HDGF. **P<0.01
vs. IgG. LINC00649, long intergenic nonprotein coding RNA 649;
HDGF, hepatoma-derived growth factor; CRC, colorectal cancer; miR,
microRNA; RT-qPCR, reverse transcription-quantitative PCR; RIP, RNA
immunoprecipitation; si-, small interfering; NC, negative control;
Ago2, Argonaute-2. |
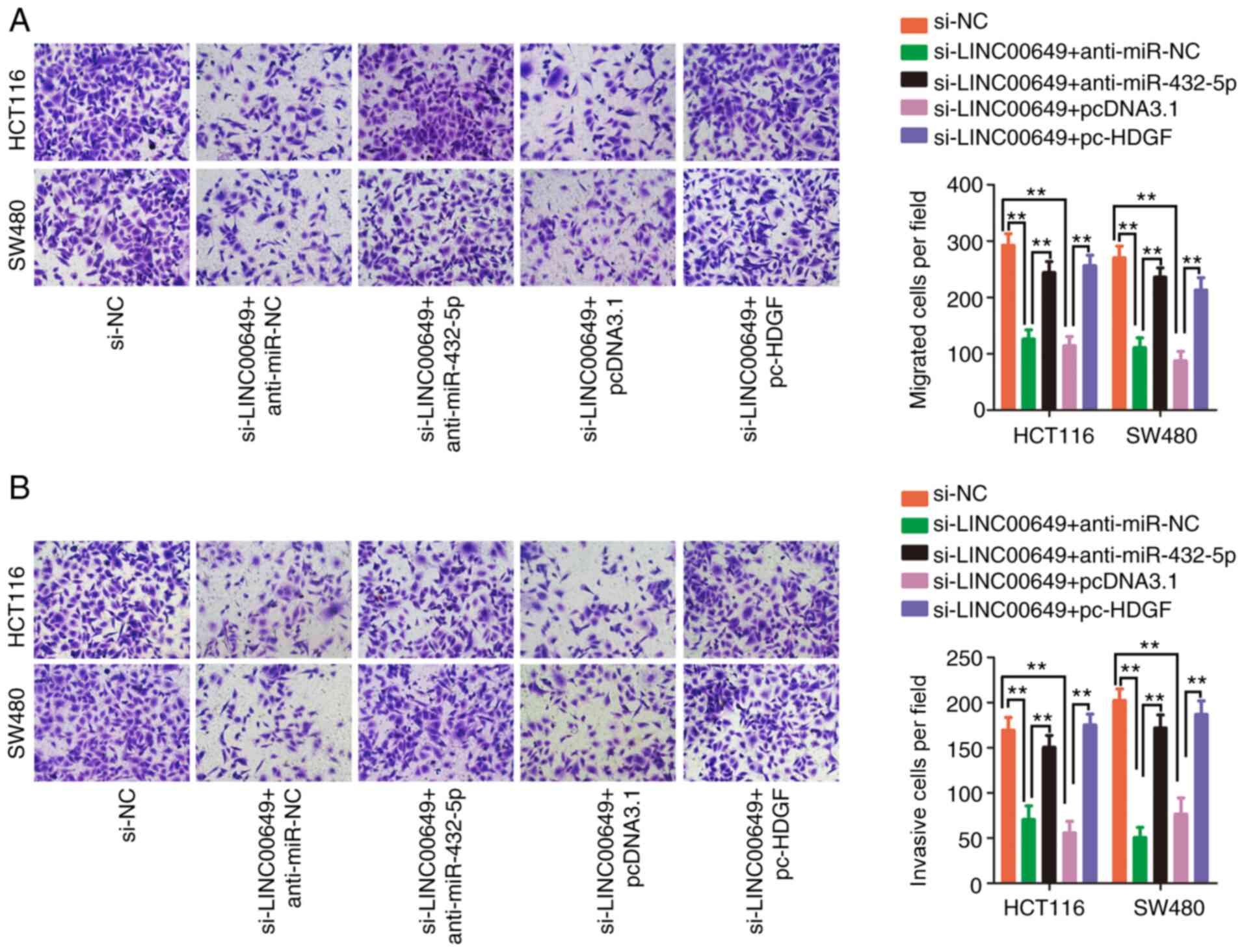
Given primarily the obtained data, it was
hypothesized that the miR-432-5p/HDGF axis is necessary for the
effects of LINC00649 in CRC cells. Rescue experiments were
performed to corroborate this theory. Prior to transfection, the
efficiency of anti-miR-432-5p and pc-HDGF transfection was
confirmed (Fig. 7A and B). The
proliferative activity of CRC cells was inhibited by LINC00649
downregulation, which was mitigated by miR-432-5p inhibition or
HDGF upregulation (Fig. 7C).
Additionally, the si-LINC00649-induced effect on cell apoptosis was
counteracted by co-transfection of anti-miR-432-5p or pc-HDGF
(Fig. 7D). Furthermore,
interference with LINC00649 hindered cell migration (Fig. 8A) and invasion (Fig. 8B) properties, whereas these effects
were abated by the addition of anti-miR-432-5p or pc-HDGF. In
summary, LINC00649 played a carcinogenic role in CRC cells via the
miR-432-5p/HDGF axis.
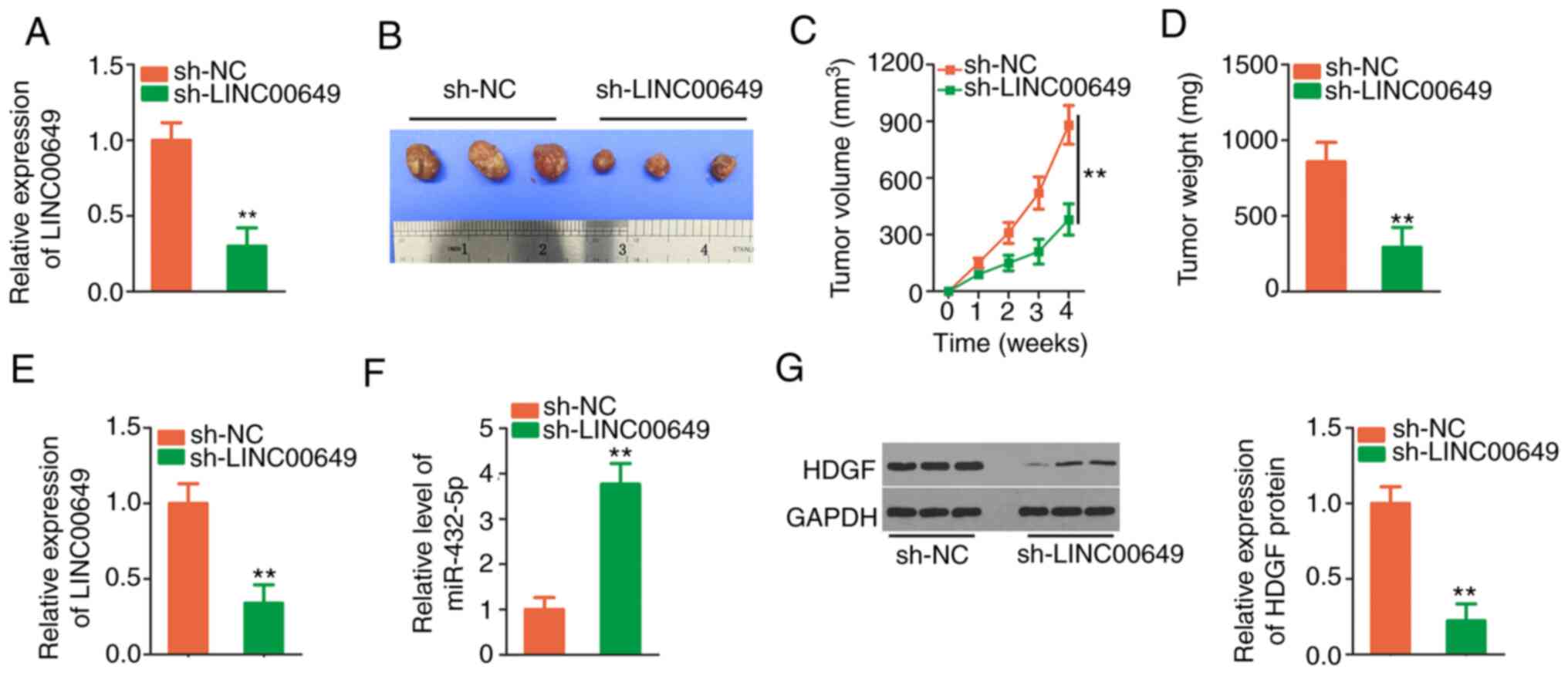
Depletion of LINC00649 attenuates
tumour growth in vivo
To demonstrate the effect of LINC00649 on in
vivo tumour growth, sh-LINC00649 was designed, and HCT 116
cells stably expressing sh-LINC00649 were established (Fig. 9A). The tumour diameter (Fig. 9B) and volume (Fig. 9C) in the sh-LINC00649 group were
evidently lower than sh-NC group. Additionally, the weight of
tumours in sh-LINC00649 group was decreased (Fig. 9D). Moreover, sh-LINC00649-injected
tumours presented decreased LINC00649 (Fig. 9E) and increased miR-432-5p levels
(Fig. 9F). Furthermore, HDGF
protein was reduced in tumours obtained from the sh-LINC00649 group
(Fig. 9G). Collectively,
downregulation of LINC00649 inhibited CRC growth in
vivo.
Discussion
As tumour molecular biology progresses, extensive
studies have confirmed that lncRNAs have a significant effect on
the oncogenicity of CRC (21–23).
With the universal application of high-throughput sequencing,
various lncRNAs have been identified to be differentially expressed
in CRC (10). Nevertheless,
numerous lncRNAs have yet to be investigated, and the mechanisms
involving lncRNAs in the pathogenesis of CRC are not completely
understood. Thus, the present study was initiated to offer
conclusive evidence regarding the involvement of LINC00649 in CRC
and the crosstalk between LINC00649 and miR-432-5p.
LINC00649 is expressed at a low level in acute
myeloid leukaemia (17). Patients
with acute myeloid leukaemia manifesting a low LINC00649 level have
shorter progression-free survival and overall survival than
patients with a high LINC00649 level (17). However, the expression and
contribution of LINC00649 in CRC have yet to be described.
According to TCGA database data, it was confirmed that LINC00649
was overexpressed in both COAD and READ relative to normal tissues.
In line with this tendency, the expression level of LINC00649 was
revealed to be increased in CRC tissues of our own cohort. More
importantly, the aberrant expression of LINC00649 exhibited an
inverse association with the prognosis of patients with CRC.
Functionally, loss of LINC00649 hindered the aggressive properties
of CRC cells. Therefore, the expression status and functions of
LINC00649 in human cancers exhibited tissue specificity.
Mechanistically, lncRNAs exercise their biological
tasks by working with different molecules (24). At the transcriptional level, lncRNAs
can epigenetically silence mRNAs or degrade proteins (25). At the post-transcriptional level,
ceRNA crosstalk among lncRNAs, miRNAs and mRNAs has been recognized
and has attracted increasing interest (26). The cytoplasmic localization of
lncRNAs can operate as ‘miRNA sponges’ and competitively bind to
certain miRNAs, thereby decreasing the targets of miRNAs from
silencing or translational inhibition (27). Clarification of the subcellular
localization of LINC00649 revealed that it was located in the
cytosol of CRC cells. Accordingly, LINC00649 may function as a
ceRNA in CRC.
Predictably, it was revealed that miR-432-5p may
play a role as a miRNA sponge of LINC00649. Additionally, the
present study revealed that the silencing of LINC00649 distinctly
improved miR-432-5p expression in CRC cells. Next, luciferase
reporter assays and Ago2 RIP assays confirmed that LINC00649
physically interacted and directly bound with miR-432-5p in CRC,
thus sequestering miR-432-5p at the molecular level. Subsequent
experiments were performed to determine the target mRNA of
miR-432-5p to construct a complete lncRNA/miRNA/mRNA pathway. The
mechanistic investigation identified HDGF as a direct downstream
functional target of miR-432-5p. In CRC, a negative expression
correlation was confirmed between LINC00649 and miR-432-5p, while
LINC00649 and HDGF levels presented a positive correlation.
Furthermore, HDGF was under the positive control of LINC00649 in
CRC cells, which occurred through sponging miR-432-5p.
Collectively, a novel ceRNA theory consisting of LINC00649,
miR-432-5p and HDGF was identified in CRC.
To date, multiple studies have highlighted the
involvement of miR-432-5p in several human cancers (28–30).
Our present study also attempted to reveal the importance of
miR-432-5p in CRC. A notable downregulation of miR-432-5p in CRC
was revealed in both the TCGA database and our own cohort, which
was in line with a recent study (31). miR-432-5p has anti-oncogenic
activities in CRC, and HDGF was confirmed as a downstream target of
miR-432-5p. HDGF was validated as an essential contributor to the
malignancy of CRC (32–35). Numerous miRNAs, including miR-93-5p
(36), miR-511 (37), and miR-610 (38), have been revealed to participate in
the regulation of HDGF in CRC, but it remains uncertain whether
HDGF can be regulated by certain lncRNAs in CRC. In particular,
compelling evidence has been presented to support that LINC00649
positively affects the expression pattern of HDGF. Furthermore, a
functional rescue assay was employed and revealed that the
exogenous introduction of miR-432-5p inhibitor or HDGF
overexpression plasmid partially abated the anticarcinogenic
actions of LINC00649 knockdown in CRC cells. All of the
aforementioned observations indicated that LINC00649 modulated the
miR-432-5p/HDGF axis and thereby aggravated the oncogenicity of CRC
cells.
The present study had several limitations. First,
only 52 pairs of CRC tissues and adjacent normal tissues were
collected, thus the sample size was too small. Furthermore, the
regulatory actions of LINC00649 silencing on CRC cell
proliferation, apoptosis, migration and invasion were explored
in vitro; however, the effects of LINC00649 overexpression
on these aggressive phenotypes were not examined. Finally, this
study did not investigate the influence of LINC00649 on metastasis
in vivo.
In summary, the present research demonstrated that
LINC00649 played a carcinogenic role in CRC by competitively
binding miR-432-5p, thus inducing increased HDGF expression.
Jointly, the LINC00649/miR-432-5p/HDGF pathway may serve as an
attractive target for CRC therapy.
Acknowledgements
Not applicable.
Funding
The present study was supported by The First People's Hospital
of Chongqing Liangjiang New District (Chongqing, China).
Availability of data and materials
All datasets used and/or analysed during the current
study are available from the corresponding author on reasonable
request.
Authors' contributions
JW and XL designed the research. JB, XB, JW and XL
conducted the experiments. XL analysed the obtained data. JW and XL
wrote the manuscript. XL supervised the junior staff. All authors
read and approved the final manuscript. JW and XL confirm the
authenticity of all raw data.
Ethics approval and consent to
participate
The present study was authorized (approval no.
EC-FPHCQLND-20150815) by the Ethics Committee of The First People's
Hospital of Chongqing Liangjiang (Chongqing, China). Written
informed consent was provided by all participants. The animal study
was authorized (approval no. IACUC-FPHCQLND-20190601) by the
Institutional Animal Care and Use Committee of The First People's
Hospital of Chongqing Liangjiang.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Bray F, Ferlay J, Soerjomataram I, Siegel
RL, Torre LA and Jemal A: Global cancer statistics 2018: GLOBOCAN
estimates of incidence and mortality worldwide for 36 cancers in
185 countries. CA Cancer J Clin. 68:394–424. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Siegel RL, Miller KD, Goding Sauer A,
Fedewa SA, Butterly LF, Anderson JC, Cercek A, Smith RA and Jemal
A: Colorectal cancer statistics, 2020. CA Cancer J Clin.
70:145–164. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Mitry E, Guiu B, Cosconea S, Jooste V,
Faivre J and Bouvier AM: Epidemiology, management and prognosis of
colorectal cancer with lung metastases: A 30-year population-based
study. Gut. 59:1383–1388. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Dekker E and Rex DK: Advances in CRC
prevention: Screening and surveillance. Gastroenterology.
154:1970–1984. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Tol J, Koopman M, Cats A, Rodenburg CJ,
Creemers GJ, Schrama JG, Erdkamp FL, Vos AH, van Groeningen CJ,
Sinnige HA, et al: Chemotherapy, bevacizumab, and cetuximab in
metastatic colorectal cancer. N Engl J Med. 360:563–572. 2009.
View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Ponting CP, Oliver PL and Reik W:
Evolution and functions of long noncoding RNAs. Cell. 136:629–641.
2009. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Kung JT, Colognori D and Lee JT: Long
noncoding RNAs: Past, present, and future. Genetics. 193:651–669.
2013. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Quinn JJ and Chang HY: Unique features of
long non-coding RNA biogenesis and function. Nat Rev Genet.
17:47–62. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Schwarzmueller L, Bril O, Vermeulen L and
Léveillé N: Emerging role and therapeutic potential of lncRNAs in
colorectal cancer. Cancers (Basel). 12:38432020. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Talebi A, Azizpour M, Agah S, Masoodi M,
Mobini GR and Akbari A: The relevance of long noncoding RNAs in
colorectal cancer biology and clinical settings. J Cancer Res Ther.
16 (Suppl):S22–S33. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Srinivasan S, Selvan ST, Archunan G,
Gulyas B and Padmanabhan P: MicroRNAs -the next generation
therapeutic targets in human diseases. Theranostics. 3:930–942.
2013. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Shukla GC, Singh J and Barik S: MicroRNAs:
Processing, maturation, target recognition and regulatory
functions. Mol Cell Pharmacol. 3:83–92. 2011.PubMed/NCBI
|
|
13
|
Wen XQ, Qian XL, Sun HK, Zheng LL, Zhu WQ,
Li TY and Hu JP: MicroRNAs: multifaceted regulators of colorectal
cancer metastasis and clinical applications. Onco Targets Ther.
13:10851–10866. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Ahadi A: The significance of microRNA
deregulation in colorectal cancer development and the clinical uses
as a diagnostic and prognostic biomarker and therapeutic agent.
Noncoding RNA Res. 5:125–134. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Weidle UH, Brinkmann U and Auslaender S:
microRNAs and corresponding targets involved in metastasis of
colorectal cancer in preclinical in vivo models. Cancer Genomics
Proteomics. 17:453–468. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Wang L, Cho KB, Li Y, Tao G, Xie Z and Guo
B: Long noncoding RNA (lncRNA)-mediated competing endogenous RNA
networks provide novel potential biomarkers and therapeutic targets
for colorectal cancer. Int J Mol Sci. 20:57582019. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Guo C, Gao YY, Ju QQ, Zhang CX, Gong M and
Li ZL: LINC00649 underexpression is an adverse prognostic marker in
acute myeloid leukemia. BMC Cancer. 20:8412020. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
He M, Lin Y and Xu Y: Identification of
prognostic biomarkers in colorectal cancer using a long non-coding
RNA-mediated competitive endogenous RNA network. Oncol Lett.
17:2687–2694. 2019.PubMed/NCBI
|
|
19
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(−Delta Delta C(T)) method. Methods. 25:402–408. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Deng M, Bragelmann J, Schultze JL and
Perner S: Web-TCGA: An online platform for integrated analysis of
molecular cancer data sets. BMC Bioinformatics. 17:722016.
View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Hon KW, Abu N, Ab Mutalib NS and Jamal R:
miRNAs and lncRNAs as predictive biomarkers of response to FOLFOX
therapy in colorectal cancer. Front Pharmacol. 9:8462018.
View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Rivandi M, Pasdar A, Hamzezadeh L,
Tajbakhsh A, Seifi S, Moetamani-Ahmadi M, Ferns GA and Avan A: The
prognostic and therapeutic values of long noncoding RNA PANDAR in
colorectal cancer. J Cell Physiol. 234:1230–1236. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Wei L, Wang X, Lv L, Zheng Y, Zhang N and
Yang M: The emerging role of noncoding RNAs in colorectal cancer
chemoresistance. Cell Oncol (Dordr). 42:757–768. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Gao N, Li Y, Li J, Gao Z, Yang Z, Li Y,
Liu H and Fan T: Long non-coding RNAs: The regulatory mechanisms,
research strategies, and future directions in cancers. Front Oncol.
10:5988172020. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Samimi H, Sajjadi-Jazi SM, Seifirad S,
Atlasi R, Mahmoodzadeh H, Faghihi MA and Haghpanah V: Molecular
mechanisms of long non-coding RNAs in anaplastic thyroid cancer: A
systematic review. Cancer Cell International. 20:3522020.
View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Qi X, Zhang DH, Wu N, Xiao JH, Wang X and
Ma W: ceRNA in cancer: Possible functions and clinical
implications. J Med Genet. 52:710–718. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Ergun S and Oztuzcu S: Oncocers:
ceRNA-mediated cross-talk by sponging miRNAs in oncogenic pathways.
Tumour Biol. 36:3129–3136. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Das S and Bhattacharyya NP: Heat shock
factor 1 regulates hsa-miR-432 expression in human cervical cancer
cell line. Biochem Biophys Res Commun. 453:461–466. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Jiang N, Chen WJ, Zhang JW, Xu C, Zeng XC,
Zhang T, Li Y and Wang GY: Downregulation of miR-432 activates
Wnt/β-catenin signaling and promotes human hepatocellular carcinoma
proliferation. Oncotarget. 6:7866–7879. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Chen L, Kong G, Zhang C, Dong H, Yang C,
Song G, Guo C, Wang L and Yu H: MicroRNA-432 functions as a tumor
suppressor gene through targeting E2F3 and AXL in lung
adenocarcinoma. Oncotarget. 7:20041–20053. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Luo M, Hu Z, Kong Y and Li L:
MicroRNA-432-5p inhibits cell migration and invasion by targeting
CXCL5 in colorectal cancer. Exp Ther Med. 21:3012021. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Lian J, Tang J, Shi H, Li H, Zhen T, Xie
W, Zhang F, Yang Y and Han A: Positive feedback loop of
hepatoma-derived growth factor and beta-catenin promotes
carcinogenesis of colorectal cancer. Oncotarget. 6:29357–29374.
2015. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Liao F, Liu M, Lv L and Dong W:
Hepatoma-derived growth factor promotes the resistance to
anti-tumor effects of nordihydroguaiaretic acid in colorectal
cancer cells. Eur J Pharmacol. 645:55–62. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Hu TH, Lin JW, Chen HH, Liu LF, Chuah SK
and Tai MH: The expression and prognostic role of hepatoma-derived
growth factor in colorectal stromal tumors. Dis Colon Rectum.
52:319–326. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Liao F, Dong W and Fan L: Apoptosis of
human colorectal carcinoma cells is induced by blocking
hepatoma-derived growth factor. Med Oncol. 27:1219–1226. 2010.
View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Hong YG, Huang ZP, Liu QZ, E JF, Gao XH,
Xin C, Zhang W, Li P and Hao LQ: MicroRNA-95-3p inhibits cell
proliferation and metastasis in colorectal carcinoma by HDGF.
Biomed J. 43:163–173. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
He S, Wang G, Ni J, Zhuang J, Zhuang S,
Wang G, Ye Y and Xia W: MicroRNA-511 inhibits cellular
proliferation and invasion in colorectal cancer by directly
targeting hepatoma-derived growth factor. Oncol Res. 26:1355–1363.
2018. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Sun B, Gu X, Chen Z and Xiang J: MiR-610
inhibits cell proliferation and invasion in colorectal cancer by
repressing hepatoma-derived growth factor. Am J Cancer Res.
5:3635–3644. 2015.PubMed/NCBI
|