Muscular atrophy refers to the progressive loss of
skeletal muscle mass (SMM) and strength and is a disease that has
profound effects on the general health and quality of life of
patients (1,2). Muscular atrophy is associated with
various factors, including aging, immobility, long-term illness,
poor nutrition and genetic diseases (3,4).
Muscular atrophy involves several intricate molecular mechanisms,
an understanding of which is of importance when developing
effective treatment strategies. MicroRNAs (miRNAs/miRs) are small
RNA molecules, usually 19–23 nucleotides in size, which
post-transcriptionally regulate gene expression. miRNAs regulate
signaling associated with protein synthesis within cells, thereby
controlling a variety of cellular processes, including muscle
development and maintenance (5,6). In
the context of muscular atrophy, miRNAs can either promote or
alleviate muscle loss (7); thus,
the genes and pathways involved in muscular atrophy can be targeted
to either enhance or reduce muscle wasting. Skeletal muscular
atrophy presents as changes, including muscle fiber contraction, as
well as the loss of muscle cytoplasm, organelles and all cellular
proteins (8). Biomarkers are
measurable indicators of biological or pathological processes, and
are valuable tools for monitoring, diagnosing and predicting
treatment responses for diseases (9). Physiological and pathological changes
accompanying skeletal muscular atrophy can be biomarkers. Atrophy
occurs when the rate of proteolysis exceeds the rate of protein
synthesis. This atrophy of skeletal muscle is closely related to
high-fat and high-sugar consumption, which are commonly associated
with Western diets, as well as aging and long-term illnesses, such
as diabetes, obesity, heart failure, Alzheimer's disease and
cachexia (10). Understanding why
muscular atrophy occurs under various conditions is essential for
its prevention and treatment.
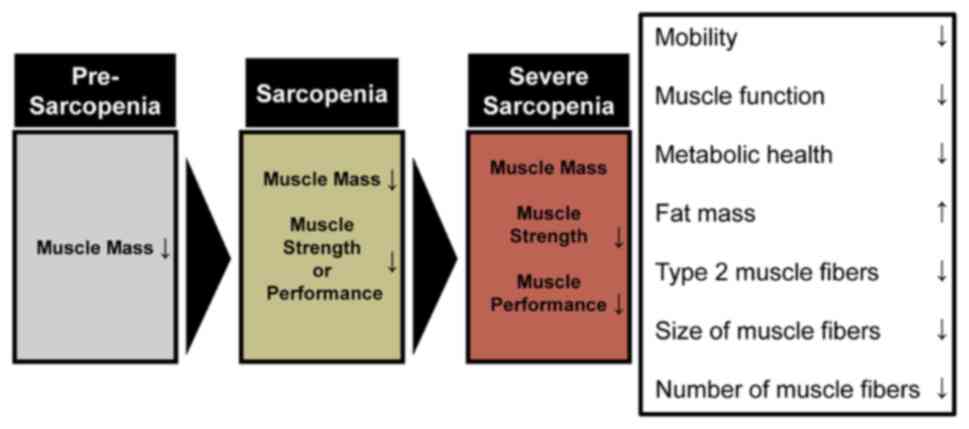
Sarcopenia is the most representative disease
associated with symptoms of muscular atrophy. In 2010, the European
Working Group on Sarcopenia in Older People (EWGSOP) developed
three diagnostic criteria for sarcopenia: Changes in muscle mass,
muscle strength and physical performance (11). A diagnosis of sarcopenia requires
both low muscle mass (LMM), and either low muscle strength (LMS) or
low physical performance (LPP) (12). LMM is identified by a SMM index of
<8.90 kg/m2; LMS is identified by a hand-grip
strength of <30 kg in men and <20 kg in women; LPP is
identified by a gait speed of ≤0.8 m/sec (13). According to the EWGSOP, sarcopenia
can be categorized into three subgroups: Pre-sarcopenia, sarcopenia
and severe sarcopenia, based on LMM status and the presence or
absence of functional impairment (LMS and LPP) (11).
In 2018, a new consensus was reached in terms of
both the definition and diagnosis of sarcopenia. The EWGSOP2
criteria were defined based on sarcopenia research conducted after
2010. The operational definitions of pre-sarcopenia, sarcopenia and
severe sarcopenia are generally defined by LMS, LMM and LPP,
respectively (14). Several
sarcopenia tests and cut-offs have been suggested. LMS can be
tested by measuring grip strength and the ability to stand from a
seated position (15).
Appendicular SMM (ASM) is measured to evaluate muscle mass and
quality, and LPP is assessed by measuring walking speed,
determining the Short Physical Performance Battery (SPPB) score,
and conducting timed up-and-go and 400-m walk tests. Pre-sarcopenia
is diagnosed when rising from a chair takes >15 sec in five
trials, or grip strength is <27 kg for men or <16 kg for
women (14). Sarcopenia is
diagnosed when the ASM is <20 kg for men or <15 kg for women,
or when the ASM/height2 is <7.0 kg/m2 for
men or <5.5 kg/m2 for women. Severe sarcopenia is
diagnosed when the walking speed is ≤0.8 m/sec, the SPPB score is
≤8 points, the timed up-and-go test requires ≥20 sec, and the 400-m
walk test is either not completed or requires ≥6 min for completion
(16). Muscle mass can be
estimated via dual-energy X-ray absorption or bioelectrical
impedance analysis, and the results can be modified based on height
or body mass index (17). Muscle
quality can be evaluated via computed tomography or magnetic
resonance imaging; both provide comprehensive data on the SMM, ASM,
third lumbar muscle cross-sectional area and middle thigh
cross-sectional area (14,18). In addition, EWGSOP2 recommends the
use of the self-reporting SARC-F questionnaire, which is a
screening test for sarcopenia consisting of five questions; SARC-F
predicts LMS with low-to-moderate sensitivity but very high
specificity and can therefore detect even the most severe cases
(14).
Muscle cell differentiation serves a crucial role in
muscular atrophy. After exercise or damage, skeletal muscle can
effectively regenerate through the actions of versatile satellite
cells (33). Upon activation,
satellite cells transition from a state of dormancy to engage in
proliferation and differentiation, ultimately transforming into
myoblasts, which further differentiate and merge to form
multinucleated myotubes (34).
This intricate myogenic process is carefully regulated by a complex
network of genes. At the core of this network are basic
helix-loop-helix transcription factors known as myogenic regulatory
factors (MRFs), which include myogenic factor 5, myoblast
determination protein 1, myogenin and MRF4 (35,36).
miRNAs, which do not code for proteins, are vital components of
this network, targeting specific mRNAs to finely adjust gene
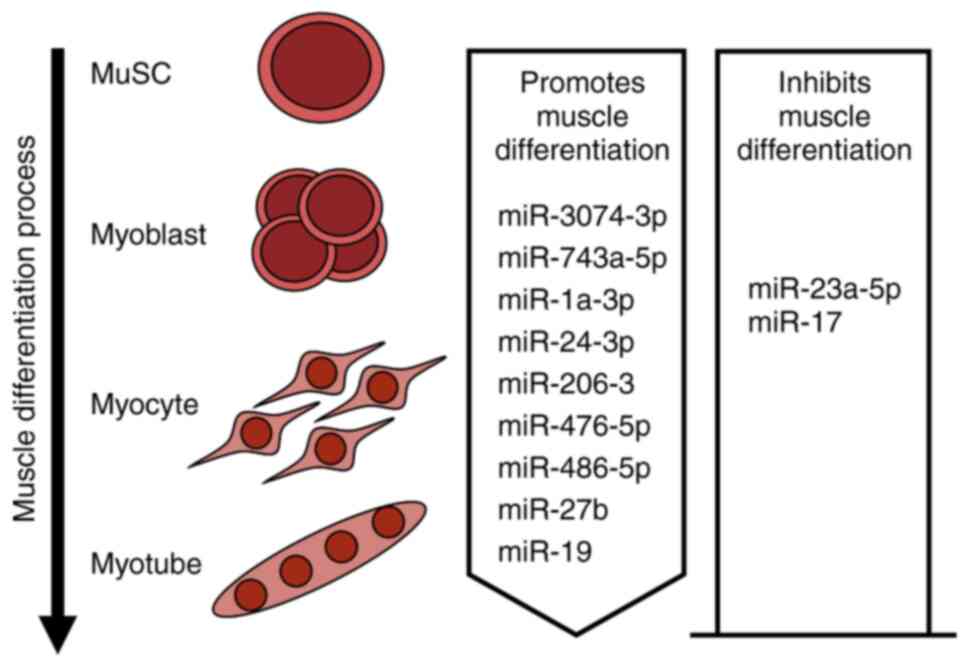
expression (37–40). Various miRNAs regulate muscle
differentiation by targeting genes involved in muscle cell
differentiation (Fig. 3).
Skeletal muscle differentiation involves various
miRNAs, including miR-24, miR-3074, miR-743a, miR-1a/206, miR-486,
miR-23a, miR-27, miR-19 and miR-17. In neonatal mice, miR-24-3p
leads to increased numbers of actively dividing PAX7-positive
muscle stem cells, which is attributable to decreased inhibition of
muscle differentiation and regeneration by Hmga1 and Id3 (41). A conserved miRNA termed miR-3074-3p
is involved in the regulation of Cav1 expression in both C2C12
cells and human skeletal muscle myoblasts, which ultimately
enhances myogenesis (42). The
modulation of miR-743a-5p activity is key to the regulation of
myoblast differentiation, and is achieved by targeting Mob1b,
another key player in skeletal muscle development and regeneration.
Notably, elevated levels of miR-743a actively promote the
differentiation of C2C12 myoblasts (43). miR-1a-3p, miR-206-3p, miR-24-3p and
miR-486-5p act as regulators of myoblast differentiation that
repress MRTF-A synthesis via the MRTF-A 3′-untranslated region
(UTR). Upregulation of these miRNAs during myogenesis inhibits the
translation of MRTF-A, allowing progression to late stages of
differentiation. The inhibitory effect of MRTF-A on muscle
differentiation is reduced upon the binding of miR-1a-3p, miR-24-3p
and miR-486-5p to the MRTF-A 3′-UTR (44). miR-23a-5p enhances the
proliferation of C2C12 myoblasts while simultaneously inhibiting
their differentiation, thereby influencing muscle fiber composition
(45). Myogenic differentiation is
facilitated by the downregulation of PAX3 protein levels under the
control of miR-27b, which targets the 3′-UTR of Pax3 mRNA. This
process ensures the strong, rapid initiation of differentiation
(46). miR-17 influences cell
proliferation to some extent by directly affecting Ccnd2 and Jak1
and reduces cell motility and cell fusion by targeting Rhoc.
Notably, treatment of C2C12 myoblast cells with miR-19 has been
shown to counteract the harmful effects of miR-17 and to aid
myotube development (47).
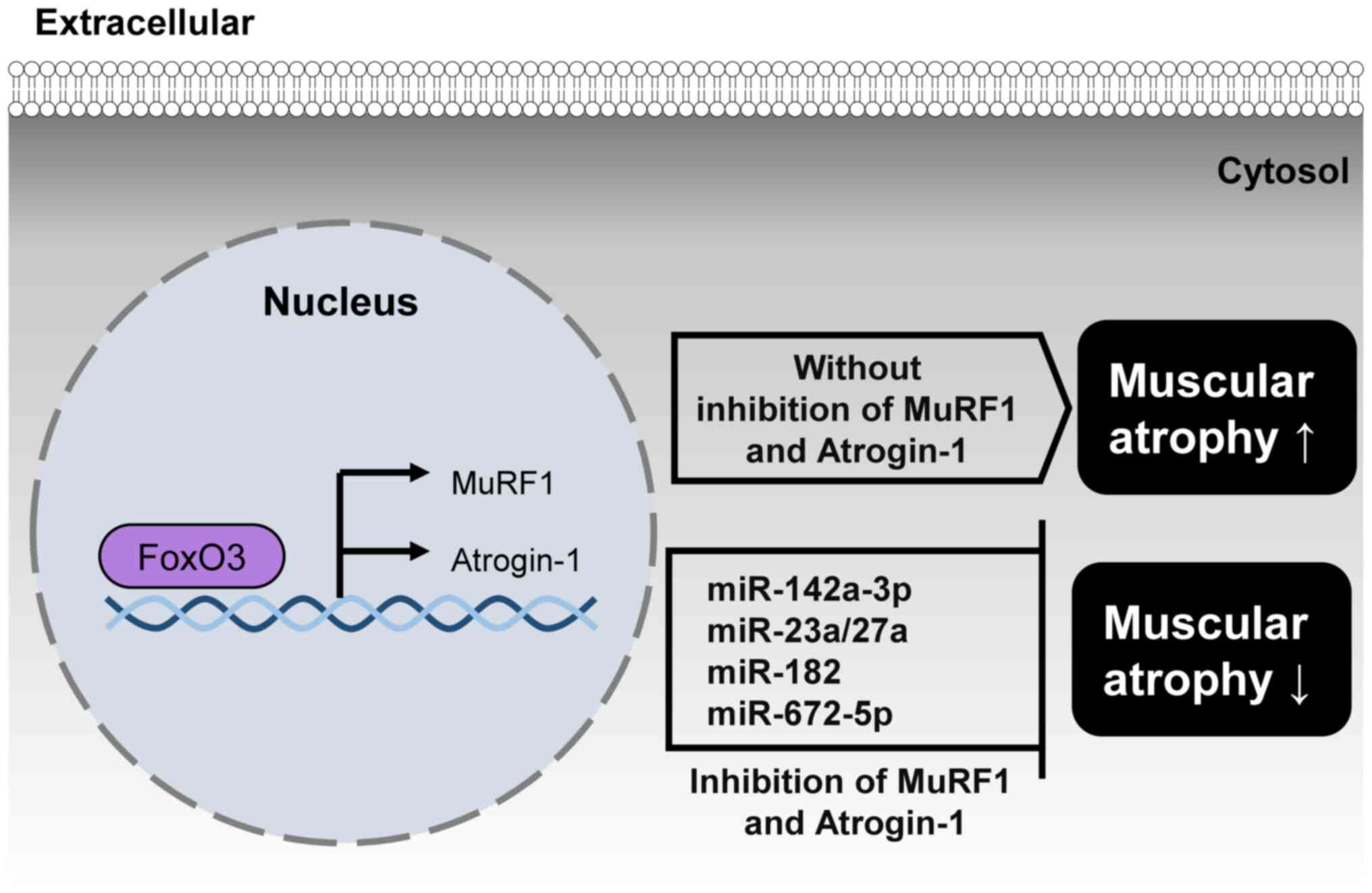
Muscular atrophy, or the wasting of muscle tissue,
is primarily caused by abnormal protein degradation (48). The ubiquitin-proteasome pathway
serves a crucial role in this process, through the identification
and breakdown of poly-ubiquitinated proteins (49). The key players in protein
ubiquitination are E3 ligases, with muscle RING finger 1 (MuRF1)
and Atrogin-1 being specific to muscle tissue (50). In patients with muscular atrophy,
both MuRF1 and Atrogin-1 are overexpressed; inhibition of the
activity of these proteins effectively prevents muscle loss and
mitigates the effects of muscular atrophy (51,52).
FOXO1, Atrogin1, and MuRF1 regulate muscular atrophy by promoting
the breakdown of muscle proteins through the ubiquitin-proteasome
pathway. Several miRNAs regulate muscular atrophy by targeting
FoxO1, MuRF1, and Atrogin-1, which induce muscle protein
degradation. (Fig. 4).
A previous study indicated that suppression of
miR-142a-3p decreases the expression levels of Atrogin-1, MuRF1 and
Nedd4, potentially hindering activation of the ubiquitin-proteasome
system and other pathways associated with muscular atrophy
(53). In skeletal muscle,
miR-23a/27a reduces atrophy by lowering the levels of the E3
ubiquitin ligases TRIM63/MuRF1 and FBXO32/Atrogin-1, which
contribute to muscle wasting (54). When miR-182 is introduced into
C2C12 myotubes treated with dexamethasone, it interacts
specifically with the 3′-UTR of FoxO3, decreasing the expression of
various genes controlled by FoxO3, such as Atrogin-1 (7). Additionally, miR-672-5p treatment
reduces ovariectomy-induced increased expression by targeting
Atrogin-1 and MuRF1 (55).
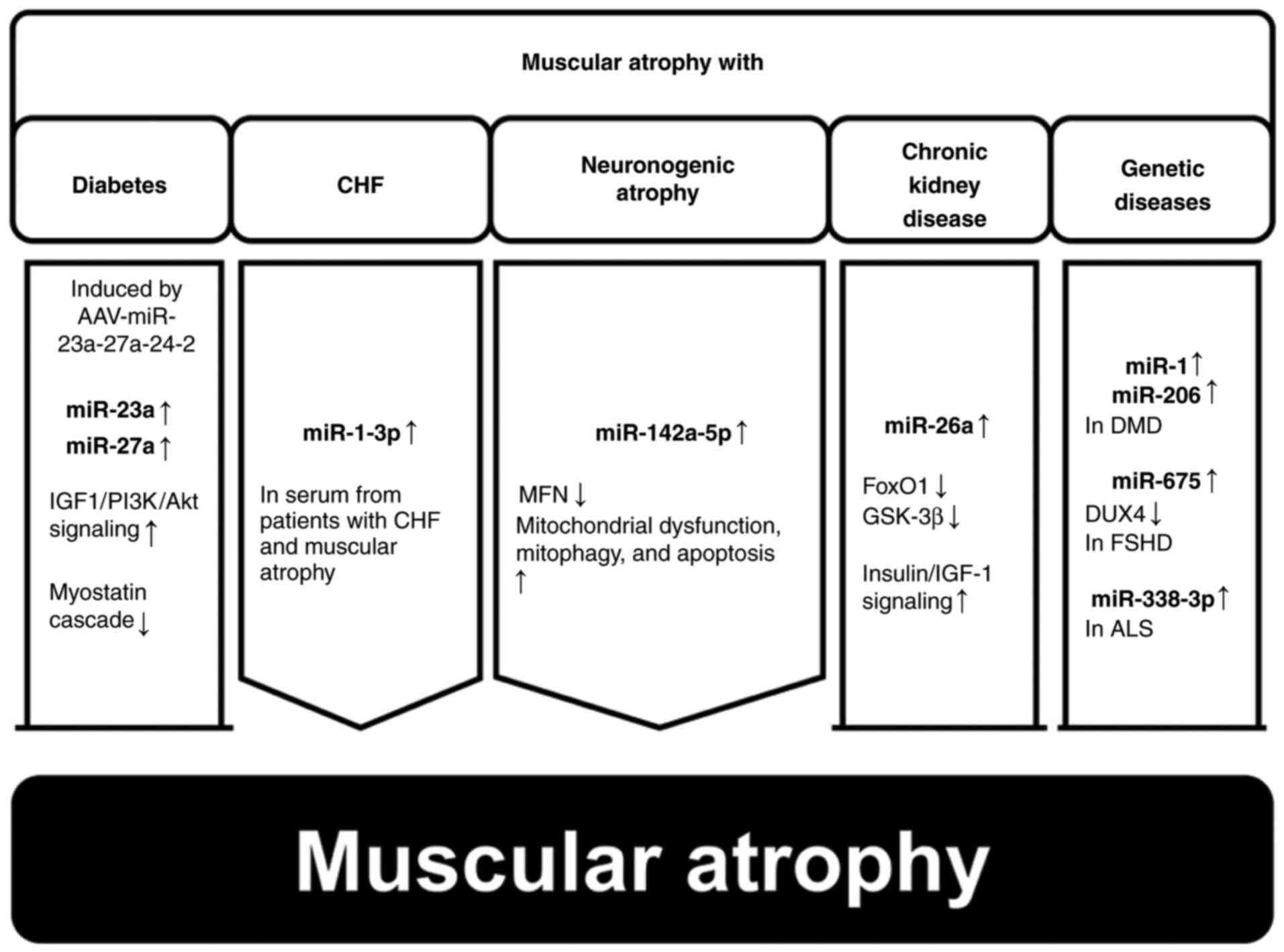
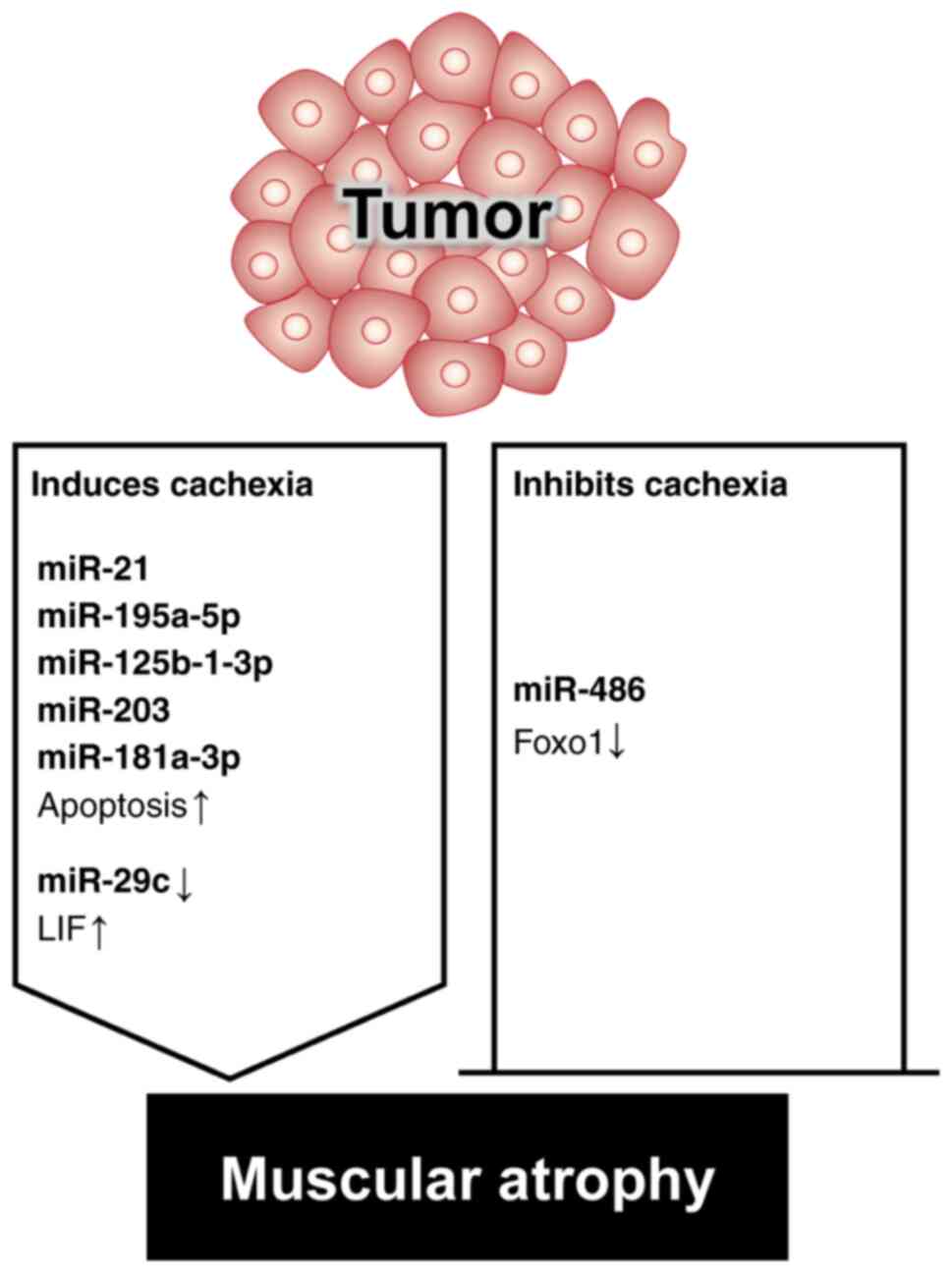
Cachexia is a medical condition associated with the
unintended loss of muscle and fat tissue in individuals with cancer
or chronic inflammatory diseases; this disease significantly
compromises patient outcomes and increases mortality. However, few
established interventions or treatments are available for cachexia
(56). The pathogenesis of cancer
cachexia is marked by an imbalance between protein and energy
levels, caused by various factors, including decreased metabolism
and diminished appetite (57–61).
Various miRNAs are involved in cachexia-induced muscular atrophy
(Fig. 5).
The degradation of muscle protein in patients with
cachexia is usually facilitated by the ubiquitin-proteasome system
and is initiated via E3 ligase activation (62). The inhibition of FoxO
transcriptional activity has been reported to curb muscle fiber
atrophy in patients with cachexia (63). miR-486 inhibits E3 ubiquitin ligase
activity by reducing FoxO1 protein expression and increasing FoxO1
phosphorylation (64). However,
miR-21 binds to and triggers the action of Toll-like receptor 7,
resulting in muscle cell apoptosis via the c-Jun N-terminal kinase
pathway, ultimately resulting in atrophy (65). Muscle catabolism in patients with
lung cancer is attributable to the activation of the leukemia
inhibitory factor (LIF) via the downregulation of miR-29c
expression. LIF has been shown to promote muscle wasting via the
mitogen-activated protein kinase and JAK/signal transducers and
activators of transcription pathways (66). Tumor-released exosomal miRNAs, such
as miR-195a-5p and miR-125b-1-3p, target Bcl-2 and induce muscle
wasting by reversing Bcl-2-mediated inhibition of cell death in
patients with colon cancer and cachexia (67). Elevated serum miR-203 levels in
patients with colorectal cancer have also been reported to act as
independent risk factor for sarcopenia; miR-203 induces apoptosis
through the downregulation of survivin in human skeletal muscle
cells (68). Notably, miR-181a-3p
in oral squamous cell carcinoma exosomes regulates the endoplasmic
reticulum stress pathway, triggering muscular atrophy and muscle
cell apoptosis (69). Several
studies have reported that the miR-181a family targets the 3′-UTR
of Grp78, reducing its expression and increasing the susceptibility
of cancer cells and muscle cells to apoptosis (70–73).
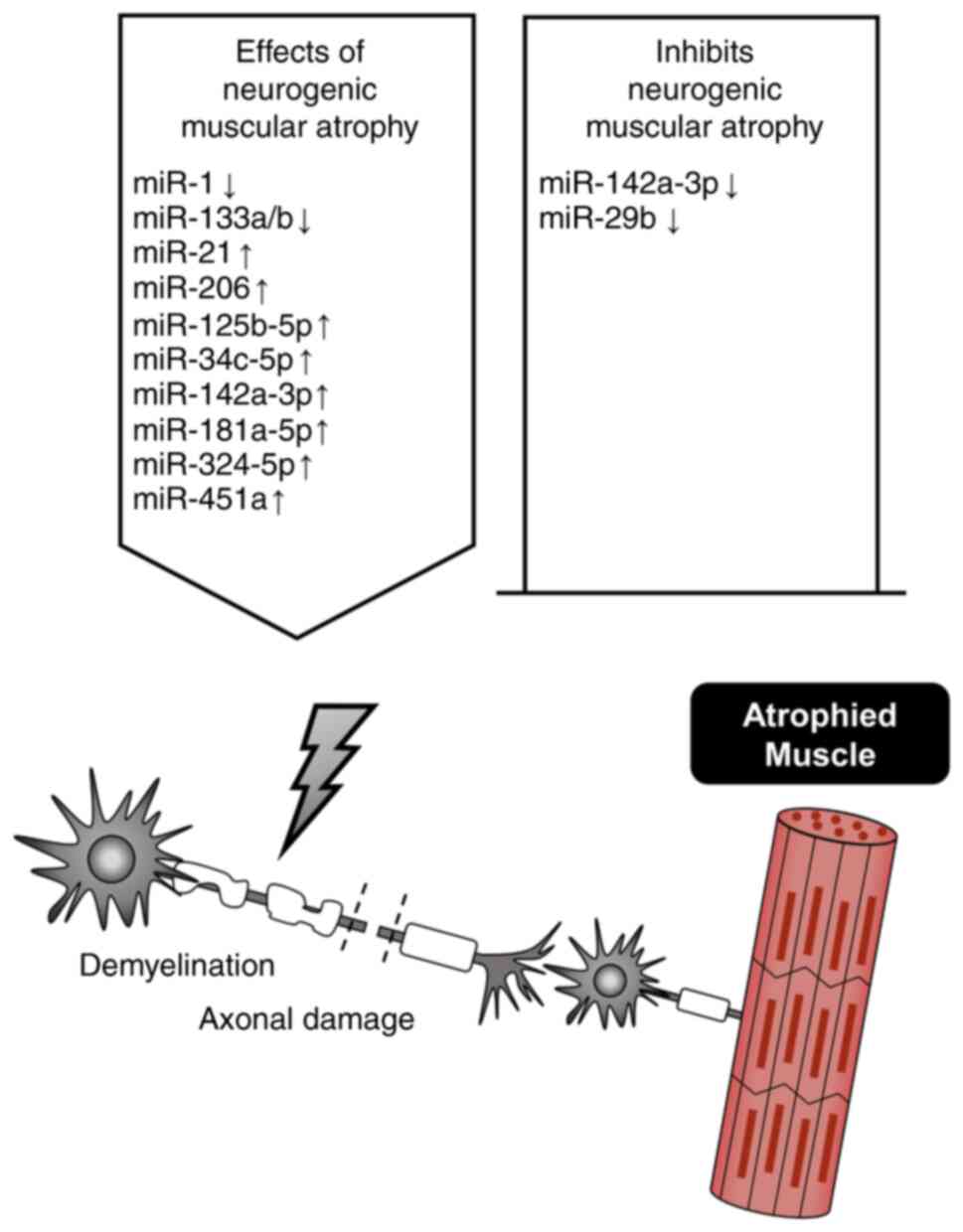
The denervation of skeletal muscle causes severe
muscular atrophy preceded by several cellular changes that increase
the permeability of the plasma membrane, decrease resting membrane
potential and accelerate protein breakdown (74). The nerves of skeletal muscles have
crucial roles in maintaining physiological muscle tone and function
(75–77). Denervation is followed by
significant muscular atrophy and weakness, accompanied by various
cellular alterations (78–80) that disrupt the ionic balance
(81–83) and accelerate protein catabolism
(84–86). Neurogenic muscular atrophy may
regulate miRNA levels, whereas the downregulation of miRNAs may
alleviate neurogenic muscular atrophy (Fig. 6).
miR-142a-3p has been reported to exhibit the most
significant differential expression among miRNAs in mouse skeletal
muscle after denervation (87).
This miRNA is considered to be a key modulator of cell fate in the
hematopoietic system (88). The
knockdown of miR-142a-3p alleviates decreases in body weight,
muscle strength and muscle fiber cross-sectional area caused by
nerve injury, and increases the number of mesenchymal stem cells,
as well as the expression levels of genes related to proliferation
and differentiation that are susceptible to Mef2a-mediated
inhibition. Additionally, nerve regeneration in areas of nerve
damage has been detected (53). In
another study, denervation was shown to trigger a significant
increase in miR-206 levels, and a decrease in the expression levels
of miR-1, miR-133a and miR-133b in muscle fiber-derived exosomes
(79). Furthermore, miR-206
overexpression attenuates denervation-induced skeletal muscular
atrophy via the inhibition of TGF-β1 and HDAC4 signaling, and the
promotion of satellite cell differentiation (89). CRISPR/Cas9-mediated editing of
miR-29b prevents denervation-induced muscular atrophy via
PKB-FoxO3A-mTOR signaling pathway activation and inhibits
angiotensin II-induced myocyte apoptosis in mice, increasing their
exercise capacity (90). Notably,
an exploration of the impact of denervation on muscle tissue miRNA
expression showed that denervation triggers significant changes in
the miRNA expression profile; specifically, miR-21 and miR-206
levels were revealed to be markedly increased after 3, 7 and 14
days. Srivastava et al (91) reported that miR-125b-5p is elevated
under similar conditions and could potentially act as a therapeutic
target in patients with denervated muscular atrophy. Both
miR-34c-5p and miR-142a-3p were significantly upregulated in
denervated skeletal muscle (92).
Abiusi et al (93) showed
that three miRNAs (miR-181a-5p, miR-324-5p and miR-451a) were
overexpressed in skeletal muscle and serum samples from patients
with spinal muscular atrophy.
Recent research on muscle miRNAs has significantly
advanced our understanding of the intricate regulatory mechanisms
governing muscle development, maintenance and disease. Explorations
of the roles played by miRNAs in muscle biology have yielded
valuable insights into the post-transcriptional control of gene
expression, highlighting the pivotal roles played by these small
RNA molecules in orchestrating various cellular processes. Despite
significant progress, a number of challenges and knowledge gaps
remain; notably, the specific mechanisms by which miRNAs exert
their effects on target genes in muscle cells remain unclear.
Further research is therefore considered necessary. Additionally,
the context-dependent functions of miRNAs and their interactions
with other non-coding RNAs remain crucial areas of investigation.
The integration of omics technologies, such as genomics,
transcriptomics and proteomics, will likely enhance our
comprehension of the regulatory networks underlying muscle biology.
The future of research on miRNA-mediated regulation of muscular
atrophy offers notable possibilities but poses significant
challenges. It is essential to improve miRNA-based therapeutic
strategies for clinical application. To effectively translate
research findings into treatments for muscle-wasting conditions, it
is essential to explore the efficacy, safety and specificity of
miRNA mimics or inhibitors in preclinical models and clinical
trials. The development of personalized medicine approaches is also
very promising. An understanding of how various miRNA levels vary
among individuals, and tailoring of interventions based on these
differences, will improve treatment efficacy and minimize potential
side effects. Furthermore, miRNAs may be useful as valuable
biomarkers for the early detection and monitoring of muscular
atrophy. The establishment of miRNA signatures associated with
different stages of muscular atrophy may aid timely intervention
and improve patient outcomes.
Not applicable.
The present study was financially supported by National Research
Foundation of Korea grants funded by the Korea Government (Ministry
of Education, Science and Technology; grant nos.
NRF-2021R1A2C1008492 and NRF-2020R1F1A1049801).
Not applicable.
WJ, UJ, SG, HN and JP contributed to the conception
and design of the study. WJ wrote the first draft of the
manuscript. WJ, UJ, SG, HN, QH, SL and JP wrote sections of the
manuscript. WJ, UJ, SG, HN, SL and BL searched the relevant
literature. SoHK, SeHK and JP critically reviewed the manuscript.
Data authentication is not applicable. All authors contributed to
manuscript revision and read and approved the final version of the
manuscript.
Not applicable.
Not applicable.
The authors declare that they have no competing
interests.
|
1
|
Duan K, Gao X and Zhu D: The clinical
relevance and mechanism of skeletal muscle wasting. Clin Nutr.
40:27–37. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Larsson L, Degens H, Li M, Salviati L, Lee
YI, Thompson W, Kirkland JL and Sandri M: Sarcopenia: Aging-Related
loss of muscle mass and function. Physiol Rev. 99:427–511. 2019.
View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Damluji AA, Alfaraidhy M, AlHajri N,
Rohant NN, Kumar M, Al Malouf C, Bahrainy S, Ji Kwak M, Batchelor
WB, Forman DE, et al: Sarcopenia and cardiovascular diseases.
Circulation. 147:1534–1553. 2023. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Nishio H, Niba ETE, Saito T, Okamoto K,
Takeshima Y and Awano H: Spinal muscular atrophy: The past,
present, and future of diagnosis and treatment. Int J Mol Sci.
24:119392023. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
O'Brien J, Hayder H, Zayed Y and Peng C:
Overview of MicroRNA biogenesis, mechanisms of actions, and
circulation. Front Endocrinol (Lausanne). 9:4022018. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Gebert LFR and MacRae IJ: Regulation of
microRNA function in animals. Nat Rev Mol Cell Biol. 20:21–37.
2019. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Brzeszczynska J, Brzeszczynski F, Hamilton
DF, McGregor R and Simpson AHRW: Role of microRNA in muscle
regeneration and diseases related to muscle dysfunction in atrophy,
cachexia, osteoporosis, and osteoarthritis. Bone Joint Res.
9:798–807. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
De Paepe B: Progressive skeletal muscle
atrophy in muscular dystrophies: A role for toll-like
receptor-signaling in disease pathogenesis. Int J Mol Sci.
21:44402020. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Vo TT, Kong G, Kim C, Juang U, Gwon S,
Jung W, Nguyen H, Kim SH and Park J: Exploring scavenger receptor
class F member 2 and the importance of scavenger receptor family in
prediagnostic diseases. Toxicol Res. 39:341–353. 2023. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Jun L, Robinson M, Geetha T, Broderick TL
and Babu JR: Prevalence and mechanisms of skeletal muscle atrophy
in metabolic conditions. Int J Mol Sci. 24:29732023. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Cruz-Jentoft AJ, Baeyens JP, Bauer JM,
Boirie Y, Cederholm T, Landi F, Martin FC, Michel JP, Rolland Y,
Schneider SM, et al: Sarcopenia: European consensus on definition
and diagnosis: Report of the European working group on sarcopenia
in older people. Age Ageing. 39:412–423. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Cho MR, Lee S and Song SK: A review of
sarcopenia pathophysiology, diagnosis, treatment and future
direction. J Korean Med Sci. 37:e1462022. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Jang JY, Kim D and Kim ND: Pathogenesis,
intervention, and current status of drug development for
sarcopenia: A review. Biomedicines. 11:16352023. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Cruz-Jentoft AJ, Bahat G, Bauer J, Boirie
Y, Bruyere O, Cederholm T, Cooper C, Landi F, Rolland Y, Sayer AA,
et al: Sarcopenia: Revised European consensus on definition and
diagnosis. Age Ageing. 48:16–31. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Guttikonda D and Smith AL: Sarcopenia
assessment techniques. Clin Liver Dis (Hoboken). 18:189–192. 2021.
View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Koo BK: Assessment of muscle quantity,
quality and function. J Obes Metab Syndr. 31:9–16. 2022. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Cheng KY, Chow SK, Hung VW, Wong CH, Wong
RM, Tsang CS, Kwok T and Cheung WH: Diagnosis of sarcopenia by
evaluating skeletal muscle mass by adjusted bioimpedance analysis
validated with dual-energy X-ray absorptiometry. J Cachexia
Sarcopenia Muscle. 12:2163–2173. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Faron A, Sprinkart AM, Kuetting DLR,
Feisst A, Isaak A, Endler C, Chang J, Nowak S, Block W, Thomas D,
et al: Body composition analysis using CT and MRI: intra-individual
intermodal comparison of muscle mass and myosteatosis. Sci Rep.
10:117652020. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Dufour AB, Hannan MT, Murabito JM, Kiel DP
and McLean RR: Sarcopenia definitions considering body size and fat
mass are associated with mobility limitations: The Framingham
Study. J Gerontol A Biol Sci Med Sci. 68:168–174. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Singh H, Kim D, Kim E, Bemben MG, Anderson
M, Seo DI and Bemben DA: Jump test performance and sarcopenia
status in men and women, 55 to 75 years of age. J Geriatr Phys
Ther. 37:76–82. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Lee J, Hong YP, Shin HJ and Lee W:
Associations of sarcopenia and sarcopenic obesity with metabolic
syndrome considering both muscle mass and muscle strength. J Prev
Med Public Health. 49:35–44. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Hunter GR, McCarthy JP and Bamman MM:
Effects of resistance training on older adults. Sports Med.
34:329–348. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Hepple RT: Sarcopenia-a critical
perspective. Sci Aging Knowledge Environ. 2003:pe312003. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Hunter GR, Singh H, Carter SJ, Bryan DR
and Fisher G: Sarcopenia and its implications for metabolic health.
J Obes. 2019:80317052019. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Zhang A, Li M, Wang B, Klein JD, Price SR
and Wang XH: miRNA-23a/27a attenuates muscle atrophy and renal
fibrosis through muscle-kidney crosstalk. J Cachexia Sarcopenia
Muscle. 9:755–770. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Xu R, Cui S, Chen L, Chen XC, Ma LL, Yang
HN and Wen FM: Circulating miRNA-1-3p as biomarker of accelerated
sarcopenia in patients diagnosed with chronic heart failure. Rev
Invest Clin. 74:276–268. 2022.PubMed/NCBI
|
|
27
|
Yang X, Xue P, Chen H, Yuan M, Kang Y,
Duscher D, Machens HG and Chen Z: Denervation drives skeletal
muscle atrophy and induces mitochondrial dysfunction, mitophagy and
apoptosis via miR-142a-5p/MFN1 axis. Theranostics. 10:1415–1432.
2020. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Wang B, Zhang A, Wang H, Klein JD, Tan L,
Wang ZM, Du J, Naqvi N, Liu BC and Wang XH: miR-26a limits muscle
wasting and cardiac fibrosis through exosome-mediated microRNA
transfer in chronic kidney disease. Theranostics. 9:1864–1877.
2019. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Oikawa S, Yuan S, Kato Y and Akimoto T:
Skeletal muscle-enriched miRNAs are highly unstable in vivo and may
be regulated in a Dicer-independent manner. FEBS J. 290:5692–5703.
2023. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Meng Q, Zhang J, Zhong J, Zeng D and Lan
D: Novel miRNA biomarkers for patients with duchenne muscular
dystrophy. Front Neurol. 13:9217852022. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Saad NY, Al-Kharsan M, Garwick-Coppens SE,
Chermahini GA, Harper MA, Palo A, Boudreau RL and Harper SQ: Human
miRNA miR-675 inhibits DUX4 expression and may be exploited as a
potential treatment for Facioscapulohumeral muscular dystrophy. Nat
Commun. 12:71282021. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
De Felice B, Annunziata A, Fiorentino G,
Borra M, Biffali E, Coppola C, Cotrufo R, Brettschneider J,
Giordana ML, Dalmay T, et al: miR-338-3p is over-expressed in
blood, CFS, serum and spinal cord from sporadic amyotrophic lateral
sclerosis patients. Neurogenetics. 15:243–253. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Dumont NA, Wang YX and Rudnicki MA:
Intrinsic and extrinsic mechanisms regulating satellite cell
function. Development. 142:1572–1581. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Feige P, Brun CE, Ritso M and Rudnicki MA:
Orienting muscle stem cells for regeneration in homeostasis, aging,
and disease. Cell Stem Cell. 23:653–664. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Yafe A, Shklover J, Weisman-Shomer P,
Bengal E and Fry M: Differential binding of quadruplex structures
of muscle-specific genes regulatory sequences by MyoD, MRF4 and
myogenin. Nucleic Acids Res. 36:3916–3925. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Gunther S, Kim J, Kostin S, Lepper C, Fan
CM and Braun T: Myf5-positive satellite cells contribute to
Pax7-dependent long-term maintenance of adult muscle stem cells.
Cell Stem Cell. 13:590–601. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Chen JF, Mandel EM, Thomson JM, Wu Q,
Callis TE, Hammond SM, Conlon FL and Wang DZ: The role of
microRNA-1 and microRNA-133 in skeletal muscle proliferation and
differentiation. Nat Genet. 38:228–233. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Lee SW, Yang J, Kim SY, Jeong HK, Lee J,
Kim WJ, Lee EJ and Kim HS: MicroRNA-26a induced by hypoxia targets
HDAC6 in myogenic differentiation of embryonic stem cells. Nucleic
Acids Res. 43:2057–2073. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Wu R, Li H, Zhai L, Zou X, Meng J, Zhong
R, Li C, Wang H, Zhang Y and Zhu D: MicroRNA-431 accelerates muscle
regeneration and ameliorates muscular dystrophy by targeting Pax7
in mice. Nat Commun. 6:77132015. View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Ma G, Wang Y, Li Y, Cui L, Zhao Y, Zhao B
and Li K: MiR-206, a key modulator of skeletal muscle development
and disease. Int J Biol Sci. 11:345–352. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Dey P, Soyer MA and Dey BK: MicroRNA-24-3p
promotes skeletal muscle differentiation and regeneration by
regulating HMGA1. Cell Mol Life Sci. 79:1702022. View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Lee B, Shin YJ, Lee SM, Son YH, Yang YR
and Lee KP: miR-3074-3p promotes myoblast differentiation by
targeting Cav1. BMB Rep. 53:278–283. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
43
|
Zhang Y, Yao Y, Wang Z, Lu D, Zhang Y,
Adetula AA, Liu S, Zhu M, Yang Y, Fan X, et al: MiR-743a-5p
regulates differentiation of myoblast by targeting Mob1b in
skeletal muscle development and regeneration. Genes Dis.
9:1038–1048. 2022. View Article : Google Scholar : PubMed/NCBI
|
|
44
|
Holstein I, Singh AK, Pohl F, Misiak D,
Braun J, Leitner L, Huttelmaier S and Posern G:
Post-transcriptional regulation of MRTF-A by miRNAs during myogenic
differentiation of myoblasts. Nucleic Acids Res. 48:8927–8942.
2020. View Article : Google Scholar : PubMed/NCBI
|
|
45
|
Zhao X, Gu H, Wang L, Zhang P, Du J, Shen
L, Jiang D, Wang J, Li X, Zhang S, et al: MicroRNA-23a-5p mediates
the proliferation and differentiation of C2C12 myoblasts. Mol Med
Rep. 22:3705–3714. 2020.PubMed/NCBI
|
|
46
|
Crist CG, Montarras D, Pallafacchina G,
Rocancourt D, Cumano A, Conway SJ and Buckingham M: Muscle stem
cell behavior is modified by microRNA-27 regulation of Pax3
expression. Proc Natl Acad Sci USA. 106:13383–13387. 2009.
View Article : Google Scholar : PubMed/NCBI
|
|
47
|
Kong D, He M, Yang L, Zhou R, Yan YQ,
Liang Y and Teng CB: MiR-17 and miR-19 cooperatively promote
skeletal muscle cell differentiation. Cell Mol Life Sci.
76:5041–5054. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
48
|
Attaix D, Combaret L, Bechet D and
Taillandier D: Role of the ubiquitin-proteasome pathway in muscle
atrophy in cachexia. Curr Opin Support Palliat Care. 2:262–266.
2008. View Article : Google Scholar : PubMed/NCBI
|
|
49
|
Hartmann-Petersen R and Gordon C: Proteins
interacting with the 26S proteasome. Cell Mol Life Sci.
61:1589–1595. 2004.PubMed/NCBI
|
|
50
|
Bodine SC, Latres E, Baumhueter S, Lai VK,
Nunez L, Clarke BA, Poueymirou WT, Panaro FJ, Na E, Dharmarajan K,
et al: Identification of ubiquitin ligases required for skeletal
muscle atrophy. Science. 294:1704–1708. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
51
|
Eddins MJ, Marblestone JG, Suresh Kumar
KG, Leach CA, Sterner DE, Mattern MR and Nicholson B: Targeting the
ubiquitin E3 ligase MuRF1 to inhibit muscle atrophy. Cell Biochem
Biophys. 60:113–118. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
52
|
Clavel S, Coldefy AS, Kurkdjian E, Salles
J, Margaritis I and Derijard B: Atrophy-related ubiquitin ligases,
atrogin-1 and MuRF1 are up-regulated in aged rat Tibialis Anterior
muscle. Mech Ageing Dev. 127:794–801. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
53
|
Gu X, Wang S, Li D, Jin B, Qi Z, Deng J,
Huang C and Yin X: MicroRNA-142a-3p regulates neurogenic skeletal
muscle atrophy by targeting Mef2a. Mol Ther Nucleic Acids.
33:191–204. 2023. View Article : Google Scholar : PubMed/NCBI
|
|
54
|
Xhuti D, Nilsson MI, Manta K, Tarnopolsky
MA and Nederveen JP: Circulating exosome-like vesicle and skeletal
muscle microRNAs are altered with age and resistance training. J
Physiol. 601:5051–5073. 2023. View Article : Google Scholar : PubMed/NCBI
|
|
55
|
Ahmad N, Kushwaha P, Karvande A, Tripathi
AK, Kothari P, Adhikary S, Khedgikar V, Mishra VK and Trivedi R:
MicroRNA-672-5p identified during weaning reverses osteopenia and
sarcopenia in ovariectomized mice. Mol Ther Nucleic Acids.
14:536–549. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
56
|
Webster JM, Kempen LJAP, Hardy RS and
Langen RCJ: Inflammation and skeletal muscle wasting during
cachexia. Front Physiol. 11:5976752020. View Article : Google Scholar : PubMed/NCBI
|
|
57
|
Emery PW, Edwards RH, Rennie MJ, Souhami
RL and Halliday D: Protein synthesis in muscle measured in vivo in
cachectic patients with cancer. Br Med J (Clin Res Ed).
289:584–586. 1984. View Article : Google Scholar : PubMed/NCBI
|
|
58
|
Warnold I, Lundholm K and Schersten T:
Energy balance and body composition in cancer patients. Cancer Res.
38:1801–1807. 1978.PubMed/NCBI
|
|
59
|
Chang VT, Xia Q and Kasimis B: The
functional assessment of anorexia/cachexia therapy (FAACT) Appetite
Scale in veteran cancer patients. J Support Oncol. 3:377–382.
2005.PubMed/NCBI
|
|
60
|
Martin L, Birdsell L, Macdonald N, Reiman
T, Clandinin MT, McCargar LJ, Murphy R, Ghosh S, Sawyer MB and
Baracos VE: Cancer cachexia in the age of obesity: Skeletal muscle
depletion is a powerful prognostic factor, independent of body mass
index. J Clin Oncol. 31:1539–1547. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
61
|
Yang W, Huang J, Wu H, Wang Y, Du Z, Ling
Y, Wang W, Wu Q and Gao W: Molecular mechanisms of cancer
cachexia-induced muscle atrophy (Review). Mol Med Rep.
22:4967–4980. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
62
|
Bilodeau PA, Coyne ES and Wing SS: The
ubiquitin proteasome system in atrophying skeletal muscle: Roles
and regulation. Am J Physiol Cell Physiol. 311:C392–C403. 2016.
View Article : Google Scholar : PubMed/NCBI
|
|
63
|
Reed SA, Sandesara PB, Senf SM and Judge
AR: Inhibition of FoxO transcriptional activity prevents muscle
fiber atrophy during cachexia and induces hypertrophy. FASEB J.
26:987–1000. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
64
|
Xu J, Li R, Workeneh B, Dong Y, Wang X and
Hu Z: Transcription factor FoxO1, the dominant mediator of muscle
wasting in chronic kidney disease, is inhibited by microRNA-486.
Kidney Int. 82:401–411. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
65
|
He WA, Calore F, Londhe P, Canella A,
Guttridge DC and Croce CM: Microvesicles containing miRNAs promote
muscle cell death in cancer cachexia via TLR7. Proc Natl Acad Sci
USA. 111:4525–4529. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
66
|
Xie K, Xiong H, Xiao W, Xiong Z, Hu W, Ye
J, Xu N, Shi J, Yuan C, Chen Z, et al: Downregulation of miR-29c
promotes muscle wasting by modulating the activity of leukemia
inhibitory factor in lung cancer cachexia. Cancer Cell Int.
21:6272021. View Article : Google Scholar : PubMed/NCBI
|
|
67
|
Miao C, Zhang W, Feng F, Gu X, Shen Q, Lu
S, Fan M, Li Y, Guo X, Ma Y, et al: Cancer-derived exosome miRNAs
induce skeletal muscle wasting by Bcl-2-mediated apoptosis in colon
cancer cachexia. Mol Ther Nucleic Acids. 24:923–938. 2021.
View Article : Google Scholar : PubMed/NCBI
|
|
68
|
Okugawa Y, Toiyama Y, Hur K, Yamamoto A,
Yin C, Ide S, Kitajima T, Fujikawa H, Yasuda H, Koike Y, et al:
Circulating miR-203 derived from metastatic tissues promotes
myopenia in colorectal cancer patients. J Cachexia Sarcopenia
Muscle. 10:536–548. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
69
|
Qiu L, Chen W, Wu C, Yuan Y and Li Y:
Exosomes of oral squamous cell carcinoma cells containing
miR-181a-3p induce muscle cell atrophy and apoptosis by
transmissible endoplasmic reticulum stress signaling. Biochem
Biophys Res Commun. 533:831–837. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
70
|
Su SF, Chang YW, Andreu-Vieyra C, Fang JY,
Yang Y, Han B, Lee AS and Liang G: miR-30d, miR-181a and
miR-199a-5p cooperatively suppress the endoplasmic reticulum
chaperone and signaling regulator GRP78 in cancer. Oncogene.
32:4694–4701. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
71
|
Liu J, Huang Y, Cai F, Dang Y, Liu C and
Wang J: MicroRNA-181a regulates endoplasmic reticulum stress in
offspring of mice following prenatal microcystin-LR exposure.
Chemosphere. 240:1249052020. View Article : Google Scholar : PubMed/NCBI
|
|
72
|
Wei Y, Tao X, Xu H, Chen Y, Zhu L, Tang G,
Li M, Jiang A, Shuai S, Ma J, et al: Role of miR-181a-5p and
endoplasmic reticulum stress in the regulation of myogenic
differentiation. Gene. 592:60–70. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
73
|
Zhang M, Zhang Q, Hu Y, Xu L, Jiang Y,
Zhang C, Ding L, Jiang R, Sun J, Sun H and Yan G: miR-181a
increases FoxO1 acetylation and promotes granulosa cell apoptosis
via SIRT1 downregulation. Cell Death Dis. 8:e30882017. View Article : Google Scholar : PubMed/NCBI
|
|
74
|
Cisterna BA, Vargas AA, Puebla C,
Fernandez P, Escamilla R, Lagos CF, Matus MF, Vilos C, Cea LA,
Barnafi E, et al: Active acetylcholine receptors prevent the
atrophy of skeletal muscles and favor reinnervation. Nat Commun.
11:10732020. View Article : Google Scholar : PubMed/NCBI
|
|
75
|
Burke RE: Sir Charles Sherrington's the
integrative action of the nervous system: A centenary appreciation.
Brain. 130:887–894. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
76
|
Dulhunty AF: Excitation-contraction
coupling from the 1950s into the new millennium. Clin Exp Pharmacol
Physiol. 33:763–772. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
77
|
Canfora I, Tarantino N and Pierno S:
Metabolic pathways and ion channels involved in skeletal muscle
atrophy: A starting point for potential therapeutic strategies.
Cells. 11:25662022. View Article : Google Scholar : PubMed/NCBI
|
|
78
|
Bruusgaard JC and Gundersen K: In vivo
time-lapse microscopy reveals no loss of murine myonuclei during
weeks of muscle atrophy. J Clin Invest. 118:1450–1457. 2008.
View Article : Google Scholar : PubMed/NCBI
|
|
79
|
De Gasperi R, Hamidi S, Harlow LM,
Ksiezak-Reding H, Bauman WA and Cardozo CP: Denervation-related
alterations and biological activity of miRNAs contained in exosomes
released by skeletal muscle fibers. Sci Rep. 7:128882017.
View Article : Google Scholar : PubMed/NCBI
|
|
80
|
Magnusson C, Svensson A, Christerson U and
Tagerud S: Denervation-induced alterations in gene expression in
mouse skeletal muscle. Eur J Neurosci. 21:577–580. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
81
|
Ehmsen JT and Hoke A: Cellular and
molecular features of neurogenic skeletal muscle atrophy. Exp
Neurol. 331:1133792020. View Article : Google Scholar : PubMed/NCBI
|
|
82
|
Daeschler SC, Feinberg K, Harhaus L,
Kneser U, Gordon T and Borschel GH: Advancing nerve regeneration:
Translational perspectives of tacrolimus (FK506). Int J Mol Sci.
24:127712023. View Article : Google Scholar : PubMed/NCBI
|
|
83
|
Zheng H, Liu X, Katsurada K and Patel KP:
Renal denervation improves sodium excretion in rats with chronic
heart failure: Effects on expression of renal ENaC and AQP2. Am J
Physiol Heart Circ Physiol. 317:H958–H968. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
84
|
Tokinoya K, Shirai T, Ota Y, Takemasa T
and Takekoshi K: Denervation-induced muscle atrophy suppression in
renalase-deficient mice via increased protein synthesis. Physiol
Rep. 8:e144752020. View Article : Google Scholar : PubMed/NCBI
|
|
85
|
Sandri M: Protein breakdown in muscle
wasting: Role of autophagy-lysosome and ubiquitin-proteasome. Int J
Biochem Cell Biol. 45:2121–2129. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
86
|
Bongers KS, Fox DK, Ebert SM, Kunkel SD,
Dyle MC, Bullard SA, Dierdorff JM and Adams CM: Skeletal muscle
denervation causes skeletal muscle atrophy through a pathway that
involves both Gadd45a and HDAC4. Am J Physiol Endocrinol Metab.
305:E907–E915. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
87
|
Weng J, Zhang P, Yin X and Jiang B: The
whole transcriptome involved in denervated muscle atrophy following
peripheral nerve injury. Front Mol Neurosci. 11:692018. View Article : Google Scholar : PubMed/NCBI
|
|
88
|
Nimmo R, Ciau-Uitz A, Ruiz-Herguido C,
Soneji S, Bigas A, Patient R and Enver T: MiR-142-3p controls the
specification of definitive hemangioblasts during ontogeny. Dev
Cell. 26:237–249. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
89
|
Huang QK, Qiao HY, Fu MH, Li G, Li WB,
Chen Z, Wei J and Liang BS: MiR-206 Attenuates Denervation-Induced
Skeletal Muscle Atrophy in Rats Through Regulation of Satellite
Cell Differentiation via TGF-beta1, Smad3, and HDAC4 Signaling. Med
Sci Monit. 22:1161–1170. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
90
|
Li J, Wang L, Hua X, Tang H, Chen R, Yang
T, Das S and Xiao J: CRISPR/Cas9-Mediated miR-29b editing as a
treatment of different types of muscle atrophy in mice. Mol Ther.
28:1359–1372. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
91
|
Srivastava S, Rathor R, Singh SN and
Suryakumar G: Emerging role of MyomiRs as biomarkers and
therapeutic targets in skeletal muscle diseases. Am J Physiol Cell
Physiol. 321:C859–C875. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
92
|
Gu XY, Jin B, Qi ZD and Yin XF: MicroRNA
is a potential target for therapies to improve the physiological
function of skeletal muscle after trauma. Neural Regen Res.
17:1617–1622. 2022. View Article : Google Scholar : PubMed/NCBI
|
|
93
|
Abiusi E, Infante P, Cagnoli C, Lospinoso
Severini L, Pane M, Coratti G, Pera MC, D'Amico A, Diano F, Novelli
A, et al: SMA-miRs (miR-181a-5p, −324-5p, and −451a) are
overexpressed in spinal muscular atrophy skeletal muscle and serum
samples. Elife. 10:e680542021. View Article : Google Scholar : PubMed/NCBI
|