Introduction
Melanin is a ubiquitous biological pigment in plants
and animals that absorbs and scatters ultraviolet radiation and
reduces free radical production, thus protecting cells from DNA,
protein and lipid damage (1,2).
Melanin is synthesized on melanosomes in melanocytes, transported
to adjacent keratinocytes through dendritic structures and further
diffused on the skin surface (3).
Hypo- and hyperpigmentation disorders cause serious skin diseases,
such as excessive melanin synthesis, which can cause chloasma,
freckles and senile plaques (4,5).
Therefore, there is significant interest in inhibiting the abnormal
synthesis of melanin to treat melanin-related diseases.
Melanogenesis is regulated by tyrosinase,
tyrosinase-related protein 1 (TRP-1), TRP-2 and
microphthalmia-associated transcription factor (MITF) (6). Tyrosinase catalyzes the hydroxylation
of tyrosine into 3,4-dihydroxyphenylalanine (L-DOPA), which is
oxidized to dopa-quinone (7).
Tyrosinase catalyzes the rate-limiting step of melanogenesis and
TRP-1 and TRP-2 act downstream in the melanin biosynthetic pathway
(8). MITF, a basic
helix-loop-helix transcription factor, plays a crucial role in
melanocyte biology and development by regulating the expression of
tyrosinase-related proteins (9,10).
MITF expression is enhanced by the activation of melanocyte
differentiation (11). Melanocytes
that overexpress MITF display high cell proliferation, reduced
apoptosis and increased melanin levels when compared with normal
melanocytes (12). These cellular
features are consistent with the hypothesis that MITF is a master
regulator of pigmentation. Expression of the MITF gene is regulated
by several signaling pathways, including the p38 MAPK pathway,
which is involved in the activation of MITF expression and the
consequent increase in tyrosinase expression (13).
Arbutin, kojic acid and ascorbic acid have been
reported to possess an inhibitory effect on tyrosinase and act as
whitening agents on the skin; however, agents such as kojic acid
usually have adverse side effects, including dermatitis and
melanocyte damage (14).
Therefore, identifying natural compounds with few adverse reactions
that target tyrosinase and inhibit melanin synthesis are desirable.
Pueraria Lobata Radix (P. Lobata Radix) belongs to
the Leguminosae family and contains various active
compounds, including flavonoids, isoflavones, proteins and
polysaccharides (15). These
compounds give P. Lobata Radix pharmacological activity,
including liver protection and anti-cancer and anti-inflammatory
activities (16–18). Proteins from P. Lobata Radix
have attracted increasing interest in recent years. Using an
extracellular screening model, it was found that a P. Lobata
Radix water-soluble total protein extract (PLP) strongly inhibited
tyrosinase activity. As this PLP inhibited tyrosinase and P.
Lobata Radix has been reported to have anti-cancer activity
(19,20), there is interest in investigating
the effect of PLP on melanin formation in melanoma.
The present study obtained a PLP by alkali
extraction and acid precipitation, characterized the physical and
chemical properties of this PLP and evaluated its effect on
melanogenesis in B16 melanoma cells. It also analyzed the effect of
the PLP on apoptosis in B16 melanoma cells to investigate the
potential of this extract as an anti-melanoma agent. The mechanisms
by which PLP functions were also investigated. The inhibition of
tyrosinase activity and reduction in melanin content by PLP and the
related melanogenesis inhibitory mechanism are described here for
the first time, to the best of the authors' knowledge.
Materials and methods
Chemicals and reagents
P. Lobata Radix was purchased from Hongjian
Pharmacy Co., Ltd. Antibodies against GAPDH (cat. no. BS65483M),
tyrosinase (cat. no. BS1484), TRP-1 (cat. no. BS91379), MITF (cat.
no. BS6666), Bcl-2 associated X protein (Bax, BS90120), succinate
dehydrogenase complex B subunit (SDHB; cat. no. BS8003),
phosphorylated (p-)p38 (cat. no. BS6383) and p38 (cat. no. BS60458)
were purchased from Biogot Technology Co., Ltd. Antibodies against
caspase-3 (cat. no. 9662), caspase-9 (cat. no. 9508), B-cell
lymphoma-2 (Bcl-2, #3498) and succinate dehydrogenase complex A
subunit (SDHA; cat. no. 11998) were purchased from Cell Signaling
Technology, Inc. The antibody against p-MITF (cat. no. PA5-104707)
was purchased from Invitrogen (Thermo Fisher Scientific, Inc.). All
other reagents and solvents used were of analytical grade. The
reactive oxygen species (ROS; cat. no. YX-181519M), superoxide
dismutase (SOD; cat. no. YX-191504M), catalase (CAT; cat. no.
YX-191518M) and glutathione (GSH; cat. no. YX-181509M) ELISA kits
were purchased from Shanghai Youyou Biotechnology Co., Ltd.
Preparation of PLP from dried P.
Lobata Radix
Dried P. Lobata Radix was ground to powder
and passed through a sieve (<0.18 mm). The powder (100 g) was
mixed with water (1:10 w/v) and the pH was adjusted to 8.0 by
adding dilute NaOH. The sample was incubated at 4°C for 4 h,
centrifuged at 7,000 × g for 10 min at 4°C and the precipitate was
discarded. The pH of the supernatant was adjusted to 3.5 by adding
dilute HCl and centrifuged at 7,000 × g for 10 min at 4°C. The
supernatant was then discarded and the remaining sediment
lyophilized.
Preparation of PLP from fresh P.
Lobata Radix
Fresh P. Lobata Radix (100 g) was dispersed
in water at a ratio of 1:10 and homogenized. The remaining steps
were the same as in the previous section.
Preparation of total flavonoids from
P. Lobata Radix
The powder (50 g) obtained from dried P.
Lobata Radix was extracted by refluxing in 70% (v/v) ethanol
(1:40). Extraction was carried out twice for 2 h at 70°C and the
ethanol was removed by evaporation to yield the dried extract.
Preparation of isoflavones from P.
Lobata Radix
The powder (50 g) obtained from dried P.
Lobata Radix was dispersed in 60% (v/v) ethanol (1:20 w/v) and
extracted with 300 W ultrasonic power for 60 min at 50°C. The
extraction process was carried out twice. The ethanol fraction was
recovered and evaporated to yield the dried extract.
Preparation of P. Lobata Radix water
extract
The powder (100 g) obtained from dried P.
Lobata Radix was extracted twice with water (1:10) for 2 and
1.5 h, respectively. The filtrate was combined and concentrated at
6,000 × g for 10 min at 4°C and concentrated to a small volume
before drying. The obtained P. Lobata Radix products were
stored in a sealed, dark, dry and cool place.
Tyrosinase inhibition assay
Tyrosine was used as the substrate to detect
tyrosinase activity in samples and 3-O-ethyl ascorbate ether was
used as the control. In the reaction, 2 mM L-tyrosine (50 µl), 110
µl phosphate buffer (pH 6.8) and 30 µl final samples were mixed and
incubated at 37°C for 10 min. Finally, 50 µl tyrosinase (300 U/ml)
was added rapidly to the reaction mixture and incubated for 5 min.
The absorbance at 475 nm was measured with a microplate reader
(Infinite 200 pro; Tecan Group, Ltd.) every 10 min for at least 1
h.
Determination of water, ash and fat
contents of PLP
Moisture determination was carried out according to
the National Food Safety Standards GB 5009.3-2016 ‘Determination of
Moisture in Food’ (direct drying method) (https://www.eshian.com/standards/36398.html). Fat
levels were determined according to the National Food Safety
Standards GB 5009.6-2016 ‘Determination of Fat in Food’ (Soxhlet
extraction method) (https://www.eshian.com/standards/37367.html). Ash
levels were determined according to the National Food Safety
Standards GB 5009.4-2016 ‘Determination of Ash in Food’ (quality
method) (https://www.eshian.com/standards/36415.html). The
carbohydrate content in samples was determined using the
colorimetric phenol-sulfuric acid method according to the
previously reported protocol (21).
Determination of surface
hydrophobicity of PLP
The surface hydrophobicity was measured using sodium
8-anilino-1-naphthalenesulfonate as the fluorescent probe. PLP
solution (1 mg/ml) was diluted with phosphate buffer to a
concentration between 0.1-1.0 mg/ml. Diluted PLP solution (4 ml)
was mixed with 20 µl 8 mmol/l ANS. The sample was then left
standing in the dark for 10 min. The fluorescence intensity of the
sample was measured by a fluorophotometer (FL6500; PerkinElmer,
Inc.).
Determination of free sulfhydryl and
disulfide bond content of PLP
The PLP solution (2 ml) was mixed with 5.0 ml
Tris-Gly buffer and 100 µl Ellman reagent (10 mM). After the
reaction, the sample was maintained at 25°C for 15 min and the
absorbance at 412 nm was measured. A solution without Ellman
reagent was used as a control and Tris-HCl (pH 8.0) was used
instead of the sample to determine the baseline value.
The total sulfhydryl content was determined by
adding the PLP solution (1.0 ml) to 4.0 ml Tris-Gly-10 mol/l urea
and 50 µl Ellman reagent (10 mmol/l). The remaining steps are the
same as described for determining the free sulfhydryl content.
Analysis of PLP amino acids
Amino acid standards were prepared by taking 200 µl
amino acid mixed standard solution and adding 0.1 mol/l HCl to give
a constant volume in a 5 ml volumetric flask. Following dilution,
the amino acid sample was used as a standard for testing. Amino
acid mixed standard products included 17 standard amino acids.
PLP (30 mg) was dissolved with 20 ml 6 M HCl in a 50
ml hydrolysis tube. The tube was sealed under nitrogen gas and
hydrolyzed. After hydrolysis, the tube was cooled to room
temperature and the volume was adjusted to 50 ml with ultrapure
water. The sample (2 ml) was placed in a vacuum drying oven at 70°C
and evaporated to dryness. The residue was washed and evaporated to
dryness twice with the same volume of ultrapure water, diluted with
2.0 ml of 0.02 M HCl, shaken well and filtered through a
microporous membrane. A ZORBAX Eclipse Plus C18 (4.6×250 mm; 5 µm;
Agilent Technologies, Inc.) column was used for high-performance
liquid chromatography analysis. The mobile phase consisted of
solvent A (0.1 M methanol) and solvent B (0.1 M sodium acetate
buffer). The gradient elution program was: 0–30 min, A 10%; 30–40
min, A 10–35%; 40–50 min, A 35–55%; 50–70 min, A 55–70%; 70–75 min,
A 70–10%; 75–78 min, A 10%. The flow rate was 1.0 ml/min, the
detection wavelength was 360 nm, the column temperature was 20°C
and the injection volume was 10 µl.
Analysis of PLP by SDS-PAGE
Lyophilized PLP (5 mg) was diluted with distilled
water to prepare 1, 3 and 5 mg/ml protein samples. The protein
solutions were centrifuged at 12,000 × g for 5 min at 4°C, the
supernatant was collected and a 2X loading buffer was added. The
samples were boiled for 5 min to denature proteins and left to cool
for later use. Protein electrophoresis was performed and the
protein was stained using Coomassie blue staining (cat. no. C8420;
Beijing Solarbio Science & Technology Co., Ltd.) and imaged
using a gel intelligent imaging system (iBright FL1000; Thermo
Fisher Scientific, Inc.).
PLP solubility determination
PLP (1 g) was dissolved in 100 ml of distilled
water. The sample was stirred for 20 min (pH set to 1.0-11.0) and
incubated at 25°C for 20 min. The sample was then centrifuged at
3,500 × g for 20 min at 4°C. The supernatant was tested with the
BCA kit (P0012, Beyotime, Shanghai, China) and the test was
repeated three times.
Measurement of water absorption
capacity (WAC) and oil absorption capacity (OAC) of PLP
PLP (500 mg) was mixed with 2.5 ml of grease in a
centrifuge tube and incubated at 25°C for 30 min. The sample was
spun at 3,500 × g for 20 min at 4°C to remove the upper layer of
grease. The weight of the centrifuge tube before and after
centrifugation was measured. Using a water-based method, 2.5 ml of
oil was required to replace 2.5 ml of distilled water. Finally, WAC
or OAC of PLP was characterized by the weight ratio of the
centrifuge tubes before and after removing the grease or
supernatant.
PLP foamability and foam stability
determination
PLP (2 g) was mixed with 200 ml of distilled water
(pH adjusted to 2.0, 3.0, 4.0, 5.0, 6.0, 7.0, 8.0, 9.0 and 10.0).
The sample was centrifuged at 14,580 × g for 2 min at 4°C. The
foaming volume V0 at the end of the stirring
period was measured. After 30 min (V30), the
foaming volume was also measured and the foaming property and
foaming stability were calculated. Foaming
(%)=V0/V ×100%. Foaming stability
(%)=V0/V30 × 100%. V is
the initial volume.
PLP emulsification and emulsification
stability determination
PLP (2 g) was mixed with 200 ml of distilled water
(pH adjusted to 2.0, 3.0, 4.0, 5.0, 6.0, 7.0, 8.0, 9.0 and 10.0).
Sample (15 ml) and corn oil (5 ml) were added to a tube and stirred
at 14,580 × g for 2 min at 4°C. After stirring, 50 µl of the sample
from the bottom of the tube was transferred to a new tube
containing 5 ml 0.1% (w/v) SDS. The absorbance at 500 nm was
measured using the microplate reader. The emulsification and
emulsification stability were calculated. Emulsifying activity
(m2/g)=(2×2.303 × A0 x
DF)/(c × Φ × 10,000). Emulsion stability
(min)=(A0 x
t)/(A0-At).
A0 is the absorbance value of the sample.
At is the absorbance value of the sample after
standing for 30 min. c is the concentration of the PLP
sample. DF is the dilution factor. Φ is the ratio of the oil
phase in the emulsion. t is the time.
UV absorption spectrum
An ultraviolet spectrum of the PLP solution was
measured over 200–450 nm with an ultraviolet-visible
spectrophotometer at room temperature.
Fourier transform infrared absorption
spectroscopy (FTIR)
After grinding and mixing the freeze-dried PLP
powder and dried potassium bromide at a ratio of 1:50, the sample
was pressed on a tablet press and scanned in the range 400~4,000
cm−1 using a VERTEX 70 infrared spectrometer (Bruker
Corporation).
Circular dichroism (CD)
spectroscopy
The sample was diluted to 0.2 mg/ml using buffer (20
mM PBS). The standard solution (ultraviolet spectrum, 180–340 nm)
and the test samples (far-UV region scan, 190–260 nm) were recorded
at room temperature (bandwidth was 1.0 nm, time-per-point was 0.5
sec, pathlength was 0.5 mm and spectra were recorded three times).
Pro-Data Viewer software (version 4.5, Applied Photophysics Ltd.)
was used to process raw data and CDNN software (version 2.1,
Applied Photophysics Ltd.) was used to fit data for calculating the
secondary structure of the test protein sample.
Endogenous fluorescence
spectroscopy
PLP lyophilized powder was formulated into a 0.1
mg/ml solution using 10 mM phosphate buffer. The pH of the solution
was adjusted to 5.0, 7.0 and 9.0 with 1.0 M HCl and 1.0 M NaOH. The
excitation wavelength was 280 nm and the fluorescence spectrum with
a wavelength range of 300–500 nm was recorded. The excitation and
emission slit width was 5 nm.
Cell culture
The mouse melanoma cell line (B16 cells) was
obtained from the Type Culture Collection of the Chinese Academy of
Sciences and cultured in RPMI 1640 medium (cat. no. SH30809.01)
supplemented with 10% fetal bovine serum (cat. no. SV30208), 1%
penicillin and streptomycin (cat. no. SV30010; all from HyClone;
Cytiva). Cells were cultured in a humidified normoxic chamber
(Thermo Fisher Scientific, Inc.) with 5% CO2 at
37°C.
Cell viability assay
B16 cells were seeded in 96-well plates
(3×103 cells/well) and cultured for 24 h. The medium was
then replaced with fresh medium containing PLP at various
concentrations (0.1, 0.3 and 0.5 mg/ml) for 48 h at 37°C. Thiazolyl
tetrazolium (MTT; 5 mg/ml) was added to each well and incubated in
darkness at 37°C for 4 h. MTT is a yellow compound that acts on the
mitochondria of living cells to produce blue formazan crystals. The
resulting formazan crystals were dissolved in 150 µl DMSO and the
absorbance at 490 nm was recorded using a microplate reader. Cell
viability
(%)=(Asample-Ablank)/(Acontrol-Ablank)
×100%. The EC50 value was calculated using GraphPad
Prism 6.0 software (Dotmatics).
Assay of melanin content
B16 cells were seeded into 6-well plates at a
density of 6×104 cells/well and treated with or without
PLP (0.1, 0.3, 0.5 mg/ml) at 37°C for 48 h. After removing the
medium, the cells were washed twice with PBS and harvested by
centrifuging at 530 × g for 5 min at room temperature. The cells
were then lysed by incubation in 1 M NaOH [10% (v/v) DMSO] at 60°C
for 1 h. The amount of melanin was determined
spectrophotometrically immediately after lysis by measuring the
absorbance at 475 nm using a microplate reader. The melanin content
in control wells was set as 100% and the results were expressed as
percentages of the control wells. The assay was repeated at least
three times.
Cellular tyrosinase activity
assay
Tyrosinase enzyme activity was estimated by
measuring L-DOPA oxidation. Briefly, the method to culture B16
cells was the same as aforementioned. The cells were washed twice
with PBS, mixed with 1% Triton X-100 and cryopreserved at −80°C for
30 min. The cells were lysed at room temperature, heated in water
at 37°C for 5 min and mixed with 200 µl of 0.1% (w/v) L-DOPA at
37°C for 2 h. The absorbance was measured at 475 nm using a
microplate reader every 10 min for at least 1 h. Tyrosinase
activity was expressed as percentages of activity in untreated
cells.
Reverse transcription-quantitative PCR
(RT-qPCR)
Total RNA was extracted from 1×106 B16
cells using a total RNA extraction kit (DP424; Tiangen Biotech Co.,
Ltd.), according to the manufacturer's instructions and reverse
transcription was performed according to Prime Script RT Reagent
kit instructions (DRR047A; Takara Bio, Inc.). Aliquots of cDNA were
subjected to qPCR analysis with SYBR Green PCR Master Mix (cat. no.
DRR420A; Takara Biotechnology) and a Real-Time PCR system equipped
with a CFX 96 Connect Optics Module (Bio-Rad Laboratories, Inc.).
Program was: 94°C pre-denaturation for 30 sec; 40 cycles (94°C
denaturation for 5 sec, 60°C annealing for 1 min). Expression
levels of target genes were normalized to the GAPDH gene expression
level using the 2−ΔΔCq method (22). All assays were performed in
triplicate. The primer sequences are listed in Table I.
 | Table I.Primer sequences used in the present
study. |
Table I.
Primer sequences used in the present
study.
| Gene | Gene ID | Accession
number | Forward
(5′-3′) | Reverse
(5′-3′) |
|---|
| GAPDH | 14433 | NM_001289726.2 |
TGCCCAGAACATCATCCCT |
TGAAGTCGCAGGAGACAACC |
| Bax | 12028 | NM_001411994.1 |
GGAGATGAACTGGATAGCAATATGG |
GTTTGCTAGCAAAGTAGAAGAGGGCG |
| Bcl-2 | 12043 | NM_009741.5 |
CTGGCATCTTCTCCTTCCAG |
GACGGTAGCGACGAGAGAAG |
| MITF | 17342 | NM_001113198.2 |
CAACGGGAACAGCAACGAGC |
CGGTGGATGGGATAAGGGAAA |
|
Tyrosinase | 22173 | NM_001317397.1 |
GAACACCTGAGGGACCAC |
CATTGGCTTCTGGGTAAA |
| TRP-1 | 22178 | NM_001282014.1 |
CATAACAGGCAATACAACAT |
GTAACAACGCAGCCACTA |
| TRP-2 | 13190 | NM_010024.3 |
CCCCTTTCGCCACAGCCCAACT |
GCGGTCCTCAGACTCTTCCACATTCC |
| p38 | 26416 | NM_001168508.1 |
GGGAGGTGCCCGAACGATAC |
TGGCGTGAATGATGGACTGAAA |
Western blotting
Cells were washed twice with PBS, centrifuged at 530
× g and 4°C for harvesting and lysed with RIPA lysis buffer
(P0013C; Beyotime). Total protein was measured using a BCA kit
(P0010S; Beyotime Institute of Biotechnology). Proteins (20
µg/lane) were separated by 12% SDS-PAGE and transferred onto a
nitrocellulose membrane by semidry blotting. Non-specific protein
binding was blocked by incubating the membranes with 5% (v/v)
non-fat dried milk (P0216; Beyotime Institute of Biotechnology) in
PBS at 25°C for 1 h and the membranes were then incubated overnight
at 4°C with primary antibodies as follows: GAPDH (1:1,000),
tyrosinase (1:1,000), TRP-1 (1:1,000), MITF (1:1,000), Bax
(1:1,000), SDHB (1:1,000), p-p38 (1:1,000), p38 (1:500), caspase-3
(1:1,000), caspase-9 (1:1,000), Bcl-2 (1:1,000), SDHA (1:1,000) and
p-MITF (1:1,000). Following washing with PBS containing 0.05% (v/v)
Tween-20 (P1033; Bejing Solarbio Science & Technology Co.,
Ltd.) (PBST), the membranes were incubated with horseradish
peroxidase-conjugated anti-mouse IgG (cat. no. BS12478; 1:5,000;
Biogot Tech) or horseradish peroxidase-conjugated anti-rabbit IgG
(cat. no. BS13278; 1:5,000; Biogot Tech) for 1 h at 25°C and bands
were visualized using Enhanced Chemiluminescence Reagent kit (cat.
no. P0018FS; Beyotime Institute of Biotechnology). Membranes were
imaged using the iBright FL1000 Imaging System. Image J software
1.53a (National Institutes of Health) was used to quantify the gray
value of western blot bands and the protein expression level was
normalized as the gray value of the target protein/loading control
protein.
Cell apoptosis analysis
The apoptosis incidence was determined by an Amnis
Flow Sight flow cytometer (Merck & Co., Inc.) using the Annexin
V-FITC Apoptosis Detection kit (cat. no. 556547; BD Biosciences).
Briefly, B16 cells were incubated with fresh medium (as control) or
medium containing PLP for 48 h at 37°C. After treatment, the cells
were collected by centrifugation at 303 × g for 5 min at 24°C,
washed twice with ice-cold PBS, suspended in 1X Binding Buffer and
incubated with 5 µl each of propidium iodide (PI) and Annexin
V-FITC solutions. Probes were incubated in darkness at 37°C for 15
min. Samples were examined with a flow cytometer and quantified
using Amnis IDEAS software v6.1 (Merck & Co., Inc.). The number
of total apoptotic cells, including early and late apoptotic cells,
was counted and represented as percentages of the total cell
count.
Mitochondrial membrane potential (MMP)
assessment
MMP changes in B16 cells were measured by
rhodamine-123 staining (cat. no. C2008S; Beyotime Institute of
Biotechnology). Briefly, treated cells were trypsinized, washed
with PBS and stained with rhodamine-123 in darkness at 37°C for 30
min. After centrifugation (430 × g at room temperature for 5 min),
the cells were resuspended in 300 µl PBS and staining was measured
by the Amnis Flow Sight flow cytometer (Merck & Co., Inc.) at
an excitation wavelength of 507 nm and a maximum emission
wavelength of 529 nm.
Mitochondrial fluorescence
photography
B16 cells were incubated with fresh medium (as
control) or medium containing PLP for 48 h at 37°C. The cell
culture medium was removed, Mito-Tracker Green (cat. no. C1048;
Beyotime Institute of Biotechnology) was added and the cells were
incubated in darkness at 37°C for 30 min. Mito-Tracker Green was
removed and 1 ml Hoechst 33342 staining solution (cat. no. C1022;
Beyotime Institute of Biotechnology) was added. The cells were
washed twice again after labeling in darkness at 37°C for 10 min
and then observed with an EVOS® FL Auto Imaging System
(Thermo Fisher Scientific, Inc.).
Measurement of oxygen consumption rate
(OCR)
OCR, as a surrogate indicator of mitochondrial
oxidative phosphorylation, were measured with the Seahorse Cell
Mitochondrial Stress Test kit (cat. no. 103010-100; Agilent
Technologies, Inc.). In brief, control or PLP-treated B16 cells
were harvested to detect OCR, including basal OCR, ATP-linked OCR,
reserve capacity OCR and proton leak OCR, after the administration
of oligomycin (Complex V inhibitor, 1 µM), carbonyl cyanide
4-(trifluoromethoxy)-phenylhydrazone (0.5 µM; an uncoupling agent
of mitochondrial respiration to achieve the maximal respiration
rate) and rotenone/antimycin A (Complex I/III inhibitor, 0.5 µM) in
a Seahorse XFp Analyzer (Agilent Technologies, Inc.). The OCR was
displayed as pmol/min.
Measurement of intracellular ATP
levels
Intracellular ATP concentrations were assayed with
an ATP assay kit (cat. no. S0026; Beyotime Institute of
Biotechnology) according to the manufacturer's instructions.
Briefly, 200 µl ice-cold ATP lysis buffer was added to cells and
the cell lysate was harvested at 12,000 × g and 4°C for 10 min. The
supernatant was taken and the reaction started at 37°C for 3 min,
by adding 100 µl ATP detection buffer to deplete background ATP and
then adding 10 µl supernatant for detection. Relative light unit
(RLU) values were recorded by the microplate reader.
ROS and oxidative stress marker
analysis
The ROS content, SOD, CAT and GSH in B16 cells was
detected by ELISA. Cells were harvested and ruptured by repeated
freeze-thaw steps. The sample was then centrifuged at 3,000 × g and
4°C for 10 min and the supernatant was taken for testing. Reagents
were added according to the kit method and samples were incubated
at 37°C for 60 min. After adding the color reagent, the samples
were incubated in darkness at 37°C for 15 min. Finally, 50 µl stop
solution was added and the absorbance at 450 nm was measured within
15 min using the microplate reader.
Statistical analysis
Data were evaluated using GraphPad software (version
9; GraphPad Software; Dotmatics). Data were presented as the mean ±
standard deviation of three independent experiments and were
analyzed by one-way ANOVA and Dunnett's post-hoc test. P<0.05
was considered to indicate a statistically significant
difference.
Results
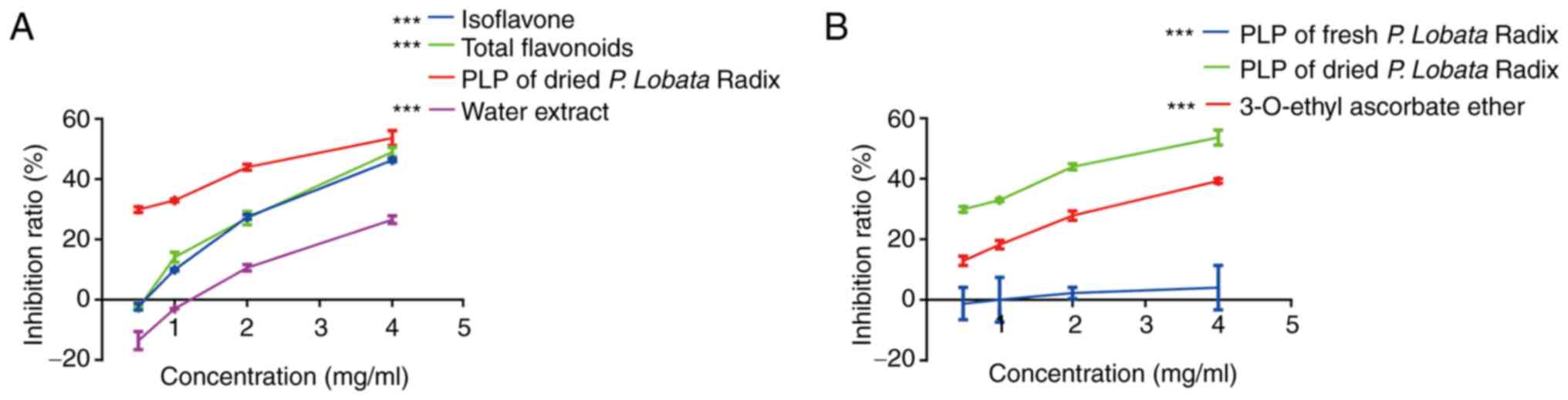
Inhibitory effect of P. Lobata Radix
extracts on tyrosinase activity
Tyrosinase is a copper-containing and rate-limiting
enzyme that regulates melanin biosynthesis (23). The present study first screened
different P. Lobata Radix extracts for inhibition of
tyrosinase activity using a spectrophotometric assay. Tyrosinase
inhibition results for different dried P. Lobata Radix
extracts showed that PLP had the strongest inhibitory effect on
tyrosinase with an IC50 of 4.0 mg/ml (Fig. 1A).
In addition, the tyrosinase inhibition rates of PLP
extracted from fresh P. Lobata Radix and dried P.
Lobata Radix were compared. The result showed that dried P.
Lobata Radix inhibition of tyrosinase was
concentration-dependent and stronger than the inhibition observed
for proteins from fresh P. Lobata Radix and the positive
control (3-O-ethyl ascorbate ether) (Fig. 1B). Thus, we used dried P.
Lobata Radix in subsequent experiments. These results indicated
that PLP is a potential tyrosinase inhibitor.
Analysis of the properties of PLP
The analysis of PLP showed that the protein content
was 90.17% (Table II). The
surface hydrophobicity was 457, the free sulfhydryl content was
6.45 µmol/g, the total sulfhydryl content was 12.65 µmol/g and the
disulfide bond content was 3.1 µmol/g (Table III). The leucine content was the
highest of the essential amino acids at 3.05/100 g, whereas the
aspartic acid content was the highest of the non-essential amino
acids at 3.51/100 g (Table
IV).
 | Table II.Water, ash, protein, carbohydrate and
fat content of PLP |
Table II.
Water, ash, protein, carbohydrate and
fat content of PLP
| Index | Content (%) |
|---|
| Moisture | 3.1±0.1 |
| Ash | 0.51±0.2 |
| protein | 90.17±0.06 |
| carbohydrate | 6.14±0.73 |
| Fat | 0.09±0.01 |
 | Table III.PLP hydrophobicity and sulfur
content |
Table III.
PLP hydrophobicity and sulfur
content
| Index | PLP |
|---|
| Surface
hydrophobicity | 457±16.77 |
| Free sulfhydryl
content (µmol/g) | 6.45±0.6 |
| Total sulfhydryl
content (µmol/g) | 12.65±0.83 |
| Disulfide bond
(µmol/g) | 3.1±0.53 |
 | Table IV.PLP amino acid content |
Table IV.
PLP amino acid content
| A, Essential amino
acids |
|---|
|
|---|
| Amino acids | PLP (g/100 g) |
|---|
| Histidine
(His) | 0.73±0.003 |
| Isoleucine
(Ile) | 1.54±0.03 |
| Leucine (Leu) | 3.05±0.07 |
| Lysine (Lys) | 1.85±0.04 |
| Methionine
(Met) | 0.29±0.01 |
| Phenylalanine
(Phe) | 1.93±0.03 |
| Threonine
(Thr) | 1.91±0.04 |
| Valine (Val) | 2.50±0.06 |
|
| B, Non-essential
amino acids |
|
| Amino
acids | PLP (g/100
g) |
|
| Aspartic (Asp) | 3.51±0.06 |
| Glutamic (Glu) | 3.09±0.07 |
| Serine (Ser) | 1.49±0.05 |
| Cystine (Cys) | 0.27±0.01 |
| Glycine (Gly) | 1.90±0.04 |
| Tyrosine (Tyr) | 1.22±0.04 |
| Arginine (Arg) | 1.53±0.04 |
| Alanine (Ala) | 1.10±0.02 |
| Proline (Pro) | 1.90±0.04 |
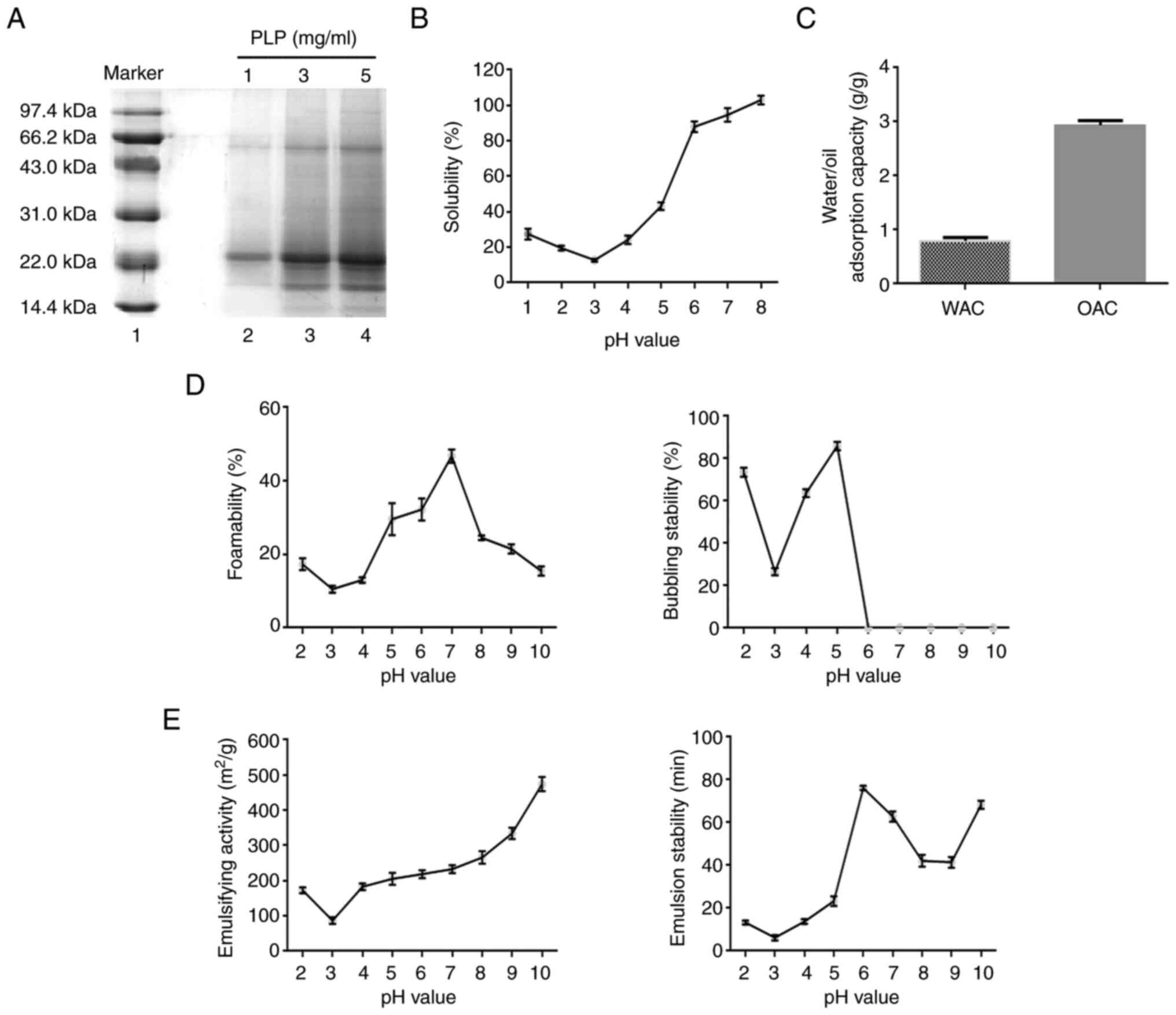
Chemical characterization of PLP
Separation by SDS-PAGE showed that PLP was composed
of 11 proteins (Fig. 2A). As shown
in Fig. 2B, at pH 3.0, the
solubility of PLP was the lowest when the isoelectric point was
reached. The solubility of PLP increased gradually with increasing
pH, with the highest solubility at pH 8.0. WAC and OAC experiments
revealed that there are more hydrophobic than hydrophilic groups in
PLP (Fig. 2C). The foaming ability
of PLP was highest when the pH was 7.0 and the foaming stability
was found to be highest at pH 5.0 (Fig. 2D). The emulsifying activity and
stability of PLP were the lowest when the pH reached the
isoelectric point (Fig. 2E). With
increasing pH, the emulsifying activity of PLP gradually increased
and the emulsifying stability reached a maximum at pH 6.0.
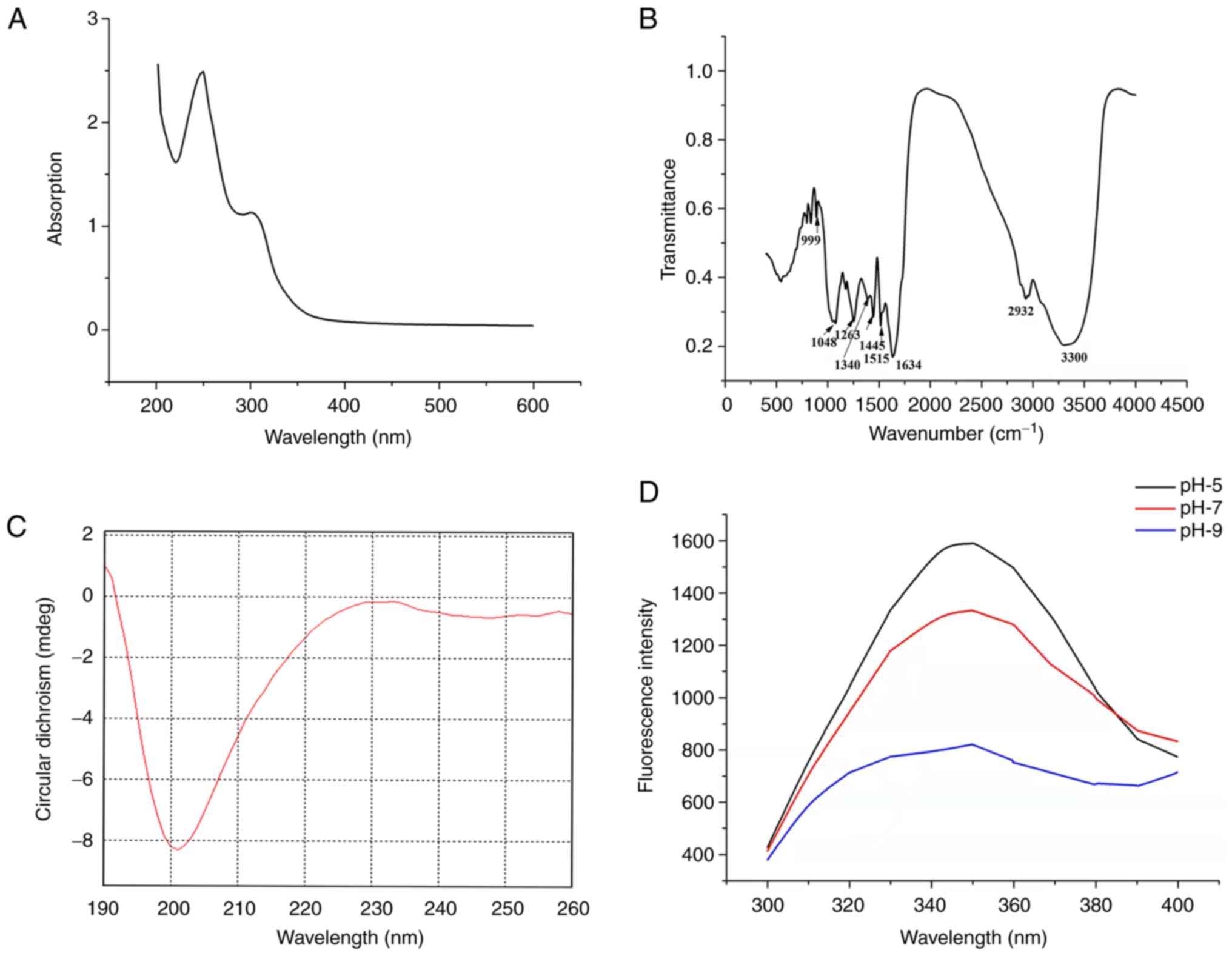
Spectral analysis of PLP
The UV spectrum of PLP had absorption peaks at ~257
and 280 nm (Fig. 3A). PLP has a
number of characteristic absorption peaks in the FTIR spectrum
(Fig. 3B). For example, the amide
I band (1,600–1,700 cm−1) contains abundant secondary
structure information on proteins, with the 1,634 cm−1
peak indicating proteins with β-sheet secondary structure and the
amide III band (1,220–1,330 cm−1) suitable for
distinguishing between α-helix secondary structure and random coil.
The peak at 1,263 cm−1 in the FTIR spectrum arises from
protein with random coil. The infrared spectroscopic results were
consistent with the CD spectrum and the secondary structure of PLP
was analyzed using this spectrum (Fig.
3C and Table V). The CD
results for PLP showed a negative peak near 198 nm and a small and
broad positive peak near 202 nm, indicative of random coil. The
endogenous fluorescence spectrum showed that the fluorescence
intensity of PLP decreased with increasing pH. In addition,
changing the pH yielded a minor redshift effect (Fig. 3D).
 | Table V.PLP secondary structure. |
Table V.
PLP secondary structure.
| Secondary
structure | 190-260 nm (%) | 195-260 nm (%) | 200-260 nm (%) | 205-260 nm (%) | 210-260 nm (%) |
|---|
| α-helix | 7.1 | 7.6 | 6.5 | 4.7 | 5.4 |
| β-sheet | 40.7 | 37.1 | 39.2 | 40.8 | 44.9 |
| β-turn | 22.7 | 23.6 | 25.3 | 23.5 | 19.4 |
| Random curl | 31.6 | 34.5 | 38.3 | 35.0 | 35.2 |
| Total | 102.2 | 102.8 | 109.4 | 104.0 | 104.9 |
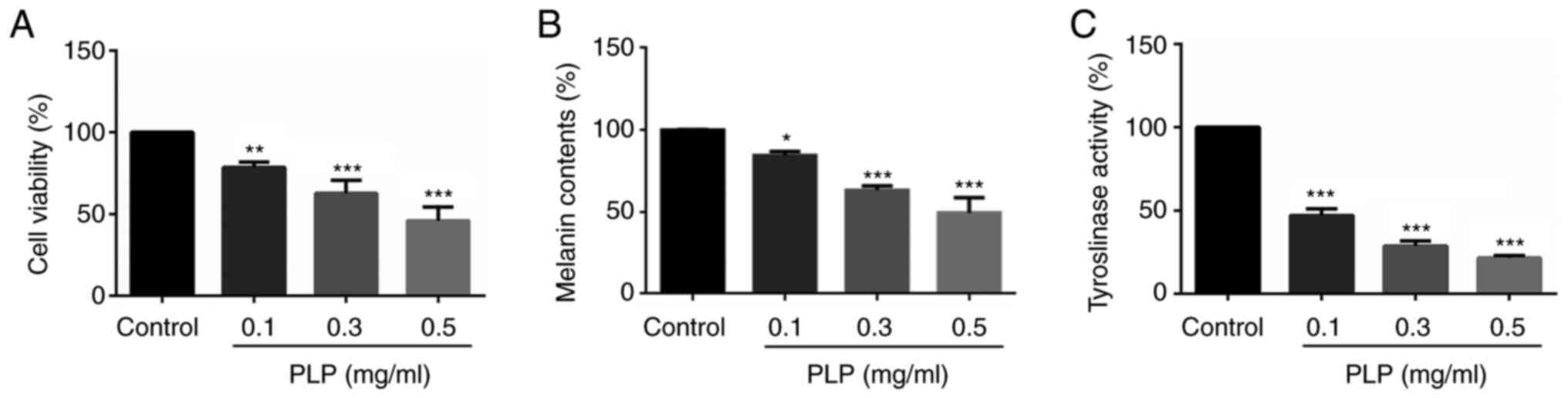
Effect of PLP on cellular melanin
synthesis in mouse melanoma B16 cells
Cell viability was first evaluated to determine the
effects of PLP on cellular melanin synthesis. The MTT assay
revealed that the proliferation of B16 melanoma cells decreased
significantly as the concentration of PLP increased (78.40±3.03,
62.56±6.81 and 45.82±6.99%) and the EC50 of PLP was 0.35
mg/ml (Fig. 4A). To examine the
effects of PLP on melanogenesis, a melanin content assay was
performed in mouse melanoma B16 cells. As shown in Fig. 4B, treatment with PLP decreased the
melanin content in B16 melanoma cells in a dose-dependent manner
(84.41±1.99, 63.22±2.11, 49.32±7.69%). In addition, cellular
tyrosinase activity was also suppressed by PLP treatment in a
dose-dependent manner (47.00±3.22, 28.70±2.46 and 21.43±1.20%;
Fig. 4C). These data indicated
that PLP inhibited tyrosinase activity and had an inhibitory effect
on melanocytes, suggesting that PLP may be a good whitening agent
at the molecular and cellular levels.
Effect of PLP on melanogenesis-related
gene expression
RT-qPCR and western blot analyses were performed to
investigate the effects of PLP on the expression of
melanogenesis-related genes. The mRNA expression levels of MITF and
its downstream genes, tyrosinase, TRP-1 and TRP-2, all decreased as
the concentration of PLP increased (Fig. 5A). In addition, treatment with PLP
markedly inhibited the protein expression levels of p-MITF, MITF,
tyrosinase and TRP-1 in B16 melanoma cells and did not affect the
p-MITF/MITF ratio (Fig. 5B-G).
These findings indicated that the inhibitory effects of PLP on
melanogenesis in B16 melanoma cells may be mediated by the
downregulation of MITF-related melanogenic genes.
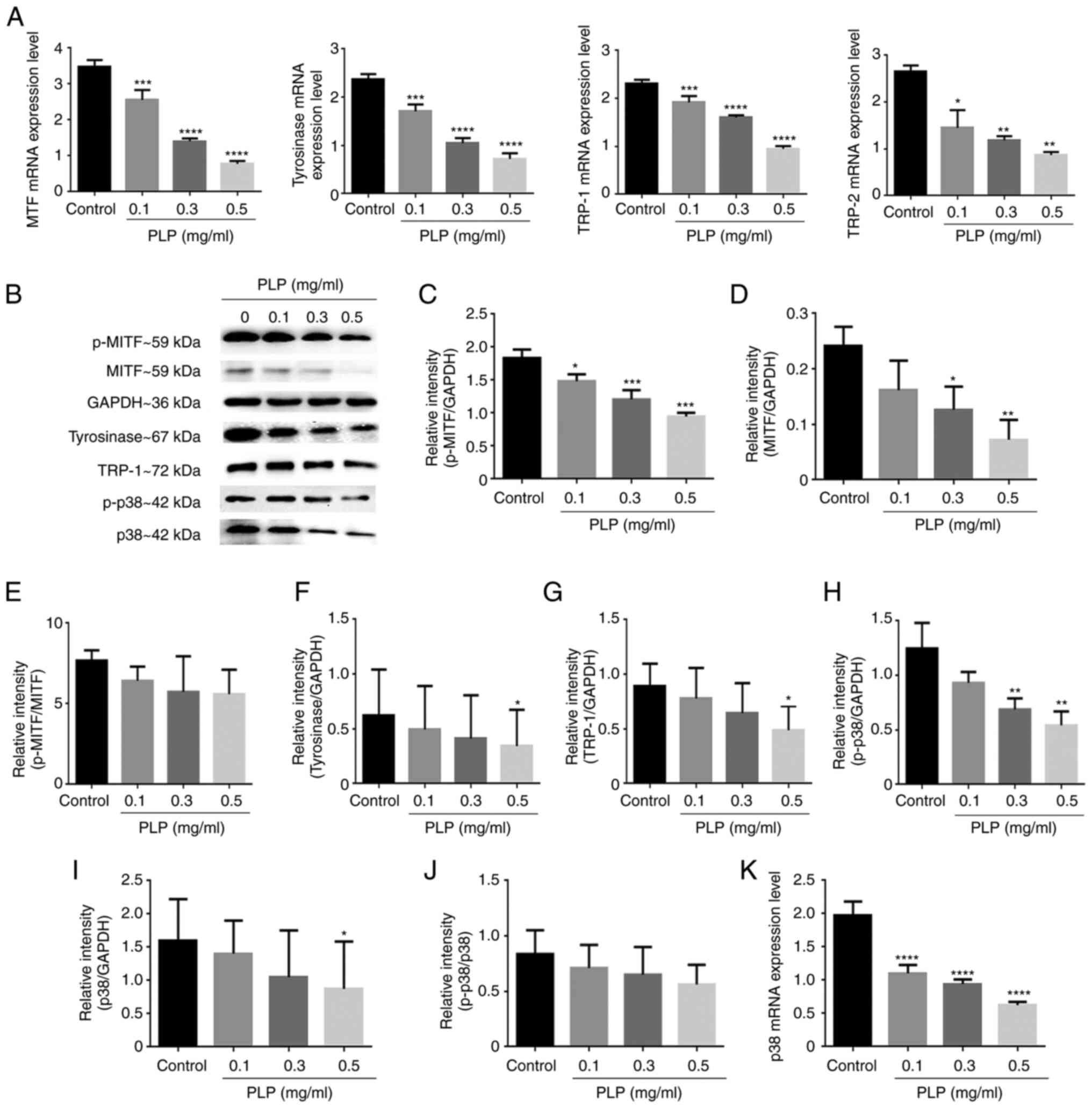 | Figure 5.Effects of PLP on melanogenic-related
and p38 gene and protein expression levels. (A) The effects of PLP
on the mRNA levels of melanogenesis-related genes (MITF,
tyrosinase, TRP-1, TRP-2) were detected by reverse
transcription-quantitative PCR analysis. (B) Effects of PLP on
p-MITF, MITF, tyrosinase, TRP-1, p-p38 and p38 expression levels
determined by western blot analysis with GAPDH as an internal
reference. (C-J) The protein expression levels of p-MITF, MITF,
p-MITF/MITF ratio, tyrosinase, TRP-1, p-p38, p38 and p-p38/p38
ratio. (K) The mRNA level of p38 were detected. Data are expressed
as the mean ± SD of three independent experiments. *P<0.05;
**P<0.01; ***P<0.001; ****P<0.0001 vs. control. PLP, P.
Lobata Radix water-soluble total protein extract; MITF,
microphthalmia-associated transcription factor; TRP-1,
tyrosinase-related protein 1; TRP-2, tyrosinase-related protein 2;
p-, phosphorylated. |
The expression of the MITF gene is regulated by p38
MAPK. For this reason, the effect of PLP on the expression of p38
was examined. As shown in Fig.
5H-K, PLP decreased the mRNA level of p38 and protein
expression levels of p-p38 and p38 significantly, but did not
affect the p-p38/p38 ratio, which were consistent with the
expression of MITF. These results suggested that p38 MAPK operates
upstream of MITF in PLP-regulated melanogenesis.
Effect of PLP on the apoptosis of
mouse melanoma B16 cells
MITF-overexpressing melanocytes showed reduced
apoptosis and increased melanin levels when compared with normal
melanocytes (24). The results of
the present study indicated that PLP inhibited the expression of
the MITF gene in melanoma B16 cells and it was hypothesized that
PLP induced cell apoptosis. Thus, apoptosis in B16 cells was
examined by Annexin V-FITC/PI staining and flow cytometry to
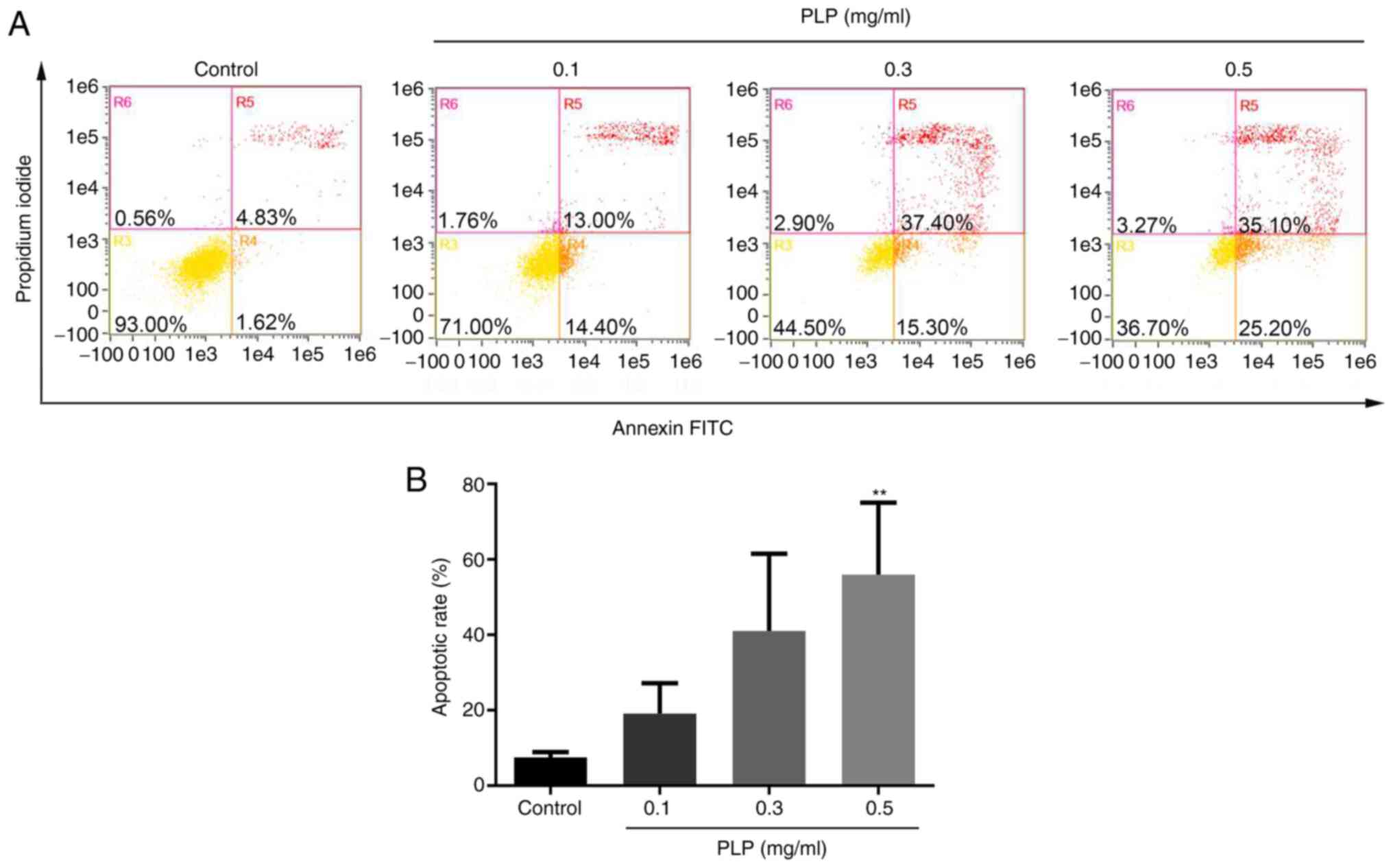
investigate the effect of PLP. As shown in Fig. 6A and B, the percentage of apoptotic
cells in the control group was 7.33±1.23%. As the concentration of
PLP increased, the apoptosis rate of B16 melanoma cells increased
gradually (17.18±3.57, 39.73±14.67 and 55.60±11.96%).
Effect of PLP on mitochondrial
apoptotic pathways
Mitochondria play a central role in the growth and
apoptosis of cells (25). To
determine whether mitochondrial apoptotic signaling pathways are
potentially involved in PLP-induced apoptosis, the expression
levels of a pro-apoptotic protein (Bax), anti-apoptotic protein
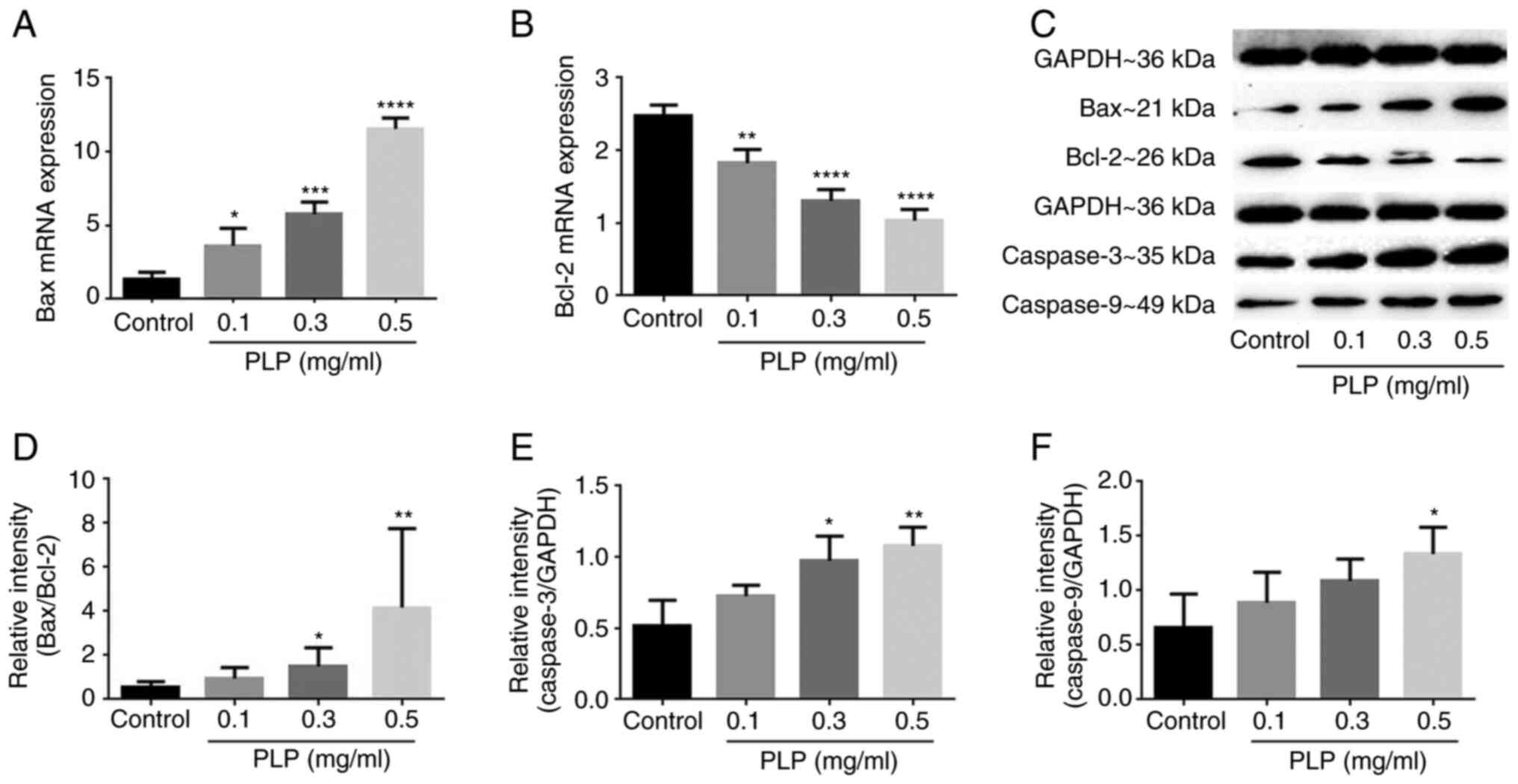
(Bcl-2), caspase-3 and caspase-9 were evaluated. The results showed
that B16 cells treated with PLP had higher Bax expression and lower
Bcl-2 expression (Fig. 7A-D) and
the expression levels of procaspase-3 (Fig. 7E) and procaspase-9 were also
upregulated significantly when compared with untreated cells
(Fig. 7F). Thus, PLP-induced
apoptosis might be related to mitochondrial apoptotic signaling
pathways.
PLP-induced mitochondrial dysfunction
in mouse melanoma B16 cells
The role of mitochondria in PLP-induced apoptosis of
melanoma B16 cells was further elucidated by detecting the MMP and
mitochondria of live cells by flow cytometry and Mito-Tracker
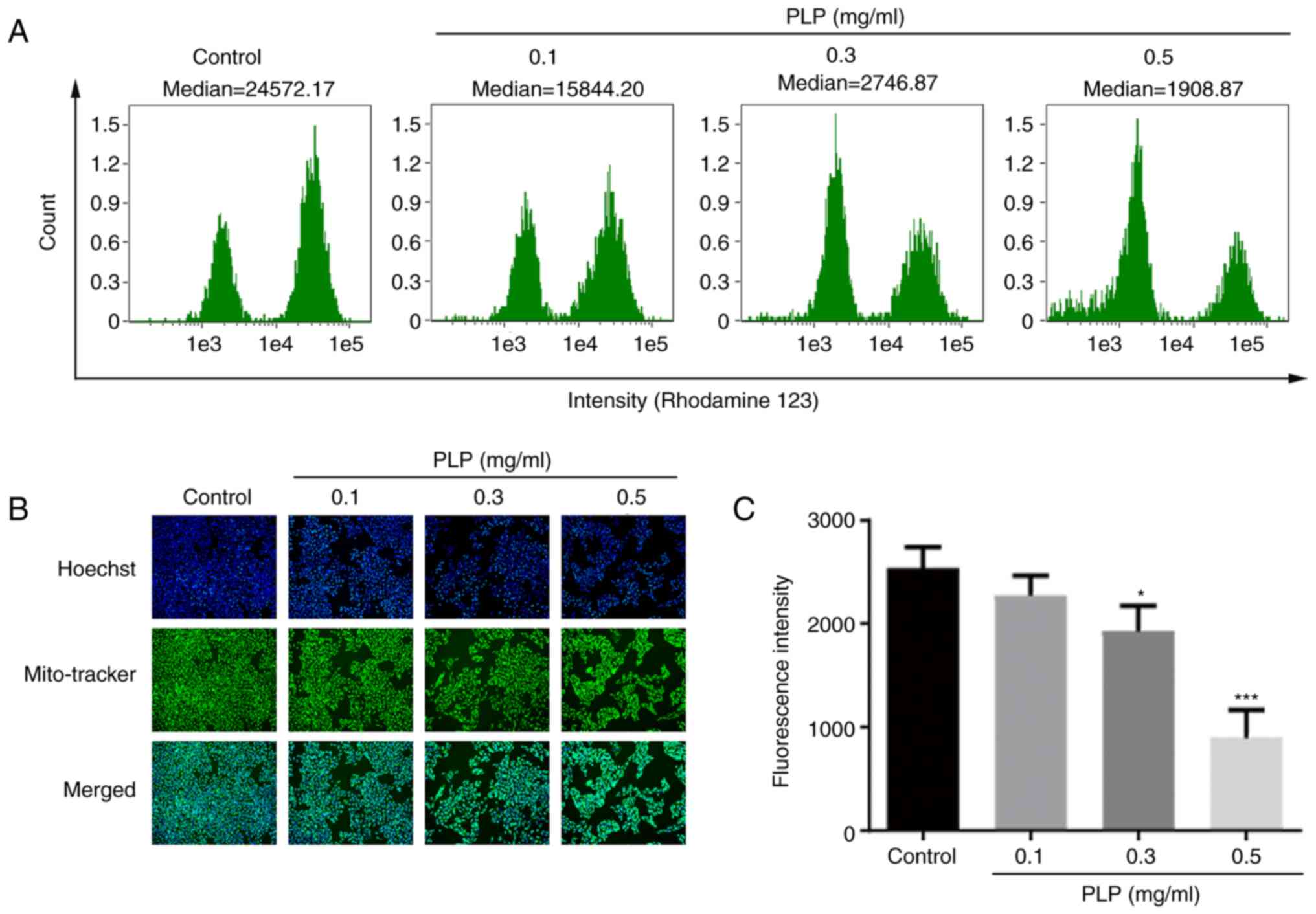
probes, respectively. Compared with control cells, the MMP of cells
treated with PLP was reduced significantly and the MMP decreased
gradually as the concentration of PLP increased (Fig. 8A). In addition, Hoechst was used to
locate cells and Mito-Tracker was used to stain the mitochondria of
live cells specifically. The results showed that the green
fluorescence gradually weakened (Fig.
8B and C).
Effect of PLP on mitochondrial energy
metabolism and ATP generation
Mitochondrial energy metabolism is essential for the
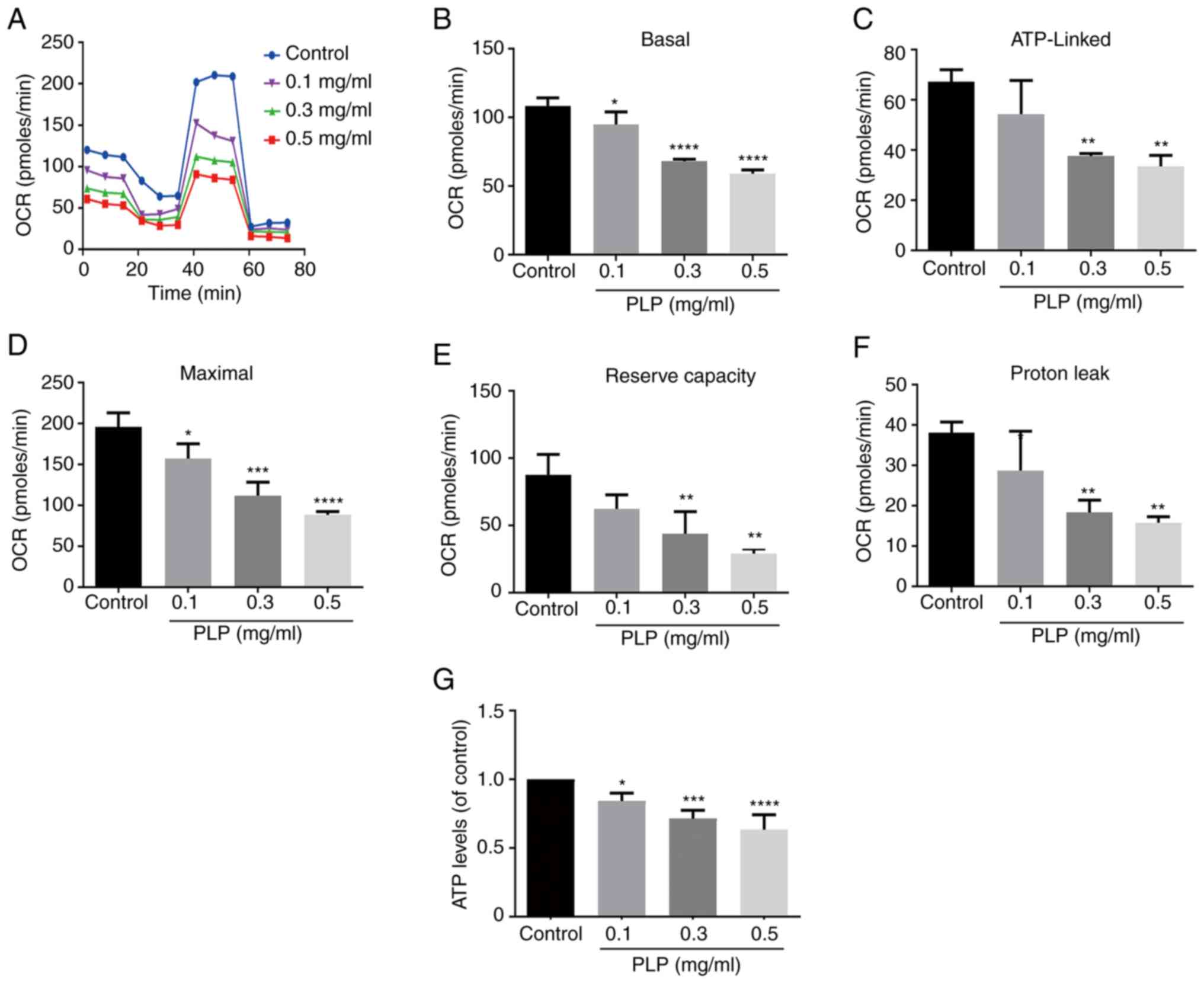
ability of cells to maintain the MMP (4). A Seahorse XFp extracellular flux
analyzer was used to study the effect of PLP on mitochondrial
energy metabolism. Experiments monitoring mitochondrial energy
metabolism evaluated mitochondrial oxygen utilization buffer
capacity, oxygen consumption and other functions (26). Under normal conditions, melanoma
cells display a high rate of mitochondrial energy metabolism
(27). The rate of tumor cell
metabolism decreased after treatment with PLP and a concentration
of 0.5 mg/ml PLP had the strongest effect (Fig. 9A and B). PLP treatment reduced OCR
significantly, which was linked to ATP production (Fig. 9C-F). Similarly, PLP also decreased
the levels of ATP in treated cells (Fig. 9G).
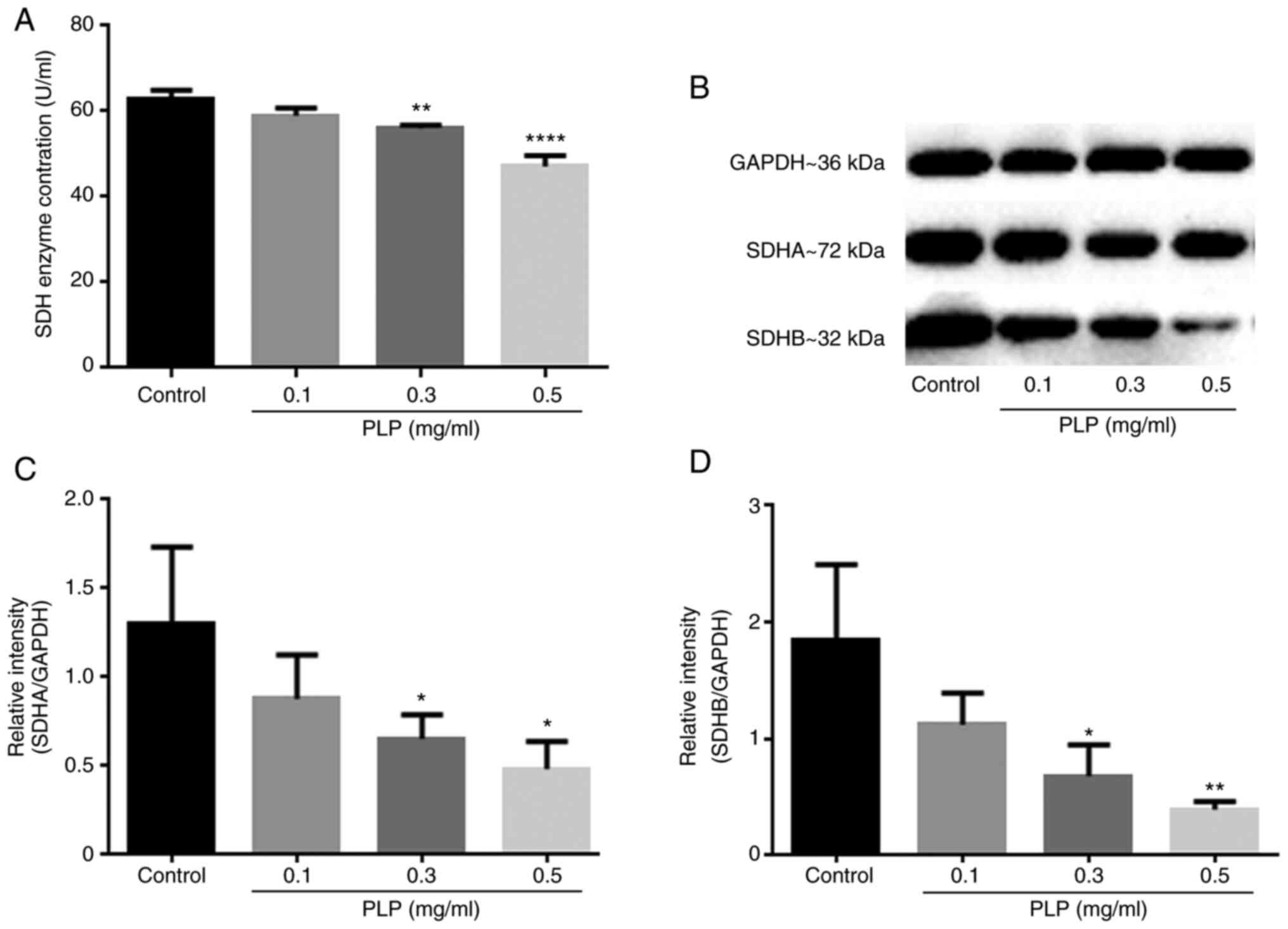
Effect of PLP on succinate
dehydrogenase
Mitochondrial energy metabolism and MMP have been
shown previously to decrease in a concentration-dependent manner
after treatment with PLP. However, the cause of the decrease has
remained unresolved. Therefore, an ELISA assay was used to detect
changes in SDH and protein levels of its two related subunits. The
results showed that the content of SDH decreased after treatment
with PLP (Fig. 10A) and the
levels of its two related subunit proteins also decreased
simultaneously (Fig. 10B-D).
Treatment with 0.5 mg/ml PLP had the strongest effect.
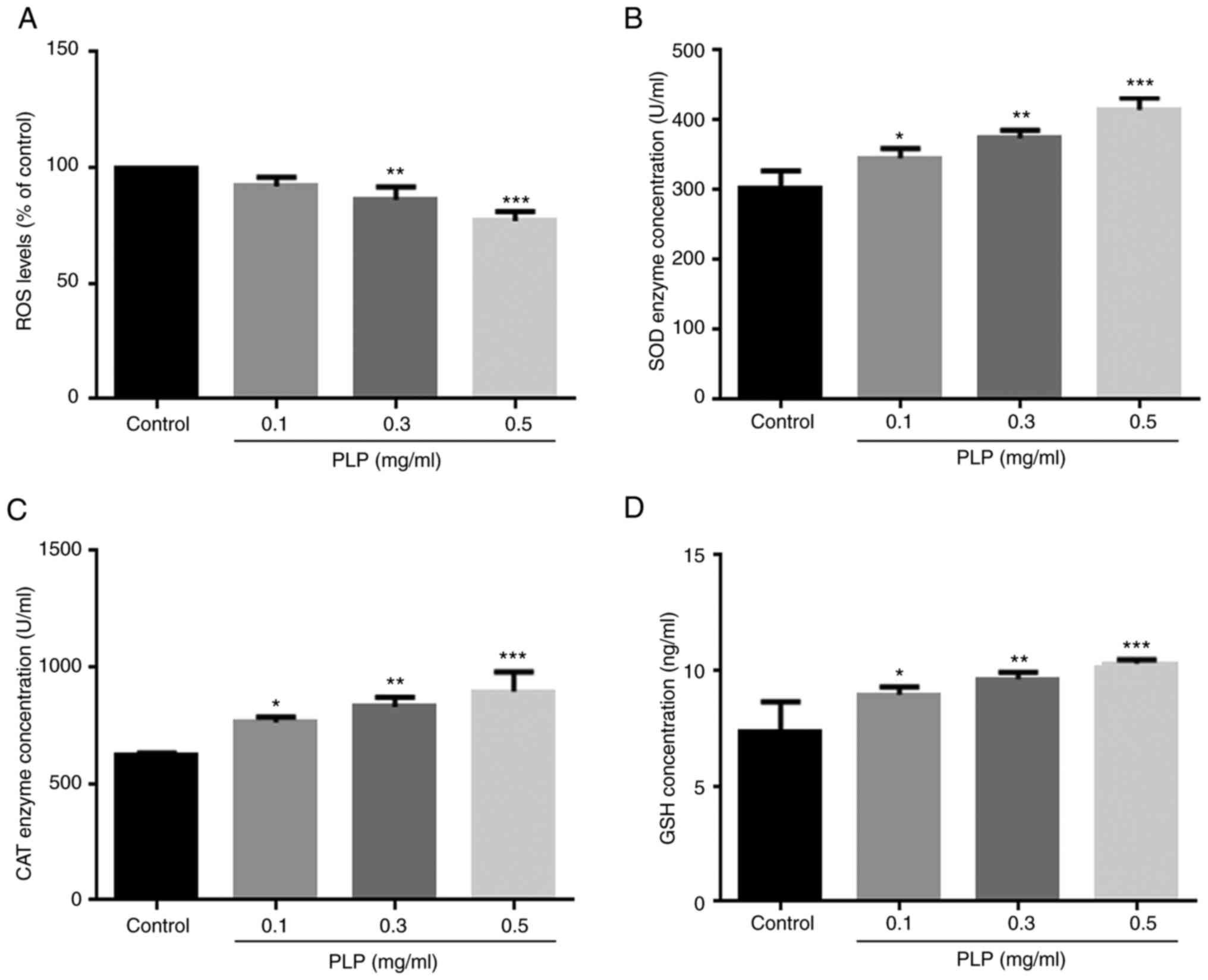
Effect of PLP on intracellular ROS
levels and oxidative stress
Melanin synthesis involves an oxidation reaction and
is important for ROS generation and hyperpigmentation. The present
study investigated the correlation between the inhibition of
melanogenesis and the antioxidant activity of PLP. The level of ROS
in cells decreased significantly when cells were incubated with PLP
(Fig. 11A) and the activities of
SOD (Fig. 11B), CAT (Fig. 11C) and GSH (Fig. 11D) increased significantly. The
results suggested that the inhibitory effect of PLP on melanin
synthesis may also be related to an antioxidant effect.
Discussion
PLP was shown to inhibit melanin synthesis in B16
melanoma cells. PLP is rich in amino acids, including eight
essential amino acids: His, Ile, Leu, Lys, Met, Phe, Thr and Val,
and nine non-essential amino acids: Asp, Glu, Ser, Cys, Gly, Tyr,
Arg, Ala and Pro. PLP exhibited good solubility, foamability,
foaming stability, emulsification activity and emulsification
stability under different pH conditions. In addition, spectroscopic
analysis provided structural information on PLP. The effect of PLP
on melanogenesis in B16 melanoma cells was attributed to the
inhibition of cellular tyrosinase activity. PLP inhibited
melanogenesis by decreasing the expression of tyrosinase, TRP-1 and
TRP-2 through downregulation of the MITF gene, which was mediated
by inhibition of p38 MAPK signaling. In addition, PLP significantly
inhibited cell viability and induced apoptosis in B16 melanoma
cells. Exposure to PLP altered the mitochondrial function in B16
melanoma cells, leading to mitochondria-related apoptosis. Cell
death induced by PLP was associated with the regulation of
mitochondrial energy metabolism, which may explain why MMP
collapsed and a reduction in ATP were observed in B16 melanoma
cells treated with PLP. Finally, it was demonstrated that the
inhibition of melanin synthesis in B16 cells by PLP was correlated
with the regulation of antioxidant enzymes to reduce ROS
levels.
Melanogenesis is the process of synthesizing melanin
within melanosomes and is a major function of both differentiated
normal melanocytes and malignant melanoma cells (28,29).
The present study used mouse melanoma cells, as this cell line has
been used widely in a number of studies on melanin synthesis
(30). It was demonstrated that
PLP significantly inhibited the melanin content in B16 cells. A
previous study indicated that humans and mice share a number of
common genetic features and mice and murine cultured cells are
typically used instead of human equivalents in biological
experiments (31). A number of
reagents have been found to have similar effects on mouse melanoma
B16 cells and human epidermal melanocytes (32–35).
It was hypothesized that the effects of PLP on melanogenesis in
mouse melanoma cells and human melanocytes are likely to be
similar, but further research is necessary as animal models provide
only an approximation of the situation in humans.
Tyrosinase is the rate-limiting enzyme of melanin
biosynthesis in the body (36).
Inhibiting the synthesis of tyrosinase and reducing the activity of
tyrosinase are effective approaches to reduce melanin synthesis.
The extracellular tyrosinase inhibition results for different
fractions of P. Lobata Radix showed that PLP had the
strongest inhibitory effect. The inhibition of tyrosinase by PLP
was also studied in melanoma B16 cells and the DOPA-oxidation rate
was used as the indicator of inhibition. It was found that
treatment with PLP inhibited tyrosinase activity and melanin
synthesis in a dose-dependent manner. TRP-1 and TRP-2 are two other
key enzymes downstream of tyrosinase that affect melanin
biosynthesis (8). The expression
of these melanogenic enzymes is affected by MITF (37,38).
Therefore, the effect of PLP on the expression of p-MITF, MITF,
tyrosinase, TRP-1 and TRP-2 was examined but p-tyrosinase was not
evaluated by western blotting which is a limitation of the present
study. The expression levels of p-MITF, MITF, tyrosinase, TRP-1 and
TRP-2 were inhibited by PLP, indicating that the anti-melanogenesis
activity of PLP was produced through downregulation of MITF, which
affected the related melanogenic enzymes in melanoma B16 cells.
Melanogenesis is regulated by several melanogenic signaling
pathways, including p38 MAPK signaling (39). Activation of p38 MAPK increases the
expression of MITF and tyrosinase, leading to the induction of
melanin synthesis (13). The
present study found that p-p38 and p38 were inhibited by PLP, which
indicated that the inhibitory effect of PLP on MITF expression was
related to p38 MAPK.
MITF is a major regulatory protein for melanocyte
growth, differentiation and pigment synthesis and also plays an
important role in the malignant transformation of melanocytes and
the development and apoptosis of melanocytes. Low levels of MITF
can promote cell senescence and ultimately death (40,41).
As PLP inhibited MITF expression, whether PLP induced apoptosis of
B16 cells was investigated. The results demonstrated that
increasing concentrations of PLP enabled stronger cell apoptosis
responses. Mitochondria play a key role in the induction of cell
apoptosis (42). The
mitochondria-related apoptosis pathway is regulated mainly by
proteins from the Bcl-2 family, including the anti-apoptotic
protein Bcl-2 and the pro-apoptotic protein Bax (43,44).
Accumulation of Bax on the mitochondrial outer membrane results in
loss of MMP and activation of caspase-9, which activates caspase-3
and causes apoptosis of cells (45,46).
Changes in these proteins and decreased MMP were observed in the
present study, which indicated that the mitochondrial apoptosis
pathway was activated in the PLP-induced apoptosis of melanoma B16
cells. In addition, it was found that the reduction in the MMP was
related to a change in the level of SDH on the membrane. SDH
directly affects electron transfer in the mitochondrial respiratory
chain and thus affects mitochondrial function (47). The activity of SDH and its two
subunits, SDHA and SDHB, was determined at the protein level. The
content of SDH decreased after treatment with PLP and the levels of
the two related subunit proteins decreased simultaneously.
Mitochondrial energy metabolism was also evaluated. The results
showed that after treatment with PLP, the mitochondrial utilization
of oxygen and oxygen consumption were significantly reduced. These
results indicated that PLP mediated the mitochondrial apoptotic
pathway to inhibit melanoma cell proliferation, thereby reducing
melanin synthesis. However, the direct role of MITF in PLP-induced
mitochondrial-associated melanoma cell apoptosis induced by PLP
needs further characterization.
The over-synthesis of melanin is also related to
increases in oxidative stress caused by external stimuli (48,49).
ROS have been shown to increase pigmentation (50). Thus, ROS scavengers and inhibitors
of ROS production may suppress melanogenesis (51). PLP contains a variety of amino
acids, which may be natural antioxidants. PLP reduced the levels of
ROS and increased the expression of SOD, CAT and GSH, which
indicated a potential antioxidant capacity of PLP. Mitochondria are
both the source and target of ROS and excessive or low levels of
ROS might disrupt the MMP and induce apoptosis (52–55).
Notably, in the present study, mitochondria-associated melanoma B16
cell apoptosis induced by PLP may have been related to a decrease
in ROS rather than an increase.
Acknowledgments
Not applicable.
Funding
The present study was supported by the Major Science and
Technology Program of Jilin Province (grant no. 20210304002YY).
Availability of data and materials
The datasets used and/or analyzed during the current
study are available from the corresponding authors on reasonable
request.
Authors' contributions
ML and SW conceived and designed the study; YZ and
SY performed experiments; YW, YC and JC collected the data and
confirmed the authenticity of all original data; YZ and JW analyzed
the data and prepared the figures; SW, SY and ML drafted and
revised the manuscript. All authors read and approved the final
manuscript.
Ethics approval and consent to
participate
Not applicable.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
Glossary
Abbreviations
Abbreviations:
|
TRP-1
|
tyrosinase-related protein 1
|
|
TRP-2
|
tyrosinase-related protein 2
|
|
L-DOPA
|
3,4-dihydroxyphenylalanine
|
|
MITF
|
microphthalmia-associated
transcription factor
|
|
PLP
|
P. Lobata Radix water-soluble
total protein
|
|
SDHA
|
succinate dehydrogenase complex A
subunit
|
|
SDHB
|
succinate dehydrogenase complex B
subunit
|
|
WAC
|
water absorption capacity
|
|
OAC
|
oil absorption capacity
|
|
FTIR
|
Fourier transform infrared absorption
spectroscopy
|
|
CD
|
circular dichroism
|
|
MTT
|
thiazolyl tetrazolium
|
|
MMP
|
mitochondrial membrane potential
|
|
OCR
|
oxygen consumption rate
|
|
ROS
|
reactive oxygen species
|
|
SDH
|
succinate dehydrogenase
|
|
SOD
|
superoxide dismutase
|
|
CAT
|
catalase
|
|
GSH
|
glutathione
|
References
|
1
|
Bonaventure J, Domingues MJ and Larue L:
Cellular and molecular mechanisms controlling the migration of
melanocytes and melanoma cells. Pigment Cell Melanoma Res.
26:316–325. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Wang HD, Chen CC, Huynh P and Chang JS:
Exploring the potential of using algae in cosmetics. Bioresour
Technol. 184:355–362. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Otręba M, Rok J, Buszman E and Wrześniok
D: Regulation of melanogenesis: The role of cAMP and MITF. Postepy
Hig Med Dosw (Online). 66:33–40. 2012.(In Polish). PubMed/NCBI
|
|
4
|
Otręba M, Miliński M, Buszman E, Wrześniok
D and Beberok A: Hereditary hypomelanocytoses: The role of PAX3,
SOX10, MITF, SNAI2, KIT, EDN3 and EDNRB genes. Postepy Hig Med Dosw
(Online). 67:1109–1118. 2013.(In Polish). View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Ahn SJ, Koketsu M, Ishihara H, Lee SM, Ha
SK, Lee KH, Kang TH and Kima SY: Regulation of melanin synthesis by
selenium-containing carbohydrates. Chem Pharm Bull (Tokyo).
54:281–286. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
D'Mello SA, Finlay GJ, Baguley BC and
Askarian-Amiri ME: Signaling pathways in melanogenesis. Int J Mol
Sci. 17:11442016. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Buitrago E, Hardré R, Haudecoeur R, Jamet
H, Belle C, Boumendjel A, Bubacco L and Réglier M: Are human
tyrosinase and related proteins suitable targets for melanoma
therapy? Curr Top Med Chem. 16:3033–3047. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Pillaiyar T, Manickam M and Namasivayam V:
Skin whitening agents: Medicinal chemistry perspective of
tyrosinase inhibitors. J Enzyme Inhib Med Chem. 32:403–425. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Wang Y, Li SM, Huang J, Chen SY and Liu
YP: Mutations of TYR and MITF genes are associated with plumage
colour phenotypes in geese. Asian-Australas J Anim Sci. 27:778–783.
2014. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Sun L, Guo Y, Zhang Y and Zhuang Y:
Antioxidant and anti-tyrosinase activities of phenolic extracts
from rape bee pollen and inhibitory melanogenesis by cAMP/MITF/TYR
pathway in B16 mouse melanoma cells. Front Pharmacol. 8:1042017.
View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Raja DA, Gotherwal V, Burse SA,
Subramaniam YJ, Sultan F, Vats A, Gautam H, Sharma B, Sharma S,
Singh A, et al: pH-controlled histone acetylation amplifies
melanocyte differentiation downstream of MITF. EMBO Rep.
21:e483332020. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Pathria G, Garg B, Borgdorff V, Garg K,
Wagner C, Superti-Furga G and Wagner SN: Overcoming MITF-conferred
drug resistance through dual AURKA/MAPK targeting in human melanoma
cells. Cell Death Dis. 7:e21352016. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Jung E, Kim JH, Kim MO, Jang S, Kang M, Oh
SW, Nho YH, Kang SH, Kim MH, Park SH and Lee J: Afzelin positively
regulates melanogenesis through the p38 MAPK pathway. Chem Biol
Interact. 254:167–172. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Wang DD, Jin Y, Wang C, Kim YJ, Perez ZEJ,
Baek NI, Mathiyalagan R, Markus J and Yang DC: Rare ginsenoside Ia
synthesized from F1 by cloning and overexpression of the
UDP-glycosyltransferase gene from bacillus subtilis: Synthesis,
characterization, and in vitro melanogenesis inhibition activity in
BL6B16 cells. J Ginseng Res. 42:42–49. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Wang S, Zhang S, Wang S, Gao P and Dai L:
A comprehensive review on Pueraria: Insights on its chemistry and
medicinal value. Biomed Pharmacother. 131:1107342020. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Hien TT, Kim HG, Han EH, Kang KW and Jeong
HG: Molecular mechanism of suppression of MDR1 by puerarin from
Pueraria lobata via NF-kappaB pathway and cAMP-responsive
element transcriptional activity-dependent up-regulation of
AMP-activated protein kinase in breast cancer MCF-7/adr cells. Mol
Nutr Food Res. 54:918–928. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Xu L, Zheng N, He Q, Li R, Zhang K and
Liang T: Puerarin, isolated from Pueraria lobata (Willd.),
protects against hepatotoxicity via specific inhibition of the
TGF-β1/Smad signaling pathway, thereby leading to anti-fibrotic
effect. Phytomedicine. 20:1172–1179. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Jeon YD, Lee JH, Lee YM and Kim DK:
Puerarin inhibits inflammation and oxidative stress in dextran
sulfate sodium-induced colitis mice model. Biomed Pharmacother.
124:1098472020. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Ye Y, Gao Y, Fang Y, Xu L and He F:
Anticancer effect of puerarin on ovarian cancer progression
contributes to the tumor suppressor gene expression and gut
microbiota modulation. J Immunol Res. 2022:44725092022. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Li Y, Zhang C, Ma X, Yang L and Ren H:
Identification of the potential mechanism of Radix pueraria in
colon cancer based on network pharmacology. Sci Rep. 12:37652022.
View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Zhu J, Xu J, Wang Y, Li C, Chen Z, Song L,
Gao J and Yu R: Purification and structural characterization of a
novel anti-tumor protein from Arca inflata. Int J Biol Macromol.
105:103–110. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(−Delta Delta C(T)) method. Methods. 25:402–408. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Noh H, Lee SJ, Jo HJ, Choi HW, Hong S and
Kong KH: Histidine residues at the copper-binding site in human
tyrosinase are essential for its catalytic activities. J Enzyme
Inhib Med Chem. 35:726–732. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Shen XF, Teng Y, Sha KH, Wang XY, Yang XL,
Guo XJ, Ren LB, Wang XY, Li J and Huang N: Dietary flavonoid
luteolin attenuates uropathogenic Escherichia. Coli invasion of the
urinary bladder. Biofactors. 42:674–685. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Bock FJ and Tait SWG: Mitochondria as
multifaceted regulators of cell death. Nat Rev Mol Cell Biol.
21:85–100. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Tian L, Zhu C, Yang H, Li Y and Liu Y:
Protective effect of mitochondrial ND2 C5178A gene mutation on cell
and mitochondrial functions. Oxid Med Cell Longev.
2021:47287142021. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Li L, Meng Y, Wu X, Li J and Sun Y:
Bromodomain-containing protein 4 inhibitor JQ1 promotes melanoma
cell apoptosis by regulating mitochondrial dynamics. Cancer Sci.
112:4013–4025. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Kleszczyński K, Kim TK, Bilska B, Sarna M,
Mokrzynski K, Stegemann A, Pyza E, Reiter RJ, Steinbrink K, Böhm M
and Slominski AT: Melatonin exerts oncostatic capacity and
decreases melanogenesis in human MNT-1 melanoma cells. J Pineal
Res. 67:e126102019. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Orhan IE and Deniz FSS: Inhibition of
melanogenesis by some well-known polyphenolics: A review. Curr
Pharm Biotechnol. 22:1412–1423. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Sarna M, Krzykawska-Serda M, Jakubowska M,
Zadlo A and Urbanska K: Melanin presence inhibits melanoma cell
spread in mice in a unique mechanical fashion. Sci Rep. 9:92802019.
View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Dutta S and Sengupta P: Men and mice:
Relating their ages. Life Sci. 152:244–248. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Villareal MO, Kume S, Neffati M and Isoda
H: Upregulation of Mitf by phenolic compounds-rich cymbopogon
schoenanthus treatment promotes melanogenesis in B16 melanoma cells
and human epidermal melanocytes. Biomed Res Int. 2017:83036712017.
View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Lee CS, Nam GB and Park JS:
Protopanaxatriol inhibits melanin synthesis through inactivation of
the pCREB-MITF-tyrosinase signalling pathway in melanocytes. Clin
Exp Dermatol. 44:295–299. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Hu Y, Huang J, Li Y, Jiang L, Ouyang Y, Li
Y, Yang L, Zhao X, Huang L, Xiang H, et al: Cistanche deserticola
polysaccharide induces melanogenesis in melanocytes and reduces
oxidative stress via activating NRF2/HO-1 pathway. J Cell Mol Med.
24:4023–4035. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Lim HY, Kim E, Park SH, Hwang KH, Kim D,
Jung YJ, Kopalli SR, Hong YD, Sung GH and Cho JY: Antimelanogenesis
effects of theasinensin A. Int J Mol Sci. 22:74532021. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Song W, Zhao YY, Ren YJ, Liu LL, Wei SD
and Yang HB: Proanthocyanidins isolated from the leaves of Photinia
× fraseri block the cell cycle and induce apoptosis by inhibiting
tyrosinase activity in melanoma cells. Food Funct. 12:3978–3991.
2021. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Tsatmali M, Ancans J and Thody AJ:
Melanocyte function and its control by melanocortin peptides. J
Histochem Cytochem. 50:125–133. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Imokawa G: Autocrine and paracrine
regulation of melanocytes in human skin and in pigmentary
disorders. Pigment Cell Res. 17:96–110. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Sun L, Guo C, Yan L, Li H, Sun J, Huo X,
Xie X and Hu J: Syntenin regulates melanogenesis via the p38 MAPK
pathway. Mol Med Rep. 22:733–738. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Giuliano S, Ohanna M, Ballotti R and
Bertolotto C: Advances in melanoma senescence and potential
clinical application. Pigment Cell Melanoma Res. 24:295–308. 2011.
View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Strub T, Giuliano S, Ye T, Bonet C, Keime
C, Kobi D, Le Gras S, Cormont M, Ballotti R, Bertolotto C and
Davidson I: Essential role of microphthalmia transcription factor
for DNA replication, mitosis and genomic stability in melanoma.
Oncogene. 30:2319–2332. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Liu WK, Ho JC, Cheung FW, Liu BP, Ye WC
and Che CT: Apoptotic activity of betulinic acid derivatives on
murine melanoma B16 cell line. Eur J Pharmacol. 498:71–78. 2004.
View Article : Google Scholar : PubMed/NCBI
|
|
43
|
Yang F, Yu Y, Lei Q, Zeng A, Li Y, Xie Y,
Ye T and Wei Y: Lobaplatin arrests cell cycle progression, induces
apoptosis and impairs migration and invasion in B16-F10 melanoma
cell line in vitro. Biomed Pharmacother. 69:402–408. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
44
|
Li J, Li J, Aipire A, Gao L, Huo S, Luo J
and Zhang F: Phenylethanoid glycosides from cistanche tubulosa
inhibits the growth of B16-F10 cells both in vitro and in vivo by
induction of apoptosis via mitochondria-dependent pathway. J
Cancer. 7:1877–1887. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
45
|
Cartron PF, Bellot G, Oliver L,
Grandier-Vazeille X, Manon S and Vallette FM: Bax inserts into the
mitochondrial outer membrane by different mechanisms. FEBS Lett.
582:3045–3051. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
46
|
Yang CC, Hung CF and Chen BH: Preparation
of coffee oil-algae oil-based nanoemulsions and the study of their
inhibition effect on UVA-induced skin damage in mice and melanoma
cell growth. Int J Nanomedicine. 12:6559–6580. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
47
|
Dalla Pozza E, Dando I, Pacchiana R, Liboi
E, Scupoli MT, Donadelli M and Palmieri M: Regulation of succinate
dehydrogenase and role of succinate in cancer. Semin Cell Dev Biol.
98:4–14. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
48
|
Zhang BB, Wang DG, Guo FF and Xuan C:
Mitochondrial membrane potential and reactive oxygen species in
cancer stem cells. Fam Cancer. 14:19–23. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
49
|
Wang LX, Qian J, Zhao LN and Zhao SH:
Effects of volatile oil from ginger on the murine B16 melanoma
cells and its mechanism. Food Funct. 9:1058–1069. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
50
|
Alfadda AA and Sallam RM: Reactive oxygen
species in health and disease. J Biomed Biotechnol.
2012:9364862012. View Article : Google Scholar : PubMed/NCBI
|
|
51
|
Wang J and Yi J: Cancer cell killing via
ROS: To increase or decrease, that is the question. Cancer Biol
Ther. 7:1875–1884. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
52
|
Yang S, Zhang Y, Luo Y, Xu B, Yao Y, Deng
Y, Yang F, Ye T, Wang G, Cheng Z, et al: Hinokiflavone induces
apoptosis in melanoma cells through the ROS-mitochondrial apoptotic
pathway and impairs cell migration and invasion. Biomed
Pharmacother. 103:101–110. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
53
|
Lu L, Zhang H, Liu J, Liu Y, Wang Y, Xu S,
Zhu Z and Xu J: Synthesis, biological evaluation and mechanism
studies of C-23 modified 23-hydroxybetulinic acid derivatives as
anticancer agents. Eur J Med Chem. 182:1116592019. View Article : Google Scholar : PubMed/NCBI
|
|
54
|
Rizwan H, Pal S, Sabnam S and Pal A: High
glucose augments ROS generation regulates mitochondrial dysfunction
and apoptosis via stress signalling cascades in keratinocytes. Life
Sci. 241:1171482020. View Article : Google Scholar : PubMed/NCBI
|
|
55
|
Yang N, Guan QW, Chen FH, Xia QX, Yin XX,
Zhou HH and Mao XY: Antioxidants targeting mitochondrial oxidative
stress: Promising neuroprotectants for epilepsy. Oxid Med Cell
Longev. 2020:66871852020. View Article : Google Scholar : PubMed/NCBI
|