Introduction
Type 2 diabetes mellitus (T2DM), one of the most
common and fastest growing diseases worldwide, is an endocrine and
metabolic disease. It is estimated that 693 million adults will
have T2DM by 2045 (1). The
vascular complications of T2DM are some of the most important and
pressing concerns in patients (2). In addition, the leading cause of
death in diabetic patients is cardiovascular disease.
Atherosclerosis constitutes the primary pathological outcome
following the development of macrovascular complications, and it
causes the thickening and hardening of the arterial wall and
narrowing of the vascular lumen (3). Inflammation underlies the
pathogenesis of atherosclerosis, which is the most common cause of
cardiovascular disease (CVD) (4).
The association between oxidative stress and inflammation has
garnered growing interest in the study of the progression of the
disease (5). Inflammation leads
to increased ROS levels, which can induce oxidative stress
(6). However, when the
physiological antioxidant defense system is overwhelmed, excessive
levels of ROS can lead to oxidative stress (7). Therefore, there is an urgent need to
discover novel treatments to prevent and treat the
diabetes-associated macrovascular diseases.
High concentrations of glucose can promote apoptosis
of endothelial cells, which is closely related to vascular
complications. In addition, high glucose conditions can not only
cause metabolic disorders, but also produce excess quantities of
oxygen-free radicals, resulting in oxidative stress, which in-turn
results in toxic effects on endothelial cells, and this process
plays an important role in the development of atherosclerosis, a
vascular complication of diabetes. Therefore, in the present study
HUVECs cultured under high glucose conditions were used as a model
to study the antioxidant effect of 3,4-DHAP.
Cellular antioxidant defense plays a crucial role in
protecting against oxidative stress (8). Nuclear factor E2-related factor 2
(Nrf2) is the major transcriptional regulator of antioxidant gene
expression (9). Nrf2 participates
in the pathogenesis of several diseases (10–13). Under physiological conditions,
Nrf2 forms a complex with Keap1, thereby mediating Nrf2 proteasomal
degradation and ubiquitination (14). However, when subjected to
oxidative stress or other physiological stimuli, Nrf2 cannot
interact with Keap1, resulting in Nrf2 activation, nuclear
translocation and transcription of downstream genes of the Nrf2
transcription factor, including heme oxygenase-1 (HO-1) (15). Increased production of ROS
disassociates Nrf2 from Keap1, and Nrf2 translocates to the nucleus
in the dissociated form, where it results in transcription of
several genes (16). Nrf2
protects cells against oxidative stress by activating several
signaling pathways. HO-1 is a critical antioxidant enzyme regulated
by Nrf2 (17). Zhang et al
(18) indicated that the
Nrf2/HO-1 signaling pathway was activated under conditions of
increased ROS. Ci et al (19) demonstrated that farrerol decreased
oxidative stress through activation of Nrf2 to induce HO-1
expression. Mohammad et al (20) also showed that HO-1 was
upregulated in response to oxidative stress. Thus, the Nrf2/HO-1
pathway has become a research hotspot in recent years.
Similar to the Nrf2 pathway, autophagy plays a role
in cell homeostasis when stimulated by oxidative stress (21). Autophagy, which includes
macro-autophagy, micro-autophagy and chaperone-mediated autophagy,
is a process of regulated cellular degradation (22). During autophagy, autophagosomes
‘swallow’ cytoplasmic proteins or organelles and fuse with
lysosomes to form autophagic lysosomes, and the components of the
autophagosome are degraded by the contained lysosomal hydrolases
(23). Microtubule-associated
protein 1A/1B-light chain 3 (LC3) is a marker of autophagy. It is
primarily involved in the formation of autophagosomes. LC3 also
plays a role in mitochondrial autophagy, regulating the quantity of
mitochondria by eliminating them to minimal levels required to meet
the immediate energy demands of the cell and prevent excessive ROS
production (24). The autophagy
of mitochondria is primarily initiated by PTEN-induced putative
kinase 1 (PINK1). PINK proteins degrade cellular components through
the actions of presenilin-associated rhomboid-like (PARL) under
physiological conditions, whereas the function of PARL is inhibited
when the mitochondria are damaged, and in this situation, PINK
stabilizes and recruits Parkin, the E3 ligase, to initiate
autophagy (25). Concurrently,
the cytoplasmic form of LC3 (LC3-I) binds to
phosphatidylethanolamine to form LC3 phosphatidylethanolamine
conjugate (LC3-II), which is recruited to the autophagic membrane.
The formation of autophagosomes is the basis of autophagy and
affects the transformation of LC3-I to LC3-II (26). Hence, LC3 expression is accepted
as a marker for autophagy. When the autophagic process is
initiated, LC3-I is transformed into LC3-II and this commits the
cell to undergoing autophagy (27).
DNA, the genetic material of eukaryotic cells, is
damaged every day by a variety of internal and external factors.
When the DNA is damaged, DNA damage repair pathways are activated
to ensure the stability of the genome. Reactive oxygen species
(ROS) are a factor that can cause DNA damage. PARP is a DNA damage
response sensor. PARP-1, the cleavage substrate of caspase, is also
involved in DNA repair, gene expression regulation, genomic
stability and apoptosis (28,29). A previous study has shown that
PARP-1 plays crucial roles in DNA cell repair and survival using
PARP-1 knockout mice (30).
Another study has also shown that PARP-1 regulates DNA repair
factor availability, and this is an attractive target in the study
of cancer therapeutics (31).
Pazzaglia and Pioli (32) showed
that PARP exerted a protective role in DNA repair and regulated
inflammatory processes. Moreover, it has been shown that autophagy
may be cytoprotective in response to DNA repair, via regulation of
PARP-1 activation (33). Wang
et al (34) determined
that farrerol could ameliorate hepatotoxicity induced by PARP-1,
and this was achieved through activation of Nrf2 and induction of
autophagy. Therefore, whether 3,4-dihydroxyacetophenone (3,4-DHAP)
could protect HUVECs against high glucose-induced damage via
regulating PARP-1 was assessed in the present study.
3,4-DHAP is an active ingredient from Ilex
glauca leaves and has a variety of beneficial biological
activities, including anti-inflammatory, antioxidative and
cardioprotective properties (35), and has been shown to suppress
melanin production (36), inhibit
platelet aggregation, promote coronary artery dilation and improve
blood circulation (37). In our
previous study, it was shown that 3,4-DHAP reduced the levels TNF-α
secretion from RAW264.7 cells, thus exhibiting an anti-inflammatory
effect. It was also shown that 3,4-DHAP decreased the levels of
inflammation-related indicators in a rabbit model of
atherosclerosis induced by hypercholesterolemia (38). However, the effects of 3,4-DHAP on
oxidative stress and its underlying mechanism remain to be
assessed. Therefore, the aim of the present study was to
investigate whether 3,4-DHAP could protect HUVECs against oxidative
stress via regulation of the Nrf2/HO-1 signaling pathway, and the
effects on autophagy and DNA damage repair in this process.
Materials and methods
Reagents
3,4-DHAP was purchased from Jinan Luxin Chemical
Technology Co., Ltd. Sulforaphane (SFN), an antioxidant reagent
that was used as a positive control, was obtained from
Sigma-Aldrich; Merck KGaA. ML385, a novel and specific Nrf2
inhibitor, was purchased from Selleck Chemicals. ML385 is an
inhibitor of Nrf2 activity and nuclear translocation, which has
been confirmed to affect the expression of downstream genes. The
ROS assay kit, Nuclear and Cytoplasmic Protein Extraction Kit,
double antibiotics, MTT kit, DAPI and Fluorescent Mounting Media
were obtained from Beijing Solarbio Science & Technology Co.,
Ltd.
Cell culture
HUVECs were obtained from Shanghai Baili
Biotechnology (produced by ATCC). The HUVECs used were an
immortalized cell line. The cells were cultured in DMEM (Gibco;
Thermo Fisher Scientific, Inc.) supplemented with 15% FBS (EVERY
GREEN; Zhejiang Tianhang Biotechnology, Co., Ltd.) and 1%
penicillin/streptomycin (Beijing Solarbio Science & Technology
Co., Ltd.) at 37°C, in a humidified incubator supplied with 5%
CO2. HUVECs were randomly grouped according to the
experimental design as follows: Control group, high glucose group,
SFN group (SFN + high glucose), 3,4-DHAP group (3,4-DHAP + high
glucose), and 3,4-DHAP + ML385 group (3,4-DHAP + ML385 + high
glucose). The cells were pretreated with SFN (20 µmol/l), 3,4-DHAP
(10 µmol/l) or ML385 (0.25 µmol/l) for 6 h, then exposed to high
glucose conditions (33.3 mmol/l) for 12 h.
MTT assay
For assessment of cell viability and cytotoxicity,
an MTT assay was performed. The HUVECs were evenly plated on a
96-well cell culture plate with ~5,000 cells/well and cultured at
37°C for 24 h. HUVECs were pretreated with 1, 10, 20, 50 or 100
µmol/l 3,4-DHAP, after which, the OD values were measured. The
cells were pretreated with 20 µmol/l SFN and 10 µmol/l 3,4-DHAP for
6 h and then cultured at 37°C with 33.3 mmol/l glucose for 12 h.
Subsequently, to each well, 10 µl MTT solution was added (5 mg/ml),
and cells were incubated for a further 4 h. Finally, the
supernatant was discarded, the resulting blue-purple crystals were
dissolved using 150 µl DMSO with shaking for 10 min. Using a
microplate reader, the absorbance of each well was measured at 490
nm. Cell viability was calculated, and a histogram was created.
ROS activity
The ROS levels are the most commonly detected
indicator of oxidative stress. HUVECs were plated in a 6-well plate
at a density of 1×105 cells/well. After treatment as
described above, the cells were washed with PBS. DCFH-DA was added
to each well, and incubated at 37°C for 30 min in dark. The ROS
levels were measured using a fluorescent enzyme label instrument
(Spectra Max M5; Molecular Devices LLC). Images were obtained using
an Inverted Fluorescence Microscope (Olympus Corporation).
Western blot analysis
Total protein from cells in each group was collected
using RIPA lysis buffer (Beijing Solarbio Science & Technology
Co., Ltd.). The cytoplasmic and the nuclear proteins were
separately acquired using a Nuclear and Cytoplasmic Protein
Extraction Kit according to the manufacturer's instructions. A BCA
assay kit was used to measure the protein concentration. According
to the protein concentration, the amount of sample protein (40 µg)
was calculated and separated by 10 or 12% SDS-PAGE. When the
electrophoresis had finished, a strip of gel was cut and this was
used to transfer proteins to a PVDF film. The PVDF film containing
the protein of interest was immersed in 5% skimmed milk powder at
37°C for 2 h, washed three times with TBST (0.05% Tween-20; 10 min
each), and incubated with one of the following primary antibodies:
Nrf2 (1:1,000; cat. no. SAB4501984; Sigma-Aldrich; Merck KGaA),
HO-1 (1:25,000; cat. no. ab68477; Abcam), LC3 (1:1,000; cat. no.
ab192890; Abcam) and PARP-1 (1:2,500; cat. no. ab32138; Abcam) at
4°C overnight. Then the PVDF film was washed with TBST and
incubated with a secondary antibody: GAPDH (1:7,000; cat. no.
AF1186; Beyotime Institute of Biotechnology) or H3 (1:2,000; cat.
no. ab32356; Abcam) at 37°C for 1 h. The PVDF film was treated with
an enhanced chemiluminescence reagent (Beijing Solarbio Science
& Technology Co., Ltd.), and the signals were visualized using
a chemiluminescence detection system (FluorChem E; Protein Simple
Ltd.). Quantitative expression of proteins was calculated using
ImageJ software (v1.8.0; National Institutes of Health).
Reverse transcription-quantitative PCR
(RT-qPCR)
According to the manufacturer's instructions, the
RNA of HUVECs was extracted by lysing cells on ice using
TRIzol® reagent (Beijing ComWin Biotech Co., Ltd.), and
then reverse transcribed into cDNA using the ReverTra Ace qPCR RT
Kit [cat. no. FSQ-101; Toyobo (Shanghai) Biotech, Co., Ltd.]. qPCR
was performed using SYBR® Green Real-time PCR Master Mix
(cat. no. QPK-201; Toyobo (Shanghai) Biotech, Co., Ltd.) in a 7500
Sequence Detection System. The thermocycling conditions were as
follows: Initial denaturation at 95°C for 60 sec; followed by 40
cycles at 95°C for 15 sec, 60°C for 15 sec and 72°C for 45 sec. The
primer sequences used were: Nrf2 forward,
5′-CCCAGCACATCCAGTCAGAAACC-3′ and reverse,
5′-AGCCGAAGAAACCTCATTGTCATCTAC-3′; HO-1 forward,
5′-TGCCAGTGCCACCAAGTTCAAG-3′ and reverse,
5′-TGTTGAGCAGGAACGCAGTCTTG-3′; and GAPDH forward,
5′-CAGGAGGCATTGCTGATGAT-3′ and reverse, 5′-GAAGGCTGGGGCTCATTT-3′.
All mRNA expression levels were normalized to the housekeeping gene
GAPDH. Relative expression was calculated using the
2−ΔΔCq method (39).
Cellular immunofluorescence
After discarding the culture medium, the cells were
washed with PBS three times (5 min each). Formaldehyde (2%) was
added and cells were fixed at 37°C for 30 min, after which, the
solution was removed, cells were washed with PBS three times (5 min
each) permeabilized using 0.3% Triton X-100 at 37°C for 15 min,
washed as above, blocked using 10% goat serum at 37°C for 2 h,
incubated with the Nrf2 primary antibody (1:100; cat. no.
SAB4501984; Sigma-Aldrich) overnight at 4°C, washed, incubated with
the secondary FITC-conjugated antibody (1:100; cat. no. ZF-0311;
ZSGB-BIO; OriGene Technologies, Inc.) in the dark at 37°C for 1 h,
washed, stained with DAPI at 37°C for 5 min, then washed again. The
cell climbing piece (Thermo Fisher Scientific, Inc.) and coverslip
were removed and sealed using the Fluorescent Mounting Media.
Images were obtained using a fluorescence microscope
(magnification, ×10; Olympus Corporation).
Assessment of autophagosome
formation
After treating cells as described above, the cells
were collected, centrifuged at 1,006.2 × g for 10 min, fixed with
3% glutaraldehyde at 4°C for 12 h, fixed with 1% osmic acid at 37°C
for 2 h, and embedded using pure embedding solution for 2 h. The
samples were dehydrated in a series of increasing ethanol
solutions, embedded and set in epoxy resin at different
temperatures and for different lengths of times (37°C for 12 h,
45°C for 12 h and 60°C for 24 h). The embedded samples were cut
into ultra-thin sections (70 nm), and then stained (3% uranium
acetate for 15–30 min and lead citrate for 5–10 min, at 37°C).
Finally, the images were captured using a transmission electron
microscope (magnification, ×5,000).
Statistical analysis
All data were analyzed using GraphPad Prism version
7.0 (GraphPad Software, Inc.). Results are presented as the mean ±
SD. Unpaired Student's t-tests were used for comparisons between
two groups. Comparisons among multiple groups were analyzed using
one-way ANOVA followed by Tukey's post hoc test. P<0.05 was
considered to indicate a statistically significant difference.
Results
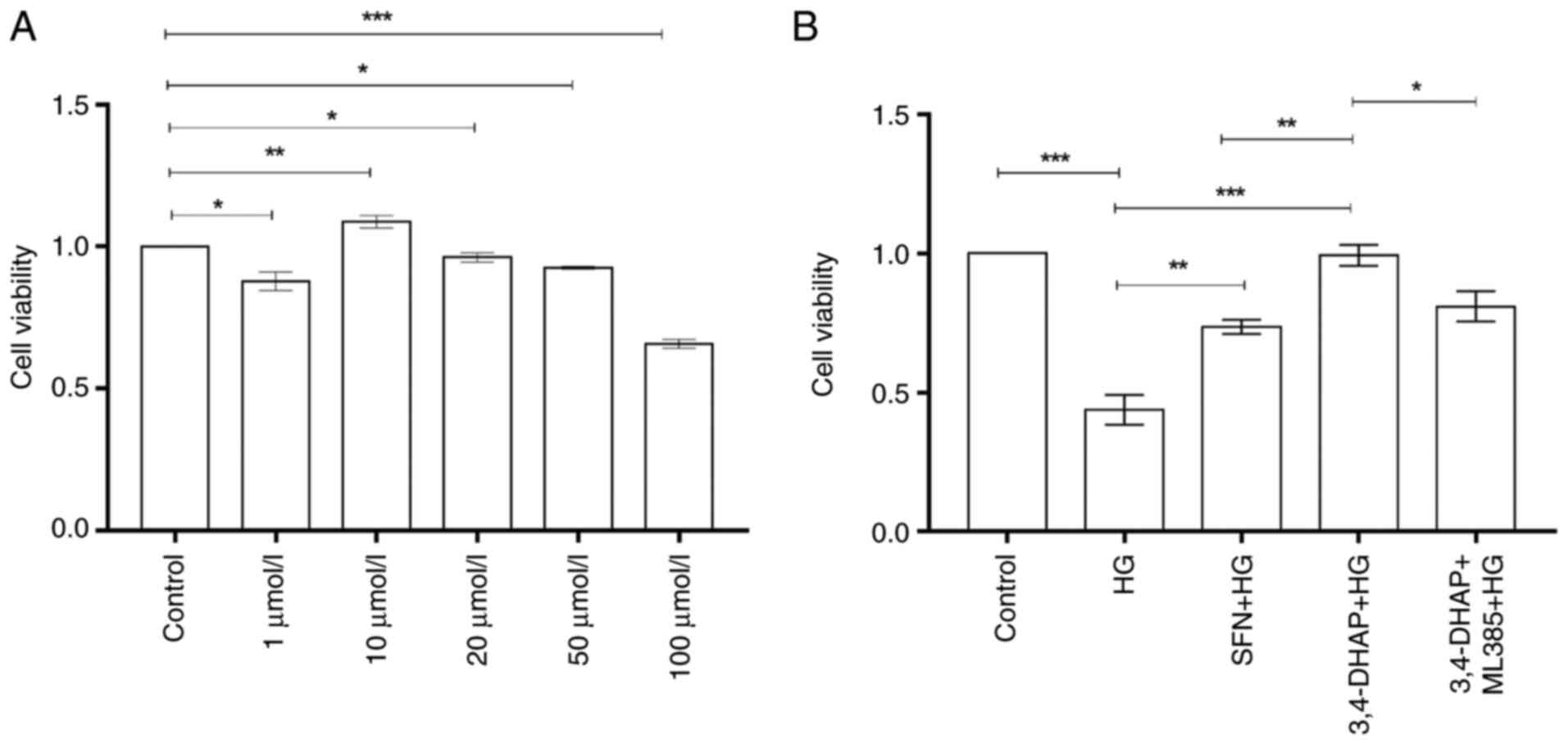
3,4-DHAP increases cell viability and
reduces cytotoxicity
The cell viability of HUVECs was assessed using an
MTT assay. The basic principle is that the amber dehydrogenase in
the mitochondria of living cells can reduce the exogenous MTT,
causing it to crystallize and deposit the blue-purple formazan,
which is difficult to dissolve in water in living cells, while the
dead cells have no such function. In Fig. 1A, compared with the control group,
in cells treated with 10 µmol/l 3,4-DHAP, the cell viability was
increased (P<0.01); the cell viability of 3,4-DHAP when treated
with 1, 20 and 50 µmol/l 3,4 DHAP was reduced (P<0.05); and the
cell viability when treated with 100 µmol/l 3,4-DHAP was also
reduced (P<0.001). Thus, 10 µmol/l 3,4-DHAP was selected for
subsequent experiments. In Fig.
1B, the cell viability of the high glucose group was
significantly reduced compared with the control group (P<0.001),
and this demonstrated that a successful in vitro model of
diabetes had been established. The cell viability was significantly
increased after pretreatment with SFN (P<0.01) and 3,4-DHAP
(P<0.001), and there was significant difference between the SFN
group and the 3,4-DHAP group (P<0.01). Compared to the
3,4-DHAP group, the cell viability in the 3,4-DHAP + ML385 group
was reduced (P<0.05). These results indicated that 3,4-DHAP
could protect HUVECs from high glucose-induced cell death.
3,4-DHAP reduces ROS levels in
HUVECs
Intracellular ROS levels were determined using a
fluorescent enzyme labeling instrument. ROS levels were detected
using the fluorescent probe DCFH-DA. DCFH-DA does not fluoresce
itself and can pass through the cell membrane freely. After
entering the cell, DCFH can be hydrolyzed by esterases in the cell
to generate DCFH. DCFH cannot penetrate a cell membrane, making it
easy to probe in loaded cells. Intracellular ROS can oxidize
non-fluorescent DCFH to generate fluorescent DCF, and the
fluorescence of DCF can be detected to determine the levels of
intracellular ROS (40). As shown
in Fig. 2A, compared with the
control group, the high glucose group exhibited significantly
increased ROS production (P<0.001); compared with the high
glucose group, SFN and 3,4-DHAP group significantly reduced the
production of ROS (P<0.001), but there was no significant
difference between the SFN group and the 3,4-DHAP group
(P>0.05). Compared with the 3,4-DHAP group, the ROS levels in
the 3,4-DHAP + ML385 group were increased (P<0.01). In Fig. 2B, compared with the control group,
the high glucose group significantly increased fluorescence
intensity (P<0.001); compared with the high glucose group, the
SFN and 3,4-DHAP groups significantly reduced the fluorescence
intensity (P<0.01), but there was no significant difference
between the SFN group and the 3,4-DHAP group (P>0.05). Compared
with the 3,4-DHAP group, fluorescence intensity in the 3,4-DHAP +
ML385 group was increased (P<0.05). The fluorescent images also
showed corroborating results (Fig.
2C). These findings suggested that 3,4-DHAP could attenuate the
oxidative stress induced by high glucose conditions in HUVECs.
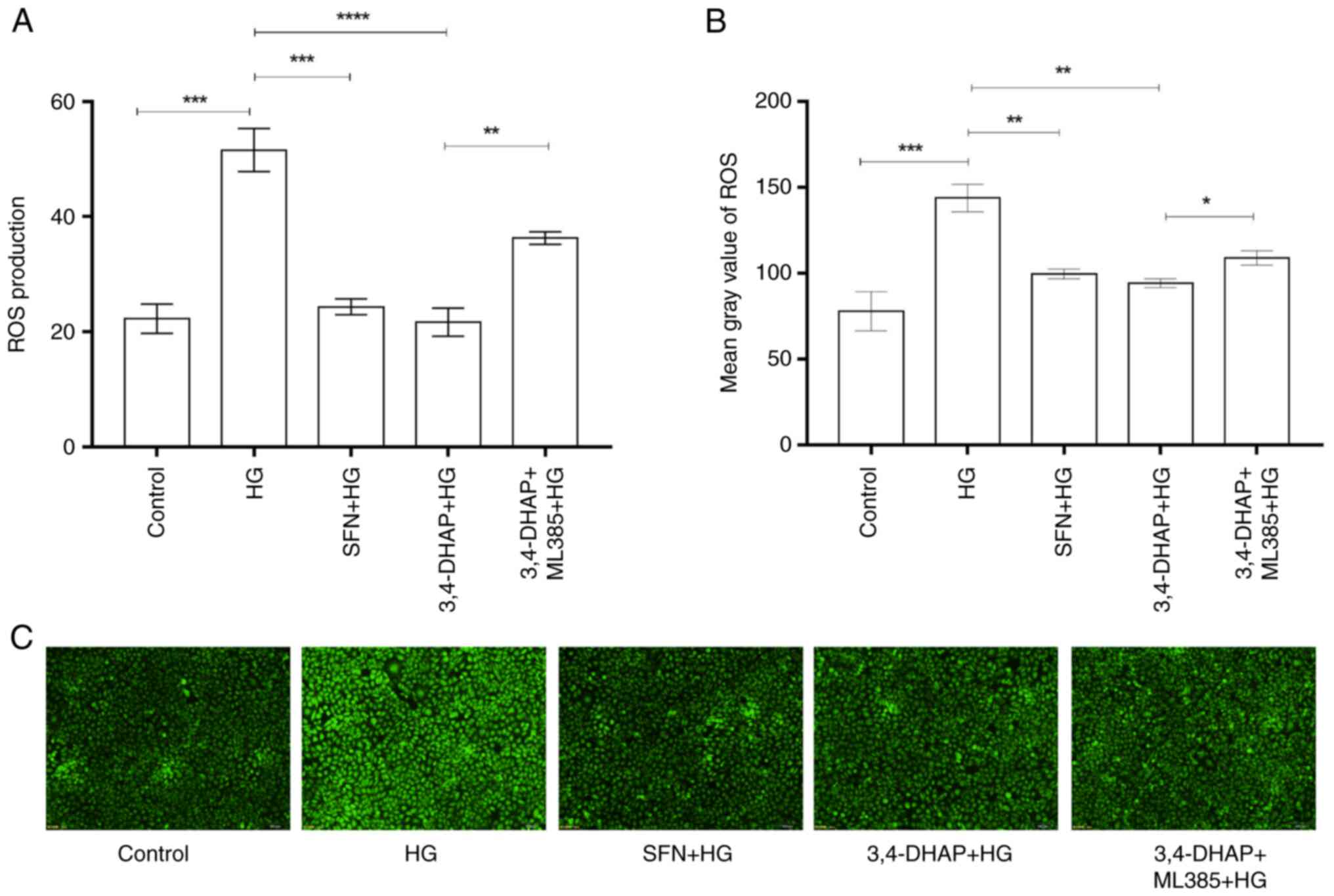 | Figure 2.3,4-DHAP reduces ROS activity in
HUVECs. (A) ROS levels were measured using a fluorescent enzyme
label instrument. (B) Mean gray values of ROS were measured using
ImageJ software. (C) ROS were imaged using an inverted fluorescence
microscope (magnification, ×10). Data are presented as the mean ±
SD of three repeats. *P<0.05, **P<0.01, ***P<0.001 and
****P<0.0001. 3,4-DHAP, 3,4-dihydroxyacetophenone; ROS, reactive
oxygen species; HUVECs, human umbilical vein endothelial cells; HG,
high glucose; SFN, sulforaphane. |
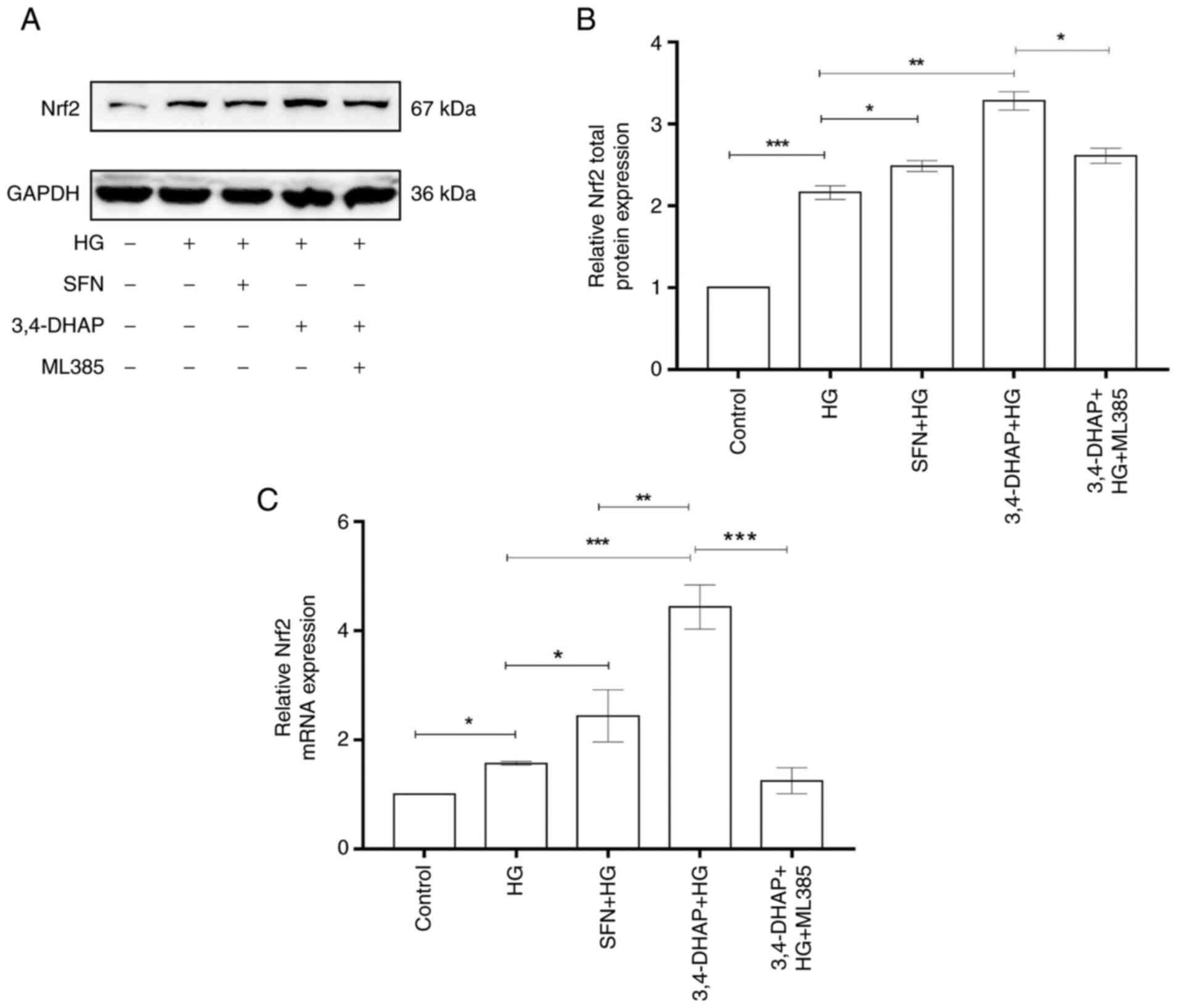
3,4-DHAP upregulates Nrf2 protein and
mRNA expression levels
As shown in Fig. 3A
and B, Nrf2 total protein expression in the high glucose group
was increased compared with the control group (P<0.001).
Compared to the high glucose group, Nrf2 total protein expression
was increased in the SFN group (P<0.05). Nrf2 total
protein expression in the 3,4-DHAP group was also significantly
increased compared to the high glucose group (P<0.01).
The total Nrf2 protein expression in the 3,4-DHAP group was higher
than that in the SFN group, but there was no significant difference
between these groups (P>0.05). Compared with the 3,4-DHAP
group, the total Nrf2 protein expression in the 3,4-DHAP + ML385
group was significantly reduced (P<0.05). In Fig. 3C, the mRNA expression level of
Nrf2 in the high glucose group was increased compared with the
control group (P<0.05). Compared with the high glucose
group, Nrf2 mRNA expression was increased in the SFN group
(P<0.05). Nrf2 mRNA expression in the 3,4-DHAP group was
also significantly increased compared with the high glucose group
(P<0.001). Nrf2 mRNA expression in the 3,4-DHAP group was
significantly higher than that in the SFN group, and the difference
was significant (P<0.01). Compared to the 3,4-DHAP group,
the mRNA expression level of Nrf2 in the 3,4-DHAP + ML385 group was
significantly reduced (P<0.001). These findings indicated
that 3,4-DHAP could protect HUVECs against oxidative stress, and
this may have been regulated by the Nrf2 pathway.
3,4-DHAP increases Nrf2 nuclear
translocation
Nrf2 nuclear protein expression is shown in Fig. 4A. The nuclear translocation of
Nrf2 was reduced when cells were exposed to oxidative stress. As
shown in Fig. 4B, compared with
the control group, the high glucose group exhibited slightly
increased Nrf2 nuclear protein expression (P<0.05). Nrf2 nuclear
protein expression increased when pretreated with SFN (P>0.05)
and 3,4-DHAP (P<0.01) compared with the high glucose group, and
the difference between the SFN and the 3,4-DHAP group was also
significant (P<0.05). Compared with the 3,4-DHAP group,
the nuclear expression of Nrf2 in the 3,4-DHAP + ML385 group was
decreased (P<0.001). In Fig. 4C
and D, the fluorescence intensity of the high glucose group was
higher than that of control group (P<0.001). Compared with the
high glucose group, the fluorescence intensity of the SFN group was
increased (P<0.05), the fluorescence intensity of the 3,4-DHAP
group was significantly also increased (P<0.01), and the
difference between the SFN and the 3,4-DHAP group was also
significant (P<0.05). Compared with the 3,4-DHAP group,
the fluorescence intensity in the 3,4-DHAP + ML385 group was
decreased (P<0.0001). These results showed that 3,4-DHAP exerted
antioxidant effects by regulating Nrf2 nuclear translocation.
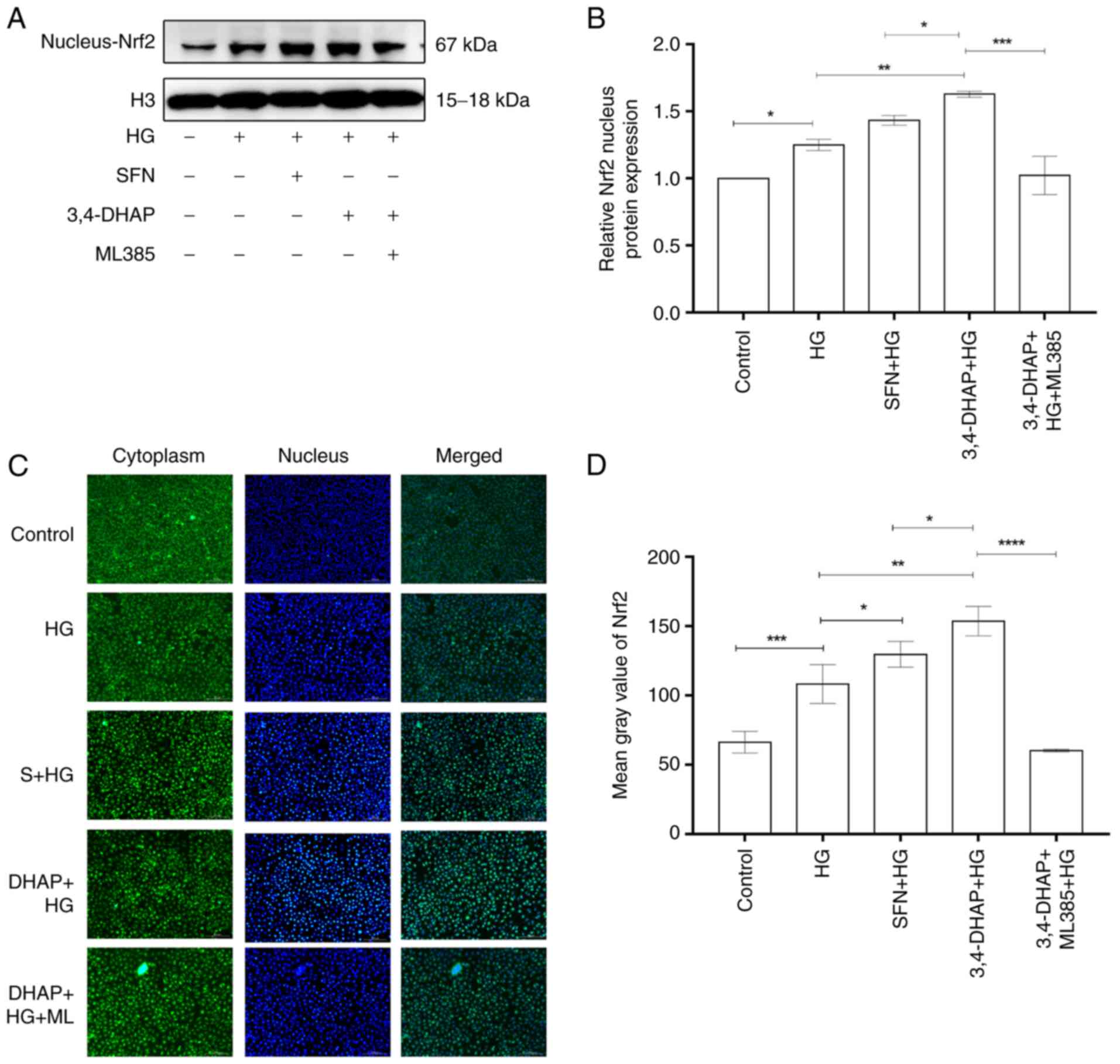 | Figure 4.3,4-DHAP promotes Nrf2 nuclear
translocation. (A) Nrf2 nuclear protein expression levels were
measured by western blotting and (B) quantified using ImageJ
software. (C) Nrf2 protein expression levels in the cytoplasm and
nucleus were imaged using an inverted fluorescence microscope
(magnification, ×10). (D) Mean gray values of ROS were measured
using ImageJ software. Data are presented as the mean ± SD of three
repeats. *P<0.05, **P<0.01, ***P<0.001 and
****P<0.0001. 3,4-DHAP, 3,4-dihydroxyacetophenone; Nrf2, nuclear
factor E2-related factor 2; ROS, reactive oxygen species; HG, high
glucose; SFN, sulforaphane. |
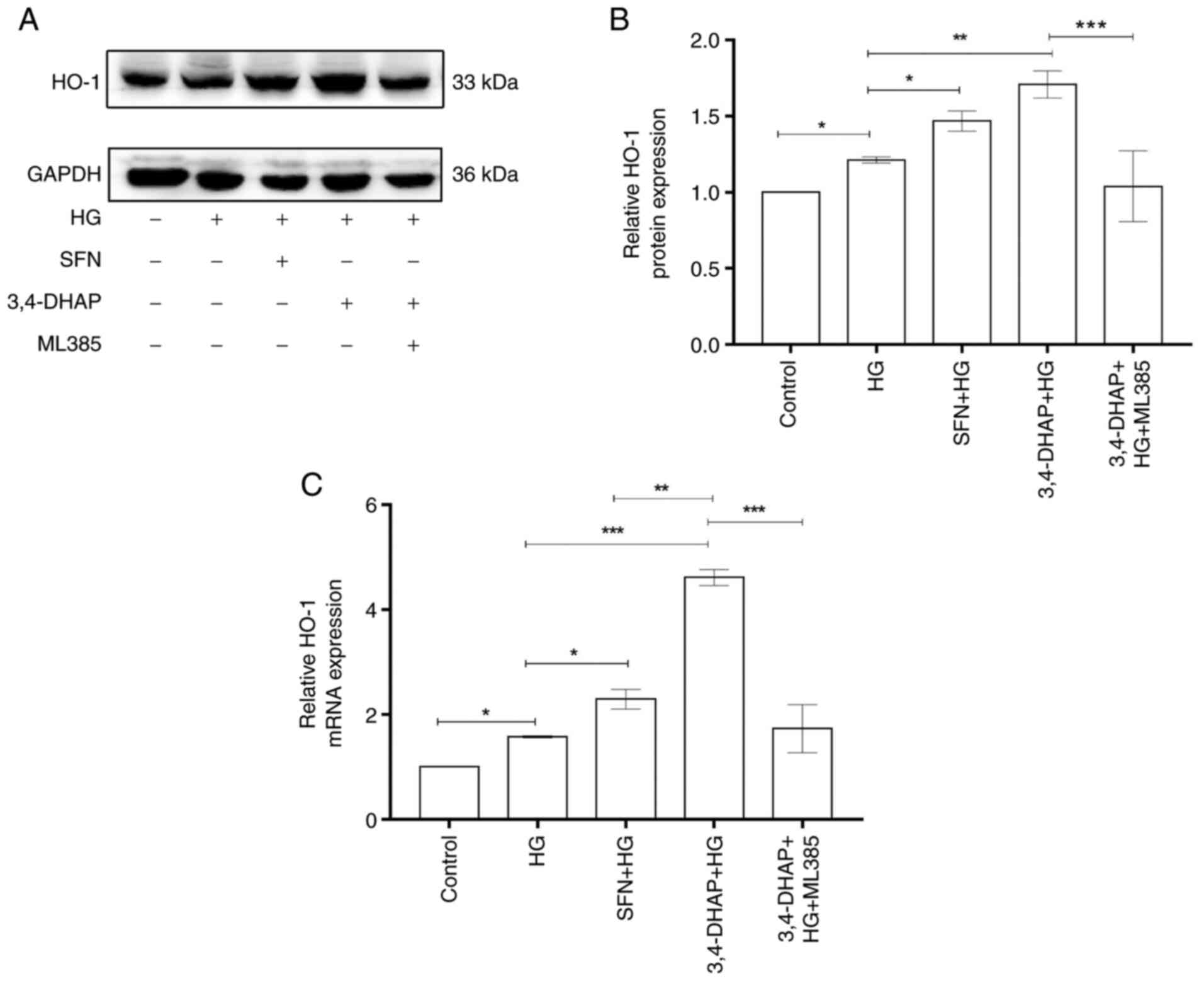
3,4-DHAP enhances HO-1 expression
Fig. 5A shows the
protein expression levels of HO-1. As shown in Fig. 5B, HO-1 expression at the protein
level in the high glucose group was slightly increased compared
with the control group (P<0.05). Compared with the high
glucose group, HO-1 protein expression was enhanced in the SFN
group (P<0.05). Compared with the high glucose group,
3,4-DHAP group also exhibited significantly increased expression of
HO-1 (P<0.01). HO-1 expression in the 3,4-DHAP group was
higher than that in the SFN group, and there was no significant
difference between the SFN and 3,4-DHAP groups (P>0.05).
HO-1 expression in the 3,4-DHAP + ML385 group was reduced compared
with 3,4-DHAP group (P<0.001). As shown in Fig. 5C, the mRNA expression level of
HO-1 in the high glucose group was slightly increased compared with
the control group (P<0.05). Compared with the high
glucose group, HO-1 mRNA expression levels were significantly
increased in the SFN group (P<0.05) and in the 3,4-DHAP
group (P<0.001). HO-1 mRNA expression in the 3,4-DHAP
group was higher than that in the SFN group, and there was a
significant difference between the SFN and 3,4-DHAP groups
(P<0.01). Compared to the 3,4-DHAP group, the mRNA
expression levels of HO-1 in the 3,4-DHAP + ML385 group was
decreased (P<0.001). These findings further indicated
that 3,4-DHAP could protect HUVECs from oxidative stress by
regulating the Nrf2/HO-1 pathway.
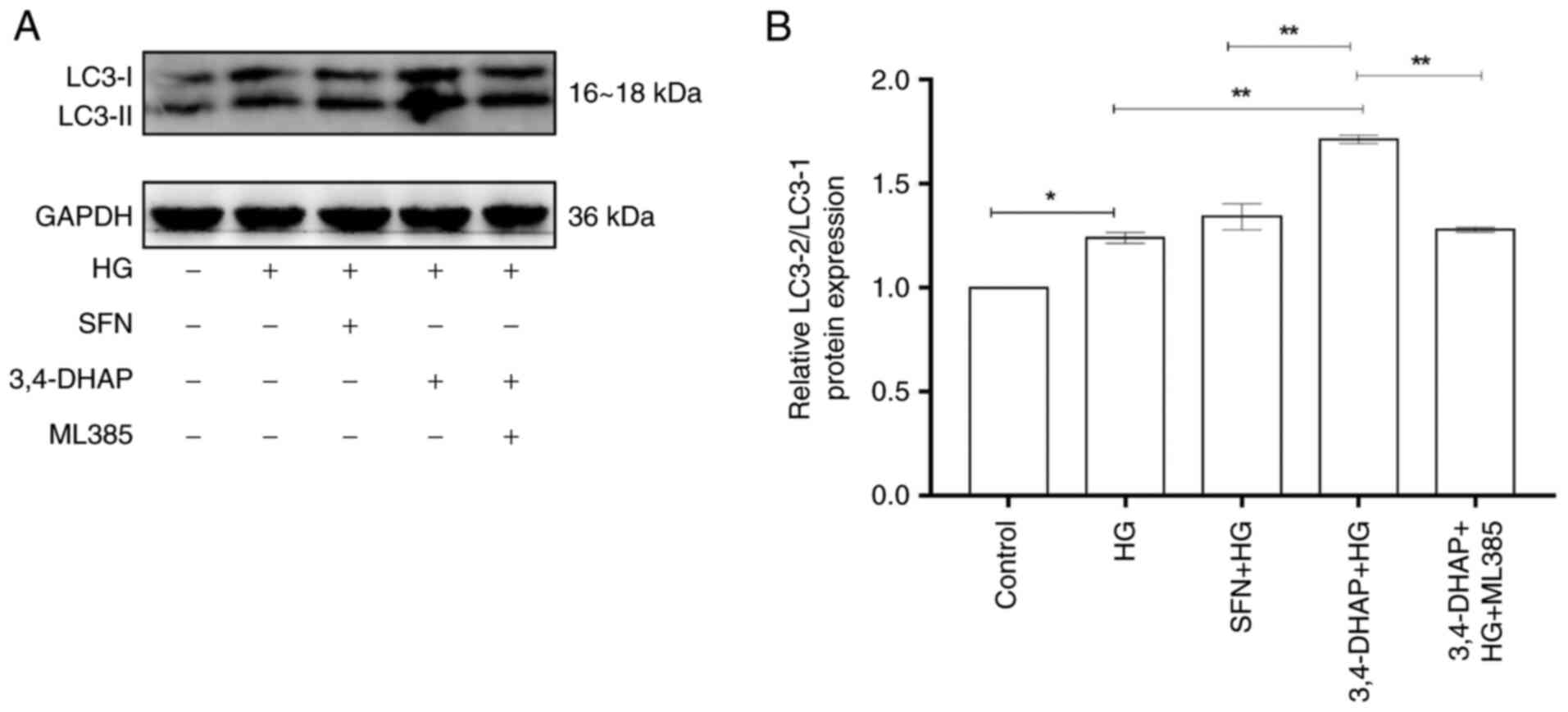
3,4-DHAP upregulates LC3 protein
expression
LC3 is considered to the primary indicator of
autophagy (41). Fig. 6A shows LC3 protein expression. As
shown in Fig. 6B, the protein
expression ratio of LC3-II/LC3-I in the high glucose group was
increased compared with the control group (P<0.05).
Compared with the high glucose group, the LC3-II/LC3-ratio was
increased in the SFN group (P>0.05). Compared with the
high glucose group, 3,4-DHAP markedly increased the LC3-II/LC3-I
protein ratio (P<0.01). The LC3-II/LC3-I ratio in the
3,4-DHAP group was higher than that in the SFN group, and the
difference was significant (P<0.01). Compared with the
3,4-DHAP group, the 3,4-DHAP + ML385 group exhibited a reduced
LC3-II/LC3-I ratio (P<0.01). These findings suggest that
3,4-DHAP can promote autophagy in response to oxidative stress
induced by high glucose treatment in HUVECs.
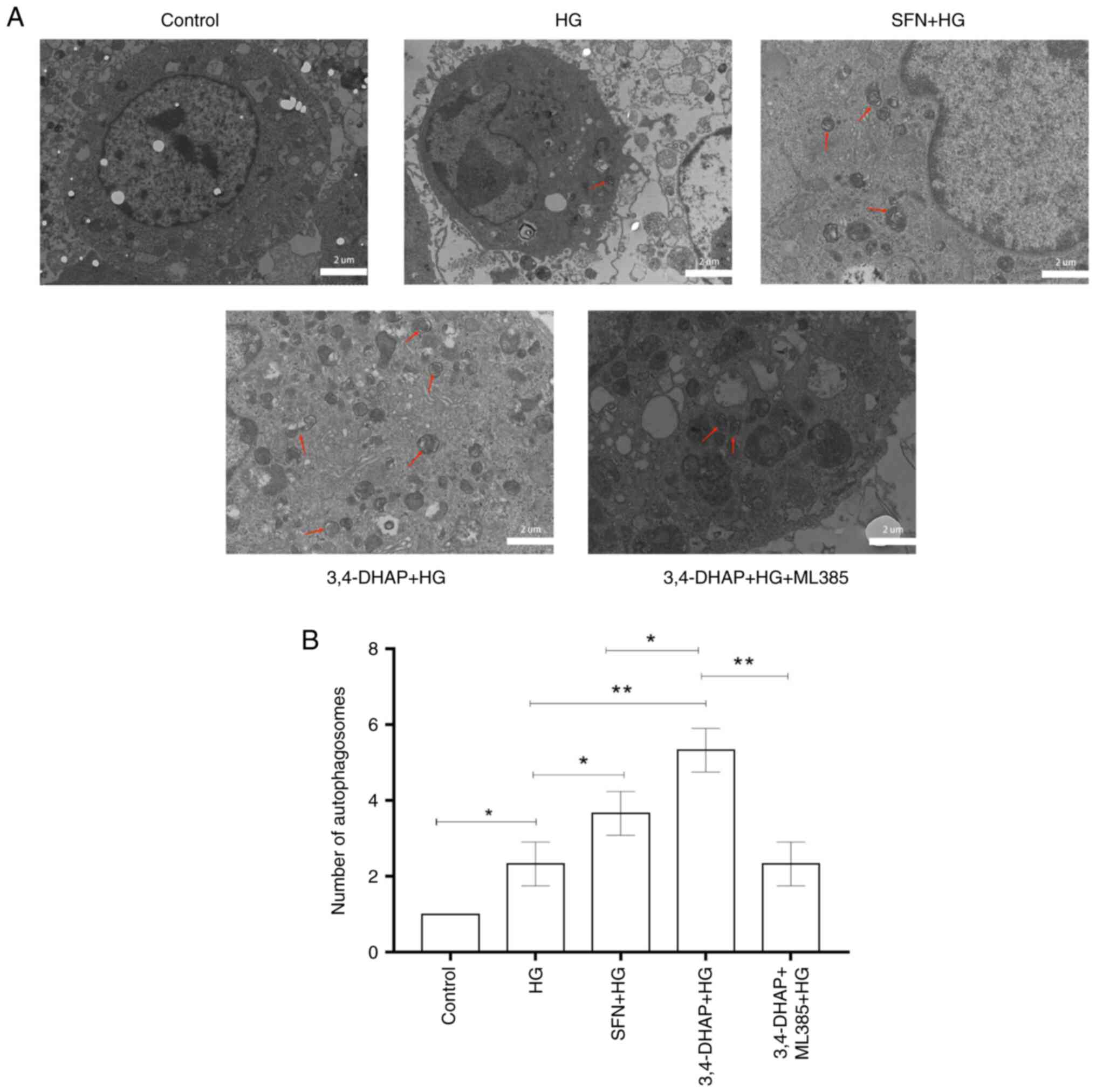
3,4-DHAP promotes the formation of
autophagosomes
Formation of autophagosomes was detected by TEM.
Compared with the control group, the high glucose group exhibited a
slight increase in autophagosome formation (P<0.05).
However, the formation of autophagosomes was evidently increased
after SFN (P<0.05) and 3,4-DHAP (P<0.01)
treatment compared with high glucose group, and there was a
significant difference between the SFN and 3,4-DHAP groups
(P<0.05). Compared with the 3,4-DHAP group, the 3,4-DHAP
+ ML385 group exhibited decreased formation of autophagosomes
(P<0.01) (Fig. 7). The
results further showed that 3,4-DHAP could promote autophagy in
response to oxidative stress.
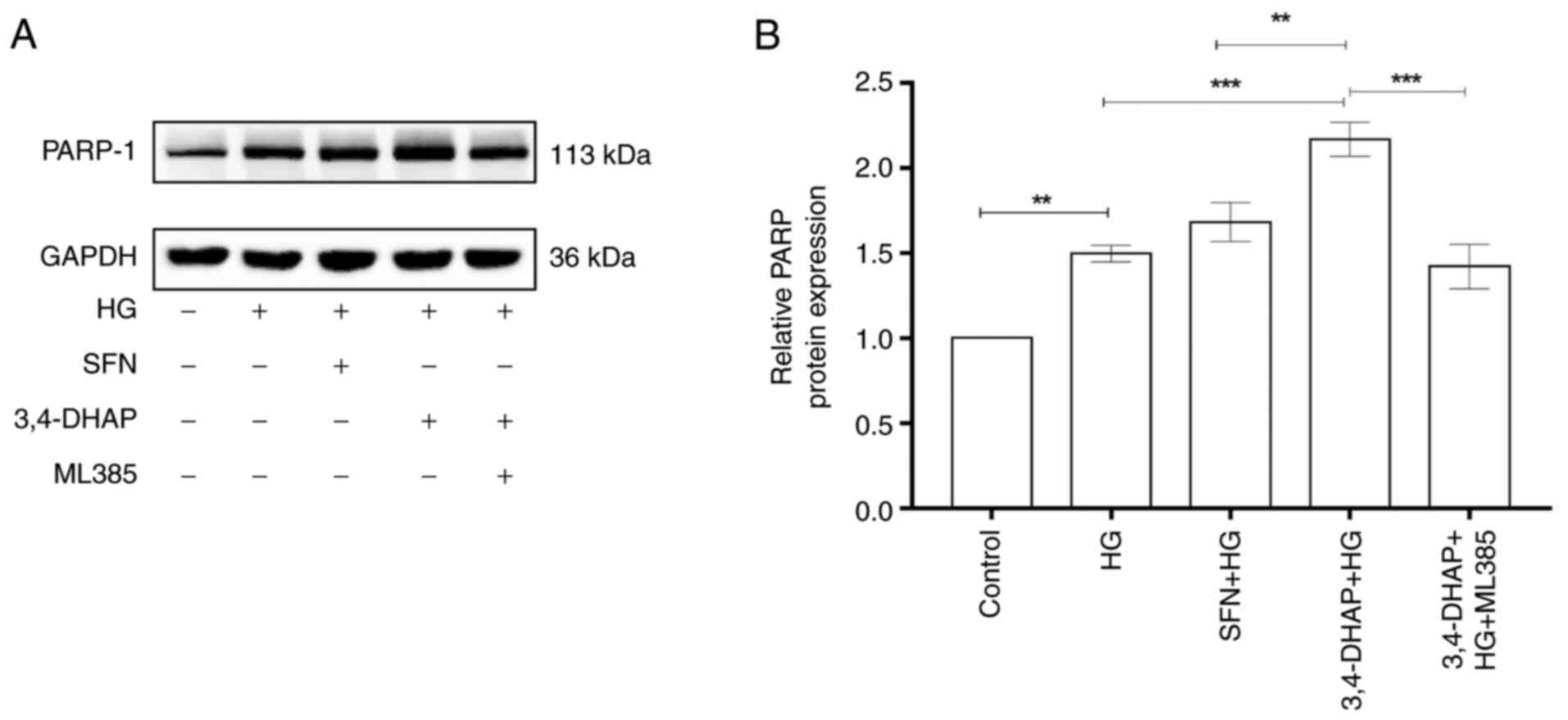
3,4-DHAP enhances PARP-1 protein
expression
PARP-1 is a receptor for DNA damage response
(42). Fig. 8A shows PARP-1 protein expression.
As shown in Fig. 8B, the protein
expression levels of PARP-1 in the high glucose group were
increased compared with the control group (P<0.01).
Compared with the high glucose group, PARP-1 protein expression was
increased in the SFN group (P>0.05); 3,4-DHAP also
significantly increased PARP-1 protein expression levels
(P<0.001). PARP-1 levels in the 3,4-DHAP group were
higher than those in the SFN group, and the difference was
significant (P<0.01). Compared with the 3,4-DHAP group,
the protein expression levels of PARP-1 in the 3,4-DHAP + ML385
group were reduced (P<0.001). These findings further
demonstrated that 3,4-DHAP could promote the response to cell
damage via regulation of PARP-1 expression.
Discussion
With the increasing adoption of unhealthy diets and
improvements in living standards, the incidence of T2DM is
increasing annually, posing a significant burden to the health and
quality of life of individuals (43). Atherosclerosis, the most common
complication of T2DM, is a chronic inflammatory disease (44). Endothelial dysfunction, the
initial link in the early stage of atherosclerosis, is an early
manifestation that occurs prior to the formation of atherosclerosis
and affects the occurrence and development of atherosclerosis.
Endothelial cells can be damaged by several factors, such as
oxidative stress, proinflammatory factors, hyperglycemia,
hyperlipidemia and hypertension (45). Oxidative stress is a state that
arises following an imbalance between oxidation and antioxidation.
Vascular oxidative stress promotes endothelial dysfunction and
atherosclerotic progression (46). Oxidative stress induced by high
glucose levels is a major factor in diabetic macroangiopathy
(47). Therefore, a model of
diabetes was established using high glucose medium. The results
also showed that there was a significant reduction in cell
viability in cells cultured under high glucose conditions.
ROS homeostasis in the majority of organisms is
maintained through the balance between ROS production and ROS
scavenging (48). When there is
an imbalance between generation and reduction of ROS, ROS levels
are increased. Increased ROS levels leads to a disorder of the
antioxidant system and accumulation of ROS, resulting in an
oxidative stress (49). Excessive
ROS levels notably alter the function of endothelial cells, such as
causing metabolic imbalances and oxidative stress (50), affecting mitochondrial morphology
and function (51) and inducing
autophagy and cell death (52).
There are numerous studies that have suggested that the levels of
ROS exert a potent effect in the occurrence of oxidative stress
(53–55). Similarly, in the present study,
ROS production was markedly increased in the high glucose group
compared with the control group.
ROS can be eliminated through antioxidant effects.
Zhong et al (56) showed
that the defense function of the cell was activated by the increase
in ROS levels when cells were damaged, thus cellular antioxidant
mechanisms are activated and upregulated to scavenge the ROS. Das
et al (57) reported that
persistent hyperglycemia impaired the pro-oxidant and antioxidant
balance, which in-turn reduced antioxidant levels and increased ROS
production under diabetic conditions. Wang et al (58) indicated that baicalein (BL)
increased production of ROS or decreased the expression of
antioxidant proteins mediated by ROS, to promote cancer cell death.
3,4-DHAP, a compound extracted from Ilex glauca leaves, has
physiological activities and beneficial effects for cardiovascular
diseases with relatively little toxicity and few side effects
(38). It also has a therapeutic
effect on coronary heart diseases, angina pectoris and pregnancy
hypertension (59), and has been
clinically adopted as a novel treatment for coronary heart diseases
and angina pectoris. Wu et al (60) suggested that 3,4-DHAP exerted
anti-inflammatory function on LPS-activated macrophages. Lu and
Chen (37) showed that 3,4-DHAP
could eliminate free radicals and increase resistance to lipid
peroxidation to protect the function of the brain. The present
study showed that 3,4-DHAP increased cell viability and markedly
reduced ROS levels compared with the high glucose group. Thus,
3,4-DHAP was hypothesized to exhibit a potent antioxidant effect
and to attenuate oxidative stress induced by high glucose in
HUVECs.
It is well established that Nrf2 is the primary
defense mechanism against cellular oxidative stress (61). When an antioxidant stimulates a
cell, Nrf2 becomes decoupled from the cytoplasmic protein chaperone
molecule Keap1 and enters the nucleus, where it binds to the
antioxidant reaction element (ARE) to eliminate ROS (62). HO-1, an important antioxidant
enzyme, primarily catalyzes hemoglobin into ferrous iron, carbon
monoxide and biliverdin. The degradation of the heme group is
conducive in preventing its oxidative promotion. When Nrf2 is
present in the nucleus, HO-1 promoter activity is regulated by
Nrf2. Furthermore, the activation of the Nrf2/HO-1 pathway exerts a
potent effect on cells, especially when under conditions of
oxidative stress (63). Martinez
et al (64) demonstrated
that BML-111 could increase Nrf2, HO-1 and NQO1 expression levels
to lower oxidative stress induced by ultraviolet radiation B (UVB).
Piao et al (65) also
showed that the Nrf2 and HO-1 levels were upregulated after
treatment with mangiferin (MF), suggesting that the antioxidant
effects of MF were regulated by Nrf2/HO-1. A previous study has
shown that Nrf2 expression is increased as well as its
translocation to the nucleus, and this plays a pivotal role in its
antioxidant effects (66). In the
present study, Nrf2 and HO-1 protein and mRNA expression levels in
the 3,4-DHAP group were higher than that in the high glucose group.
The Nrf2 nuclear levels were enhanced in cells pretreated with
3,4-DHAP when compared with the high glucose group. Thus, it was
considered that 3,4-DHAP may exert an antioxidant role by
regulating the Nrf2/HO-1 pathway to eliminate excessive ROS.
In addition, ML385, the Nrf2 activity inhibitor,
could directly interact with the Nrf2 protein, binding to the Neh1
binding region of Nrf2, thus preventing the establishment of the
Nrf2-mafg complex at ARE promoter sequences and in-turn reducing
transcriptional activity. In lung cancer cells, ML385 targeted Nrf2
signaling, affected colony formation ability and the growth of
cells, as well as Nrf2-mediated functions (67). Liu et al (68) confirmed that the protective
effects of isoliquiritigenin on acute pancreatitis in mice were
mediated through inhibition of oxidative stress and modulation of
the Nrf2/HO-1 pathway. Thus, in order to determine whether 3,4-DHAP
exerted its antioxidant effects through the Nrf2/HO-1 pathway,
ML385 was used to inhibit Nrf2. The results showed that compared
with the 3,4-DHAP group, the Nrf2 total and nuclear protein levels
in the 3,4-DHAP + ML385 group were decreased, the mRNA expression
levels of Nrf2 in the 3,4-DHAP + ML385 group were reduced, and the
HO-1 protein and mRNA expression levels in the 3,4-DHAP + ML385
group were also reduced. These findings further suggested that
3,4-DHAP protected HUVECs from oxidative stress induced by high
glucose through the Nrf2/HO-1 pathway. However, HO-1 is one of
numerous downstream genes regulated by Nrf2; other downstream genes
such as NQO1, GCLC, GCLM may have also participated in the
oxidative protective mechanisms in the HUVECs treated with high
glucose (69). The involvement of
other genes will be assessed in future studies.
Autophagy is a biological process in which damaged
organelles, misfolded proteins and invading pathogens are enveloped
in an intracellular double-membraned structure and degraded. When
stimulated by stressors such as starvation, hypoxia, infection and
DNA damage (70), autophagy is
further activated and plays a protective role by removing abnormal
organelles (71). Growing
evidence supports the notion that a series of biological factors
and compounds can induce vascular endothelial cells to undergo
autophagy to resist stress responses and protect the cell. Xu et
al (72) found that rapamycin
activated autophagy and reduced DNA radiation damage of bone marrow
blood cells through the STAT3 signaling pathway. However, when
autophagy was over-activated, it resulted in autophagic death
(73). Studies have shown that
treatment of endothelial cells with endostatin, an endogenous
angiogenic inhibitor, induced autophagy and cell death (74,75). LC3 is a specific marker for
autophagosome formation (76).
LC3 was previously considered to be involved in the regulation of
microtubule assembly and disassembly. Subsequently, LC3 was found
to exert specific autophagic effects. During the formation of
autophagosomes, LC3-I is transformed into LC3-II by binding to
phosphatidylethanolamine. When the LC3-II/LC3-I ratio increases,
this is indicative that autophagy has occurred. Qiao et al
(77) found that the anticancer
effects of TRAIL were increased following azithromycin treatment,
which may be related to LC3-mediated autophagy. A previous study
has shown that aminoguanidine (AG) reduces the LC3-II/LC3-I ratio
and ROS production to inhibit autophagy (78). The autophagosome, a key structure
in the process of autophagy, envelops damaged organelles or
proteins and combines with lysosomes to generate an
autophagolysosome. Liang et al (79) observed that autophagosomes were
formed in the early stages of autophagy and matured in the later
stages of autophagy. In the present study, the protein expression
levels of LC3-II/LC3-I and the formation of autophagosomes in the
high glucose group was slightly increased compared with the control
group. This demonstrated that autophagy was activated when
stimulated by oxidation. Furthermore, 3,4-DHAP markedly enhanced
LC-3II/LC3-I protein levels and the formation of autophagosomes
compared with the high glucose group. Therefore, it was suggested
that 3,4-DHAP could protect HUVECs from oxidative stress by
enhancing autophagy levels.
The stability of genomic DNA is vital for the
survival of individuals and the long-term reproduction of species.
When DNA damage occurs, cells need to activate the DNA damage
repair mechanisms, and the cell cycle is arrested, preventing the
cell in question from continuing mitosis. After the DNA damage is
repaired, the cell cycle is resumed and the cell begins to undergo
mitosis again (80). The working
system of DNA damage repair includes sensors, mediators, signal
transmitters and effectors (81).
The receptors primarily involved in this process are PARP1/2, the
9-1-1 complex and the RAD17-RFC complex (82,83). PARP-1 is a multifunctional protein
that post-translationally modifies enzymes already present in the
majority eukaryotic cells. PARP-1 can sense DNA damage and is
activated by identifying DNA fragments with structural damage.
Studies have shown that PARP inhibitors cause an increase in DNA
damage and prevent cells from repairing single-stranded DNA breaks
(84). Therefore, PARP-1 plays a
significant role in DNA damage repair and transcriptional
regulation (85). Isakoff et
al (86) discovered that DNA
damage repair was blocked by PARP inhibitors, and the clinical
activity of DNA-damaging chemotherapy was enhanced when combined
with these inhibitors. The results of the present study showed that
3,4-DHAP markedly enhanced PARP-1 protein expression levels
compared with the high glucose group. In short, these findings
demonstrated that 3,4-DHAP could promote DNA repair in HUVECs by
regulating the expression of PARP-1.
An increasing number of studies have found that the
mutual regulation between autophagy and Nrf2 is involved in
modulation of ROS and other factors. ROS plays a critical role in
autophagy. Pajares et al (87) found that Nrf2 regulated autophagy
gene transcription in a mouse model of Alzheimer's disease. Feng
et al (88) showed that
activating the Nrf2 pathway could upregulate autophagy to protect
LPS-induced HK-2 cell injury. In the present study, a possible
association between Nrf2 and autophagy was identified. Compared
with the 3,4-DHAP group, the protein expression levels of
LC3-II/LC3-I were reduced in the 3,4-DHAP + ML385 group. Therefore,
it was hypothesized that Nrf2 may induce autophagy to repair
damaged endothelial cells. Furthermore, autophagy was involved in
the repair of damaged DNA in cells (89). Studies have suggested that PARP-1
has a profound effect on autophagy and is an important indicator in
the formation and maturation of autophagosomes (90). Rodríguez-Vargas et al
(91) showed that the production
of ROS led to DNA damage and excessive activation of PARP-1 when
autophagy was induced by starvation. Thus, in the present study, it
was hypothesized that there was a link between autophagy and DNA
damage. The protein expression levels of PARP were reduced in the
3,4-DHAP + ML385 group compared with the 3,4-DHAP group. However,
the mechanisms linking autophagy and Nrf2, and linking autophagy
and DNA damage repair require further study to elucidate.
In the present study it was determined that 3,4-DHAP
treatment reduced ROS production, upregulated Nrf2 protein and mRNA
expression, increased HO-1 protein and mRNA expression, promoted
Nrf2 nuclear translocation, increased LC3-II/LC3-I and PARP-1
protein expression and promoted the formation of autophagosomes. At
present, the prevention and treatment of T2DM and its complications
are still a major problem to be solved in China. Clinically, drugs
for the comprehensive treatment of diabetes and AS are still in
short supply. The present study found that 3,4-DHAP has an
inhibitory effect on inflammatory response and oxidative stress.
Therefore, 3,4-DHAP may be the drug of choice for treatment.
However, the current experimental study has limitations. We are
currently investigating the protective effect of 3,4-DHAp on HUVECs
and its molecular mechanism, which needs to be verified in
vivo. Regarding autophagy and DNA damage repair, only 3,4-DHAP
has been studied to regulate autophagy and DNA damage repair, and
its mechanism needs to be further studied. In addition, the manner
in which 3,4-DHAP regulates Nrf2, autophagy and DNA repair is also
a problem to be solved in the next experimental stage. In
conclusion, the results suggest that 3,4-DHAP possesses
antioxidative properties and was thus able to protect HUVECs from
oxidative stress via regulation of the Nrf2/HO-1 pathway, enhancing
autophagy and promoting DNA damage repair.
Acknowledgements
Not applicable.
Funding
This work was supported by the Shandong Natural Science
Foundation (grant nos. ZR2020MH415 and ZR2021LZY033).
Availability of data and materials
The datasets used and/or analyzed during the current
study are available from the corresponding author on reasonable
request.
Authors' contributions
DZ, JL and DC conceived and designed the study. DC
and YW performed the experiments. DC drafted the manuscript. WL,
YW, JJ, and JG performed data analysis. DZ and JL were responsible
for data interpretation. WL, JJ and JG revised the manuscript
critically for important intellectual content. DZ, JL and DC
confirm the authenticity of all the raw data. All authors have read
and approved the final manuscript.
Ethics approval and consent to
participate
Not applicable.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Cho NH, Shaw JE, Karuranga S, Huang Y, da
Rocha Fernandes JD, Ohlrogge AW and Malanda B: IDF diabetes atlas:
Global estimates of diabetes prevalence for 2017 and projections
for 2045. Diabetes Res Clin Pract. 138:271–281. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Newman JD, Schwartzbard AZ, Weintraub HS,
Goldberg IJ and Berger JS: Primary prevention of cardiovascular
disease in diabetes mellitus. J Am Coll Cardiol. 70:883–893. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Sheng B, Truong K, Spitler H, Zhang L,
Tong X and Chen L: The long-term effects of bariatric surgery on
type 2 diabetes remission, microvascular and macrovascular
complications, and mortality: A systematic review and
meta-analysis. Obes Surg. 27:2724–2732. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
GBD 2016 Causes of Death Collaborators, .
Global, regional, and national age-sex specific mortality for 264
causes of death, 1980-2016: A systematic analysis for the Global
Burden of Disease Study 2016. Lancet. 390:1151–1210. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Luo JF, Shen XY, Lio CK, Dai Y, Cheng CS,
Liu JX, Yao YD, Yu Y, Xie Y, Luo P, et al: Activation of Nrf2/HO-1
pathway by nardochinoid C inhibits inflammation and oxidative
stress in lipopolysaccharide-stimulated macrophages. Front
Pharmacol. 9:9112018. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Zuo L, Prather ER, Stetskiv M, Garrison
DE, Meade JR, Peace TI and Zhou T: Inflammaging and oxidative
stress in human diseases: From molecular mechanisms to novel
treatments. Int J Mol Sci. 20:44722019. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Förstermann U, Xia N and Li H: Roles of
vascular oxidative stress and nitric oxide in the pathogenesis of
atherosclerosis. Circ Res. 120:713–735. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Costa JG, Saraiva N, Batinic-Haberle I,
Castro M, Oliveira NG and Fernandes AS: The SOD Mimic
MnTnHex-2-PyP5+ reduces the viability and migration of
786-O human renal cancer cells. Antioxidants (Basel). 8:4902019.
View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Ge C, Tan J, Zhong S, Lai L, Chen G, Zhao
J, Yi C, Wang L, Zhou L, Tang T, et al: Nrf2 mitigates prolonged
PM2.5 exposure-triggered liver inflammation by positively
regulating SIKE activity: Protection by Juglanin. Redox Biol.
36:1016452020. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Rojo de la Vega M, Chapman E and Zhang DD:
NRF2 and the hallmarks of cancer. Cancer Cell. 34:21–43. 2018.
View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Suzuki T, Motohashi H and Yamamoto M:
Toward clinical application of the Keap1-Nrf2 pathway. Trends
Pharmacol Sci. 34:340–346. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Leinonen HM, Kansanen E, Pölönen P,
Heinäniemi M and Levonen AL: Role of the Keap1-Nrf2 pathway in
cancer. Adv Cancer Res. 122:281–320. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Cuadrado A, Rojo AI, Wells G, Hayes JD,
Cousin SP, Rumsey WL, Attucks OC, Franklin S, Levonen AL, Kensler
TW and Dinkova-Kostova AT: Therapeutic targeting of the NRF2 and
KEAP1 partnership in chronic diseases. Nat Rev Drug Discov.
18:295–317. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Kobayashi A, Kang MI, Okawa H, Ohtsuji M,
Zenke Y, Chiba T, Igarashi K and Yamamoto M: Oxidative stress
sensor Keap1 functions as an adaptor for Cul3-based E3 ligase to
regulate proteasomal degradation of Nrf2. Mol Cell Biol.
24:7130–7139. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Warabi E, Takabe W, Minami T, Inoue K,
Itoh K, Yamamoto M, Ishii T, Kodama T and Noguchi N: Shear stress
stabilizes NF-E2-related factor 2 and induces antioxidant genes in
endothelial cells: Role of reactive oxygen/nitrogen species. Free
Radic Biol Med. 42:260–269. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Farkhondeh T, Pourbagher-Shahri AM,
Azimi-Nezhad M, Forouzanfar F, Brockmueller A, Ashrafizadeh M,
Talebi M, Shakibaei M and Samarghandian S: Roles of Nrf2 in gastric
cancer: Targeting for therapeutic strategies. Molecules.
26:31572021. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Su L, Cao P and Wang H: Tetrandrine
mediates renal function and redox homeostasis in a
streptozotocin-induced diabetic nephropathy rat model through
Nrf2/HO-1 reactivation. Ann Transl Med. 8:9902020. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Zhang S, Li T, Zhang L, Wang X, Dong H, Li
L, Fu D, Li Y, Zi X, Liu HM, et al: A novel chalcone derivative S17
induces apoptosis through ROS dependent DR5 up-regulation in
gastric cancer cells. Sci Rep. 7:98732017. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Ci X, Lv H, Wang L, Wang X, Peng L, Qin FX
and Cheng G: The antioxidative potential of farrerol occurs via the
activation of Nrf2 mediated HO-1 signaling in RAW 264.7 cells. Chem
Biol Interact. 239:192–199. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Mohammad J, Singh RR, Riggle C, Haugrud B,
Abdalla MY and Reindl KM: JNK inhibition blocks
piperlongumine-induced cell death and transcriptional activation of
heme oxygenase-1 in pancreatic cancer cells. Apoptosis. 24:730–744.
2019. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Levine B and Kroemer G: Autophagy in the
pathogenesis of disease. Cell. 132:27–42. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Hu B, Zhang Y, Jia L, Wu H, Fan C, Sun Y,
Ye C, Liao M and Zhou J: Binding of the pathogen receptor HSP90AA1
to avibirnavirus VP2 induces autophagy by inactivating the AKT-MTOR
pathway. Autophagy. 11:503–515. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Zhao YG, Codogno P and Zhang H: Machinery,
regulation and pathophysiological implications of autophagosome
maturation. Nat Rev Mol Cell Biol. 22:733–750. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Laverdure S, Wang Z, Yang J, Yamamoto T,
Thomas T, Sato T, Nagashima K and Imamichi T: Interleukin-27
promotes autophagy in human serum-induced primary macrophages via
an mTOR- and LC3-independent pathway. Sci Rep. 11:148982021.
View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Rakovic A, Shurkewitsch K, Seibler P,
Grünewald A, Zanon A, Hagenah J, Krainc D and Klein C: Phosphatase
and tensin homolog (PTEN)-induced putative kinase 1
(PINK1)-dependent ubiquitination of endogenous Parkin attenuates
mitophagy: Study in human primary fibroblasts and induced
pluripotent stem cell-derived neurons. J Biol Chem. 288:2223–2237.
2013. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Yang A, Pantoom S and Wu YW: Elucidation
of the anti-autophagy mechanism of the Legionella effector RavZ
using semisynthetic LC3 proteins. Elife. 6:e239052017. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Han X, Liu JX and Li XZ: Salvianolic acid
B inhibits autophagy and protects starving cardiac myocytes. Acta
Pharmacol Sin. 32:38–44. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Mikolaskova B, Jurcik M, Cipakova I,
Kretova M, Chovanec M and Cipak L: Maintenance of genome stability:
The unifying role of interconnections between the DNA damage
response and RNA-processing pathways. Curr Genet. 64:971–983. 2018.
View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Wang Y, Luo W and Wang Y: PARP-1 and its
associated nucleases in DNA damage response. DNA Repair (Amst).
81:1026512019. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Zhou Y, Tang S, Chen T and Niu MM:
Structure-based pharmacophore modeling, virtual screening,
molecular docking and biological evaluation for identification of
potential poly (ADP-Ribose) Polymerase-1 (PARP-1) Inhibitors.
Molecules. 24:42582019. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Schiewer MJ, Mandigo AC, Gordon N, Huang
F, Gaur S, de Leeuw R, Zhao SG, Evans J, Han S, Parsons T, et al:
PARP-1 regulates DNA repair factor availability. EMBO Mol Med.
10:e88162018. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Pazzaglia S and Pioli C: Multifaceted Role
of PARP-1 in DNA repair and inflammation: Pathological and
therapeutic implications in cancer and non-cancer diseases. Cells.
9:412019. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Liu Y, Song H, Song H, Feng X, Zhou C and
Huo Z: Targeting autophagy potentiates the anti-tumor effect of
PARP inhibitor in pediatric chronic myeloid leukemia. AMB Express.
9:1082019. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Wang L, Wei W, Xiao Q, Yang H and Ci X:
Farrerol Ameliorates APAP-induced hepatotoxicity via activation of
Nrf2 and autophagy. Int J Biol Sci. 15:788–799. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Fan SF, Zhou NH, Hu SL and Xu SG: Effects
of 3,4-dihydroxyacetophenone in shortening of action potential
duration of cardiac cells (author's transl). Zhongguo Yao Li Xue
Bao. 2:107–110. 1981.(In Chinese). PubMed/NCBI
|
|
36
|
Kim YJ, No JK, Lee JS, Kim MS and Chung
HY: Antimelanogenic activity of 3,4-dihydroxyacetophenone:
Inhibition of tyrosinase and MITF. Biosci Biotechnol Biochem.
70:532–534. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Lu XY and Chen WC: Effect of 3,
4-dihydroxyacetophenone on Na+, K+-ATPase
activity of injured mitochondria and the oxygen consumption of
brain cells of rat. Yao Xue Xue Bao. 40:13–16. 2005.PubMed/NCBI
|
|
38
|
Zhang D, Liu J, Wang L, Wang J, Li W,
Zhuang B, Hou J and Liu T: Effects of 3,4-dihydroxyacetophenone on
the hypercholesterolemia-induced atherosclerotic rabbits. Biol
Pharm Bull. 36:733–740. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(−Delta Delta C(T)) method. Methods. 25:402–408. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Aranda A, Sequedo L, Tolosa L, Quintas G,
Burello E, Castell JV and Gombau L: Dichloro-dihydro-fluorescein
diacetate (DCFH-DA) assay: A quantitative method for oxidative
stress assessment of nanoparticle-treated cells. Toxicol In Vitro.
27:954–963. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Arakawa S, Honda S, Yamaguchi H and
Shimizu S: Molecular mechanisms and physiological roles of
Atg5/Atg7-independent alternative autophagy. Proc Jpn Acad Ser B
Phys Biol Sci. 93:378–385. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Dunphy G, Flannery SM, Almine JF, Connolly
DJ, Paulus C, Jønsson KL, Jakobsen MR, Nevels MM, Bowie AG and
Unterholzner L: Non-canonical activation of the DNA sensing adaptor
STING by ATM and IFI16 mediates NF-κB signaling after nuclear DNA
damage. Mol Cell. 71:745–760.e5. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
43
|
Hemmingsen B, Gimenez-Perez G, Mauricio D,
Roqué IFM, Metzendorf MI and Richter B: Diet, physical activity or
both for prevention or delay of type 2 diabetes mellitus and its
associated complications in people at increased risk of developing
type 2 diabetes mellitus. Cochrane Database Syst Rev. Dec
4–2017.(Epub ahead of print). View Article : Google Scholar
|
|
44
|
Wu Y, Song F, Li Y, Li J, Cui Y, Hong Y,
Han W, Wu W, Lakhani I, Li G and Wang Y: Acacetin exerts
antioxidant potential against atherosclerosis through Nrf2 pathway
in apoE(−/-) Mice. J Cell Mol Med. 25:521–534. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
45
|
Kaur R, Kaur M and Singh J: Endothelial
dysfunction and platelet hyperactivity in type 2 diabetes mellitus:
Molecular insights and therapeutic strategies. Cardiovasc Diabetol.
17:1212018. View Article : Google Scholar : PubMed/NCBI
|
|
46
|
Sena CM, Leandro A, Azul L, Seiça R and
Perry G: Vascular oxidative stress: Impact and therapeutic
approaches. Front Physiol. 9:16682018. View Article : Google Scholar : PubMed/NCBI
|
|
47
|
Jiao B, Wang YS, Cheng YN, Gao JJ and
Zhang QZ: Valsartan attenuated oxidative stress, decreased MCP-1
and TGF-β1 expression in glomerular mesangial and epithelial cells
induced by high-glucose levels. Biosci Trends. 5:173–181. 2011.
View Article : Google Scholar : PubMed/NCBI
|
|
48
|
Wang M, Sun X, Yu D, Xu J, Chung K and Li
H: Genomic and transcriptomic analyses of the tangerine pathotype
of Alternaria alternata in response to oxidative stress. Sci Rep.
6:324372016. View Article : Google Scholar : PubMed/NCBI
|
|
49
|
Guo J, Zhao MH, Shin KT, Niu YJ, Ahn YD,
Kim NH and Cui XS: The possible molecular mechanisms of bisphenol A
action on porcine early embryonic development. Sci Rep. 7:86322017.
View Article : Google Scholar : PubMed/NCBI
|
|
50
|
An J, Li Q, Yang J, Zhao Z, Wu Y, Wang Y
and Wang W: Wheat F-box protein TaFBA1 positively regulates plant
drought tolerance but negatively regulates stomatal closure. Front
Plant Sci. 10:12422019. View Article : Google Scholar : PubMed/NCBI
|
|
51
|
Zhang R, Liu B, Fan X, Wang W, Xu T, Wei
S, Zheng W, Yuan Q, Gao L, Yin X, et al: Aldehyde dehydrogenase 2
protects against post-cardiac arrest myocardial dysfunction through
a novel mechanism of suppressing mitochondrial reactive oxygen
species production. Front Pharmacol. 11:3732020. View Article : Google Scholar : PubMed/NCBI
|
|
52
|
Bazhin AV, Philippov PP and Karakhanova S:
Reactive oxygen species in cancer biology and anticancer therapy.
Oxid Med Cell Longev. 2016:41978152016. View Article : Google Scholar : PubMed/NCBI
|
|
53
|
Senoner T and Dichtl W: Oxidative stress
in cardiovascular diseases: Still a therapeutic target? Nutrients.
11:20902019. View Article : Google Scholar : PubMed/NCBI
|
|
54
|
Kang Q and Yang C: Oxidative stress and
diabetic retinopathy: Molecular mechanisms, pathogenetic role and
therapeutic implications. Redox Biol. 37:1017992020. View Article : Google Scholar : PubMed/NCBI
|
|
55
|
Shah MS and Brownlee M: Molecular and
cellular mechanisms of cardiovascular disorders in diabetes. Circ
Res. 118:1808–1829. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
56
|
Zhong Z, Fu X, Li H, Chen J, Wang M, Gao
S, Zhang L, Cheng C, Zhang Y, Li P, et al: Citicoline protects
auditory hair cells against neomycin-induced damage. Front Cell Dev
Biol. 8:7122020. View Article : Google Scholar : PubMed/NCBI
|
|
57
|
Das SK, Prusty A, Samantaray D, Hasan M,
Jena S, Patra JK, Samanta L and Thatoi H: Effect of Xylocarpus
granatum bark extract on amelioration of hyperglycaemia and
oxidative stress associated complications in STZ-induced diabetic
mice. Evid Based Complement Alternat Med. 2019:84931902019.
View Article : Google Scholar : PubMed/NCBI
|
|
58
|
Wang SX, Wen X, Bell C and Appiah S:
Liposome-delivered baicalein induction of myeloid leukemia K562
cell death via reactive oxygen species generation. Mol Med Rep.
17:4524–4530. 2018.PubMed/NCBI
|
|
59
|
Yang DS, Xi-Rui W and Ting-Yuan M: Effects
of 3,4-dihydroxyacetophenone on the biosynthesis of TXA2 and PGI2
in human placental villus and umbilical artery segments in vitro.
Prostaglandins. 38:497–504. 1989. View Article : Google Scholar : PubMed/NCBI
|
|
60
|
Wu P, Ye D, Zhang D, Zhang L, Wan J and
Pan Q: Dual effect of 3,4-dihydroxyacetophenone on LPS-induced
apoptosis in RAW264.7 cells by modulating the production of
TNF-alpha. J Huazhong Univ Sci Technolog Med Sci. 25:131–134. 2005.
View Article : Google Scholar : PubMed/NCBI
|
|
61
|
Fan X, Wei W, Huang J, Liu X and Ci X:
Isoorientin attenuates cisplatin-induced nephrotoxicity through the
inhibition of oxidative stress and apoptosis via activating the
SIRT1/SIRT6/Nrf-2 pathway. Front Pharmacol. 11:2642020. View Article : Google Scholar : PubMed/NCBI
|
|
62
|
Qin JJ, Cheng XD, Zhang J and Zhang WD:
Dual roles and therapeutic potential of Keap1-Nrf2 pathway in
pancreatic cancer: A systematic review. Cell Commun Signal.
17:1212019. View Article : Google Scholar : PubMed/NCBI
|
|
63
|
Li T, Chen B, Du M, Song J, Cheng X, Wang
X and Mao X: Casein glycomacropeptide hydrolysates exert
cytoprotective effect against cellular oxidative stress by
Up-Regulating HO-1 expression in HepG2 cells. Nutrients. 9:312017.
View Article : Google Scholar : PubMed/NCBI
|
|
64
|
Martinez RM, Fattori V, Saito P, Pinto IC,
Rodrigues CCA, Melo CPB, Bussmann AJC, Staurengo-Ferrari L, Bezerra
JR, Vignoli JA, et al: The lipoxin Receptor/FPR2 agonist BML-111
protects mouse skin against ultraviolet B radiation. Molecules.
25:29532020. View Article : Google Scholar : PubMed/NCBI
|
|
65
|
Piao CH, Fan YJ, Nguyen TV, Song CH and
Chai OH: Mangiferin alleviates ovalbumin-induced allergic rhinitis
via Nrf2/HO-1/NF-κB signaling pathways. Int J Mol Sci. 21:34152020.
View Article : Google Scholar : PubMed/NCBI
|
|
66
|
Probst BL, McCauley L, Trevino I, Wigley
WC and Ferguson DA: Cancer cell growth is differentially affected
by constitutive activation of NRF2 by KEAP1 deletion and
pharmacological activation of NRF2 by the synthetic triterpenoid,
RTA 405. PLoS One. 10:e01352572015. View Article : Google Scholar : PubMed/NCBI
|
|
67
|
Singh A, Venkannagari S, Oh KH, Zhang YQ,
Rohde JM, Liu L, Nimmagadda S, Sudini K, Brimacombe KR, Gajghate S,
et al: Small molecule inhibitor of NRF2 selectively intervenes
therapeutic resistance in KEAP1-deficient NSCLC tumors. ACS Chem
Biol. 11:3214–3225. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
68
|
Liu X, Zhu Q, Zhang M, Yin T, Xu R, Xiao
W, Wu J, Deng B, Gao X, Gong W, et al: Isoliquiritigenin
ameliorates acute pancreatitis in mice via inhibition of oxidative
stress and modulation of the Nrf2/HO-1 pathway. Oxid Med Cell
Longev. 2018:71615922018. View Article : Google Scholar : PubMed/NCBI
|
|
69
|
Zhao Y, Sun Y, Wang G, Ge S and Liu H:
Dendrobium officinale polysaccharides protect against MNNG-Induced
PLGC in rats via activating the NRF2 and antioxidant enzymes HO-1
and NQO-1. Oxid Med Cell Longev. 2019:93102452019. View Article : Google Scholar : PubMed/NCBI
|
|
70
|
Gomes LR, Menck CFM and Leandro GS:
Autophagy roles in the modulation of DNA repair pathways. Int J Mol
Sci. 18:23512017. View Article : Google Scholar : PubMed/NCBI
|
|
71
|
Liao SX, Sun PP, Gu YH, Rao XM, Zhang LY
and Ou-Yang Y: Autophagy and pulmonary disease. Ther Adv Respir
Dis. 13:17534666198905382019. View Article : Google Scholar : PubMed/NCBI
|
|
72
|
Xu F, Li X, Yan L, Yuan N, Fang Y, Cao Y,
Xu L, Zhang X, Xu L, Ge C, et al: Autophagy promotes the repair of
Radiation-Induced DNA damage in bone marrow hematopoietic cells via
enhanced STAT3 signaling. Radiat Res. 187:382–396. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
73
|
Galati S, Boni C, Gerra MC, Lazzaretti M
and Buschini A: Autophagy: A player in response to oxidative stress
and DNA damage. Oxid Med Cell Longev. 2019:56929582019. View Article : Google Scholar : PubMed/NCBI
|
|
74
|
Poluzzi C, Iozzo RV and Schaefer L:
Endostatin and endorepellin: A common route of action for similar
angiostatic cancer avengers. Adv Drug Deliv Rev. 97:156–173. 2016.
View Article : Google Scholar : PubMed/NCBI
|
|
75
|
Nguyen TM, Subramanian IV, Xiao X, Ghosh
G, Nguyen P, Kelekar A and Ramakrishnan S: Endostatin induces
autophagy in endothelial cells by modulating Beclin 1 and
beta-catenin levels. J Cell Mol Med. 13:3687–3698. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
76
|
Deng W, Long Q, Zeng J, Li P, Yang W, Chen
X and Xie J: Mycobacterium tuberculosis PE_PGRS41 enhances the
intracellular survival of M. smegmatis within macrophages via
blocking innate immunity and inhibition of host defense. Sci Rep.
7:467162017. View Article : Google Scholar : PubMed/NCBI
|
|
77
|
Qiao X, Wang X, Shang Y, Li Y and Chen SZ:
Azithromycin enhances anticancer activity of TRAIL by inhibiting
autophagy and up-regulating the protein levels of DR4/5 in colon
cancer cells in vitro and in vivo. Cancer Commun (Lond). 38:432018.
View Article : Google Scholar : PubMed/NCBI
|
|
78
|
Lee JH, Parveen A, Do MH, Kang MC, Yumnam
S and Kim SY: Molecular mechanisms of methylglyoxal-induced aortic
endothelial dysfunction in human vascular endothelial cells. Cell
Death Dis. 11:4032020. View Article : Google Scholar : PubMed/NCBI
|
|
79
|
Liang C, Lee JS, Inn KS, Gack MU, Li Q,
Roberts EA, Vergne I, Deretic V, Feng P, Akazawa C and Jung JU:
Beclin1-binding UVRAG targets the class C Vps complex to coordinate
autophagosome maturation and endocytic trafficking. Nat Cell Biol.
10:776–787. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
80
|
Shaltiel IA, Krenning L, Bruinsma W and
Medema RH: The same, only different-DNA damage checkpoints and
their reversal throughout the cell cycle. J Cell Sci. 128:607–620.
2015.PubMed/NCBI
|
|
81
|
Ivy SP, de Bono J and Kohn EC: The
‘Pushmi-Pullyu’ of DNA REPAIR: Clinical synthetic lethality. Trends
Cancer. 2:646–656. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
82
|
He G, Siddik ZH, Huang Z, Koomen J,
Kobayashi R, Khokhar AR and Kuang J: Induction of p21 by p53
following DNA damage inhibits both Cdk4 and Cdk2 activities.
Oncogene. 24:2929–2943. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
83
|
Bartek J and Lukas J: Mammalian G1- and
S-phase checkpoints in response to DNA damage. Curr Opin Cell Biol.
13:738–747. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
84
|
Eckert MA, Orozco C, Xiao J, Javellana M
and Lengyel E: The Effects of chemotherapeutics on the ovarian
cancer microenvironment. Cancers (Basel). 13:31362021. View Article : Google Scholar : PubMed/NCBI
|
|
85
|
Min X, Heng H, Yu HL, Dan M, Jie C, Zeng
Y, Ning H, Liu ZG, Wang ZY and Lin W: Anticancer effects of
10-hydroxycamptothecin induce apoptosis of human osteosarcoma
through activating caspase-3, p53 and cytochrome c pathways. Oncol
Lett. 15:2459–2464. 2018.PubMed/NCBI
|
|
86
|
Isakoff SJ, Puhalla S, Domchek SM,
Friedlander M, Kaufman B, Robson M, Telli ML, Diéras V, Han HS,
Garber JE, et al: A randomized Phase II study of veliparib with
temozolomide or carboplatin/paclitaxel versus placebo with
carboplatin/paclitaxel in BRCA1/2 metastatic breast cancer: Design
and rationale. Future Oncol. 13:307–320. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
87
|
Pajares M, Jiménez-Moreno N, García-Yagüe
ÁJ, Escoll M, de Ceballos ML, Van Leuven F, Rábano A, Yamamoto M,
Rojo AI and Cuadrado A: Transcription factor NFE2L2/NRF2 is a
regulator of macroautophagy genes. Autophagy. 12:1902–1916. 2016.
View Article : Google Scholar : PubMed/NCBI
|
|
88
|
Feng LX, Zhao F, Liu Q, Peng JC, Duan XJ,
Yan P, Wu X, Wang HS, Deng YH and Duan SB: Role of Nrf2 in
lipopolysaccharide-induced acute kidney injury: Protection by human
umbilical cord blood mononuclear cells. Oxid Med Cell Longev.
2020:61234592020. View Article : Google Scholar : PubMed/NCBI
|
|
89
|
Yang J, Yu J, Li D, Yu S, Ke J, Wang L,
Wang Y, Qiu Y, Gao X, Zhang J and Huang L: Store-operated calcium
entry-activated autophagy protects EPC proliferation via the
CAMKK2-MTOR pathway in ox-LDL exposure. Autophagy. 13:82–98. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
90
|
Lin W, Yin CY, Yu Q, Zhou SH, Chai L, Fan
J and Wang WD: Expression of glucose transporter-1, hypoxia
inducible factor-1α and beclin-1 in head and neck cancer and their
implication. Int J Clin Exp Pathol. 11:3708–3717. 2018.PubMed/NCBI
|
|
91
|
Rodríguez-Vargas JM, Ruiz-Magaña MJ,
Ruiz-Ruiz C, Majuelos-Melguizo J, Peralta-Leal A, Rodríguez MI,
Muñoz-Gámez JA, de Almodóvar MR, Siles E, Rivas AL, et al:
ROS-induced DNA damage and PARP-1 are required for optimal
induction of starvation-induced autophagy. Cell Res. 22:1181–1198.
2012. View Article : Google Scholar : PubMed/NCBI
|