Introduction
The unexpected outbreak of severe acute respiratory
syndrome coronavirus 2 (SARS-CoV-2) has highlighted that the world
was unprepared for a global pandemic of such a catastrophic nature.
The emergence and recurrence of viral outbreaks, including severe
acute respiratory syndrome coronavirus (SARS-CoV), Middle East
respiratory syndrome corona virus, and SARS-CoV-2, have
demonstrated the need to develop diagnostics and effective
antivirals that can be used to alleviate the disease burden.
Current diagnostic and antiviral development approaches are
insufficient to control the onset of diseases at an early stage. In
particular, in current technologies, the limitations include cost,
processing time and the requirement for sophisticated equipment. In
addition, diagnostic reagents, testing infrastructure and trained
staff are under pressure when demand is high during the peak of a
global pandemic (1–3). In the future, more outbreaks of human
pathogens are expected due to the increasing access to wild
habitats through man-made encroachments. Owing to the increased
contact between wildlife and humans, the likelihood of future virus
outbreaks is very high (4). There
are only ~25 approved vaccines and effective antivirals to treat
known viruses (2). Therefore,
without developing efficient diagnostic approaches to detect
emerging and recurrent viruses, and effective antiviral agents for
early treatment in the initial stages of outbreaks, viruses will
continue to pose a significant threat to human health (2).
The presence of numerous viruses as reservoirs in
wild animal species, such as bats with pandemic potential,
complicates preparation. The advent and success of modern genomics
have accelerated the identification of new microbes, including
viruses, in animal or insect species. A number of these viruses
have pandemic or epidemic potential and can cause human diseases
while infecting others (4–7). There is high sequence diversity among
viruses; for example, there are three types of influenza (A, B and
C). Within Influenza A, the virus has subtypes, including 18
hemagglutinins and 11 neuraminidases (8,9).
Viruses have evolved at an increased rate because of higher
mutation acquisition rates than that in humans. Mutation
acquisition can render a virus tolerant to its host, cause
acquisition of changes in diagnostic targets or dropout and
development of resistance to therapy, as seen in SARS-CoV-2 variant
B1.1.7, with the 69/70 mutation, which originated in the United
Kingdom (2,10,11).
Consequently, there is uncertainty regarding the virus species and
variants in circulation during pandemics or epidemics. It is
necessary that diagnostics and therapy do not require in-depth
knowledge of viral biology and that the technologies for detecting
viral mutations are robust and can be easily modified if
necessary.
Molecular diagnostic tools based on the polymerase
chain reaction (PCR) are considered the gold standard in
traditional diagnostics. Programmability and usability are marked
concerns associated with the traditional PCR-based diagnostic
approaches. Although PCR-based assays require only sequence
information to design primers or probes, laboratory infrastructure
and requirements for trained personnel limit their use outside a
well-equipped laboratory. Antigen-based tests offer alternatives
that are easy to use but have lower specificity and sensitivity
compared with the gold standard of PCR-based tests and are much
slower to design and develop (1).
The development of effective antiviral drugs requires an
understanding of the viral and host target proteins. The utility of
numerous small-molecule inhibitors has led to the approval of ~90
antiviral drugs for human use; however, identifying small molecules
and reusing previously used inhibitors is a lengthy and
time-consuming process (12).
Recent global proteomics studies have reported novel SARS-CoV-2
targets that could be used to develop new treatment regimens
against the virus. These studies provide new targets that can also
be used to identify potent small molecules or to reuse known
molecules with potent antiviral activity without in-depth knowledge
of SARS-CoV-2 biology (13–16).
Numerous nucleic acid-based antiviral drugs, including small
interfering RNAs, have been successfully tested in cell cultures
and animal models. Unfortunately, none of these drugs have been
approved for treating viral diseases (17,18).
Therefore, the development of technologies that only
require sequence information will improve future virus detection.
One of the reported components of the bacterial adaptive immune
system, CRISPR-Cas systems, offers opportunities for the rapid
development of virus diagnostics. The successful use of CRISPR-Cas
systems in genetic engineering and the treatment of certain
hereditary human diseases has been previously reported (19). Furthermore, the continuous
detection of new CRISPR-Cas systems and orthologous Cas proteins,
and their in-depth molecular insights have expanded the scope of
CRISPR-Cas systems in the urgent development of modern diagnostics
(19,20). CRISPR-Cas systems have been used to
develop virus diagnostics, including for the recently emerged
SARS-CoV-2 (21). A number of
these have been used on an emergency basis and may, in the future,
be developed as point-of-care (POC) devices and as easy-to-use
diagnostic tests for numerous human diseases (2).
CRISPR-Cas systems provide tools for modern
diagnostics
In modern diagnostics, viral nucleic acids are
detected using PCR. PCR can be either qualitative or quantitative,
and is considered the gold standard (22). Reverse transcription-quantitative
PCR (RT-qPCR) is commonly used when quantification is required.
RT-qPCR only requires knowledge of the viral genome sequence.
However, the cost, time from sample to reaction, need for trained
personnel and expensive laboratory equipment are significant
barriers to using RT-qPCR for virus diagnostics (23).
In addition, certain other methods have been
developed with advantages and limitations in performance,
sensitivity, specificity, multiplexing ability, readouts and test
throughput, which eliminating the need for costly thermal cycling
or PCR, such as isothermal amplification which can be used instead
of PCR (24,25). The most commonly used isothermal
amplification methods are loop-mediated isothermal amplification
(LAMP), recombinase polymerase amplification (RPA), nucleic acid
sequence-based amplification (NASBA) and nicking enzyme
amplification reaction (26–30).
CRISPR-based diagnostic tests are versatile and can
detect viruses that contain DNA or RNA genomes. Several Cas
proteins, reaction conditions, amplification methods and readouts
have been used to demonstrate their versatility for diagnosing
viral diseases (31–35) (Tables
I and II). CRISPR-based
diagnostics complement current diagnostic technologies because
their sensitivity and specificity are similar to those of PCR
methods. CRISPR-based tests are also easily adapted for
fluorescence-based detection or visual displays using lateral flow
assays (36–38).
 | Table I.Cas12 and Cas13 orthologues in
clustered regularly interspaced short palindromic
repeats-diagnostics. |
Table I.
Cas12 and Cas13 orthologues in
clustered regularly interspaced short palindromic
repeats-diagnostics.
| Essential
features | Cas12 | Cas13 |
|---|
| Requirement of
PAM | Required | Not required |
| Identity of
PAM | TTTV | Not applicable |
| Cleavage type | Single staggered
cut | Multiple cut
sites |
| Type of target | ssDNA, dsDNA | ssRNA |
| Collateral cleavage
activity | Present | Present |
 | Table II.Comparative features of Cas13 and
Cas12 orthologs. |
Table II.
Comparative features of Cas13 and
Cas12 orthologs.
| Origin of Cas
enzymes and associated essential features | Cas enzymes |
|---|
|
|---|
| LwaCas13a | LbaCas13a | CcaCas13b | PsmCas13b | AsCas12a |
|---|
| Organism | Leptotrichia
wadei | Lachnospiraceae
bacterium NK4A179 | Capnocytophaga
canimorsus | Prevotella sp.
MA2016 |
Acidaminococcus sp. BV3L6 |
| Target | ssRNA | ssRNA | ssRNA | ssRNA | ssDNA/dsDNA |
| Direct repeat
orientation | 5′ | 5′ | 3′ | 3′ | 5′ |
| Motif
preference | Poly U/AU | Poly A/AC | Poly U/UA/UC | Poly A/GA | Not applicable |
| Spacer length,
nt | 28 | 28 | 30 | 30 | 20 |
| Sensitivity,
aM |
~5×105 |
~1×109 |
~5×106 |
~5×108 |
~5×1010 |
Cas proteins recognize target
double-stranded DNA (dsDNA) and RNA mediated by single-stranded RNA
(ssRNA)
CRISPR is organized into CRISPR arrays containing
direct repeats (DRs) and spacer sequences (short sequences of
viruses or plasmids) (39,40). The Cas proteins responsible for
enzymatic activity are located near CRISPR arrays. CRISPR-Cas
systems are divided into two classes, six types and numerous
sub-types. Only class 2 Cas proteins are used in viral diagnostics
or therapy because a single subunit protein with multiple domains
acts as the effector required for nucleic acid cleavage, whereas
multiple subunits are required in class 1. The Cas effectors Cas9,
Cas12, and Cas13 interfere with target nucleic acids when complexed
with mature CRISPR RNA (crRNA), also called guide RNA (gRNA)
(Fig. 1). Each Cas effector
protein possesses a different biochemical or catalytic activity
that recognizes different nucleic acids for cleavage, which also
determines their potential applications (Fig. 1; Tables I and II) (2).
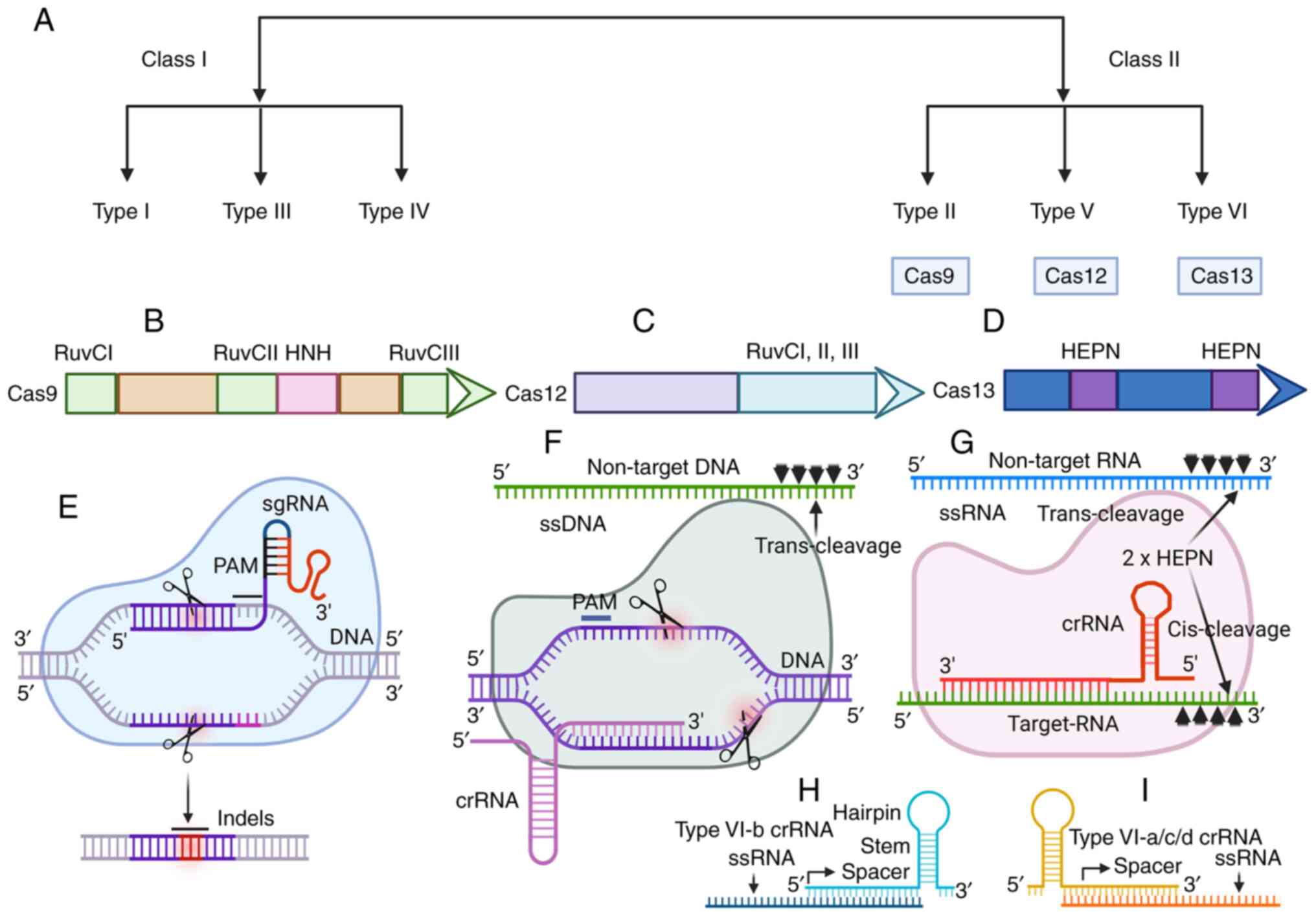 | Figure 1.Cas12 and Cas13 Cas orthologs
recognize dsDNA and ssRNA possessing ssDNA and ssRNA cleaving
trans-collateral activity. (A) CRISPR-Cas systems can be divided
into two classes and six types. The class II system encodes single
subunit enzymes, such as Cas9, Cas12 and Cas13, that target nucleic
acids for modifications. Schematic representations of (B) Cas9, (C)
Cas12 and (D) Cas13. Cas9 contains three RuvC (I, II and III) and
one catalytically active HNH domain required to induce DNA
cleavage. Cas12 contains three RuvC (I, II and III) endonuclease
domains. Cas13 has two HEPN-binding domains, as presented in the
figure. (E) Cas9 is presented in complex with sgRNA and target DNA.
The position of PAM, target binding and cleavage position are
marked in the figure. (F) Cas12 is presented in complex with crRNA
and a double-stranded DNA target. The position of PAM and the
cleavage sites are also presented. Cas12 also possesses promiscuous
ssDNA degrading activity. (G) Cas13 is presented in complex with
crRNA and ssRNA targets. Cas13 also possesses ssRNA-degrading
collateral activity when complexed with target RNA. (H) The crRNA
structure demonstrated in type VI b-Cas orthologs. The pre-crRNA
processing occurs at the 3′ ends, which create spacers. (I) The
most common crRNA structure reported in type a/c/d Cas orthologs in
which processing of pre-crRNA occurs at the 5′ end, creating
spacers. This figure was generated using BioRender (www.biorender.com). CRISPR, clustered regularly
interspaced short palindromic repeats; Cas, CRISPR-associated; PAM,
protospacer-associated motif; Indels, insertions or deletions; ss,
single-stranded; sgRNA, single guide RNA; crRNA, CRISPR RNA; HEPN,
higher eukaryotic and prokaryotic nucleotide; RuvC, an endonuclease
domain named for an Escherichia coli protein involved in DNA
repair; HNH, an endonuclease domain with catalytic histidine and
asparagine residues. |
Cas9 is the best-characterized Cas protein, and
requires crRNA and trans-activating RNA (tracrRNA) for enzymatic
activity. Although these ssRNAs are produced separately, they can
be engineered into a single RNA molecule known as a single gRNA
(Fig. 1). Cas9 proteins are used
in epigenome and genome editing, among numerous other applications,
which have been described previously (41,42).
Cas9 enzymes are modular proteins, which comprise RuvC (an
endonuclease domain named for an Escherichia coli protein
involved in DNA repair)-like endonuclease and HNH (an endonuclease
domain with catalytic histidine and asparagine residues) nuclease
domains, which are critical for target cleavage (Fig. 1B). Cas9 requires a 30 nucleotide
(nt) long protospacer sequence flanked by a protospacer-associated
motif (PAM) (5′-NGG-3′). Cas9 cleaves dsDNA 3-nt upstream of the
conserved PAM sequence. Consequently, host DNA repair enzymes
repair the breakpoints, which leads to the incorporation of
insertions or deletions (Fig. 1E)
(42). CRISPR arrays and Cas
proteins recognize and degrade foreign DNA, RNA and plasmids. All
characterized Cas proteins identify target nucleic acids when
guided by crRNAs (37,43–45).
The single subunit C2c2, or Cas13a, is an RNA
endonuclease (RNase). The function of Cas13a is regulated by ssRNAs
(37,43,46).
Cas13a possesses dual RNase activity, which is required for crRNA
processing and RNA-dependent RNA degradation (37,47).
The two unique RNase activities of Cas13a provide flexibility for
multiplex processing of pre-crRNA into crRNA and the loading of
gRNA or a spacer sequence into the target sequence for cleavage,
which is necessary for the sensitive detection of cellular RNAs or
any RNA substrates (37). Cas13a
proteins encompass two higher eukaryotic and prokaryotic nucleotide
(HEPN) binding domains that are critical for RNA degradation
(Fig. 1D) (43). Unlike Cas9 and Cas12, Cas13 does
not require a conserved PAM (Table
II). Cas13 proteins cleave ssRNA at multiple sites that do not
depend on the crRNA position but on ssRNA secondary structures such
as stems and loops known as hairpins (37,43,48).
Initiation of CRISPR-mediated immunity requires processing of
pre-crRNA into individual mature crRNAs composed of a single
spacer. Pre-crRNAs harbor multiple spacers flanked by palindromic
DRs (37,49–51).
Three possible steps are used alone or in
combination by different CRISPR-Cas systems in the production of
mature crRNAs, including: i) A dedicated endonuclease being
required for crRNA processing or target cleavage (37); ii) integration of a host
endonuclease with CRISPR-Cas proteins (37,52)
or iii) an intrinsic RNase activity for effector function (37,47).
The Cas13a homologs from three distinct branches of the Cas13
protein family including Leptotrichia shahii Cas13a
(LshCas13a), Leptotrichia buccalis Cas13a (LbuCas13a), and
Listeria seeligeri Cas13a cleave at the 5′ end of the
pre-crRNA, which comprises consensus DRs and a conserved 20 nt
spacer sequence (Fig. 1I).
LshCas13a and LbuCas13a process crRNA three or five nucleotides
upstream or downstream of the DRs and form hairpin structures
depending on the Cas13a homolog (37,43).
Changing the stem and reducing or inverting the hairpin in the DRs
attenuates LbuCas13a processing of pre-crRNA. Four contiguous
nucleotide mutations in a single-stranded region near or including
the cleavable bond completely abolishes the activity of LshCas13a
and LbuCas13a (37,49,50,53).
Similarly, LshCas13a requires 3′ flanking sequences
for DRs, stems and secondary hairpin structures in crRNA processing
(43). Target identification using
LshCas13a is sensitive to mutations in the protospacer region
(target RNA sequence). The processing activity of LbuCas13a is
independent of divalent cations (37,47).
The mature crRNA binds to Cas proteins, generating an RNA-directed
RNA-degrading complex for highly sequence-specific recognition and
cleavage of target RNA (Fig. 1F and
G) (37,42,44,45).
LshCas13a and LbuCas13a have demonstrated non-specific promiscuous
RNA-dependent RNA trans-degradation activity (Fig. 1G). LshCas13a has been reported to
preferentially cleave uracil (U) residues in the ssRNA region of
crRNA (37,43). crRNA processing and ssRNA target
cleavage are two independent activities of the Cas13a proteins.
ssRNA cleavage is ~80 times faster compared with crRNA processing.
Cas13a proteins, including LshCas13a and LbuCas13a, contained two
HEPN domains (Fig. 1D). Mutations
in conserved arginine (R) or histidine residues abolish
crRNA-directed target cleavage, while retaining crRNA processing
and binding activity (37,43). In addition, mutational studies with
LbuCas13a have reported a conserved R1079, which is required for
crRNA processing; however, crRNA-driven RNA cleavage and binding
activities remain unaffected. These observations indicated that
Cas13a proteins possess distinct pre-crRNA processing and
crRNA-directed cleavage activities (Fig. 1D) (37).
ssRNA or dsDNA target recognition-dependent
collateral activity of Cas13 or Cas12 proteins
Cas12 and Cas13 proteins have demonstrated
single-stranded DNA (ssDNA) and ssRNA-degrading collateral activity
(cleavage of nonspecific/nontarget ssDNA or ssRNA molecules)
(32,36) (Fig. 1F
and G). The identification of CRISPR-Cas systems in bacteria
and the development of Cas12- and Cas13-based diagnostics for the
detection of viral diseases and other applications occurred over a
~35 year period (Fig. 2). ssRNA-
and ssDNA-degrading collateral activities were used to detect viral
RNA or DNA in a readable format when combined in a CRISPR reaction
using a synthetic ssRNA or ssDNA molecule flanked by a fluorophore
and quencher. Collateral cleavage of the fluorophore and
quencher-labeled synthetic ssRNA or DNA physically separated the
quencher from the fluorophore, which generated a fluorescent signal
that could be assessed with a fluorimeter or fluorescence detector
(Fig. 3) (37). In the second approach, synthetic
reporters were flanked by fluorescein amidite (FAM) and biotin
molecules. The FAMs were labeled with gold nanoparticles (AuNPs)
for visualization. A streptavidin-coated lateral flow strip and
anti-FAM antibody were used to visualize the cleavage of the target
molecule. If the target was present in the sample, the lateral flow
strip would present two bands at the streptavidin and anti-FAM
antibody positions (Fig. 3)
(43). LshCas13a and LbuCas13a
comprise the trans-RNA-stimulated cleavage activity of ssRNA
substrates. This is an essential feature of Cas13a proteins, which
recognize specific RNAs from a pool or mixture of RNA molecules.
Although numerous polymerase-based methods for nucleic acid
amplification and subsequent detection are available, only a few,
including NASBA, LAMP and RPA, can directly detect the target RNA
without further improvements (37,54–57).
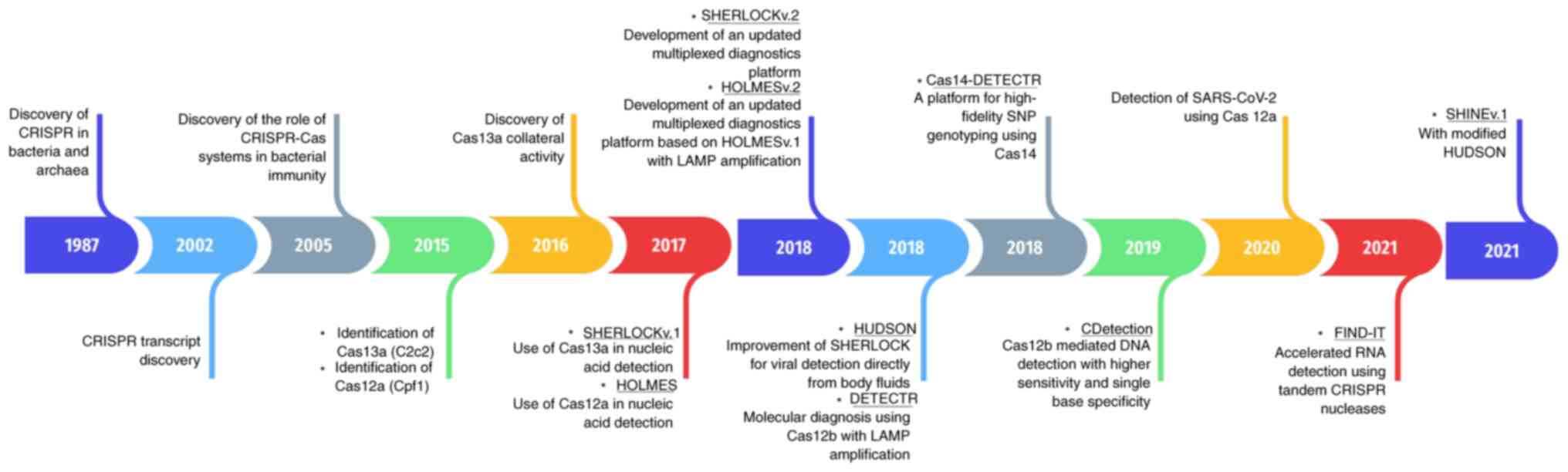 | Figure 2.The discovery of Cas12 and Cas13 Cas
orthologs has revolutionized the development of CRISPR-based
diagnostics. Timeline of the major discoveries for CRISPR-Cas
systems in nucleic acid detection. The discovery of CRISPR-Cas
systems in bacteria and archaea, together with biochemical
characterization of Cas12 and Cas13 ortholog enzymatic properties,
has revolutionized the development of CRISPR-based diagnostics. The
nonspecific trans-collateral activity of Cas12 and Cas13 in
cleaving single-stranded DNA and single-stranded RNA has been
exploited in the design and development of CRISPR-based diagnostic
approaches for nucleic acid detection. CRISPR, clustered regularly
interspaced short palindromic repeats; Cas, CRISPR-associated;
SHERLOCK, specific high-sensitivity enzymatic reporter unlocking;
v.1, version 1; HOLMES, 1-h low-cost multipurpose highly efficient
system; LAMP, loop-mediated isotherm amplification; HUDSON, heating
unextracted diagnostic samples to obliterate nucleases; DETECTR,
deoxyribonucleic acid endonuclease targeted CRISPR trans reporter;
SNP, single nucleotide polymorphism; SARS-CoV-2, severe acute
respiratory syndrome corona virus 2; FIND-IT, fast integrated
nuclease detection in tandem; SHINE, streamlined highlighting of
infections to navigate epidemics. |
 | Figure 3.Exploitation of collateral RNA
cleavage activity of Cas13a in CRISPR-based diagnostic approaches.
Schematic representation of Cas13a-mediated detection of any target
RNA or DNA molecule. The clinical sample is processed using
methods, such as HUDSON or chemical and heat treatment, followed by
extraction of nucleic acids. Target nucleic acids are pre-amplified
using isothermal amplification RPA or LAMP. RNA targets are first
reverse transcribed and amplified with RPA or LAMP. DNA targets are
amplified directly. After amplification, the amplified targets must
be transcribed using T7 RNA polymerase in case of Cas13-mediated
detection. After amplification, detection is performed by adding
CRISPR RNA, target and appropriate Cas enzymes. The signal is
detected using either visual indicators in lateral flow strips or
fluorescence monitoring in reaction tubes. Fluorescence signals can
also be read with a fluorimeter for quantification. Numerous
methods have been developed using clinical samples in which
reaction reagents are simultaneously used in target amplification
and detection. This figure was generated using BioRender
(www.biorender.com). CRISPR, clustered
regularly interspaced short palindromic repeats; Cas,
CRISPR-associated; LAMP, loop-mediated isotherm amplification;
HUDSON, heating unextracted diagnostic samples to obliterate
nucleases; FAM, fluorescein amidites; NP, nanoparticle; RT, reverse
transcription; RPA, recombinase polymerase amplification, -ve,
negative; +ve, positive. |
It has been reported that the RNA-directed
trans-endonuclease activity of LbuCas13a can be used to cleave
fluorophore quencher-tagged reporter RNA. Target RNA-induced
trans-RNase activity resulted in enhanced fluorescence intensity
within 30 min (37). Promiscuous
trans-RNase cleavage activity could be detected by adding 1–10 pM
of target RNA. The results indicated that LbuCas13a was a robust
RNA-cleaving enzyme capable of 104 turnovers per recognized target.
In the presence of ~0.02% target RNA, LbuCas13a demonstrated
~2,550% cleavage of the labeled reporter RNA substrate compared
with the level of crRNA-directed cleavage. These results indicate
that LbuCas13a has potent trans-RNA cleavage activity that can be
used to detect ssRNA molecules present as genetic material in
numerous viruses (37). The
presence of two different RNase activities can also be exploited in
the multiplex detection of RNA molecules with markedly increased
signal intensity using a fluorescence-quencher-labeled RNA reporter
for the easy detection of ssRNA viruses without target
amplification (37).
Diagnostic technologies which exploit the
collateral activity of Cas12 and Cas13-Cas proteins
Trans-RNA cleaving collateral activity
of Cas12 and Cas13, has been exploited in the development of
CRISPR-based diagnostics
The identification of the collateral activity of
Cas13 proteins has led to the emergence of CRISPR-Cas-based
diagnostic approaches to detect viral and human genomic variations.
Characterization of Cas13 collateral activity led to the
development of the specific high-sensitivity enzymatic reporter
unlocking (SHERLOCK) version 1 (v.1) (32,34).
LbuCas13a collateral activity is activated after crRNA binding and
target recognition. Collateral activity has been detected using
fluorophore-quencher-labeled ssRNA cleavage and the subsequent
generation of a fluorescent signal (Fig. 3). A fluorescence reader or
fluorimeter can be used for fluorescence signal detection. The
collateral activity of Cas12a was revealed after discovering the
collateral activity of Cas13a. Cas12a recognizes dsDNA and
possesses ssDNA-cleaving trans-collateral activity (Fig. 1C and F). The discovery of Cas12
ended the T7-mediated in vitro transcription of the samples
(32,36,58).
Two methods have been previously reported that
utilize the collateral activity of Cas12, known as DNA
endonuclease-targeted CRISPR trans reporter (DETECTR) (36) and 1 h low-cost, multipurpose,
highly efficient system (HOLMES) (59). The dsDNA-activated collateral
activity of Cas12a cleaves fluorescence-quencher-labeled ssDNA,
which produces a fluorescence signal that is read with a
fluorescence reader. The two approaches use different
preamplification steps: DETECTR uses RPA, whereas HOLMES uses PCR
(Table III) (32,34,36,38,60–71).
CRISPR-Cas-based approaches detect viruses, single nucleotide
polymorphisms (SNPs) and human genome variations in DNA samples
isolated from patients with cancer (72).
 | Table III.Characteristics of CRISPR-based
diagnostic approaches. |
Table III.
Characteristics of CRISPR-based
diagnostic approaches.
| Name | Cas-enzyme |
Preamplification | Assay-time | Preparation of
sample | Readout | Advantages | Disadvantages | Applications | Load of detection,
mol/l | (Refs.) |
|---|
| DETECTR | Cas12a. | RPA. | RPA for 10 min and
CRISPR for 60–120 min. | Crude
extraction. | Fluorescence | Highly specific,
fast, portable, low occurrence of false positive results,
multiplexing can be used and can differentiate viral subtypes. | Off-target effects,
limited scope and narrow target range. | HPV16 and HPV18
detection in human samples. |
1.0×10−18 | (36,60) |
| Cas14-DETECTR | Cas14
(Cas121). | PCR. | The assay time for
PCR is NS and for CRISPR it is 120 min. | Crude
extraction. | Fluorescence | High sensitivity,
simple design, high specificity, multiplexing can be used, no PAM
requirements, cost effective detection of pathogenic mutations,
user friendly, uses less complicated sample processing, has a high
fidelity in SNP detection. | Limited scope,
narrow target range, requires extensive validation. | HECT and RLD domain
containing E3 ubiquitin protein ligase 2 SNPs detection in human
samples. | NS | (61,62) |
| HOLMESv.1 | Cas12a. | PCR. | PCR for 88 min and
CRISPR for 15 min. | Column based. | Fluorescence | High sensitivity,
rapid PCR amplification, multiplexing can be used, high efficiency,
rapid results and no in vitro transcription required | Limited scope and
high cost. | Discrimination of
SNPs in cell lines and human samples; Pseudorabies and JEV
detection; and discrimination of viral strains |
1.0×10−17 | (34,63–65) |
| CDetection | Cas12b. | RPA. | RPA for 10 min and
CRISPR for 60–180 min. | Synthetic targets
or crude extraction | Fluorescence | High sensitivity,
single-base specificity, rapid results and cost-effective. | Limited scope,
off-target effects, complex sample preparation and narrow target
range. | HPV16 detection;
ABO blood genotyping in human; and SNPs detection of breast cancer
1 gene and tumor protein p53. |
1.0×10−18 | (66) |
| HOLMESv.2 | Cas12b. | LAMP. | LAMP for 40 min and
CRISPR for 35 min or one pot for 120 min. | NS. | Fluorescence | High sensitivity
and specificity, one-pot detection, enhanced multiplexing
capabilities compared with HOLMESv.1., rapid results and is
portable. The use of Cas12b in HOLMESv2 provides greater
flexibility in target selection and the detection of multiple
targets in a single reaction, which are important advantages over
HOLMESv.1 | Sample preparation
(temperature gradients were required for target amplification),
off-target effects and high costs | Discrimination of
SNP in cell lines; JEV detection; detection of human mRNA and
circular RNA; and detection of DNA methylation |
1.0×10−17 | (34,63,65) |
| SHER-LOCKv.1 | Cas13. | NASBA or RPA. | NASBA for 132 min
or RPA for 120 min and CRISPR for 60–180 min. | Crude extraction or
column based | Fluorescence | Ultra-sensitive and
specific (2×10−18 M), single molecule detection, can
detect both DNA and RNA, reagents can be lyophilized without
impacting specificity and sensitivity, specificity of Cas13 can be
enhanced by introducing synthetic mismatch, it is rapid, and it has
a low cost. | Sample preparation
requires expertise in protein purification and RNA biology,
multistep nucleic acid amplification which may affect precise
target quantification, it is less useful for precise gene
expression profiling, and the absolute digital quantification of
the target is not possible | ZIKV and DENV
detection; bacterial detection (such as E. coli, K. pneumoniae,
P. aeruginosa, M. tuberculosis and S. aureus); viral
strain discrimination; and SNPs detection |
2.0×10−18 | (32,60) |
| SHER-LOCKv.2 | Cas13. | RPA. | RPA for 60 min and
CRISPR for 60–180 min or one pot for 60–180 min. | Crude extraction or
column based | Lateral flow or
fluorescence | High specificity
for single nucleotides, high sensitivity (8×10−21 M) and
flexible detection using fluorescence and lateral flow assays. | More
time-consuming, multistep nucleic acid amplification process, which
may affect the precise target quantification and it is less
applicable for precise gene expression profiling. | ZIKV and DENV
detection; bacterial detection (such as Escherichia coli,
Klebsiella pneumoniae, Pseudomonas aeruginosa Mycobacterium
tuberculosis and Staphylococcus aureus); viral strain
discrimination; and SNPs detection. |
8.0×10−21 | (34,38,60) |
| SHINE. | Cas13. | RPA. | One pot for 50
min. | Crude
extraction. | Lateral flow or
fluorescence. | High throughput,
high sensitivity, high specificity, it is easy to use, it is less
time consuming, it is simple (easy to use lyophilized reagents
reduce assay time, target amplification and detection are performed
in a single tube), portable and has versatile detection
methods | Requires
preparation of multiple reaction mixtures and the handling of
multiple samples | SARS-CoV-2
detection |
8.0×10−18 | (96) |
| STOPcovid | Cas12b. | LAMP. | One pot for 60
min. | Crude
extraction. | Lateral flow or
fluorescence. | High sensitivity,
it is rapid, cost-effective and has a one- step reaction
assay. | Limited to
SARS-CoV-2 detection, has a limited sensitivity, there is a limited
validation and requires development of simple and efficient sample
processing steps | SARS-CoV-2
detection |
3.3×10−18 | (68,69) |
| CARMEN | Cas13. | PCR or RPA. | RPA for 20 min and
CRISPR for 180 min. | Column based | Fluorescence | High specificity,
easy to use (single array can detect more than 100 viruses), high
multiplexing capability and can be used for multiplex detection of
multiple pathogens | Limited to Cas13a
targets, limited validation and off-target effects | Detection of
viruses; influenza A strain subtyping; and drug-resistant mutation
detection in HIV |
9.0×10−19 | (70,71) |
Target pre-amplification coupled with
CRISPR-based, highly sensitive detection of viruses using
SHERLOCKv.1
Bacterial CRISPR-Cas enzymes, such as LshCas13a and
LbuCas13a, opened a new area of application for CRISPR-Cas in
diagnostics, particularly in the detection of viruses with an ssRNA
genome (37,43). LbuCas13a recognizes target RNA at
the pM level (1–10 pM) (37). In
addition, the newly characterized Leptotrichia wadei Cas13a
(LwaCas13a) has been reported to exhibit potent collateral RNA
cleavage activity, capable of detecting ~50 fM of target RNA
without amplification (32).
However, aM sensitivity is required, particularly for the in
vitro detection of pathogens containing RNA or DNA (73–75).
The two commonly used PCR-based technologies, droplet digital and
RT-qPCR, have a high sensitivity (in aM). Achieving similar, aM,
sensitivity using CRISPR-based approaches requires
pre-amplification of the target substrate, which can be achieved
with isothermal RPA (Fig. 3)
(28).
RPA offers the highest sensitivity when coupled with
T7 transcription. In RPA, RNA targets are first converted into DNA
using reverse transcriptase and DNA targets are directly amplified.
T7-mediated transcription of amplified DNA into RNA enables
detection by LwaCas13a using a fluorescence-quencher-labeled RNA
reporter, generating an amplified signal, which is referred to as
SHERLOCKv.1 (32). SHERLOCKv.1
achieves aM sensitivity in the detection of DNA and RNA, similar to
droplet digital PCR and RT-qPCR, with less inter-sample
variability, and can be easily performed in a single reaction mix
(32).
In addition, the utility of SHERLOCKv.1 for the
detection of Zika virus (ZIKV) and Dengue virus (DENV) has been
previously reported. SHERLOCKv.1 recognized ZIKV and DENV with a
sensitivity of 2 aM. The lyophilized and rehydrated components of
SHERLOCKv.1 detected unamplified target RNA at 20 fM. The
lyophilized and rehydrated SHERLOCKv.1 component detected target
RNA in an aqueous reaction with a sensitivity of 3 aM in
combination with a pre-amplification step. However, it was reported
that paper spotting and lyophilization of SHERLOCKv.1 reagents
slightly reduced the sensitivity, to 20 aM. These features of
SHERLOCKv.1 are ideal for the development of POC devices to
diagnose various human-borne pathogens with high accuracy within 1
h. SHERLOCKv.1 can detect ZIKV RNA in clinical samples, such as
urine, serum and saliva. SHERLOCKv.1 can also detect bacterial
pathogens and can be used to identify clinical isolates (34). SHERLOCKv.1 has been reported to
also identify clinical ZIKV strains harboring a single-nucleotide
mutation with high specificity and sensitivity (Table III) (32). Cas13a-based tests can detect a
single copy of the viral nucleic acid in 1 µl of sample. These
diagnostic tests can easily be adapted for virus detection in
remote areas without complex, laborious and costly instrumentation
(76–78).
HUDSON enables the development of
SHERLOCKv.2 for the multiplex detection of viruses in a single
reaction tube
To directly detect viral RNA in samples from
patients in a readable colorimetric or fluorescent format, heating
unextracted diagnostic samples to obliterate nucleases (HUDSON) was
developed for sample processing. SHERLOCKv.1, coupled with HUDSON
[SHERLOCK version 2 (SHERLOCKv.2)], enabled the specific detection
of multiple circulating isolates of DENV and ZIKV serotypes.
SHERLOCKv.2 recognizes a single copy/µl of ZIKV and DENV
reverse-transcribed cDNA from the samples of patients with 100%
specificity and sensitivity, similar to the standard RT-qPCR tests
explicitly designed for ZIKV and DENV viruses (79).
In addition, the utility of SHERLOCKv.2 in detecting
ZIKV and DENV RNA in the samples or bodily fluids of patients
without purification has been demonstrated (79). ZIKV and DENV viruses are excreted
in the saliva and urine. Urine and saliva samples can be collected
quickly, without invasive procedures. In SHERLOCKv.2,
HUDSON-treated samples are added directly to the RPA reaction
mixture without dilution or purification. Using HUDSON in
SHERLOCKv.2 does not influence downstream amplification and
detection of the target RNA. SHERLOCKv.2 easily detects spiked ZIKV
RNA in blood, plasma, serum, saliva and urine samples with aM
sensitivity. ZIKV particles been reported to have been spiked into
urine, saliva, blood, plasma and serum to mimic clinical
infections, and the total turnaround time was <2 h when
colorimetric and fluorescent reading formats were combined. The
sensitivity of SHERLOCKv.2 is comparable with that of ZIKV RNA in
the samples of patients, that is, 1–1,000 copies/ml. The
SHERLOCKv.2 also detects DENV RNA with high sensitivity and
specificity in <1 h in both the saliva and serum. SHERLOCKv.2,
adapted in a lateral flow assay format, can be easily used in a
simple pipeline with minimal equipment requirements for diagnosing
ZIKV and DENV viruses (79). In
addition, the utility of SHERLOCKv.2 in identifying ZIKV and DENV
serotypes has also been demonstrated (79) (Table
III). SHERLOCKv.2 enables precise and sensitive detection of
ZIKV and DENV serotypes due to the development of serotype-specific
crRNAs (79). In addition,
SHERLOCKv.2 can also target multiple flaviviruses (ZIKV, DENV, West
Nile virus and Yellow fever virus) in a single reaction tube using
engineered universal RPA primers and crRNAs (79). Numerous advances have been made
using orthogonal Cas proteins to further increase the versatility
of SHERLOCKv.2. New advances include the use of multiple
fluorophores in a single reaction, quantification, visible reading
in lateral flow format and a three-and-a-half-fold increase in
sensitivity through the combination of the additional Cas protein,
Csm6 (38).
The discovery and biochemical
characterization of tandem-acting Cas enzymes and orthologs enables
the development of highly sensitive and multiplex CRISPR-based
diagnostics
The multiplexing-compatible Cas proteins were
identified by biochemical characterization of Cas13a and Cas13b
family members (Fig. 4B). The
cleavage preferences of these proteins have been studied using
homopolymer reporters, and the majority of these proteins preferred
uridine (U), adenine (A) or a combination of bases (Table II) (38). Cleavage preferences were further
improved through the optimization of buffers and design of crRNAs.
Cas13b from Prevotella sp. MA2016 (PsmCas13b) is more
sensitive to A during cleavage of homopolymer A compared with
Cas13a from the Lachnospiraceae bacterium (LbaCas13a). The
cleavage preferences of other Cas proteins were refined using
dinucleotide motifs. Cleavage of RNA sequence motifs and
dinucleotide recognition preferences confirmed that the activities
of LwaCas13a, Cas13b from Capnocytophaga canimorsus
(CcaCas13b), LbaCas13a and PsmCas13b could be measured
independently using four dinucleotide reporters, namely, AU, UC, AC
and GA, respectively. In addition, the cleavage preference of Cas
proteins was further refined using a random library of RNA motifs.
It was demonstrated that Cas proteins can efficiently recognize RNA
oligomers of 6 nt, which further enhanced orthogonal usage and
provided flexibility for multiplexing in a single reaction. Using
these cleavage preferences, synthetic ssRNAs from ZIKV and DENV
were detected in a single reaction using hexachlorofluorescein
(HEX) and FAM channels (38).
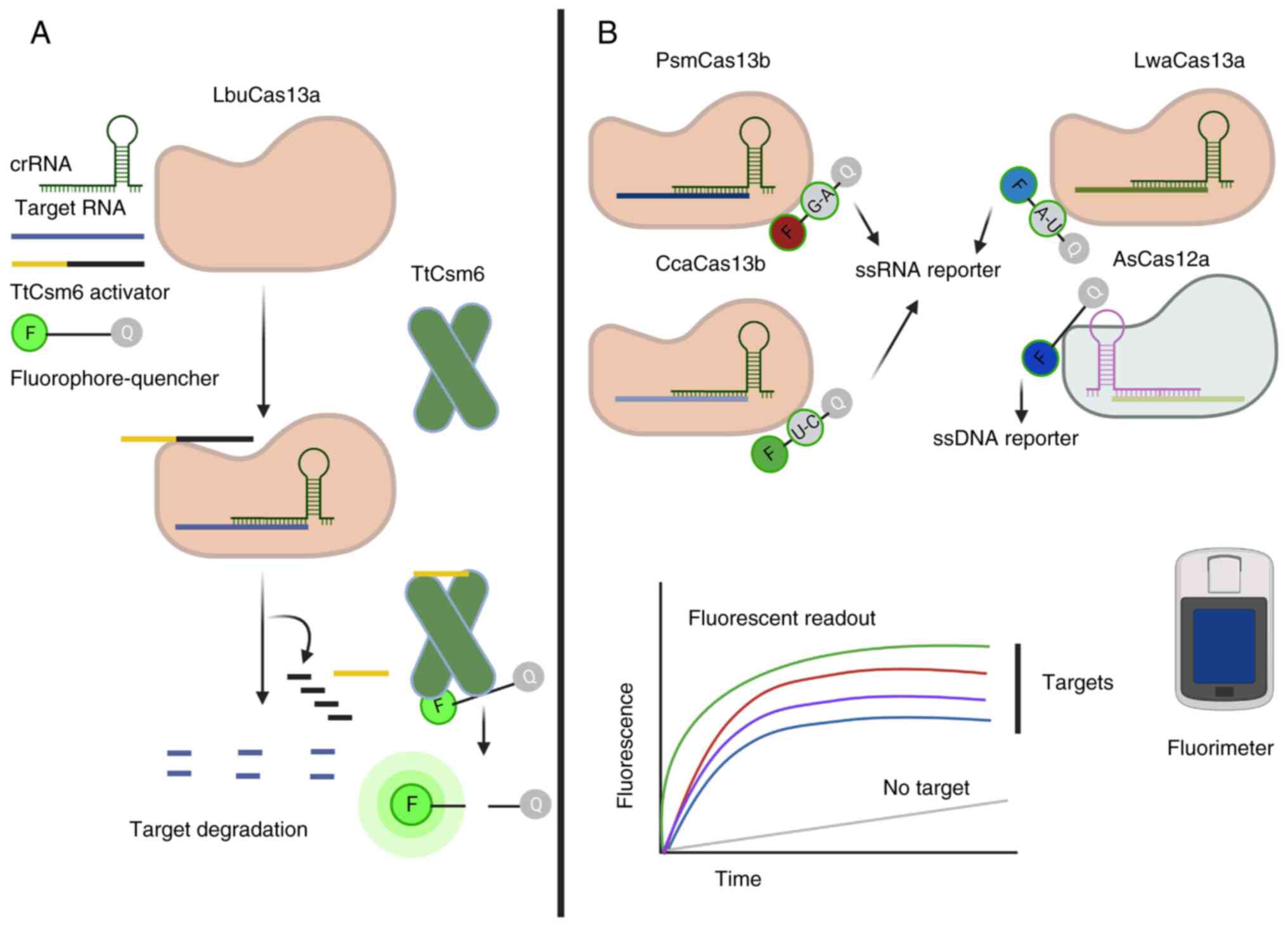 | Figure 4.The discovery and characterization of
tandem acting and orthogonal Cas enzymes have enhanced the
multiplexing of CRISPR-based diagnostic approaches. (A) Orthogonal
Cas enzymes can be used simultaneously, where target recognition by
one enzyme activates its collateral activity, which produces
byproducts that act as an activator for the second Cas enzyme.
Activation produces a stable and robust signal that can be easily
detected without a pre-amplification reaction. For example,
LbuCas13a and TtCsm6 can be used to detect any RNA target. The
collateral activity of LbuCas13a produces 2′-3′ cyclic phosphates
or linear oligonucleotides that act as an activator for TtCsm6.
Stable activation of TtCsm6 produces a strong signal easily
detected with a fluorimeter. (B) Orthologous Cas enzymes can be
used together for multiplex detection of pathogen RNA and DNA,
saving time and cost. Different Cas orthologs demonstrate different
collateral activity on oligonucleotides, including dinucleotides
and hexanucleotides. The figure presents four orthologous Cas
enzymes used together in the presence of defined di- or
oligonucleotide motifs that separate the quencher molecule.
Recognition of these targets by the enzymes activates the
collateral activity, which results in the generation of fluorescent
signals that can be detected with a fluorimeter. This figure was
generated using BioRender (www.biorender.com). Cas, clustered regularly
interspaced short palindromic repeats-associated; crRNA, CRISPR
RNA; ss, single-stranded; LbuCas13a, Leptotrichia buccalis
Cas13a; TtCsm6, Csm6 from Thermus thermophilus; PsmCas13b,
Prevotella sp. MA2016 Cas13b; CcaCas13b, Capnocytophaga
canimorsus Cas13b; LwaCas13a, Leptotrichia wadei Cas13a;
AsCas12a, Acidaminococcus sp. Cas12a. |
By extending the utility of SHERLOCKv.2 to multiplex
detection in a single reaction, the collateral cleavage activity of
Cas12a was exploited. Cas12a isolated from Acidaminococcus
sp. BV3L6 (AsCas12a) demonstrated weak collateral activity and
required an input concentration >10 nanomoles (nM). Therefore,
the combination of RPA pre-amplification with AsCas12a enabled the
detection of a single molecule with a detection limit of 2 aM.
Triplex detection was achieved using LwaCas13a [U reporter in the
cyanine-5 (Cy5) channel], PsmCas13b (A reporter in the FAM channel)
and AsCas12a (ssDNA reporter in the HEX channel). Combining these
enzymes and reporters enabled recognition of three targets in a
single reaction: A ssDNA target, ZIKV ssRNA and DENV ssRNA
(38).
Furthermore, the recognition of four targets in a
single reaction was achieved using the reporter cleavage activities
of the orthogonal dinucleotide motifs of AsCas12a, LwaCas13a,
PsmCas13b and CcaCas13b in HEX, FAM, Cy5 and TEX channels,
respectively (Fig. 4B) (38,69,80).
By combining RPA, two DNA targets (Pseudomonas aeruginosa
acyltransferase and Staphylococcus aureus thermonuclease
genes) were detected in a single reaction with an aM sensitivity
(38). In addition, multiplex
SHERLOCKv.2 detects aM concentrations of ZIKV and DENV RNA diluted
with PsmCas13b and LwaCas13a. The identification and
characterization of orthogonal Cas proteins, RNA and dinucleotide
cleavage preferences, enabled multiplex detection of DNA and ssRNA
in a single reaction with aM sensitivity combined with
pre-amplification using RPA (34,38,81).
Other improvements have also been incorporated to make SHERLOCKv.2
more versatile, quantitative and sensitive. RPA primer
concentrations were optimized, and it was reported that a primer
concentration of 240 nM demonstrated correlation between signal and
input and could detect sample concentrations as low as a single
molecule in the aM range. A number of nucleic acid detection
applications require a detection limit of one molecule/ml, for
example, for human immunodeficiency virus (HIV) (38,82).
Using LwaCas13a, PsmCas13b and the RPA reaction, a detection limit
of 200 zM was achieved. Collateral RNase activity of Csm6 was
exploited to further increase the sensitivity of SHERLOCKv.2
(38). Csm6 from Enterococcus
italicus (EiCsm6) and Lactobacillus salivarius Csm6
(LsCsm6) requires RNA with 2′,3′-cyclic phosphate ends (38). The collateral activity of LwaCas13a
and PsmCas13b produces RNA with 2′,3′-cyclic phosphate ends, which
suggests that EiCsm6 and LsCsm6 can be used to increase the
sensitivity of SHERLOCKv.2. The cleavage preference of Thermus
thermophilus Csm6 (TtCsm6), EiCsm6, LsCsm6 and Csm6 over A- and
C-rich reporters allowed measurement of LwaCas13a and Csm6 cleavage
activity using two distinct fluorophores in different channels (FAM
and HEX) of a fluorescent plate reader. LwaCas13a recognizes and
cleaves poly U oligomer reporters, whereas Csm6 cleaves
oligo-reporters containing poly-A. The combination of poly-A and
poly-U produced a reporter recognized by Cas13a, which produced a
reporter for Csm6 (Fig. 4A). Both
enzymes were used in a single reaction to achieve enhanced signal
generation, which could be easily detected using single-channel
fluorescence (38). Increased
addition of a Csm6-specific reporter correlated directly with
increased signal intensity (Fig.
4A) (38).
Cas13 orthologs have been used for the
targeting, degradation, and subsequent detection of viruses
LbaCas13a and PsmCas13b activities were used for
coupled ssRNA virus targeting, degradation and subsequent detection
(83), this study suggested the
possibility of using Cas13 proteins in viral RNA degradation with
subsequent detection as a viral treatment option. It has been
reported that the majority of human-associated viruses (HAVs) that
infect humans or related animals contain genomic regions recognized
by Cas13-specific crRNAs (83).
The efficiency and feasibility of using Cas13a and Cas13b for
detection and degradation have been previously demonstrated against
three ssRNA viruses, namely human stomatitis virus, influenza virus
A and lymphocytic choriomeningitis virus (LCMV) (83). Both enzymes can attack these
viruses in a cell culture model and can be detected with
SHERLOCKv.2 by combining HUDSON, RPA-mediated pre-amplification and
crRNA. The combination of viral targeting and detection based on
Cas13 enzymes is called the Cas13-assisted restriction of viral
expression and readout (83).
These results suggested that the majority of HAVs can be targeted
for degradation by incorporating specific crRNAs and Cas
proteins.
Cas12a comprises ssDNA-cleaving
trans-collateral activity used for the specific and sensitive
detection of HPV16 and HPV18 viruses
Type V family proteins, such as Cas12a, harbor
RNA-directed deoxyribonuclease (DNase) activity. The
Lachnospiraceae bacterium ND2006 Cas12a (LbCas12a)
recognizes dsDNA using a 20 nt gRNA. LbCas12a cleaves dsDNA using a
single catalytic RuvC endonuclease domain, which generates 5′ and
3′-staggered ends (Fig. 1C and F).
Recognition of target DNA activates ssDNase activity and
indiscriminately degrades ss-linear or circular DNA molecules
(36,84). Cas12a uses a T-rich neighboring PAM
motif to induce DNA cleavage and catalyzes the processing of its
pre-crRNA to produce mature crRNA (Fig. 1F) (36,47,85–90).
Using fluorophore quencher-labeled non-specific ssDNA,
Cas12a-specific crRNA and the dsDNA target of Cas12a, Cas12a
exhibited non-specific trans-ssDNA cleavage activity (Fig. 1F). Cas12a requires 15 nt of target
complementary to the crRNA (36).
Cas12a bound to the crRNA-DNA complex can degrade ~1,250 ssDNA
molecules/s, which is equivalent to the diffusion limit. A 2-nt
mismatch mutation in the gRNA and dsDNA target region results in an
~100-fold increase in the trans-degrading activity of LbCas12a
(36). The majority of Cas12a
orthologs contain a single RuvC catalytic domain (Fig. 1C). It has been previously
demonstrated that Cas12a orthologs AsCas12a, Francisella
novicida Cas12a and Cas12b from Alicyclobacillus
acidiphilus (AaCas12b) possess ssDNA-degrading activity when
assembled with crRNA and double-stranded activator DNA (36).
The use of LbCas12a for the detection of human
papillomavirus 16 (HPV16) and HPV18 DNA has been previously
demonstrated (36). LbCas12a
detects HPV16 and HPV18 DNA with pM sensitivity. LbCas12a combined
with RPA pre-amplification detected HPV16 and HPV18 DNA with aM
sensitivity. LbCas12a efficiently discriminates HPV16 and HPV18 DNA
from complex DNA samples purified from anal swabs with similar
specificity and sensitivity to the, gold-standard, PCR assays
(36).
SARS-CoV-2 detection using orthologous
CRISPR-Cas proteins
RT-qPCR is considered to be the gold standard for
SARS-CoV-2 detection. This requires trained personnel and special
equipment to perform the tests. However, RT-qPCR cannot be
performed under normal testing conditions, such as at room
temperature or 37°C (91). To
overcome these limitations, SHERLOCK was validated for the
diagnosis of SARS-CoV-2. In SHERLOCK, detection is performed using
either fluorescence readout or lateral flow assays. SHERLOCK was
validated using 154 nasopharyngeal and throat swab samples, which
indicated 100% specificity and sensitivity in the fluorescence
reading and 100% specificity and 97% sensitivity in the lateral
flow assay (92). Future advances
in CRISPR-based diagnostics are expected to increase the
sensitivity of lateral flow assays (92). In particular, the utility of
SHERLOCK for SARS-CoV-2 testing was further validated using 380
preoperative SARS-CoV-2 negative samples, in which SHERLOCK
indicated 100% concordance with RT-qPCR (92). These results suggested that
CRISPR-based diagnostics can successfully diagnose SARS-CoV-2
infection (92).
SHERLOCK uses two steps to detect bacterial or viral
pathogens including SARS-CoV-2 (31,32,36).
This involves RNA extraction, followed by CRISPR-Cas-based
detection. The SHERLOCK test in one pot (STOP) streamlined and
simplified RNA extraction and concentration using magnets, which
can be easily combined with detection reactions. STOP coronavirus
disease (covid) v.1 (STOPcovidv.1) used RNA extraction, reverse
transcription and LAMP-mediated isothermal amplification of the
target RNA (26). LAMP functions
optimally at 55–70°C; therefore, a thermostable Cas enzyme is
required for detection. AaCas12b was combined with RT-LAMP to
detect SARS-CoV-2 (93).
STOPcovidv.1 used fluorescence or lateral flow strip-based display
formats. It was validated with 202 positive and 200 negative
nasopharyngeal swab specimens and demonstrated a sensitivity of
93.1% and a specificity of 98.5%, consistent with RT-qPCR (68). STOPcovidv.2 detected viral loads at
1/30 the level that can be detected by the Centers for Disease
Control and Prevention (CDC) RT-qPCR, which is a standard test for
detecting SARS-CoV-2 (68). The
SARS-CoV-2 DETECTR was validated for the detection of SARS-CoV-2 in
patients and artificial samples (31). RNA extracted from samples from
patients was reverse-transcribed and amplified using LAMP at 60°C.
Detection was performed at 37°C with LbCas12a using fluorescence or
lateral flow strip-based detection, with 95% agreement with RT-qPCR
results. DETECTR demonstrated good agreement with CDC-approved
RT-qPCR and could be performed within 45 min. This technology can
also be combined with microfluidic cartridges and freeze-dried
reagents for POC applications outside clinical laboratories, such
as in airports, public spaces, private and government clinics, and
local emergency rooms (31).
LwaCas13a and EiCsm6 were used to detect, DENV, ZIKV
and SARS-CoV-2 viruses. LwaCas13a and EiCsm6, in tandem with RPA or
RT-LAMP pre-amplification, demonstrated sensitivities in the aM
range (2 aM). Without pre-amplification, the combined use of
LwaCas13a and EiCsm6 demonstrated sensitivity of 1 µM (38,94).
To increase the sensitivity and speed of CRISPR-based diagnostics,
which made the methods amenable to POC development, thermostable
TtCsm6 and LbuCas13a were characterized, along with multiple crRNAs
and linear artificial and stable A4U6 (four contiguous adenine and
six uridine) activators of TtCsm6 (Fig. 4A) (94). The utility of LbuCas13a with
TtCsm6, crRNA and an A4U6 activator and reporter was demonstrated
using purified SARS-CoV-2 RNA and RNA extracted from nasal swab
specimens. LbuCas13a and TtCsm6 detected SARS-CoV-2 at ~30
copies/µl in 20 min. A microfluidic cartridge with a light-emitting
diode light source and detector was developed to make the method
easier to use and more versatile. This device was called
fast-integrated nuclease detection in tandem (FIND-IT). FIND-IT
could detect SARS-COV-2 in samples with an RT-qPCR-determined cycle
threshold value of ~33 (94).
During the peak of pandemics, continuous viral
surveillance and monitoring is essential and requires an on-site
POC device that can provide results within 1 h. Diagnostics with
coronavirus enzymatic reporting (DISCoVER) was developed to test
for SARS-CoV-2 using saliva as a sample. DISCoVER utilizes sample
denaturation and inactivation (lysis and reduction) with simple
reagents, such as tris(2-carboxyethyl)phosphine and
ethylenediaminetetraacetic acid. In addition, DISCoVER uses LAMP
followed by detection using LbuCas13a. DISCoVER contains a
gravity-driven microfluidic device with sample inactivation,
isothermal amplification and separate fluorescence detection of the
control and positive samples. DISCoVER indicated ~95% sensitivity
and 100% specificity for RT-qPCR-validated samples (95). Furthermore, DISCoVER has been
reported to have detected ~40 copies/µl of the SARS-CoV-2 genome
and demonstrated 100% positive predictive value and ~93% negative
predictive value (95).
To develop a POC device that can be used outside
well-equipped laboratory conditions, streamlined highlighting of
infections to navigate epidemics v.1 (SHINEv.1) was developed
(96). In SHINEv.1, a modified
HUDSON is used for nasal swab and saliva processing. The modified
HUDSON could inactivate the sample within 10 min. Isothermal
amplification is performed in a single tube using RPA- and
Cas13a-based detection methods. The sample values are recorded in a
sealed tube using a smartphone. SHINEv.1, validated on 60
nasopharyngeal samples, demonstrated 90% sensitivity and 100%
specificity for samples validated using the gold standard RT-qPCR,
with a sample response time of 50 min (96).
Additionally, SHINEv.2 was designed to test for
SARS-CoV-2 and different variants of concern, including α, β, γ, δ
and ο (67). A commercially
available FastAmp virus and cell solution is used for isothermal
amplification to effectively inactivate SARS-CoV-2 in combination
with RNase inhibitors at ambient temperature. The need for
refrigerated storage of reagents is eliminated using SHINEv.2.
Optimized lyophilized reagents, including mannitol and sucrose, are
used in the reaction. The reaction is performed at 37°C using a
heating block or body heat without compromising the sensitivity and
specificity within 90 min. SHINEv.2, validated on nasopharyngeal
specimens, demonstrated ~90% sensitivity and 100% specificity in a
paper-based lateral flow strip, and different variants of concern
could be identified visually (67).
In addition, a Cas12a-based test procedure was
developed to distinguish between the α, β, γ and δ variants of
concern (67). LbCas12a and
AsCas12a are used for target recognition and signal generation,
followed by detection of fluorescence signals. The assay
distinguished the open reading frame 8a-L/S mutation in SARS-CoV-2.
The CRISPR-Cas12a assay also efficiently detects all characteristic
spike protein mutations, including (K-417-N/T, L-452-R/Q, T-478-K,
E-484-K/Q, N-501-Y and D-614-G) to discriminate between the α, β,
γ, δ, κ, λ and ε variants of SARS-CoV-2. The assay was validated
using 32 positive samples of SARS-CoV-2 and demonstrated 100%
agreement with sequencing results. This Cas12a-based multiplex
assay can be easily adapted for genotyping different variants of
SARS-CoV-2 in laboratories performing SARS-CoV-2 testing (67).
Multiplexed high-throughput CRISPR-Cas-based
virus detection
Multiplexing has additional benefits, as it can
process multiple samples and targets in parallel. Multiplexing can
save time and money, and is beneficial for monitoring virus
outbreaks by distinguishing multiple strains and virus types
(Figs. 4 and 5). It can distinguish the dominant
circulating variants of viruses at any time point using
variant-specific crRNAs (79).
CRISPR-based multiplex assays have been developed for flaviviruses
to distinguish between the four serotypes of DENV (79). Multiplexing of CRISPR assays
requires the separate amplification of targets, followed by
detection using Cas13 and crRNA (79). Microfluidic devices have been
integrated into CRISPR diagnostics to increase the versatility and
ease of use of CRISPR-based testing, while handling thousands of
targets or samples. The handheld microfluidic device was first used
to detect Ebola virus RNA in up to 24 samples without amplification
(97). In addition, microwell
arrays have been combined with CRISPR-based assays for the highly
multiplexed detection of thousands of samples using a method known
as combinatorial array reactions for the multiplex evaluation of
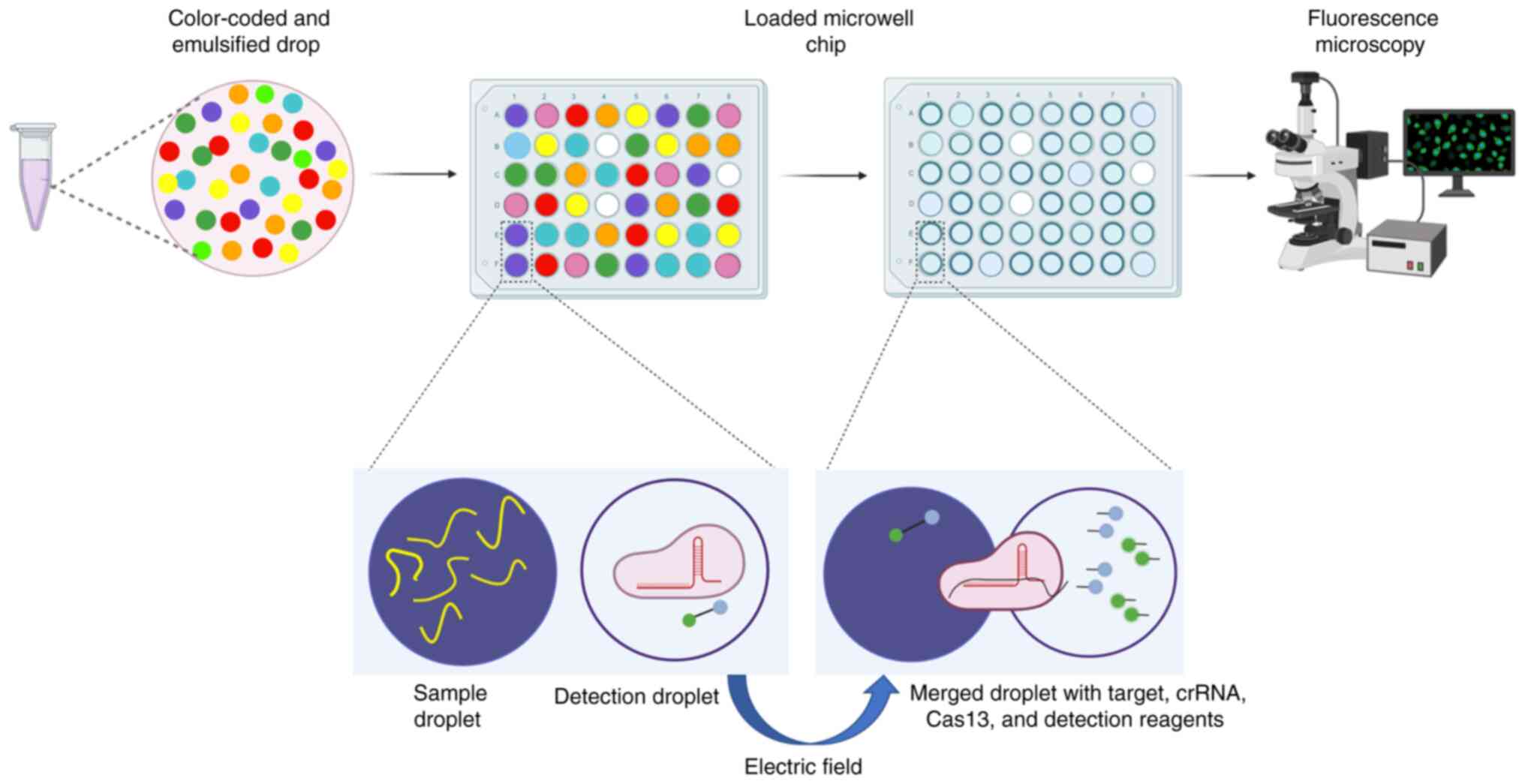
nucleic acids (CARMEN) (70).
CARMEN can process up to 5,000 samples simultaneously. In addition,
it is modular in design, allowing the processing of thousands of
samples for multiple targets or a few samples for all viral targets
simultaneously. In CARMEN, re-amplified samples are used to produce
nl volume droplets, and a pair is produced, which includes a sample
droplet with the target and a detection droplet comprising crRNA,
Cas enzyme, and detection reagents. Detection is performed by
fusing the target and crRNA with a reagent on a microfluidic chip
(Fig. 5). CARMEN-Cas can detect
all known viruses or viruses suspected to infect humans in a single
sample using a pan-viral detection test. Additionally, CARMEN can
distinguish all known haemagglutinin 1–6 and neuraminidase 1–9
subtypes of Influenza A virus (FLUAV) (70). Multiplexing CRISPR-Cas-based
detection technologies with highly automated microfluidic and
microchip-based assays could improve high-throughput viral
screening and surveillance.
Utilization of Cas13 and Cas12 for the
specific and sensitive detection of a single virus
Several approaches have been developed to detect
viral pathogens since validation of the SHERLOCK and DETECTR
technologies (Table III)
(98–102). The sudden appearance of
SARS-CoV-2 in 2019 and the first publication of its genome sequence
led to the development of several new methods for its detection
(2,103). The Food and Drug Administration
issued an emergency authorization for the use of CRISPR-based
diagnostic technologies such as SHERLOCK and DETECTR to diagnose
SARS-CoV-2 (2). The precise and
sensitive detection of viral nucleic acids and their dependence on
the complementarity between crRNA and target sequences has led to
the development of Cas13- and Cas12-based assays that can detect a
single nucleotide difference between viral isolates (Table III). A single-nucleotide mismatch
between the crRNA and the target sequence does not entirely abolish
target cleavage or collateral activity. However, introducing a
single mismatch near a mutant site markedly abolishes the target
and ssRNA cleavage activity (32).
This property of Cas13 and Cas12 has been exploited for the
detection of ZIKV isolates, DENV serotypes and single-nucleotide
mutations in HPV16 and HPV18 DNA (32,79).
Cas13 has been used to identify two isolates of ZIKV and two common
serotypes of DENV circulating in Asia and America (79). Cas12 has been used to identify
HPV16 and HPV18 DNA SNP (66). In
addition, Cas13 has been used to identify drug resistance mutations
in several HIV genes. For example, 27 common mutations in HIV drug
resistance have been verified, including six reverse transcriptase
inhibitors and 21 integrase inhibitor mutations (70). The development of Cas13- and
Cas12-based methods beyond the detection of viral nucleic acids has
expanded the scope of CRISPR-Cas-based technologies. This will
provide further impetus for developing CRISPR-Cas-based
technologies for clinical use to identify various human diseases
caused by bacteria, viruses and fungi (32,79).
Application of CRISPR-based diagnostics
beyond the laboratory
To further expand the utility of CRISPR-based
diagnostics as a POC device, numerous sample processing methods
have been developed that are easier to use than previously
developed CRISPR technologies, such as SHERLOCK and DETECTR, and do
not require trained personnel or laboratory equipment. These
methods utilize optimized lysis buffers, heat inactivation and
chemical treatments for subsequent applications. Numerous samples
such as saliva, nasal swabs, urine, blood, blood plasma and serum
have been successfully processed (35). Modified HUDSON has been reported to
inactivate the LSV virus and its viral inactivation efficiency has
been demonstrated using a plaque assay (102).
During the processing of samples containing HPV
DNA, a simple chemical (proteinase K) and heat inactivation method
was effectively combined with the DETECTR assay to minimize the
complex sample processing and ensure the safety of working
personnel (36). Simple and
easy-to-use room-temperature lysis buffers compatible with reverse
transcription and LAMP-based pre-amplification of target RNA
combined with magnetic bead-based RNA extraction containing
concentrated SARS-CoV-2 nucleic acids are easy to use with the
current detection methods, including STOPcovidv.2 (68).
Several approaches have been used for the minimal
handling of processed samples, such as streamlined magnetic
bead-based nucleic acid extraction, easy-to-use chemical reagents,
and reverse transcription, pre-amplification and detection in a
single tube. This is important to prevent contamination during
sample handling after the initial processing. This can be
accomplished in two ways: i) Physical separation of reaction
components that can be mixed in a single tube; and ii) standardized
reaction conditions that can eliminate pre-amplification or the
identification of reaction conditions in which amplification and
CRISPR-based cleavage can be performed together. With the former,
there is still a need to manipulate the reaction tubes without
opening them. Several modifications have also been used to separate
amplification and CRISPR-based cleavage, such as using mineral oil
(104), placing the reagents on
the side of the tubes (105) or
capping the reagent (106)
followed by shaking after the initial amplification is complete.
Cas12 orthologs of thermophilic origin were used in HOLMESv.2 and
STOPcovidv.1and STOPcovidv.2, combining RT-LAMP amplification and
cleavage in a single tube to detect Japanese encephalitis virus
(JEV) and SAR-CoV-2 (63,68). In addition, SHERLOCK was combined
with RT-RPA and Cas13a to enable amplification, cleavage and
detection in a single reaction for the detection of SARS-CoV-2.
Performing amplification and cleavage reactions in a single step
requires optimization of the reaction conditions, reporter and
enzymatic components (96).
Finally, it has been demonstrated that the combination of crRNA and
hyperactive LbuCas13a eliminated the need for amplification
(107). These approaches
efficiently detect viruses without manipulating the reaction
conditions after being set up in a single tube, thus reducing the
risk of contamination of the starting sample (107).
A portable and inexpensive fluorimeter and
fluorescence reader were used to collect signals in either a
lateral flow paper strip or colorimetric assay (Fig. 3). When detecting SARS-CoV-2 with
Cas12a- or Cas13a-based methods, a green-fluorescent signal with a
blue light-emitting diode has been reported (96,106–109). Colorimetric detection is easier
to use because it does not require expensive laboratory equipment
and is also easy to use at home. Lateral flow strips were combined
with Cas13a cleavage and AuNP-tagged FAM-biotin using SHERLOCKv.2.
The tests and control lines could be visualized with the naked eye
on the lateral flow strip (Fig. 3)
(38,79). A similar method was used to
identify the African swine flu virus (ASFV) using Cas12a-based
detection with pre-amplified samples (110). Compared with strip-based lateral
flow tests, colorimetric or fluorescence-based detection in
reaction vessels is more convenient and scalable and has a lower
risk of contamination. An isothermal amplification-based assay that
requires only nucleic acid purification and heat inactivation, such
as LAMP, can detect pH-based, in-tube, color changes (111). However, LAMP-based colorimetric
assays have not been reported as being used for Cas13-based
detection. AuNP-tagged amplified ASFV DNA was detected by
Cas12-based cleavage in a reaction tube, where cleavage of the
target by Cas12 resulted in a color change in the disaggregated
AuNPs, whereas the aggregated AuNPs were colorless. AuNP color is
determined by proximity, which presents a visual color change when
Cas12-based target cleavage occurs. Although not used for
Cas13-based detection of ASFV, AuNP-based colorimetric assays can
also be used for Cas13 (112).
Inhibiting viruses with CRISPR
CRISPR-Cas systems have evolved in half of bacteria
and almost all archaea to protect them from invading bacteriophages
and plasmids (113). Effective
antiviral agents, antibodies, or other therapeutics are needed for
effective treatment of viral diseases. Developing an effective
treatment against viral diseases requires detailed mechanistic
insights into viral biology and is time consuming. In contrast,
CRISPR-based interventions require only genomic sequence
information as the basis from which to inhibit viruses as a
treatment option. CRISPR-Cas systems can target the genomes or
genomic intermediates of viruses. Several approaches have been used
to inhibit viruses by using different Cas orthologs, such as
targeting viral mRNA or RNA genomes (2).
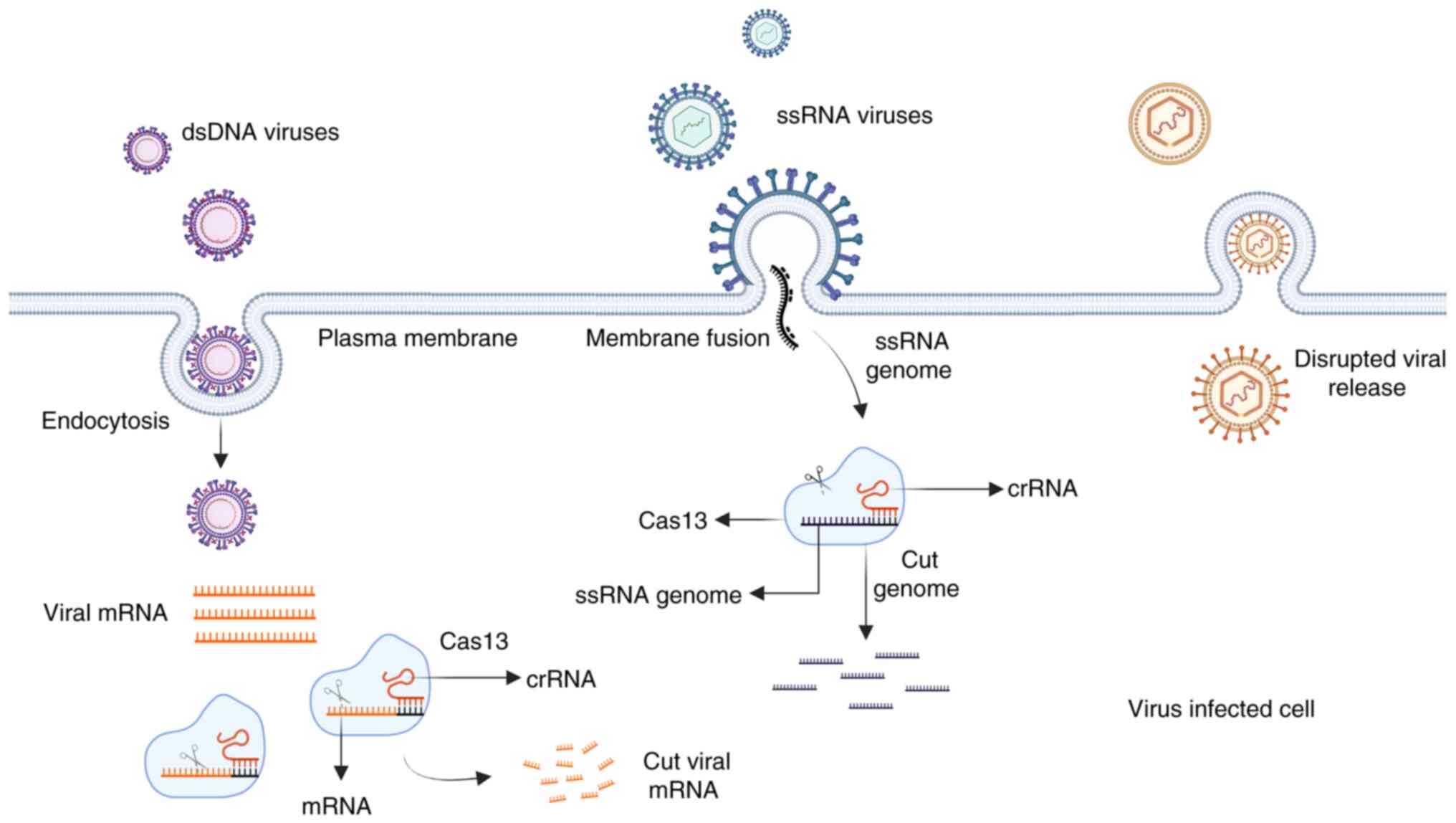
Cas13-based targeting of viral mRNA or RNA
genome
Several viruses contain ssRNA in their genome,
making Cas13 a powerful tool to treat them. Cas13a can be used to
slow down replication and growth (Fig.
6). Previous studies have demonstrated the utility of Cas13a
and Cas13b for reducing host mRNA using mammalian cells (114,115). These studies prompted research
into the investigation of the reduction of ssRNA expression in
LCMV, FLUAV and vesicular stomatitis viruses by Cas13. The study
indicated that Cas13 efficiently inhibited RNA expression (83). These aforementioned studies
identified the optimal crRNA design for effective targeting of
ssRNA in mammalian cells: i) The secondary structure of crRNAs
should be limited; ii) several cytosines should be present nearby;
and iii) several Us or preferred nucleotides should be present
upstream of the target (116).
Further studies have reported that Cas13a can
inhibit the expression and infectivity of viruses, such as HIV-1
and DENV (Fig. 6). Cas13a inhibits
the production of HIV-1 RNA and viral particles, RNA expression
from reactivated proviruses and integrated DNA. Cas13a can also
inhibit the infectivity and expression of DENV and its RNA when
expressed with non-structural 3-targeted crRNA (2,117,118). Detailed characterization of Cas13
revealed that it could be used to combat viruses containing ssRNA
genomes for which the sequence information is known (Fig. 6). SARS-CoV-2 emerged in December
2019 and its first genomic information was published in January
2020. The first Cas13-based method was reported in April 2020 to
inhibit the virus both in vitro and in vivo.
Prophylactic antiviral CRISPR in human cells (PACMAN) was developed
to inhibit SARS-CoV-2 in human cells using smaller Cas13 orthologs
known as Cas13d or CasRx (engineered ribonuclease effector derived
from Ruminococcus flavefaciens XPD3002), which can be
rapidly packaged into adeno-associated viruses (AAVs) (119–121). PACMAN has also been used to
decrease FLUAV levels in human lung epithelial cells. In addition,
crRNA was constructed against the conserved regions of
coronaviruses capable of infecting humans, including SARS-CoV-2,
and Cas13d activity was analyzed using the SARS-CoV-2 reporter
assay system to assess the efficiency of crRNA to target and
degrade SARS-CoV-2 sequences in human cells (119). In addition, further work has
demonstrated that Cas13a, mRNA and virus-specific crRNAs packaged
with a poly(β-amino ester)-based polymer can be delivered to in
vivo mouse and hamster models using a nebulizer. The
effectiveness of this approach was demonstrated using hamster and
mouse models of SARS-CoV-2 and FLUAV infection. It was revealed
that delivery of Cas13a and crRNAs resulted in decreased weight
loss and viral load in the lung tissue, even though the effects
observed were not pronounced (122).
The collateral cleavage activity of Cas13 orthologs
has been widely used in viral diagnostics; however, its effect on
viral RNA or cellular mRNA has not been well explored. Cas13-based
cellular mRNA and viral RNA targeting has little or no collateral
or off-target effects. In addition, it has also been reported that
Cas13- and crRNA-mediated targeting of non-cytotoxic viruses does
not affect cell viability (83,114,115,120,122). Additionally, overexpression of
Cas13a using a lentiviral transduction system and Cas13a-mediated
targeting of overexpressed RNA in glioma cells demonstrated
collateral activity (123).
Transplanting these cells into a mouse model of intracranial
glioblastoma resulted in effective tumor inhibition. The
aforementioned studies indicated that characterization of the
collateral activity of Cas13 against host mRNA and viral RNA still
requires further development. Comprehensive insight into the
collateral activity of Cas13 orthologs, their expression conditions
and the concentration of the target RNA would be beneficial for the
characterization of the therapeutic utility of Cas13.
Cas13-based antiviral approaches are potent and
effective agents for combating viral infections. However, concerted
efforts are needed to study their efficacy and safety requirements
before realizing their clinical potential. Detailed molecular
insights are needed to provide a way to design crRNAs that are less
likely to promote the development of resistance in the target. This
could be achieved by identifying the most conserved regions in the
viral genomes. The enigmatic properties of Cas13 proteins,
including their inherent quality of crRNA processing and
maturation, facilitate multiplexing, which is critical for reducing
the development of resistance during infection and preventing the
inhibition of Cas13 enzymatic activity (46). A previous study indicated that no
mutations in crRNAs were detected in cells treated with Cas13 and
infected with LCMV or DENV2 viruses (117). However, insertions and deletions
flanking crRNA-binding sites were observed in DENV2 isolated from
the supernatant (83,117). Similar studies should be
performed in vitro and in vivo to investigate the
efficacy of Cas13-based treatments that would benefit from
stringent crRNA design. The effectiveness of Cas13-based antiviral
therapies will depend on the further investigations of the optimal
delivery system and minimal or no off-target effects due to the
expression of Cas13 and crRNAs in mammalian cells. The majority of
experiments have been performed with Cas13 and its crRNA before
viral infection, and the in vivo results indicated very low
potency (2). Therefore, more
detailed experiments and insights are required to demonstrate the
effectiveness of Cas13-based treatment approaches for viral
diseases. Furthermore, expression and inhibition by Cas13 should be
equally effective at different stages of viral replication.
Additionally, the potency of Cas13-based therapeutics should
outperform newer antiviral approaches (2).
Conclusions and future perspectives
The identification of CRISPR-Cas proteins, such as
Cas12 and Cas13, has provided a significant impetus for developing
CRISPR-Cas-based diagnostics. Additional molecular insights into
the ssRNA- and ssDNA-degrading collateral activity of these
proteins have prompted the rapid development of CRISPR-based
diagnostics for disease detection (34,36).
Multiplexing, ease of use and use outside well-equipped
laboratories have made these tests increasingly popular.
Furthermore, the development of buffers, reagents and effective
methods for inactivating viral samples that are compatible with
pre-amplification and detection by Cas13 or Cas12 in a single tube
makes CRISPR-based diagnostics viable as a POC device (2,35,68,96).
Biochemical characterization of Cas13 and Cas12 orthologs with
specific and robust fluorescence-quencher reporters have enabled
multiplexing (Fig. 4B). The
identification and characterization of Csm6-Cas type III proteins
makes tandem utilization of Cas orthologs possible (Fig. 4A) (38). Detailed molecular knowledge of
hyperactive LbuCas13a has increased the sensitivity of CRISPR-based
diagnostics from aM to zM (107).
The present review hypothesizes that CRISPR-based diagnostic tests
will remain at the forefront of diagnosing viral and
human-associated diseases in the coming decades.
The detection of smaller Cas enzymes would support
the development of CRISPR-based diagnostics and therapeutics. The
previously reported Cas14a (CasX) proteins are small and contain
~400–700 amino acids (61). CasX
proteins contain a single RuvC endonuclease domain that is required
for ssDNA cleavage. It recognizes ssDNA, which is directed by
ssRNA, to induce ssDNA cleavage. This requires crRNAs and tracrRNAs
for target recognition and cleavage. CasX recognizes and binds to
the 20 nt target ssDNA. CasX does not require PAM; variations in
the middle of the target sequence (seed) are the most sensitive,
and changes in this region abolish cleavage. Recognition of the
target by CasX gRNA on ssDNA activates ssDNA-cleaving collateral
activity (61). The utility of
Cas14a in the efficient detection of SNP was validated against
Cas12a using a quencher-labeled ssDNA probe (61). In addition, finding CRISPR-Cas
systems in bacteriophages could lead to the detection of the
smaller counterparts of the Cas enzymes that could be quickly
packaged in AAV particles, which could improve the scope of
CRISPR-based diagnostic and therapeutic approaches to treat a
variety of human genetic disorders (124). LbuCas13a has recently been
demonstrated to manipulate phage genomes, and multiple phage genes
can be simultaneously edited. Advances in the detection and
in-depth characterization of these enzymes have expanded the scope
of CRISPR-based diagnostics for the detection of human diseases
(125,126).
Acknowledgements
Not applicable.
Funding
Funding: No funding was received.
Availability of data and materials
Not applicable.
Authors' contributions
MS and MK conceptualized the project. MS reviewed
the literature. MS and MK prepared the manuscript. MS, RS and NA
designed and created the schematic representation. MS, MK, NA, RS,
RR and PK edited the manuscript. All authors have read and approved
the final version of the manuscript. Data authentication is not
applicable.
Ethics approval and consent to
participate
Not applicable.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
Glossary
Abbreviations
Abbreviations:
|
CRISPR
|
clustered regularly interspaced short
palindromic repeats
|
|
Cas
|
CRISPR-associated
|
|
AaCas12b
|
Alicyclobacillus acidiphilus
Cas12b
|
|
AAVs
|
adeno-associated viruses
|
|
AsCas12a
|
Acidaminococcus sp. Cas12a
|
|
ASFV
|
African swine flu virus
|
|
AuNPs
|
gold nanoparticles
|
|
CARMEN
|
combinatorial array reactions for the
multiplex evaluation of nucleic acids
|
|
CcaCas13b
|
Capnocytophaga canimorsus
Cas13b
|
|
CDC
|
Centers for Disease Control and
Prevention
|
|
crRNA
|
CRISPR RNA
|
|
gRNA
|
guide RNA
|
|
DENV
|
dengue virus
|
|
DETECTR
|
DNA endonuclease targeted CRISPR
trans reporter
|
|
DISCoVER
|
diagnostics with coronavirus
enzymatic reporting
|
|
DRs
|
direct repeats
|
|
dsDNA
|
double-stranded DNA
|
|
EiCsm6
|
Csm6 from Enterococcus
italicus
|
|
FAM
|
fluorescein amidites
|
|
FIND-IT
|
fast integrated nuclease detection in
tandem
|
|
FLUAV
|
Influenza A virus
|
|
HAVs
|
human-associated viruses
|
|
HEPN
|
higher eukaryotic and prokaryotic
nucleotide
|
|
HEX
|
hexachloro-fluorescein
|
|
HIV
|
human immune deficiency virus
|
|
HOLMES
|
1-h low-cost multipurpose highly
efficient system
|
|
HPV
|
human papillomavirus
|
|
HUDSON
|
heating unextracted diagnostic
samples to obliterate nucleases
|
|
JEV
|
Japanese encephalitis virus
|
|
LAMP
|
loop-mediated isotherm
amplification
|
|
LbaCas13a
|
Lachnospiraceae bacterium
Cas13a
|
|
LbCas12a
|
Lachnospiraceae bacterium
Cas12a
|
|
LbuCas13a
|
Leptotrichia buccalis
Cas13a
|
|
LCMV
|
lymphocytic choriomeningitis
virus
|
|
LsCsm6
|
Csm6 from Lactobacillus
salivarius
|
|
LshCas13a
|
Leptotrichia shahii Cas13a
|
|
LwaCas13a
|
Leptotrichia wadei Cas13a
|
|
NASBA
|
nucleic acid sequence-based
amplification
|
|
nt
|
nucleotide
|
|
PACMAN
|
prophylactic antiviral CRISPR in
human cells
|
|
PAM
|
protospacer-associated motif
|
|
POC
|
point-of-care
|
|
PsmCas13b
|
Prevotella sp. MA2016
Cas13b
|
|
RNase
|
RNA endonuclease
|
|
RPA
|
recombinase polymerase
amplification
|
|
RT-qPCR
|
reverse transcription-quantitative
PCR
|
|
SARS-CoV
|
severe acute respiratory syndrome
corona virus
|
|
ssDNA
|
single-stranded DNA
|
|
SHERLOCK
|
specific high-sensitivity enzymatic
reporter unlocking
|
|
SHINE
|
streamlined highlighting of
infections to navigate epidemics
|
|
SNP
|
single nucleotide polymorphism
|
|
ssRNA
|
single-stranded RNA
|
|
STOP
|
SHERLOCK test in one pot
|
|
tracrRNA
|
trans-activating CRISPR RNA
|
|
TtCsm6
|
Csm6 from Thermus
thermophilus
|
|
ZIKV
|
zika virus
|
References
|
1
|
Jayamohan H, Lambert CJ, Sant HJ, Jafek A,
Patel D, Feng H, Beeman M, Mahmood T, Nze U and Gale BK: SARS-CoV-2
pandemic: A review of molecular diagnostic tools including sample
collection and commercial response with associated advantages and
limitations. Anal Bioanal Chem. 413:49–71. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Freije CA and Sabeti PC: Detect and
destroy: CRISPR-based technologies for the response against
viruses. Cell Host Microbe. 29:689–703. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
da Costa VG, Moreli ML and Saivish MV: The
emergence of SARS, MERS and novel SARS-2 coronaviruses in the 21st
century. Arch Virol. 165:1517–1526. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Johnson CK, Hitchens PL, Pandit PS,
Rushmore J, Evans TS, Young CCW and Doyle MM: Global shifts in
mammalian population trends reveal key predictors of virus
spillover risk. Proc Biol Sci. 287:201927362020.PubMed/NCBI
|
|
5
|
Morse SS, Mazet JA, Woolhouse M, Parrish
CR, Carroll D, Karesh WB, Zambrana-Torrelio C, Lipkin WI and Daszak
P: Prediction and prevention of the next pandemic zoonosis. Lancet.
380:1956–1965. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Nieto-Rabiela F, Wiratsudakul A, Suzan G
and Rico-Chavez O: Viral networks and detection of potential
zoonotic viruses in bats and rodents: A worldwide analysis.
Zoonoses Public Health. 66:655–666. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Olival KJ, Hosseini PR, Zambrana-Torrelio
C, Ross N, Bogich TL and Daszak P: Host and viral traits predict
zoonotic spillover from mammals. Nature. 546:646–650. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Bouvier NM and Palese P: The biology of
influenza viruses. Vaccine. 26 (Suppl 4):D49–D53. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Tong S, Zhu X, Li Y, Shi M, Zhang J,
Bourgeois M, Yang H, Chen X, Recuenco S, Gomez J, et al: New world
bats harbor diverse influenza A viruses. PLoS Pathog.
9:e10036572013. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Elena SF, Bedhomme S, Carrasco P, Cuevas
JM, de la Iglesia F, Lafforgue G, Lalić J, Pròsper A, Tromas N and
Zwart MP: The evolutionary genetics of emerging plant RNA viruses.
Mol Plant Microbe Interact. 24:287–293. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Sanjuan R, Nebot MR, Chirico N, Mansky LM
and Belshaw R: Viral mutation rates. J Virol. 84:9733–9748. 2010.
View Article : Google Scholar : PubMed/NCBI
|
|
12
|
De Clercq E and Li G: Approved antiviral
drugs over the past 50 years. Clin Microbiol Rev. 29:695–747. 2016.
View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Gordon DE, Hiatt J, Bouhaddou M, Rezelj
VV, Ulferts S, Braberg H, Jureka AS, Obernier K, Guo JZ, Batra J,
et al: Comparative host-coronavirus protein interaction networks
reveal pan-viral disease mechanisms. Science. 370:eabe94032020.
View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Gordon DE, Jang GM, Bouhaddou M, Xu J,
Obernier K, O'Meara MJ, Guo JZ, Swaney DL, Tummino TA, Hüttenhain
R, et al: A SARS-CoV-2 protein interaction map reveals targets for
drug repurposing. Nature. 583:459–468. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Zhou Y, Liu Y, Gupta S, Paramo MI, Hou Y,
Mao C, Luo Y, Judd J, Wierbowski S, Bertolotti M, et al: A
comprehensive SARS-CoV-2-human protein-protein interactome reveals
COVID-19 pathobiology and potential host therapeutic targets. Nat
Biotechnol. 41:128–139. 2023. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Bojkova D, Klann K, Koch B, Widera M,
Krause D, Ciesek S, Cinatl J and Münch C: Proteomics of
SARS-CoV-2-infected host cells reveals therapy targets. Nature.
583:469–472. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Kole R, Krainer AR and Altman S: RNA
therapeutics: Beyond RNA interference and antisense
oligonucleotides. Nat Rev Drug Discov. 11:125–140. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Warren TK, Warfield KL, Wells J, Swenson
DL, Donner KS, Van Tongeren SA, Garza NL, Dong L, Mourich DV,
Crumley S, et al: Advanced antisense therapies for postexposure
protection against lethal filovirus infections. Nat Med.
16:991–994. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Xu Y and Li Z: CRISPR-Cas systems:
Overview, innovations and applications in human disease research
and gene therapy. Comput Struct Biotechnol J. 18:2401–2415. 2020.
View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Kostyusheva A, Brezgin S, Babin Y,
Vasilyeva I, Glebe D, Kostyushev D and Chulanov V: CRISPR-Cas
systems for diagnosing infectious diseases. Methods. 203:431–446.
2022. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Ebrahimi S, Khanbabaei H, Abbasi S, Fani
M, Soltani S, Zandi M and Najafimemar Z: CRISPR-Cas System: A
promising diagnostic tool for Covid-19. Avicenna J Med Biotechnol.
14:3–9. 2022.PubMed/NCBI
|
|
22
|
Yang S and Rothman RE: PCR-based
diagnostics for infectious diseases: Uses, limitations, and future
applications in acute-care settings. Lancet Infect Dis. 4:337–348.
2004. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Mackay IM, Arden KE and Nitsche A:
Real-time PCR in virology. Nucleic Acids Res. 30:1292–1305. 2002.
View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Zanoli LM and Spoto G: Isothermal
amplification methods for the detection of nucleic acids in
microfluidic devices. Biosensors (Basel). 3:18–43. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Cassedy A, Parle-McDermott A and O'Kennedy
R: Virus Detection: A review of the current and emerging molecular
and immunological methods. Front Mol Biosci. 8:6375592021.
View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Notomi T, Okayama H, Masubuchi H, Yonekawa
T, Watanabe K, Amino N and Hase T: Loop-mediated isothermal
amplification of DNA. Nucleic Acids Res. 28:E632000. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Compton J: Nucleic acid sequence-based
amplification. Nature. 350:91–92. 1991. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Piepenburg O, Williams CH, Stemple DL and
Armes NA: DNA detection using recombination proteins. PLoS Biol.
4:e2042006. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Kurosaki Y, Martins DBG, Kimura M, Catena
ADS, Borba MACSM, Mattos SDS, Abe H, Yoshikawa R, de Lima Filho JL
and Yasuda J: Development and evaluation of a rapid molecular
diagnostic test for Zika virus infection by reverse transcription
loop-mediated isothermal amplification. Sci Rep. 7:135032017.
View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Patel P, Abd El Wahed A, Faye O, Prüger P,
Kaiser M, Thaloengsok S, Ubol S, Sakuntabhai A, Leparc-Goffart I,
Hufert FT, et al: A field-deployable reverse transcription
recombinase polymerase amplification assay for rapid detection of
the chikungunya virus. PLoS Negl Trop Dis. 10:e00049532016.
View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Broughton JP, Deng X, Yu G, Fasching CL,
Servellita V, Singh J, Miao X, Streithorst JA, Granados A,
Sotomayor-Gonzalez A, et al: CRISPR-Cas12-based detection of
SARS-CoV-2. Nat Biotechnol. 38:870–874. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Gootenberg JS, Abudayyeh OO, Lee JW,
Essletzbichler P, Dy AJ, Joung J, Verdine V, Donghia N, Daringer
NM, Freije CA, et al: Nucleic acid detection with
CRISPR-Cas13a/C2c2. Science. 356:438–442. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Liang Y, Lin H, Zou L, Zhao J, Li B, Wang
H, Lu J, Sun J, Yang X, Deng X and Tang S: CRISPR-Cas12a-based
detection for the major SARS-CoV-2 variants of concern. Microbiol
Spectr. 9:e01017212021. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Kellner MJ, Koob JG, Gootenberg JS,
Abudayyeh OO and Zhang F: SHERLOCK: Nucleic acid detection with
CRISPR nucleases. Nat Protoc. 14:2986–3012. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Kaminski MM, Abudayyeh OO, Gootenberg JS,
Zhang F and Collins JJ: CRISPR-based diagnostics. Nat Biomed Eng.
5:643–656. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Chen JS, Ma E, Harrington LB, Da Costa M,
Tian X, Palefsky JM and Doudna JA: CRISPR-Cas12a target binding
unleashes indiscriminate single-stranded DNase activity. Science.
360:436–439. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
East-Seletsky A, O'Connell MR, Knight SC,
Burstein D, Cate JH, Tjian R and Doudna JA: Two distinct RNase
activities of CRISPR-C2c2 enable guide-RNA processing and RNA
detection. Nature. 538:270–273. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Gootenberg JS, Abudayyeh OO, Kellner MJ,
Joung J, Collins JJ and Zhang F: Multiplexed and portable nucleic
acid detection platform with Cas13, Cas12a, and Csm6. Science.
360:439–444. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Marraffini LA and Sontheimer EJ: CRISPR
interference limits horizontal gene transfer in staphylococci by
targeting DNA. Science. 322:1843–1845. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Barrangou R, Fremaux C, Deveau H, Richards
M, Boyaval P, Moineau S, Romero DA and Horvath P: CRISPR provides
acquired resistance against viruses in prokaryotes. Science.
315:1709–1712. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Pickar-Oliver A and Gersbach CA: The next
generation of CRISPR-Cas technologies and applications. Nat Rev Mol
Cell Biol. 20:490–507. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Jinek M, Chylinski K, Fonfara I, Hauer M,
Doudna JA and Charpentier E: A programmable dual-RNA-guided DNA
endonuclease in adaptive bacterial immunity. Science. 337:816–821.
2012. View Article : Google Scholar : PubMed/NCBI
|
|
43
|
Abudayyeh OO, Gootenberg JS, Konermann S,
Joung J, Slaymaker IM, Cox DB, Shmakov S, Makarova KS, Semenova E,
Minakhin L, et al: C2c2 is a single-component programmable
RNA-guided RNA-targeting CRISPR effector. Science. 353:aaf55732016.
View Article : Google Scholar : PubMed/NCBI
|
|
44
|
van der Oost J, Westra ER, Jackson RN and
Wiedenheft B: Unravelling the structural and mechanistic basis of
CRISPR-Cas systems. Nat Rev Microbiol. 12:479–492. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
45
|
Wright AV, Nunez JK and Doudna JA: Biology
and applications of CRISPR systems: Harnessing Nature's toolbox for
genome engineering. Cell. 164:29–44. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
46
|
Shmakov S, Abudayyeh OO, Makarova KS, Wolf
YI, Gootenberg JS, Semenova E, Minakhin L, Joung J, Konermann S,
Severinov K, et al: Discovery and functional characterization of
diverse class 2 CRISPR-cas systems. Mol Cell. 60:385–397. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
47
|
Fonfara I, Richter H, Bratovic M, Le Rhun
A and Charpentier E: The CRISPR-associated DNA-cleaving enzyme Cpf1
also processes precursor CRISPR RNA. Nature. 532:517–521. 2016.
View Article : Google Scholar : PubMed/NCBI
|
|
48
|
East-Seletsky A, O'Connell MR, Burstein D,
Knott GJ and Doudna JA: RNA targeting by functionally orthogonal
type VI-A CRISPR-cas enzymes. Mol Cell. 66:373–383. e3732017.
View Article : Google Scholar : PubMed/NCBI
|
|
49
|
Li H: Structural principles of CRISPR RNA
processing. Structure. 23:13–20. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
50
|
Charpentier E, Richter H, van der Oost J
and White MF: Biogenesis pathways of RNA guides in archaeal and
bacterial CRISPR-Cas adaptive immunity. FEMS Microbiol Rev.
39:428–441. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
51
|
Hochstrasser ML and Doudna JA: Cutting it
close: CRISPR-associated endoribonuclease structure and function.
Trends Biochem Sci. 40:58–66. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
52
|
Deltcheva E, Chylinski K, Sharma CM,
Gonzales K, Chao Y, Pirzada ZA, Eckert MR, Vogel J and Charpentier
E: CRISPR RNA maturation by trans-encoded small RNA and host factor
RNase III. Nature. 471:602–607. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
53
|
Fonfara I, Le Rhun A, Chylinski K,
Makarova KS, Lécrivain AL, Bzdrenga J, Koonin EV and Charpentier E:
Phylogeny of Cas9 determines functional exchangeability of dual-RNA
and Cas9 among orthologous type II CRISPR-Cas systems. Nucleic
Acids Res. 42:2577–2590. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
54
|
Yang W: Nucleases: Diversity of structure,
function and mechanism. Q Rev Biophys. 44:1–93. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
55
|
Cordray MS and Richards-Kortum RR:
Emerging nucleic acid-based tests for point-of-care detection of
malaria. Am J Trop Med Hyg. 87:223–230. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
56
|
Rohrman BA, Leautaud V, Molyneux E and
Richards-Kortum RR: A lateral flow assay for quantitative detection
of amplified HIV-1 RNA. PLoS One. 7:e456112012. View Article : Google Scholar : PubMed/NCBI
|
|
57
|
Yan L, Zhou J, Zheng Y, Gamson AS, Roembke
BT, Nakayama S and Sintim HO: Isothermal amplified detection of DNA
and RNA. Mol Biosyst. 10:970–1003. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
58
|
Ortiz-Cartagena C, Fernandez-Garcia L,
Blasco L, Pacios O, Bleriot I, López M, Cantón R and Tomás M:
Reverse Transcription-loop-mediated isothermal
Amplification-CRISPR-Cas13a technology as a promising diagnostic
tool for SARS-CoV-2. Microbiol Spectr. 10:e02398222022. View Article : Google Scholar : PubMed/NCBI
|
|
59
|
Li SY, Cheng QX, Wang JM, Li XY, Zhang ZL,
Gao S, Cao RB, Zhao GP and Wang J: CRISPR-Cas12a-assisted nucleic
acid detection. Cell Discov. 4:202018. View Article : Google Scholar : PubMed/NCBI
|
|
60
|
Mustafa MI and Makhawi AM: SHERLOCK and
DETECTR: CRISPR-Cas systems as potential rapid diagnostic tools for
emerging infectious diseases. J Clin Microbiol. 59:e00745–20. 2021.
View Article : Google Scholar : PubMed/NCBI
|
|
61
|
Harrington LB, Burstein D, Chen JS,
Paez-Espino D, Ma E, Witte IP, Cofsky JC, Kyrpides NC, Banfield JF
and Doudna JA: Programmed DNA destruction by miniature CRISPR-Cas14
enzymes. Science. 362:839–842. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
62
|
Hillary VE and Ceasar SA: A review on the
mechanism and applications of CRISPR/Cas9/Cas12/Cas13/Cas14
proteins utilized for genome engineering. Mol Biotechnol.
65:311–325. 2023. View Article : Google Scholar : PubMed/NCBI
|
|
63
|
Li L, Li S, Wu N, Wu J, Wang G, Zhao G and
Wang J: HOLMESv2: A CRISPR-Cas12b-assisted platform for nucleic
acid detection and DNA methylation quantitation. ACS Synth Biol.
8:2228–2237. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
64
|
Li SY, Cheng QX, Liu JK, Nie XQ, Zhao GP
and Wang J: CRISPR-Cas12a has both cis- and trans-cleavage
activities on single-stranded DNA. Cell Res. 28:491–493. 2018.
View Article : Google Scholar : PubMed/NCBI
|
|
65
|
Safari F, Afarid M, Rastegari B,
Borhani-Haghighi A, Barekati-Mowahed M and Behzad-Behbahani A:
CRISPR systems: Novel approaches for detection and combating
COVID-19. Virus Res. 294:1982822021. View Article : Google Scholar : PubMed/NCBI
|
|
66
|
Teng F, Guo L, Cui T, Wang XG, Xu K, Gao
Q, Zhou Q and Li W: CDetection: CRISPR-Cas12b-based DNA detection
with sub-attomolar sensitivity and single-base specificity. Genome
Biol. 20:1322019. View Article : Google Scholar : PubMed/NCBI
|
|
67
|
Arizti-Sanz J, Bradley A, Zhang YB, Boehm
CK, Freije CA, Grunberg ME, Kosoko-Thoroddsen TF, Welch NL, Pillai
PP, Mantena S, et al: Simplified Cas13-based assays for the fast
identification of SARS-CoV-2 and its variants. Nat Biomed Eng.
6:932–943. 2022. View Article : Google Scholar : PubMed/NCBI
|
|
68
|
Joung J, Ladha A, Saito M, Kim NG, Woolley
AE, Segel M, Barretto RPJ, Ranu A, Macrae RK, Faure G, et al:
Detection of SARS-CoV-2 with SHERLOCK One-Pot Testing. N Engl J
Med. 383:1492–1494. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
69
|
Soh JH, Balleza E, Abdul Rahim MN, Chan
HM, Mohd Ali S, Chuah JKC, Edris S, Atef A, Bahieldin A, Ying JY
and Sabir JSM: CRISPR-based systems for sensitive and rapid on-site
COVID-19 diagnostics. Trends Biotechnol. 40:1346–1360. 2022.
View Article : Google Scholar : PubMed/NCBI
|
|
70
|
Ackerman CM, Myhrvold C, Thakku SG, Freije
CA, Metsky HC, Yang DK, Ye SH, Boehm CK, Kosoko-Thoroddsen TF, Kehe
J, et al: Massively multiplexed nucleic acid detection with Cas13.
Nature. 582:277–282. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
71
|
Yu TC: Beginnings and development of
nursing education in Republic of China. Hu Li Za Zhi. 24:39–42.
1977.(In Chinese). PubMed/NCBI
|
|
72
|
Wang M, Zhang R and Li J: CRISPR/cas
systems redefine nucleic acid detection: Principles and methods.
Biosens Bioelectron. 165:1124302020. View Article : Google Scholar : PubMed/NCBI
|
|
73
|
Emmadi R, Boonyaratanakornkit JB,
Selvarangan R, Shyamala V, Zimmer BL, Williams L, Bryant B,
Schutzbank T, Schoonmaker MM, Amos Wilson JA, et al: Molecular
methods and platforms for infectious diseases testing a review of
FDA-approved and cleared assays. J Mol Diagn. 13:583–604. 2011.
View Article : Google Scholar : PubMed/NCBI
|
|
74
|
Song L, Shan D, Zhao M, Pink BA, Minnehan
KA, York L, Gardel M, Sullivan S, Phillips AF, Hayman RB, et al:
Direct detection of bacterial genomic DNA at sub-femtomolar
concentrations using single molecule arrays. Anal Chem.
85:1932–1939. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
75
|
Barletta JM, Edelman DC and Constantine
NT: Lowering the detection limits of HIV-1 viral load using
real-time immuno-PCR for HIV-1 p24 antigen. Am J Clin Pathol.
122:20–27. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
76
|
Faye O, Faye O, Dupressoir A, Weidmann M,
Ndiaye M and Alpha Sall A: One-step RT-PCR for detection of Zika
virus. J Clin Virol. 43:96–101. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
77
|
Faye O, Faye O, Diallo D, Diallo M,
Weidmann M and Sall AA: Quantitative real-time PCR detection of
Zika virus and evaluation with field-caught mosquitoes. Virol J.
10:3112013. View Article : Google Scholar : PubMed/NCBI
|
|
78
|
Waggoner JJ, Abeynayake J, Sahoo MK, Gresh
L, Tellez Y, Gonzalez K, Ballesteros G, Guo FP, Balmaseda A,
Karunaratne K, et al: Comparison of the FDA-approved CDC DENV-1-4
real-time reverse transcription-PCR with a laboratory-developed
assay for dengue virus detection and serotyping. J Clin Microbiol.
51:3418–3420. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
79
|
Myhrvold C, Freije CA, Gootenberg JS,
Abudayyeh OO, Metsky HC, Durbin AF, Kellner MJ, Tan AL, Paul LM,
Parham LA, et al: Field-deployable viral diagnostics using
CRISPR-Cas13. Science. 360:444–448. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
80
|
Javalkote VS, Kancharla N, Bhadra B,
Shukla M, Soni B, Sapre A, Goodin M, Bandyopadhyay A and Dasgupta
S: CRISPR-based assays for rapid detection of SARS-CoV-2. Methods.
203:594–603. 2022. View Article : Google Scholar : PubMed/NCBI
|
|
81
|
Huang Z, Fang J, Zhou M, Gong Z and Xiang
T: CRISPR-Cas13: A new technology for the rapid detection of
pathogenic microorganisms. Front Microbiol. 13:10113992022.
View Article : Google Scholar : PubMed/NCBI
|
|
82
|
Shen F, Sun B, Kreutz JE, Davydova EK, Du
W, Reddy PL, Joseph LJ and Ismagilov RF: Multiplexed quantification
of nucleic acids with large dynamic range using multivolume digital
RT-PCR on a rotational SlipChip tested with HIV and hepatitis C
viral load. J Am Chem Soc. 133:17705–17712. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
83
|
Freije CA, Myhrvold C, Boehm CK, Lin AE,
Welch NL, Carter A, Metsky HC, Luo CY, Abudayyeh OO, Gootenberg JS,
et al: Programmable inhibition and detection of RNA viruses using
Cas13. Mol Cell. 76:826–837. e8112019. View Article : Google Scholar : PubMed/NCBI
|
|
84
|
Tang Y and Fu Y: Class 2 CRISPR/Cas: An
expanding biotechnology toolbox for and beyond genome editing. Cell
Biosci. 8:592018. View Article : Google Scholar : PubMed/NCBI
|
|
85
|
Zetsche B, Gootenberg JS, Abudayyeh OO,
Slaymaker IM, Makarova KS, Essletzbichler P, Volz SE, Joung J, van
der Oost J, Regev A, et al: Cpf1 is a single RNA-guided
endonuclease of a class 2 CRISPR-Cas system. Cell. 163:759–771.
2015. View Article : Google Scholar : PubMed/NCBI
|
|
86
|
Swarts DC, van der Oost J and Jinek M:
Structural basis for guide RNA processing and seed-dependent DNA
targeting by CRISPR-Cas12a. Mol Cell. 66:221–233.e4. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
87
|
Dong D, Ren K, Qiu X, Zheng J, Guo M, Guan
X, Liu H, Li N, Zhang B, Yang D, et al: The crystal structure of
Cpf1 in complex with CRISPR RNA. Nature. 532:522–526. 2016.
View Article : Google Scholar : PubMed/NCBI
|
|
88
|
Gao P, Yang H, Rajashankar KR, Huang Z and
Patel DJ: Type V CRISPR-Cas Cpf1 endonuclease employs a unique
mechanism for crRNA-mediated target DNA recognition. Cell Res.
26:901–913. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
89
|
Stella S, Alcon P and Montoya G: Structure
of the Cpf1 endonuclease R-loop complex after target DNA cleavage.
Nature. 546:559–563. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
90
|
Yamano T, Nishimasu H, Zetsche B, Hirano
H, Slaymaker IM, Li Y, Fedorova I, Nakane T, Makarova KS, Koonin
EV, et al: Crystal structure of Cpf1 in complex with Guide RNA and
target DNA. Cell. 165:949–962. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
91
|
Drame M, Tabue Teguo M, Proye E, Hequet F,
Hentzien M, Kanagaratnam L and Godaert L: Should RT-PCR be
considered a gold standard in the diagnosis of COVID-19? J Med
Virol. 92:2312–2313. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
92
|
Patchsung M, Jantarug K, Pattama A,
Aphicho K, Suraritdechachai S, Meesawat P, Sappakhaw K, Leelahakorn
N, Ruenkam T, Wongsatit T, et al: Clinical validation of a
Cas13-based assay for the detection of SARS-CoV-2 RNA. Nat Biomed
Eng. 4:1140–1149. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
93
|
Teng F, Cui T, Feng G, Guo L, Xu K, Gao Q,
Li T, Li J, Zhou Q and Li W: Repurposing CRISPR-Cas12b for
mammalian genome engineering. Cell Discov. 4:632018. View Article : Google Scholar : PubMed/NCBI
|
|
94
|
Liu TY, Knott GJ, Smock DCJ, Desmarais JJ,
Son S, Bhuiya A, Jakhanwal S, Prywes N, Agrawal S, Díaz de León
Derby M, et al: Accelerated RNA detection using tandem CRISPR
nucleases. Nat Chem Biol. 17:982–988. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
95
|
Chandrasekaran SS, Agrawal S, Fanton A,
Jangid AR, Charrez B, Escajeda AM, Son S, Mcintosh R, Tran H,
Bhuiya A, et al: Rapid detection of SARS-CoV-2 RNA in saliva via
Cas13. Nat Biomed Eng. 6:944–956. 2022. View Article : Google Scholar : PubMed/NCBI
|
|
96
|
Arizti-Sanz J, Freije CA, Stanton AC,
Petros BA, Boehm CK, Siddiqui S, Shaw BM, Adams G,
Kosoko-Thoroddsen TF, Kemball ME, et al: Streamlined inactivation,
amplification, and Cas13-based detection of SARS-CoV-2. Nat Commun.
11:59212020. View Article : Google Scholar : PubMed/NCBI
|
|
97
|
Qin P, Park M, Alfson KJ, Tamhankar M,
Carrion R, Patterson JL, Griffiths A, He Q, Yildiz A, Mathies R and
Du K: Rapid and fully microfluidic ebola virus detection with
CRISPR-Cas13a. ACS Sens. 4:1048–1054. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
98
|
Wu Y, Liu SX, Wang F and Zeng MS: Room
temperature detection of plasma Epstein-barr virus DNA with
CRISPR-Cas13. Clin Chem. 65:591–592. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
99
|
Normandin E, Solomon IH, Zamirpour S,
Lemieux J, Freije CA, Mukerji SS, Tomkins-Tinch C, Park D, Sabeti
PC and Piantadosi A: Powassan Virus neuropathology and genomic
diversity in patients with fatal encephalitis. Open Forum Infect
Dis. 7:ofaa3922020. View Article : Google Scholar : PubMed/NCBI
|
|
100
|
Liu Y, Xu H, Liu C, Peng L, Khan H, Cui L,
Huang R, Wu C, Shen S, Wang S, et al: CRISPR-Cas13a nanomachine
based simple technology for avian Influenza A (H7N9) Virus On-Site
detection. J Biomed Nanotechnol. 15:790–798. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
101
|
Curti LA, Pereyra-Bonnet F, Repizo GD, Fay
JV, Salvatierra K, Blariza MJ, Ibañez-Alegre D, Rinflerch AR,
Miretti M and Gimenez CA: CRISPR-based platform for carbapenemases
and emerging viruses detection using Cas12a (Cpf1) effector
nuclease. Emerg Microbes Infect. 9:1140–1148. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
102
|
Barnes KG, Lachenauer AE, Nitido A,
Siddiqui S, Gross R, Beitzel B, Siddle KJ, Freije CA, Dighero-Kemp
B, Mehta SB, et al: Deployable CRISPR-Cas13a diagnostic tools to
detect and report Ebola and Lassa virus cases in real-time. Nat
Commun. 11:41312020. View Article : Google Scholar : PubMed/NCBI
|
|
103
|
Broughton JP, Deng X, Yu G, Fasching CL,
Singh J, Streithorst J, Granados A, Sotomayor-Gonzalez A, Zorn K,
Gopez A, et al: Rapid detection of 2019 Novel coronavirus
SARS-CoV-2 using a CRISPR-based DETECTR lateral flow assay.
medRxiv. Mar 27–2020.doi: 10.1101/2020.03.06.20032334. PubMed/NCBI
|
|
104
|
Chen Y, Shi Y, Chen Y, Yang Z, Wu H, Zhou
Z, Li J, Ping J, He L, Shen H, et al: Contamination-free visual
detection of SARS-CoV-2 with CRISPR/Cas12a: A promising method in
the point-of-care detection. Biosens Bioelectron. 169:1126422020.
View Article : Google Scholar : PubMed/NCBI
|
|
105
|
Ali Z, Aman R, Mahas A, Rao GS, Tehseen M,
Marsic T, Salunke R, Subudhi AK, Hala SM, Hamdan SM, et al: iSCAN:
An RT-LAMP-coupled CRISPR-Cas12 module for rapid, sensitive
detection of SARS-CoV-2. Virus Res. 288:1981292020. View Article : Google Scholar : PubMed/NCBI
|
|
106
|
Guo L, Sun X, Wang X, Liang C, Jiang H,
Gao Q, Dai M, Qu B, Fang S, Mao Y, et al: SARS-CoV-2 detection with
CRISPR diagnostics. Cell Discov. 6:342020. View Article : Google Scholar : PubMed/NCBI
|
|
107
|
Fozouni P, Son S, Diaz de Leon Derby M,
Knott GJ, Gray CN, D'Ambrosio MV, Zhao C, Switz NA, Kumar GR,
Stephens SI, et al: Amplification-free detection of SARS-CoV-2 with
CRISPR-Cas13a and mobile phone microscopy. Cell. 184:323–333.e9.
2021. View Article : Google Scholar : PubMed/NCBI
|
|
108
|
Hale CR, Zhao P, Olson S, Duff MO,
Graveley BR, Wells L, Terns RM and Terns MP: RNA-guided RNA
cleavage by a CRISPR RNA-Cas protein complex. Cell. 139:945–956.
2009. View Article : Google Scholar : PubMed/NCBI
|
|
109
|
Wang X, Zhong M, Liu Y, Ma P, Dang L, Meng
Q, Wan W, Ma X, Liu J, Yang G, et al: Rapid and sensitive detection
of COVID-19 using CRISPR/Cas12a-based detection with naked eye
readout, CRISPR/Cas12a-NER. Sci Bull (Beijing). 65:1436–1439. 2020.
View Article : Google Scholar : PubMed/NCBI
|
|
110
|
Lu S, Li F, Chen Q, Wu J, Duan J, Lei X,
Zhang Y, Zhao D, Bu Z and Yin H: Rapid detection of African swine
fever virus using Cas12a-based portable paper diagnostics. Cell
Discov. 6:182020. View Article : Google Scholar : PubMed/NCBI
|
|
111
|
Poole CB, Li Z, Alhassan A, Guelig D,
Diesburg S, Tanner NA, Zhang Y, Evans TC Jr, LaBarre P, Wanji S, et
al: Colorimetric tests for diagnosis of filarial infection and
vector surveillance using non-instrumented nucleic acid
loop-mediated isothermal amplification (NINA-LAMP). PLoS One.
12:e01690112017. View Article : Google Scholar : PubMed/NCBI
|
|
112
|
Yuan C, Tian T, Sun J, Hu M, Wang X, Xiong
E, Cheng M, Bao Y, Lin W, Jiang J, et al: Universal and Naked-Eye
Gene detection platform based on the clustered regularly
interspaced short palindromic Repeats/Cas12a/13a system. Anal Chem.
92:4029–4037. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
113
|
Barrangou R and Marraffini LA: CRISPR-Cas
systems: Prokaryotes upgrade to adaptive immunity. Mol Cell.
54:234–244. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
114
|
Abudayyeh OO, Gootenberg JS,
Essletzbichler P, Han S, Joung J, Belanto JJ, Verdine V, Cox DBT,
Kellner MJ, Regev A, et al: RNA targeting with CRISPR-Cas13.
Nature. 550:280–284. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
115
|
Cox DBT, Gootenberg JS, Abudayyeh OO,
Franklin B, Kellner MJ, Joung J and Zhang F: RNA editing with
CRISPR-Cas13. Science. 358:1019–1027. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
116
|
Wessels HH, Mendez-Mancilla A, Guo X,
Legut M, Daniloski Z and Sanjana NE: Massively parallel Cas13
screens reveal principles for guide RNA design. Nat Biotechnol.
38:722–727. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
117
|
Li H, Wang S, Dong X, Li Q, Li M, Li J,
Guo Y, Jin X, Zhou Y, Song H and Kou Z: CRISPR-Cas13a cleavage of
dengue Virus NS3 gene efficiently inhibits viral replication. Mol
Ther Nucleic Acids. 19:1460–1469. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
118
|
Yin L, Zhao F, Sun H, Wang Z, Huang Y, Zhu
W, Xu F, Mei S, Liu X, Zhang D, et al: CRISPR-Cas13a inhibits HIV-1
infection. Mol Ther Nucleic Acids. 21:147–155. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
119
|
Abbott TR, Dhamdhere G, Liu Y, Lin X,
Goudy L, Zeng L, Chemparathy A, Chmura S, Heaton NS, Debs R, et al:
Development of CRISPR as an antiviral strategy to combat SARS-CoV-2
and influenza. Cell. 181:865–876. e8122020. View Article : Google Scholar : PubMed/NCBI
|
|
120
|
Konermann S, Lotfy P, Brideau NJ, Oki J,
Shokhirev MN and Hsu PD: Transcriptome Engineering with
RNA-Targeting Type VI-D CRISPR Effectors. Cell. 173:665–676.e14.
2018. View Article : Google Scholar : PubMed/NCBI
|
|
121
|
Zhu N, Zhang D, Wang W, Li X, Yang B, Song
J, Zhao X, Huang B, Shi W, Lu R, et al: A Novel coronavirus from
patients with pneumonia in China, 2019. N Engl J Med. 382:727–733.
2020. View Article : Google Scholar : PubMed/NCBI
|
|
122
|
Blanchard EL, Vanover D, Bawage SS, Tiwari
PM, Rotolo L, Beyersdorf J, Peck HE, Bruno NC, Hincapie R, Michel
F, et al: Treatment of influenza and SARS-CoV-2 infections via
mRNA-encoded Cas13a in rodents. Nat Biotechnol. 39:717–726. 2021.
View Article : Google Scholar : PubMed/NCBI
|
|
123
|
Wang Q, Liu X, Zhou J, Yang C, Wang G, Tan
Y, Wu Y, Zhang S, Yi K and Kang C: The CRISPR-Cas13a Gene-editing
system induces collateral cleavage of RNA in glioma cells. Adv Sci
(Weinh). 6:19012992019. View Article : Google Scholar : PubMed/NCBI
|
|
124
|
Al-Shayeb B, Skopintsev P, Soczek KM,
Stahl EC, Li Z, Groover E, Smock D, Eggers AR, Pausch P, Cress BF,
et al: Diverse virus-encoded CRISPR-Cas systems include streamlined
genome editors. Cell. 185:4574–4586.e16. 2022. View Article : Google Scholar : PubMed/NCBI
|
|
125
|
Guan J, Oromi-Bosch A, Mendoza SD,
Karambelkar S, Berry JD and Bondy-Denomy J: Bacteriophage genome
engineering with CRISPR-Cas13a. Nat Microbiol. 7:1956–1966. 2022.
View Article : Google Scholar : PubMed/NCBI
|
|
126
|
Adler BA, Hessler T, Cress BF, Lahiri A,
Mutalik VK, Barrangou R, Banfield J and Doudna JA: Broad-spectrum
CRISPR-Cas13a enables efficient phage genome editing. Nat
Microbiol. 7:1967–1979. 2022. View Article : Google Scholar : PubMed/NCBI
|