Introduction
Mesenchymal stem cells (MSCs) are stromal cells with
self-renewal and multilineage differentiation ability isolated from
tissue. Adipose tissue-derived MSCs (ADMSCs) are the preferred SCs
for clinical applications due to their convenient acquisition and
easy culture (1). Studies have
shown that ADMSCs can differentiate into cardiomyocyte and are an
ideal cell source for myocardial regenerative medicine (2,3).
Ghrelin, a gastric-secreted peptide hormone, was
first isolated and identified in the rat stomach in 1999 and is an
endogenous ligand for the growth hormone secretagogue receptor
(4,5). Ghrelin is involved in and regulates a
variety of physiological processes, including energy balance and
weight maintenance and studies have shown strong associations
between ghrelin and the cardiovascular system (6–8). In
addition, ghrelin is involved in the multilineage differentiation
of MSCs. Our previous study showed that ghrelin serves a critical
role in the promotion of neural differentiation of ADMSCs (9). Studies have shown that ghrelin
inhibits cardiomyocyte apoptosis and improves myocardial infarction
(10,11). Pretreatment of MSCs with ghrelin
exerts a protective effect in aged hearts (12). Studies have also found that ghrelin
can promote differentiation of embryonic SCs into cardiomyocytes
(13,14). The aforementioned studies suggest
that ghrelin serves a key role in the differentiation of MSCs.
However, it is still unknown whether ghrelin affects the
differentiation of ADMSCs into cardiomyocyte.
Studies have reported the essential role of
Wnt/β-catenin signaling pathway in embryonic heart development and
in vitro cardiomyocyte differentiation (15,16).
Ghrelin could regulate cell apoptosis, proliferation and function
via activation of Wnt/β-catenin pathway in a variety of cells and
tissue (17). Secreted
frizzled-related protein 4 (SFRP4) inhibits Wnt signaling by
competing with frizzle-protein receptor, a specific receptor of the
Wnt signaling pathway (18,19).
It has been reported that SFRP4 promotes adipogenic differentiation
of ADMSCs and inhibits the osteogenic differentiation of human
periodontal ligament MSCs (20,21).
SFRP4 inhibits the expression of cardiac-specific genes during
differentiation of P19CL6 cells, a clonal derivative of murine P19
cells, into cardiomyocytes (22).
Therefore, it was hypothesized that ghrelin may promote ADMSC
cardiomyocyte differentiation by regulating the SFRP4/Wnt/β-catenin
axis. To explore the regulatory mechanism of ghrelin in regulating
SFRP4/Wnt/β-catenin axis, the present used RNA sequencing (seq) of
cultured ADMSCs to identify potential genes regulated by
ghrelin.
Accordingly, the aim of the present study was to
investigate the critical role of ghrelin in cardiomyocyte
differentiation of ADMSCs and the potential downstream regulatory
mechanisms.
Materials and methods
Cell culture
Human ADMSCs were purchased from Cyagen Biosciences,
Inc. (cat. no. HUXMD-01001). Cells were cultured in ADMSC complete
medium (cat. no. HUXMD-90011; Cyagen Biosciences, Inc.) in an
incubator at 37°C with 5% CO2.
Immunophenotyping analysis
Cell surface markers were identified by flow
cytometry. Cells were collected by centrifugation (90 × g; 5 min;
4°C). The cells of the 3rd to 5th generation were trypsinized (cat.
no. T4799; MilliporeSigma). When the cells became round,
FBS-containing ADMSC complete medium (cat. no. HUXMD-90011; Cyagen
Biosciences, Inc.) was added to stop the reaction, cells were
collected by centrifugation (90 × g; 5 min; 4°C) and the density
was adjusted to 1×106 cells/ml. Cells were added to
labeled antibodies, including the control, to a final volume of 100
µl. After being incubated at 4°C for 30 min, 1 ml buffer was added
to each tube. The cells were collected by centrifugation (90 × g; 5
min; 4°C) and resuspended in 500 µl buffer before testing. Analysis
was performed using the NovoExpress software (version 1.4.1) based
on a NovoCyte flow cytometer (ACEA Bioscience, Inc.; Agilent
Technologies, Inc.). Antibodies were as follows: CD29 (cat. no.
CL488-65191; mouse; Proteintech Group, Inc.), CD90-FITC (cat. no.
328107; mouse; Biolegend, Inc.), CD34-FITC (cat. no. FITC-65183;
mouse; Proteintech Group, Inc.), CD45-FITC (cat. no. AH04501;
mouse; Multi Sciences), CD11b-FITC (cat. no. AH011B01; rat; Multi
Sciences), CD44-FITC (cat. no. AH04401; rat; Multi Sciences).
Multilineage differentiation of
ADMSCs
To evaluate the multilineage differentiation
potential of ADMSCs, cells were exposed to osteogenic and
adipogenic differentiation medium, as previously described
(23). For osteogenic
differentiation, cells were cultured in high-glucose Dulbecco's
Modified Eagle Medium (DMEM; cat. no. G4510, Wuhan Servicebio
Technology Co., Ltd.) containing 10% fetal bovine serum (FBS; cat.
no. 11011; Zhejiang Tianhang Biotechnology Co., Ltd.), 200 µM
L-ascorbic acid (cat. no. A8101; Beijing Solarbio Science &
Technology Co., Ltd.), 0.1 µM dexamethasone (cat. no. D137736;
Shanghai Aladdin Biochemical Technology Co., Ltd.) and 10 mM
β-glycerol phosphate disodium pentahydrate (cat. no. G9140; Beijing
Solarbio Science & Technology Co., Ltd.) at 37°C for 21 days.
Control cells were cultured in high-glucose DMEM containing 10%
FBS. Cells were fixed with 4% paraformaldehyde (cat. no. C104188;
Shanghai Aladdin Biochemical Technology Co., Ltd.) for 15 min at
room temperature. Alizarin red staining solution at 1%
concentration (cat. no. G1452; Beijing Solarbio Science &
Technology Co., Ltd.) was added for 20 min at room temperature.
For adipogenic differentiation, Oil Red O Stain kit
(cat. no. G1262; Beijing Solarbio Science & Technology Co.,
Ltd.) was used according to the manufacturer's instructions. Cells
were cultured in low-glucose DMEM (cat. no. G4520; Wuhan Servicebio
Technology Co., Ltd.) containing 10% fetal bovine serum (FBS; cat.
no. 11011; Zhejiang Tianhang Biotechnology Co., Ltd.), 100 µM
L-ascorbic acid, 1 µM dexamethasone, 0.5 mM 1-methyl-3-isobutyl
xanthine (cat. no. I106812; Shanghai Aladdin Biochemical Technology
Co., Ltd.), 100 µM indomethacin (cat. no. HY-14397, MedChemExpress)
and 10 µg/ml human recombinant insulin (cat. no. INS-100MG; Bioing
Biotech, China) at 37°C for 21 days. Control cells were cultured in
low-glucose DMEM containing 10% FBS at 37°C for 21 days. The newly
prepared Oil red O staining solution was added for 20 min at room
temperature. Isopropyl alcohol (60%; cat. no. I112011; Shanghai
Aladdin Biochemical Technology Co., Ltd.) was used for washing.
Finally, cells were soaked in Oil red O buffer for 1 min at room
temperature. Images were captured using a fluorescence microscope
(200×, cat. no. IX-53; Olympus Corporation).
Cell treatment
Cells were treated with DMEM/F12 (cat. no. BL305A;
Biosharp Life Sciences) containing 10 µM 5-Azacytidine (5-Aza; cat.
no. A100625; Shanghai Aladdin Biochemical Technology Co., Ltd.) for
24 h at 37°C. Treatment 1: After washing twice with PBS (cat. no.
B548117; Sangon Biotech Co., Ltd.), cells were cultured in DMEM/F12
without 5-Aza for 14 days at 37°C. Ghrelin at different
concentrations (1, 5, 10, 50 and 100 nM; cat. no. G111460; Shanghai
Aladdin Biochemical Technology Co., Ltd.) was added to some group.
Treatment 2: After washing twice with PBS, cells were cultured in
5-Aza-free DMEM/F12 for 7 days at 37°C. In the ghrelin group, 10 nM
ghrelin was added when the medium was changed to 5-Aza-free.
Thereafter, cells were cultured for another 7 days at 37°C.
Treatment 3: After washing twice with PBS, cells were cultured in
DMEM/F12 without 5-Aza for 14 days at 37°C. To examine the effect
of ghrelin and SFRP4, ghrelin (10 nM) and recombinant human SFRP4
(100 ng/ml; cat. no. 120-50; PeproTech, Inc.) were added when the
medium was changed to 5-Aza-free.
RNA extraction and sequencing
(seq)
RNA-seq was performed 14 days later. Paired-end
sequencing was performed. Total RNA was extracted from cells using
TRIzol® (cat. no. 15596026; Invitrogen; Thermo Fisher
Scientific, Inc.) according to the manufacturer's protocol. RNA
quality was determined using a Nanodrop (Micro-Drop; Shanghai
Baoyude Scientific Instrument Co., Ltd.) and the integrity was
assessed using Fragment Analyzer 5400 (Agilent Technologies, Inc.).
cDNA fragments of 250–300 bp were selected preferentially and the
sequencing was directed from 5′ to 3′ end. Then, sequencing
libraries were generated using NEBNext®
Ultra™ RNA Library Prep kit for Illumina®
(cat. no. E7530L; New England Biolabs) according to the
manufacturer's instructions. The loading concentration of final
library in different cell sample was different (2 to 4 nM). After
library construction, the concentration was quantified by CFX96
Touch Real-Time PCR Detection System (Bio-Rad Laboratories, Inc.).
Finally, libraries were sequenced by NovaSeq 6000 S4 Reagent kit
v1.5 (300 cycles; cat. no. 20028312; Illumina, Inc.) based on an
Illumina Novaseq 6000 platform and 150 bp paired-end reads were
generated. The quality of raw sequencing data was assessed by Fastp
(version 0.23.1; http://github.com/OpenGene/fastp). Trimmed sequence
reads were mapped to human reference genome (GRCh38/hg38;
http://genome.ucsc.edu/) using hisat2
(daehwankimlab.github.io/hisat2/) with default parameters.
Bioinformatics analysis
Principal component analysis (PCA) and volcano plot
were analyzed by ggplot2 package (Version: 3.4.1) in R (http://www.R-project.org/). Hierarchical clustering
was analyzed by pheatmap package (Version: 1.0.12) in R. Gene
Ontology (GO; http://geneontology.org/) and Kyoto Encyclopedia of
Genes and Genomes (KEGG; http://www.kegg.jp/) enrichment analysis of
differentially expressed genes (DEGs) were analyzed using
ClusterProfiler package (Version 4.2.2, Bioconductor platform;
http://bioconductor.org/packages/release/bioc/html/clusterProfiler.html)
in R. DEGs were selected by Log2 |fold-change|>1 and
P<0.05.
Lentivirus infection
Coding sequence of the human DDX17 gene (NM_006386;
National Institutes of Health) was inserted into pLJM1-EGFP (cat.
no. ZT101; Hunan Fenghui Biotechnology Co., Ltd.) plasmid at the
NheI and AgeI restriction sites. pLJM1-EGFP-DDX17 or
its empty vector plasmid was transfected into 293T cells [cat. no.
iCell-h237 Saibaikang (Shanghai) Biotechnology Co., Ltd.] using
Lipofectamine® 3000 (cat. no. L3000015; Invitrogen;
Thermo Fisher Scientific, Inc.) to produce lentivirus particles,
according to the manufacturer's protocols. The second-generation
lentiviral packaging system was used. Briefly, two solutions were
prepared containing 24 µl Lipofectamine 3000 and 500 µl Opti-MEM
(cat. no. 31985070; Invitrogen, Thermo Fisher Scientific, Inc.) or
8 µg lentiviral plasmid, 6 µg packaging vector (psPAX2; cat. no.
12260, Addgene, Inc.), 2 µg envelope (pMD2.G; cat. no. 12259,
Addgene, Inc.), 28 µl Lipofectamine 3000 and 500 µl Opti-MEM. The
solutions were mixed and placed at room temperature for 15 min. The
mixture was added to 293T cells (37°C; 6 h) and lentiviral
particles were collected using a 0.45-µm filter membrane (Tianjin
Jinteng Experimental Equipment Co., Ltd.). ADMSCs were then
infected with the packaged lentiviral particles (MOI=100).
Polybrene (cat. no. BL628A; Biosharp Life Sciences) at 5 µg/ml was
also added. Cells were cultured in a 5% CO2 incubator at
37°C for 24 h. After a further 48 h (5% CO2, 37°C), the
cells were collected for subsequent experiments.
Western blotting
Total protein was extracted from cells by radio
immunoprecipitation assay lysis buffer (cat. no. P0013B) and 1%
PMSF (cat. no. ST506; both Beyotime Institute of Biotechnology) and
the product was collected by centrifugation (4°C, 1,000 × g, 3
min). Nuclear protein was extracted using a nuclear protein
extraction kit (cat. no. P0028; Beyotime Institute of
Biotechnology). Protein quantification was performed using BCA
detection kit (cat. no. P0009; Beyotime Institute of
Biotechnology). Proteins (15 µl/lane, containing 15–30 µg protein)
were separated by sodium dodecyl sulfate polyacrylamide gel
electrophoresis (8% for proteins in 35 to 50 kDa, 10% for proteins
in 50 to 75 kDa and 12% for proteins over 75 kDa; cat. no. P0015;
Beyotime Institute of Biotechnology) and transferred to
polyvinylidene fluoride membrane (cat. no. LC2005; Thermo Fisher
Scientific, Inc.). The non-specific sites were blocked using 5%
bovine serum albumin (BSA; cat. no. BS043; Biosharp Life Sciences)
to avoid non-specific adsorption (1 h, room temperature). The
membrane was incubated with the primary antibody at 4°C overnight,
followed by incubation with horseradish peroxidase-labeled goat
anti rabbit (cat. no. SA00001-2) or mouse (both 1:10,000; cat. no.
SA00001-1; both Wuhan Sanying Biotechnology) IgG at 37°C for 40
min. Enhanced chemiluminescence reagent (cat. no. E003; 7 Sea
Biotech) was added to the membrane for 5 min. The gel was scanned
and optical density of the target band was analyzed by gel image
processing system (GEL-Pro-Analyzer software, version 4.0, Roper
Technologies, Inc.). Antibodies were as follows: Rabbit GATA
binding protein 4 (GATA4; 1:1,000; cat. no. A13756; ABclonal
Biotech Co., Ltd.), α-myosin heavy chain (α-MHC; 1:20,000; cat. no.
22281-1-AP; Wuhan Sanying Biotechnology), ISL LIM homeobox 1 (ISL1;
1:1,000; cat. no. A0871; ABclonal Biotech Co., Ltd.), NK2 homeobox
5 (Nkx2.5; 1:1,000; cat. no. A5651; ABclonal Biotech Co., Ltd.),
Troponin T2, cardiac type (TNNT2; 1:1,000; cat. no. 15513-1-AP;
Wuhan Sanying Biotechnology), secreted frizzled-related protein 4
(SFRP4; 1:1,000; cat. no. A4189; ABclonal Biotech Co., Ltd.),
Dead-box helicase 17 (DDX17; 1:1,000; cat. no. DF12935; Affinity
Biosciences), β-catenin (1:1,000; cat. no. A19657; ABclonal Biotech
Co., Ltd.), Histone H3 (1:500; cat. no. 17168-1-AP; Wuhan Sanying
Biotechnology) and mouse β-actin (1:2,000; cat. no. 60008-1-Ig;
Wuhan Sanying Biotechnology).
Immunofluorescence (IF) staining
Cells were fixed in 4% paraformaldehyde (cat. no.
80096618; Sinopharm Chemical Reagent Co., Ltd.) for 15 min at room
temperature. Then, 0.1% triton X-100 (cat. no. ST795; Beyotime
Institute of Biotechnology) was added for 30 min at room
temperature. BSA (cat. no. A602440-0050; Sangon Biotech Co., Ltd.)
was added as the blocking buffer at room temperature for 15 min.
The primary antibody was diluted in PBS and incubated overnight at
4°C. Cy3-labeled goat anti-rabbit IgG (1:200; cat. no. A27039;
Invitrogen; Thermo Fisher Scientific, Inc.) was added for 60 min at
room temperature in the dark. For double IF staining, Cy3-labeled
goat anti-rabbit (red; 1:200; cat. no. A27039; Invitrogen; Thermo
Fisher Scientific, Inc.) and FITC-labeled goat anti-mouse IgG
(green; 1:200; cat. no. ab6785; Abcam) were used as the secondary
antibody. DAPI (cat. no. D106471; Shanghai Aladdin Biochemical
Technology Co., Ltd.) was used for nuclear counterstaining (5 min,
room temperature). After adding anti-fluorescence quencher (cat.
no. S2100; Beijing Solarbio Science & Technology Co., Ltd.),
images were captured under a fluorescence microscope
(magnification, ×400; DP73 system; Olympus Corporation). Antibodies
were as follows: Mouse GATA4 (1:50; cat. no. sc-25310; Santa Cruz
Biotechnology, Inc.) and rabbit α-MHC (1:100; cat. no. 22281-1-AP;
Wuhan Sanying Biotechnology) and β-catenin (1:100; cat. no. A19657;
ABclonal Biotech Co., Ltd.).
Reverse transcription-quantitative PCR
(RT-qPCR)
Total RNA was extracted using TRIpure isolation
reagent (cat. no. RP1001; BioTeke Corporation) and the product was
collected by centrifugation (4°C, 10,000 × g, 10 min). RNA
concentration was measured by a UV spectrophotometer (Nano 2000;
Thermo Fisher Scientific, Inc.). The RNA samples were
reverse-transcribed to obtain the corresponding cDNA by BeyoRT II
M-MLV reverse transcriptase (cat. no. D7160L; Beyotime Institute of
Biotechnology) and RNase inhibiter (cat. no. B600478; Sangon
Biotech Co., Ltd.). RT-qPCR was performed using SYBR GREEN (cat.
no. SY1020; Beijing Solarbio Science & Technology Co., Ltd.)
and 2XTaq PCR MasterMix (cat. no. PC1105; Beijing Solarbio Science
& Technology Co., Ltd.) with the following thermocycling
conditions: 95°C for 5 min, followed by 40 cycles of 95°C for 10
sec, 60°C for 10 sec and 72°C for 15 sec. Exicycler TM 96
fluorimeter (Bioneer Corporation) was used for fluorescence
quantitative analysis. Results were calculated using
2−ΔΔCq method (24).
β-actin was the internal control. Primers were as follows: secreted
frizzled-related protein 4 (SFRP4) forward, 5′-CAGCACGCAGGAGAACG-3′
and reverse, 5′-GCACGGCTTGATAGGGTC-3′; thyroid hormone receptor
interactor 6 (TRIP6) forward, 5′-GCTGGATAGGCTGACGAAGA-3′ and
reverse, 5′-GCGATCAAGGGCCACAAC-3′; Dead-box helicase 17 (DDX17)
forward, 5′-GTGTTTGCCTTCCATCAT-3′ and reverse,
5′-TCTTCCCAGAGCCAGTC-3′; cellular communication network factor 1
(CCN1) forward, 5′-AGTGGGTCTGTGACGAGGATAG-3′ and reverse,
5′-AACAGGGAGCCGCTTCAGT-3′; SRY-box transcription factor 30 (SOX30)
forward, 5′-AAAGAAACCCTATTACGATG-3′ and reverse
5′-GGCTTGGGCTCTGGACT-3′; tripartite motif containing 36 (TRIM36)
forward, 5′-CTTGATAAATTGGCACCAT-3′ and reverse,
5′-GGACTACAGATTGAACCCT-3′; WNK lysine deficient protein kinase 1
(WNK1) forward, 5′-AAGCAGCCCTCCTAATG-3′ and reverse,
5′-TGTGATACTTGAAACTACGC-3′; transcription factor 3 (TCF3) forward,
5′-GGAGCAGAGGTGAACGG-3′ and reverse, 5′-AAGGGCTGGACGAGAAGT-3′ and
β-actin forward, 5′-CACTGTGCCCATCTACGAGG-3′ and reverse,
5′-TAATGTCACGCACGATTTCC-3′.
Statistical analysis
Data are presented as the mean ± SD from three
independent experiments. Statistical analysis was performed using
Graphpad Prism 8.0 software (GraphPad Software, Inc.; Dotmatics).
Unpaired t test (Fig. 5) and
one-way ANOVA followed by Dunnett's multiple comparisons test
(Figs. 1B, 2B and C) or Tukey's multiple comparisons
test (Figs. 6A, B, C, E, 7A and C) were performed. P<0.05 was
considered to indicate a statistically significant difference.
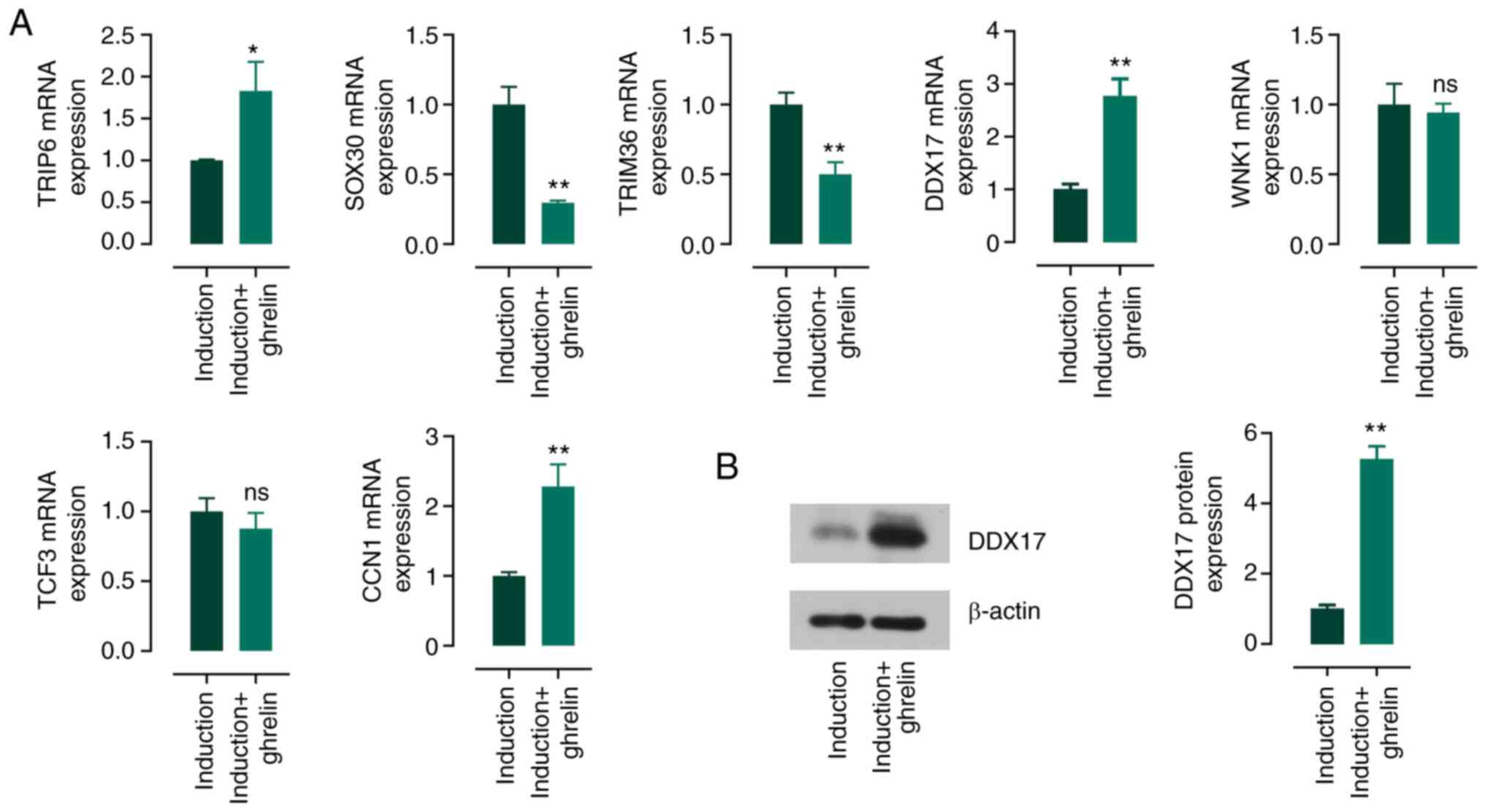 | Figure 5.Potential genes that regulated by
ghrelin in the process of the cardiomyocyte differentiation of
ADMSCs. (A) The mRNA expression of TRIP6, SOX30, TRIM36, DDX17,
WNK1, TCF3 and CCN1 in the induction medium-cultured ADMSCs in the
presence or absence of ghrelin. (B) Protein expression of DDX17 in
ADMSCs. Data are expressed as mean ± SD (n=3). *P<0.05,
**P<0.01 vs. induction. ADMSCs, adipose tissue-derived
mesenchymal stem cells; TRIP6, thyroid hormone receptor interactor
6; SOX30, SRY-box transcription factor 30; TRIM36, tripartite motif
containing 36; DDX17, Dead-box helicase 17; WNK1, WNK lysine
deficient protein kinase 1; TCF3, transcription factor 3; CCN1,
cellular communication network factor 1; ADMSC, adipose
tissue-derived mesenchymal stem cell. |
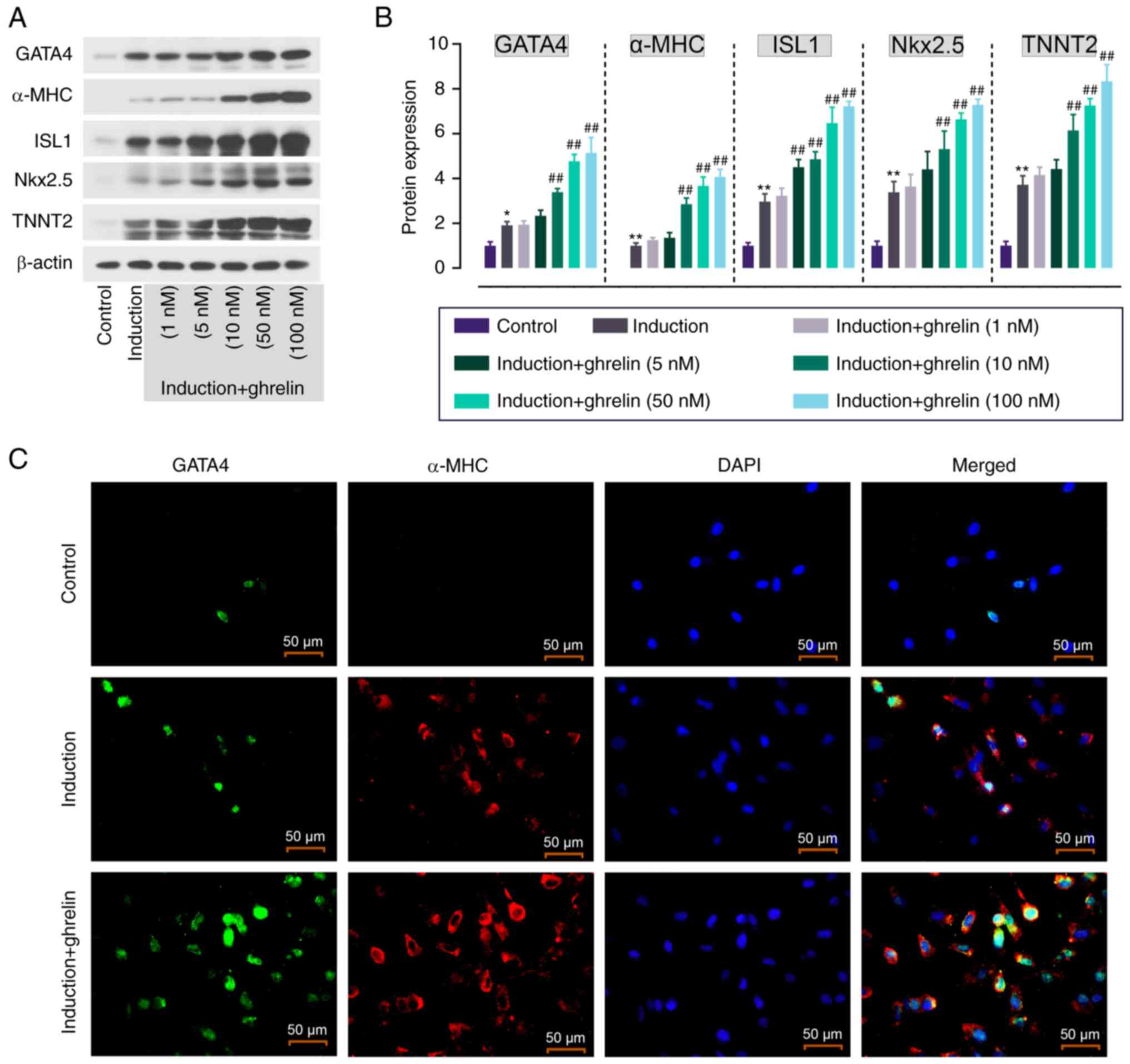 | Figure 1.Ghrelin promotes cardiomyocyte
differentiation of ADMSCs. (A) Protein expression and (B)
quantification of cardiomyocyte markers in ghrelin (1, 5, 10, 50
and 100 nM)-treated ADMSCs. (C) Immunofluorescence staining of
GATA4 (green) and α-MHC (red) in ADMSCs. Scale bar, 50 µm. Data are
expressed as mean ± SD (n=3). *P<0.05, **P<0.01 vs. control;
##P<0.01 vs. induction. ADMSC, adipose tissue-derived
mesenchymal stem cell; GATA4, GATA binding protein 4; MHC, myosin
heavy chain; ISL1, ISL LIM homeobox 1; Nkx2.5, NK2 homeobox 5;
TNNT2, Troponin T2, cardiac type. |
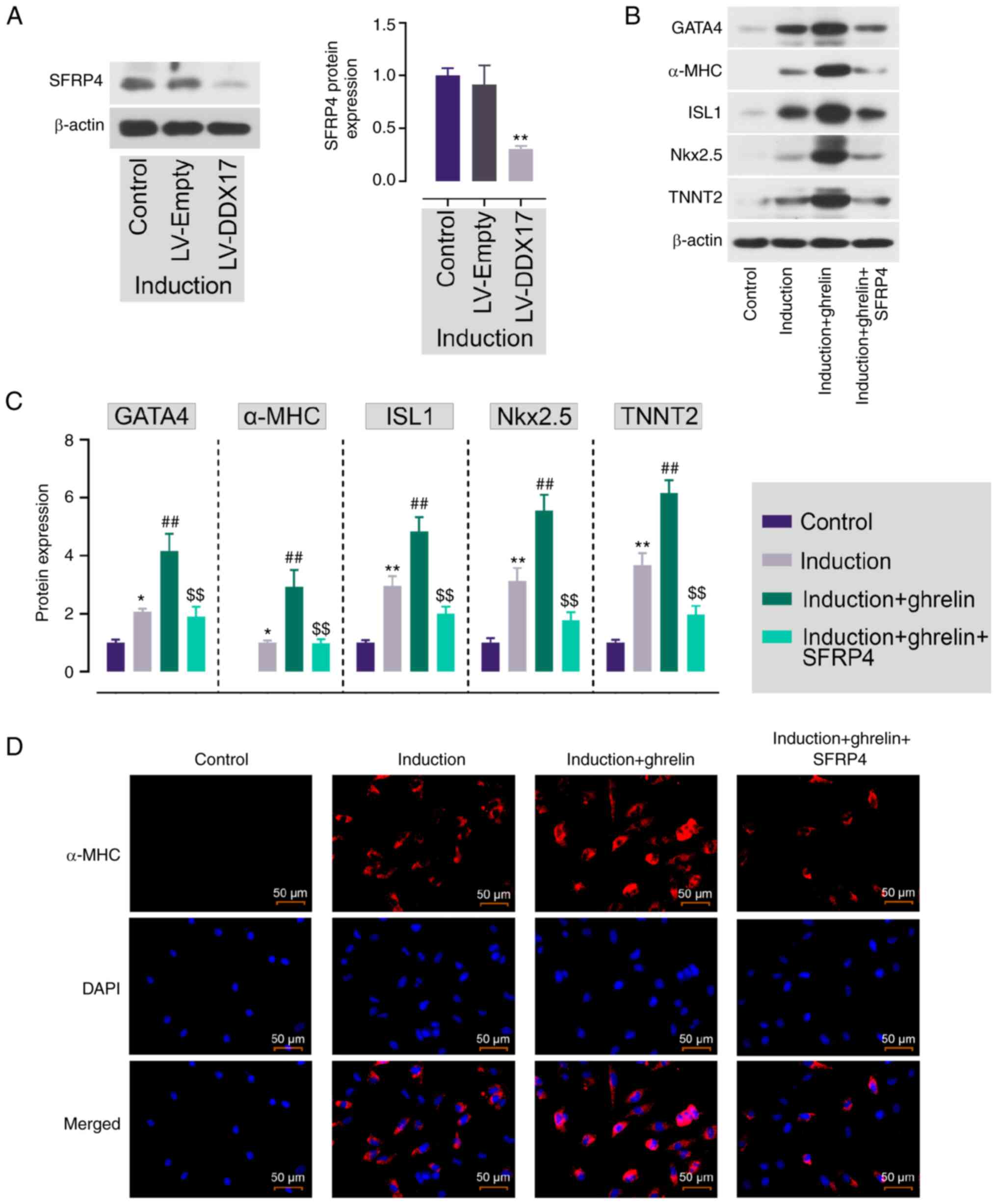 | Figure 7.DDX17, upregulated by ghrelin,
promotes ADMSC cardiomyocyte differentiation by downregulating
SFRP4. (A) Protein expression of SFRP4 in DDX17-overexpressing
ADMSCs. **P<0.01 vs. LV-Empty. (B) Protein expression and (C)
quantification of GATA4, α-MHC, ISL1, Nkx2.5 and TNNT2. (D)
Immunofluorescence staining of α-MHC in ADMSCs. Scale bar, 50 µm.
Data are expressed as mean ± SD (n=3). *P<0.05, **P<0.01 vs.
control; ##P<0.01 vs. induction;
$$P<0.01 vs. induction + ghrelin. DDX17, Dead-box
helicase 17; ADMSC, Adipose tissue-derived mesenchymal stem cell;
SFRP4, secreted frizzled-related protein 4; LV, lentivirus; GATA4,
GATA binding protein 4; MHC, myosin heavy chain; ISL1, ISL LIM
homeobox 1; Nkx2.5, NK2 homeobox 5; TNNT2, Troponin T2, cardiac
type. |
Results
Ghrelin promotes cardiomyocyte
differentiation of ADMSCs
ADMSCs were characterized by flow cytometry and
multilineage differentiation. ADMSCs were positive for CD90, CD44
and CD29 and negative for CD45, CD34 and CD11 (Fig. S1A). Osteogenic (Fig. S1B) and adipogenic (Fig. S1C) induction were used to confirm
the multilineage differentiation of ADMSCs. The differentiation
ability of ADMSCs into cardiomyocyte was assessed by measuring the
expression of cardiomyocyte markers such as GATA4, α-MHC, ISL1,
Nkx2.5 and TNNT2. Induction medium enhanced the expression of these
markers in ADMSCs compared with the control and the addition of
ghrelin further enhanced this in a concentration-dependent manner
(Fig. 1A and B). Based on this,
ghrelin at 10 nM was selected for subsequent experiments.
IF staining was used to show the expression and
localization of GATA4 and α-MHC in ADMSCs. Following ghrelin
treatment, GATA4 (green fluorescence) showed increased nuclear
localization and notable expression of α-MHC (red fluorescence) was
observed in induced ADMSCs (Fig.
1C). By contrast, GATA4 and α-MHC were not notably expressed in
control cells. This indicated that ghrelin promoted cardiomyocyte
differentiation of ADMSCs.
Ghrelin promotes cardiomyocyte
differentiation of ADMSCs via regulation of SFRP4/Wnt/β-catenin
axis
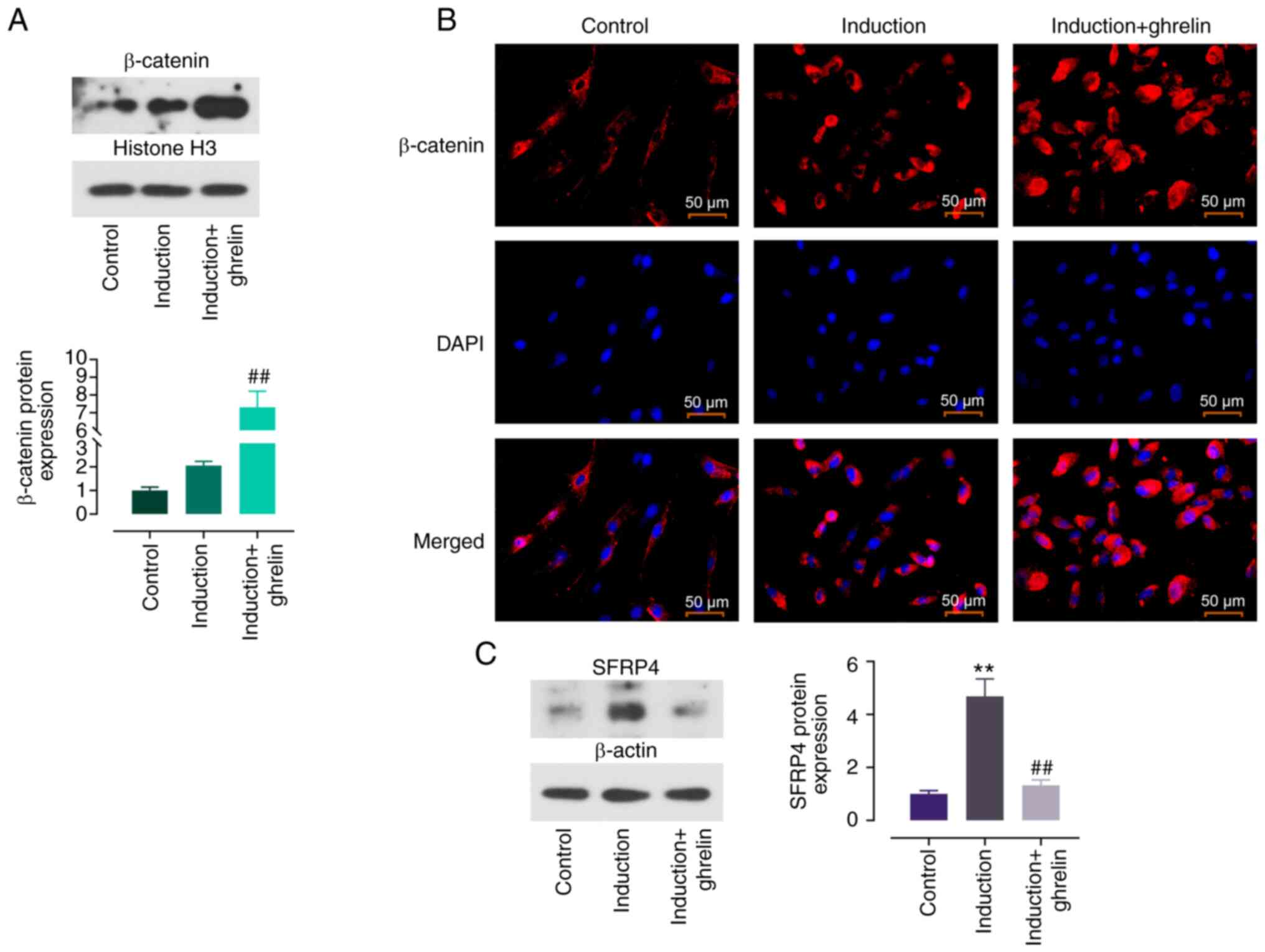
β-catenin expression was enhanced in induced ADMSCs
compared with control cells and ghrelin further increased this
trend (Fig. 2A). IF staining was
performed to determine localization of β-catenin. The red
fluorescence of β-catenin was enhanced following induction
(Fig. 2B). This was further
enhanced by ghrelin and primarily localized in the nucleus,
evidenced by the overlapping blue fluorescence with DAPI. Protein
expression of SFRP4, an inhibiter of Wnt signaling pathway, was
assessed. SFRP4 showed increased expression in induced cells
compared with control and the addition of ghrelin partially
reversed this effect (Fig. 2C).
These results showed that ghrelin promoted activation of the
Wnt/β-catenin signaling pathway in ADMSC cardiomyocyte
differentiation by inhibiting SFRP4.
DEGs and functional enrichment
analysis of RNA-seq
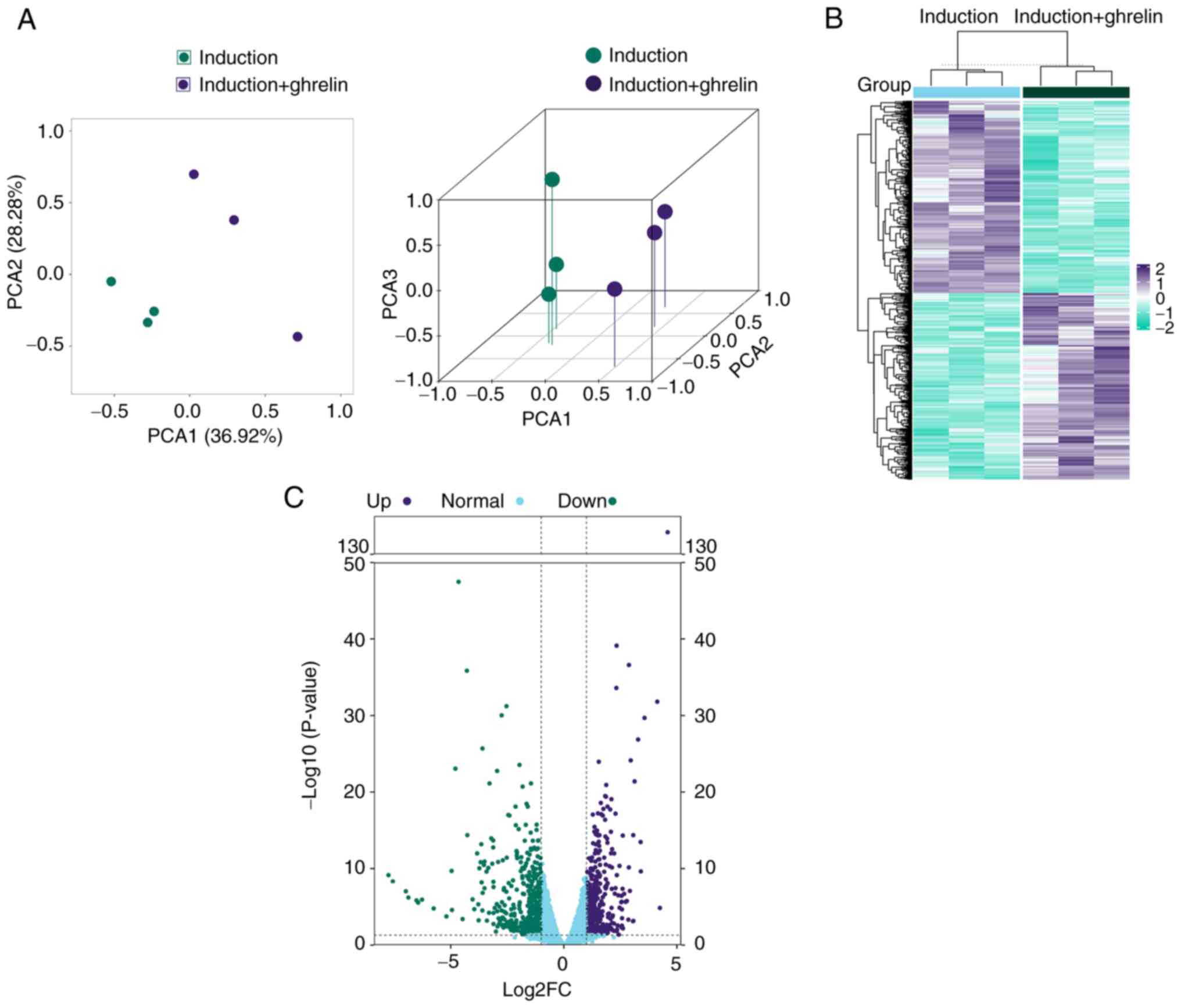
To confirm the role of ghrelin in the cardiomyocyte
differentiation process of ADMSCs, RNA-seq of cultured ADMSCs was
performed. To assess the consistency and variance of cell samples,
PCA and hierarchical clustering were performed. PCA showed clear
clustering in the induction and induction + ghrelin groups
(Fig. 3A), which was also
supported by hierarchical clustering results (Fig. 3B). Up- and downregulated DEGs
(Fig. 3C) were annotated with GO
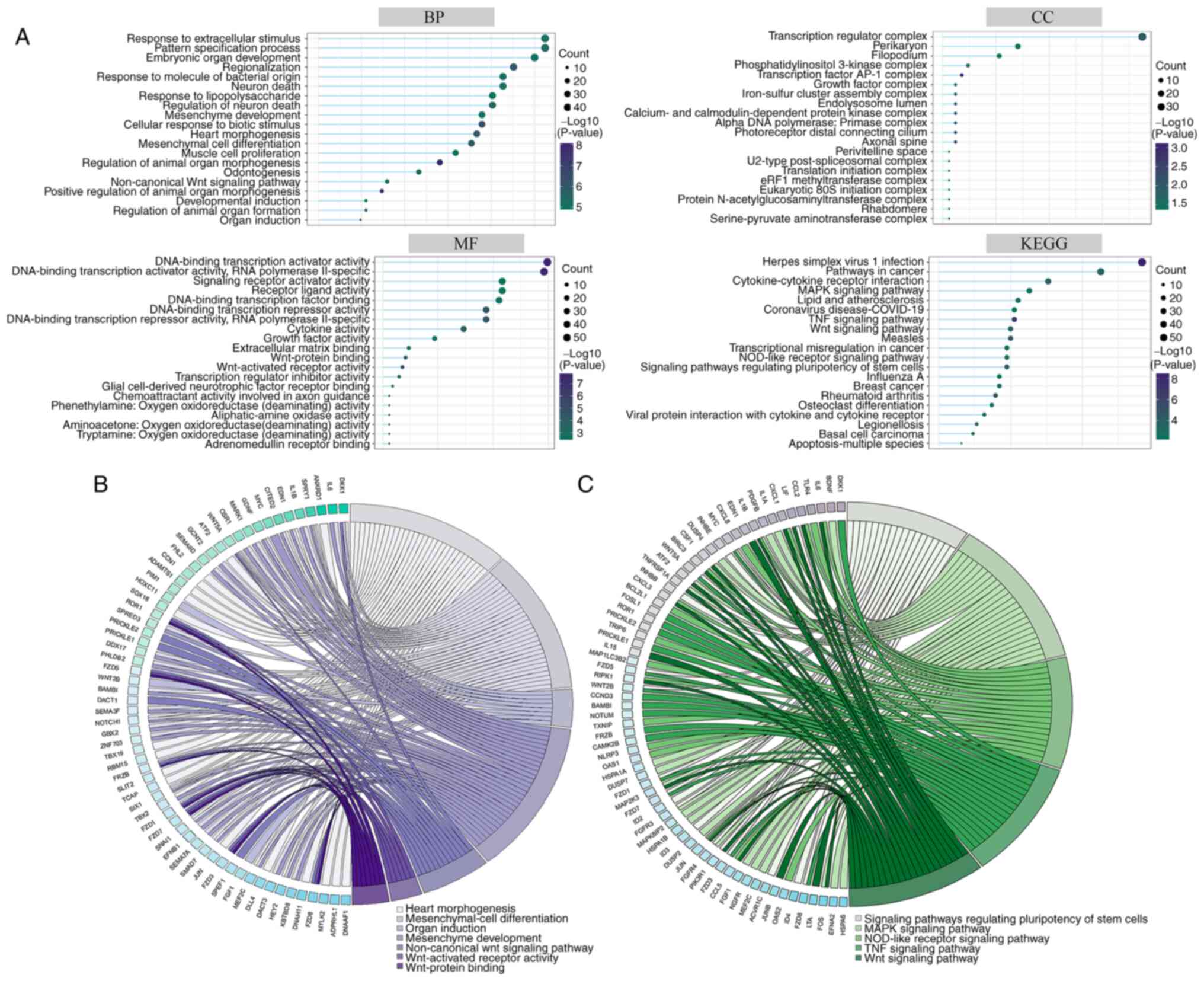
and KEGG for functional analysis (Fig.
4A). GO was sorted into three primary functional categories:
Biological process (BP), cellular component (CC) and molecular
function (MF). Top 20 terms of BP were associated with ‘mesenchyme
development’, ‘heart morphogenesis’, ‘mesenchymal cell
differentiation’ and ‘muscle cell proliferation’; CC was associated
with ‘transcription regulator complex’ and ‘perikaryon’; MF was
associated with ‘Wnt-protein binding’, ‘Wnt-activated receptor
activity’ and ‘transcription regulator inhibitor activity’. Top 20
terms of KEGG were primarily associated with ‘pathways in cancer’,
‘cytokine-cytokine receptor interaction’, ‘MAPK signaling pathway’.
Key GO terms (Fig. 4B) and KEGG
pathways (Fig. 4C) associated with
mesenchymal cell differentiation and Wnt pathways are
displayed.
Potential genes that regulated by
ghrelin in the process of the cardiomyocyte differentiation of
ADMSCs
Based on the results of RNA-seq, the present study
investigated genes that were associated with the Wnt/β-catenin
signaling pathway, including TRIP6 (25,26),
SOX30 (27,28), TRIM36 (29,30),
DDX17 (31–33), WNK1 (34,35),
TCF3 (36) and CCN1 (37). All of these genes have been
reported to be associated with activation or inhibition of the Wnt
signaling pathway in previously reported studies. For verification,
mRNA levels of these genes in cultured cells were measured. Ghrelin
enhanced mRNA expression of TRIP6, DDX17 and CCN1 and suppressed
the levels of SOX30 and TRIM36 (Fig.
5A). WNK1 and TCF3 exhibited no significant difference. Among
these five changed genes, DDX17 has been reported to promote the
expression of myocyte enhancer factor 2C (MEF2C) and TNNT1 in
myoblast cells (38). Therefore,
the present study detected the protein expression of DDX17 in
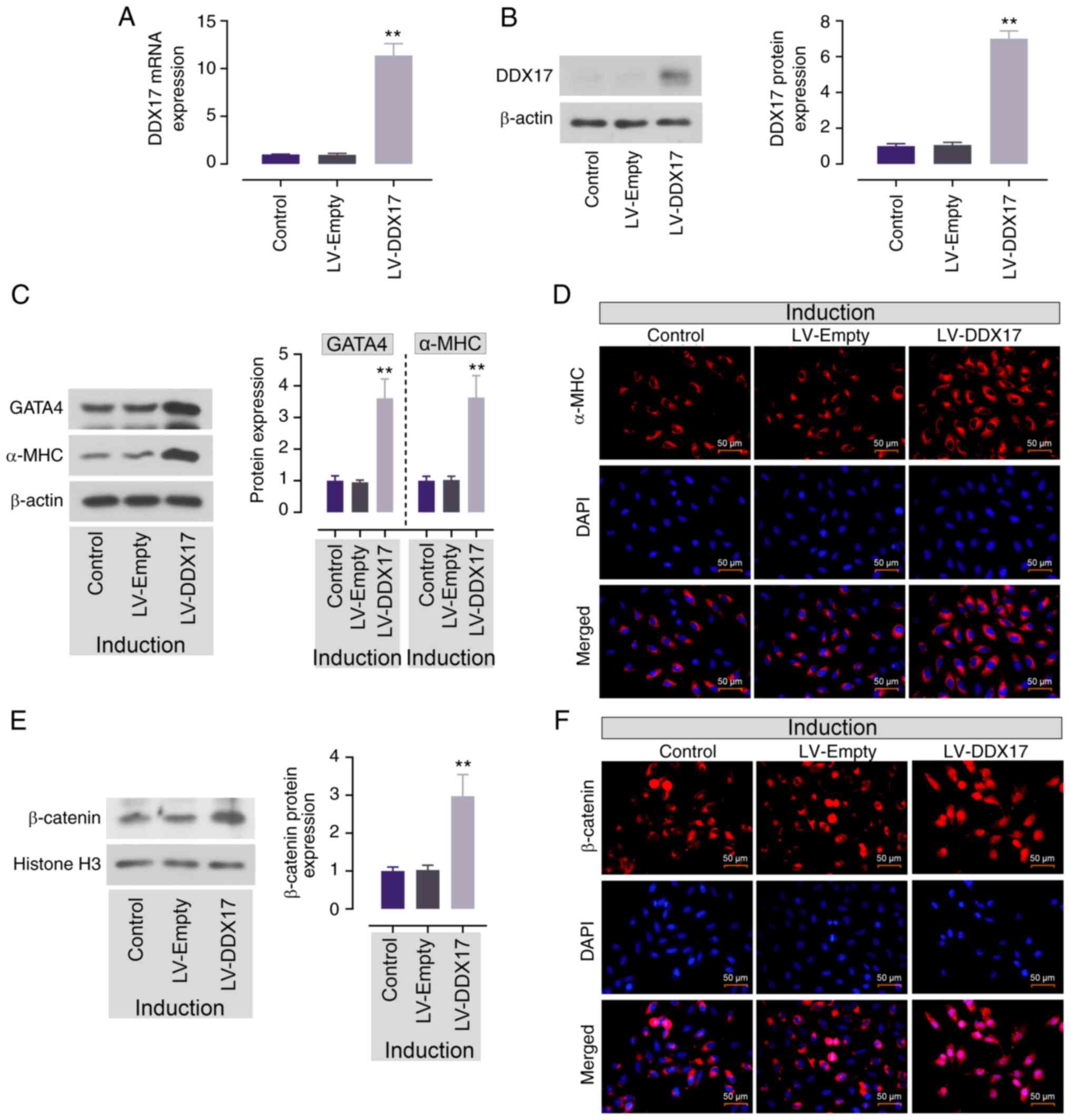
ADMSCs. Ghrelin enhanced the protein expression of DDX17 (Fig. 5B). Accordingly, it was hypothesized
that DDX17 may be involved in ghrelin-induced cardiomyocyte
differentiation of ADMSCs.
DDX17 promotes ADMSC cardiomyocyte
differentiation and β-catenin nuclear accumulation
To explore whether DDX17 was involved in ADMSC
cardiomyocyte differentiation, DDX17 was overexpressed in ADMSCs
and confirmed by RT-qPCR and western blotting (Fig. 6A and B). Then, protein expression
of GATA4 and α-MHC was assessed in ADMSCs to verify the effect of
DDX17. DDX17 overexpression enhanced the expression of GATA4 and
α-MHC in induced ADMSCs (Fig. 6C).
Consistently, IF staining of α-MHC also showed same trend, DDX17
overexpression enhanced the red fluorescence in ADMSCs (Fig. 6D). This demonstrated that enhanced
expression of DDX17 promoted cardiomyocyte differentiation of
ADMSCs.
The present study detected the expression of
β-catenin to verify the effect of DDX17 on Wnt/β-catenin signaling
pathway activation. DDX17 upregulation caused enhanced protein
expression and nuclear accumulation of β-catenin in ADMSCs
(Fig. 6E and F). This demonstrated
that enhanced expression of DDX17 may participate in
ghrelin-induced Wnt/β-catenin signaling in cardiomyocyte
differentiation of ADMSCs.
DDX17, upregulated by ghrelin,
promotes ADMSC cardiomyocyte differentiation by downregulating
SFRP4
The present study investigated the potential
regulatory mechanism between DDX17 and Wnt/β-catenin signaling
pathway activation. Protein expression of SFRP4 in ADMSCs was
assessed; DDX17 overexpression decreased protein expression levels
of SFRP4 (Fig. 7A). This
demonstrated that DDX17 downregulated protein expression of
SFRP4.
To determine the participation of DDX17 and SFRP4 in
ghrelin-induced cardiomyocyte differentiation of ADMSCs, protein
expression of cardiomyocyte markers following SFRP4 addition was
detected. Ghrelin promoted the protein expression levels of GATA4,
α-MHC, ISL1, Nkx2.5 and TNNT2 (Fig. 7B
and C). However, the addition of SFRP4 reversed this effect and
the expression levels of GATA4, α-MHC, ISL1, Nkx2.5 and TNNT2 were
significantly decreased. IF staining of α-MHC also showed that
SFRP4 abolished the enhanced expression of α-MHC (Fig. 7D). This indicated that DDX17 was
involved in ghrelin-induced ADMSC cardiomyocyte differentiation by
downregulating SFRP4.
Discussion
MSCs serve key roles in tissue repair and
regeneration and have been studied in cardiovascular disease
(39,40), including myocardial infarction (MI)
(41,42). ADMSCs differentiate into
cardiomyocyte-like cells and are one of the ideal cell sources for
myocardial regenerative medicine (2); compared with other types of MSC,
ADMSCs are abundant, easy to obtain (43) and free from ethical concerns
(44). These advantages make
ADMSCs a candidate in clinical application. Several studies have
reported that ADMSCs injected into animal myocardium post-MI
results in improved cardiac function compared with untreated rats
(45,46) or mice (47), as well as less fibrosis and wall
thinning (48,49). However, the therapeutic efficacy of
transplanted MSCs is limited by their poor engraftment and low
survival rate in the injured tissues (50–52).
Therefore, accelerating the differentiation of MSCs into
cardiomyocytes may help to facilitate their clinical application.
In the present study, ghrelin promoted differentiation of ADMSCs
into cardiomyocyte via activation of the Wnt/β-catenin pathway.
DDX17, an upregulated DEG following ghrelin addition, promoted the
protein expression of cardiac-specific markers and β-catenin and
inhibited expression of SFRP4. The present study showed that DDX17
was involved in ghrelin-induced cardiomyocyte differentiation of
ADMSCs, which was associated with regulation of the
SFRP4/Wnt/β-catenin axis (Fig. 8).
This provides a scientific foundation for the effect of ghrelin in
cardiac differentiation of ADMSCs and application in clinical
treatment.
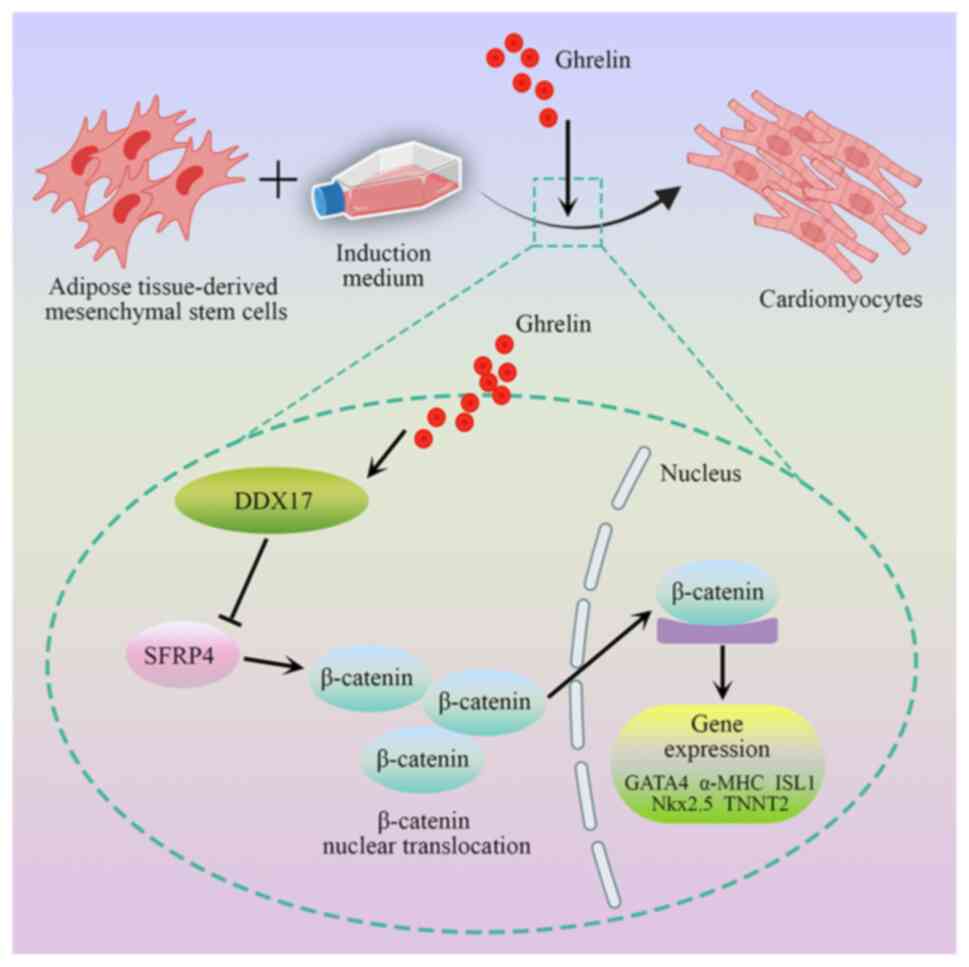 | Figure 8.Role of ghrelin in promotion of
ADMSCs cardiomyocyte differentiation. Ghrelin promotes
differentiation of ADMSCs into cardiomyocyte via activation of the
Wnt/β-catenin pathway. DDX17, an upregulated differentially
expressed gene after ghrelin treatment, promotes nuclear
translocation of β-catenin and inhibits protein expression of
SFRP4. DDX17 is involved in ghrelin-induced cardiomyocyte
differentiation of ADMSCs, which is associated with regulation of
the SFRP4/Wnt/β-catenin axis. ADMSC, adipose tissue-derived
mesenchymal stem cell; DDX17, Dead-box helicase 17; SFRP4, Secreted
frizzled-related protein 4; GATA4, GATA binding protein 4; MHC,
myosin heavy chain; Nkx2.5, NK2 homeobox 5; ISL1, ISL LIM homeobox
1; TNNT2, Troponin T2, cardiac type. |
Ghrelin, a gastric-secreted peptide hormone, is
involved in multilineage differentiation of MSCs (12,53,54),
including bone marrow-derived MSCs (55). Our previous study showed that
ghrelin serves a critical role in the promotion of neural
differentiation of ADMSCs (9).
Another study has showed that intramyocardial injection of ADMSCs
combined with ghrelin inhibits host cardiomyocyte apoptosis and
improves cardiac function, in a mice myocardial infarction model
(56). Therefore, it was
hypothesized that ghrelin may also participate in cardiomyocyte
differentiation of ADMSCs. The present results showed that ghrelin
promoted expression of cardiac-specific markers GATA4, α-MHC, ISL1,
Nkx2.5 and TNNT2. This result implied that ghrelin promoted
cardiomyocyte differentiation of ADMSCs. After confirming the
positive effect of ghrelin on differentiation of ADMSCs into
cardiomyocyte, the present study explored the underlying mechanism.
In previously reported studies, Wnt pathway was confirmed to serve
an important role in cardiomyocyte differentiation and cardiac
formation (57,58). A previous study suggested that
Wnt/β-catenin regulates the crucial transcription factors as
Baf60c, Nkx2.5 and ISL1 at an early stage of progenitor formation
(59). The aforementioned results
demonstrate that the regulation of Wnt/β-catenin pathway is key
during cardiomyocyte differentiation. Similarly, ghrelin promotes
neural SC differentiation via the Wnt/β-catenin pathway (60). Ghrelin can regulate apoptosis in
cells and tissue by regulating the Wnt/β-catenin pathway (61,62).
Therefore, it was hypothesized that the positive effect of ghrelin
on cardiomyocyte differentiation of ADMSCs may be associated with
the Wnt/β-catenin pathway. The present study found that ghrelin
significantly promoted expression of nuclear β-catenin, thus
enhancing the activity of the aforementioned signaling pathway. In
addition, studies have reported that SFRP4, an inhibiter of the
Wnt/β-catenin pathway, inhibits the expression of cardiac-specific
genes during cardiomyocyte differentiation of P19CL6 cells
(22). Knockdown of SFRP4
attenuates apoptosis to protect against myocardial
ischemia/reperfusion injury (63).
Therefore, the expression of SFRP4 was also tested; ghrelin
inhibited the elevated SFRP4 protein expression in ADMSCs.
Therefore, it was hypothesized that ghrelin may promote
cardiomyocyte differentiation of ADMSCs by regulating the
SFRP4/Wnt/β-catenin axis.
To determine the role of ghrelin in ADMSCs, cells
were subjected to RNA-seq to determine the genes regulated by
ghrelin. Expression of numerous genes changed following ghrelin
treatment. Following screening, TRIP6, SOX30, TRIM36, DDX17, WNK1,
TCF3 and CCN1 were selected for further study; these genes were
associated with the Wnt/β-catenin signaling pathway. Among these
genes, TRIP6 (25,26), DDX17 (31–33),
WNK1 (34,35) and CCN1 (37) are associated with the activation of
the Wnt/β-catenin pathway and SOX30 (27,28),
TRIM36 (29,30) and TCF3 (36) are associated with its inhibition.
DDX17 has been reported to promote mRNA expression of myocyte
enhancer factor 2C (MEF2C) and TNNT1 in myoblast cells (38) and serve a protective role in
doxorubicin-induced cardiomyocyte injury (64). Therefore, DDX17 was selected for
further study. Here, DDX17 overexpression showed a similar effect
to ghrelin in promoting the cardiomyocyte differentiation of ADMSCs
and Wnt/β-catenin signaling pathway activation. More importantly,
the present results showed that DDX17 downregulated SFRP4
expression. Rescue assay was performed to determine the role of
SFRP4. SFRP4 reversed the promoting effect of ghrelin on
cardiomyocyte differentiation of ADMSCs, which demonstrated the
participation of DDX17 in this process.
The present study demonstrated that ghrelin promoted
cardiomyocyte differentiation of ADMSCs. However, there are
limitations. First, RNA-seq indicated that the promotive effect of
ghrelin may be related to DDX17-mediated regulation of the
SFRP4/Wnt/β-catenin axis. However, the expression of numerous genes
was significantly changed following ghrelin treatment, including
456 up- and 470 downregulated genes. Therefore, ghrelin may promote
cardiomyocyte differentiation of ADMSCs by regulating other genes.
Second, based on the critical role of the Wnt/β-catenin pathway in
cardiomyocyte differentiation, the present study focused on the
connection between ghrelin and activation of the Wnt/β-catenin
pathway. However, TGF-β (65) and
Notch (66,67) signaling pathways also serve an
important role in cardiomyocyte differentiation. Therefore, the
promoting effect of ghrelin on cardiomyocyte differentiation of
ADMSCs may not be solely dependent on the Wnt/β-catenin pathway.
Third, the present study mainly focused on the effect of ghrelin on
cardiomyocyte differentiation of ADMSCs in vitro; effects of
ghrelin in vivo need to be determined.
In conclusion, the present study indicated that
ghrelin promoted differentiation of ADMSCs into cardiomyocytes by
DDX17-mediated regulation of the SFRP4/Wnt/β-catenin axis.
Specifically, ghrelin promoted cardiomyocyte differentiation of
ADMSCs via activation of the Wnt/β-catenin pathway. DDX17, an
upregulated DEG, enhanced protein expression of cardiac-specific
markers and β-catenin and inhibited the expression of SFRP4. The
present results provide scientific foundation for the effect of
ghrelin in cardiac-differentiation of ADMSCs and may assist to
promote the application of ghrelin in the ADMSCs mediated clinical
treatment of cardiac-associated injury.
Supplementary Material
Supporting Data
Acknowledgements
Not applicable.
Funding
The present study was supported by the Joint Guidance Project of
Natural Science Foundation of Heilongjiang Province (grant no.
LH2020H074) and the Scientific Research Project of Basic Scientific
Research Business Expenses of Provincial Colleges and Universities
in Heilongjiang Province (grant number 2020-KYYWF-0780).
Availability of data and materials
The datasets generated and analyzed during the
current study are available in the NCBI Gene Expression Omnibus
repository, ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE232876
(accession no. GSE232876).
Authors' contributions
GBL conceived the study, designed the experiments,
collected data and wrote the manuscript. YXC conceived the study,
designed the experiments and wrote the manuscript. HML and YL
collected, analyzed and clarified the data. LXS and QW designed the
experiments and performed the literature review. SFG and TTL
analyzed and interpreted data and performed the literature review.
CLD collected data and performed the literature review. GS
conceived and supervised the study, designed the experiments and
reviewed the manuscript. GBL and GS confirm the authenticity of all
the raw data. All authors have read and approved the final
manuscript.
Ethics approval and consent to
participate
Not applicable.
Patient consent for publication
Not applicable.
Competing interests
The authors declare they have no competing
interests.
Glossary
Abbreviations
Abbreviations:
|
ADMSC
|
adipose tissue-derived mesenchymal
stem cell
|
|
GATA4
|
GATA binding protein 4
|
|
α-MHC
|
α-myosin heavy chain
|
|
ISL1
|
ISL LIM homeobox 1
|
|
Nkx2.5
|
NK2 homeobox 5
|
|
TNNT2
|
troponin T2, cardiac type
|
|
SFRP4
|
secreted frizzled-related protein
4
|
|
DEG
|
differentially expressed gene
|
|
DDX17
|
dead-box helicase 17
|
|
FBS
|
fetal bovine serum
|
|
BSA
|
bovine serum albumin
|
|
IF
|
immunofluorescence
|
|
RT-q
|
reverse transcription-quantitative
|
|
seq
|
sequencing
|
|
PCA
|
principal component analysis
|
|
GO
|
Gene Ontology
|
|
KEGG
|
Kyoto Encyclopedia of Genes and
Genomes
|
|
BP
|
biological process
|
|
CC
|
cellular component
|
|
MF
|
molecular function
|
References
|
1
|
Zhou Z, Chen Y, Zhang H, Min S, Yu B, He B
and Jin A: Comparison of mesenchymal stromal cells from human bone
marrow and adipose tissue for the treatment of spinal cord injury.
Cytotherapy. 15:434–448. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Joo HJ, Kim JH and Hong SJ: Adipose
tissue-derived stem cells for myocardial regeneration. Korean Circ
J. 47:151–159. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Miyahara Y, Nagaya N, Kataoka M, Yanagawa
B, Tanaka K, Hao H, Ishino K, Ishida H, Shimizu T, Kangawa K, et
al: Monolayered mesenchymal stem cells repair scarred myocardium
after myocardial infarction. Nat Med. 12:459–465. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Kojima M, Hosoda H, Date Y, Nakazato M,
Matsuo H and Kangawa K: Ghrelin is a growth-hormone-releasing
acylated peptide from stomach. Nature. 402:656–660. 1999.
View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Yuan MJ, Li W and Zhong P: Research
progress of ghrelin on cardiovascular disease. Biosci Rep.
41:BSR202033872021. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Khatib MN, Shankar A, Kirubakaran R, Agho
K, Simkhada P, Gaidhane S, Saxena D B U, Gode D, Gaidhane A and
Zahiruddin SQ: Effect of ghrelin on mortality and cardiovascular
outcomes in experimental rat and mice models of heart failure: A
systematic review and meta-analysis. PLoS One. 10:e01266972015.
View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Tokudome T, Otani K, Miyazato M and
Kangawa K: Ghrelin and the heart. Peptides. 111:42–46. 2019.
View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Tokudome T and Kangawa K: Physiological
significance of ghrelin in the cardiovascular system. Proc Jpn Acad
Ser B Phys Biol Sci. 95:459–467. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Liu GB, Pan YM, Liu YS, Hu JH, Zhang XD,
Zhang DW, Wang Y, Feng YK, Yu JB and Cheng YX: Ghrelin promotes
neural differentiation of adipose tissue-derived mesenchymal stem
cell via AKT/mTOR and β-catenin signaling pathways. Kaohsiung J Med
Sci. 36:405–416. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Eid RA, Alkhateeb MA, Al-Shraim M, Eleawa
SM, Shatoor AS, El-Kott AF, Zaki MSA, Shatoor KA, Bin-Jaliah I and
Al-Hashem FH: Ghrelin prevents cardiac cell apoptosis during
cardiac remodelling post experimentally induced myocardial
infarction in rats via activation of Raf-MEK1/2-ERK1/2 signalling.
Arch Physiol Biochem. 125:93–103. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Eid RA, Alkhateeb MA, Eleawa S, Al-Hashem
FH, Al-Shraim M, El-Kott AF, Zaki MSA, Dallak MA and Aldera H:
Cardioprotective effect of ghrelin against myocardial
infarction-induced left ventricular injury via inhibition of SOCS3
and activation of JAK2/STAT3 signaling. Basic Res Cardiol.
113:132018. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Sun L and Zhang W: Preconditioning of
mesenchymal stem cells with ghrelin exerts superior
cardioprotection in aged heart through boosting mitochondrial
function and autophagy flux. Eur J Pharmacol. 903:1741422021.
View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Gao M, Yang J, Wei R, Liu G, Zhang L, Wang
H, Wang G, Gao H, Chen G and Hong T: Ghrelin induces cardiac
lineage differentiation of human embryonic stem cells through
ERK1/2 pathway. Int J Cardiol. 167:2724–2733. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Yang J, Liu GQ, Wei R, Hou WF, Gao MJ, Zhu
MX, Wang HN, Chen GA and Hong TP: Ghrelin promotes differentiation
of human embryonic stem cells into cardiomyocytes. Acta Pharmacol
Sin. 32:1239–1245. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Jiang M, Liu T, Zhang J, Gao S, Tao B, Cao
R, Qiu Y, Liu J, Li Y, Wang Y and Cao F: Rapamycin promotes
cardiomyocyte differentiation of human induced pluripotent stem
cells in a stage-dependent manner. Stem Cells Dev. 29:1229–1239.
2020. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Ueno S, Weidinger G, Osugi T, Kohn AD,
Golob JL, Pabon L, Reinecke H, Moon RT and Murry CE: Biphasic role
for Wnt/beta-catenin signaling in cardiac specification in
zebrafish and embryonic stem cells. Proc Natl Acad Sci USA.
104:9685–9690. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Li B, Zeng M, He W, Huang X, Luo L, Zhang
H and Deng DY: Ghrelin protects alveolar macrophages against
lipopolysaccharide-induced apoptosis through growth hormone
secretagogue receptor 1a-dependent c-Jun N-terminal kinase and
Wnt/β-catenin signaling and suppresses lung inflammation.
Endocrinology. 156:203–217. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Gay D, Ghinatti G, Guerrero-Juarez CF,
Ferrer RA, Ferri F, Lim CH, Murakami S, Gault N, Barroca V, Rombeau
I, et al: Phagocytosis of Wnt inhibitor SFRP4 by late wound
macrophages drives chronic Wnt activity for fibrotic skin healing.
Sci Adv. 6:eaay37042020. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Zhang Y, Guan H, Fu Y, Wang X, Bai L, Zhao
S and Liu E: Effects of SFRP4 overexpression on the production of
adipokines in transgenic mice. Adipocyte. 9:374–383. 2020.
View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Visweswaran M, Schiefer L, Arfuso F,
Dilley RJ, Newsholme P and Dharmarajan A: Wnt antagonist secreted
frizzled-related protein 4 upregulates adipogenic differentiation
in human adipose tissue-derived mesenchymal stem cells. PLoS One.
10:e01180052015. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Yamada A, Iwata T, Yamato M, Okano T and
Izumi Y: Diverse functions of secreted frizzled-related proteins in
the osteoblastogenesis of human multipotent mesenchymal stromal
cells. Biomaterials. 34:3270–3278. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Tian Y, Wang W, Lu Q, Chen P, Ma K, Jia Z
and Zhou C: Cross-talk of SFRP4, integrin α1β1, and Notch1 inhibits
cardiac differentiation of P19CL6 cells. Cell Signal. 28:1806–1815.
2016. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Qazi RE, Naeem N, Khan I, Qadeer Q,
Shaheen F and Salim A: Effect of a dianthin G analogue in the
differentiation of rat bone marrow mesenchymal stem cells into
cardiomyocytes. Mol Cell Biochem. 475:27–39. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(−Delta Delta C(T)) method. Methods. 25:402–408. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Zhao X, Jiang C, Xu R, Liu Q, Liu G and
Zhang Y: TRIP6 enhances stemness property of breast cancer cells
through activation of Wnt/β-catenin. Cancer Cell Int. 20:512020.
View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Gou H, Liang JQ, Zhang L, Chen H, Zhang Y,
Li R, Wang X, Ji J, Tong JH, To KF, et al: TTPAL promotes
colorectal tumorigenesis by stabilizing TRIP6 to activate
Wnt/β-catenin signaling. Cancer Res. 79:3332–3346. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Han F, Liu WB, Shi XY, Yang JT, Zhang X,
Li ZM, Jiang X, Yin L, Li JJ, Huang CS, et al: SOX30 inhibits tumor
metastasis through attenuating Wnt-signaling via transcriptional
and posttranslational regulation of β-catenin in lung cancer.
EBioMedicine. 31:253–266. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Fu Q, Sun Z, Yang F, Mao T, Gao Y and Wang
H: SOX30, a target gene of miR-653-5p, represses the proliferation
and invasion of prostate cancer cells through inhibition of
Wnt/β-catenin signaling. Cell Mol Biol Lett. 24:712019. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Tong Q, Yi M, Kong P, Xu L, Huang W, Niu
Y, Gan X, Zhan H, Tian R and Yan D: TRIM36 inhibits tumorigenesis
through the Wnt/β-catenin pathway and promotes caspase-dependent
apoptosis in hepatocellular carcinoma. Cancer Cell Int. 22:2782022.
View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Zhao B, Qiao G, Li J, Wang Y, Li X, Zhang
H and Zhang L: TRIM36 suppresses cell growth and promotes apoptosis
in human esophageal squamous cell carcinoma cells by inhibiting
Wnt/β-catenin signaling pathway. Hum Cell. 35:1487–1498. 2022.
View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Li K, Mo C, Gong D, Chen Y, Huang Z, Li Y,
Zhang J, Huang L, Li Y, Fuller-Pace FV, et al: DDX17
nucleocytoplasmic shuttling promotes acquired gefitinib resistance
in non-small cell lung cancer cells via activation of β-catenin.
Cancer Lett. 400:194–202. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Shin S, Rossow KL, Grande JP and Janknecht
R: Involvement of RNA helicases p68 and p72 in colon cancer. Cancer
Res. 67:7572–7578. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Germann S, Gratadou L, Zonta E, Dardenne
E, Gaudineau B, Fougère M, Samaan S, Dutertre M, Jauliac S and
Auboeuf D: Dual role of the ddx5/ddx17 RNA helicases in the control
of the pro-migratory NFAT5 transcription factor. Oncogene.
31:4536–4549. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Serysheva E, Berhane H, Grumolato L, Demir
K, Balmer S, Bodak M, Boutros M, Aaronson S, Mlodzik M and Jenny A:
Wnk kinases are positive regulators of canonical Wnt/β-catenin
signalling. EMBO Rep. 14:718–725. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Sato A, Shimizu M, Goto T, Masuno H,
Kagechika H, Tanaka N and Shibuya H: WNK regulates Wnt signalling
and β-Catenin levels by interfering with the interaction between
β-Catenin and GID. Commun Biol. 3:6662020. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Kuwahara A, Sakai H, Xu Y, Itoh Y,
Hirabayashi Y and Gotoh Y: Tcf3 represses Wnt-β-catenin signaling
and maintains neural stem cell population during neocortical
development. PLoS One. 9:e944082014. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Yang Y, Qi Q, Wang Y, Shi Y, Yang W, Cen
Y, Zhu E, Li X, Chen D and Wang B: Cysteine-rich protein 61
regulates adipocyte differentiation from mesenchymal stem cells
through mammalian target of rapamycin complex 1 and canonical Wnt
signaling. FASEB J. 32:3096–3107. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Caretti G, Schiltz RL, Dilworth FJ, Di
Padova M, Zhao P, Ogryzko V, Fuller-Pace FV, Hoffman EP, Tapscott
SJ and Sartorelli V: The RNA helicases p68/p72 and the noncoding
RNA SRA are coregulators of MyoD and skeletal muscle
differentiation. Dev Cell. 11:547–560. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Choi YH, Kurtz A and Stamm C: Mesenchymal
stem cells for cardiac cell therapy. Hum Gene Ther. 22:3–17. 2011.
View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Gupta S, Sharma A S A and Verma RS:
Mesenchymal stem cells for cardiac regeneration: From
differentiation to cell delivery. Stem Cell Rev Rep. 17:1666–1694.
2021. View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Neshati V, Mollazadeh S, Fazly Bazzaz BS,
de Vries AA, Mojarrad M, Naderi-Meshkin H, Neshati Z and Kerachian
MA: Cardiomyogenic differentiation of human adipose-derived
mesenchymal stem cells transduced with Tbx20-encoding lentiviral
vectors. J Cell Biochem. 119:6146–6153. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Shafei AE, Ali MA, Ghanem HG, Shehata AI,
Abdelgawad AA, Handal HR, Talaat KA, Ashaal AE and El-Shal AS:
Mesenchymal stem cell therapy: A promising cell-based therapy for
treatment of myocardial infarction. J Gene Med. 19:2017. View Article : Google Scholar : PubMed/NCBI
|
|
43
|
Casteilla L, Planat-Benard V, Laharrague P
and Cousin B: Adipose-derived stromal cells: Their identity and
uses in clinical trials, an update. World J Stem Cells. 3:25–33.
2011. View Article : Google Scholar : PubMed/NCBI
|
|
44
|
Fraser JK, Wulur I, Alfonso Z and Hedrick
MH: Fat tissue: An underappreciated source of stem cells for
biotechnology. Trends Biotechnol. 24:150–154. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
45
|
Otto Beitnes J, Oie E, Shahdadfar A,
Karlsen T, Müller RM, Aakhus S, Reinholt FP and Brinchmann JE:
Intramyocardial injections of human mesenchymal stem cells
following acute myocardial infarction modulate scar formation and
improve left ventricular function. Cell Transplant. 21:1697–1709.
2012. View Article : Google Scholar : PubMed/NCBI
|
|
46
|
Irion CI, Martins EL, Christie MLA, de
Andrade CBV, de Moraes ACN, Ferreira RP, Pimentel CF, Suhett GD, de
Carvalho ACC, Lindoso RS, et al: Acute myocardial infarction
reduces respiration in rat cardiac fibers, despite adipose tissue
mesenchymal stromal cell transplant. Stem Cells Int.
2020:43279652020. View Article : Google Scholar : PubMed/NCBI
|
|
47
|
Davy PM, Lye KD, Mathews J, Owens JB, Chow
AY, Wong L, Moisyadi S and Allsopp RC: Human adipose stem cell and
ASC-derived cardiac progenitor cellular therapy improves outcomes
in a murine model of myocardial infarction. Stem Cells Cloning.
8:135–148. 2015.PubMed/NCBI
|
|
48
|
Cai L, Johnstone BH, Cook TG, Tan J,
Fishbein MC, Chen PS and March KL: IFATS collection: Human adipose
tissue-derived stem cells induce angiogenesis and nerve sprouting
following myocardial infarction, in conjunction with potent
preservation of cardiac function. Stem Cells. 27:230–237. 2009.
View Article : Google Scholar : PubMed/NCBI
|
|
49
|
Cho JW, Seo MS, Kang KK and Sung SE:
Effect of human thymus adipose tissue-derived mesenchymal stem
cells on myocardial infarction in rat model. Regen Ther.
11:192–198. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
50
|
Kuraitis D, Ruel M and Suuronen EJ:
Mesenchymal stem cells for cardiovascular regeneration. Cardiovasc
Drugs Ther. 25:349–362. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
51
|
Penicka M, Widimsky P, Kobylka P, Kozak T
and Lang O: Images in cardiovascular medicine. Early tissue
distribution of bone marrow mononuclear cells after transcoronary
transplantation in a patient with acute myocardial infarction.
Circulation. 112:e63–e65. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
52
|
Müller-Ehmsen J, Krausgrill B, Burst V,
Schenk K, Neisen UC, Fries JW, Fleischmann BK, Hescheler J and
Schwinger RH: Effective engraftment but poor mid-term persistence
of mononuclear and mesenchymal bone marrow cells in acute and
chronic rat myocardial infarction. J Mol Cell Cardiol. 41:876–884.
2006. View Article : Google Scholar : PubMed/NCBI
|
|
53
|
Russo C, Mannino G, Patanè M, Parrinello
NL, Pellitteri R, Stanzani S, Giuffrida R, Lo Furno D and Russo A:
Ghrelin peptide improves glial conditioned medium effects on
neuronal differentiation of human adipose mesenchymal stem cells.
Histochem Cell Biol. 156:35–46. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
54
|
Li K, Fan L, Lin J, Heng BC, Deng Z, Zheng
Q, Zhang J, Jiang Y and Ge Z: Nanosecond pulsed electric fields
prime mesenchymal stem cells to peptide ghrelin and enhance
chondrogenesis and osteochondral defect repair in vivo. Sci China
Life Sci. 65:927–939. 2022. View Article : Google Scholar : PubMed/NCBI
|
|
55
|
Ge S, He W, Zhang L, Lin S, Luo Y, Chen Q
and Zeng M: Ghrelin pretreatment enhanced the protective effect of
bone marrow-derived mesenchymal stem cell-conditioned medium on
lipopolysaccharide-induced endothelial cell injury. Mol Cell
Endocrinol. 548:1116122022. View Article : Google Scholar : PubMed/NCBI
|
|
56
|
Han D, Huang W, Ma S, Chen J, Gao L, Liu
T, Zhang R, Li X, Li C, Fan M, et al: Ghrelin improves functional
survival of engrafted adipose-derived mesenchymal stem cells in
ischemic heart through PI3K/Akt signaling pathway. Biomed Res Int.
2015:8583492015. View Article : Google Scholar : PubMed/NCBI
|
|
57
|
Lian X, Zhang J, Azarin SM, Zhu K,
Hazeltine LB, Bao X, Hsiao C, Kamp TJ and Palecek SP: Directed
cardiomyocyte differentiation from human pluripotent stem cells by
modulating Wnt/β-catenin signaling under fully defined conditions.
Nat Protoc. 8:162–175. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
58
|
Lian X, Hsiao C, Wilson G, Zhu K,
Hazeltine LB, Azarin SM, Raval KK, Zhang J, Kamp TJ and Palecek SP:
Robust cardiomyocyte differentiation from human pluripotent stem
cells via temporal modulation of canonical Wnt signaling. Proc Natl
Acad Sci USA. 109:E1848–E1857. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
59
|
Klaus A, Müller M, Schulz H, Saga Y,
Martin JF and Birchmeier W: Wnt/β-catenin and Bmp signals control
distinct sets of transcription factors in cardiac progenitor cells.
Proc Natl Acad Sci USA. 109:10921–10926. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
60
|
Gong B, Jiao L, Du X, Li Y, Bi M, Jiao Q
and Jiang H: Ghrelin promotes midbrain neural stem cells
differentiation to dopaminergic neurons through Wnt/β-catenin
pathway. J Cell Physiol. 235:8558–8570. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
61
|
Liu X, Chen D, Wu Z, Li J, Li J, Zhao H
and Liu T: Ghrelin inhibits high glucose-induced 16HBE cells
apoptosis by regulating Wnt/β-catenin pathway. Biochem Biophys Res
Commun. 477:902–907. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
62
|
Qu R, Chen X, Yuan Y, Wang W, Qiu C, Liu
L, Li P, Zhang Z, Vasilev K, Liu L, et al: Ghrelin fights against
Titanium particle-induced inflammatory osteolysis through
activation of β-catenin signaling pathway. Inflammation.
42:1652–1665. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
63
|
Zeng W, Cao Y, Jiang W, Kang G, Huang J
and Xie S: Knockdown of Sfrp4 attenuates apoptosis to protect
against myocardial ischemia/reperfusion injury. J Pharmacol Sci.
140:14–19. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
64
|
Lin B, Wang F, Wang J, Xu C, Zhao H, Cheng
Z and Feng D: The protective role of p72 in doxorubicin-induced
cardiomyocytes injury in vitro. Mol Med Rep. 14:3376–3380.
2016. View Article : Google Scholar : PubMed/NCBI
|
|
65
|
Wang MK, Sun HQ, Xiang YC, Jiang F, Su YP
and Zou ZM: Different roles of TGF-β in the multi-lineage
differentiation of stem cells. World J Stem Cells. 4:28–34. 2012.
View Article : Google Scholar : PubMed/NCBI
|
|
66
|
MacGrogan D, Münch J and de la Pompa JL:
Notch and interacting signalling pathways in cardiac development,
disease, and regeneration. Nat Rev Cardiol. 15:685–704. 2018.
View Article : Google Scholar : PubMed/NCBI
|
|
67
|
Fang X, Miao S, Yu Y, Ding F, Han X, Wu H,
Zhao ZA, Wang Y, Hu S and Lei W: MIR148A family regulates
cardiomyocyte differentiation of human embryonic stem cells by
inhibiting the DLL1-mediated NOTCH signaling pathway. J Mol Cell
Cardiol. 134:1–12. 2019. View Article : Google Scholar : PubMed/NCBI
|