Construction of ceRNA regulatory networks for osteoporosis
- Authors:
- Published online on: June 13, 2023 https://doi.org/10.3892/mmr.2023.13033
- Article Number: 145
-
Copyright: © Chen et al. This is an open access article distributed under the terms of Creative Commons Attribution License.
Abstract
Introduction
Osteoporosis is defined as a systemic skeletal disorder characterized by low bone mass and microarchitectural deterioration of bone tissue, leading to increased bone fragility and increased susceptibility to fracture (1). Osteoporosis is a highly prevalent disease that is estimated to affect 200 million women and men worldwide, mainly those over 60 years of age (2). Osteoporosis imposes a huge public health and medical burden on society and causes significant economic losses to countries (3). Population aging, urbanization and menopause have contributed to the marked increase in osteoporotic fractures (4,5). Both estrogen deficiency and immunoinflammatory conditions lead to decreased bone density and osteoporosis due to an altered balance of bone remodeling processes that maintain healthy bone (6,7). In addition, genetic predisposition, lifestyle, and nutrition are factors associated with their pathogenesis (8).
A bone mineral density (BMD) T score of −2.5 or less can be used as a diagnostic criterion of osteoporosis (9). However, since the determination of osteoporosis requires a comprehensive assessment of the risk of fracture of a patient, it is noted that the BMD test is a limited clinical indicator for diagnosis (10,11). Therefore, in addition to BMD, it is necessary to develop new clinical predictors to detect and diagnose osteoporosis early. On the basis of understanding the pathogenesis of osteoporosis, an effective method for appropriate treatment is provided.
Circular RNAs (circRNAs) do not have 3′ and 5′ ends but form a closed loop different from linear RNA. circRNAs are described as having common characteristics such as stability and conservation and can become biomarkers in clinical practice and research (12). CircRNAs can regulate the growth, regeneration and function of osteoblasts, chondrocytes and osteoclasts (13). In osteoporosis, circRNAs are involved in proliferation, differentiation and apoptosis (14). Studies have shown that microRNAs (miRNAs) can provide valuable information concerning bone status and can be used as biomarkers of osteoporosis (15,16). MiRNAs are small noncoding RNAs with a length of 21–25 nucleotides that regulate protein expression through post-transcriptional gene silencing (17). CircRNAs affect the differentiation of osteoblasts and osteoclasts by acting as miRNA sponges, which will facilitate the analysis of pathogenesis and the development of new drugs to treat osteoporosis (18). However, research on osteoporosis is still very limited.
In the present study, osteoporosis-related gene expression data was analyzed from public datasets. Bioinformatics tools were used to explore the potential molecular imbalance mechanism of osteoporosis. In addition, circRNAs and their regulatory interaction networks were constructed. These findings were further validated in in vitro experiments with patient tissues and human mesenchymal stem cells. These findings aid in clarifying the pathogenesis and provide new suggestions for the diagnosis and treatment of senile osteoporosis.
Materials and methods
Data collection
The GSE35958 dataset (19) included mRNA expression profiles of mesenchymal stem cells (MSCs) from bone marrow in 5 primary patients with osteoporosis and 4 age-matched, nonosteoporotic donors based on the GPL570 platform. The GSE56815 dataset (20) included mRNA expression profiles of blood monocytes from 40 females with low hip BMD and 40 females with high BMD based on the GPL96 platform. GSE159121 included miRNA and circRNA expression profiles of peripheral blood samples from 3 healthy controls and 3 patients with osteoporosis based on the GPL16791 platform.
Difference analysis
Differential expression analysis for mRNAs between osteoporosis and controls in GSE35958 and GSE56815 was performed using the limma R package (21). Differential expression analysis for miRNAs and circRNA between osteoporosis and controls in GSE159121 was performed using the DEseq2 R package (22). The genes with a P-value <0.05 when comparing between patients with osteoporosis and controls were assigned as differentially expressed mRNAs, miRNAs, and circRNAs (DEmRs, DEmiRs, and DEcircRs).
Enrichment analysis
The enrichment analysis of Gene Ontology (GO) analysis and Kyoto Encyclopedia of Genes and Genomes (KEGG) pathways for DEmRs were performed using the clusterProfiler R package (23). A P-value <0.05 was set to screen the significant enrichment results.
Target prediction of DEmiRs
The target circRNAs of DEmiRs were predicted using the circBank database (http://www.circbank.cn/) and miRanda database (http://www.microrna.org/microrna/home.do). The target mRNAs of DEmiRs were predicted using the miRanda database. According to the predicted negative regulation of miRNAs, a network of competing endogenous RNAs (ceRNAs) was constructed.
Logistic regression analysis
A nomogram was built using logistic regression analysis with the rms R package (v3.5.3) (24) based on DEmRs to evaluate the risk assessment of osteoporosis. Forest plots were also constructed using logistic regression analysis with the forestplot R package (v4.0.3) (25). Receiver operating characteristic (ROC) curve analysis was performed using the pROC package (v1.16.2) (26).
Patients and bone marrow collection
Bone marrow tissue from 5 patients (2 males) with osteoporosis and 5 patients (2 males) with accidental fracture (without osteoporosis) were collected using surgery from August 2020 to November 2021 in the Sixth Affiliated Hospital of Xinjiang Medicine University (Urumqi, China). The age of the osteoporotic patients ranged from 31 to 96 years, with a mean age of 72.34±12.47 years, and that of patients without osteoporosis ranged from 31 to 95 years, with a mean age of 70.50±12.34 years. All participants had normal levels of parathyroid hormone with a mean level of 48.05±12.76 (normal range, 9–90 pg/ml), thyroid stimulating hormone with a mean level of 2.31±0.69 (normal range, 0.3-4.5 mIU/l). Inclusion criteria for osteoporosis were as follows: T-score of <-2.5 as detected by X-ray absorptiometry. Exclusion criteria were as follows: i) Patients with a history of brittle fractures within the past 12 months; ii) patients with bone diseases; iii) patients with a previous history of external bone irradiation or implanted radiation therapy; and iv) patients who had suffered from any type of cancer within the past 5 years. The present study was approved by the Ethics Committee of the Sixth Affiliated Hospital of Xinjiang Medicine University (approval no. LFYLLSC20191231-06). Patients were informed about the study and provided written informed consent.
Cell culture and transfection
Primary human bone marrow mesenchymal stem cells (BMSCs) were obtained from Pricella (https://www.procell.com.cn/view/2301.html). The use of BMSCs was approved by the Ethics Committee of the Sixth Affiliated Hospital of Xinjiang Medicine University (approval no. LFYLLSC20191231-06). BMSCs were cultured in DMEM medium (Hyclone; Cytiva) containing 10% FBS (Beijing Solarbio Science & Technology Co., Ltd.) and 100 U/ml penicillin and streptomycin at 37°C with 5% CO2. The recombinant plasmid pcDNA3/circ_0070304 for circ_0070304 expression and the negative control plasmid (NC) were purchased from Shanghai Gemma Medical Technоlоgy Cо., Ltd. These plasmids (500 ng/ml) were transfected with Lipofectamine 2000 (Invitrogen; Thermo Fisher Scientific, Inc.) into BMSCs at 37°C for 48 h, then immediately used for subsequent experiments.
Osteogenic induction and Alizarin Red staining
For osteogenic differentiation, BMSCs were cultured in osteogenic induction medium (50 mM ascorbic acid, 100 nM dexamethasone, 10 mM β-glycerophosphate, and 15% FBS) for 2 weeks.
Subsequently, BMSCs (3×105/well) were fixed in 60% isopropanol for 60 sec at 37°C and washed twice with PBS. These cells were then stained with 1% Alizarin Red (Beijing Solarbio Science & Technology Co., Ltd.) staining for 3 min at 37°C. After washing the cells with PBS twice, the cells were observed and imaged using a fluorescence inverted microscope (Leica Microsystems GmbH). The type of microscope was a fluorescence inverted microscope, however fluorescence was not used.
Dual-luciferase reporter assay
For the dual luciferase reporter assay, circ_0070304 and the ring finger and CCCH-type domains 2 (RC3H2) 3′ untranslated region (3′UTR) were synthesized and cloned into psiCHECK-2 vectors, termed circRNA-wild-type (WT) or RC3H2-WT. The mutant (Mut) sequences of circ_0070304 and RC3H2 3′UTR were also synthesized and cloned into the same vectors to obtain circRNA-Mut or RC3H2-Mut. Then, these vectors (Shanghai Gemma Medical Technоlоgy Cо., Ltd.) were cotransfected with miR-183-5p mimics (forward, 5′-UAUGGCACUGGUAGAAUUCACU-3′ and reverse, 5′-UGAAUUCUACCAGUGCCAUAUU-3′) or miR-183-5p control (forward, 5′-UUCUCCGAACGUGUCACGUTT-3′ and reverse, 5′-ACGUGACACGUUCGGAGAATT-3′) into BMSCs using Lipofectamine 2000 (Invitrogen; Thermo Fisher Scientific, Inc.). The sequences were designed by Shanghai Gemma Medical Technоlоgy Cо., Ltd., according to the sequence in the miRbase database. To detect the fluorescence in each well, the Dual Luciferase Reporter Assay System (Thermo Fisher Scientific, Inc.) was applied after 36 h of transfection. Relative Luciferase activity is the ratio of Renilla luciferase activity normalized to firefly fluorescence activity.
Reverse transcription-quantitative PCR (RT-qPCR)
Total RNA was extracted from bone marrow tissue or BMSCs using TRIzol (Invitrogen; Thermo Fisher Scientific, Inc.). Reverse transcription of RNA into cDNA was performed using Superscript II reverse transcriptase (Invitrogen; Thermo Fisher Scientific, Inc.) according to the maufacturer's instructions. RT-qPCR was then performed with a SYBR Green PCR kit (Invitrogen; Thermo Fisher Scientific, Inc.). The reaction conditions were as follows: 95°C for 10 min; 95°C for 10 sec, 40 cycles; 60°C for 15 sec; 72°C for 20 sec; 72°C for 10 min. GAPDH and U6 were used as internal reference genes to normalize the expression of target genes using the 2−ΔΔCq method (27). Primer sequences were designed by Primer Premier 5 software (Premier Biosoft International) and are shown in Table SI.
Western blotting (WB)
Proteins were extracted from bone marrow tissue or BMSCs with RIPA lysis buffer (Beyotime Institute of Biotechnology) on ice. Quantification was performed using BCA Protein Assay Kit (Beyotime Institute of Biotechnology). A total of 50 µg proteins were segregated using 10% SDS-PAGE and then transferred onto PVDF membranes (MilliporeSigma). After blocking with 5% nonfat dry milk for 2 h at 37°C, the membranes were incubated with primary antibodies against ZFP36L1 (1:1,000; cat. no. ab230507; Abcam), P4HB (1:5,000; cat. no. ab137119; Abcam), RC3H2 (1:1,000; cat. no. ab241406; Abcam), TRIM28 (1:5,000; cat. no. ab109287; Abcam), WNK1 (1:2,500; cat. no. ab181029; Abcam), GAPDH (1:5,000; cat. no. ab181602; Abcam) overnight at 4°C. The membranes were then incubated with HRP-conjugated secondary antibody (anti-rabbit IgG; 1:2,500; cat. no. ab6734; Abcam) for 2 h at room temperature. GAPDH was used as an internal reference protein. Finally, ECL chemiluminescent liquid (Beijing Solarbio Science & Technology Co., Ltd.) was employed to identify the protein bands in the chemiluminescence using ChemiDoc MP imaging system (Bio-Rad Laboratories, Inc.). Densitometric analysis was performed using Image J software (v3.0; National Instiutes of Health).
Statistical analyses
Statistical significance between groups was analyzed by unpaired two-tailed Student's t-test using SPSS 20.0 software (IBM Corp.). At least three independent experiments were performed for all assays. All data are shown as the mean ± standard deviation (SD). Correlation analysis was performed using Pearson's correlation. P<0.05 was considered to indicate a statistically significant difference.
Results
Differentially expressed mRNAs in osteoporosis
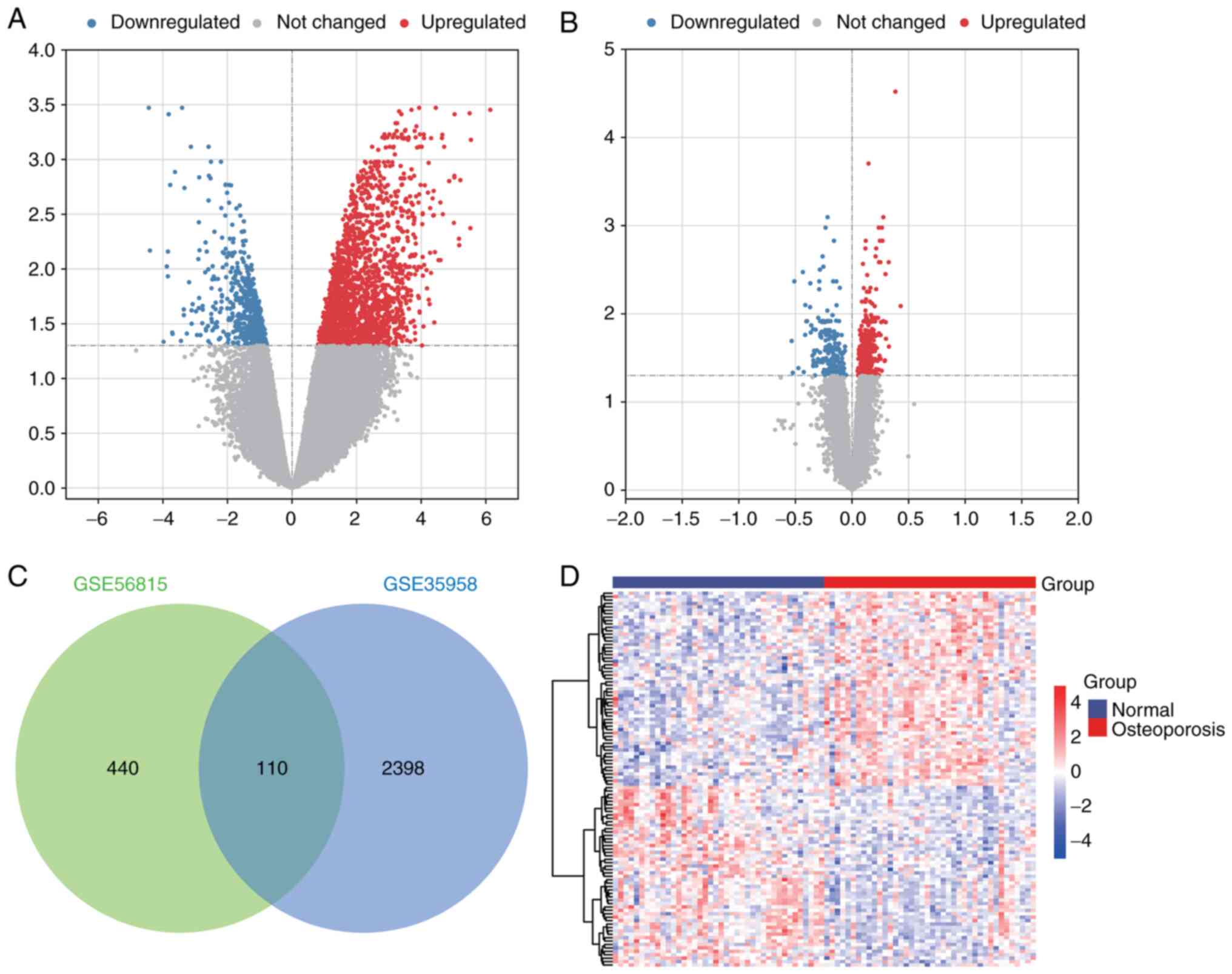
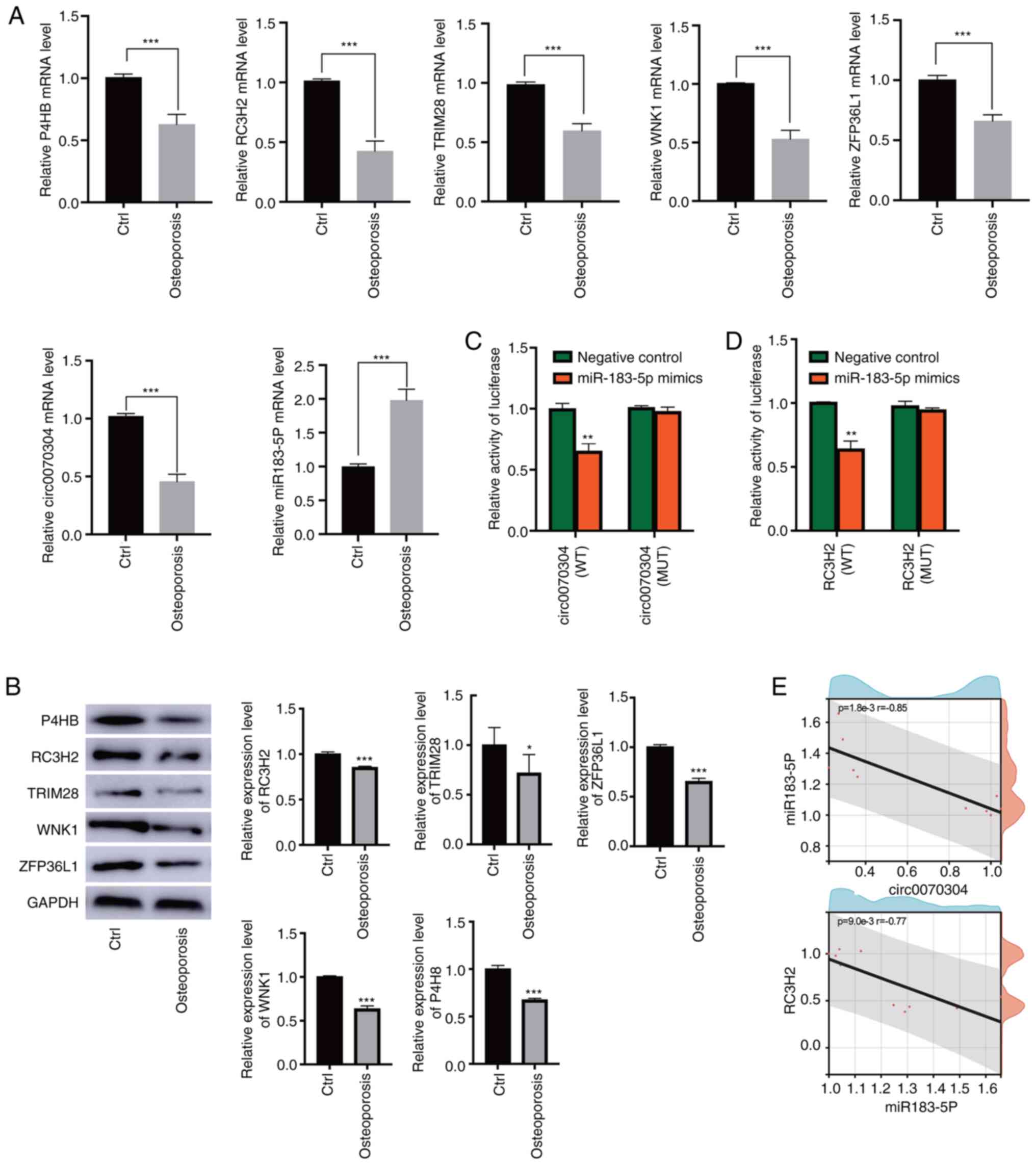
To identify mRNAs potentially associated with osteoporosis, differential analysis of mRNA expression was performed. In GSE35958, a total of 2,508 DEmRs were identified in patients with osteoporosis compared with controls (Fig. 1A). In GSE56815, 550 DEmRs (Fig. 1B) were identified. Finally, through intersection analysis, 110 DEmRs that were present in both GSE35958 and GSE56815 (Fig. 1C) were identified. There was a clear difference in the expression of these intersectional DEmRs between osteoporotic patients and controls (Fig. 1D). These DEmRs may be associated with osteoporosis.
Enrichment analysis of intersectional DEmRs
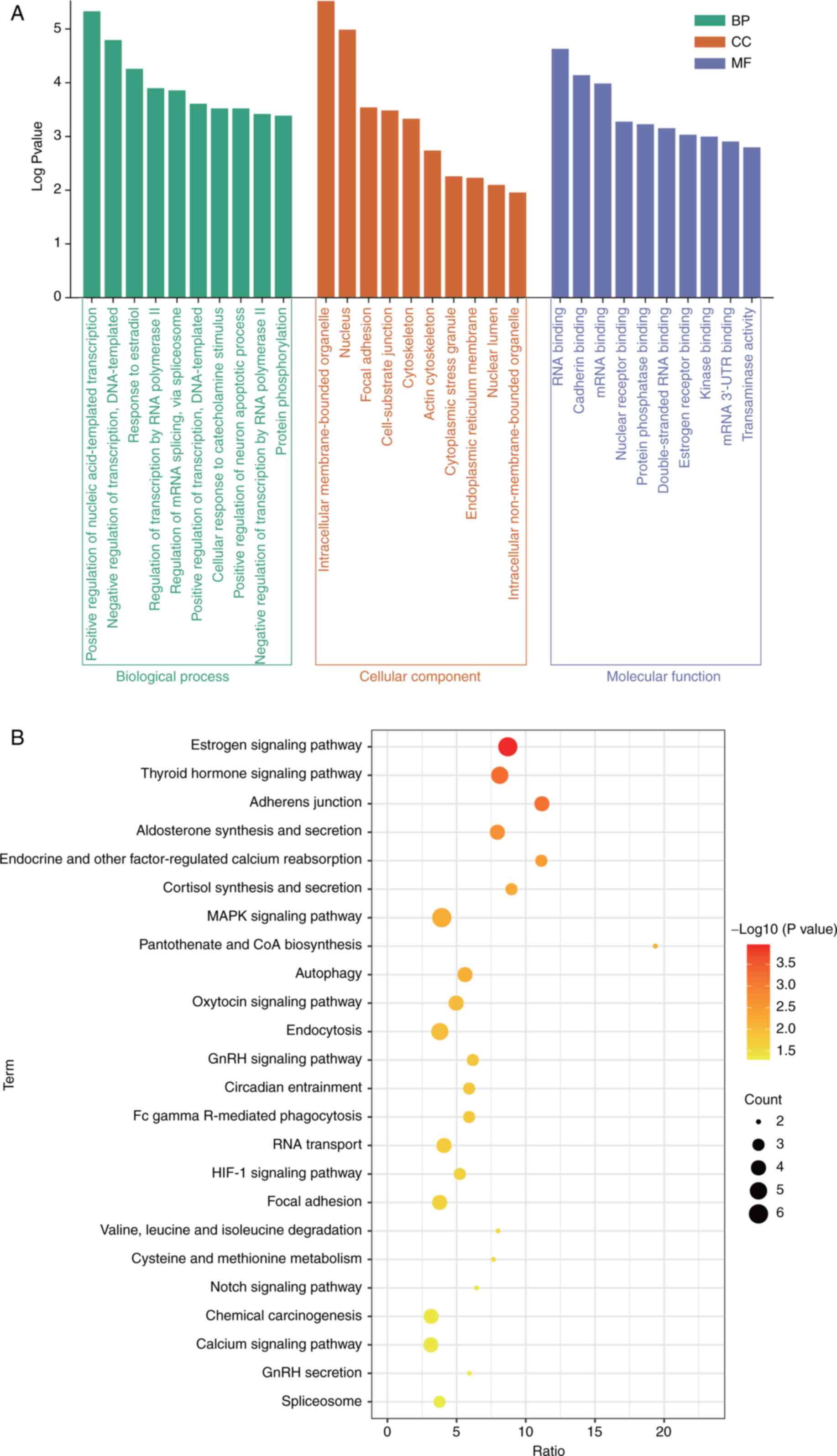
To explore the potential pathological mechanism of osteoporosis, biofunction enrichment analysis of intersectional DEmRs was performed. The results of GO analysis for intersectional DEmRs included biological processes (BP), cellular component (CC), and molecular function (MF) (Fig. 2A). In the BP category, it was identified that ‘positive regulation of nucleic acid-templated transcription’, ‘negative regulation of transcription, DNA-templated transcription’, and ‘response to estradiol’ were mainly enriched. For CC, ‘intracellular membrane-bound organelle’, ‘nucleus’, and ‘focal adhesion’ were significantly enriched. ‘RNA binding’, ‘cadherin binding’, and ‘mRNA binding’ were significantly enriched in MF. In addition, the ‘estrogen signaling pathway’, ‘thyroid hormone signaling pathway’, and ‘adherens junction’ were significantly enriched in KEGG pathways (Fig. 2B).
Construction of ceRNA networks
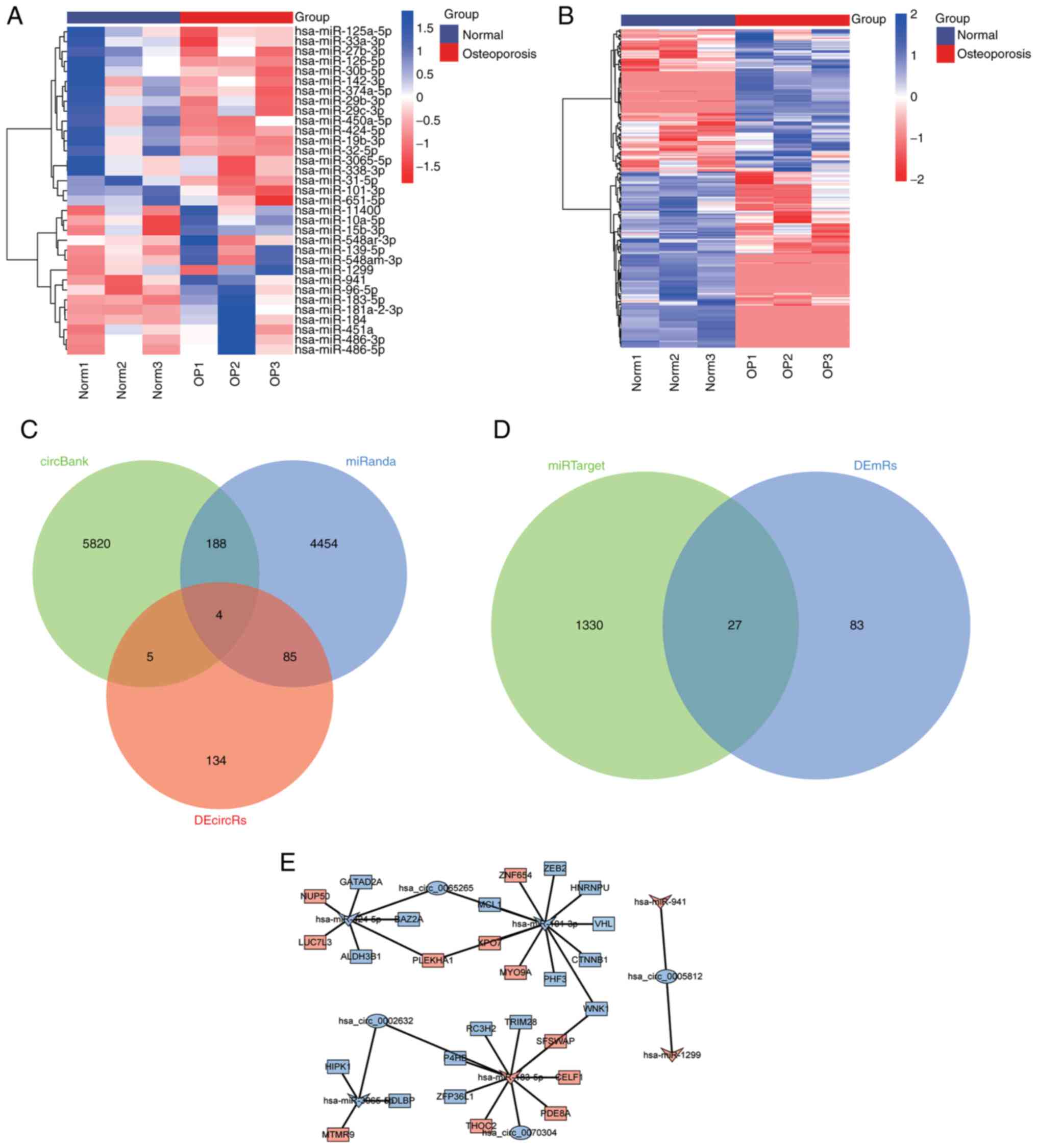
To identify the regulatory network associated with osteoporosis, 33 DEmiRs (Fig. 3A) and 228 DEcircRs (Fig. 3B) in GSE159121 were identified. The circRNAs targeted by the DEmiRs were then predicted using the circBank database and miRanda database, of which four overlapped with DEcircRs: circ_0065265, circ_0002632, circ_0005812, and circ_0070304 (Fig. 3C). Next, the mRNAs targeted by the DEmiRs were predicted using the miRanda database, and 27 downstream mRNAs overlapped with DEmRs (Fig. 3D). The circRNAs, miRNAs and mRNAs were synthesized into ceRNA networks (Fig. 3E). It was then determined that circRNAs were all downregulated in the network. According to the negatively regulated sponge relationship, the targeted DEcircRs and DEmRs that are negatively regulated by miRNAs were identified. Finally, miR-183-5p (upregulation) was identified as a key DEmiR, circ_0002632 and circ_0070304 (downregulation) were identified as key DEcircRs, and ZFP36L1, P4HB, RC3H2, TRIM28, and WNK1 (downregulation) were identified as key DEmRs.
Evaluation of key DEmRs
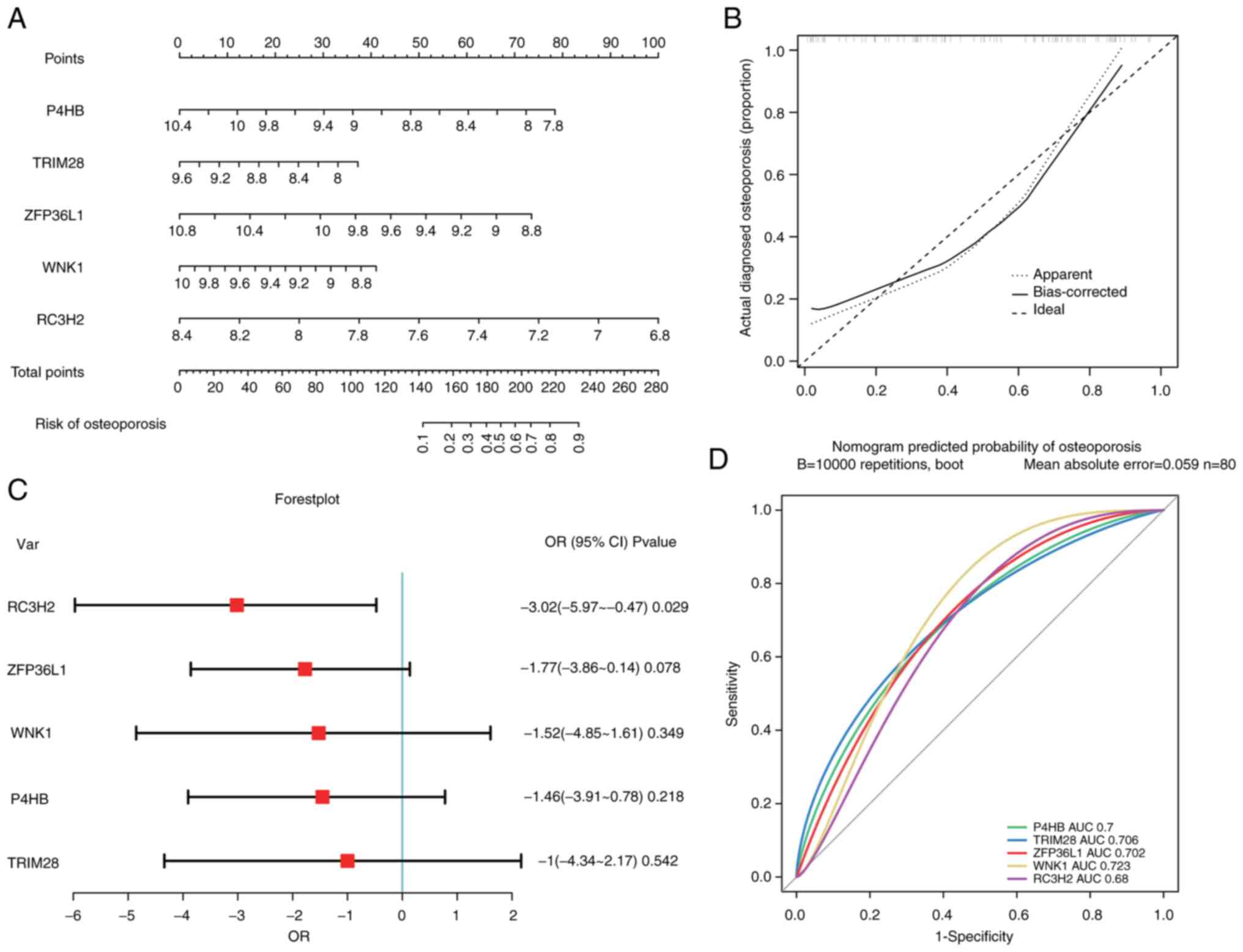
To evaluate the key DEmRs in the ceRNA network, a nomogram (Fig. 4A) was first constructed. P4HB, ZFP36L1, and RC3H2 contributed the most to the total score. The predictive values were consistent with the observed values according to the calibration curve (Fig. 4B). Furthermore, the distribution of key DEmRs was visualized using a forest plot (Fig. 4C). RC3H2 was a protective factor for osteoporosis. The area under the ROC (AUC) value of RC3H2 was 0.68, which had predictive diagnostic power (Fig. 4D). Therefore, it was identified that circ_0002632 and circ_0070304 may compete with RC3H2 for binding to miR-183-5p and may serve as promising targets for the diagnosis and treatment of osteoporosis.
Experimental validation of significant genes
To verify the aberrant expression of key genes, bone marrow tissue from patients was collected. In the RT-qPCR results, key DEmRs were all downregulated in osteoporosis compared to controls, circ_0070304 was significantly downregulated, and miR-183-5p was upregulated (Fig. 5A). According to the results of WB, the protein levels of ZFP36L1, P4HB, RC3H2, TRIM28, and WNK1 were all downregulated in osteoporosis (Fig. 5B).
Dual luciferase reporter assays were then performed to determine the specific 3′UTR binding between genes within the ceRNA network (circ_0070304/miR-183-5p/RC3H2). Notably, circRNA-WT or circRNA-Mut was cotransfected with miR-183-5p mimics or miR-183-5p control into hBMSCs. The luciferase activity of circRNA-WT was significantly inhibited by miR-183-5p mimics, while the luciferase activity of circRNA-Mut was not significantly changed by miR-183-5p mimics (Fig. 5C). The luciferase activity of RC3H2-WT was significantly inhibited by miR-183-5p mimics, and that of RC3H2-Mut was not significantly changed by miR-183-5p mimics (Fig. 5D). The results of correlation analysis showed a significant negative correlation between circ_0070304 and miR-183-5p with a correlation coefficient of −0.85 or between miR-183-5p and RC3H2 with a correlation coefficient of −0.77 (Fig. 5E).
Circ_0070304 promotes osteoblastic differentiation
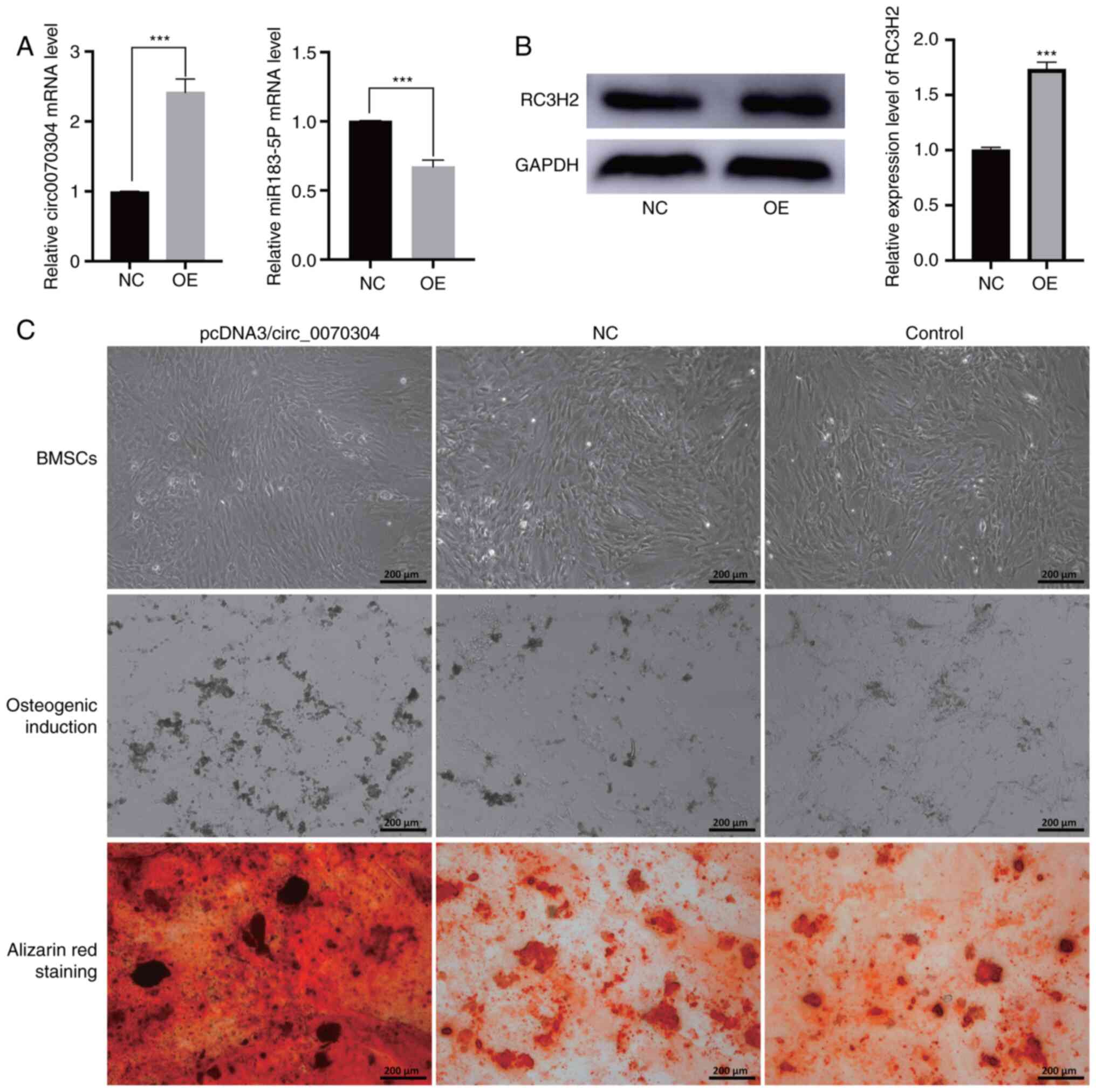
To elucidate the function of circ_0070304 in osteoporosis, the plasmid pcDNA3/circ_0070304 was transfected into BMSCs to overexpress circ_0070304. The expression of circ_0070304 was significantly increased in the overexpression group compared to the NC group (Fig. 6A). In the overexpression group, the RNA level of miR-183-5p was reduced (Fig. 6A). WB demonstrated that RC3H2 protein expression was significantly increased in the circ_0070304 overexpression group compared to the NC group (Fig. 6B).
Subsequently, osteogenic induction was performed and BMSCs transfected with plasmid pcDNA3/circ_0070304 and NC were stained with Alizarin Red. Increased calcium deposition in the circ_0070304 overexpression group was revealed compared to the NC group (Fig. 6C). These results all suggested that circ_0070304 promotes osteoblastic differentiation.
Discussion
Diagnostic screening for osteoporosis is essential for primary fracture prevention, as it is the gateway to treatment (28). In the present study, using gene expression data from osteoporotic patients, mRNAs with diagnostic value as well as ceRNA regulatory networks were identified. It was demonstrated that circ_0070304 exhibited significantly lower expression in osteoporotic patients than in controls and had a good discriminating effect. Furthermore, circ_0070304 promoted osteoblast differentiation by upregulating RC3H2 via miR-183-5p in vitro. These findings suggest that the regulation of the ceRNA network (circ_0070304/miR-183-5p/RC3H2) may be a useful therapy for the treatment and prevention of osteoporosis, as well as a potential biomarker for detecting osteoporosis.
Currently, much research has been devoted to exploring the pathogenesis of osteoporosis to develop effective targeted therapeutics. In the present study, important pathways that may be associated with osteoporosis were identified by analyzing the biological functions and signaling pathways in which DEmRs participate. The decrease in estrogen levels caused by menopause or aging leads to loss of bone mass and physical strength and contributes to the development of osteoporosis (29). Estrogen slows the rate of bone remodeling by decreasing the birth rate of osteoclasts and osteoblasts and is an important therapeutic target for osteoporosis (30). Thyroid hormones are required for skeletal development and are important regulators of adult bone maintenance (31). Abnormal thyroid hormone signaling is considered a well-established cause of secondary osteoporosis, which leads to increased osteoclast number and activity (32). Adherens junctions are considered to be involved in osteogenesis, bone formation and resorption (33).
Multiple circRNAs and miRNAs have important post-transcriptional regulatory effects on gene expression during bone formation (34). In the present study, a ceRNA network that is upregulated in osteoporosis was identified. miR-183-5p was found to increase with age, inhibit bone marrow stromal cell proliferation, and induce stem cell senescence and was considered a potential therapeutic target for osteoporosis (35,36). During osteoclastogenesis of bone marrow-derived macrophages, it was revealed that miR-183-5p expression is upregulated, and heme oxygenase-1 expression is inhibited (37).
Decreased expression of circ_0070304 in osteoporosis was verified by molecular experiments. To the best of our knowledge, no study has reported the circ_relationship between circ_0070304 and osteoporosis. The occurrence and development of osteoporosis are mainly due to the imbalance between osteoblast and osteoclast activity (38). Alizarin Red staining revealed that circ_0070304 overexpression in BMSCs significantly promoted osteoblast differentiation. The results of the present study suggest for the first time that circ_0070304 may be a potential diagnostic and therapeutic target for osteoporotic patients.
The results of the present study suggested that among the key DEmRs in the ceRNA network, upregulated expression of RC3H2 in osteoporosis may have a significant protective effect on disease. RC3H2 represses the mRNA of genes that control T helper cell development, thereby participating in the regulation of the adaptive immune system (39). In addition, RC3H2 acts as an E3 ligase, regulates the turnover of apoptosis signal regulating kinase 1 (ASK1), inhibits the sustained activation of JNK and p38 MAPK, and impedes cell death (40,41). JNK and MAPK signaling activation leads to the expression of proinflammatory cytokines and chemokines, resulting in osteoporosis (42). Therefore, these data suggest that RC3H2-mediated ubiquitination activity is a regulatory mechanism involved in the development of osteoporosis and may serve as an effective therapeutic target.
The present study has limitations. Due to the small sample size used in this study, which may affect the accuracy of the experimental results, the sample size should be expanded to aid clinical applicability in the future. Also, the role of ceRNA network was only validated in vitro, and since no in vivo studies were conducted in this study, it is difficult to accurately evaluate whether this ceRNA network can improve bone quality.
In conclusion, circ_0070304 and RC3H2 in osteoporosis were downregulated, and miR-183-5p was upregulated. Upregulated circ_0070304 upregulated RC3H2 by sponging miR-183-5p, promoting osteoblast differentiation. It is suggested that the ceRNA network may be a new therapeutic target for osteoporosis, and the mechanism may be associated with the imbalance of osteoblast differentiation.
Supplementary Material
Supporting Data
Acknowledgements
Not applicable.
Funding
The present study was supported by the Natural Science Foundation of Xinjiang Uygur Autonomous Region (grant no. 2020D01C198).
Availability of data and materials
The datasets used during the present study are available from the corresponding author upon reasonable request.
Authors' contributions
HC designed experiments, analyzed the data, and wrote the manuscript. HW and XL performed the experiments and analyzed data. LL and YA contributed to the experimental design and manuscript editing. YT conceived and supervised the study. HC and HW confirm the authenticity of all the raw data. All authors read and approved the manuscript and agree to be accountable for all aspects of the research in ensuring that the accuracy or integrity of any part of the work are appropriately investigated and resolved.
Ethics approval and consent to participate
The present study was approved by the Ethics Committee of the Sixth Affiliated Hospital of Xinjiang Medicine University, Urumqi, China (approval no. LFYLLSC20191231-06). Patients were informed about the study and provided written informed consent.
Patients consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
References
|
Kanis JA, Cooper C, Rizzoli R and Reginster JY; Scientific Advisory Board of the European Society for Clinical and Economic Aspects of Osteoporosis (ESCEO) and the Committees of Scientific Advisors and National Societies of the International Osteoporosis Foundation (IOF), : European guidance for the diagnosis and management of osteoporosis in postmenopausal women. Osteoporos Int. 30:3–44. 2019. View Article : Google Scholar : PubMed/NCBI | |
|
Wright NC, Looker AC, Saag KG, Curtis JR, Delzell ES, Randall S and Dawson-Hughes B: The recent prevalence of osteoporosis and low bone mass in the United States based on bone mineral density at the femoral neck or lumbar spine. J Bone Miner Res. 29:2520–2526. 2014. View Article : Google Scholar : PubMed/NCBI | |
|
Liu T, Huang J, Xu D and Li Y: Identifying a possible new target for diagnosis and treatment of postmenopausal osteoporosis through bioinformatics and clinical sample analysis. Ann Transl Med. 9:11542021. View Article : Google Scholar : PubMed/NCBI | |
|
Chandran M, Mitchell PJ, Amphansap T, Bhadada SK, Chadha M, Chan DC, Chung YS, Ebeling P, Gilchrist N, Habib Khan A, et al: Development of the Asia Pacific consortium on osteoporosis (APCO) framework: Clinical standards of care for the screening, diagnosis, and management of osteoporosis in the Asia-Pacific region. Osteoporos Int. 32:1249–1275. 2021. View Article : Google Scholar : PubMed/NCBI | |
|
Zhang H, Wang A, Shen G, Wang X, Liu G, Yang F, Chen B, Wang M and Xu Y: Hepcidin-induced reduction in iron content and PGC-1β expression negatively regulates osteoclast differentiation to play a protective role in postmenopausal osteoporosis. Aging (Albany NY). 13:11296–11314. 2021. View Article : Google Scholar : PubMed/NCBI | |
|
D'Amelio P and Sassi F: Gut microbiota, immune system, and bone. Calcif Tissue Int. 102:415–425. 2018. View Article : Google Scholar : PubMed/NCBI | |
|
Cosman F, de Beur SJ, LeBoff MS, Lewiecki EM, Tanner B, Randall S and Lindsay R; National Osteoporosis Foundation, : Clinician's guide to prevention and treatment of osteoporosis. Osteoporos Int. 25:2359–2381. 2014. View Article : Google Scholar : PubMed/NCBI | |
|
De Martinis M, Ginaldi L, Allegra A, Sirufo MM, Pioggia G, Tonacci A and Gangemi S: The osteoporosis/microbiota linkage: The role of miRNA. Int J Mol Sci. 21:88872020. View Article : Google Scholar : PubMed/NCBI | |
|
No authors listed. Assessment of fracture risk and its application to screening for postmenopausal osteoporosis. Report of a WHO study group. World Health Organ Tech Rep Ser. 843:1–129. 1994.PubMed/NCBI | |
|
Compston JE, McClung MR and Leslie WD: Osteoporosis. Lancet. 393:364–376. 2019. View Article : Google Scholar : PubMed/NCBI | |
|
Li R, Zhu X, Zhang M, Zong G and Zhang K: Association of serum periostin level with classical bone turnover markers and bone mineral density in shanghai chinese postmenopausal women with osteoporosis. Int J Gen Med. 14:7639–7646. 2021. View Article : Google Scholar : PubMed/NCBI | |
|
Pan X, Cen X, Zhang B, Pei F, Huang W, Huang X and Zhao Z: Circular RNAs as potential regulators in bone remodeling: A narrative review. Ann Transl Med. 9:15052021. View Article : Google Scholar : PubMed/NCBI | |
|
Fu M, Fang L, Xiang X, Fan X, Wu J and Wang J: Microarray analysis of circRNAs sequencing profile in exosomes derived from bone marrow mesenchymal stem cells in postmenopausal osteoporosis patients. J Clin Lab Anal. 36:e239162022. View Article : Google Scholar : PubMed/NCBI | |
|
Luo Y, Qiu G, Liu Y, Li S, Xu Y, Zhang Y, Cao Y and Wang Y: Circular RNAs in osteoporosis: Expression, functions and roles. Cell Death Discov. 7:2312021. View Article : Google Scholar : PubMed/NCBI | |
|
Cui Q, Xing J, Yu M, Wang Y, Xu J, Gu Y, Nan X, Ma W, Liu H and Zhao H: Mmu-miR-185 depletion promotes osteogenic differentiation and suppresses bone loss in osteoporosis through the Bgn-mediated BMP/Smad pathway. Cell Death Dis. 10:1722019. View Article : Google Scholar : PubMed/NCBI | |
|
Kerschan-Schindl K, Hackl M, Boschitsch E, Foger-Samwald U, Nägele O, Skalicky S, Weigl M, Grillari J and Pietschmann P: Diagnostic performance of a panel of miRNAs (OsteomiR) for osteoporosis in a cohort of postmenopausal women. Calcif Tissue Int. 108:725–737. 2021. View Article : Google Scholar : PubMed/NCBI | |
|
Alberti C, Manzenreither RA, Sowemimo I, Burkard TR, Wang J, Mahofsky K, Ameres SL and Cochella L: Cell-type specific sequencing of microRNAs from complex animal tissues. Nat Methods. 15:283–289. 2018. View Article : Google Scholar : PubMed/NCBI | |
|
Liang B, Burley G, Lin S and Shi YC: Osteoporosis pathogenesis and treatment: Existing and emerging avenues. Cell Mol Biol Lett. 27:722022. View Article : Google Scholar : PubMed/NCBI | |
|
Benisch P, Schilling T, Klein-Hitpass L, Frey SP, Seefried L, Raaijmakers N, Krug M, Regensburger M, Zeck S, Schinke T, et al: The transcriptional profile of mesenchymal stem cell populations in primary osteoporosis is distinct and shows overexpression of osteogenic inhibitors. PLoS One. 7:e451422012. View Article : Google Scholar : PubMed/NCBI | |
|
Zhou Y, Gao Y, Xu C, Shen H, Tian Q and Deng HW: A novel approach for correction of crosstalk effects in pathway analysis and its application in osteoporosis research. Sci Rep. 8:6682018. View Article : Google Scholar : PubMed/NCBI | |
|
Ritchie ME, Phipson B, Wu D, Hu Y, Law CW, Shi W and Smyth GK: limma powers differential expression analyses for RNA-sequencing and microarray studies. Nucleic Acids Res. 43:e472015. View Article : Google Scholar : PubMed/NCBI | |
|
Love MI, Huber W and Anders S: Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 15:5502014. View Article : Google Scholar : PubMed/NCBI | |
|
Yu G, Wang LG, Han Y and He QY: clusterProfiler: An R package for comparing biological themes among gene clusters. OMICS. 16:284–287. 2012. View Article : Google Scholar : PubMed/NCBI | |
|
Gao Z, Yao K, Cai P, Long N, Cao Y, He F, Liu L and Xiao C: Establishment of a survival risk prediction model for adolescent and adult osteosarcoma patients. Biomed Res Int. 2022:95594962022. View Article : Google Scholar : PubMed/NCBI | |
|
Liao X, Wang W, Yu B and Tan S: Thrombospondin-2 acts as a bridge between tumor extracellular matrix and immune infiltration in pancreatic and stomach adenocarcinomas: An integrative pan-cancer analysis. Cancer Cell Int. 22:2132022. View Article : Google Scholar : PubMed/NCBI | |
|
Robin X, Turck N, Hainard A, Tiberti N, Lisacek F, Sanchez JC and Müller M: pROC: An open-source package for R and S+ to analyze and compare ROC curves. BMC Bioinformatics. 12:772011. View Article : Google Scholar : PubMed/NCBI | |
|
Livak KJ and Schmittgen TD: Analysis of relative gene expression data using real-time quantitative PCR and the 2(−Delta Delta C(T)) method. Methods. 25:402–408. 2001. View Article : Google Scholar : PubMed/NCBI | |
|
Lin C, Yu S, Jin R, Xiao Y, Pan M, Pei F, Zhu X, Huang H, Zhang Z, Chen S, et al: Circulating miR-338 Cluster activities on osteoblast differentiation: Potential diagnostic and therapeutic targets for postmenopausal osteoporosis. Theranostics. 9:3780–3797. 2019. View Article : Google Scholar : PubMed/NCBI | |
|
Manolagas SC: The quest for osteoporosis mechanisms and rational therapies: How far we've come, how much further we need to go. J Bone Miner Res. 33:371–385. 2018. View Article : Google Scholar : PubMed/NCBI | |
|
Almeida M, Laurent MR, Dubois V, Claessens F, O'Brien CA, Bouillon R, Vanderschueren D and Manolagas SC: Estrogens and androgens in skeletal physiology and pathophysiology. Physiol Rev. 97:135–187. 2017. View Article : Google Scholar : PubMed/NCBI | |
|
Williams GR and Bassett JHD: Thyroid diseases and bone health. J Endocrinol Invest. 41:99–109. 2018. View Article : Google Scholar : PubMed/NCBI | |
|
Bassett JH and Williams GR: Role of thyroid hormones in skeletal development and bone maintenance. Endocr Rev. 37:135–187. 2016. View Article : Google Scholar : PubMed/NCBI | |
|
Zeng Y, Zhang L, Zhu W, He H, Sheng H, Tian Q, Deng FY, Zhang LS, Hu HG and Deng HW: Network based subcellular proteomics in monocyte membrane revealed novel candidate genes involved in osteoporosis. Osteoporos Int. 28:3033–3042. 2017. View Article : Google Scholar : PubMed/NCBI | |
|
Jin D, Wu X, Yu H, Jiang L, Zhou P, Yao X, Meng J, Wang L, Zhang M and Zhang Y: Systematic analysis of lncRNAs, mRNAs, circRNAs and miRNAs in patients with postmenopausal osteoporosis. Am J Transl Res. 10:1498–1510. 2018.PubMed/NCBI | |
|
Davis C, Dukes A, Drewry M, Helwa I, Johnson MH, Isales CM, Hill WD, Liu Y, Shi X, Fulzele S and Hamrick MW: MicroRNA-183-5p increases with age in bone-derived extracellular vesicles, suppresses bone marrow stromal (stem) cell proliferation, and induces stem cell senescence. Tissue Eng Part A. 23:1231–1240. 2017. View Article : Google Scholar : PubMed/NCBI | |
|
Weigl M, Kocijan R, Ferguson J, Leinfellner G, Heimel P, Feichtinger X, Pietschmann P, Grillari J, Zwerina J, Redl H and Hackl M: Longitudinal changes of circulating miRNAs during bisphosphonate and teriparatide treatment in an animal model of postmenopausal osteoporosis. J Bone Miner Res. 36:1131–1144. 2021. View Article : Google Scholar : PubMed/NCBI | |
|
Ji X, Chen X and Yu X: MicroRNAs in osteoclastogenesis and function: Potential therapeutic targets for osteoporosis. Int J Mol Sci. 17:3492016. View Article : Google Scholar : PubMed/NCBI | |
|
Wang G, Zhang L, Yan C, Wang F and Zhang Y: Overexpression of miR125b promotes osteoporosis through miR-125b-TRAF6 pathway in postmenopausal ovariectomized rats. Diabetes Metab Syndr Obes. 14:671–682. 2021. View Article : Google Scholar : PubMed/NCBI | |
|
Hoefig KP, Reim A, Gallus C, Wong EH, Behrens G, Conrad C, Xu M, Kifinger L, Ito-Kureha T, Defourny KAY, et al: Defining the RBPome of primary T helper cells to elucidate higher-order Roquin-mediated mRNA regulation. Nat Commun. 12:52082021. View Article : Google Scholar : PubMed/NCBI | |
|
Maruyama T, Araki T, Kawarazaki Y, Naguro I, Heynen S, Aza-Blanc P, Ronai Z, Matsuzawa A and Ichijo H: Roquin-2 promotes ubiquitin-mediated degradation of ASK1 to regulate stress responses. Sci Signal. 7:ra82014. View Article : Google Scholar : PubMed/NCBI | |
|
Zhang Q, Fan L, Hou F, Dong A, Wang YX and Tong Y: New insights into the RNA-binding and E3 ubiquitin ligase activities of Roquins. Sci Rep. 5:156602015. View Article : Google Scholar : PubMed/NCBI | |
|
De Martinis M, Sirufo MM, Suppa M and Ginaldi L: IL-33/IL-31 axis in osteoporosis. Int J Mol Sci. 21:12392020. View Article : Google Scholar : PubMed/NCBI |