Introduction
Osteosarcoma (OS) is the commonest primary malignant
bone tumor in young patients. Although, with the development of
surgical skills and the application of neoadjuvant chemotherapy,
the 5-year survival rate of patients has increased to 70%, there
are still a number of challenges that physicians face in OS,
including the chemoresistance, local recurrence and pulmonary
metastasis (1,2). The most important prognostic
indicator for OS patients is the necrosis of OS after chemotherapy
before operations. Patients with <90% tumor necrosis, who are
considered as chemoresistant to the agents, will have poor outcomes
in the future (3). Unfortunately,
>70% of patients are insensitive to chemotherapeutic agents and
the chemoresistance appears to be mediated by a variety of
mechanisms (4).
Exosomes are a class of 30- to 150-nm extracellular
vesicles (EVs) generated by almost all cell types, including cancer
cells (5). Exosomes, with lipid
bilayer membranes, can be taken up by neighboring or distant
recipient cells and exosomal contents, including small RNA and
protein, can exhibit biological activities such as immunomodulation
(6), autophagy (7), stem cell differentiation (8) and intercellular communication
(9), which is known as the third
method of cellular communication. Exosomal miRNAs, protected by
lipid membranes of exosomes from being digested by RNases, can be
stably transferred between two different tumor cells and
participate in tumor progression (9). Some related research about the
function of cell-secreted exosomal miRNAs (exo-miRNAs) in growth,
metastasis, angiogenesis and multidrug resistance (MDR) for
malignant tumors has been reported (10). However, the importance of exosomes
and related exo-miRNAs in the pathogenesis of the OS cells has yet
to be established. The present study investigated the potential
influence of exosomes from doxorubicin-resistant OS cells in the
proliferation, migration, invasion and MDR on the
doxorubicin-sensitive OS cells and searched for potential
differentially expressed exo-miRNAs that would predict the
different response to chemotherapy.
Methods and materials
Cell culture and reagents
A total of four OS cell lines, MG63 cells
(doxorubicin-sensitive OS cells), doxorubicin-resistant MG63 cells
(MG63/DXR), KHOS cells (doxorubicin-sensitive OS cells) and
doxorubicin-resistant KHOS cells (KHOS/DXR), were chosen in the
present study and purchased from the American Type Culture
Collection. All four cell lines were cultured in DMEM (Gibco;
Thermo Fisher Scientific, Inc.) containing 10% FBS depleted of
exosomes (FDE; ScienCell Research Laboratories, Inc.) and 1%
penicillin/streptomycin (Gibco; Thermo Fisher Scientific, Inc.).
All of the cells were cultured in a humidified incubator with 5%
CO2 at 37°C. When the cell density reached 70–80%, the
conditioned medium was collected.
Exosome isolation
After the collection of the conditioned medium from
OS cells, the supernatant was collected and centrifuged as follows:
300 × g for 10 min to remove cells, 2,000 × g for 10 min to remove
dead cells, 10,000 × g for 30 min to remove cell debris, 100,000 ×
g for 70 min to collect pellets, washed with PBS and 100,000 × g
for 70 min to collect exosomes all at 4°C. NanoSight particle
tracking analysis (NTA) was conducted to identify the concentration
and number of exosomes and the bicinchoninic acid (BCA) method
(Thermo Fisher Scientific, Inc.) was applied to examine the
exosomal protein concentration.
Transmission electron microscopy
(TEM)
For TEM observation, 10 µl of exosome solution was
placed onto copper mesh and incubated at room temperature for 10
min. After that, the copper mesh was washed with sterile distilled
water and 10 µl of 2% uranyl acetate was pipetted on the copper
mesh for negative staining for 1 min, the excess fluid was removed
and the mesh was dried under an incandescent lamp for 2 min.
Finally, the copper mesh was observed under a transmission electron
microscope at 80 KV (JEOL, Ltd.).
NTA
After the isolation of the exosomes from OS cells,
PBS was used to dilute at a factor of 100 or 1,000 for exosomes to
obtain an approximate number of vesicles prior to NTA. ZetaView PMX
110 (Particle Metrix GmbH) and its corresponding software (ZetaView
8.02.28) were applied to analyze the size and concentration of the
exosomes from OS cells.
Western blot analysis
Total protein was isolated using cell lysis buffer
and the bicinchoninic acid (BCA) method (Thermo Scientific, Inc.)
was applied to examine the exosomal protein concentration as
described before. The densitometry of the protein was detected by
NanoDrop-1000 (Thermo Scientific, Inc. USA) with the software
Nanodrop 3.3.0 by the BCA method. The equal amounts of protein (50
µg per lane) were loaded for western blot analysis. To identify the
three specific proteins, CD9 (23 kDa), CD63 (26 kDa), and TSG-101
(72 kDa) were positive experssed, exosomes were collected and lysed
using RIPA protein extraction reagent (Beyotime Institute of
Biotechnology) supplemented with a protease inhibitor cocktail
(Roche Applied Science). Proteins were loaded onto 10% SDS-PAGE
gels for electrophoresis, transferred to PVDF membranes and blocked
in 5% milk for 1 h at 4°C overnight prior to incubation with the
indicated primary antibodies for 3 h at room temperature. After
washing with trisbuffered saline with 0.1% Tween 20 (TBST) four
times, the secondary antibodies were incubated with the membrane at
room temperature for 1.5 h. The antibodies used in the experiments
included anti-CD63 (1:1,000; Santa Cruz Biotechnology, Inc.),
anti-CD9 (1:1,000; Santa Cruz Biotechnology, Inc.), anti-TSG101
(1:1,000; Santa Cruz Biotechnology, Inc.) and anti-β-actin
(1:1,000; Santa Cruz Biotechnology, Inc.). All experiments were
repeated three times. The bound antibodies were visualized with
Pierce ECL Western Blotting Substrate (Thermo Fisher Scientific).
Image pro-Plus 6.0 (Media Cybernetics, Inc.) was used for the
densitometry of the brands.
Exosome labeling and uptake
MG63 cells were stained with the CellTrace CFSE Cell
Proliferation kit (Invitrogen; Thermo Fisher Scientific, Inc.)
according to the manufacturer's instructions and exosomes were
labeled using the PKH67 Blue Fluorescent Cell Linker kit
(MINI67-1KT; MilliporeSigma). PKH67 dye solution (1 ml; 1:1,000)
was mixed with 20 µg of exosomes for 20 min at 36.5°C, washed with
PBS and centrifuged at 100,000 × g for 70 min at 4°C. PKH67-labeled
exosomes (4 µg) were resuspended in IMDM supplemented with 10% FDE
and added to MG63 cells at 36.5°C. The cells were washed and fixed
in 3.7% PFA for 10 min to stop the process of uptake with different
time of incubation. Then, the cells were stained with fluorescein
isothiocyanate (FITC)-conjugated phalloidin (MilliporeSigma) and
the uptake of exosomes at different time points was observed under
a confocal fluorescence microscope (Nikon Eclipse E800M; Nikon
Corporation).
Cell viability
Cell viability was evaluated by the CellTiter-Glo
2.0 Reagent (cat. no. G7572; Promega Corporation). MG63 cells were
seeded at a density of 2×103 cells/well in 96-well
flat-bottomed tissue culture plates in the presence of DMEM +10%
FDE and test compound at room temperature for approximately 30 min.
Then, 100 µl of CellTiter-Glo 2.0 Reagent was added and the content
was mixed for 2 min on an orbital shaker. After incubating the
plate at room temperature for 10 min, the luminescence was
recorded.
Cell migration assay
Transwell experiment was conducted for the migration
assay. OS cells were seeded at a density of 1.2×105
cells/well in 24-well plates with 500 µl of cell culture medium for
48 h. Then the MG63 cells were fixed with 500 µl of 4%
paraformaldehyde at room temperature for 15 min. After washing with
deionization water for 3 to 5 times, the OS cells were observed
under a microscope. Experiments were performed at least three times
and the results were recorded as the mean of these experiments.
Cell invasion assay
MG63 cells were seeded at the destiny of
2.0×105 cells/ml in 24-well plates and incubated with
exosomes from MG63/DXR (Exo-MG63-DXR; 100 µl/ml) for 48 h at
36.5°C. Matrigel (cat. no. 356235; Corning, Inc.) was thawed on a
shaker at 4°C for 2 h. Matrigel (10 µl) was added the cell pellet
in an Eppendorf tube and mixed gently. Cells that were mixed with
cold Matrigel were gently pipetted into the middle of a well in a
24-well plate into a drop-like shape. The Matrigel drops are
solidified in a 37°C incubator with 5% CO2 injection for
20 min. The cells were digested and centrifuged with the speed of
200 × g for 5 min at room temperature. After termination of
digestion, the cells washed twice with PBS, and resuspended in 10
g/l BSA. The cells were seeded at a density of 1.2×105
cells/well in 24-well plates. Doxorubicin (100 ng/ml) was added
into the treated MG63 cells for 24 h and the cells were fixed with
500 µl of 4% paraformaldehyde at room temperature for 15 min. The
chamber in the control group was treated with 500 µl crystal violet
staining solution for 20 min at room temperature (control group).
After washing with deionized water 3–5 times, the tumor cells were
observed under a confocal microscope (LSM 900; Carl Zeiss AG) with
magnification ×400. Experiments were performed at least three times
and the results were recorded as the mean of these experiments.
RNA sequence and reverse
transcription-quantitative (RT-q) PCR
Exosomal RNAs were extracted using
TRIzol® (Thermo Fisher Scientific, Inc.) according to
the manufacturer's instructions and miRNAs were extracted by the
RNeasy/miRNeasy Mini kit (Qiagen, Inc.). The amount and quality of
small RNAs in the total RNAs were tested by Heyuan Biotechnology
(Shanghai) Co., Ltd. Small RNA library construction and sequencing
were performed by Heyuan Biotechnology (Shanghai) Co., Ltd. Then,
the cDNA library was sequenced on an Illumina Hiseq 2500 (Illumina,
Inc.). Raw reads were collected using related Illumina analysis
software and RT-qPCR was performed on a CFX96 Real-Time System
(Bio-Rad Laboratories, Inc.) using iTaq Universal One-Step RT-qPCR
kits (Bio-Rad Laboratories, Inc.). The PCR cycling conditions were:
94°C for 5 min, 94°C for 1 min, 55°C for 40 sec, 72°C for 50 sec,
72°C for 7 min and the temperature lowered to 4°C at the end of
each cycles. The cycles were repeated 29 times. The probes and
primers by a web based assay design software (Probe Finder
http://www.roche-applied-science.com)
to identify the expression of MDR-1 (MDR-1-f
5′-GCCATCAGTCCTGTTCTTGG-3′; MDR-1-r 5′-GCTTTTGCATACGCTAAGAGTTC-3′)
and the results were expressed as the ratio between the MDR-1 and
GAPDH according to the 2−ΔΔCq method. RT-qPCR was
repeated three times (11).
Micro (mi)RNA mimic and inhibitor
transfection
MG63 cells and KHOS cells (4×104 cell/ml)
were transfected with 100 nM of the exosomal miRNA mimic (0.16
µM/µl) [Heyuan Biotechnology (Shanghai) Co., Ltd.] and MG63/DXR and
KHOS/DXR were transfected with 100 nM of the exosomal miRNA
inhibitor (0.16 µM/µl) [Heyuan Biotechnology (Shanghai) Co., Ltd.]
using Lipofectamine® 2000 (Invitrogen; Thermo Fisher
Scientific, Inc.). PBS was used as the negative control (NC). The
negative control inhibitor and mimic of the exosomal miRNAs were
used for corresponding negative controls (NC; Table I). The OS cells (4×104
cell/ml) were seeded in 96-well, flat, clear-bottomed, opaque wall
microplates and treated with miRNA mimic or inhibitor (0.16 µM/µl)
for 48 h at the temperature of 36.5°C. After 48 h of transfection,
the four OS cells (4×104 cell/ml) were treated with
doxorubicin (100 ng/ml) for another 24 h at 36.5°C. The effect of
exosomal RNAs on the viability of OS cells was determined using
CellTiter-Glo Luminescent Cell Viability Assay (Promega
Corporation) according to the manufacturer's instructions. The
total ATP content as an estimate of total number of viable cells
was measured on an automatic Fluoroskan Luminometer (Thermo Fisher
Scientific, Inc.).
 | Table I.Sequences of all miRNA mimics, mimic
NC, miRNA inhibitors and inhibitor NC. |
Table I.
Sequences of all miRNA mimics, mimic
NC, miRNA inhibitors and inhibitor NC.
| miRNA | Sequence |
|---|
| miRNA inhibitor
NC | CAG UAC UUU UGU GUA
GUA CAA |
| miRNA mimic NC | UUC UUC GAA CGU GUC
ACG UTT |
|
| ACG UGA CAC GUU CGG
AGA ATT |
| miRNA-143-3p
inhibitor | GAG CUA CAG UGC UUC
AUC UCA |
| miRNA-143-3p
mimic | UGA GAU GAA GCA CUG
UAG CUC |
|
| GCU ACA GUG CUU CAU
CUC AUU |
| miRNA-493-5p
inhibitor | AAU GAA AGC CUA CCA
UGU ACAA |
| miRNA-493-5p
mimic | UUG UAC AUG GUA GGC
UUU CAUU |
|
| UGA AAG CCU ACC AUG
UAC AAUU |
| miRNA-494-3p
inhibitor | UGA AAC AUA CAC GGG
AAA CCU UCU |
| miRNA-494-3p
mimics | UGA AAC AUA CAC GGG
AAA CCU CU |
|
| AGG UUU CCC GUG UAU
GUU UCA UU |
Bioinformatic analysis of the exosomal
miRNAs
According to the exosomal miRNA sequences of MG63
and MG63/DXR, Gene Ontology (GO) analysis of target genes were
conducted using the DAVID database (https://david.ncifcrf.gov/) and the pathway enrichment
analysis for related.
Statistical analysis
The expression levels of exosomal miRNA from RNA
sequence were analyzed by SDS software version 2.2.2 (Applied
Biosystems; Thermo Fisher Scientific, Inc.). R software for Windows
4.1.2 (https://www.r-project.org/) and RStudio
(https://www.rsrudio.com/products/rstudio/download)
were used to draw a heatmap of differentially expressed exosomal
miRNAs and between-group statistical analysis. The results are
presented as the mean ± standard deviation. Statistical
significance between two groups was determined using a two-tailed
Student's t test. P-values were either listed or represented by the
following number of asterisks: *P<5×10−2;
**P<1×10-2; ***P<1×10-3. P<5×10-2 was considered to
indicate a statistically significant difference.
Results
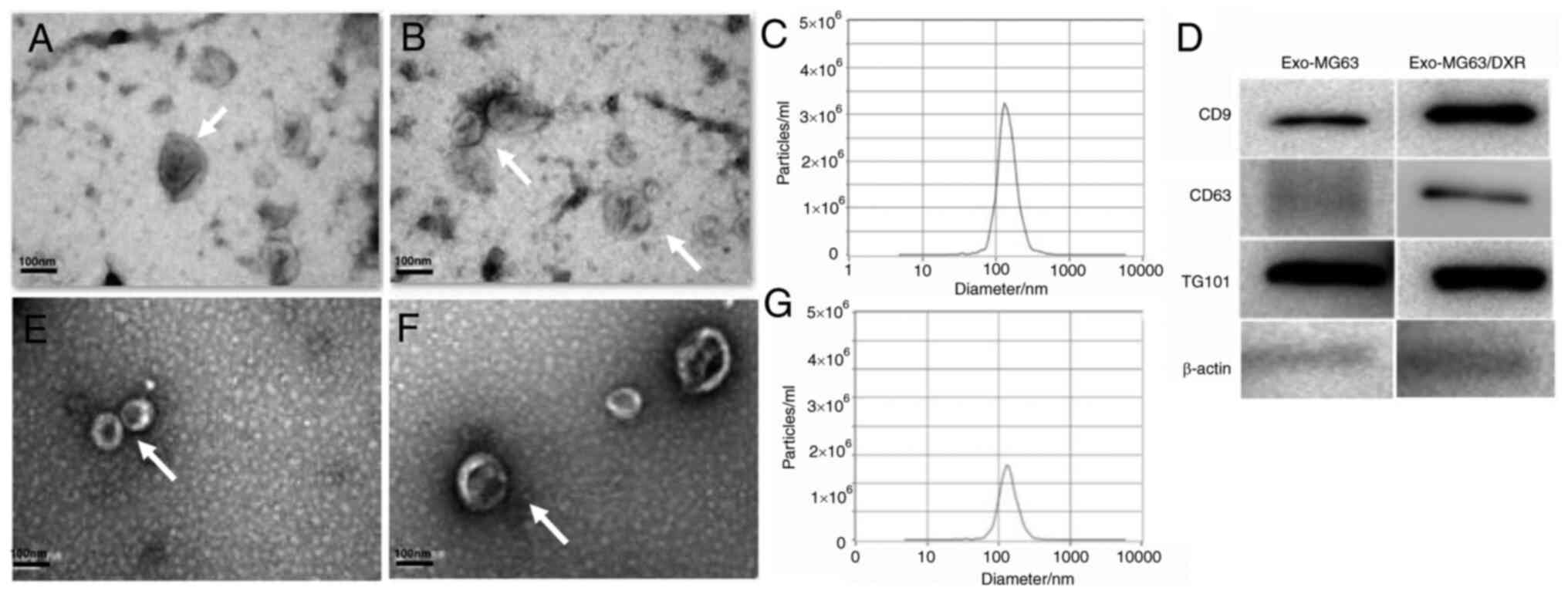
Isolation and identification of
exosomes
First, the exosomes of the MG63 cells and MG63/DXR
cells were isolated and under the TEM, the vesicles from the OS
cells exhibited a cup shape with bilayered membranes and a diameter
ranging from 30–150 nm (Fig. 1A and B,
E and F). The main size of the vesicles was 132.4 and 136.3 nm
and the concentrations were 2.6×106 particles/ml and
3.1×106 particles/ml for MG63 and MG63/DXR cells
respectively as identified by NTA (Fig. 1C and G). In addition, the exosomal
markers CD9, CD63 and TSG101 were detected by western blot analysis
(Fig. 1D). Vesicles isolated from
MG63 cells and MG63/DXR cells displayed typical characteristics of
exosomes and MG63/DXR cells secreted more exosomes than MG63
cells.
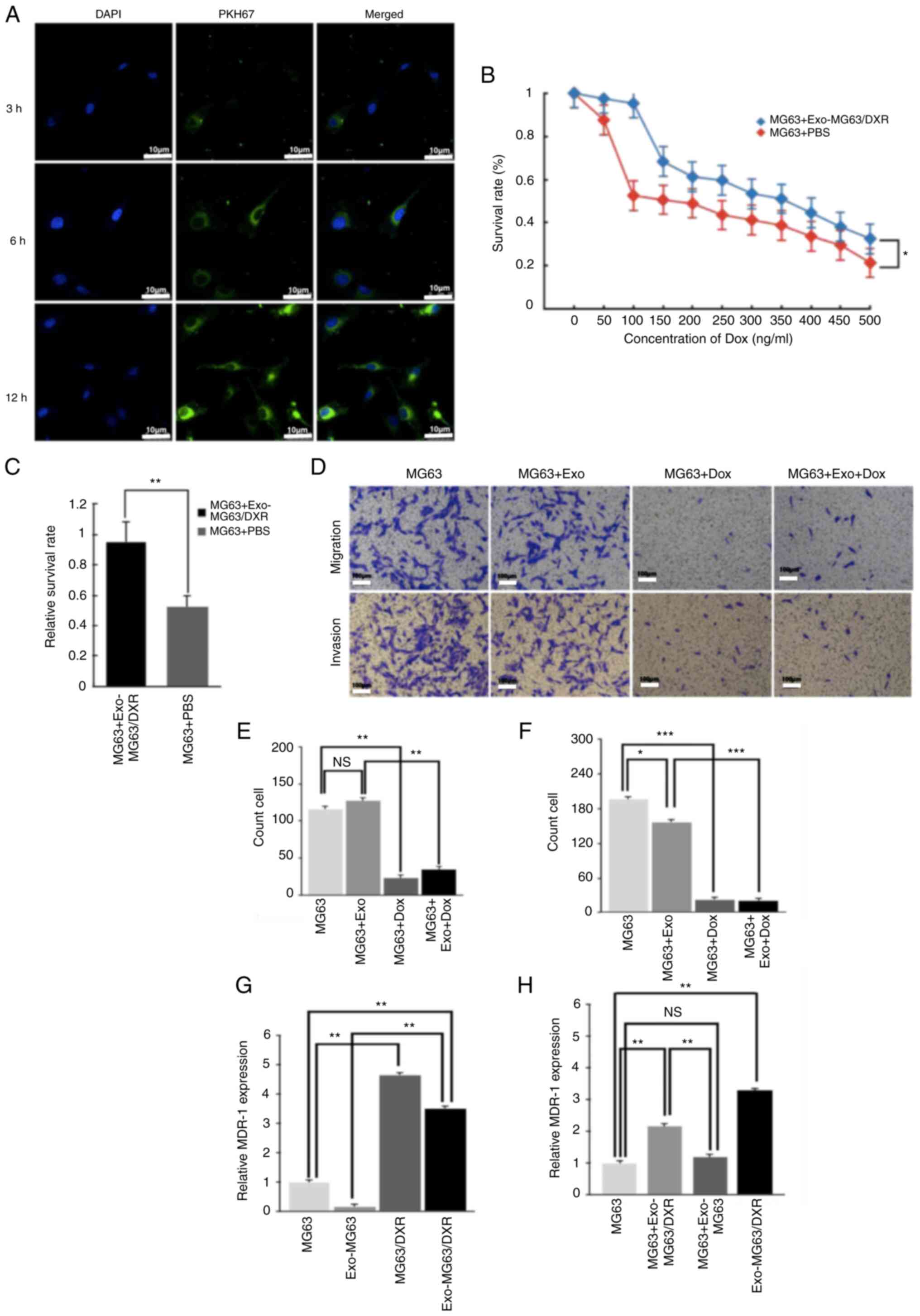
Exosomes labelling and uptake
To examine whether exosomes from MG63/DXR
(Exo-MG63/DXR) could be taken up in MG63 cells, PKH67-labeled
Exo-MG63/DXR were incubated with MG63 cells and examined using
fluorescence microscopy. After 3 h of incubation, the PKH67 signal
was detected in the perinuclear region and an increasing PKH67
signal was observed in the perinuclear region of MG63 cells 6 and
12 h later (Fig. 2A). The results
suggested that the Exo-MG63/DXR was assimilated and internalized by
MG63 cells following incubation.
Influence of Exo-MG63/DXR for MG63
cells on the proliferation, invasion and migration after treatment
with doxorubicin
To investigate the influence of Exo-MG63/DXR on the
proliferation of MG63 cells to doxorubicin, cell viability was
examined in MG63 cells in the presence of increasing concentrations
of doxorubicin (1–1,000 ng/ml) for 24 h after incubation with
Exo-MG63/DXR (100 µg/ml) for 48 h. MG63 viability was affected by
doxorubicin in a dose-dependent way and the resistance of
doxorubicin for MG63 cells was increased following incubation with
Exo-MG63/DXR (Fig. 2B). In
particular, MG63 cells showed a significant increase in viability
after exposed to 100 ng/ml of doxorubicin compared with other
concentration of doxorubicin following incubation with Exo-MG63/DXR
(Fig. 2C). However, the invasion
of MG63 cells was significantly inhibited and the migration of MG63
cells was not affected by the Dox-MG63-Exo while the invasion and
migration of MG63 cells was significantly inhibited at the same
time following treatment with doxorubicin (Fig. 2D-F).
MG63 expressed MDR-1 mRNA after
incubation with Exo-MG63/DXR
The expression of MDR-1 was evaluated by RT-qPCR
after extracting total RNA from Exo-MG63/DXR, exosomes of MG63
(Exo-MG63) and their cells of origin. Exo-MG63/DXR expressed higher
levels of MDR-1 mRNA compared with Exo-MG63. In addition, the
expression of MDR-1 mRNA in Exo-MG63/DXR was significantly higher
compared with that in MG63 cells (Fig.
2G). In addition, following incubation with Exo-MG63/DXR, the
treated MG63 cells expressed higher levels of MDR-1 mRNA compared
with the untreated MG63 cells (Fig.
2H). As expected, there was no difference in the expression of
MDR-1 mRNA in the MG63 cells incubated with Exo-MG63.
Sequence and bioinformatic analysis of
exosomal miRNAs
According to the analysis of the miRNA sequence of
exosomes from MG63 cells and MG63/DXR cells, 2864 differentially
expressed exo-miRNAs were detected and 456 miRNAs were upregulated
and 98 miRNAs were downregulated significantly (fold-change>2.0,
P<5×10−2 and FDR<0.05; Fig. 3A-C). The 10 most up- and
downregulated exo-miRNAs are shown in Table II. The related exo-miRNAs and
pathways involved in the doxorubicin resistance for OS cells were
identified by bioinformatic analysis (Fig. 3D-E). As a result, 20 high-risk
pathways were found according to the KEGG analysis. Among them,
pathways in cancer (P=1.77×10−9), PI3K-Akt signaling
pathway (P=1.94×10−5), proteoglycans in cancer
(P=2.06×10−9), Rap1 signaling pathway
(P=4.86×10−3), Ras signaling pathway
(P=1.71×10−7) and regulation of actin cytoskeleton
(P=1.26×10−5) were the most prominent pathways enriched
in quantiles with different exo-miRNAs in MG63/DXR cells. In
addition, according to the analysis of GO enrichment, protein
binding, membrane, cytosol and cytoplasm were the prominent GO
terms for the differentially expressed exo-miRNAs in MG63/DXR and
MG63 cells.
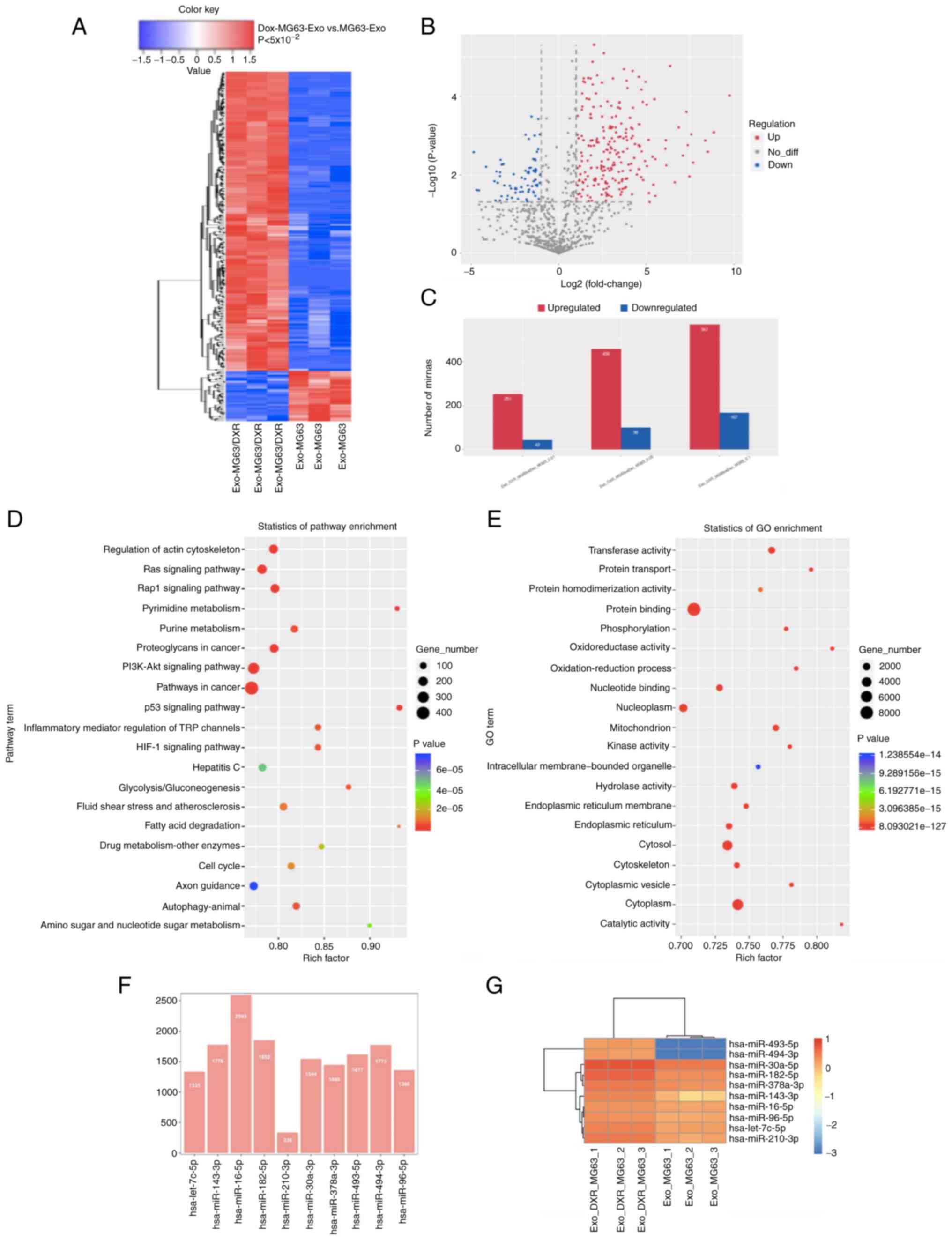 | Figure 3.Exosomal miRNA sequence and
bioinformatic analysis of exosomal miRNAs. (A) Heatmap of different
exosomal miRNA profiles in MG63/DXR and MG63 cell lines. (B)
Volcano map for exosomal miRNAs of the two cell lines according to
the results of exo-miRNA sequence. (C) Among the exosomal miRNAs,
456 were upregulated and 98 were downregulated significantly
(fold-change>2.0, P<0.05 and FDR<0.05) in exosomes. (D and
E) Bioinformatic analysis of the exo-miRNAs by KEGG and GO
enrichment. Pathways in cancer, PI3K-Akt signaling pathway,
Proteoglycans in cancer, Rap1 signaling pathway, Ras signaling
pathway and Regulation of actin cytoskeleton were the most
prominent pathways enriched in quantiles with different exo-miRNAs
in MG63/DXR cells. The protein binding, membrane, cytosol and
cytoplasm were the prominent GO terms for the different expressed
exo-miRNAs in MG63/DXR and MG63 cells. (F) Number of the target
genes and (G) heatmap for the 10 randomly selected exosomal miRNAs.
miRNA, microRNA; KEGG, Kyoto Encyclopedia of Genes and Genomes; GO,
Gene Ontology; Exo, exosomal. |
 | Table II.Ten most up- and downregulated
exo-miRNAs. |
Table II.
Ten most up- and downregulated
exo-miRNAs.
| miR name | miR sequence | Regulation | Fold change | P-values |
|---|
| hsa-miR-494-3p |
TGAAACATACACGGGAAACCTCT |
Up | inf |
8.53×10−4 |
| hsa-miR-493-5p |
TTGTACATGGTAGGCTTTCATT |
Up | inf |
1.06×10−3 |
| hsa-let-7c-5p |
TGAGGTAGTAGGTTGTATGGTT |
Up | 4.05 |
4.86×10−6 |
| hsa-miR-210-3p |
CTGTGCGTGTGACAGCGGCTGA |
Up | 8.57 |
3.84×10−5 |
| hsa-miR-182-5p |
TTTGGCAATGGTAGAACTCACACCG |
Up | 8.04 |
1.76×10−4 |
| hsa-miR-30a-5p |
TGTAAACATCCTCGACTGGAAGCT |
Up | 5.30 |
1.79×10−4 |
| hsa-miR-25-3p |
CATTGCACTTGTCTCGGTCTGA |
Up | 5.58 |
3.27×10−4 |
| hsa-miR-143-3p |
TGAGATGAAGCACTGTAGCTC |
Up | 24.20 |
5.21×10−4 |
| hsa-miR-183-5p |
TATGGCACTGGTAGAATTCACT |
Up | 7.53 |
6.43×10−4 |
| hsa-miR-34c-5p |
AGGCAGTGTAGTTAGCTGATTGC |
Up | 190.42 |
9.40×10−4 |
|
hsa-miR-199a-5p_R-1 |
CCCAGTGTTCAGACTACCTGTT | Down | 0.44 |
2.19×10−3 |
| hsa-miR-152-3p |
TCAGTGCATGACAGAACTTGG | Down | 0.42 |
2.59×10−3 |
| hsa-miR-185-5p |
TGGAGAGAAAGGCAGTTCCTGA | Down | 0.39 |
4.00×10−3 |
|
hsa-miR-23a-3p_R-1 |
ATCACATTGCCAGGGATTTC | Down | 0.33 |
6.58×10−3 |
|
hsa-miR-16-2-3p_L+1R-1 |
ACCAATATTACTGTGCTGCTTT | Down | 0.30 |
8.30×10−3 |
|
hsa-miR-27a-3p_R-1 |
TTCACAGTGGCTAAGTTCCG | Down | 0.31 |
8.95×10−3 |
|
hsa-miR-24-3p_R-2 |
TGGCTCAGTTCAGCAGGAAC | Down | 0.49 |
9.61×10−3 |
|
hsa-miR-146a-5p |
TGAGAACTGAATTCCATGGGTT | Down | 0.26 |
1.25×10−2 |
|
hsa-miR-148a-3p |
TCAGTGCACTACAGAACTTTGT | Down | 0.08 |
1.44×10−2 |
| hsa-miR-423-5p |
TGAGGGGCAGAGAGCGAGACTTT | Down | 0.43 |
1.79×10−2 |
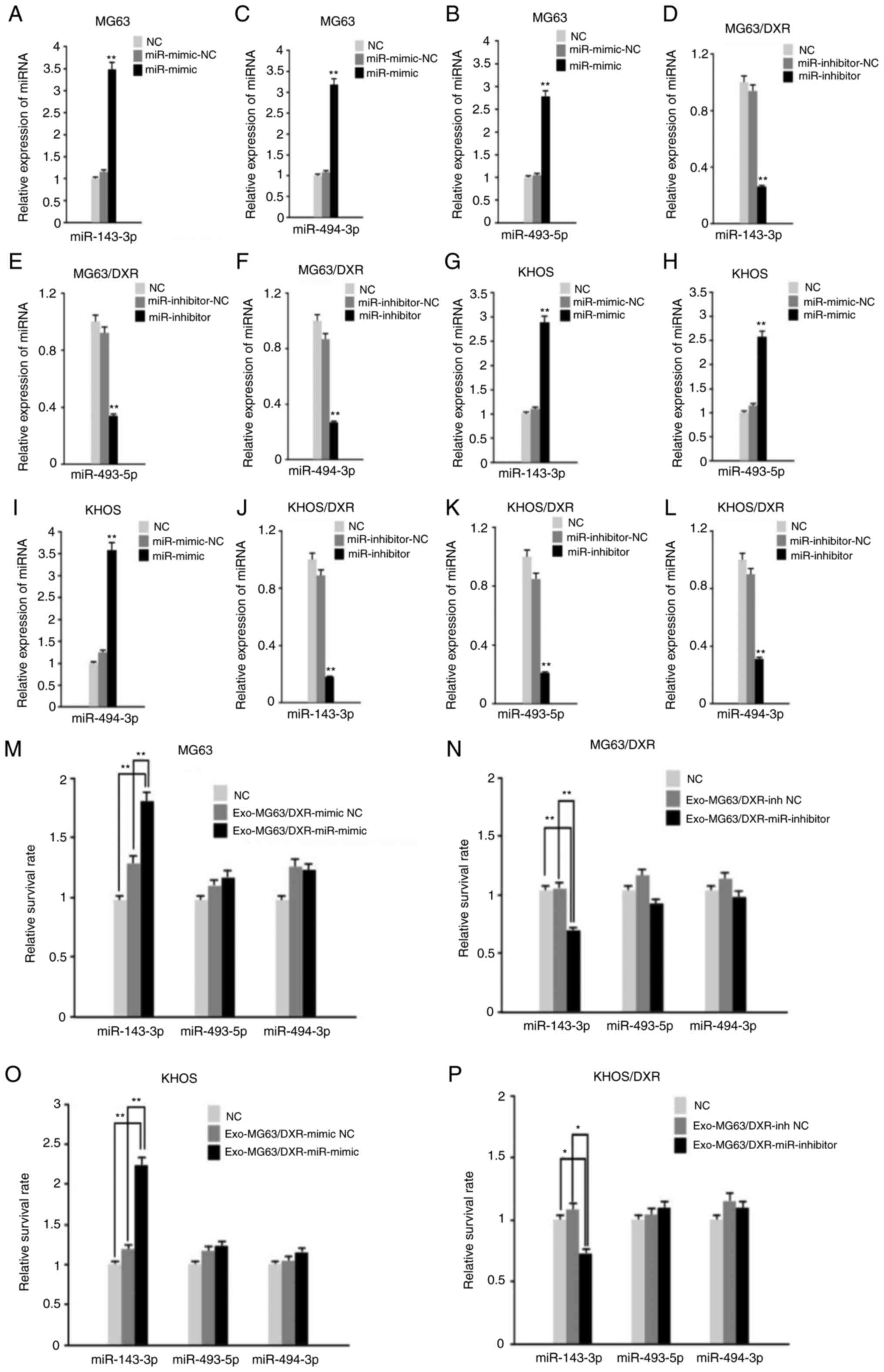
Validation of different exosomal
miRNAs confer doxorubicin resistance to OS cells
Based on the statistical significance and biological
plausibility, 10 miRNAs, including miR-30a-5p, miR-16-5p,
miR-96-5p, let-7c-5p, miR-182-5p, miR-210-3P, miR-378a-3P,
miR-493-5p, miR-494-3p and miR-143-3p, were selected for validation
from the exo-miRNA sequence by RT-qPCR in four OS cells including
MG63 cells, MG63/DXR cells, KHOS cells and KHOS/DXR cells (Table III). The heatmap and the number
of the target genes of the exo-miRNAs are shown in Fig. 3F and G. A total of 10 randomly
exosomal miRNAs, including miR-30a-5p, miR-16-5p, miR-96-5p,
let-7c-5p, miR-182-5p, miR-210-3P, miR-378a-3P, miR-493-5p,
miR-494-3p and miR-143-3p, were selected for validation by TaqMan
RT-qPCR in four OS cells. According to the result of RT-qPCR, the
expression of miR-143-3p was significantly increased in exosomes
from MG63/DXR cells and KHOS/DXR cells compared with MG63 cells and
KHOS cells and miR-493-5p and miR-494-3p were the two most
significantly differentially expressed miRNAs between MG63 cells
and MG63/DXR cells (Fig. 4A-J). To
investigate the function of the three exo-miRNAs in OS cells, MG63
cells and KHOS cells were infected with lentiviral vectors
expressing mimic of exosomal miR-143-3p, miR-493-5p and miR-494-3p
and the MG63/DXR cells and KHOS/DXR cells were infected with
lentiviral vectors expressing inhibitor of the three miRNAs. The
negative control mimic and inhibitor of the three miRNAs were used
as negative controls (NC). The transfection for each of the three
miRNA mimics in MG63 cells and KHOS cells and inhibitors in
MG63/DXR cells and KHOS/DXR cells are shown separately in Fig. 5A-L. After 48 h of incubation of the
infected exosomes, the four OS cells were treated with doxorubicin
(100 ng/ml) for another 24 h (Fig.
5M-P). The results of cell viability evaluated by CellTiter-Glo
indicated that exosomal miR-143-3p mimic significantly increased
the doxorubicin resistance for the MG63 cells and KHOS cells
compared with the NC group while the exosomal miR-493-5p mimic and
exosomal miR-494-3p mimic did not perform a similar function in the
two OS cell lines. In addition, the exosomal miR-143-3p inhibitor
reduced the doxorubicin resistance for the MG63/DXR cells and
KHOS/DXR cells according to the result of cell viability compared
with the NC group (Fig. 5M-P). As
a result, the present study found upregulation of exosomal
miR-143-3p led to poor chemotherapeutic response to osteosarcoma,
highlighting the importance of miR-143-3p as an oncogene in
chemotherapy for osteosarcoma.
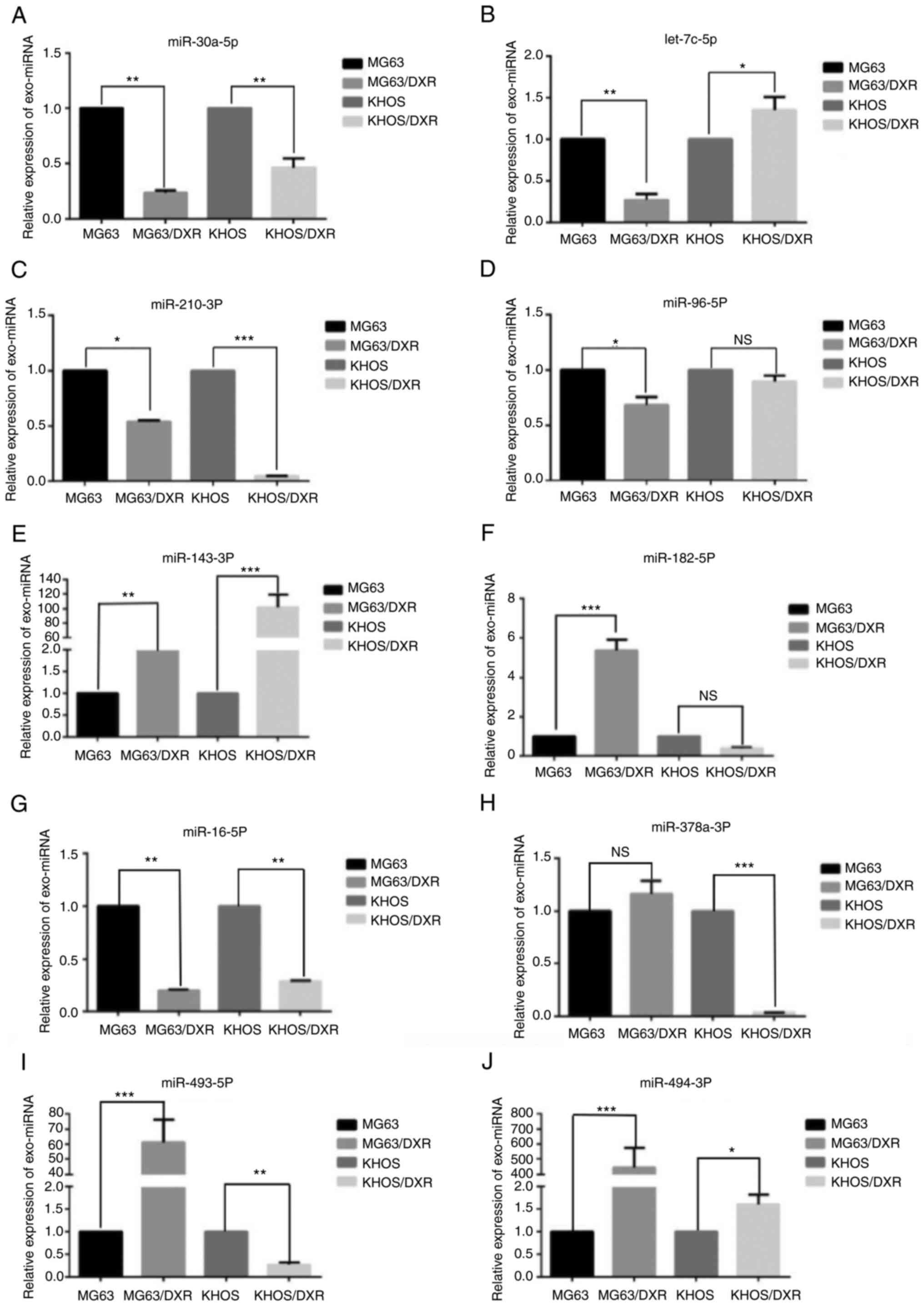 | Figure 4.Validation of different exo-miRNAs
confer doxorubicin resistance to OS cells by RT-qPCR. A total of 10
randomly exosomal miRNAs, including (A) miR-30a-5p, (B) let-7c-5p,
(C) miR-210-3P, (D) miR-96-5p, (E) miR-143-3p, (F) miR-182-5p, (G)
miR-16-5p, (H) miR-378a-3P, (I) miR-493-5p and (J) miR-494-3p were
selected for validation by TaqMan RT-qPCR in four OS cells.
*P<0.05, **P<0.01 and ***P<0.001. miRNA, microRNA; OS,
osteosarcoma; RT-qPCR, reverse transcription-quantitative PCR; NS,
not significant. |
 | Table III.Ten randomly selected miRNAs for
validation from exo-miRNA sequence by TaqMan reverse
transcription-quantitative PCR. |
Table III.
Ten randomly selected miRNAs for
validation from exo-miRNA sequence by TaqMan reverse
transcription-quantitative PCR.
| miR name | miR sequence | Regulation | Fold change | P-values | KEGG name | GO name | GO function |
|---|
| miR-30a-5p |
TGTAAACATCCTCGACTGGAAGCT |
Up | 5.30 |
1.79×10−4 | Pathways in
cancer | Nucleoplasm | Cellular
component |
| miR-16-5p |
TAGCAGCACGTAAATATTGGCG |
Up | 2.55 |
1.83×10−4 | MicroRNAs in
cancer | Nucleotide
binding | Cellular
component |
| miR-96-5p |
TTTGGCACTAGCACATTTTTGCT |
Up | 3.83 |
1.32×10−5 | MicroRNAs in
cancer | Nucleoplasm | Cellular
component |
| let-7c-5p |
TGAGGTAGTAGGTTGTATGGTT |
Up | 4.05 |
4.86×10−6 | MicroRNAs in
cancer | Nucleoplasm | Cellular
component |
| miR-182-5p |
TTTGGCAATGGTAGAACTCACACCG |
Up | 8.04 |
1.76×10−4 | Hippo signaling
pathway | Nucleoplasm | Cellular
component |
| miR-210-3P |
CTGTGCGTGTGACAGCGGCTGA |
Up | 8.57 |
3.84×10−5 | Ras signaling
pathway | Nucleoplasm | Cellular
component |
| miR-378a-3P |
ACTGGACTTGGAGTCAGAAGGC |
Up | 3.56 |
3.47×10−5 | Ras signaling
pathway | Protein
binding | Molecular
function |
| miR-493-5p |
TTGTACATGGTAGGCTTTCATT |
Up | inf |
1.06×10−3 | p53 signaling
pathway | Protein
binding | Molecular
function |
| miR-494-3p |
TGAAACATACACGGGAAACCTCT |
Up | inf |
8.53×10−4 | Proteoglycans in
cancer | Protein
binding | Molecular
function |
| miR-143-3p |
TGAGATGAAGCACTGTAGCTC |
Up | 24.20 |
5.21×10−4 | Pathways in
cancer | Protein
binding | Molecular
function |
Discussion
Multidrug resistance (MDR) is a major concern
regarding the clinical management of osteosarcoma patients and a
key issue in the failure of current treatment (4). Exosomes have been reported to serve
an increasingly important role in different stages of tumor
progression and chemotherapy resistance (5–7). The
present study reported that exosomes derived from
doxorubicin-resistant OS cells are able to transfer phenotypic
characteristics to doxorubicin-sensitive OS cells. According to the
results of exo-miRNA sequence and bioinformatic analysis of the
differentially expressed exo-miRNAs from doxorubicin-resistant OS
cells and doxorubicin-sensitive OS cells, the present study found a
substantial profile of exo-miRNAs was differentially expressed in
OS cell lines with different chemotherapeutic response. Notably,
the results for validation of different exosomal miRNAs by RT-qPCR
indicated that the expression level of miR-143-3p was significantly
different in the exosomes of OS cell lines with different
chemotherapeutic responses and upregulation of exosomal
miRNA-143-3p abundance can induce a poor chemotherapeutic response
in OS cells. These results could help monitor or predict disease
progression during chemotherapeutic treatment of OS.
Exosomes facilitate cell-cell crosstalk within the
tumor environment, which serves a crucial role in augmenting MDR
pathways (12,13). Santos et al (14) report that breast cancer stem cells
and doxorubicin- and paclitaxel-resistant breast cells can secrete
more exosomes than parental cells. Fang et al (15) found that liver cancer cells with
high metastatic potential secreted more exosomes than those with
low metastatic potential. The above findings are consistent with
the findings of the present study that doxorubicin-resistant OS
cells release more exosomes than doxorubicin-sensitive OS cells.
The phenomenon that exosomes released by drug resistant cells can
mediate the acquired MDR of drug-sensitive cancer cells is observed
in ovarian cancer (16), prostate
cancer (17), breast cancer
(18) and melanoma (19). In the present study, this
phenomenon was also observed in OS cells, including the capacity
for doxorubicin resistance and the presence of selective MDR-1
mRNA, except for the invasion and migration of OS cells, which need
further investigation in the future.
A number of studies have identified that exosomes
have the ability to transport molecular information such as
proteins, mRNAs and miRNAs from one cell to another to induce
chemoresistance and malignant phenotypic traits (20–23).
Exosomal miR-126a, miR-222-3p, miR-32-5p and miR-222 are reported
to be involved in the MDR of malignant tumors (20–23).
The present study provided the first results of the miRNA sequence
for different exosomal miRNAs expression in doxorubicin-sensitive
and doxorubicin-resistant OS cells. In addition, according to the
results of bioinformatic analysis and validation of differential
exosomal miRNAs, a substantial profile of different expressed
exosomal miRNAs was found in OS cell lines with different
chemotherapeutic response.
miR-143 is located on human chromosome 5. Depending
on the different site of cleavage, the stem-loop structure of
miR-143 precursor can form miR-143-3p and miR-143-5p (24). Previous reports indicate that the
expression levels of miR-143 is associated with clinical stage,
disease grade and lymph node metastasis (25,26).
Regulating the expression of miR-143-3p inhibits or promotes the
cell proliferation, migration and invasion in hepatocellular
carcinoma and triple negative breast cancer (27,28).
The expression level of miR-143-3p is significantly decreased for
OS cells and serum of OS patients (29). The present study was the first, to
the best of the authors' knowledge, to report on the expression of
exosomal miR-143-3p for OS cells with different chemotherapeutic
response. It was found that miR-143-3p was highly expressed in
exosomes from doxorubicin-resistant OS cells and that upregulation
of exosomal miR-143-3p abundance can induce a poor chemotherapeutic
response to osteosarcoma, highlighting the importance of miR-143-3p
as an oncogene in osteosarcoma and providing new insights into
chemotherapy for osteosarcoma. In 2005, Zhang et al
(30) reported that forced miR-143
expression significantly reversed chemoresistance, which was
different from the result of the present study. However, the
present study investigated the miRNAs which could be stably present
in exosomes and the expression level of miRNA-143 may be different
in the exosomes and OS cells; the present study focused on the
function of exosomal miRNA in chemoresistance in OS cells.
Furthermore, depending on the different site of cleavage, the
stem-loop structure of miR-143 precursor can form miR-143-3p and
miR-143-5p (29). The present
study studied the function of exosomal miRNA143-3p in the
chemoresistance of OS cells and exosomal miR-143-3p induced the
doxorubicin-resistant phenotype according to the PI3K/mTOR pathway
and the key protein in this pathway will be reported in the
future.
There are some limitations to the present study.
First, multivariate analysis, such as Fisher discriminant analysis,
could be further applied to explore the differential role of miRNAs
in the chemotherapeutic response. Second, animal experiments need
to be conducted to support the conclusion of the present study.
Last but not least, the investigation of the different samples from
different OS patients, such as serum of the patients and the
specimen of the osteosarcoma, need to be performed to prove the
results of the present study.
To conclude, the present study corroborated the
evidence that exosomes from doxorubicin-resistant osteosarcoma
cells are capable of transferring chemoresistant phenotypic traits
and MDR-1, a specific mRNA of chemoresistance. In addition, it
revealed a substantial abundance of differentially expressed miRNAs
present in the exosomes from OS cells with the different
chemotherapeutic response. Importantly, the present study found
that the upregulation of exosomal miR-143-3p abundance was
associated with poor chemotherapeutic response to osteosarcoma
cells, which was highly expressed in doxorubicin-resistant OS
cells, highlighting the importance of miR-143-3p as an oncogene in
osteosarcoma; this may provide new insights into chemotherapy of
osteosarcoma.
Acknowledgements
Not applicable.
Funding
The present study was supported by grants from the National
Natural Science Foundation of China (grant nos. 81872174 and
82072963), Program of Shanghai Academic Research Leader (grant no.
19XD1402900) and Program of Shanghai Sailing Program (grant no.
20YF1437700).
Availability of data and materials
The datasets used and/or analyzed during the current
study are available from the corresponding author on reasonable
request.
Authors' contributions
TC contributed to the conception and design of the
study, analysis and interpretetion of the data. TZ contributed to
the drafting of the manuscript and acquisition of data. CZ
contributed to the design of the study, gave final approval of the
version to be published and agreed to be accountable for all aspect
of work. TC and CZ confirm the authenticity of all the raw data.
All authors read and approved the final manuscript.
Ethics approval and consent to
participate
The the study was approved by the Institutional
Review Board of Tongji University (Shanghai, China) and was
performed in accordance with the ethical standards prescribed by
the Helsinki Declaration.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
Authors' information
Tao Cai was an undergraduate medical student in the
Department of Orthopedic Surgery, Shanghai Tenth People's Hospital
Affiliated To Tongji University when the article was submitted.
Now, Dr Tao Cai is a surgeon of the Department of Orthopedic
Surgery, Tongji Hospital Affiliated to Tongji University.
References
|
1
|
Miller RW: Contrasting epidemiology of
childhood osteosarcoma, Ewing's tumor, and rhabdomyosarcoma. Natl
Cancer Inst Monogr. 56:9–15. 1981.PubMed/NCBI
|
|
2
|
Ottaviani G and Jaffe N: The Epidemiology
of Osteosarcoma. Cancer Treat Res. 152:3–13. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Provisor AJ, Ettinger LJ, Nachman JB,
Krailo MD, Makley JT, Yunis EJ, Huvos AG, Betcher DL, Baum ES,
Kisker CT and Miser JS: Treatment of nonmetastatic osteosarcoma of
the extremity with preoperative and postoperative chemotherapy: A
report from the Children's Cancer Group. J Clin Oncol. 15:76–84.
1997. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Gianferante DM, Mirabello L and Savage SA:
Germline and somatic genetics of osteosarcoma-connecting aetiology,
biology and therapy. Nat Rev Endocrinol. 13:480–491. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Milane L, Singh A, Mattheolabakis G,
Suresh M and Amiji MM: Exosome mediated communication within the
tumor microenvironment. J Control Release. 219:278–294. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Teng X, Chen L, Chen W, Yang J, Yang Z and
Shen Z: Mesenchymal stem cell-derived exosomes improve the
microenvironment of infarcted myocardium contributing to
angiogenesis and anti-inflammation. Cell Physiol Biochem.
37:2415–2424. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Baixauli F, López-Otín C and Mittelbrunn
M: Exosomes and autophagy: Coordinated mechanisms for the
maintenance of cellular fitness. Front Immunol. 5:4032014.
View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Nair R, Santos L, Awasthi S, von Erlach T,
Chow LW, Bertazzo S and Stevens MM: Extracellular vesicles derived
from preosteoblasts influence embryonic stem cell differentiation.
Stem Cells Dev. 23:1625–1635. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Tkach M and Théry C: Communication by
extracellular vesicles: Where we are and where we need to go. Cell.
164:1226–1232. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Zhao L, Liu W, Xiao J and Cao B: The role
of exosomes and ‘exosomal shuttle microRNA’ in tumorigenesis and
drug resistance. Cancer Lett. 356((2 Pt B)): 339–346. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(−Delta Delta C(T)) method. Methods. 25:402–408. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Chen WX, Liu XM, Lv MM, Chen L, Zhao JH,
Zhong SL, Ji MH, Hu Q, Luo Z, Wu JZ and Tang JH: Exosomes from
drug-resistant breast cancer cells transmit chemoresistance by a
horizontal transfer of microRNAs. PLoS One. 9:e952402014.
View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Robbins PD and Morelli AE: Regulation of
immune responses by extracellular vesicles. Nat Rev Immunol.
14:195–208. 2014. View
Article : Google Scholar : PubMed/NCBI
|
|
14
|
Santos JC, Lima NDS, Sarian LO, Matheu A,
Ribeiro ML and Derchain SFM: Exosome-mediated breast cancer
chemoresistance via miR-155 transfer. Sci Rep. 8:8292018.
View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Fang T, Lv H, Lv G, Li T, Wang C, Han Q,
Yu L, Su B, Guo L, Huang S, et al: Tumor-derived exosomal
miR-1247-3p induces cancer-associated fibroblast activation to
foster lung metastasis of liver cancer. Nat Commun. 9:1912018.
View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Safaei R, Larson BJ, Cheng TC, Gibson MA,
Otani S, Naerdemann W and Howell SB: Abnormal lysosomal trafficking
and enhanced exosomal export of cisplatin in drug-resistant human
ovarian carcinoma cells. Mol Cancer Ther. 4:1595–1604. 2005.
View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Corcoran C, Rani S and O'Driscoll L:
miR-34a is an intracellular and exosomal predictive biomarker for
response to docetaxel with clinical relevance to prostate cancer
progression. Prostate. 74:1320–1334. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Lv MM, Zhu XY, Chen WX, Zhong SL, Hu Q, Ma
TF, Zhang J, Chen L, Tang JH and Zhao JH: Exosomes mediate drug
resistance transfer in MCF-7 breast cancer cells and a probable
mechanism is delivery of P-glycoprotein. Tumour Biol.
35:10773–10779. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Federici C, Petrucci F, Caimi S, Cesolini
A, Logozzi M, Borghi M, D'Ilio S, Lugini L, Violante N, Azzarito T,
et al: Exosome release and low pH belong to a framework of
resistance of human melanoma cells to cisplatin. PLoS One.
9:e881932014. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Yu D, Wu Y, Zhang X, Lv MM, Chen WX, Chen
X, Yang SJ, Shen H, Zhong SL, Tang JH and Zhao JH: Exosomes from
adriamycin-resistant breast cancer cells transmit drug resistance
partly by delivering miR-222. Tumour Biol. 37:3227–3235. 2016.
View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Deng Z, Rong Y, Teng Y, Zhuang X,
Samykutty A, Mu J, Zhang L, Cao P, Yan J, Miller D and Zhang HG:
Exosomes miR-126a released from MDSC induced by DOX treatment
promotes lung metastasis. Oncogene. 36:639–651. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Wei F, Ma C, Zhou T, Dong X, Luo Q, Geng
L, Ding L, Zhang Y, Zhang L, Li N, et al: Exosomes derived from
gemcitabine-resistant cells transfer malignant phenotypic traits
via delivery of miRNA-222-3p. Mol Cancer. 16:1322017. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Fu X, Liu M, Qu S, Ma J, Zhang Y, Shi T,
Wen H, Yang Y, Wang S, Wang J, et al: Exosomal microRNA-32-5p
induces multidrug resistance in hepatocellular carcinoma via the
PI3K/Akt pathway. J Exp Clin Cancer Res. 37:522018. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Sun X, Dai G, Yu L, Hu Q, Chen J and Guo
W: miR-143-3p inhibits the proliferation, migration and invasion in
osteosarcoma by targeting FOSL2. Sci Rep. 8:6062018. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Wang H, Li Q, Niu X, Wang G, Zheng S, Fu G
and Wang Z: miR-143 inhibits bladder cancer cell proliferation and
enhances their sensitivity to gemcitabine by repressing IGF-1R
signaling. Oncol Lett. 13:435–440. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Wang L, He J, Xu H, Xu L and Li N: MiR-143
targets CTGF and exerts tumour-suppressing functions in epithelial
ovarian cancer. Am J Transl Res. 8:2716–2726. 2016.PubMed/NCBI
|
|
27
|
Chen L, Yao H, Wang K and Liu X: Long
non-coding RNA MALAT1 regulates ZEB1 expression by sponging
miR-143-3p and promotes hepatocellular carcinoma progression. J
Cell Biochem. 118:4836–4843. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Li D, Hu J, Song H, Xu H, Wu C, Zhao B,
Xie D, Wu T, Zhao J and Fang L: miR-143-3p targeting LIM domain
kinase 1 suppresses the progression of triple-negative breast
cancer cells. Am J Transl Res. 9:2276–2285. 2017.PubMed/NCBI
|
|
29
|
Yang L, Li H and Huang A: MiR-429 and
MiR-143-3p function as diagnostic and prognostic markers for
osteosarcoma. Clin Lab. 66:2020. View Article : Google Scholar
|
|
30
|
Zhou J, Wu S, Chen Y, Zhao J, Zhang K,
Wang J and Chen S: microRNA-143 is associated with the survival of
ALDH1+CD133+ osteosarcoma cells and the chemoresistance of
osteosarcoma. Exp Biol Med (Maywood). 240:867–875. 2015. View Article : Google Scholar : PubMed/NCBI
|