Introduction
The liver is a crucial digestive organ of humans and
in charge of detoxification. Acute hepatic injury is an aggressive
type of hepatic disease (1), and
its harmful effect cannot be ignored. Acute hepatic injury refers
to acute damage or necrosis of hepatocytes (2), causing abnormal hepatic function.
Patients with acute hepatic injury exhibit a series of symptoms of
acute hepatitis, such as yellowing of the skin and sclera, loss of
appetite, and pain in the hepatic area. Generally, drug-induced
hepatic injury is the most widespread cause (3,4).
In addition, alcohol (5),
hepatitis B (6) and appendicitis
are also principal factors. Endoplasmic reticulum stress (ERS) is a
signal response pathway of the cellular self-protection mechanism,
which participates in the physiological and pathological processes
of numerous diseases (7–11). Without exception, ERS is also
involved in the occurrence and development of a variety of hepatic
injuries (12). By studying the
effect of drugs on hepatic injury in mice, the protective effect of
drugs against ERS-induced acute hepatic injury has been revealed
(13). Through the establishment
of alcoholic hepatic injury in mice, it has been demonstrated that
alcohol may induce ERS and cause hepatic damage (14). The aforementioned mechanisms
suggest that hepatic damage is relevant to ERS.
Stress-associated endoplasmic reticulum protein 1
(SERP1) is involved in the regulation of ERS (15). SERP1 stimulates the generation of
β-amyloid in cells which undergo ERS, as is observed in diabetes
(16). Additionally, increased
ERS in the pancreas and other organs of mice lacking SERP1
indicated that SERP1 can alleviate ERS (17). The levels of ERS increases during
early acute hepatic injury, and it can activate GSK3β signaling
(18,19). GSK3β, a serine/threonine kinase,
was originally reported to be a key enzyme in the process of
glycogen synthesis. It acts on numerous signal proteins and
transcription factors, and regulates cell survival,
differentiation, proliferation and apoptosis. Furthermore, using a
D-galactosamine (D-GalN)/lipopolysaccharide (LPS)-induced acute
hepatic failure mouse model, it has been revealed that ERS is
activated in these mice. Inhibition of activation may have a
protective effect on hepatic injury, and the mechanism of action
involves reducing inflammation, expression of apoptosis factors and
signal transduction (20).
In the present study, D-GalN/LPS induction was used
to construct a model of early acute hepatic injury. Furthermore, a
series of assays was performed to explore whether the
overexpression of SERP1 could reduce ERS by inhibiting the activity
of GSK3β signaling, thereby reducing hepatocyte apoptosis.
Materials and methods
Cell culture and mouse model
establishment
Normal rat hepatocytes (BRL) were purchased from the
American Type Culture Collection. Cells were cultured in DMEM
(Gibco; Thermo Fisher Scientific, Inc.) supplemented with 10% FBS
(Gibco; Thermo Fisher Scientific, Inc.), 100 U/ml
penicillin/streptomycin (Thermo Fisher Scientific, Inc.) and
maintained in an incubator with 5% CO2 at 37°C. LPS (1
mg/l) was used to induce the formation of damaged hepatocytes.
Sodium phenylbutyrate (4-PBA; Sigma-Aldrich), an ERS inhibitor, was
dissolved using sterile PBS.
The present study was performed following the
National Institutes of Health Guide for the Care and Use of
Laboratory Animals. The study was approved by the Huaihe Hospital
of Henan University Animal Ethics Committee (approval no.
HHIACUC-201956; Kaifeng, China). A total of 16 male wild-type
(C57BL/6) mice (age, 6–8 weeks; weight, 20–22 g; Guangdong Medical
Laboratory Animal Center) were used in the present study. Half of
the mice were injected intraperitoneally with D-GalN (400 mg/kg)
and LPS (0.02 mg/kg; Sigma-Aldrich). The control mice were injected
with the same amount of normal saline. Throughout the experiment,
mice were kept at a room temperature of 25°C with 12 h light/12 h
dark cycle and a relative humidity of 55%. They had free access to
water and food. For blood collection, the mice were anesthetized
using sodium pentobarbital (80 mg/kg) and the abdominal cavity was
then opened through an incision along the midline of the abdomen to
expose the abdominal aorta. A total of 0.6 ml of blood was
collected using a syringe. Subsequently, the mice were euthanized
by cervical dislocation. All efforts were made to minimize animal
suffering.
Cell transfection
BRL cells (5×105 cells/well) were seeded
into 6-well plates until they reached 70–80% confluence. The coding
sequences of SERP1 (Gene ID: 80881, Accession Number: NM_030835.2)
and GSK3β (Gene ID: 84027, NM_032080.1) were cloned into pcDNA3.1
(Hunan YouBio Co., Ltd.). pcDNA3.1-SERP1, pcDNA3.1-GSK3β and empty
vector as a negative control (NC) were transfected into BRL cells
at 37°C for 6 h using Lipofectamine® 3000 reagent
(Thermo Fisher Scientific, Inc.) according to the manufacturer's
instructions. Cells were induced using LPS as requested (1 mg/l)
after 48 h of transfection.
H&E staining
The hepatic tissue was obtained after lavage, fixed
in 4% paraformaldehyde at room temperature overnight, dehydrated
and embedded in paraffin. Subsequently, it was cut into 4 µm
sections and stained with H&E for 10 min at room temperature.
Following color separation, rinsing and dehydration, the tissue was
pressed flat and fixed with a new cover glass. After drying,
physiological changes of the tissue were observed under a light
microscope (magnification, ×200; Olympus Corporation).
Immunohistochemistry
Tissues were pretreated as aforementioned for
H&E staining. Subsequently, paraffin sections were washed with
xylene, rehydrated with descending alcohol and antigen retrieval
was performed at 95°C for 20 min. Then, 5% goat serum (Beijing
Solarbio Technology Co. Ltd.) was used for blocking for 10 min at
room temperature and sections were then incubated with the SERP1
primary antibody (1:200; cat. no. DF9873; Affinity Biosciences) at
4°C overnight. After washing with PBS, biotin-labeled goat
anti-rabbit secondary antibody (1:2,000; cat. no. 31820; Thermo
Fisher Scientific, Inc.) was added. Streptavidin Biotin-peroxidase
Complex (Beyotime Institute of Biotechnology) was diluted and added
after incubation at 37°C for 0.5 h. Subsequently, the sections were
developed using 3,3′-diaminobenzidine staining solution,
counterstained with hematoxylin for 25 sec at room temperature,
dehydrated and mounted. The differences were observed under a light
microscope (magnification, ×400; Olympus Corporation).
Detection of alanine aminotransferase
(ALT) and aspartate aminotransferase (AST)
The mouse abdominal aorta blood was drawn and the
upper serum was separated by centrifugation (1,000 × g) for 10 min
at 4°C. The sample was added to the well and mixed with the matrix
solution at 37°C, and the color developing agent was added. The
stop solution was added after 20 min in a 37°C water bath. After 15
min at room temperature, the microplate reader was used to detect
the optical density (OD) value at 510 nm. ALT and AST levels were
calculated using the standard curve.
Western blotting
Protein was extracted from BRL cells and homogenized
in RIPA lysis buffer (Beyotime Institute of Biotechnology). A BCA
protein assay kit (Beyotime Institute of Biotechnology) was used
for protein quantification. The proteins (20 µg/lane), which were
separated using an 10% SDS-polyacrylamide gel, were transferred to
a PVDF membrane. The membranes were first incubated in 5% skimmed
milk for 1 h at room temperature. Blots were incubated with primary
antibodies [glucose-regulated protein 78 (GRP78; product code
ab108615; 1:1,000), glucose-regulated protein 94 (GRP94; product
code ab238126; 1:1,000), SERP1 (product code ab254839; 1:1,000),
NLR family pyrin domain containing 3 (NLRP3; product code ab263899;
1:1,000), apoptosis-associated speck-like protein containing a CARD
(ASC; product code ab180799; 1:1,000), GSK3β (cat. no. ab227208;
1:1,000), phosphorylated (p-)GSK3β (product code ab75745; 1:1,000),
β-catenin (product code ab32572; 1:5,000), Bcl-2 (product code
ab196495; 1:1,000), Bad (product code ab32445; 1:1,000), Bax
(product code ab182733; 1:2,000), all from Abcam; cleaved-caspase3
(cat. no. AC033; 1:1,000) and caspase3 (cat. no. AF1213; 1:1,000),
caspase-1 (cat. no. AF1681; 1:1,000), CHOP (AF6684; 1:1,000) and
GAPDH (AF1186; 1:2,000) from Beyotime Institute of Biotechnology]
at 4°C overnight after washing with Tris-buffered saline with 0.01%
Tween-20. Subsequently, they were washed again, and strips were
incubated with HRP-conjugated anti-rabbit antibody (cat. no. 31460;
1:50,000; Thermo Fisher Scientific, Inc.) at room temperature for
1.5 h. ECL (Shanghai Yeasen Technology Co. Ltd.) was added. GAPDH
was used as a control and Image Lab software (v4.0; Bio-Rad
Laboratories, Inc.) was used to analyze data.
Reverse transcription-quantitative PCR
(RT-qPCR)
Total RNA was isolated from BRL cells using
TRIzol® reagent (Invitrogen; Thermo Fisher Scientific,
Inc.) and cDNA was primed using a Reverse Transcriptase kit
(Invitrogen; Thermo Fisher Scientific, Inc.) according to the
manufacturer's instructions. RT-qPCR was performed using a
QuantiTect SYBR Green PCR Kit (Qiagen, Inc.). The conditions of PCR
amplification were as follows: 95°C for 3 min, followed by 40
cycles at 95°C for 10 sec and 60°C for 30 sec. The
2−ΔΔCq method (21)
was applied for data analysis with normalization to GAPDH. The
sequences of the primers were as follows: SERP1 forward,
5′-CGGGCGGAGTCCATACTTC-3′ and reverse, 5′CCGGGAGGAAAGAGTCCAAC-3′;
GSK3β forward, 5′-CACTGTGTAGCCGTCTCCTG-3′ and reverse,
5′-GAAGAGGGCAGGTGTGTCTC-3′; and GAPDH forward,
5′-TCTCTGCTCCTCCCTGTTCT-3′ and reverse,
5′-TACGGCCAAATCCGTTCACA-3′.
ELISA
Expression levels of inflammatory factors: TNF-α
(product no. PT516; Beyotime Institute of Biotechnology), IL-18
(cat. no. E-EL-R0567c; Wuhan Elabscience Co. Ltd.), IL-6 (product
no. PI328; Beyotime Institute of Biotechnology) and IL-1β (product
no. PI303; Beyotime Institute of Biotechnology) in BRL cells were
detected using ELISA kits according to the manufacturer's protocol.
A microplate reader was used to detect the OD value at 450 nm.
TUNEL assay
Cells (1×104/well) were seeded in a
24-well plate and cultured to occupy 80% of the well in an
incubator with 5% CO2 at 37°C. Subsequently, cells were
fixed with 4% paraformaldehyde for 1 h at 4°C, stained using the
TUNEL test kit for 1 h at 37°C (Beyotime Institute of
Biotechnology) according to the manufacturer's protocol. DAPI (2
µg/ml) was used for nuclear staining for 5 min at room temperature.
Antifade mounting medium (Beyotime Institute of Biotechnology) was
added on the slide and cells were observed from five field of view
under a fluorescence microscope (magnification, ×200; Olympus
Corporation).
Luciferase assay
BRL cells (5×105/well) were seeded into
6-well plates and co-transfected with 1 µg reporter gene TOP Flash
and FOP Flash (Beyotime Institute of Biotechnology), and
overexpression vector. After transfection with
Lipofectamine® 3000 reagent (Thermo Fisher Scientific,
Inc.) for 18 h, cells were lysed in passive phenylbenzothiazole
buffer and sample lysates were measured using a Mithras LB 940
Multimode Microplate Reader (Berthold Technologies GmbH & Co.
KG). The activity was measured using a dual-luciferase reporter
gene assay kit (Beyotime Institute of Biotechnology) and normalized
to Renilla luciferase. The results are presented as a normalized
TOP Flash/FOP Flash value.
Statistical analysis
Data are presented as the mean ± SD and were
analyzed using GraphPad Prism 8.0 software (GraphPad Software,
Inc.). Statistical significance was determined by one-way ANOVA
with Tukey's post hoc test for multiple groups and unpaired
Student's t-test for two groups. Each cell experiment was performed
at least three times. P<0.05 was considered to indicate a
statistically significant difference.
Results
SERP1 expression in acute hepatic
injury tissues and cells
The mice were injected intraperitoneally with
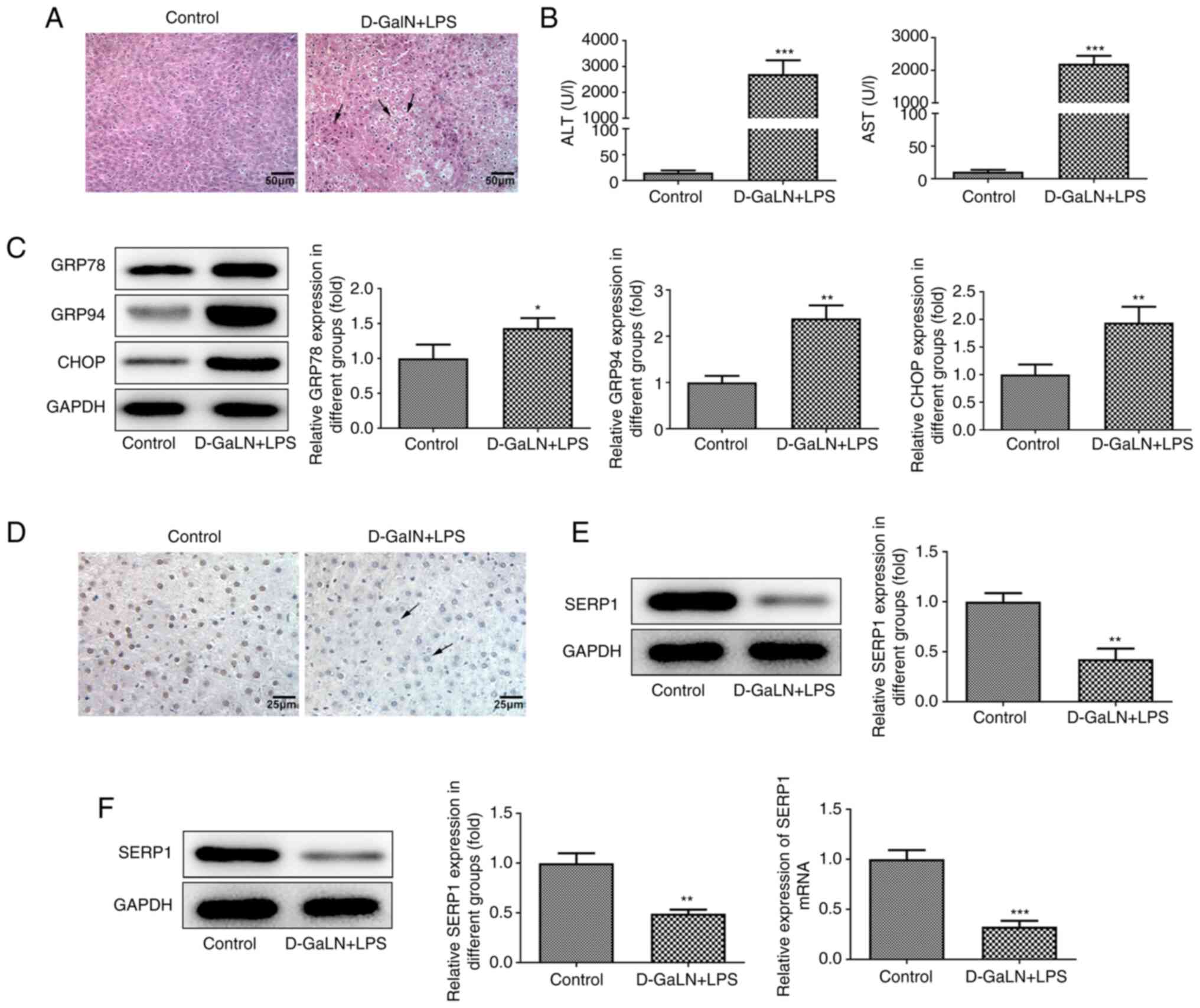
D-GalN/LPS. H&E staining was performed to examine pathological
changes of the tissues of mice in the model group. The hepatic
structure of the control group was normal and the structure of the
LPS/D-GalN group exhibited a mass of vacuoles. The hepatic cells
were disintegrated, with diffuse necrosis, and normal structure was
lost, accompanied by inflammatory cell infiltration (Fig. 1A). ALT and AST are both biomarkers
of acute hepatic injury in the serum (22). The value of the LPS/D-GalN group
was significantly increased compared with that of the control group
(Fig. 1B). The expression levels
of ERS-related proteins were determined using western blotting. The
expression levels of GRP78, GRP94 and CHOP were all significantly
increased (Fig. 1C).
The expression levels of SERP1 in acute hepatic
injury tissues were subsequently assessed using
immunohistochemistry and western blotting. Compared with that of
the control, the brownish yellow color of the cytoplasm in the
LPS/D-GalN group was less intense (Fig. 1D). The SERP1 expression in the
LPS/D-GalN group was significantly decreased according to the
results of western blotting (Fig.
1E). In addition, SERP1 expression in acute hepatic injury
cells was determined by western blotting and RT-qPCR. In these two
assays, the expression levels in the induced group were
significantly decreased (Fig.
1F).
Effect of SERP1 overexpression on the
expression of inflammatory factors and cell apoptosis
The overexpression plasmid of SERP1 was constructed,
and overexpression was confirmed using RT-qPCR and western
blotting. SERP1 expression in the overexpression-SERP1 (OE-SERP1)
group was higher than that in the NC group (Fig. 2A and B). The expression levels of
inflammatory factors (TNF-α, IL-18, IL-6 and IL-1β) were
subsequently determined by ELISAs. It was observed from the
histograms that the levels of inflammatory factors of hepatocytes
in the LPS-induced group were increased; however, when SERP1 was
overexpressed, the levels of inflammatory factors were
significantly decreased. The effect of its overexpression on
inflammatory factors was similar to the effect of 4-PBA, which is
an ERS inhibitor (Fig. 2C). In
addition, NLRP3 inflammasome (NLRP3, ASC and caspase-1) expression
was determined by western blotting. The expression levels were
increased when cells were induced by LPS, and SERP1 overexpression
contributed to the decreased inflammasome expression. Although the
inhibitory effect on inflammasome expression was not as distinct as
that of 4-PBA, it was still notable (Fig. 2D).
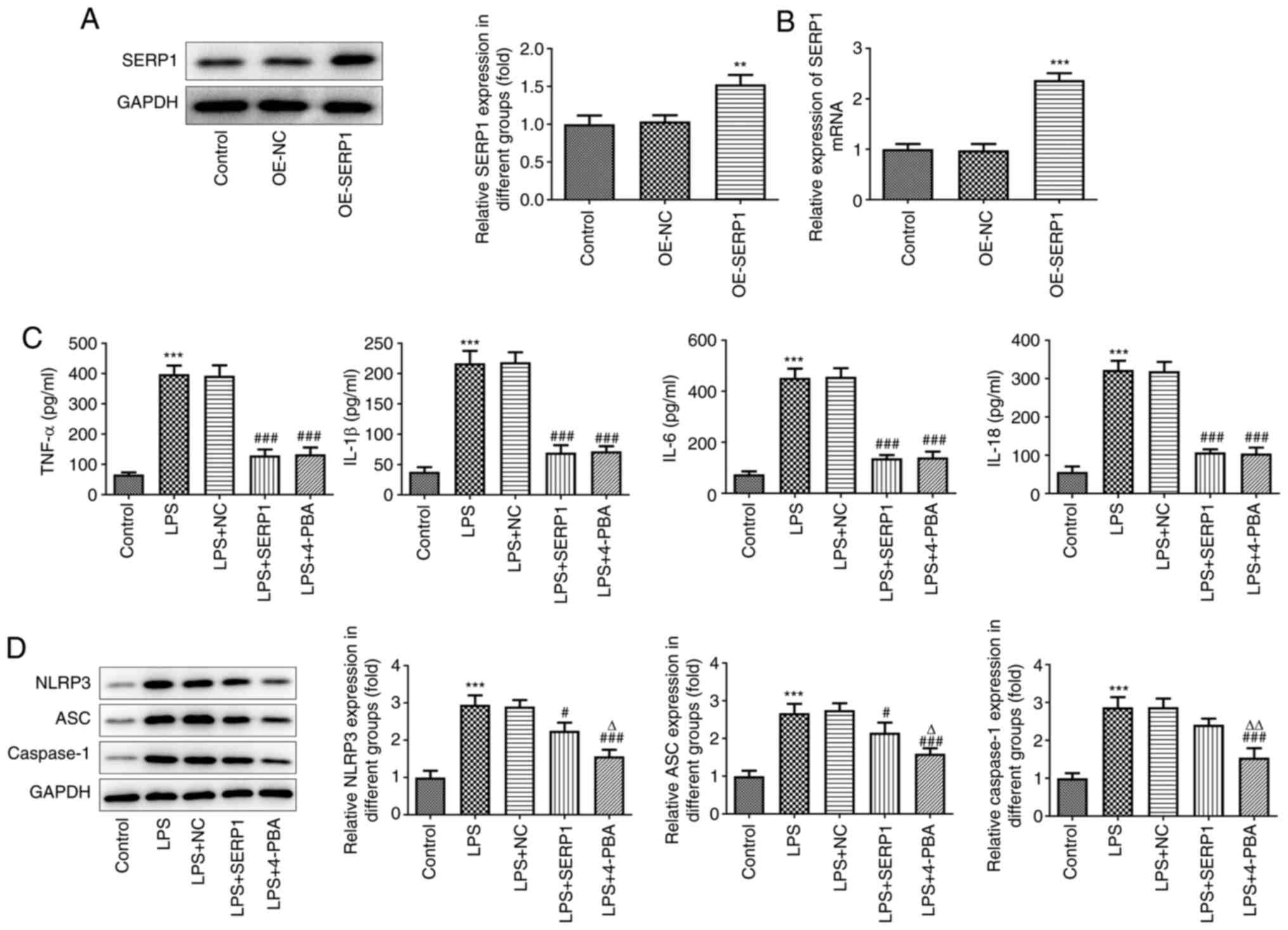 | Figure 2.Effect of SERP1 overexpression on the
expression of inflammatory factors in hepatocytes. Expression
levels of SERP1 following transfection with SERP1 plasmid detected
by (A) western blotting and (B) reverse transcription-quantitative
PCR. **P<0.01, ***P<0.001 vs. OE-NC. (C) Expression levels of
inflammatory factors (TNF-α, IL-18, IL-6 and IL-1β) detected by
ELISA. (D) NLRP3 inflammasome (NLRP3, ASC and caspase-1) expression
detected by western blotting. ***P<0.001 vs. the control;
#P<0.05, ###P<0.001 vs. LPS + NC;
ΔP<0.05, ΔΔP<0.01 vs. LPS + SERP1; n≥3.
SERP1, stress-associated endoplasmic reticulum protein 1; OE,
overexpression; NC, negative control; NLRP3, NLR family pyrin
domain containing 3; ASC, apoptosis-associated speck-like protein
containing a CARD; LPS, lipopolysaccharide. |
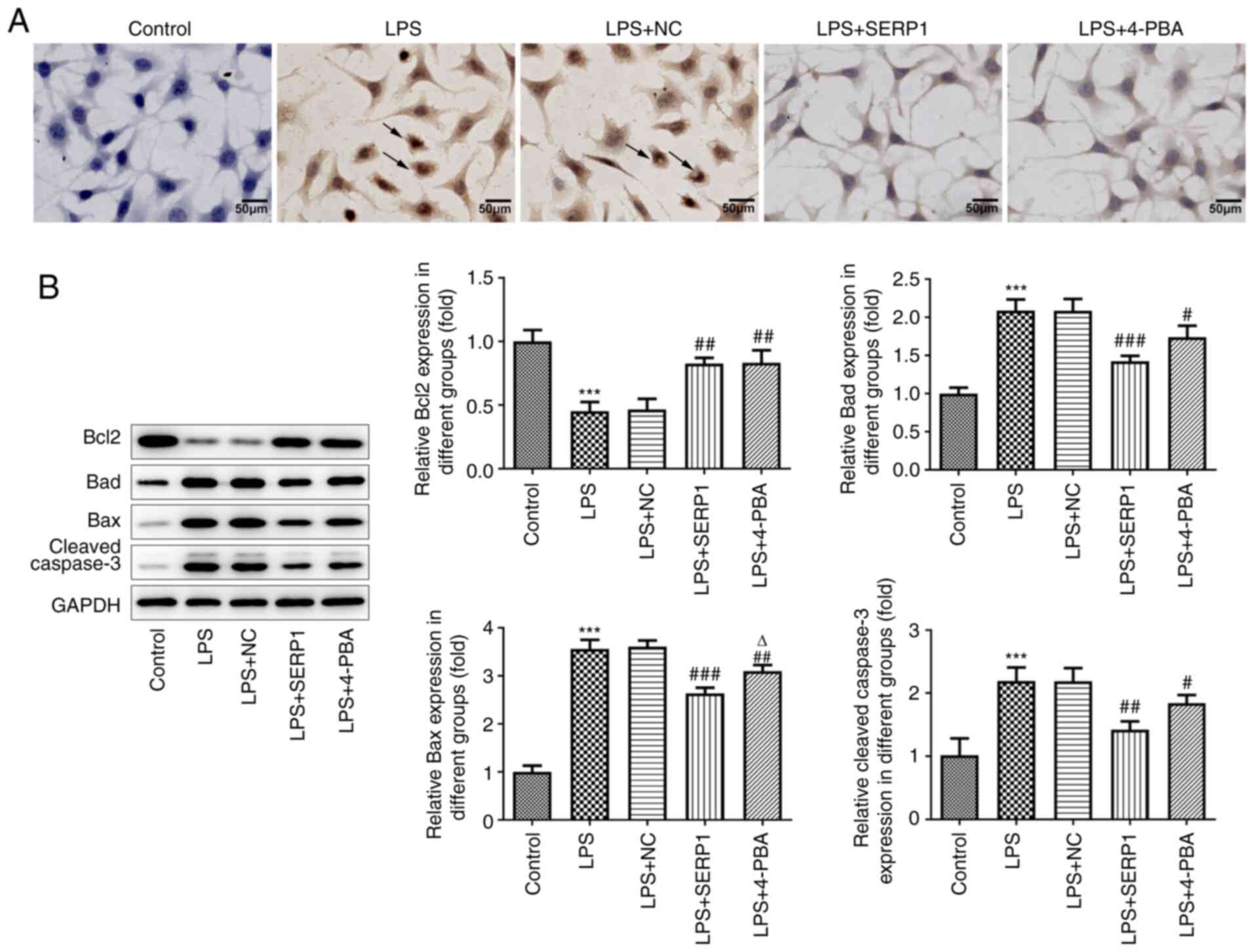
A TUNEL assay was performed to assess cell
apoptosis. Normal cells will not be stained because there is no DNA
break. It was observed from the images that the cytoplasm of
LPS-induced cells was stained, but not that of the control group.
Reduced staining was observed following SERP1 overexpression, which
was consistent with the results observed for the 4-PBA group
(Fig. 3A). The expression levels
of apoptosis-related proteins were detected by western blotting.
The expression levels of Bad, Bax and cleaved caspase-3 were
increased when cells were induced by LPS and decreased when SERP1
was overexpressed. The opposite trend was observed for Bcl-2
(Fig. 3B).
Effect of SERP1 overexpression on the
expression levels of ERS-related proteins and
GSK3β/β-catenin/T-cell factor (TCF)/lymphoid enhancing factor (LEF)
signaling in hepatocytes
Western blotting was performed to assess the
expression levels of GRP78, GRP94 and CHOP in hepatocytes. The
expression levels were all increased when cells were induced by LPS
and decreased following SERP1 overexpression or addition of 4-PBA
(Fig. 4A). This suggested that
SERP1 overexpression alleviated ERS caused by LPS. To investigate
the effect of SERP1 overexpression on signaling, a TOP Flash/FOP
Flash fluorescent gene reporter assay was used to detect TCF/LEF
activity. TOP Flash/FOP Flash luciferase activity ratios in the
control, LPS, LPS with NC, LPS with OE-SERP1 and LPS with 4-PBA
groups were investigated. FOP Flash was used to correct for
β-catenin-independent expression and transfection efficiency. In
the LPS combined with OE-SERP1 group, the TOP Flash/FOP Flash
luciferase activity ratio was decreased compared with that of the
LPS + NC group (Fig. 4B).
Additionally, GSK3β/β-catenin expression was detected by western
blotting. The expression levels of β-catenin and GSK3β in the LPS
group were increased, and this was reversed by SERP1
overexpression. Conversely, the levels of p-GSK3β in different
groups revealed the opposite trend (Fig. 4C).
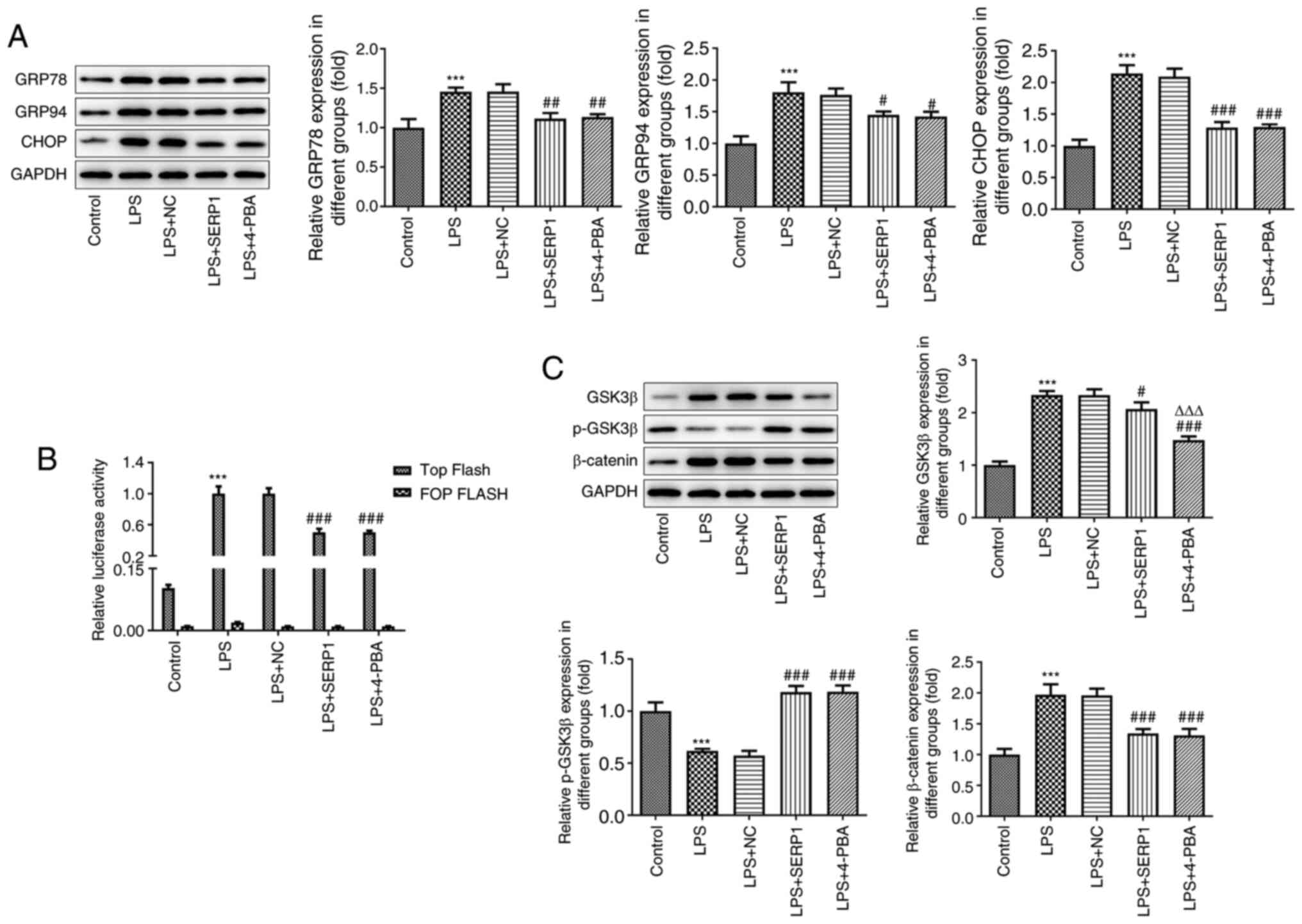 | Figure 4.Effect of SERP1 overexpression on
endoplasmic reticulum stress-related protein expression and
GSK3β/β-catenin/TCF/LEF signaling in hepatocytes. (A) Expression
levels of GRP78, GRP94 and CHOP in hepatocytes detected by western
blotting. (B) TOP Flash/FOP Flash fluorescent gene reporter assay
to detect TCF/LEF activity. (C) GSK3β/β-catenin expression was
detected by western blotting. ***P<0.001 vs. the control;
#P<0.05, ##P<0.01,
###P<0.001 vs. LPS + NC; ΔΔΔP<0.001 vs.
LPS + SERP1; n≥3. SERP1, stress-associated endoplasmic reticulum
protein 1; GRP78, glucose-regulated protein 78; GRP94,
glucose-regulated protein 94; TCF, T-cell factor; LEF, lymphoid
enhancing factor; LPS, lipopolysaccharide; NC, negative
control. |
GSK3β/β-catenin signaling activation
reverses the effect of SERP1 overexpression on ERS and cell
apoptosis
The GSK3β overexpression plasmid was constructed and
transfected into hepatocytes, and the overexpression was verified
by RT-qPCR and western blotting. The results demonstrated that
GSK3β was successfully overexpressed (Fig. 5A). Additionally, GSK3β/β-catenin
expression was detected by western blotting and TCF/LEF activity
was detected using a luciferase assay. The results of the TOP
Flash/FOP Flash luciferase assay were reversed following GSK3β
overexpression compared with the results following SERP1
overexpression (Fig. 5B).
Additionally, based on the results regarding GSK3β and β-catenin
expression, GSK3β overexpression reversed the inhibitory effect of
SERP1 overexpression (Fig. 5C).
These results indicated that GSK3β/β-catenin signaling activation
reversed the role of SERP1 overexpression in GSK3β/β-catenin
signaling expression. In addition to the aforementioned
experiments, western blotting was performed to detect the
expression levels of proteins related to ERS and apoptosis, and a
TUNEL assay was employed to detect cell apoptosis. It was revealed
that GSK3β overexpression reversed the effect of SERP1
overexpression (Fig. 5D-F).
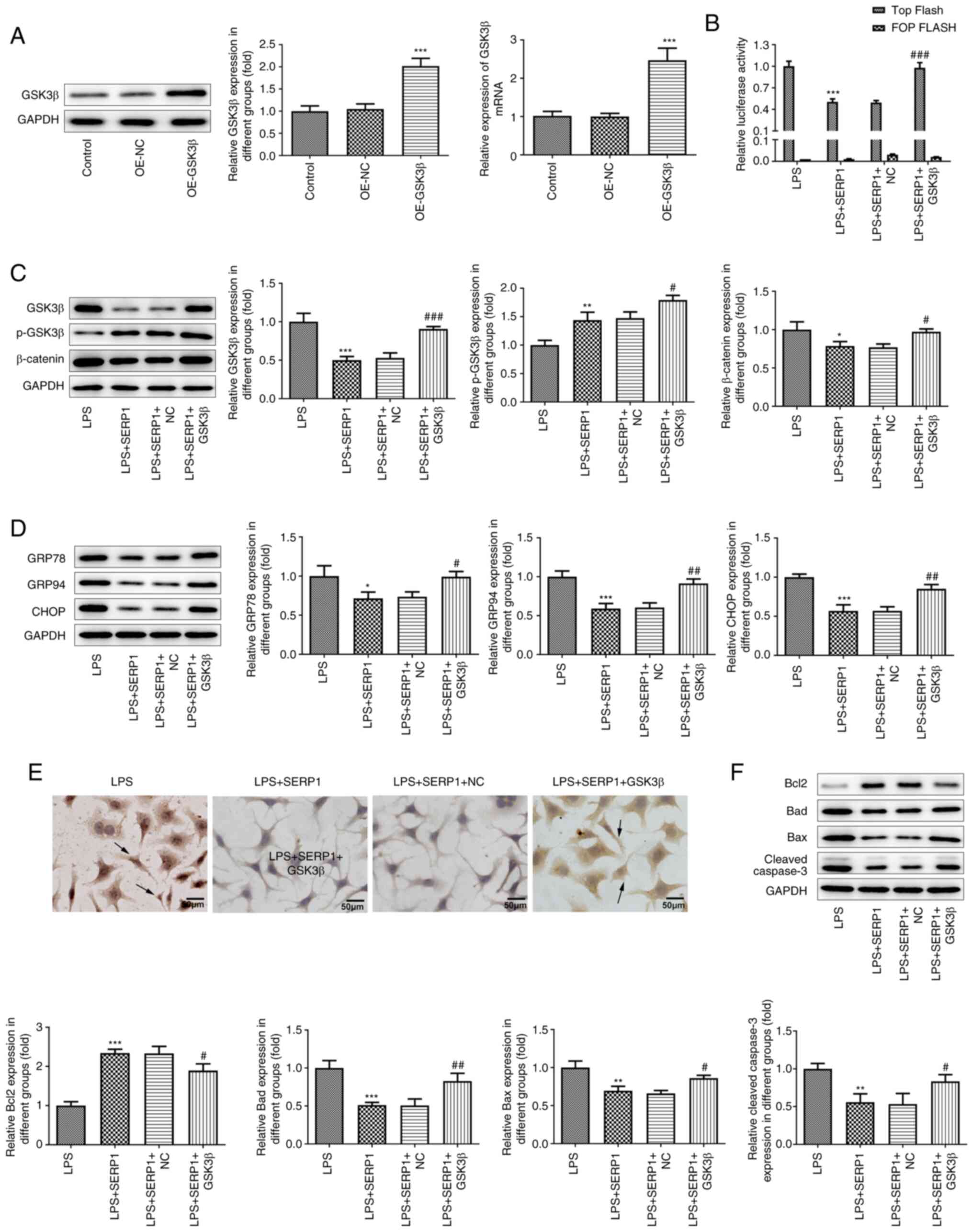 | Figure 5.GSK3β/β-catenin signaling activation
reverses the effect of SERP1 overexpression on ERS and cell
apoptosis. (A) Overexpression was verified by reverse
transcription-quantitative PCR and western blotting. ***P<0.001
vs. OE-NC; n≥3. (B) TCF/LEF activity was detected using a
luciferase assay. (C) GSK3β/β-catenin expression was detected by
western blotting. (D) Western blotting was applied to detect
ERS-related protein expression. (E) A TUNEL assay was employed to
detect cell apoptosis. (F) Apoptosis-related protein expression was
detected by western blotting. *P<0.05, **P<0.01,
***P<0.001 vs. LPS; #P<0.05,
##P<0.01, ###P<0.001 vs. LPS + SERP1 +
NC; n≥3. SERP1, stress-associated endoplasmic reticulum protein 1;
ERS, endoplasmic reticulum stress; OE, overexpression; TCF, T-cell
factor; LEF, lymphoid enhancing factor; LPS, lipopolysaccharide;
NC, negative control. |
Discussion
The liver serves a central role in immune
homeostasis and metabolism. The liver is responsible for numerous
functions, such as blood coagulation, energy production, hormone
balance and detoxification (23).
Additionally, it contains numerous macrophages, which can detect,
capture and eliminate bacteria and viruses that enter the human
body. At the same time, the biotransformation process of the human
body is mainly carried out in the liver, and thus, it is vulnerable
to numerous factors (24).
Alcoholism, excessive drug use and viral infections can all cause
acute hepatic damage (25,26).
D-GalN can cause hepatic degeneration and necrosis through
competitive depletion of uridine phosphate. As a uridine
phosphate-interfering agent, D-GalN is converted into uridine
galactose diphosphate in the body, which affects the dysfunction of
hepatocyte synthesis, causing the liver to produce an increased
amount of reactive oxygen species, and thus causing hepatic damage
in the body (27). LPS is one of
the main components of the cell wall of gram-negative bacteria and
is also called endotoxin. It can produce numerous inflammatory
factors. When LPS and D-GalN are used in combination, D-GalN can
strengthen the hepatic toxicity caused by LPS. The combined use is
widely used as a method of modeling acute hepatic injury (28).
In the present study, following intraperitoneal
injection of LPS/D-GalN in mice, by examining pathological sections
obtained from the mice, it was revealed that hepatocytes were
disintegrated and necrotic, and inflammatory cell infiltration was
observed, which is consistent with the pathological manifestations
of acute hepatic injury. The activities of ALT and AST in the
serum, which are related to liver function, were significantly
increased compared with those of the control group.
Glucose-regulated proteins GRP78 and GRP94, which are endoplasmic
reticulum chaperones, are expressed after being induced by ERS
(29). In addition, induction of
CCAAT/enhancer binding protein-homologous protein CHOP expression
is one of the main mechanisms of ERS (30). Therefore, their expression can be
detected as a marker of ERS.
SERP1 is a protein-coding gene. When the SERP1
protein was discovered, histological analysis of rabbits infected
with the myxoma virus demonstrated that the absence of the protein
was associated with increased inflammation (31). Furthermore, mice with SERP1 gene
deletion exhibit growth retardation, increased mortality and
impaired glucose tolerance (17).
The results of the aforementioned study indicated that subtle
changes of SERP1 serve a vital role in regulating ERS. The results
of these two studies based on animal models revealed that SERP1 is
obligatory for the normal growth of rabbits and mice. Additionally,
a recent study revealed that microRNA-1-3p improves cell
permeability and causes membrane damage by targeting SERP1, leading
to endothelial cell dysfunction, which means that SERP1 serves an
essential role in sepsis-induced lung injury (32). The present study detected SERP1
expression in acute liver injury tissues and cells, and the results
revealed low expression in tissues as well as cells. On this basis,
a SERP1-overexpression plasmid was established, and the effect of
its overexpression was further studied.
ERS is associated with inflammation and cell
apoptosis. In terms of inflammation, quercetin protects the
intestinal barrier destruction and inflammation in acute
necrotizing pancreatitis by inhibiting Toll-like receptor 4/MYD88
innate immune signal transduction adaptor/p38 MAPK and ERS
activation (33). Additionally,
the NLRP3 inflammasome is a complex composed of a variety of
proteins, which activates the secretion of the pro-inflammatory
cytokine IL-1β in a caspase-1-dependent manner, thereby regulating
inflammation (34). The reduced
expression levels of inflammatory factors and inflammasomes in the
present study indicated the role of SERP1 upregulation in
inflammation. In terms of apoptosis, in the case of persistently
high ERS, activation of the unfolded protein response can trigger
cell death, which has already been demonstrated by numerous studies
(34–36). ERS induced by cyclic mechanical
stretching can give rise to caspase-12 lysis and myoblast apoptosis
(37). Additionally, in
cardiomyopathy (38), nephropathy
(39) and osteosarcoma (40), ERS has been found to be induced
and results in cell apoptosis. In the present study, the reduced
expression levels of apoptosis-related proteins demonstrated that
SERP1 overexpression reduced hepatocyte apoptosis. This finding
extends the potential role of SERP1 in alleviating liver disease
through regulating ERS.
Based on the confirmed role of SERP1 overexpression,
the present study explored its mechanism. In the research of cancer
and neuropsychiatric diseases, GSK3β has been selected as a
therapeutic target and has attracted increasing attention.
Astragaloside IV protects nerve cells from ERS by inactivating
GSK3β (41). Additionally, GSK3β
regulated by ERS participates in the pathogenesis of Alzheimer's
disease (42). TCF/LEF is a type
of transcription factor with a dual regulatory role in the nucleus,
and when combined with β-catenin, it promotes the transcription of
downstream target genes. Subsequently, the activity of TCF/LEF and
GSK3β/β-catenin expression was detected in the present study, and
the effect of SERP1 overexpression was similar to that of the ERS
inhibitor. By constructing a GSK3β overexpression plasmid, it was
revealed that GSK3β overexpression reversed the effect of SERP1
overexpression. This demonstrated that SERP1 acts on ERS via the
GSK3β signaling pathway. In addition, the present study
demonstrated that when ERS was inhibited, TCF/LEF activity was also
inhibited, however, the specific mechanism of their mutual
influence has not been revealed yet.
The present study revealed that SERP1 overexpression
alleviated ERS via the GSK3β/β-catenin/TCF/LEF signaling pathway
and reduced hepatocyte apoptosis. This enables the public to have a
more comprehensive understanding of the mechanism of hepatic injury
and provides a novel direction for its therapeutic targets.
Acknowledgements
Not applicable.
Funding
Funding: No funding was received.
Availability of data and materials
The datasets used and/or analyzed during the current
study are available from the corresponding author on reasonable
request.
Authors' contributions
JC designed and performed experiments and made
considerable contributions to manuscript writing. ZS and LZ
performed the experiments and analyzed the data. HX performed the
experiments and provided critical opinions on the manuscript. JC
and HX confirm the authenticity of data. All authors have read and
approved the final manuscript.
Ethics approval and consent to
participate
This research followed the National Institutes of
Health Guide for the Care and Use of Laboratory Animals. The
present study was approved by the Animal Ethics Committee of Huaihe
Hospital of Henan University (approval no. HHIACUC-201956; Kaifeng,
China).
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Wu G, Win S, Than TA, Chen P and Kaplowitz
N: Gut microbiota and liver injury (I)-acute liver injury. Adv Exp
Med Biol. 1238:23–37. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
He GW, Günther C, Kremer AE, Thonn V,
Amann K, Poremba C, Neurath MF, Wirtz S and Becker C:
PGAM5-mediated programmed necrosis of hepatocytes drives acute
liver injury. Gut. 66:716–723. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Hassan A and Fontana RJ: The diagnosis and
management of idiosyncratic drug-induced liver injury. Liver Int.
39:31–41. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Katarey D and Verma S: Drug-induced liver
injury. Clin Med (Lond). 16 (Suppl 6):s104–s109. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Hosseini N, Shor J and Szabo G: Alcoholic
hepatitis: A review. Alcohol Alcohol. 54:408–16. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Zhao RH, Shi Y, Zhao H, Wu W and Sheng JF:
Acute-on-chronic liver failure in chronic hepatitis B: An update.
Expert Rev Gastroenterol Hepatol. 12:341–350. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Guzel E, Arlier S, Guzeloglu-Kayisli O,
Tabak MS, Ekiz T, Semerci N, Larsen K, Schatz F, Lockwood CJ and
Kayisli UA: Endoplasmic reticulum stress and homeostasis in
reproductive physiology and pathology. Int J Mol Sci. 18:7922017.
View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Fernández A, Ordóñez R, Reiter RJ,
González-Gallego J and Mauriz JL: Melatonin and endoplasmic
reticulum stress: Relation to autophagy and apoptosis. J Pineal
Res. 59:292–307. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Oakes SA and Papa FR: The role of
endoplasmic reticulum stress in human pathology. Annu Rev Pathol.
10:173–194. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Song M and Cubillos-Ruiz JR: Endoplasmic
reticulum stress responses in intratumoral immune cells:
Implications for cancer immunotherapy. Trends Immunol. 40:128–141.
2019. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Yilmaz E: Endoplasmic reticulum stress and
obesity. Adv Exp Med Biol. 960:261–276. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Lebeaupin C, Vallée D, Hazari Y, Hetz C,
Chevet E and Bailly-Maitre B: Endoplasmic reticulum stress
signalling and the pathogenesis of non-alcoholic fatty liver
disease. J Hepatol. 69:927–947. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Zhao YF, Gao Y, Wu XF, Wang JG and Duan
LX: Protective effect of earthworm active ingredients against
endoplasmic reticulum stress-induced acute liver injury in mice.
Zhongguo Zhong Yao Za Zhi. 42:1183–1188. 2017.(In Chinese).
PubMed/NCBI
|
|
14
|
Wang S, Luan J and Lv X: Inhibition of
endoplasmic reticulum stress attenuated ethanol-induced exosomal
miR-122 and Acute liver injury in mice. Alcohol Alcohol.
54:465–471. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Faria D, Lentze N, Almaça J, Luz S,
Alessio L, Tian Y, Martins JP, Cruz P, Schreiber R, Rezwan M, et
al: Regulation of ENaC biogenesis by the stress response protein
SERP1. Pflugers Arch. 463:819–827. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Jung S, Hyun J, Nah J, Han J, Kim SH, Park
J, Oh Y, Gwon Y, Moon S, Jo DG and Jung YK: SERP1 is an assembly
regulator of γ-secretase in metabolic stress conditions. Sci
Signal. 13:eaax89492020. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Hori O, Miyazaki M, Tamatani T, Ozawa K,
Takano K, Okabe M, Ikawa M, Hartmann E, Mai P, Stern DM, et al:
Deletion of SERP1/RAMP4, a component of the endoplasmic reticulum
(ER) translocation sites, leads to ER stress. Mol Cell Biol.
26:4257–4267. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Zhang L, Ren F, Zhang X, Wang X, Shi H,
Zhou L, Zheng S, Chen Y, Chen D, Li L, et al: Peroxisome
proliferator-activated receptor alpha acts as a mediator of
endoplasmic reticulum stress-induced hepatocyte apoptosis in acute
liver failure. Dis Model Mech. 9:799–809. 2016.PubMed/NCBI
|
|
19
|
Ren F, Zhou L, Zhang X, Wen T, Shi H, Xie
B, Li Z, Chen D, Wang Z and Duan Z: Endoplasmic reticulum
stress-activated glycogen synthase kinase 3β aggravates liver
inflammation and hepatotoxicity in mice with acute liver failure.
Inflammation. 38:1151–1165. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Ren F, Yang B, Zhang X, Wen T, Wang X, Yin
J, Piao Z, Zheng S, Zhang J, Chen Y, et al: Role of endoplasmic
reticulum stress in D-GalN/LPS-induced acute liver failure.
Zhonghua Gan Zang Bing Za Zhi. 22:364–368. 2014.(In Chinese).
PubMed/NCBI
|
|
21
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(−Delta Delta C(T)) method. Methods. 25:402–408. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Cao P, Sun J, Sullivan MA, Huang X, Wang
H, Zhang Y, Wang N and Wang K: Angelica sinensis polysaccharide
protects against acetaminophen-induced acute liver injury and cell
death by suppressing oxidative stress and hepatic apoptosis in vivo
and in vitro. Int J Biol Macromol. 111:1133–1139. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Trefts E, Gannon M and Wasserman DH: The
liver. Curr Biol. 27:R1147–R1151. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Lupberger J, Croonenborghs T, Roca Suarez
AA, Van Renne N, Jühling F, Oudot MA, Virzì A, Bandiera S, Jamey C,
Meszaros G, et al: Combined analysis of metabolomes, proteomes, and
transcriptomes of hepatitis c virus-infected cells and liver to
identify pathways associated with disease development.
Gastroenterology. 157:537–551.e9. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Kleiner DE: Drug-induced liver injury: The
hepatic pathologist's approach. Gastroenterol Clin North Am.
46:273–296. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Jothimani D, Venugopal R, Abedin MF,
Kaliamoorthy I and Rela M: COVID-19 and the liver. J Hepatol.
73:1231–1240. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Li R, Yang W, Yin Y, Zhang P, Wang Y and
Tao K: Protective role of 4-Octyl itaconate in murine
LPS/D-GalN-induced acute liver failure via inhibiting inflammation,
oxidative stress, and apoptosis. Oxid Med Cell Longev.
2021:99320992021.PubMed/NCBI
|
|
28
|
Pan CW, Yang SX, Pan ZZ, Zheng B, Wang JZ,
Lu GR, Xue ZX and Xu CL: Andrographolide ameliorates
d-galactosamine/lipopolysaccharide-induced acute liver injury by
activating Nrf2 signaling pathway. Oncotarget. 8:41202–41210. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Ghiasi SM, Dahlby T, Hede Andersen C,
Haataja L, Petersen S, Omar-Hmeadi M, Yang M, Pihl C, Bresson SE,
Khilji MS, et al: Endoplasmic reticulum chaperone glucose-regulated
protein 94 is essential for proinsulin handling. Diabetes.
68:747–760. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Yi S, Chen K, Zhang L, Shi W, Zhang Y, Niu
S, Jia M, Cong B and Li Y: Endoplasmic reticulum stress is involved
in stress-induced hypothalamic neuronal injury in rats via the
PERK-ATF4-CHOP and IRE1-ASK1-JNK pathways. Front Cell Neurosci.
13:1902019. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Macen JL, Upton C, Nation N and McFadden
G: SERP1, a serine proteinase inhibitor encoded by myxoma virus, is
a secreted glycoprotein that interferes with inflammation.
Virology. 195:348–363. 1993. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Gao M, Yu T, Liu D, Shi Y, Yang P, Zhang
J, Wang J, Liu Y and Zhang X: Sepsis plasma-derived exosomal
miR-1-3p induces endothelial cell dysfunction by targeting SERP1.
Clin Sci (Lond). 135:347–365. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Junyuan Z, Hui X, Chunlan H, Junjie F,
Qixiang M, Yingying L, Lihong L, Xingpeng W and Yue Z: Quercetin
protects against intestinal barrier disruption and inflammation in
acute necrotizing pancreatitis through TLR4/MyD88/p38 MAPK and ERS
inhibition. Pancreatology. 18:742–752. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Li W, Cao T, Luo C, Cai J, Zhou X, Xiao X
and Liu S: Crosstalk between ER stress, NLRP3 inflammasome, and
inflammation. Appl Microbiol Biotechnol. 104:6129–6140. 2020.
View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Sankrityayan H, Oza MJ, Kulkarni YA, Mulay
SR and Gaikwad AB: ER stress response mediates diabetic
microvascular complications. Drug Discov Today. 24:2247–2257. 2019.
View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Wu Z, Wang H, Fang S and Xu C: Roles of
endoplasmic reticulum stress and autophagy on H2O2-induced
oxidative stress injury in HepG2 cells. Mol Med Rep. 18:4163–4174.
2018.PubMed/NCBI
|
|
37
|
Song J, Zhang Q, Wang S, Yang F, Chen Z,
Dong Q, Ji Q, Yuan X and Ren D: Cleavage of caspase-12 at Asp94,
mediated by endoplasmic reticulum stress (ERS), contributes to
stretch-induced apoptosis of myoblasts. J Cell Physiol.
233:9473–9487. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Li W, Li W, Leng Y, Xiong Y and Xia Z:
Ferroptosis is involved in diabetes myocardial ischemia/reperfusion
injury through endoplasmic reticulum stress. DNA Cell Biol.
39:210–225. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Shen H, Ming Y, Xu C, Xu Y, Zhao S and
Zhang Q: Deregulation of long noncoding RNA (TUG1) contributes to
excessive podocytes apoptosis by activating endoplasmic reticulum
stress in the development of diabetic nephropathy. J Cell Physiol.
Jan 22–2019.(Epub ahead of print). View Article : Google Scholar
|
|
40
|
Wang Z, Yin F, Xu J, Zhang T, Wang G, Mao
M, Wang Z, Sun W, Han J, Yang M, et al: CYT997(Lexibulin) induces
apoptosis and autophagy through the activation of mutually
reinforced ER stress and ROS in osteosarcoma. J Exp Clin Cancer
Res. 38:442019. View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Fu Y, Cai J, Xi M and He Y, Zhao Y, Zheng
Y, Zhang Y, Xi J and He Y: Neuroprotection effect of astragaloside
IV from 2-DG-induced endoplasmic reticulum stress. Oxid Med Cell
Longev. 2020:97820622020. View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Liu XJ, Wei J, Shang YH, Huang HC and Lao
FX: Modulation of AβPP and GSK3β by endoplasmic reticulum stress
and involvement in Alzheimer's disease. J Alzheimers Dis.
57:1157–1170. 2017. View Article : Google Scholar : PubMed/NCBI
|