Introduction
Intervertebral disc degeneration (IDD) is a major
cause of degenerative spine diseases, such as cervical spondylosis,
spinal column stenosis and intervertebral disc herniation (1,2).
Current treatments for such diseases include conservative treatment
and surgery. Conservative treatments for this condition include
rest, administration of anti-inflammatory and analgesic drugs, or
combined physiotherapy (3–5)
However, their disadvantages include the need for early
intervention, recurrent symptoms and failure of radical treatment.
Surgical treatments include nucleus pulposus removal, discectomy
and fusion (6,7). Although short- and medium-term
efficacy is satisfactory after surgery, complications, such as
adjacent segment degeneration, may occur during long-term follow-up
(8). Therefore, there is
currently no effective treatment for this disease. Although
conventional methods may relieve local symptoms, they are
insufficient to properly treat IDD; thus, the normal physiological
structure and function of the intervertebral disc are not recovered
(9). Therefore, novel treatments
are urgently needed to treat IDD.
At present, the pathogenesis of IDD remains unclear.
The major pathological changes of the disease are characterized by
a decrease in the number and function of nucleus pulposus cells, as
well as extracellular matrix degradation (1,10).
Under normal conditions, nucleus pulposus cells secrete a large
amounts of proteoglycans, collagen-II and other extracellular
matrix proteins, to maintain the elasticity and the load-bearing
balance of the intervertebral disc (11). Under pathological conditions,
fewer nucleus pulposus cells, decreased matrix secretion and stress
changes in the intervertebral disc result in degradation,
ultimately contributing to intervertebral disc protrusion, spinal
column stenosis and spinal instability (12,13).
Among the biological therapies for IDD, mesenchymal
stem cells show great potential in IDD recovery due to their
ability to self-replicate and differentiate into numerous cell
lineages (14). In previous
studies (15,16), it was observed that nucleus
pulposus-derived stem cells (NPSCs) are important for maintaining
the normal function and homeostasis of the nucleus pulposus.
Another study reported that the differentiation of NPSCs is
weakened in the elderly and individuals with a degenerative
condition (17). Therefore, an
ideal therapeutic approach would be to activate NPSCs to
differentiate into nucleus pulposus-like cells for the self-healing
of tissues. However, little is known about the influencing factors
and specific mechanisms of NPSC degeneration or the functional
inhibition of the intervertebral disc microenvironment. Therefore,
clarifying the influencing factors and mechanism of degeneration of
NSPCs in the intervertebral disc microenvironment, in order to
restore their biological performance, is important for IDD
treatment.
The degenerating intervertebral disc is considered
to be a complex and harsh microenvironment, characterized by
hypoxia, inflammation, high osmotic pressure and high mechanical
stress. These complex factors in the microenvironment affect the
activity of cells in the disc (18). The injured intervertebral disc can
secrete various cytokines or chemokines, induce endogenous stem
cells in the surrounding tissue stem cell nests to migrate to the
lesion site, and promote endogenous repair and regeneration
(19,20). Previous studies on the locally
expressed cytokines in IDD have mainly focused on TGF-β, TNF-α,
IL-1β and monocyte chemoattractant protein-1 (MCP-1) (21–23). In certain studies, IDD has been
reported to be associated with high local expression levels of
MCP-1, which may be associated with the degree of IDD (24,25). However, relevant reports outlining
the role of MCP-1 in IDD are rare.
Current studies regarding mesenchymal stem cells and
MCP-1 have focused on promoting the migration of endogenous neural
stem cells to improve neurofunctional repair and to promote the
homing of bone marrow stem cells post-myocardial infarction to
enhance recovery. These studies have also investigated their roles
in human immunoregulation and the promotion of wound healing
(26–28). The functions of MCP-1 were mainly
realized via binding to its specific receptor, C-C chemokine
receptor type 2 (CCR2) (29). The
MCP-1/CCR2 signaling axis has extensive biological effects and has
been reported to be important in immunodeficiency diseases,
implant-related immune responses and in tumors (17). However, there are few reports on
the local role of MCP-1 in IDD (30). The present study aimed to
determine whether the upregulation of MCP-1 in IDD could enhance
endogenous regeneration in the intervertebral disc by regulating
the differentiation of NPSCs to IDD stem niches. To the best of our
knowledge, the present study was the first to reveal the role of
the MCP-1/CCR2 axis in the chondrogenic NPSC differentiation at the
injury site of IDD.
Materials and methods
NPSC isolation from primary
cultures
The NPSCs used in the present study were collected
from a 16-year-old female patient with trauma-induced
intervertebral disc protrusion. The patient's parents provided
written consent for the collection and use of her vertebral tissue.
This research was approved by the Ethics Committee of the Second
School of Clinical Medicine, Southern Medical University
(Guangzhou, China; approval no. ICE2017058). Under sterile
conditions, the isolated human nucleus pulposus was washed twice
with PBS, separated carefully from the surrounding tissue, cut into
0.5×0.5×0.5-mm cubes and placed into a 0.02% collagenase II
solution. The tissue was digested in an incubator at 37°C with 5%
CO2 for 4 h. A sterile steel wire mesh (200 µm) was used
to filter the tissue fragments. The resulting cell suspension was
centrifuged at 800 × g for 5 min at 4°C. The supernatant was
discarded and the cells were collected. The cells were then
cultured in DMEM/F12 (cat. no. 12400024; Thermo Fisher Scientific,
Inc.) medium containing 10% fetal bovine serum (FBS; cat. no.
16140071; Thermo Fisher Scientific, Inc.) and 1%
penicillin/streptomycin in an incubator at 37°C with an atmosphere
of 5% CO2. After 2 weeks of static culture, the cells
adhered to the culture vessel and the spent culture medium was
replenished every 2–3 days. Subculturing was performed when the
cells reached 80–90% confluence. During subculturing, a trypsin
solution (cat. no. C0204; Beyotime Institute of Biotechnology) was
used to dissociate the cells for 3 min. The cells were then seeded
into culture flasks at a density of 5×103
cells/cm2. After the cells reached P2 generation with
80–90% confluence, NPSCs were obtained.
NPSC sorting via flow cytometry
Adherent NPSC cultures at the P2 generation were
dissociated using a trypsin solution when they reached 80–90%
confluence. The cell suspensions were centrifuged at 800 × g for 5
min at 4°C to collect the cells. After pelleting the cells,
phycoerythrin (PE)-CD73 (1:500; cat. no. ab157335), PE-CD34 (1:500;
cat. no. ab223930), peridinin-chlorophyll-protein-CD45 (1:500; cat.
no. ab210221), allophycocyanin-CD90 (1:500; cat. no. ab272351) and
FITC-CD105 (1:500; cat. no. ab53318) antibodies (all purchased from
Abcam) were added and the cell suspension was incubated for 30 min
at 4°C in the dark. Subsequently, the cells were centrifuged again
at the aforementioned parameters and washed twice with PBS
containing 4% FBS. A MoFlo High-Speed Flow Cytometer (Beckman
Coulter, Inc.) with Summit software (version 62; Beckman Coulter,
Inc.) was used to sort CD34+, CD45+,
CD73+, CD90+ and CD105+ cells. The
sorted NPSCs were seeded into culture dishes at a density of
5×103 cells/cm2 for further culture. The
cells were cultured in an incubator with 5% CO2 at 37°C
for 2 weeks before subsequent experimentation.
Observation of cell morphology
Sorted NPSCs were seeded into a culture flask at a
density of 5×103 cells/cm2 to observe the
cell morphology using a light microscope (Olympus Corporation). The
morphological characteristics of the sorted NPSCs were recorded 4–6
h after seeding.
Induced differentiation potential of
isolated NPSCs
Osteogenic (cat. no. Rasmx-90021.), adipogenic (cat.
no. Rasmx-90031) or chondrogenic (cat. no. Rasmx-90041) (all Cyagen
Biosciences, Inc.) medium was applied to induce three different
differentiated lines of NPSCs for ~2 weeks at 37°C. After fixing
cells with 4% neutral formaldehyde solution for 30 min at room
temperature, Alizarin Red (3–5 min; room temperature), Oil Red O
(30 min; room temperature) and Alcian Blue (30 min; room
temperature) staining were used to determine the osteogenic,
adipogenic and chondrogenic differentiation capacities of the three
NPSC lines, respectively. Images were captured using a light
microscope (Olympus Corporation).
Synthesis and expression of MCP-1 in a
pro-inflammatory environment
NPSCs (1×105 cells/well) were plated in
6-well plates and cultured in complete medium for 1 day. To
simulate the proinflammatory and poor nutrition microenvironment of
the degenerative disc, the medium was then replaced with serum-free
medium (SFM) comprising low glucose-DMEM and 10 ng/ml interleukin
(IL)-1β (PeproTech, Inc.) or 50 ng/ml tumor necrosis factor-α
(TNF-α; PeproTech, Inc.). After 48 h of incubation at 37°C, the
protein levels of MCP-1 secreted in the culture medium were
quantified using an enzyme-linked immunosorbent assay (ELISA) kit
for monocyte chemotactic protein 1 (MCP1; cat. no. SEA087Hu; Wuhan
USCN Business Co., Ltd.) according to the manufacturer's
instructions. Total RNA was extracted using TRIzol®
reagent (Invitrogen; Thermo Fisher Scientific, Inc.) followed by
the quantification of MCP-1 mRNA expression levels via reverse
transcription-quantitative PCR (RT-qPCR).
Cell proliferation assay
The Cell Counting Kit-8 kit (Dojindo Molecular
Technologies, Inc.) was applied for the detection of cell
proliferation. NPSCs in the P3 generation growing in the
logarithmic phase were seeded into a 96-well plate at a density of
1×104 cells/ml. With triplicate wells for each group,
100 µl cell suspension was added to each well and incubated
overnight in a thermostatic incubator with 5% CO2 at
37°C. The next day, the spent medium was replaced with a medium
containing different concentrations of MCP-1 (0, 10, 50 and 100
ng/ml; cat. no. SRP3109; Merck KGaA) with or without RS504393
treatment for 1, 3, 5 and 7 days of stimulation at 37°C.
Subsequently, 10 µl CCK-8 solution was added into each well and an
equivalent amount of CCK-8 solution was added into the cell-free
well. Cells were then incubated for 2 h at 37°C in the dark. A
microplate reader was used to measure the optical density (OD) at
450 nm. The final OD value was obtained by subtracting the OD value
of the blank well from that of each experimental well. Air bubbles
were carefully avoided while adding liquids to the culture as they
could affect the OD measurement.
Wound healing assay
NPSCs were seeded into 6-well plates at a density of
5×104 cells/ml and were cultured to ~100% confluence. A
200-µl pipette tip was used to create a straight wound by
disrupting the monolayer. After the cells were washed with PBS
twice, SFM containing 0, 10, 50 and 100 ng/ml MCP-1 was added and
the cells were incubated for another 12 h at 37°C to allow them to
migrate back into the wound area. For the inhibition experiment,
the cells were pre-treated with 10 µg/ml RS504393 (cat. no.
SML0711; Merck KGaA) for 2 h at 37°C, followed by incubation with
50 or 100 ng/ml MCP-1 at 37°C. To assess the mean number of
migrated cells, low-power fields were randomly selected from each
group for cell counting. Images were captured by using a light
microscope (Olympus Corporation).
Transwell migration assay
Transwell 24-well chamber plates (Costar; Corning,
Inc.) with a pore size of 8 µm, were employed for the migration
assay. In each well, 200 µl SFM containing 1×105 cells
and 600 µl SFM containing 10% serum supplemented with 0, 10, 50 or
100 ng/ml MCP-1 were placed into the upper and lower chambers,
respectively. For the inhibition experiment, the cells were
pre-treated with 10 µg/ml RS504393 for 2 h at 37°C before being
added to the upper chamber. After 12 h of incubation at 37°C, the
cells that adhered on the upper side of the membrane were gently
removed with a cotton swab and cells on the underside were fixed
with 4% paraformaldehyde fix solution (cat. no. P0099; Beyotime
Institute of Biotechnology) for 0.5–1 h at room temperature and
stained with 0.1% crystal violet for 0.5–1 h at room temperature.
The mean number of cells from three random high-power fields under
a light microscope (Olympus Corporation) was assessed to measure
cell migration.
Cell cycle detection via flow
cytometry
NPSCs in the P3 generation growing in the
logarithmic phase were seeded into 6-well plates at a density of
5×105 cells/ml. Cells were cultured in a thermostatic
incubator for 24 h to synchronize their cell cycles. The spent
medium was replaced with SFM to create a starved culture and the
cells were cultured overnight. After 72 h, complete medium
containing different concentrations (0, 10, 50 or 100 ng/ml) of
MCP-1 was added for 24–48 h at 37°C. To the collected cells, 1 ml
pre-cooled 70% ethanol solution was added into each tube and after
mixing, the solution was stored in the refrigerator overnight at
4°C. Following centrifugation (300 × g, 10 min, 4°C) and careful
removal of the supernatant, 1 ml pre-cooled PBS was added and the
cells were re-suspended, followed by further centrifugation (1,000
× g, 4°C, 10 min) and removal of the supernatant under direct
visualization. Then RNase A (100 µl; cat. no. EN0531; Thermo Fisher
Scientific, Inc.) was used to treat cells. Freshly prepared
propidium iodide (PI) staining solution (500 µl) weas used to
re-suspend the cells. After soaking in a water bath at 37°C for 30
min in the dark, the cell suspension was rapidly stored on ice for
flow cytometry. Finally, the flow cytometer (BD FACSAria III Flow
Cytometer; BD Biosciences) was used to detect the red fluorescence
intensity at an excitation wavelength of 488 nm and light
scattering was detected for the analysis of cell DNA content using
CELL QUEST analysis software (version 3.3; BD Biosciences).
Annexin V-FITC/PI staining analysis
via flow cytometry
The Annexin V-FITC/PI apoptosis detection kit (cat.
no. 40302ES20; Shanghai Yeasen Biotechnology Co., Ltd.) was used.
NPSCs in the P3 generation were seeded (1×106) into a
culture flask and the spent medium was replaced after overnight
incubation at 37°C. Different concentrations (0, 10, 50 or 100
ng/ml) of MCP-1 were then added to the cells, which were incubated
for 72 h at 37°C. The cells in each group were then collected,
washed with 1 ml binding buffer, centrifuged at 800 × g for 10 min
at 4°C and the supernatant was discarded. After the cells were
resuspended using 100 µl binding buffer, 10 µl Annexin V-FITC was
added, followed by 15 min of incubation at room temperature
(20–28°C) in the dark. After washing with 1 ml binding buffer, the
cell suspension was centrifuged at 800 × g for 10 min at 4°C and
the supernatant was discarded. Binding buffer (500 µl) was then
added to resuspend the cells, followed by the addition of 5 µl PI
solution for 10–15 min at room temperature. The cells were
immediately subjected to flow cytometry (BD FACSAria III Flow
Cytometer; BD Biosciences). Annexin V-positive cells represented
early apoptosis, PI-positive NPSC cells represented late apoptosis,
and Annexin V- and PI-positive cells represented necrosis. CELL
QUEST analysis software (version 3.3; BD Biosciences) were used to
analyze cell apoptosis.
MCP-1-induced chondrogenic
differentiation of NPSCs
NPSCs were cultured for 72 h in a complete culture
medium containing MCP-1 (0, 50 and 100 ng/ml) with or without 2 h
of pre-treatment with 10 µg/ml RS504393 at 37°C for 24–48 h. Prior
to western blotting, total protein was extracted using RIPA lysis
buffer (Applygen Technologies, Inc.) containing PMSF. Total RNA was
extracted using TRIzol reagent. The aggregate chondrogenesis model
was constructed by centrifuging NPSCs (300 × g, 4°C, 5–10 min) into
cell micromasses and inducing chondrogenic medium. MCP-1 was added
to complete chondrogenic medium (Cyagen Biosciences, Inc.) to
determine whether MCP-1 contributes to TGF-β1-induced NPSC
chondrogenesis. An inhibition assay was then performed via
treatment with 10 µg/ml RS504393 (a CCR2 antagonist) at 37°C for
24–48 h. Aggregates were collected for histological analysis after
28 days.
Immunohistochemical analysis
Cartilage samples were fixed with 4%
paraformaldehyde fix solution (cat. no. P0099; Beyotime Institute
of Biotechnology) for 0.5-1 h at room temperature.
Paraffin-embedded samples were then serially prepared into
5-µm-thick sagittal disc sections and then deparaffinized in
dimethylbenzene (three times; each 8 min) followed by rehydration
with an alcohol gradient. Subsequently, samples were incubated
overnight at 4°C with primary antibodies targeted against:
Collagen-II (1:200; cat. no. ab34712; Abcam) and aggrecan (1:400;
cat. no. bs-11655R; BIOSS). After washing with PBS, samples were
incubated with a horseradish peroxidase-conjugated goat anti-rabbit
IgG secondary antibody (1:200; cat. no. ab6721; Abcam) for 20 min
at 37°C. Color development was then performed using DAB (Beyotime
Institute of Biotechnology) for 5 min at room temperature.
Subsequently, the sections were counterstained with hematoxylin at
room temperature for 2 min (Beyotime Institute of Biotechnology),
mounted on coverslips and observed under a light microscope (Nikon
Corporation). ImageJ software (version 1.8.0; National Institutes
of Health) was used for quantification.
RT-qPCR
Total RNA was isolated from cultured cells using
TRIzol® reagent (Invitrogen; Thermo Fisher Scientific,
Inc.). cDNA was synthesized from 1 µg total RNA using the GoScript™
Reverse Transcription System (Promega Corporation) according to the
manufacturer's instructions. qPCR was performed using a
LightCycler® 480 Real-Time PCR System with SYBR Green
Master I (Roche Diagnostics). With a 20-µl reaction volume, the
reactions were run on a Peltier Thermal Cycler (Bio-Rad,
Laboratories, Inc.). The following thermocycling conditions were
used for qPCR: Initial denaturation at 95°C for 5 min; followed by
40 cycles at 95°C for 10 sec of denaturation, 60°C for 20 sec of
annealing and elongation; and 72°C for 20 sec of final extension.
The custom-made primers (Sangon Biotech, Shanghai, China) are
presented in Table I. Relative
mRNA expression levels were quantified using the 2−ΔΔCq
method and normalized to the internal reference gene, GAPDH
(31).
 | Table I.Sequences of primers used for reverse
transcription-quantitative PCR. |
Table I.
Sequences of primers used for reverse
transcription-quantitative PCR.
| Gene | Sequence
(5′-3′) |
|---|
| CCR2 | F:
TGCTCCCTGTCATAAA |
|
| R:
AGATGAGGACGACCAGCAT |
| Collagen-II | F:
TGGACGATCAGGCGAAACC |
|
| R:
GCTGCGGATGCTCTCAATCT |
| Aggrecan | F:
GTGCCTATCAGGACAAGGTCT |
|
| R:
GATGCCTTTCACCACGACTTC |
| SOX-9 | F:
AGCGAACGCACATCAAGAC |
|
| R:
CTGTAGGCGATCTGTTGGGG |
| MCP-1 | F:
CAGCCAGATGCAATCAATGCC |
|
| R:
TGGAATCCTGAACCCACTTCT |
| GAPDH | F:
GGAGCGAGATCCCTCCAAAAT |
|
| R:
GGCTGTTGTCATACTTCTCATGG |
Western blotting
Protein concentrations were determined using the BCA
Protein Assay kit (Beyotime Institute of Biotechnology). Total
protein samples (40 µg protein/lane) were separated via SDS-PAGE on
8 or 10% gels and then transferred onto polyvinylidene fluoride
membranes (MilliporeSigma). The membranes were then blocked using
5% skimmed milk in TBS containing 0.05% Tween-20 for 2 h at 37°C.
Subsequently, membranes were incubated with primary antibodies
against CCR2 (1:1,000; cat. no. ab203128; Abcam), GAPDH (1:10,000;
cat. no. ab8245; Abcam), aggrecan (1:1,000; cat. no. 13880-1-AP;
ProteinTech Group, Inc.), SOX-9 (1:1,000; cat. no. ab185966; Abcam)
and collagen-II (1:1,000; cat. no. ab34712; Abcam) overnight at
4°C. The membranes were then incubated for 1 h with horseradish
peroxidase-conjugated secondary antibodies (1:1,000; cat. nos.
ab6789 and ab6721; Abcam) at room temperature. Using Pierce ECL
Western Blotting Substrate (Thermo Fisher Scientific, Inc.) the
membranes were visualized using ImageQuant LAS4000 (GE Healthcare).
Image-Pro Plus software (version 6.0; Media Cybernetics, Inc.) was
used to semi-quantify relative protein expression levels, with
GAPDH serving as the loading control.
Statistical analysis
All experiments were repeated at least three times.
Data are presented as the mean ± SD. All statistical analyses were
conducted using SPSS 22.0 software (IBM Corp.). For normally
distributed data, the statistical difference among multiple groups
(including the control, IL-1β, TNF-α and IL-1β + TNF-α; control,
10, 50 and 100 ng/ml; and control, 50, 100 and 100 ng/ml + RS504393
groups) was evaluated using one-way ANOVA followed by Tukey's post
hoc test. For non-normally distributed data, the statistical
difference was evaluated using the Kruskal-Wallis test followed by
the post hoc Dunn test. P<0.05 was considered to indicate a
statistically significant difference.
Results
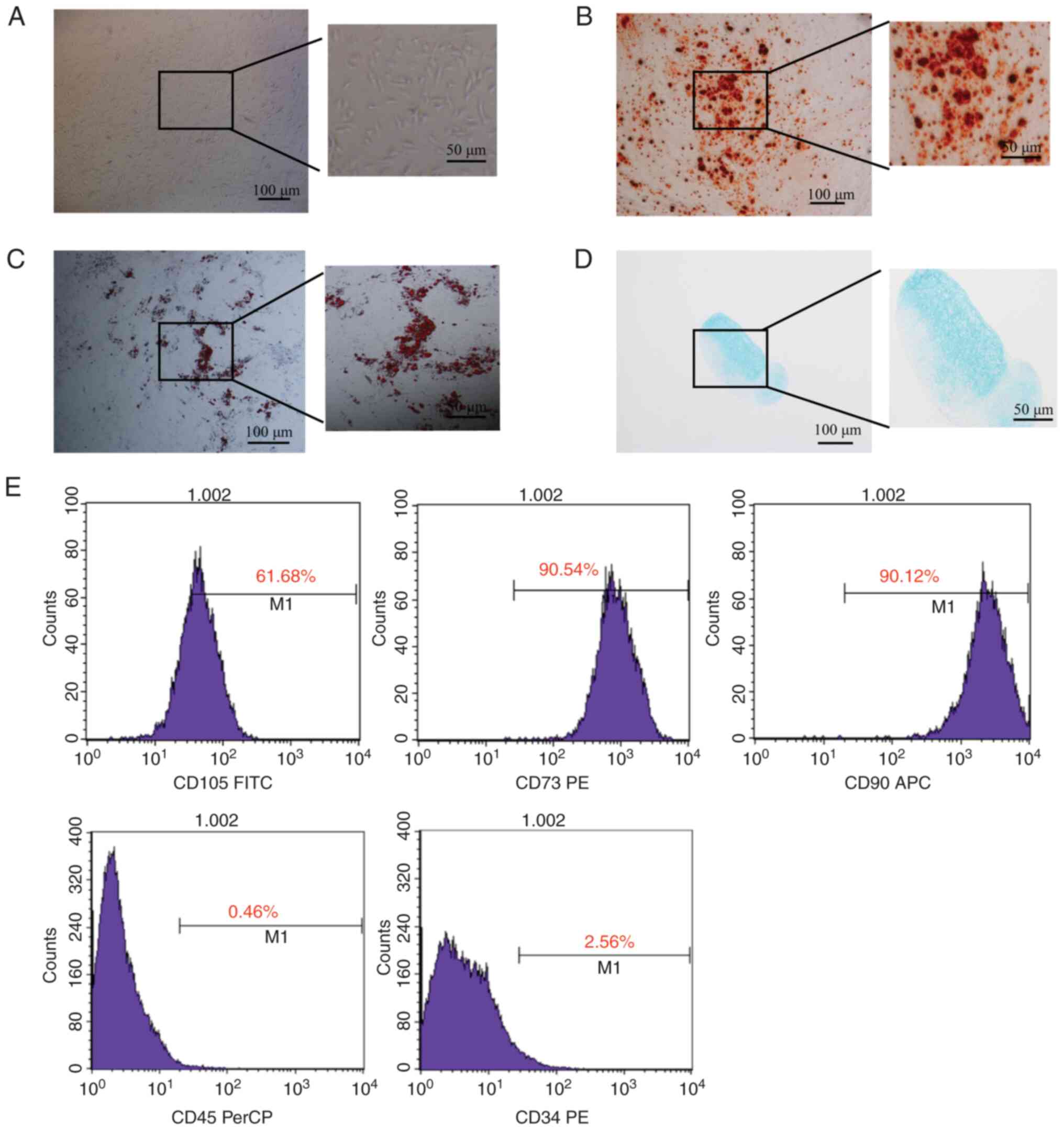
Characterization of NPSCs
Primary cells exhibited a representative
fibroblast-like morphology within 24 h of incubation (Fig. 1A). The multilineage
differentiation assays demonstrated that after specific
differentiation induction, isolated cells smoothly differentiated
into adipogenic, chondrogenic and osteogenic lineages (Fig. 1B-D). Subsequently, specific cell
surface markers commonly used for MSC identification were detected
via flow cytometry. Positive results were observed for the commonly
used MSC markers CD73 and CD90, as well as CD105, which is
expressed in certain MSCs as an auxiliary receptor for the TGF-β
receptor complex. Negative results were observed for the
hematopoietic markers CD34 and CD45 (Fig. 1E).
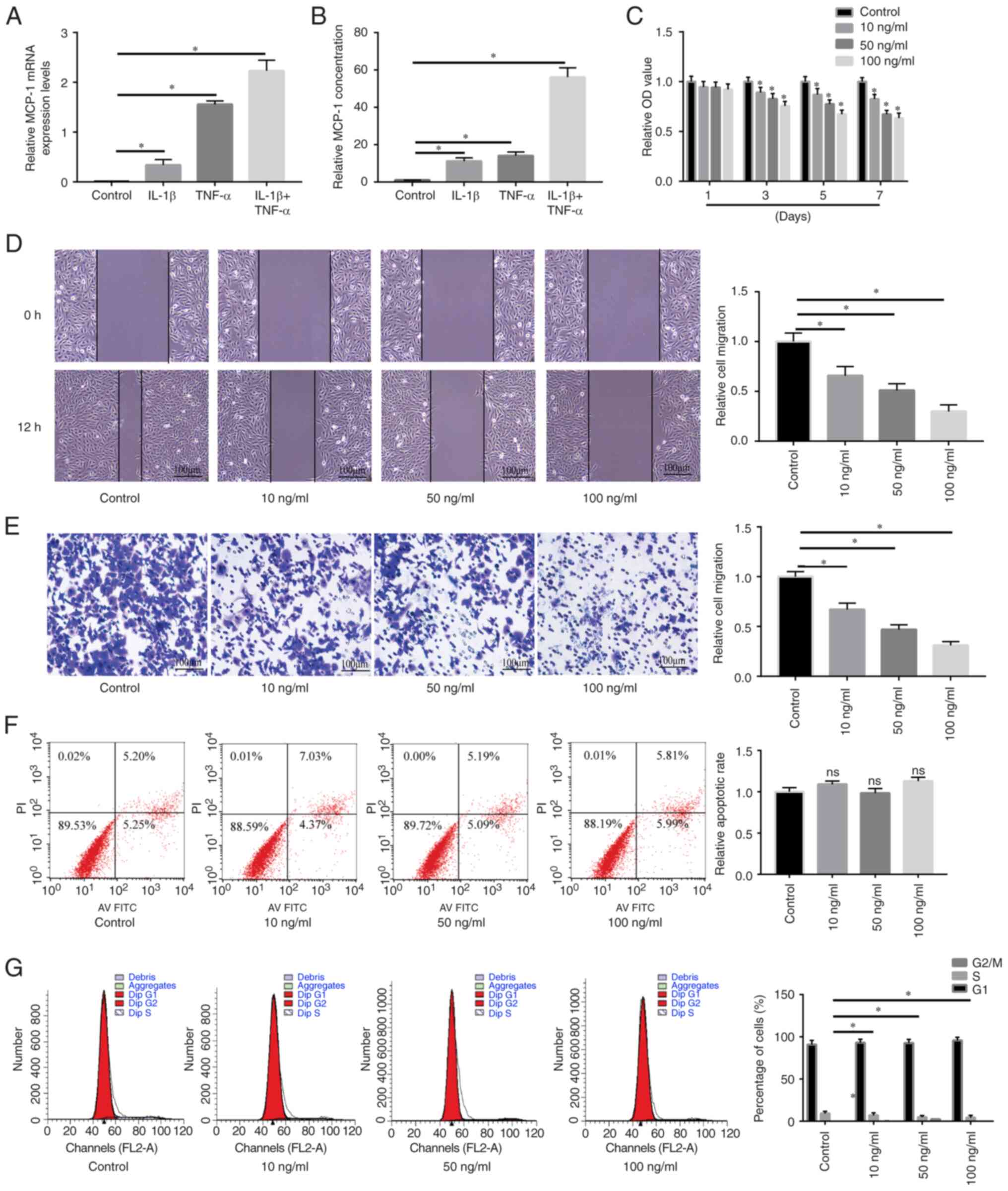
MCP-1 expression levels and secretion
in an IDD microenvironment
Following treatment with pro-inflammatory cytokines,
the mRNA expression levels and protein levels of MCP-1 in NSPCs
were examined using RT-qPCR and ELISA, respectively. As Fig. 2A and B shown, MCP-1 was
upregulated after being treated with IL-1β or TNF-α and was more
upregulated when treated with IL-1β + TNF-α To explore the effect
of MCP-1 on NPSC migration and proliferation in vitro,
Transwell and wound healing and CCK-8 assays were carried out. NPSC
migration and proliferation were significantly reduced via MCP-1
treatment in a dose-dependent manner (Fig. 2C-E). Flow cytometry results
determined that MCP-1 treatment inhibited the cell cycle in a
dose-dependent manner, whereas apoptosis of NPSCs was not affected
(Fig. 2F and G).
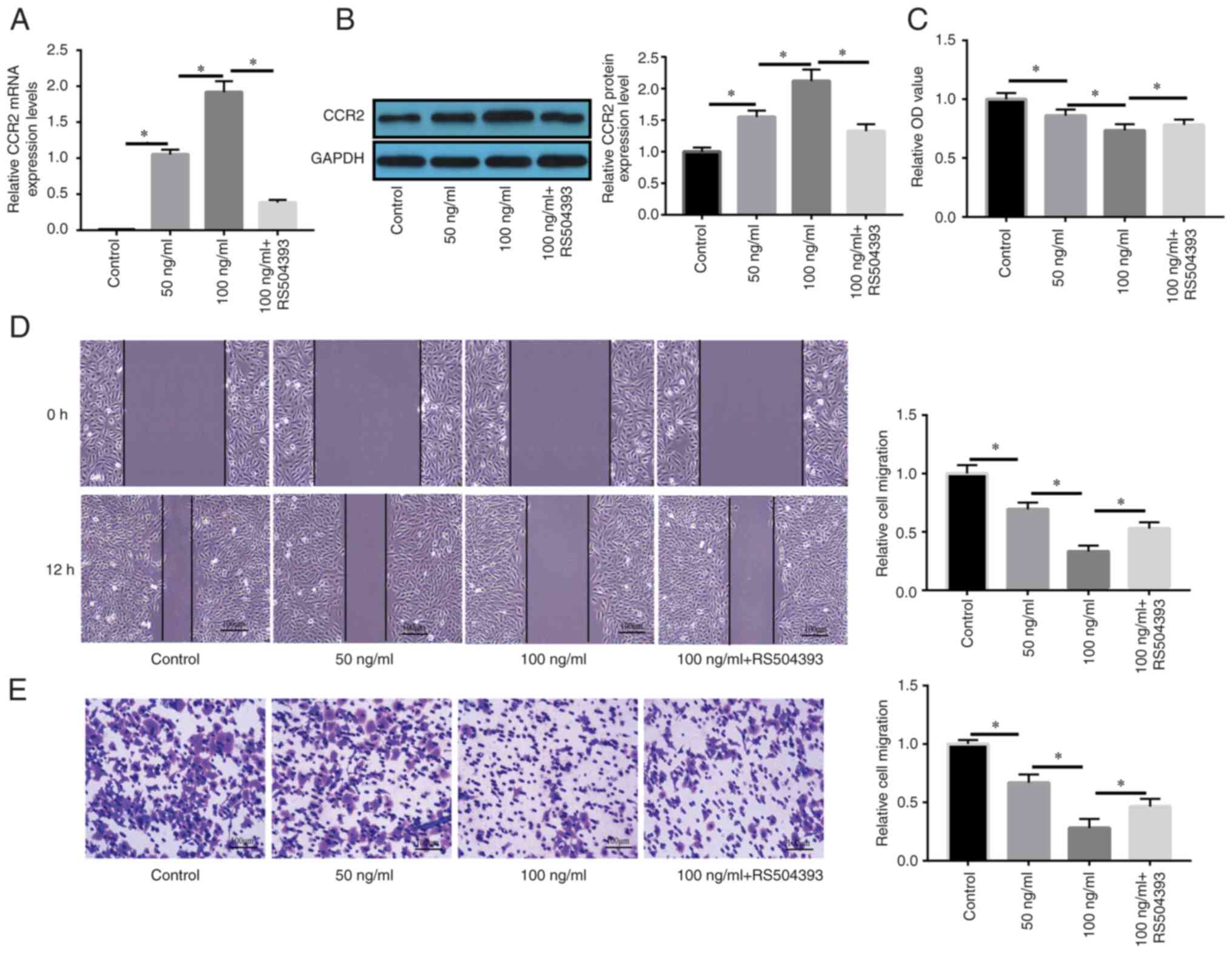
MCP-1/CCR2 axis regulates the
proliferation and migration of NPSCs
As indicated by the results of RT-qPCR and western
blotting, MCP-1 significantly enhanced CCR2 mRNA and protein
expression levels compared with in the control group; notably, CCR2
upregulation was significantly reversed by the CCR2-specific
inhibitor RS504393 compared with the 100 ng/ml MCP-1 group
(Fig. 3A and B). As presented in
Fig. 3C, the suppressive effect
of MCP-1 on cell proliferation was significantly recovered by the
CCR2-specific inhibitor RS504393 compared with in the 100 ng/ml
MCP-1 group. Similarly, wound healing and Transwell migration
assays also revealed that the suppressive effects of MCP-1 on cell
migration were significantly reversed by RS504393 compared with in
the 100 ng/ml MCP-1 group (Fig. 3D
and E).
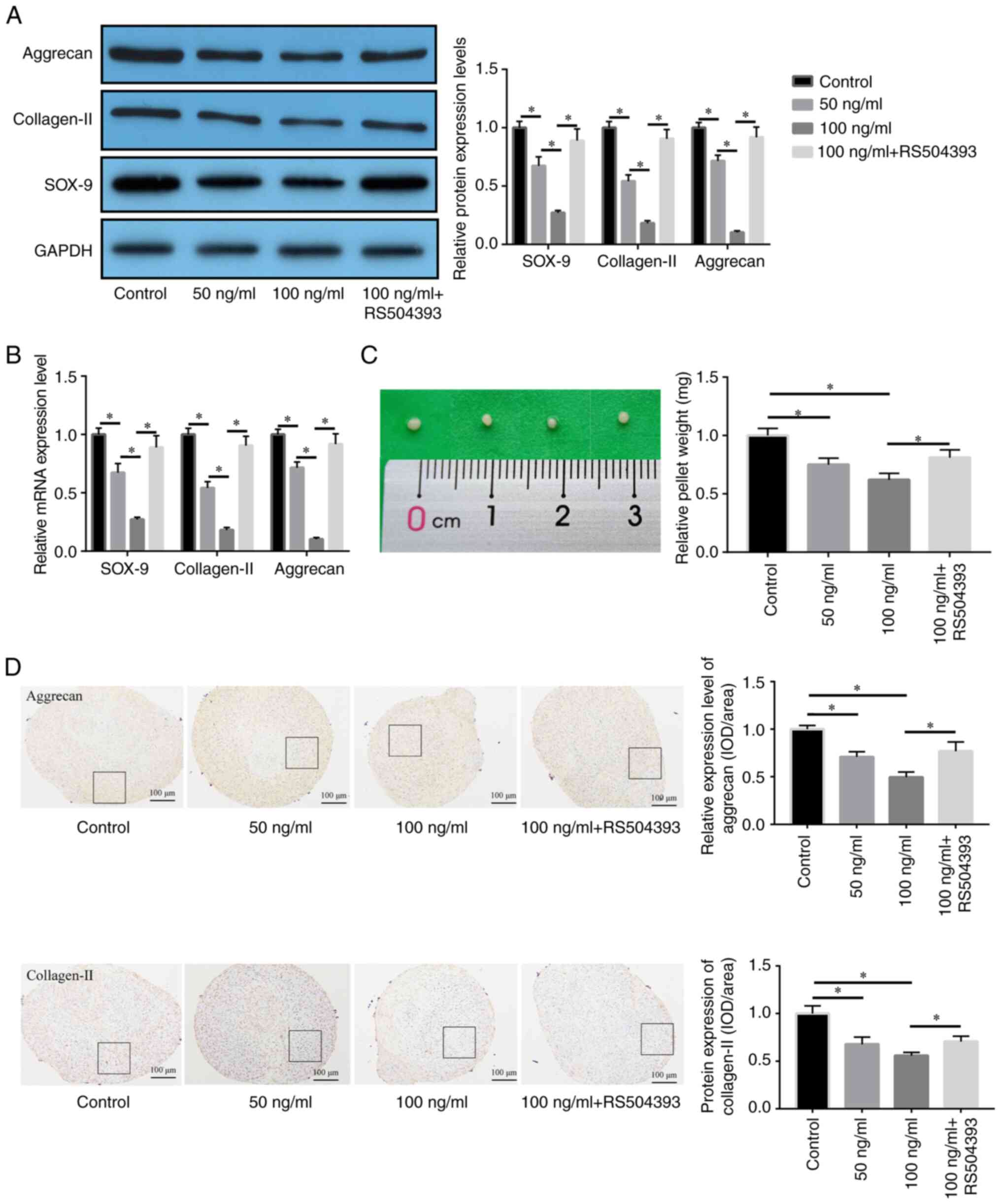
MCP-1/CCR2 axis regulates the
chondrogenic differentiation of NPSCs
To analyze the role of the MCP-1/CCR2 axis in
chondrogenic NPSC differentiation in vitro, the effect of a
CCR2 antagonist on the regulation of cartilage differentiation by
MCP-1 at the mRNA and protein levels was determined. Western
blotting indicated that MCP-1 suppressed chondrogenic NPSC
differentiation, which was manifested through the significantly
lower protein expression levels of aggrecan, collagen-II and SOX-9
compared with the control. Furthermore RT-qPCR demonstrated that
MCP-1 significantly reduced the mRNA expression levels of aggrecan,
collagen-II and SOX-9 in NPSCs compared with the control, whereas
pre-treatment with RS504393 significantly recovered this reduction
compared with the 100 ng/ml MCP-1 group (Fig. 4A and B). Through micromass
culture, the effect of MCP-1 on chondrogenic NPSC differentiation
induced by TGF-β1 was investigated. The weight of the mass
decreased significantly upon treatment with MCP-1 compared with the
control; however, this effect was significantly reversed following
RS504393 treatment compared with the 100 ng/ml MCP-1 group
(Fig. 4C). The protein expression
levels of aggrecan and collagen-II were significantly decreased
following MCP-1 treatment compared with the control, which was
significantly reversed by RS504393 compared with the 100 ng/ml
MCP-1 group, as confirmed via immunohistochemical analysis
(Fig. 4D). These results
therefore suggested that MCP-1 may synergistically inhibit
chondrogenic NPSC differentiation induced by TGF-β1 via the
MCP-1/CCR2 axis.
Discussion
Stem cell therapy has been considered a promising
option for disc regeneration (32,33); however, the potential for the
long-term survival and biological function of transplanted cells in
a harsh microenvironment remains unclear (34). To solve the possible problems of
stem cell transplantation, an alternative method is to mobilize
endogenous progenitor and stem cells to the damaged sites. As
indicated in numerous studies, progenitor cells in stem cell niches
in IDD move toward the annular fibrosus and inner parts of the IDD,
which may promote IDD repair in situ (35,36). During the natural process of
healing, affected cells and tissues release a number of cytokines
and chemokines to trigger dormant progenitor cells and activate
their migration to the injured sites (37). However, endogenous tissue repair
in degenerative IDD by targeting of the local microenvironment or
the IDD mechanism is poorly understood. The present study explored
the role of MCP-1 in the regulation of the migration and
differentiation of endogenous stem cells in IDD to investigate IDD
regeneration.
A previous study indicated that IDD releases
numerous chemokines and cytokines to effectively recruit endogenous
stem cells during the repair process (38). As a pair of ligand receptors, the
interaction between MCP-1 and CCR2 is important for the migration
and homing of stem cells. MCP-1 may mediate the recruitment of
monocytes, and regulate the phenotypes of monocytes and
lymphocytes, as well as the accumulation of fibrous tissue and
vasculogenesis, with extensive biological effects. For example, the
chemotactic effect of HConFs mediated by the MCP-1/CCR2 axis was
decreased after transdifferentiation into myofibroblasts (39). The MCP-1/CCR-2 axis activates the
PI3K/AKT/mTOR signaling pathway leading to HIF-1α-mediated VEGF-A
expression (29). The
upregulation of MCP-1 promotes a CCR2-dependent profibrotic and
inflammatory state, and accelerates the AKI-to-CKD transition
(40). Previous studies on MCP-1
in the intervertebral disc have often focused on its role in
attracting macrophages to herniated sites or areas with annular
tears (41,42). For example, in murine
intervertebral discs, MCP-1 was reported to induce macrophage
migration, which was abrogated by the addition of anti-MCP-1
neutralizing antibodies, indicating that MCP-1 may have an effect
on the pathogenesis of IDD (25).
A previous study demonstrated that MCP-1 may be detected in culture
medium after 48 h of culture with mesenchymal stem cells and could
enhance their migration (43).
However, the roles of MCP-1 in IDD are still poorly understood and
whether MCP-1 can promote the repair of IDD remains unclear.
The mechanism of tissue repair is crucial for the
normal functioning and integrity of the body, in which tissues and
organs are damaged through frequent exposure to mechanical stress.
Endogenous mesenchymal stem cells may be mobilized and activated to
promote tissue repair as a regenerative cell population (44,45). Mesenchymal stem cells may mutually
communicate with other cells in the body and move to damaged sites,
which mainly depends on the response to the cell injury signal
(46). Mesenchymal stem cells may
generate a large amount of cytokines to provide feedback for the
transduction of regeneration signals, allowing their mobilization
from each site, migration to the target sites and their support of
tissue repair (47). The present
study demonstrated significantly high expression levels and the
release of MCP-1 in native cells in vitro under the
influence of pro-inflammatory mediators IL-1β and TNF-α. In in
vitro cell migration and proliferation assays, MCP-1
demonstrated a significant dose-dependent inhibitory effect on the
migration and proliferation of NPSCs, which indicated that MCP-1
had a regulatory effect on NPSCs. Furthermore, by neutralizing CCR2
with the pharmaceutical inhibitor RS504393, MCP-1 significantly
inhibited the proliferation and migration of NPSCs.
Mesenchymal stem cells migrate to inflammatory
tissues; however, the specific mechanism of their migration needs
to be elucidated further for successful application in clinical
settings (48). The recruitment
of mesenchymal stem cells is induced via chemotaxis and the
directional migration is generated by a concentration gradient.
Previous studies have reported that mesenchymal stem cell surfaces
express various chemokines and differences in chemokine receptors
is most likely caused by differences in isolation techniques and
culture conditions (49,50).
The present study demonstrated that MCP-1
significantly upregulated the protein expression levels of its
transmembrane receptor CCR2 in NPSCs, possibly to modulate
chondrogenic NPSC differentiation upon treatment with TGF-β1.
During histological analysis, following micromass culture of NPSCs
for chondrogenesis in vitro, MCP-1 significantly reduced the
weight and size of chondrogenic pellets but also dose-dependently
significantly decreased the mRNA and protein expression levels of
aggrecan and collagen-II in NPSCs. Conversely, the CCR2 antagonist
RS504393 neutralized the inhibitory effect of MCP-1 on chondrogenic
NPSC differentiation, as demonstrated by the higher protein
expression levels of the chondrogenic markers, SOX-9, collagen-II
and aggrecan. These findings suggested that the MCP-1/CCR2 axis may
be closely associated with the proliferation and migration of
NPSCs, which is essential for the endogenous recruitment of NPSCs.
Therefore, the MCP-1/CCR2 axis could be a potential target for the
treatment of IDD.
In conclusion, the present study demonstrated a
novel mechanism of MCP-1, which accumulated in damaged or
degenerative IDD to effectively inhibit chondrogenic NPSC
differentiation and migration towards damaged sites via the
MCP-1/CCR2 axis. MCP-1 may therefore represent a novel therapeutic
target for the regeneration of IDD in situ and for
endogenous cell repair by targeted inhibition of MCP-1 expression.
However, there are still certain limitations to the present study.
The downstream signaling pathways of the MCP-1/CCR2 axis remain to
be clarified further and in vivo experiments are needed to
determine the role of MCP-1 in the regulation of numerous
biological behaviors of NPSCs in animal systems.
Acknowledgements
Not applicable.
Funding
This work was supported by the National Natural Science
Foundation of China (grant no. 81772399).
Availability of data and materials
The datasets used and/or analyzed during the present
study are available from the corresponding author on reasonable
request.
Authors' contributions
XO and TW performed the experiments and generated
data. DR made substantial contributions to the conception and
design of the present study. JY, QH and AX conducted data analysis
and interpreted the data. XO and DR confirm the authenticity of all
the raw data. All authors contributed to the drafting and revision
of the manuscript. All authors read, revised and approved the
manuscript and agreed to be accountable for all aspects of the
research, ensuring the accuracy and integrity of the work.
Ethics approval and consent to
participate
The research was approved by the Ethics Committee of
The Second School of Clinical Medicine, Southern Medical University
(Guangzhou, China; approval nos. ICE2017058 and ICE2017042). Both
human and animal ethics approval were received, and the patient's
parents provided written consent.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
Glossary
Abbreviations
Abbreviations:
|
IDD
|
intervertebral disc degeneration
|
|
MCP-1
|
monocyte chemoattractant protein-1
|
|
NPSCs
|
nucleus pulposus-derived stem
cells
|
References
|
1
|
Chan WC, Sze KL, Samartzis D, Leung VY and
Chan D: Structure and biology of the intervertebral disk in health
and disease. Orthop Clin North Am. 42:447–464. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
González Martínez E, García-Cosamalón J,
Cosamalón-Gan I, Esteban Blanco M, García-Suarez O and Vega JA:
Biology and mechanobiology of the intervertebral disc. Neurocirugia
(Astur). 28:135–140. 2017.(In Spanish). View Article : Google Scholar
|
|
3
|
Middendorp M, Vogl TJ, Kollias K,
Kafchitsas K, Khan MF and Maataoui A: Association between
intervertebral disc degeneration and the Oswestry disability index.
J Back Musculoskelet Rehabil. 30:819–823. 2017. View Article : Google Scholar
|
|
4
|
Liu Y, Li Y, Nan LP, Wang F, Zhou SF, Feng
XM, Liu H and Zhang L: Insights of stem cell-based endogenous
repair of intervertebral disc degeneration. World J Stem Cells.
12:266–276. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Vinante F and Rigo A: Heparin-binding
epidermal growth factor-like growth factor/diphtheria toxin
receptor in normal and neoplastic hematopoiesis. Toxins (Basel).
5:1180–1201. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Gunasekaran A, de Los Reyes NKM, Walters J
and Kazemi N: Clinical presentation, diagnosis, and surgical
treatment of spontaneous cervical intradural disc herniations: A
review of the literature. World Neurosurg. 109:275–284. 2018.
View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Lofrese G, Mongardi L, Cultrera F,
Trapella G and De Bonis P: Surgical treatment of
intraforaminal/extraforaminal lumbar disc herniations: Many
approaches for few surgical routes. Acta Neurochir (Wien).
159:1273–1281. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Gillet P: The fate of the adjacent motion
segments after lumbar fusion. J Spinal Disord Tech. 16:338–345.
2003. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
May M: Regenerative medicine: Rebuilding
the backbone. Nature. 503:S7–S9. 2013. View
Article : Google Scholar : PubMed/NCBI
|
|
10
|
Roberts S, Evans H, Trivedi J and Menage
J: Histology and pathology of the human intervertebral disc. J Bone
Joint Surg Am. 88 (Suppl 2):S10–S14. 2006. View Article : Google Scholar
|
|
11
|
Wang R, Luo D, Li Z and Han H: Dimethyl
fumarate ameliorates nucleus pulposus cell dysfunction through
activating the Nrf2/HO-1 pathway in intervertebral disc
degeneration. Comput Math Methods Med. 2021:60217632021.PubMed/NCBI
|
|
12
|
Colombier P, Clouet J, Hamel O, Lescaudron
L and Guicheux J: The lumbar intervertebral disc: from embryonic
development to degeneration. Joint Bone Spine. 81:125–129. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Hughes SP, Freemont AJ, Hukins DW,
McGregor AH and Roberts S: The pathogenesis of degeneration of the
intervertebral disc and emerging therapies in the management of
back pain. J Bone Joint Surg Br. 94:1298–1304. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Sivakamasundari V and Lufkin T: Stemming
the degeneration: IVD stem cells and stem cell regenerative therapy
for degenerative disc disease. Adv Stem Cells.
2013:7245472013.PubMed/NCBI
|
|
15
|
Risbud MV, Guttapalli A, Tsai TT, Lee JY,
Danielson KG, Vaccaro AR, Albert TJ, Gazit Z, Gazit D and Shapiro
IM: Evidence for skeletal progenitor cells in the degenerate human
intervertebral disc. Spine (Phila Pa 1976). 32:2537–2544. 2007.
View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Blanco JF, Graciani IF, Sanchez-Guijo FM,
Muntión S, Hernandez-Campo P, Santamaria C, Carrancio S, Barbado
MV, Cruz G, Gutierrez-Cosío S, et al: Isolation and
characterization of mesenchymal stromal cells from human
degenerated nucleus pulposus: Comparison with bone marrow
mesenchymal stromal cells from the same subjects. Spine (Phila Pa
1976). 35:2259–2265. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Zhao Y, Jia Z, Huang S, Wu Y, Liu L, Lin
L, Wang D, He Q and Ruan D: Age-related changes in nucleus pulposus
mesenchymal stem cells: An in vitro study in rats. Stem Cells Int.
2017:67615722017. View Article : Google Scholar
|
|
18
|
Urban JP: The role of the physicochemical
environment in determining disc cell behaviour. Biochem Soc Trans.
30:858–864. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Liu ZJ, Zhuge Y and Velazquez OC:
Trafficking and differentiation of mesenchymal stem cells. J Cell
Biochem. 106:984–991. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Vanden Berg-Foels WS: In situ tissue
regeneration: Chemoattractants for endogenous stem cell
recruitment. Tissue Eng Part B Rev. 20:28–39. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Wang J, Markova D, Anderson DG, Zheng Z,
Shapiro IM and Risbud MV: TNF-α and IL-1β promote a
disintegrin-like and metalloprotease with thrombospondin type I
motif-5-mediated aggrecan degradation through syndecan-4 in
intervertebral disc. J Biol Chem. 286:39738–39749. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Ou X, Ying J, Bai X, Wang C and Ruan D:
Activation of SIRT1 promotes cartilage differentiation and reduces
apoptosis of nucleus pulposus mesenchymal stem cells via the
MCP1/CCR2 axis in subjects with intervertebral disc degeneration.
Int J Mol Med. 46:1074–1084. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Ferreira JR, Teixeira GQ, Neto E,
Ribeiro-Machado C, Silva AM, Caldeira J, Pereira CL, Bidarra S,
Maia AF, Lamghari M, et al: IL-1β-pre-conditioned mesenchymal
stem/stromal cells' secretome modulates the inflammatory response
and aggrecan deposition in intervertebral disc. Eur Cell Mater.
41:431–453. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Cai WT, Guan P, Lin MX, Fu B and Wu B:
Sirt1 suppresses MCP-1 production during the intervertebral disc
degeneration by inactivating AP-1 subunits c-Fos/c-Jun. Eur Rev Med
Pharmacol Sci. 24:5895–5904. 2020.
|
|
25
|
Takayama Y, Ando T, Ichikawa J and Haro H:
Effect of thrombin-induced MCP-1 and MMP-3 production via PAR1
expression in murine intervertebral discs. Sci Rep. 8:113202018.
View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Yaghooti H, Mohammadtaghvaei N and
Mahboobnia K: Effects of palmitate and astaxanthin on cell
viability and proinflammatory characteristics of mesenchymal stem
cells. Int Immunopharmacol. 68:164–170. 2019. View Article : Google Scholar
|
|
27
|
Cao M, Liu H, Dong Y, Liu W, Yu Z, Wang Q,
Wang Q, Liang Z, Li Y and Ren H: Mesenchymal stem cells alleviate
idiopathic pneumonia syndrome by modulating T cell function through
CCR2-CCL2 axis. Stem Cell Res Ther. 12:3782021. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Ferland-McCollough D, Maselli D, Spinetti
G, Sambataro M, Sullivan N, Blom A and Madeddu P: MCP-1 feedback
loop between adipocytes and mesenchymal stromal cells causes fat
accumulation and contributes to hematopoietic stem cell rarefaction
in the bone marrow of patients with diabetes. Diabetes.
67:1380–1394. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Sun C, Li X, Guo E, Li N, Zhou B, Lu H,
Huang J, Xia M, Shan W, Wang B, et al: MCP-1/CCR-2 axis in
adipocytes and cancer cell respectively facilitates ovarian cancer
peritoneal metastasis. Oncogene. 39:1681–1695. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Yadav A, Saini V and Arora S: MCP-1:
chemoattractant with a role beyond immunity: A review. Clin Chim
Acta. 411:1570–1579. 2010. View Article : Google Scholar
|
|
31
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(−Delta Delta C(T)) method. Methods. 25:402–408. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Wang HC, Jin CH, Kong J, Yu T, Guo JW, Hu
YG and Liu Y: The research of transgenic human nucleus pulposus
cell transplantation in the treatment of lumbar disc degeneration.
Kaohsiung J Med Sci. 35:486–492. 2019.
|
|
33
|
Iwashina T, Mochida J, Sakai D, Yamamoto
Y, Miyazaki T, Ando K and Hotta T: Feasibility of using a human
nucleus pulposus cell line as a cell source in cell transplantation
therapy for intervertebral disc degeneration. Spine (Phila Pa
1976). 31:1177–1186. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Wuertz K, Godburn K, Neidlinger-Wilke C,
Urban J and Iatridis JC: Behavior of mesenchymal stem cells in the
chemical microenvironment of the intervertebral disc. Spine (Phila
Pa 1976). 33:1843–1849. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Sun K, Jing X, Guo J, Yao X and Guo F:
Mitophagy in degenerative joint diseases. Autophagy. 17:2082–2092.
2021. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Tessier S and Risbud MV: Understanding
embryonic development for cell-based therapies of intervertebral
disc degeneration: Toward an effort to treat disc degeneration
subphenotypes. Dev Dyn. 250:302–317. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Andreas K, Sittinger M and Ringe J: Toward
in situ tissue engineering: Chemokine-guided stem cell recruitment.
Trends Biotechnol. 32:483–492. 2014. View Article : Google Scholar
|
|
38
|
Li Z, Peroglio M, Alini M and Grad S:
Potential and limitations of intervertebral disc endogenous repair.
Curr Stem Cell Res Ther. 10:329–338. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Tsutsumi-Kuroda U, Inoue T, Futakuchi A,
Shobayashi K, Takahashi E, Kojima S, Inoue-Mochita M, Fujimoto T
and Tanihara H: Decreased MCP-1/CCR2 axis-mediated chemotactic
effect of conjunctival fibroblasts after transdifferentiation into
myofibroblasts. Exp Eye Res. 170:76–80. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Xu L, Sharkey D and Cantley LG: Tubular
GM-CSF promotes late MCP-1/CCR2-mediated fibrosis and inflammation
after ischemia/reperfusion injury. J Am Soc Nephrol. 30:1825–1840.
2019. View Article : Google Scholar
|
|
41
|
Schroeder M, Viezens L, Schaefer C,
Friedrichs B, Algenstaedt P, Rüther W, Wiesner L and
Hansen-Algenstaedt N: Chemokine profile of disc degeneration with
acute or chronic pain. J Neurosurg Spine. 18:496–503. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Scuderi GJ, Brusovanik GV, Anderson DG,
Dunham CJ, Vaccaro AR, Demeo RF and Hallab N: Cytokine assay of the
epidural space lavage in patients with lumbar intervertebral disk
herniation and radiculopathy. J Spinal Disord Tech. 19:266–269.
2006. View Article : Google Scholar : PubMed/NCBI
|
|
43
|
Boomsma RA and Geenen DL: Mesenchymal stem
cells secrete multiple cytokines that promote angiogenesis and have
contrasting effects on chemotaxis and apoptosis. PLoS One.
7:e356852012. View Article : Google Scholar : PubMed/NCBI
|
|
44
|
Mastrolia I, Foppiani EM, Murgia A,
Candini O, Samarelli AV, Grisendi G, Veronesi E, Horwitz EM and
Dominici M: Challenges in clinical development of mesenchymal
stromal/stem cells: Concise review. Stem Cells Transl Med.
8:1135–1148. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
45
|
Frenette PS, Pinho S, Lucas D and
Scheiermann C: Mesenchymal stem cell: Keystone of the hematopoietic
stem cell niche and a stepping-stone for regenerative medicine.
Annu Rev Immunol. 31:285–316. 2013. View Article : Google Scholar
|
|
46
|
Kang SK, Shin IS, Ko MS, Jo JY and Ra JC:
Journey of mesenchymal stem cells for homing: strategies to enhance
efficacy and safety of stem cell therapy. Stem Cells Int.
2012:3429682012. View Article : Google Scholar
|
|
47
|
Augello A, Kurth TB and De Bari C:
Mesenchymal stem cells: A perspective from in vitro cultures to in
vivo migration and niches. Eur Cell Mater. 20:121–133. 2010.
View Article : Google Scholar : PubMed/NCBI
|
|
48
|
Fan XL, Zhang Z, Ma CY and Fu QL:
Mesenchymal stem cells for inflammatory airway disorders: Promises
and challenges. Biosci Rep. 39:BSR201821602019. View Article : Google Scholar : PubMed/NCBI
|
|
49
|
Petit I, Jin D and Rafii S: The
SDF-1-CXCR4 signaling pathway: A molecular hub modulating
neo-angiogenesis. Trends Immunol. 28:299–307. 2007. View Article : Google Scholar
|
|
50
|
Humbert P, Brennan MA, Davison N, Rosset
P, Trichet V, Blanchard F and Layrolle P: Immune modulation by
transplanted calcium phosphate biomaterials and human mesenchymal
stromal cells in bone regeneration. Front Immunol. 10:6632019.
View Article : Google Scholar
|