Introduction
Alzheimer's disease (AD) is a complex neurodegenerative disorder that primarily affects elderly patients, presenting as a form of dementia (1). At present, ~55 million individuals worldwide suffer from AD (2). As the average life expectancy of the modern population increases and the population ages, the global number of patients with AD is expected to increase to 152 million by 2050, leading to a dementia epidemic (2). Notably, the development of effective treatments for AD has been slow to progress, with a failure rate of 99.6% (3). This failure rate may be due to the development of drugs that only target a single pathological manifestation of AD, such as β-amyloid deposition or Tau hyperphosphorylation (3,4). However, AD is a multifactorial disease with unclear underlying mechanisms, leading to limitations in the development of traditional single-target drugs (5). Therefore, further investigations into multi-target drugs are required for the development of a novel treatment approach.
Natural compounds extracted from plants, such as cineole, ginsenosides and resveratrol, may exhibit beneficial neuroprotective effects (6). Betulin (BT) is a lupine-type pentacyclic triterpenoid compound, and was one of the first natural substances to be isolated from plants (7). BT is mainly found in Betulaceae, with 54–82% located in the bark (8). BT exerts numerous pharmacological effects in diseases, exhibiting anti-inflammatory, antioxidant, antibacterial, antitumor and anti-HIV properties (9). Research has focused on the therapeutic potential of BT in neurodegenerative diseases due to its ability to cross the blood-brain barrier (10). A previous study demonstrated that BT reduced α-synuclein accumulation in neurons, and alleviated 6-hydroxydopamine-induced neuronal apoptosis and dopaminergic neuronal degeneration, thereby prolonging the lifespan of C. elegans, a Parkinson's disease model (11). In addition, the spatial learning and memory of streptozotocin (STZ)-induced diabetic rats were improved following treatment with BT (12). BT also alleviated STZ-induced oxidative stress and inflammation in the hippocampus by increasing the expression of heme oxygenase-1 (HO-1) and nuclear factor erythroid 2-related factor 2 (Nrf2), and inhibiting the activity of NF-κB (12). In BV-2 cells, BT was found to decrease lipopolysaccharide (LPS)-induced neuroinflammation through inhibition of inducible nitric oxide synthase, c-Jun N-terminal kinase and NF-κB activation (13). Collectively, these studies indicated that BT might exhibit potential in the treatment of neurodegenerative diseases. However, studies that focus on the effects of BT in AD are limited, and thus, the efficacy and mechanism of BT in the treatment of AD need to be further investigated.
Network pharmacology is a ‘multi-target, multi-effect’ research model that integrates high-throughput, network visualization and network analysis technology to predict the association between drugs and targets. Network pharmacology is also used to systematically explore the mechanism of action of drugs in the treatment of diseases (14). The present study aimed to determine the potential targets and molecular mechanisms of BT in the treatment of AD using network pharmacology and molecular docking technology. Furthermore, an AD cell model was established and treated with BT, and the effects of BT on the predicted signaling pathways and AD-related proteins were verified using cell viability, reactive oxygen species (ROS) probe and western blotting assays to explore the neuroprotective effects of BT and its molecular mechanisms. The study workflow is shown as a graphical summary in Fig. 1.
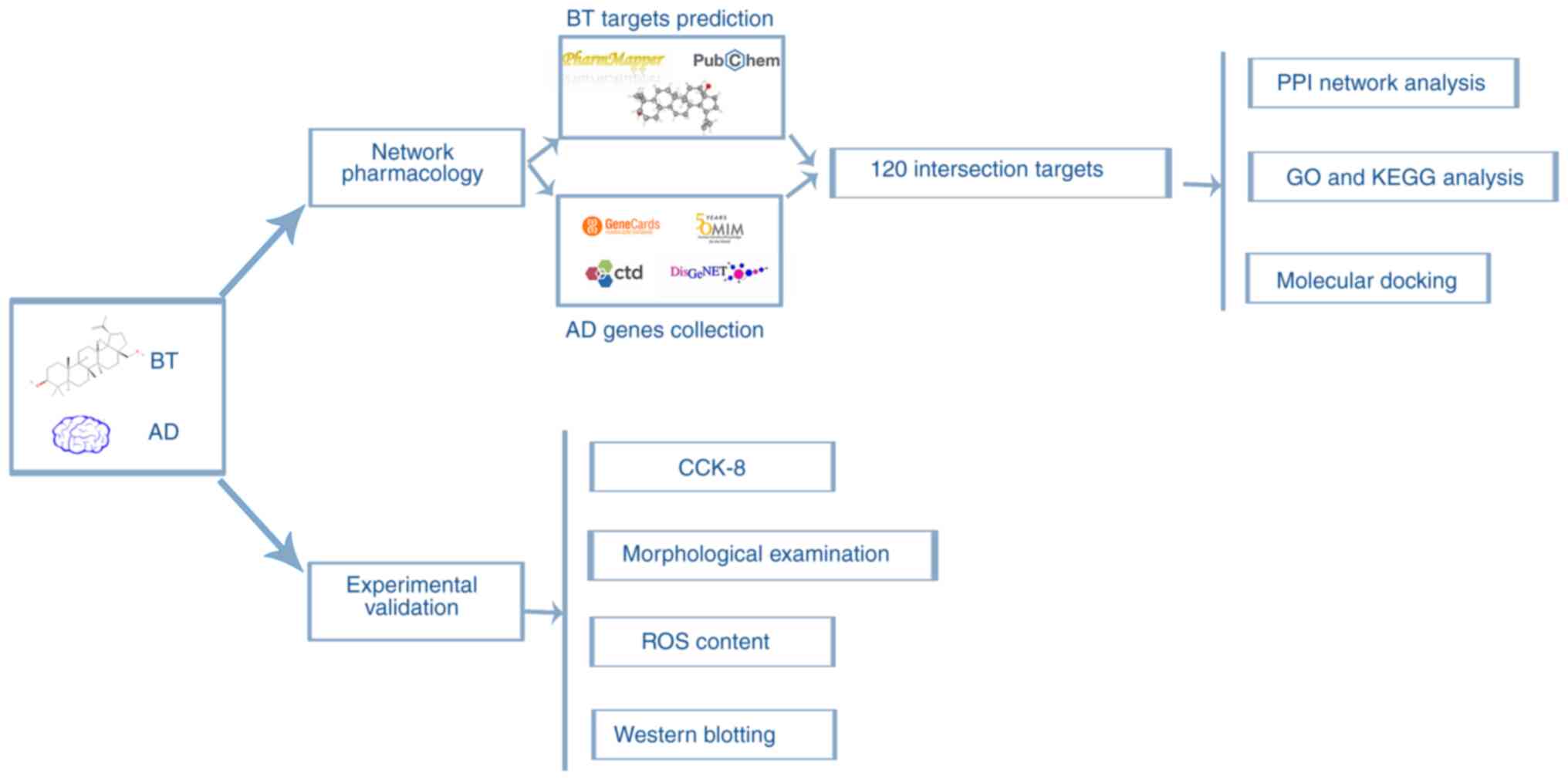 |
Figure 1.
Study concept and workflow. BT, betulin; AD, Alzheimer's disease; CCK-8, Cell Counting Kit-8; GO, Gene Oncology; KEGG, Kyoto Encyclopedia of Genes and Genomes; PPI, protein-protein interaction; ROS, reactive oxygen species.
|
Materials and methods
Acquisition and screening of drug-related target genes
The structural information of BT at the secondary and tertiary levels was acquired from the PubChem database (version; 1.8.0 beta; http://pubchem.ncbi.nlm.nih.gov/) (15), and 3D structural mapping of BT was submitted to PharmMapper (version 2017; http://lilab-ecust.cn/pharmmapper/) (16). The human-derived target protein library and pharmacophore model library were selected to fully cover the target protein, and the results obtained from the two databases were combined and de-duplicated to obtain the therapeutic target database of BT. Subsequently, the UniProt ID of BT targets was converted to a gene symbol using UniProt (version 2023_04; http://www.uniprot.org/) (17).
Acquisition of AD-related target genes
To look for AD targets, the search was conducted using the keyword ‘Alzheimer's disease’ in several databases, including the Human Gene Database and Analysis Tools (GeneCards; version 5.17; http://www.genecards.org/) (18), a database of gene-disease associations (DisGeNET; version 7.0; http://www.disgenet.org/) (19), Comparative Toxicology Database (CTD; version 17016; http://ctdbase.org/) (20) and Online Mendelian Inheritance in Man (OMIM; http://www.omim.org/) (21). The criteria for gene selection in each database were: GeneCards Inferred Functionality Score ≥50 in GeneCards, score ≥0.4 in DisGeNET and inference ≥50 in CTD. The gene selection criteria in the OMIM database were genes that exist on the gene map. The AD-related gene database was obtained by combining and de-duplicating the screened results from all databases.
Screening of drug-disease common targets and protein-protein interaction (PPI) network analysis
The intersection targets of BT and AD were determined using a Venn diagram drawn using jvenn (22) (http://www.bioinformatics.com.cn/static/others/jvenn/example.html). The PPI network was constructed using the Search Tool for the Retrieval of Interacting Genes/Proteins (STRING) database (version 12.0; http://string-db.org/) (23). Homo sapiens was selected and a minimum interaction score of 0.400 was applied. Afterwards, Cytoscape (version 3.7.2) software (24) was used for topological analysis and to draw the common targets PPI network diagram, in which nodes were sized and colored according to the degree value of the gene. The CytoHubba plugin (version 0.1) and Degree algorithm in Cytoscape were utilized to identify hub genes within the PPI network (25).
Enrichment analysis of Gene Ontology (GO) and Kyoto Encyclopedia of Genes and Genomes (KEGG) pathways
The Database for Annotation, Visualization and Integrated Discovery (DAVID; version 2021; http://david.ncifcrf.gov/home.jsp) (26) was used to input common targets of BT and AD. Briefly, the intersection targets were input into DAVID, with a focus on ‘Homo sapiens’ as the species and a choice of ‘Official Gene Symbols’ as the gene list. After submitting, the results of the GO and KEGG enrichment analysis were obtained. GO terms included biological process (BP), cellular component and molecular function terms. P<0.05 indicated a significant level of enrichment. Subsequently, SangerBox (version 1.1.3; http://vip.sangerbox.com/) (27) was used to generate bubble graphs containing the top 10 KEGG pathways and GO terms with the highest count.
Molecular docking of BT and 3D structure of hub genes
The Protein Data Bank (PDB) files of the 3D structure of hub genes were obtained from the Research Collaboration for Structural Bioinformatics PDB (RCSB PDB; version 3.48.0; http://www.rcsb.org/) (28) with the organism set as ‘Homo sapiens’. The mol2 file of BT was obtained from the Traditional Chinese Medicine Systems Pharmacology Database (TCMSP; version 2.3; http://old.tcmsp-e.com/tcmsp.php) (29). The small molecule 3D structures and protein 3D structures were imported into AutoDock Tools (version 1.5.6; The Scripps Research Institute) for the removal of water molecules and processing of hydrogen bonds, followed by molecular docking. Subsequently, the PyMOL software (version 2.5.4; Schrodinger, Inc.) was utilized to visualize the docking outcomes, while the reliability of the docking results was assessed using the binding energy and root mean square deviation (RMSD) (30). A binding energy <-1.2 kcal/mol (31) and RMSD <2 Å (32) indicate that the target protein can be bound by the compound in a spontaneous manner, and a lower binding energy indicates a stronger affinity between the ligand and the relevant target protein.
Reagents and antibodies
BT (cat. no. B35645) standards (≥98%) were purchased from Shanghai Acmec Biochemical Technology Co., Ltd. Formaldehyde (FA) aqueous solution (37% w/v; cell culture grade; F8775) was obtained from Sigma Aldrich; Merck KGaA. RIPA lysate buffer (cat. no. SB-BR040), the BCA protein assay kit (cat. no. SB-WB013) and dichlorodihydrofluorescein diacetate (DCFH-DA; ROS detection probe; cat. no. SB-R6033) were obtained from Share-bio (https://www.share-bio.com/). Special grade FBS (cat. no. C04001), DMEM/F12 (cat. no. 01-172-1ACS), penicillin-streptomycin mixture (100X; cat. no. 03-031-5B), trypsin solution (0.25%; cat. no. 03-050-1B) and phosphate buffer solution (cat. no. 02-024-1ACS) were obtained from Biological Industries; Sartorius AG. The Cell Counting Kit-8 (CCK-8; cat. no. PF00004) was purchased from Proteintech Group, Inc. Dimethyl sulfoxide (cell culture grade; cat. no. D8371) was obtained from Beijing Solarbio Science & Technology Co., Ltd. Goat anti-mouse IgG HRP-conjugated secondary antibodies (cat. no. SA00001-1) were obtained from Proteintech Group, Inc. Goat anti-rabbit IgG HRP-conjugated secondary antibodies (cat. no. SB-AB0035) and anti-β-actin (cat. no. SB-AB0101) were obtained from Share-bio (https://www.share-bio.com/). Anti-total (t-)GSK3β (cat. no. ab93926) was purchased from Abcam. Anti-phosphorylated (p-)GSK3β (Ser9; cat. no. D3A4) was obtained from Cell Signaling Technology, Inc. Anti-t-PI3K (cat. no. 251221), anti-p-PI3K (Tyr467/199; cat. no. 341468), anti-p-Tau (Thr181; cat. no. 310192), anti-t-Tau (cat. no. R25862) and anti-p-GSK3β (Tyr216; cat. no. R24515) were purchased from Chengdu Zen-Bioscience Co., Ltd. Anti-Bax (cat. no. T40051), anti-Bcl-2 (cat. no. T40056), anti-t-AKT (cat. no. T55516) and anti-p-AKT (Ser473; cat. no. T56569) were obtained from Abmart Pharmaceutical Technology, Co., Ltd. Unless stated otherwise, all analytical grade reagents were acquired from a local reagent supplier.
Cell culture
The mouse hippocampal neuronal cell line (HT22; cat. no. iCell-m020) was obtained from Cellverse Bioscience Technology Co., Ltd.,). HT22 cells were cultured in DMEM/F12 supplemented with 1% penicillin-streptomycin and 10% FBS in a humidified incubator with 5% CO2 at 37°C. When the cells grew to ~80–90% density, the cells were digested with trypsin solution and passaged.
Experimental design
HT22 cells were divided into the following groups: i) Control group (cells without any treatment); ii) FA group (FA applied at an appropriate concentration according to each experimental plan); and iii) FA + BT group (BT pre-, simultaneous- or post-treatment was performed according to each experimental plan).
Cytotoxicity assay
According to a previous study, an FA concentration of the median lethal dose (LD50; 0.5 mM) was employed to generate an AD-like cell model (33). Cell viability was determined using a CCK-8 assay according to the manufacturer's instructions. Briefly, HT22 cells in the logarithmic growth phase were digested from the dish by trypsin solution, and then were seeded at a density of 1×104 cells/well and cultured for 24 h at 37°C to ensure cell adhesion in a 96-well plate (Corning, Inc.). The medium was then substituted with FBS-free DMEM/F12 containing BT (0–100 µM), and cells were incubated with BT for 24 h at 37°C. To investigate whether BT could reduce FA-induced cytotoxicity, in 5% CO2 and at 37°C, cells were treated with BT (0.1–5 µM) for 2 h before and after exposure to FA (0.5 mM; LD50) for 4 h, as well as treated simultaneously with 0.5 mM FA for 4 h. Subsequently, the old medium was discarded and the cells were incubated with FBS-free DMEM/F12 containing CCK-8 for 2 h at 37°C. A spectrum plate reader (Bio-Rad Laboratories, Inc.) was used to measure the optical absorbance at 450 nm. Each group had at least four replicates and the experiment was repeated three times. Cell viability was presented as the percentage of viable cells compared with the control group.
To further observe cell morphology changes, HT22 cells in the logarithmic growth phase were digested by trypsin solution, and then were seeded in a 12-well plate at 1.5×105 cells/well and cultured for another 24 h at 37°C to ensure cell adhesion. Subsequently, at 37°C, cells were treated with BT (5 µM) for 2 h followed by FA (0.5 mM) exposure for 4 h. After 10 min of fixation with 4% paraformaldehyde at room temperature, cell morphology was observed using a vertical fluorescence microscopy imaging system (BXS3; Olympus Corporation). The experiment was repeated three times.
Determination of ROS content
For the detection of intracellular ROS content according to a previous study (33), HT22 cells in the logarithmic growth phase were digested from the dish by trypsin solution, and then were seeded at a density of 1.5×105 cells/well on cell slides in a 12-well plate and cultured for 24 h at 37°C for attachment. After which, cells were pre-treated with BT (5 µM) for 2 h before FA (0.5 mM) exposure for 4 h at 37°C. Subsequently, the cells were incubated with 10 µM DCFH-DA (a probe for detecting ROS green fluorescence) in the dark at 37°C for 30 min after discarding the medium. The green fluorescence intensity was measured using a vertical fluorescence microscopy imaging system (BXS3; Olympus Corporation) and analyzed using ImageJ software (version 1.49; National Institutes of Health). The assay was repeated three times with 10 cells/coverslip per replicate.
Western blotting
For western blotting, at 37°C, HT22 cells in the logarithmic growth phase were digested by trypsin solution, and then were seeded at 4×105 cells/well in a 6-well plate and were pre-treated with BT (5 µM) for 2 h before 4 h of FA (0.5 mM) at 37°C treatment, based on the literature protocols with slight modifications (33). Briefly, cells were incubated in RIPA buffer with protease inhibitors according to the manufacturer's instructions to prepare whole-cell lysates. A BCA protein assay kit was employed to determine the protein content. Protein samples (20 µg loaded/lane) were heated at 98°C for 10 min after mixing with 5X SDS-PAGE buffer. The mixture was then separated by 15% SDS-PAGE and transferred to polyvinylidene fluoride membranes (MilliporeSigma). After blocking with 5% skim milk for 1 h at room temperature, the membranes were incubated with primary antibodies, including β-actin (1:4,000), Tau (1:2,000), p-Tau (Thr181; 1:1,500), PI3K (1:1,500), p-PI3K (Tyr467; 1:1,500), AKT (1:2,000), p-AKT (Ser473; 1:2,000), GSK3β (1:2,000), p-GSK3β (Tyr216; 1:1,500), p-GSK3β (Ser9; 1:1,000), Bcl-2 (1:2,000) and Bax (1:2,000), at 37°C for 1.5 h (β-actin) or at 4°C overnight (all primary antibodies other than β-actin). The membranes were subsequently washed three times for 10 min in Tris-HCl buffered saline with 1% Tween-20 (TBST), followed by incubation with the corresponding anti-rabbit/anti-mouse IgG HRP-conjugated secondary antibodies (1:10,000) at room temperature for 1 h. The membranes were then washed three times with TBST for 10 min each. After which, the Luminol Chemiluminescent Kit (Share-bio; cat. no. SB-WB001) was added to the membrane and the optical density was measured and semi-quantified using an automatic chemiluminescence image analyzer (Shanghai Bio-Tech Co., Ltd.) and the ImageJ software package (version 1.49; National Institutes of Health), respectively. Each experiment was repeated three times, with β-actin used as an internal reference to determine the intensity of each band. Images of western blotting were processed with Adobe Photoshop CS6 (Adobe Systems Europe, Ltd.).
Statistical analysis
The statistical analysis in the present study was conducted using GraphPad Prism software (version 8.0.2; Dotmatics). Results were obtained from a minimum of three independent experiments and presented as the mean ± standard error of the mean. The normality of the data was determined using the Shapiro-Wilk test. All data were normally distributed (all P>0.05) and were analyzed using one-way ANOVA. Post hoc comparisons were performed using the Tukey test for the CCK-8 assay results and the Student-Newman-Keuls post hoc test for all other data. P<0.05 was considered to indicate a statistically significant difference.
Results
Common targets of BT in AD
In the present study, a two-dimensional chemical structure of BT was obtained from the PubChem database (Fig. 2A) and then submitted to PharmMapper. A total of 225 and 279 genes were obtained from the human target protein library and pharmacophore model library, respectively. Subsequently, and 495 therapeutic targets of BT were identified after combining the genes and removing duplicates. A total of 1,386 genes associated with AD were obtained from the OMIM, GeneCards, DisGeNET and CTD databases. Jveen was used to intersect BT therapeutic targets and AD-associated genes, leading to the identification of 120 potential therapeutic targets of BT in AD (Fig. 2B). Thus, the 120 intersection targets were submitted to the STRING website and medium confidence was selected to create a PPI network diagram. A tab separated values file was obtained from the STRING website and uploaded to Cytoscape for topology analysis. Notably, the network consisted of 120 nodes and 1,137 edges (Fig. 2C). In the PPI network diagram, nodes represent different proteins and the straight lines represent the interaction between proteins. The larger the node, the darker the color and the greater the degree value of the protein (Fig. 2C and D). The top 10 hub genes were screened according to degree values using the cytoHubba plugin in Cytoscape (Fig. 2D and E). The genes included albumin (ALB), EGFR, caspase 3 (CASP3), heat shock protein 90 α family class a member 1 (HSP90AA1), ESR1, SRC proto-oncogene, non-receptor tyrosine kinase (SRC), GSK3β, insulin-like growth factor-1 (IGF1), peroxisome proliferator-activated receptor γ (PPARG) and heat shock protein 90 α family class B member 1 (HSP90AB1) (Fig. 2E).
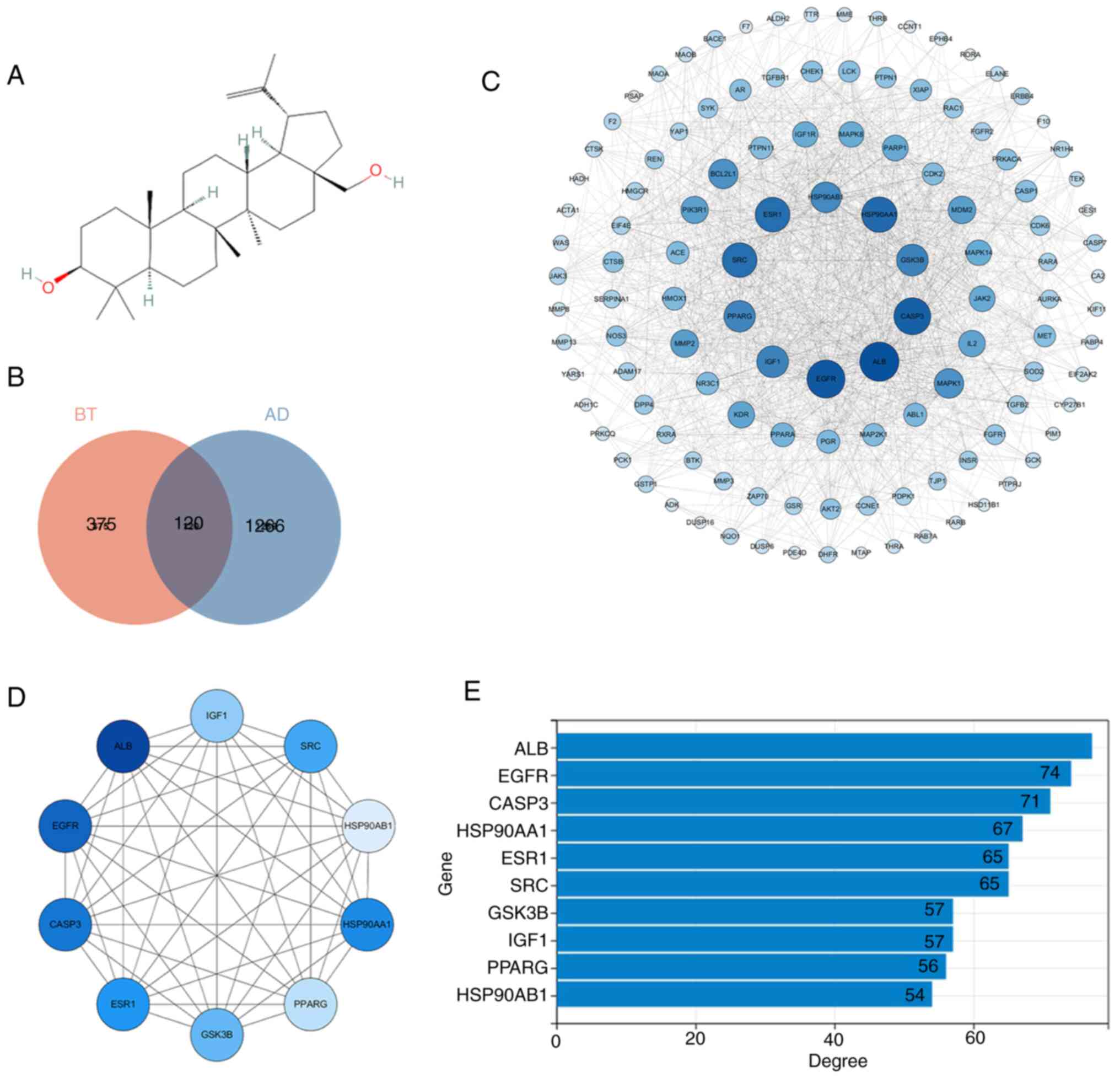 |
Figure 2.
PPI network map of the intersection targets and the top 10 hub genes. (A) Chemical structure of BT obtained from the PubChem database. Compound identifier, 72326. (B) Intersection targets of BT and AD. (C) PPI network diagram of intersection targets. Nodes represent proteins, and the larger and darker the node, the greater the degree value of the corresponding protein. (D) Top 10 hub genes according to the degree value. Nodes represent proteins, and the larger and darker the node, the greater the degree value of the corresponding protein. (E) Degree values of the 10 hub genes. AD, Alzheimer's disease; ALB, albumin; BT, betulin; CASP3, caspase 3; HSP90AA1, heat shock protein 90 α family class a member 1; HSP90AB1, heat shock protein 90 α family class B member 1; IGF1, insulin-like growth factor-1; PPARG, peroxisome proliferator-activated receptor γ; PPI, protein-protein interaction; SRC, non-receptor tyrosine kinase.
|
GO and KEGG enrichment analyses
To further clarify the potential biological functions of the 120 target genes, GO and KEGG enrichment analyses were performed. The results of the GO analysis indicated that the target genes were significantly enriched in cytosolic and nucleic functions, involving MFs such as ‘protein binding’ and ‘ATP binding’, and BPs such as ‘protein phosphorylation’ and ‘negative regulation of apoptotic process’ (Fig. 3A-C; Table I). The KEGG pathway enrichment analysis indicated that the genes were significantly enriched in signaling pathways such as ‘PI3K-Akt signaling pathway’, ‘MAPK signaling pathway’ and ‘Ras signaling pathway’ (Fig. 3D; Table II). Notably, the PI3K/AKT signaling pathway is considered to be upstream of GSK3β (34). Based on the results of the gene enrichment analysis, it was hypothesized that the mechanism of BT in the treatment of AD may be associated with changes in protein phosphorylation and negative regulation of cell apoptosis involved in the PI3K/AKT signaling pathway. Therefore, the PI3K/AKT signaling pathway and downstream proteins were further examined in vitro.
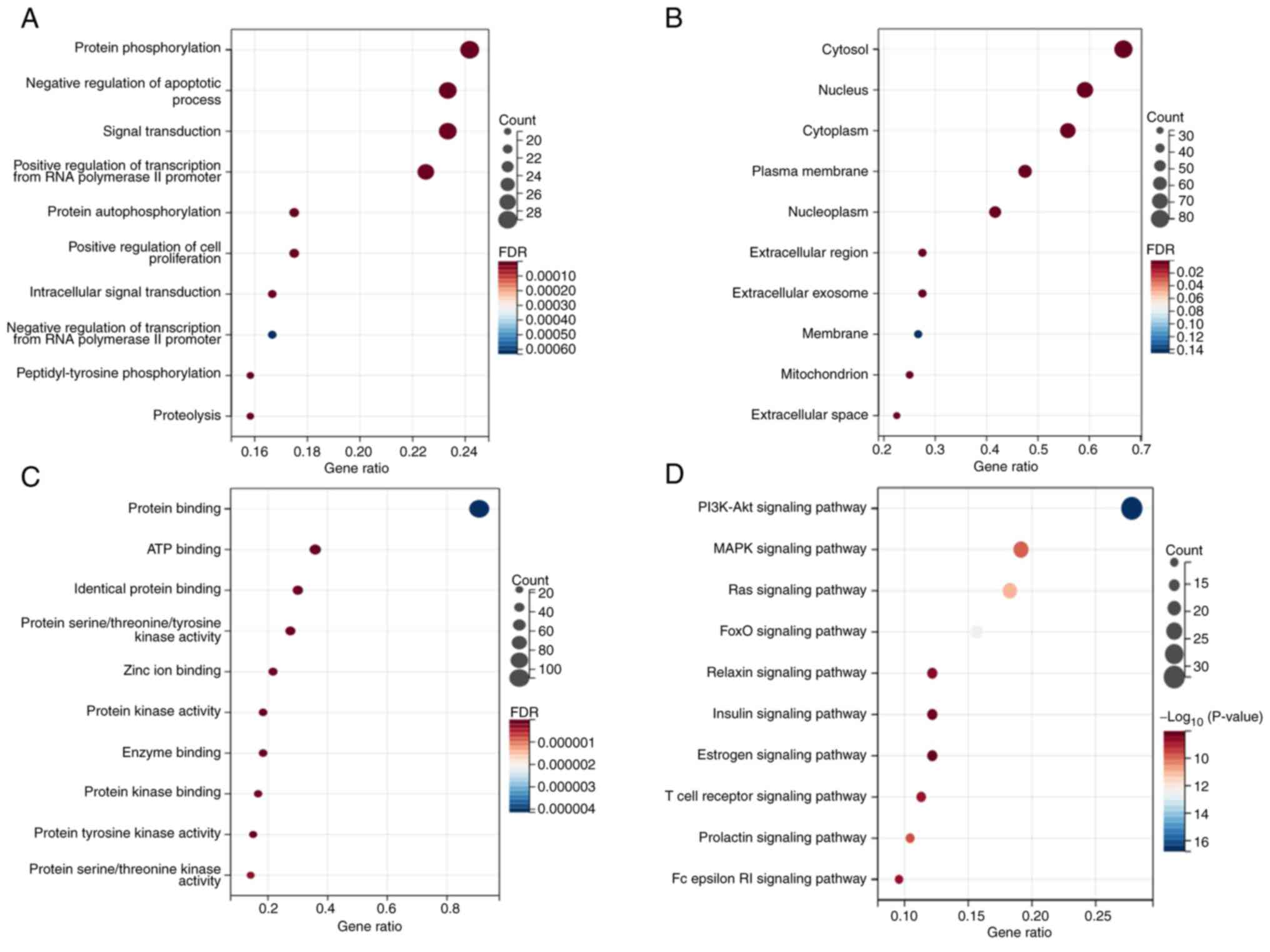 |
Figure 3.
Top 10 GO and KEGG analysis results. (A) Top 10 GO-biological process entries. (B) Top 10 GO-cellular component entries. (C) Top 10 GO-molecular function entries. (D) Top 10 KEGG signaling pathways. FDR, false discovery rate; GO, Gene Oncology; KEGG, Kyoto Encyclopedia of Genes and Genomes.
|
 |
Table I.
GO terms for the interactions of betulin and Alzheimer's disease.
|
Table I.
GO terms for the interactions of betulin and Alzheimer's disease.
| GO ID |
GO term |
Counts |
P-value |
| GOTERM_BP |
|
|
|
| GO:0006468 |
Protein phosphorylation |
29 |
4.21×10−19 |
| GO:0043066 |
Negative regulation of apoptotic process |
28 |
2.31×10−17 |
| GO:0007165 |
Signal transduction |
28 |
2.16×10−8 |
| GO:0045944 |
Positive regulation of transcription from RNA polymerase II promoter |
27 |
3.62×10−8 |
| GO:0046777 |
Protein autophosphorylation |
21 |
1.78×10−19 |
| GO:0008284 |
Positive regulation of cell proliferation |
21 |
2.41×10−10 |
| GO:0035556 |
Intracellular signal transduction |
20 |
7.60Ex10−11 |
| GO:0000122 |
Negative regulation of transcription from RNA polymerase II promoter |
20 |
1.56×10−5 |
| GO:0018108 |
Peptidyl-tyrosine phosphorylation |
19 |
8.66×10−19 |
| GO:0006508 |
Proteolysis |
19 |
1.83×10−10 |
| GOTERM_CC |
|
|
|
| GO:0005829 |
Cytosol |
80 |
6.04×10−18 |
| GO:0005634 |
Nucleus |
71 |
1.76×10−18 |
| GO:0005737 |
Cytoplasm |
67 |
9.71×10−10 |
| GO:0005886 |
Plasma membrane |
57 |
2.67×10−6 |
| GO:0005654 |
Nucleoplasm |
50 |
1.30×10−7 |
| GO:0005576 |
Extracellular region |
33 |
7.02×10−7 |
| GO:0070062 |
Extracellular exosome |
33 |
1.93×10−6 |
| GO:0016020 |
Membrane |
32 |
0.034881002 |
| GO:0005739 |
Mitochondrion |
30 |
5.95×10−9 |
| GO:0005615 |
Extracellular space |
27 |
8.16×10−5 |
| GOTERM_MF |
|
|
|
| GO:0005515 |
Protein binding |
109 |
1.71×10−7 |
| GO:0005524 |
ATP binding |
43 |
2.62×10−16 |
| GO:0042802 |
Identical protein binding |
36 |
5.15×10−10 |
| GO:0004712 |
Protein serine/threonine/tyrosine kinase activity |
33 |
1.08×10−24 |
| GO:0008270 |
Zinc ion binding |
26 |
3.58×10−10 |
| GO:0004672 |
Protein kinase activity |
22 |
5.41×10−14 |
| GO:0019899 |
Enzyme binding |
22 |
6.00×10−14 |
| GO:0019901 |
Protein kinase binding |
20 |
1.11×10−9 |
| GO:0004713 |
Protein tyrosine kinase activity |
18 |
7.63×10−19 |
| GO:0004674 |
Protein serine/threonine kinase activity |
17 |
7.24×10−9 |
 |
Table II.
Kyoto Encyclopedia of Genes and Genomes pathways for the interactions of betulin and Alzheimer's disease.
|
Table II.
Kyoto Encyclopedia of Genes and Genomes pathways for the interactions of betulin and Alzheimer's disease.
| ID |
Pathway |
Counts |
P-value |
| hsa04151 |
PI3K-Akt signaling pathway |
32 |
1.61×10−17 |
| hsa04010 |
MAPK signaling pathway |
22 |
1.68×10−10 |
| hsa04014 |
Ras signaling pathway |
21 |
1.20Ex10−11 |
| hsa04068 |
FoxO signaling pathway |
18 |
3.28×10−13 |
| hsa04926 |
Relaxin signaling pathway |
14 |
4.05×10−9 |
| hsa04910 |
Insulin signaling pathway |
14 |
8.93×10−9 |
| hsa04915 |
Estrogen signaling pathway |
14 |
9.82×10−9 |
| hsa04660 |
T cell receptor signaling pathway |
13 |
2.62×10−9 |
| hsa04917 |
Prolactin signaling pathway |
12 |
2.60×10−10 |
| hsa04664 |
Fc epsilon RI signaling pathway |
11 |
2.88×10−9 |
Molecular docking
Molecular docking simulates the binding process between a drug molecule and a protein molecule through the calculation of the interaction energy, thus providing predictions for the binding affinity, mode, position and stability of the drug-protein complex (35). The interactions and binding affinities between BT and hub genes were predicted using BT as a small molecule ligand to dock with the respective hub genes. The results of the molecular docking analysis demonstrated good binding affinity between BT and the hub genes, including ALB, EGFR, CASP3, HSP90AA1, ESR1, SRC, GSK3β, IGF1, PPARG and HSP90AB1 (Fig. 4). Specific binding energy values and the RMSD of all molecules are shown in Table III. The binding energies of BT to ten hub genes were all <-1.2 kcal/mol and the RMSD was >2 Å, which indicates that BT could regulate the physiological activity or function of these proteins by binding to them. Collectively, these results suggested that BT may exert neuroprotective effects through the aforementioned hub genes and signaling pathways that they are involved in.
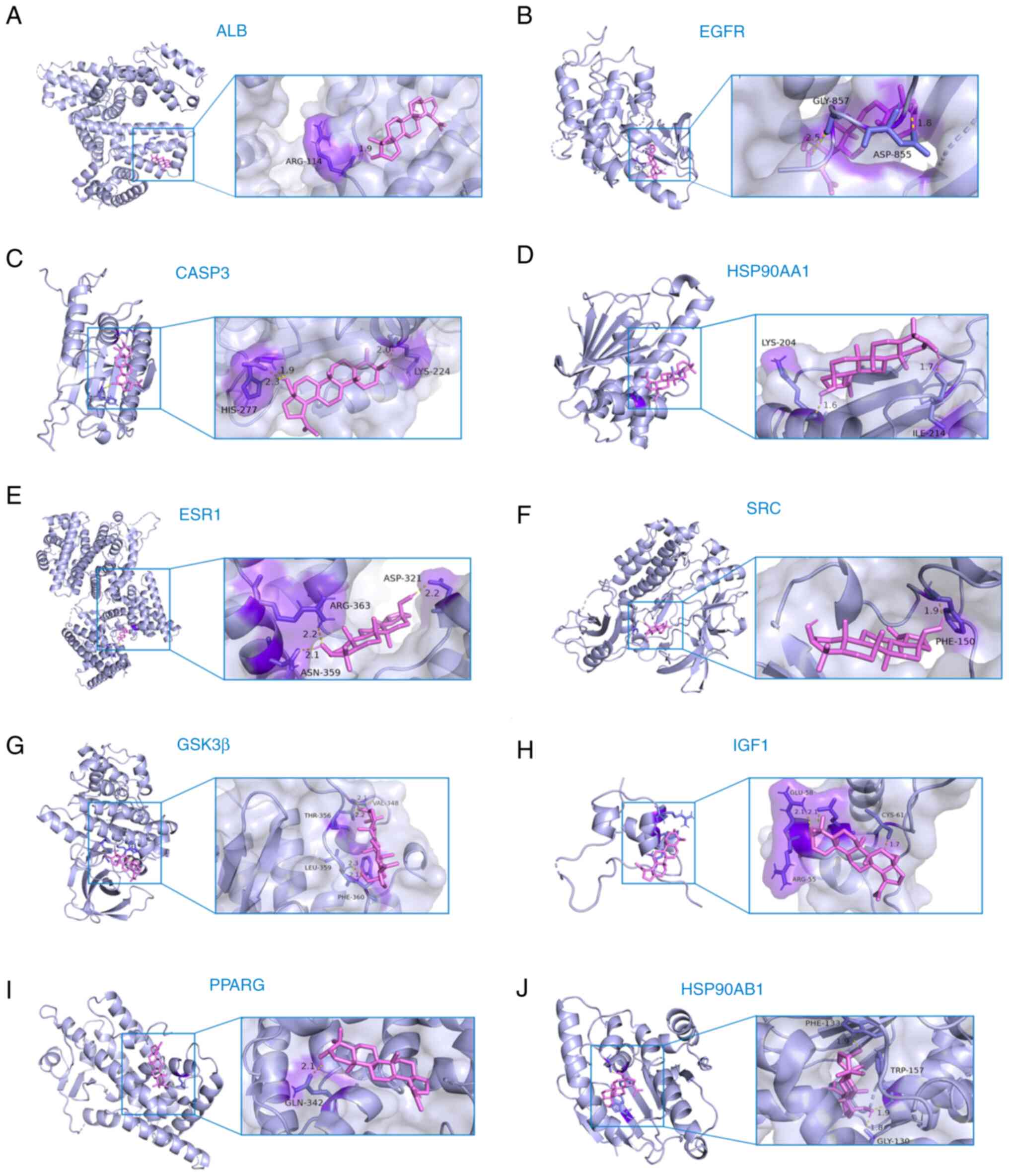 |
Figure 4.
Molecular docking results. (A) Molecular docking results of BT and ALB. (B) Molecular docking results of BT and EGFR. (C) Molecular docking results of BT and CASP3. (D) Molecular docking results of BT and HSP90AA1. (E) Molecular docking results of BT and ESR1. (F) Molecular docking results of BT and SRC. (G) Molecular docking results of BT and GSK3β. (H) Molecular docking results of BT and IGF1. (I) Molecular docking results of BT and PPARG. (J) Molecular docking results of BT and HHSP90AB1. ALB, albumin; BT, betulin; CASP3, caspase 3; HSP90AA1, heat shock protein 90 α family class a member 1; HSP90AB1, heat shock protein 90 α family class B member 1; IGF1, insulin-like growth factor-1; PPARG, peroxisome proliferator-activated receptor γ; SRC, non-receptor tyrosine kinase.
|
 |
Table III.
Molecular docking results of targets and betulin.
|
Table III.
Molecular docking results of targets and betulin.
| Target name |
PDB ID |
Binding energy, kJ/mol |
RMSD, Å |
| ALB |
6YG9 |
−6.86 |
0.69 |
| EGFR |
8A27 |
−10.36 |
0.001 |
| CASP3 |
1NME |
−8.59 |
0.10 |
| HSP90AA1 |
4BQG |
−8.06 |
0.042 |
| ESR1 |
7UJO |
−6.58 |
0.84 |
| SRC |
2SRC |
−8.29 |
0.67 |
| GSK3B |
7SXJ |
−7.78 |
0.89 |
| IGF1 |
1GZR |
−7.22 |
0.79 |
| PPARG |
6T1S |
−8.40 |
0.41 |
| HSP90AB1 |
6N8Y |
−11.57 |
0.94 |
Protective effects of BT against FA-induced cytotoxicity
In the present study, HT22 cells were used to investigate the protective effects of BT against FA-induced neurotoxicity. Data from a previous study revealed that FA induced cell death in a dose-dependent manner, with an LD50 of ~0.5 mM (33). The present study demonstrated that 24 h of treatment with BT alone (5–100 µM) dose-dependently caused loss of HT22 cell viability (Fig. 5A), with a significant effect observed from the concentration of 10 µM (cell viability, 72.63%; P<0.05). Therefore, BT at a dose of 5 µM (cell viability, 95.36%; P=0.9841) or less was used for subsequent treatments. To investigate whether BT protects cells from FA-induced cytotoxicity, the cells were treated with the following three methods and cell viability was measured separately: BT pretreatment for 2 h followed by FA treatment for 4 h; FA treatment for 4 h followed by BT post-treatment for 2 h; and simultaneous BT and FA treatment for 4 h. The CCK-8 assay demonstrated that pre-treatment with BT protected cells from FA-induced cytotoxicity in a dose-dependent manner (Fig. 5B), with a significant effect observed at 1 µM (P<0.05). The viability of HT22 cells was not influenced by post-treatment (Fig. 5C; P=0.8589) or simultaneous treatment (Fig. 5D; P=0.5501) with BT.
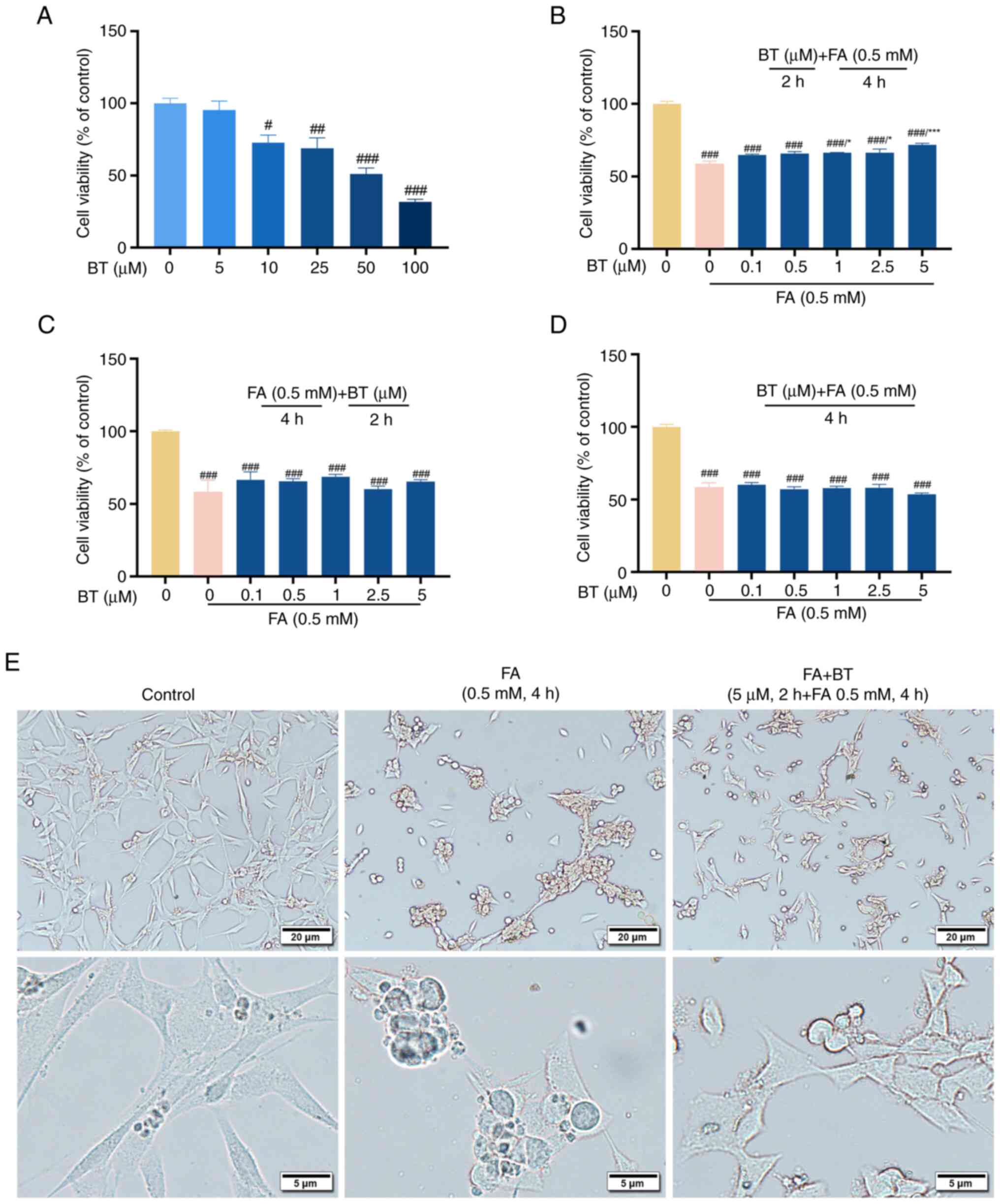 |
Figure 5.
Protective effects of BT against FA-induced cytotoxicity. (A) Viability of HT22 cells treated with BT (0–100 µM) alone for 24 h. (B) Viability of HT22 cells pretreated with BT (0.1–5 µM) for 2 h before FA (0.5 mM) treatment for 4 h. (C) Viability of HT22 cells post-treated with BT (0.1–5 µM) for 2 h after FA (0.5 mM) treatment for 4 h. (D) Viability of HT22 cells co-treated with BT (0.1–5 µM) and FA (0.5 mM) for 4 h. (E) HT22 cells were pre-treated with 5 µM BT for 2 h and then treated with 0.5 mM FA for 4 h, and cell morphology was observed with a microscope at different magnifications. Scale bar, 20 or 5 µm as indicated. All data are presented as the mean ± SEM (n=3). #P<0.05, ##P<0.01 and ###P<0.001 vs. control; *P<0.05 and ***P<0.001 vs. 0.5 mM FA group + 0 µM BT group. BT, betulin; FA, formaldehyde.
|
In addition, the morphology of HT22 cells was observed under a microscope at different magnifications. The results showed that compared with the control group, FA-treated cells were rounder and atrophied, with reduced cell processes and irregular cell boundaries. However, compared with the FA-treated group, the morphology of cells pre-treated with BT was closer to that of normal HT22 cells (Fig. 5E).
Effects of BT on FA-induced Tau protein hyperphosphorylation
As one of the crucial pathological features of AD (1), the present study revealed that the phosphorylation of Tau at the Thr181 site was significantly increased in the FA group (Fig. 6A and C; P<0.05). Pre-treatment with 5 µM BT significantly decreased the levels of p-Tau-T181 (Fig. 6A and C; P<0.01). By contrast, the expression levels of t-Tau were not significantly altered following pre-treatment with 5 µM BT (Fig. 6A and B; P=0.8646), which suggested that BT regulated the phosphorylation of Tau without affecting the expression level of total protein and thus inhibited Tau hyperphosphorylation.
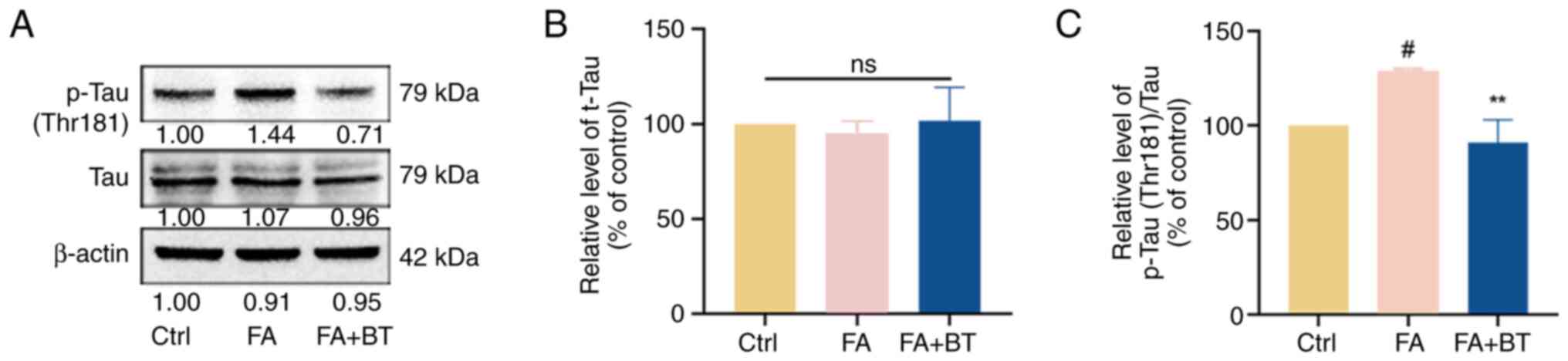 |
Figure 6.
Effects of BT on FA-induced Tau protein hyperphosphorylation. (A) Western blotting was performed to detect the protein levels of p-Tau (Thr181) and t-Tau after 4 h of FA (0.5 mM) treatment and 2 h of BT (5 µM) pre-treatment + 4 h of FA (0.5 mM) treatment. (B) Relative gray value analysis of t-Tau. (C) Relative gray value analysis of p-Tau-T181.All data are presented as the mean ± SEM (n=3). #P<0.05 vs. Ctrl; **P<0.01 vs. FA group. BT, betulin; Ctrl, control; FA, formaldehyde; ns, not significant; p-, phosphorylated; t-, total.
|
BT attenuates ROS production induced by FA
Previous studies have demonstrated that ROS are upstream of the PI3K/AKT signaling pathway, and induce apoptosis through regulation of this pathway (36,37). The results of a previous study indicated that FA induced oxidative stress and increased ROS levels in HT22 cells (33). In the present study, a significant elevation in the fluorescence intensity of intracellular ROS was observed following treatment with FA for 4 h compared with the control group (Fig. 7A and B; P<0.001). By contrast, pre-treatment with BT significantly attenuated the fluorescence intensity of intracellular ROS (Fig. 7A and B; P<0.001).
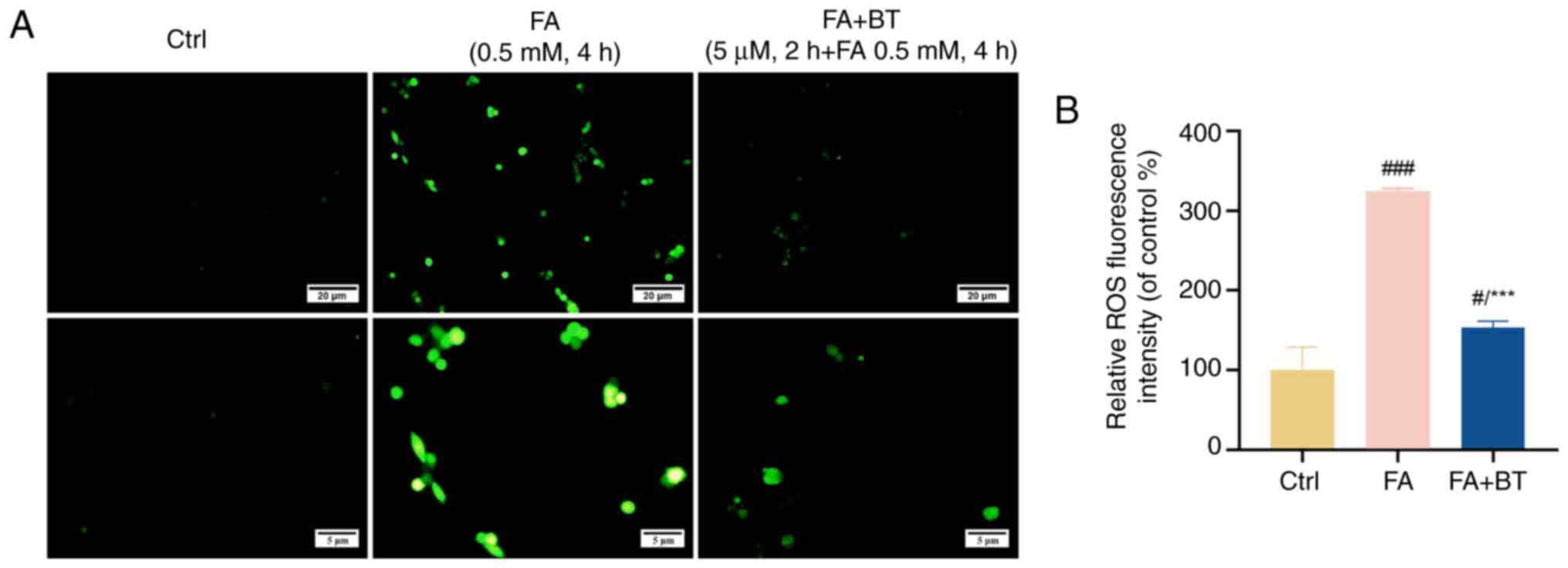 |
Figure 7.
Effects of BT on FA-induced ROS levels in HT22 cells. (A) Fluorescence intensity of ROS in HT22 cells pretreated with BT for 2 h before FA treatment for 4 h was observed by fluorescence microscopy at different magnifications. Scale bar, 20 or 5 µm as indicated. (B) Analysis of ROS fluorescence intensity. Each assay was repeated three times with 10 cells/coverslip per replicate and all data are presented as the mean ± SEM (n=3). #P<0.05 and ###P<0.001 vs. Ctrl; ***P<0.001 vs. FA group. BT, betulin; Ctrl, control; FA, formaldehyde; ROS, reactive oxygen species.
|
Effects of BT on the PI3K/AKT signaling pathway
To explore the mechanism by which BT reduced FA-induced cytotoxicity and Tau hyperphosphorylation, proteins associated with the PI3K/AKT signaling pathway were examined. The levels of p-PI3K (Fig. 8A and B; P<0.05) and p-AKT (Fig. 8A and D; P<0.001) were significantly decreased following treatment with FA for 4 h, while the levels of p-PI3K (Fig. 8A and B, P<0.05) and p-AKT (Fig. 8A and D; P<0.05) were significantly increased following pre-treatment with 5 µM BT for 2 h. As shown in Fig. 8, the expression levels of t-PI3K and t-AKT remained unaltered with (P=0.4984 for t-PI3K; P=0.9798 for t-AKT) or without (P=0.3944 for t-PI3K; P=0.9783 for t-AKT) pre-treatment with BT (Fig. 8A, C and E). These results suggested that the net effect of BT may be to alter the phosphorylation state of these proteins.
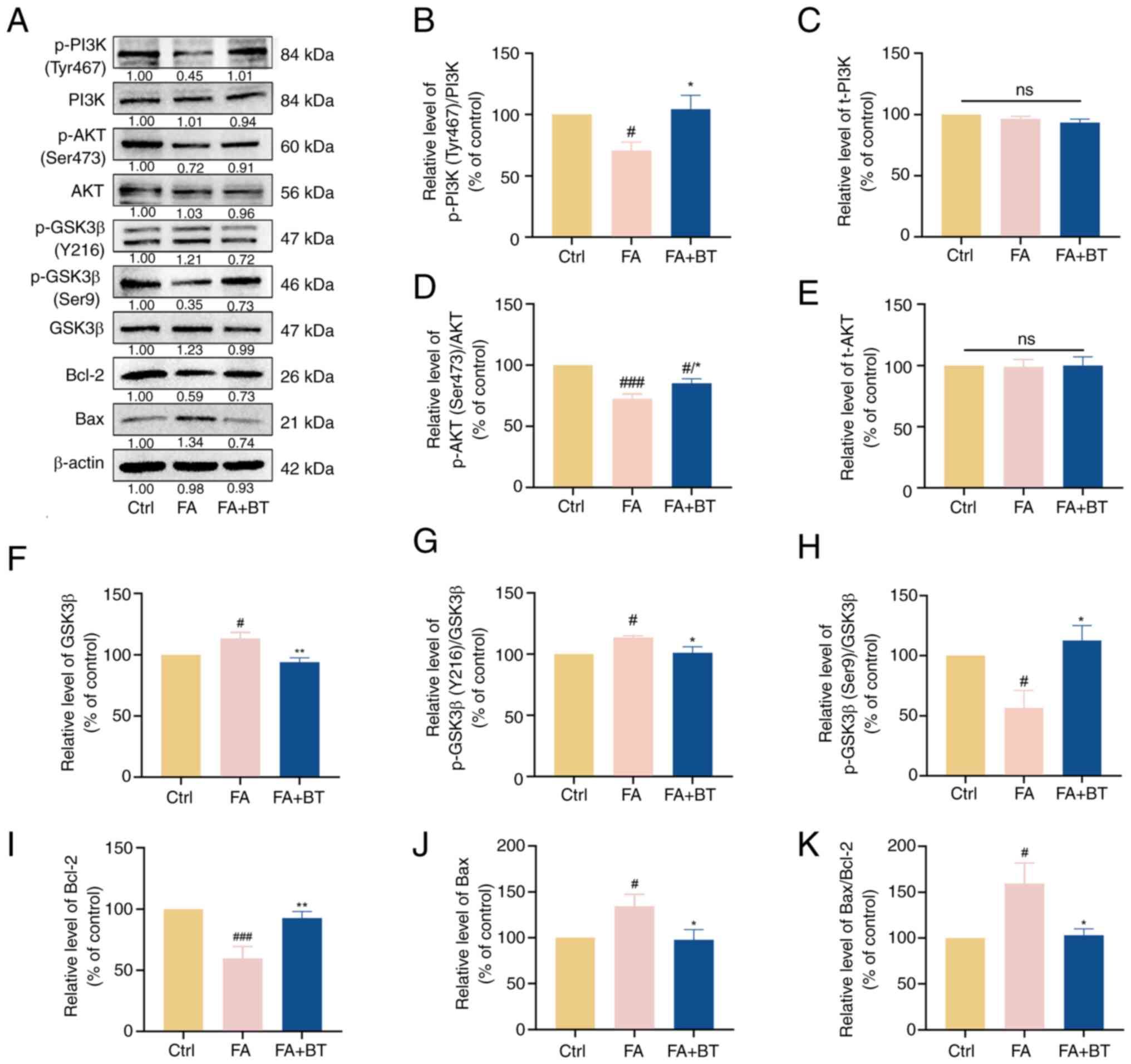 |
Figure 8.
Effects of BT on PI3K/AKT signaling pathway proteins in FA-treated HT22 cells. (A) Western blotting was performed to detect the levels of p-PI3K (Tyr467), PI3K, p-AKT (Ser473), AKT, p-GSK3β (Y216), p-GSK3β (Ser9), GSK3β, Bax and Bcl-2 after 4 h of FA (0.5 mM) treatment and 2 h of BT (5 µM) pretreatment + 4 h of FA (0.5 mM) treatment. (B) Relative gray value analysis of p-PI3K (Tyr467). (C) Relative gray value analysis of PI3K. (D) Relative gray value analysis of p-AKT (Ser473). (E) Relative gray value analysis of AKT. (F) Relative gray value analysis of GSK3β. (G) Relative gray value analysis of p-GSK3β (Y216). (H). Relative gray value analysis of p-GSK3β (Ser9). (I) Relative gray value analysis of Bcl-2. (J). Relative gray value analysis of Bax. (K) Relative gray value analysis of Bax/Bcl-2. All data are presented as the mean ± SEM (n=3). #P<0.05 and ###P<0.001 vs. Ctrl; *P<0.05 and **P<0.01 vs. FA group. BT, betulin; Ctrl, control; FA, formaldehyde; ns, not significant; p-, phosphorylated; t-, total.
|
In addition, the levels of phosphorylation of GSK3β at the activation site, namely Y216 (Fig. 8A and G; P<0.05), and t-GSK3β (Fig. 8A and F; P<0.05) were significantly increased following treatment with FA, while the levels of phosphorylation of GSK3β at the inhibition site, namely Ser9, were significantly decreased (Fig. 8A and H; P<0.05). By contrast, pre-treatment with BT significantly reduced the phosphorylation levels of GSK3β at Y216 (Fig. 8A and G; P<0.05) and t-GSK3β (Fig. 8A and F; P<0.01), and significantly increased the levels of phosphorylation of GSK3β at Ser9 (Fig. 8A and H; P<0.05). These results indicated that pre-treatment with BT inhibited the phosphorylation of GSK3β at the activation site and activated the phosphorylation of GSK3β at the inhibition site, indicating the overall decreased activity of t-GSK3β.
Bcl-2 and Bax are downstream proteins of the PI3K/AKT signaling pathway that are associated with apoptosis (38). Western blotting demonstrated that the expression levels of Bcl-2 were significantly decreased (Fig. 8A and I; P<0.001) following treatment with FA, accompanied by a significant increase in the expression levels of Bax (Fig. 8A and J; P<0.05) and the ratio of Bax/Bcl-2 (Fig. 8K; P<0.05). However, pre-treatment with BT significantly increased the expression levels of Bcl-2 (Fig. 8A and I; P<0.01), while the expression levels of Bax (Fig. 8A and J; P<0.05) and the ratio of Bax/Bcl-2 (Fig. 8K; P<0.05) were significantly decreased. Collectively, these results suggested that BT may alleviate FA-induced cytotoxicity and Tau hyperphosphorylation through modulation of the PI3K/AKT signaling pathway.
Discussion
Terrestrial plants serve a pivotal role as an indispensable source of traditional medicine, particularly in developing nations (39). The World Health Organization reported that ~80% of the global population relies on medicinal compounds derived from plants for their healthcare needs (40). Birch is used as a traditional medicine worldwide for the treatment of arthritis, gout and rheumatic disease (41,42). BT, as a key component of white birch bark, has demonstrated therapeutic potential in diseases associated with the nervous system, such as diabetic encephalopathy and Parkinson's disease (11,12). However, the therapeutic potential of BT in the treatment of AD is yet to be fully elucidated.
Using drug-disease target networks, network pharmacology illustrates complex interactions among biological systems, drugs and complex diseases (43). In the present study, network pharmacology was used to predict the therapeutic targets of BT for AD and explore the potential drug-disease target network. In addition, in vitro experiments were used to verify the network-predicted mechanisms of BT in the treatment of AD.
A single database was used to screen for BT-related targets; however, the PharmMapper database target selection covers a relatively comprehensive range, which includes 23,236 proteins covering 16,159 druggable pharmacophore models and 51,431 ligandable pharmacophore models, which can be used for comprehensive target identification (16). In the present study, 495 BT targets were predicted using the PharmMapper database, and 1,386 AD-associated genes were obtained from the OMIM, GeneCards, DisGeNET and CTD databases. Subsequently, 120 intersection targets were obtained from the 495 BT targets and 1,386 AD-associated targets, which interacted to form a complex PPI network. Topological analysis of the PPI network was performed using Cytoscape, and ALB, EGFR, CASP3, HSP90AA1, ESR1, SRC, GSK3β, IGF1, PPARG and HSP90AB were identified as hub genes of the network. Among them, GSK3β is considered a key kinase involved in the development of Tau pathology in AD (44). Previous studies indicated that GSK3β is one of the key kinases regulating Tau phosphorylation in AD (33,44,45). Further molecular docking analysis showed that BT had good binding affinity for GSK3β, indicating that BT may bind to GSK3β, regulating its physiological activity and molecular function. At the same time, based on the results of the KEGG enrichment in the PI3K/AKT signaling pathway, combined with previous studies on GSK3β in the PI3K/AKT signaling pathway (34,46,47) and the regulation of Tau phosphorylation (33,48,49), the present study focused on the effect of GSK3β on BT-mediated neuroprotection.
To further investigate the interactions and biological functions of the intersection targets, GO and KEGG enrichment analyses were performed. GO enrichment analysis revealed that the BPs associated with the targets included ‘protein phosphorylation’, ‘negative regulation of apoptotic process’ and ‘signal transduction’. KEGG analysis indicated that BT affected the development of AD via multiple pathways, including the ‘PI3K-AKT signaling pathway’, thus serving a therapeutic role. In the central nervous system, the PI3K/AKT signaling pathway serves a crucial role in a variety of physiological processes, such as cell proliferation, autophagy, apoptosis, neurogenesis and synaptic plasticity (46). Furthermore, this pathway is closely associated with the pathogenesis of AD (50). It has been reported that the levels of PI3K subunits (p85 and p110) and the phosphorylation of AKT were reduced in post-mortem brain samples of patients with AD compared with healthy individuals (51). As one of the most important intracellular signaling pathways, after binding to the growth factor receptor, PI3K activates the AKT protein via structural alterations, leading to regulation of downstream GSK3β and apoptosis-associated proteins, including Bcl-2 and Bax (46). Collectively, the results of the PPI network and pathway analysis highlighted that BT may exert neuroprotective effects on AD through regulation of the PI3K/AKT signaling pathway, thus inhibiting Tau hyperphosphorylation and apoptosis.
To further validate the predictions generated using network pharmacology, FA-induced HT22 cells were employed in the present study. FA is considered a novel pathogenic factor in AD due to the impact on cognitive impairment and pathological features (52). Thus, FA has been widely used to simulate animal and cell disease models of AD (45,53). A previous study showed that intraperitoneal injection of BT could alleviate neuropathic and inflammatory pain induced by formalin solution (0.92% FA, 20 µl; right hindpaw injection) in Swiss mice, suggesting that BT might have a protective effect against FA-induced neurotoxicity (54). The present study investigated the protective effect of BT pretreatment (2 h) against the neurotoxicity induced by FA (0.5 mM; 4 h) in HT22 cells. Notably, neuronal atrophy and death are the main causes of cognitive impairment and brain atrophy in patients with AD (55). Therefore, the present study analyzed the effects of BT on cell viability and morphology. The results showed that pre-treatment with BT for 2 h prior to 4 h of FA treatment increased cell viability in a dose-dependent manner, and a significant effect was observed at ≥1 µM. Following pre-treatment with BT, the cell density was increased, synapses were recovered and cell morphology was comparable with that of normal HT22 cells, compared with the FA group. These results indicated that BT may protect HT22 cells from FA-induced cytotoxicity.
Tau is a microtubule-associated protein, and hyperphosphorylation of Tau results in the loss of physiological function, leading to cell death, thus contributing to neuropathies (56). The levels of p-Tau-T181 in plasma and cerebrospinal fluid are associated with cognitive impairment and the pathological progression of AD, and thus, Tau is considered a highly specific marker of AD (57). A previous study revealed that FA exposure increased Tau hyperphosphorylation and ROS content (33). In the present study, BT was found to significantly reduce FA-induced Tau phosphorylation at Thr181. In addition, BT has been reported to exert antioxidant effects by increasing the protein expression levels of Nrf2 to promote the expression of HO-1 and NAD (P)H quinone oxidoreductase 1, thereby inhibiting LPS-induced oxidative stress in LPS-treated macrophages (58). Consistent with these findings, in the present study, pre-treatment with BT significantly reduced ROS generation compared with the FA group. In conclusion, pre-treatment with BT effectively ameliorated FA-induced Tau hyperphosphorylation and oxidative stress in HT22 cells and protected cells from FA-induced neurotoxicity.
Increased GSK3β activity is associated with increases in the hyperphosphorylation of Tau (48). It has been reported that inhibition of GSK3β reduced the production of neurofibrillary tangles and Tau hyperphosphorylation (48). Furthermore, BT has been found to inhibit LPS-induced oxidative stress and inflammatory responses by inhibiting the expression of GSK3β by increasing phosphorylation at Ser9, leading to the activation of Nrf2 in RAW 264.7 macrophages (58). The results of molecular docking in the present study also demonstrated that BT could regulate GSK3β. However, current docking algorithms may suffer from limited conformational sampling, leading to potential prediction errors (32). Thus, further investigation was conducted in the present study. Western blotting confirmed the molecular docking results, indicating that BT reduced the expression levels of GSK3β increased by FA. Specifically, BT inhibited the FA-induced activation of GSK3β through inhibition of the activation site at Y216 and activation of the inhibition site at Ser9, indicating an overall decreased activity of t-GSK3β, thereby alleviating FA-induced Tau hyperphosphorylation. For GSK3β, phosphorylation at its activation site Y216 is required for maximal GSK3β activity, while phosphorylation at the inhibitory site Ser9 causes the N-terminal tail of GSK3β to act as a pre-phosphorylated or pseudo-substrate, impairing its binding to the activated substrates and reducing the phosphorylation (47). Therefore, BT may exert neuroprotective effects by reducing the activity of GSK3β and consequently lowering the phosphorylation level of its substrate Tau protein.
GSK3β is considered to be a downstream effector of the PI3K/AKT signaling pathway. Following phosphorylation of AKT at Ser473, phosphorylated GSK3β is inactivated at Ser9, thereby attenuating the hyperphosphorylation of Tau and inhibiting the formation of neurofibrillary tangles (49). Therefore, activation of the PI3K/AKT signaling pathway is considered to be an effective therapeutic target for AD. Numerous studies have demonstrated that BT could mitigate the progression of various cancer types, such as metastatic colorectal cancer and ovarian cancer, by modulating the PI3K/AKT signaling pathway (59,60); however, to the best of our knowledge, there is no existing report on whether the protective effect of BT on AD is associated with the PI3K/AKT signaling pathway. In the present study, western blotting demonstrated that BT enhanced the activity of PI3K and AKT in FA-treated HT22 cells by increasing the phosphorylation levels of PI3K and AKT. These results indicated that BT may activate PI3K and subsequently induce downstream AKT phosphorylation to stimulate the PI3K/AKT signaling cascade.
Furthermore, the PI3K/AKT signaling pathway is a crucial pathway for regulating cell survival through regulating the expression of two key apoptosis-regulating proteins, Bcl-2 and Bax (38). A previous study has demonstrated that downregulation of Bcl-2 and upregulation of Bax activated caspase-3 to induce apoptosis in rat hippocampal neurons (61). In addition, the ratio of the expression levels of pro-apoptotic and anti-apoptotic proteins determines cell survival (62). In the present study, treatment with FA significantly reduced the expression levels of the anti-apoptotic protein Bcl-2, increased the production of the pro-apoptotic protein Bax and increased the ratio of Bax to Bcl-2. After pretreatment with 5 µM BT, the changes of Bax and Bcl-2 induced by FA were significantly attenuated, which was similar to a previous study in SK-NSH cells, where 2.5 µM BT and 2 µM As2O3 co-treatment for 48 h reduced the decrease in Bcl-2 and increase in Bax induced by As2O3 (63). It has been revealed that GSK3β is involved in stress-induced cell apoptosis processes, including endoplasmic reticulum stress-induced cell apoptosis (33,64). Activated GSK3β may activate or inhibit downstream targets, such as activating p53 and Bax, and inhibiting Bcl-2, thereby promoting apoptosis (64). Therefore, the regulatory effect of BT on Bax and Bcl-2 might partially be achieved by inhibiting GSK3β. Collectively, these results demonstrated that BT may exert therapeutic effects in AD through regulation of the PI3K/AKT/GSK3β signaling pathway, leading to reduced Tau hyperphosphorylation and apoptosis.
Several studies have confirmed that BT can alleviate cognitive impairment (9,12). Cho et al (9) demonstrated that BT alleviated scopolamine-induced cognitive impairment in mice and increased glutathione levels in mouse brain tissue; however, the specific molecular mechanism was not deeply analyzed. Another study indicated that BT might exhibit a protective effect on cognitive decline in STZ-induced diabetic rats via the HO-1/Nrf2/NF-κB pathway (12). To the best of our knowledge, the present study was the first to explore the potential mechanisms of BT in the treatment of AD, using compound target network construction, PPI network analysis, GO and KEGG enrichment analyses, molecular docking, and experimental validation in an FA-induced AD cell model. Briefly, the results of the present study revealed that the neuroprotective effects of BT were associated with the PI3K/AKT signaling pathway and its downstream targets, including GSK3β, Bax and Bcl-2. Thus, BT may exhibit potential for the treatment of AD, and the present study provides a theoretical basis for the development of a novel treatment option. As the PI3K/AKT signaling pathway is involved in numerous physiological functions, such as ferroptosis and inflammatory responses (65,66), further investigations are required to determine the specific molecular mechanisms underlying the effects of BT in AD. Additionally, while the HT22 cells selected in the present study have been extensively utilized (33,67,68) for investigating neurotoxicity, cell models are unable to fully replicate the cognitive decline, behavioral changes and pathological alterations associated with AD, and further animal experiments need to be performed in follow-up studies.
Acknowledgements
The authors would like to thank the Laboratory of Human Anatomy and Histoembryology, School of Basic Medicine, Dali University (Dali, China) for providing the cell lines used in the present study.
Funding
This work was supported by the Local Universities Basic Research Joint Special Project of Yunnan Province (grant no. 202301BA070001-032).
Availability of data and materials
The data generated in the present study may be requested from the corresponding author.
Authors' contributions
XH and FC designed the research idea and methodology. NW and JC wrote the manuscript and prepared images of the results. ZS and JC conducted data collection and analysis. NW and ZS carried out the experiments. XH and FC reviewed and revised the manuscript. All authors have read and approved the final version of the manuscript. NW and XH confirm the authenticity of all the raw data.
Ethics approval and consent to participate
Not applicable.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
References
|
1
|
Knopman DS, Amieva H, Petersen RC, Chételat G, Holtzman DM, Hyman BT, Nixon RA and Jones DT: Alzheimer disease. Nat Rev Dis Primers. 7:332021. View Article : Google Scholar
|
|
2
|
Scheltens P, De Strooper B, Kivipelto M, Holstege H, Chételat G, Teunissen CE, Cummings J and van der Flier WM: Alzheimer's disease. Lancet. 397:1577–1590. 2021. View Article : Google Scholar
|
|
3
|
Rajasekhar K and Govindaraju T: Current progress, challenges and future prospects of diagnostic and therapeutic interventions in Alzheimer's disease. RSC Adv. 8:23780–23804. 2018. View Article : Google Scholar
|
|
4
|
Zhang F, Zhong RJ, Cheng C, Li S and Le WD: New therapeutics beyond amyloid-β and tau for the treatment of Alzheimer's disease. Acta Pharmacol Sin. 42:1382–1389. 2021. View Article : Google Scholar
|
|
5
|
Chen YG: Research progress in the pathogenesis of Alzheimer's disease. Chin Med J (Engl). 131:1618–1624. 2018. View Article : Google Scholar
|
|
6
|
Wightman EL: Potential benefits of phytochemicals against Alzheimer's disease. Proc Nutr Soc. 76:106–112. 2017. View Article : Google Scholar
|
|
7
|
Tuli HS, Sak K, Gupta DS, Kaur G, Aggarwal D, Parashar NC, Choudhary R, Yerer MB, Kaur J, Kumar M, et al: Anti-inflammatory and anticancer properties of birch bark-derived betulin: Recent developments. Plants (Basel). 10:26632021.
|
|
8
|
Buko V, Kuzmitskaya I, Kirko S, Belonovskaya E, Naruta E, Lukivskaya O, Shlyahtun A, Ilyich T, Zakreska A and Zavodnik I: Betulin attenuated liver damage by prevention of hepatic mitochondrial dysfunction in rats with alcoholic steatohepatitis. Physiol Int. 106:323–334. 2019. View Article : Google Scholar
|
|
9
|
Cho N, Kim HW, Lee HK, Jeon BJ and Sung SH: Ameliorative effect of betulin from Betula platyphylla bark on scopolamine-induced amnesic mice. Biosci Biotechnol Biochem. 80:166–171. 2016. View Article : Google Scholar
|
|
10
|
Farzan M, Farzan M, Shahrani M, Navabi SP, Vardanjani HR, Amini-Khoei H and Shabani S: Neuroprotective properties of Betulin, Betulinic acid, and Ursolic acid as triterpenoids derivatives: A comprehensive review of mechanistic studies. Nutr Neurosci. 27:223–240. 2024. View Article : Google Scholar
|
|
11
|
Tsai CW, Tsai RT, Liu SP, Chen CS, Tsai MC, Chien SH, Hung HS, Lin SZ, Shyu WC and Fu RH: Neuroprotective effects of betulin in pharmacological and transgenic caenorhabditis elegans models of parkinson's disease. Cell Transplant. 26:1903–1918. 2017. View Article : Google Scholar
|
|
12
|
Ma C and Long H: Protective effect of betulin on cognitive decline in streptozotocin (STZ)-induced diabetic rats. Neurotoxicology. 57:104–111. 2016. View Article : Google Scholar
|
|
13
|
Liu Q, Liu JP, Mei JH, Li SJ, Shi LQ, Lin ZH, Xie BY, Sun WG, Wang ZY, Yang XL, et al: Betulin isolated from Pyrola incarnata Fisch. inhibited lipopolysaccharide (LPS)-induced neuroinflammation with the guidance of computer-aided drug design. Bioorg Med Chem Lett. 30:1271932020. View Article : Google Scholar
|
|
14
|
Nogales C, Mamdouh ZM, List M, Kiel C, Casas AI and Schmidt H: Network pharmacology: Curing causal mechanisms instead of treating symptoms. Trends Pharmacol Sci. 43:136–150. 2022. View Article : Google Scholar
|
|
15
|
Kim S, Chen J, Cheng T, Gindulyte A, He J, He S, Li Q, Shoemaker BA, Thiessen PA, Yu B, et al: PubChem 2023 update. Nucleic Acids Res. 51:D1373–D1380. 2023. View Article : Google Scholar
|
|
16
|
Wang X, Shen Y, Wang S, Li S, Zhang W, Liu X, Lai L, Pei J and Li H: PharmMapper 2017 update: A web server for potential drug target identification with a comprehensive target pharmacophore database. Nucleic Acids Res. 45:W356–W360. 2017. View Article : Google Scholar
|
|
17
|
UniProt Consortium: UniProt: The universal protein knowledgebase in 2023. Nucleic Acids Res. 51:D523–D531. 2023. View Article : Google Scholar
|
|
18
|
Stelzer G, Rosen N, Plaschkes I, Zimmerman S, Twik M, Fishilevich S, Stein TI, Nudel R, Lieder I, Mazor Y, et al: The genecards suite: From gene data mining to disease genome sequence analyses. Curr Protoc Bioinformatics. 54:1.30.31–31.30.33. 2016. View Article : Google Scholar
|
|
19
|
Pinero J, Ramirez-Anguita JM, Sauch-Pitarch J, Ronzano F, Centeno E, Sanz F and Furlong LI: The DisGeNET knowledge platform for disease genomics: 2019 update. Nucleic Acids Res. 48:D845–D855. 2020.
|
|
20
|
Davis AP, Wiegers TC, Wiegers J, Wyatt B, Johnson RJ, Sciaky D, Barkalow F, Strong M, Planchart A and Mattingly CJ: CTD tetramers: A new online tool that computationally links curated chemicals, genes, phenotypes, and diseases to inform molecular mechanisms for environmental health. Toxicol Sci. 195:155–168. 2023. View Article : Google Scholar
|
|
21
|
Amberger JS and Hamosh A: Searching online mendelian inheritance in man (OMIM): A knowledgebase of human genes and genetic phenotypes. Curr Protoc Bioinformatics. 58:1.2.1–1.2.12. 2017. View Article : Google Scholar
|
|
22
|
Bardou P, Mariette J, Escudié F, Djemiel C and Klopp C: Jvenn: An interactive Venn diagram viewer. BMC Bioinformatics. 15:2932014. View Article : Google Scholar
|
|
23
|
Szklarczyk D, Kirsch R, Koutrouli M, Nastou K, Mehryary F, Hachilif R, Gable AL, Fang T, Doncheva NT, Pyysalo S, et al: The STRING database in 2023: Protein-protein association networks and functional enrichment analyses for any sequenced genome of interest. Nucleic Acids Res. 51:D638–D646. 2023. View Article : Google Scholar
|
|
24
|
Shannon P, Markiel A, Ozier O, Baliga NS, Wang JT, Ramage D, Amin N, Schwikowski B and Ideker T: Cytoscape: A software environment for integrated models of biomolecular interaction networks. Genome Res. 13:2498–2504. 2003. View Article : Google Scholar
|
|
25
|
Chin CH, Chen SH, Wu HH, Ho CW, Ko MT and Lin CY: cytoHubba: Identifying hub objects and sub-networks from complex interactome. BMC Syst Biol. 4:S112014. View Article : Google Scholar
|
|
26
|
Sherman BT, Hao M, Qiu J, Jiao X, Baseler MW, Lane HC, Imamichi T and Chang W: DAVID: A web server for functional enrichment analysis and functional annotation of gene lists (2021 update). Nucleic Acids Res. 50:W216–W221. 2022. View Article : Google Scholar
|
|
27
|
Shen W, Song Z, Zhong X, Huang M, Shen D, Gao P, Qian X, Wang M, He X, Wang T, et al: Sangerbox: A comprehensive, interaction-friendly clinical bioinformatics analysis platform. Imeta. 1:e362022. View Article : Google Scholar
|
|
28
|
Berman HM, Battistuz T, Bhat TN, Bluhm WF, Bourne PE, Burkhardt K, Feng Z, Gilliland GL, Iype L, Jain S, et al: The protein data bank. Acta Crystallogr D Biol Crystallogr. 28:235–242. 2000.
|
|
29
|
Ru J, Li P, Wang J, Zhou W, Li B, Huang C, Li P, Guo Z, Tao W, Yang Y, et al: TCMSP: A database of systems pharmacology for drug discovery from herbal medicines. J Cheminform. 6:132014. View Article : Google Scholar
|
|
30
|
Seeliger D and de Groot BL: Ligand docking and binding site analysis with PyMOL and Autodock/Vina. J Comput Aided Mol Des. 24:417–422. 2010. View Article : Google Scholar
|
|
31
|
Goodsell DS and Olson AJ: Automated docking of substrates to proteins by simulated annealing. Proteins. 8:195–202. 1990. View Article : Google Scholar
|
|
32
|
Wang Z, Sun H, Yao X, Li D, Xu L, Li Y, Tian S and Hou T: Comprehensive evaluation of ten docking programs on a diverse set of protein-ligand complexes: The prediction accuracy of sampling power and scoring power. Phys Chem Chem Phys. 18:12964–12975. 2016. View Article : Google Scholar
|
|
33
|
Chen F, Wang N, Tian X, Su J, Qin Y, He R and He X: The protective effect of mangiferin on formaldehyde-induced HT22 cell damage and cognitive impairment. Pharmaceutics. 15:15682023. View Article : Google Scholar
|
|
34
|
Buttrick GJ and Wakefield JG: PI3-K and GSK-3: Akt-ing together with microtubules. Cell Cycle. 7:2621–2625. 2008. View Article : Google Scholar
|
|
35
|
Pinzi L and Rastelli G: Molecular docking: Shifting paradigms in drug discovery. Int J Mol Sci. 20:43312019. View Article : Google Scholar
|
|
36
|
Liu Y, Shi C, He Z, Zhu F, Wang M, He R, Zhao C, Shi X, Zhou M, Pan S, et al: Inhibition of PI3K/AKT signaling via ROS regulation is involved in Rhein-induced apoptosis and enhancement of oxaliplatin sensitivity in pancreatic cancer cells. Int J Biol Sci. 17:589–602. 2021. View Article : Google Scholar
|
|
37
|
Li H, Deng W, Yang J, Lin Y, Zhang S, Liang Z, Chen J, Hu M, Liu T, Mo G, et al: Corylifol A suppresses osteoclastogenesis and alleviates ovariectomy-induced bone loss via attenuating ROS production and impairing mitochondrial function. Biomed Pharmacother. 171:1161662024. View Article : Google Scholar
|
|
38
|
Zang G, Fang L, Chen L and Wang C: Ameliorative effect of nicergoline on cognitive function through the PI3K/AKT signaling pathway in mouse models of Alzheimer's disease. Mol Med Rep. 17:7293–7300. 2018.
|
|
39
|
Rates SM: Plants as source of drugs. Toxicon. 39:603–613. 2001. View Article : Google Scholar
|
|
40
|
Raskin I, Ribnicky DM, Komarnytsky S, Ilic N, Poulev A, Borisjuk N, Brinker A, Moreno DA, Ripoll C, Yakoby N, et al: Plants and human health in the twenty-first century. Trends Biotechnol. 20:522–531. 2002. View Article : Google Scholar
|
|
41
|
Matsuda H, Ishikado A, Nishida N, Ninomiya K, Fujiwara H, Kobayashi Y and Yoshikawa M: Hepatoprotective, superoxide scavenging, and antioxidative activities of aromatic constituents from the bark of Betula platyphylla var. japonica. Bioorg Med Chem Lett. 8:2939–2944. 1998. View Article : Google Scholar
|
|
42
|
Huh JE, Hong JM, Baek YH, Lee JD, Choi DY and Park DS: Anti-inflammatory and anti-nociceptive effect of Betula platyphylla var. japonica in human interleukin-1β-stimulated fibroblast-like synoviocytes and in experimental animal models. J Ethnopharmacol. 135:126–134. 2011. View Article : Google Scholar
|
|
43
|
Hopkins AL: Network pharmacology: The next paradigm in drug discovery. Nat Chem Biol. 4:682–690. 2008. View Article : Google Scholar
|
|
44
|
Sayas CL and Avila J: GSK-3 and tau: A key duet in Alzheimer's disease. Cells. 10:7212021. View Article : Google Scholar
|
|
45
|
He X, Li Z, Rizak JD, Wu S, Wang Z, He R, Su M, Qin D, Wang J and Hu X: Resveratrol attenuates formaldehyde induced hyperphosphorylation of tau protein and cytotoxicity in N2a cells. Front Neurosci. 10:5982016.
|
|
46
|
Long HZ, Cheng Y, Zhou ZW, Luo HY, Wen DD and Gao LC: PI3K/AKT signal pathway: A target of natural products in the prevention and treatment of Alzheimer's disease and parkinson's disease. Front Pharmacol. 12:6486362021. View Article : Google Scholar
|
|
47
|
Beurel E, Grieco SF and Jope RS: Glycogen synthase kinase-3 (GSK3): Regulation, actions, and diseases. Pharmacol Ther. 148:114–131. 2015. View Article : Google Scholar
|
|
48
|
Plattner F, Angelo M and Giese KP: The roles of cyclin-dependent kinase 5 and glycogen synthase kinase 3 in tau hyperphosphorylation. J Biol Chem. 281:25457–25465. 2006. View Article : Google Scholar
|
|
49
|
Qu ZS, Li L, Sun XJ, Zhao YW, Zhang J, Geng Z, Fu JL and Ren QG: Glycogen synthase kinase-3 regulates production of amyloid-β peptides and tau phosphorylation in diabetic rat brain. ScientificWorldJournal. 2014:8781232014. View Article : Google Scholar
|
|
50
|
Gabbouj S, Ryhänen S, Marttinen M, Wittrahm R, Takalo M, Kemppainen S, Martiskainen H, Tanila H, Haapasalo A, Hiltunen M and Natunen T: Altered insulin signaling in alzheimer's disease brain-special emphasis on PI3K-Akt pathway. Front Neurosci. 13:6292019. View Article : Google Scholar
|
|
51
|
Steen E, Terry BM, Rivera EJ, Cannon JL, Neely TR, Tavares R, Xu XJ, Wands JR and de la Monte SM: Impaired insulin and insulin-like growth factor expression and signaling mechanisms in Alzheimer's disease-is this type 3 diabetes? J Alzheimers Dis. 7:63–80. 2005. View Article : Google Scholar
|
|
52
|
Tulpule K and Dringen R: Formaldehyde in brain: An overlooked player in neurodegeneration? J Neurochem. 127:7–21. 2013. View Article : Google Scholar
|
|
53
|
Liu X, Zhang Y, Wu R, Ye M, Zhao Y, Kang J, Ma P, Li J and Yang X: Acute formaldehyde exposure induced early Alzheimer-like changes in mouse brain. Toxicol Mech Methods. 28:95–104. 2018. View Article : Google Scholar
|
|
54
|
de Souza MT, de Campos Buzzi F, Filho VC, Hess S, Monache FD and Niero R: Phytochemical and antinociceptive properties of Matayba elaeagnoides Radlk. barks. Z Naturforsch C J Biosci. 62:550–554. 2007. View Article : Google Scholar
|
|
55
|
Offen D, Elkon H and Melamed E: Apoptosis as a general cell death pathway in neurodegenerative diseases. J Neural Transm Suppl. 2000:153–166. 2000.
|
|
56
|
Gyparaki MT, Arab A, Sorokina EM, Santiago-Ruiz AN, Bohrer CH, Xiao J and Lakadamyali M: Tau forms oligomeric complexes on microtubules that are distinct from tau aggregates. Proc Natl Acad Sci USA. 118:e20214611182021. View Article : Google Scholar
|
|
57
|
Karikari TK, Pascoal TA, Ashton NJ, Janelidze S, Benedet AL, Rodriguez JL, Chamoun M, Savard M, Kang MS, Therriault J, et al: Blood phosphorylated tau 181 as a biomarker for Alzheimer's disease: A diagnostic performance and prediction modelling study using data from four prospective cohorts. Lancet Neurol. 19:422–433. 2020. View Article : Google Scholar
|
|
58
|
Ci X, Zhou J, Lv H, Yu Q, Peng L and Hua S: Betulin exhibits anti-inflammatory activity in LPS-stimulated macrophages and endotoxin-shocked mice through an AMPK/AKT/Nrf2-dependent mechanism. Cell Death Dis. 8:e27982017. View Article : Google Scholar
|
|
59
|
Yang Q, Fei Z and Huang C: Betulin terpenoid targets OVCAR-3 human ovarian carcinoma cells by inducing mitochondrial mediated apoptosis, G2/M phase cell cycle arrest, inhibition of cell migration and invasion and modulating mTOR/PI3K/AKT signalling pathway. Cell Mol Biol (Noisy-le-grand). 67:14–19. 2021. View Article : Google Scholar
|
|
60
|
Han YH, Mun JG, Jeon HD, Kee JY and Hong SH: Betulin inhibits lung metastasis by inducing cell cycle arrest, autophagy, and apoptosis of metastatic colorectal cancer cells. Nutrients. 12:662019. View Article : Google Scholar
|
|
61
|
Jin H, Wang M, Wang J, Cao H, Niu W and Du L: Paeonol attenuates isoflurane anesthesia-induced hippocampal neurotoxicity via modulation of JNK/ERK/P38MAPK pathway and regulates histone acetylation in neonatal rat. J Matern Fetal Neonatal Med. 33:81–91. 2020. View Article : Google Scholar
|
|
62
|
Korsmeyer SJ, Shutter JR, Veis DJ, Merry DE and Oltvai ZN: Bcl-2/Bax: A rheostat that regulates an anti-oxidant pathway and cell death. Semin Cancer Biol. 4:327–332. 1993.
|
|
63
|
Chen KY, Hsu WL, Hsu SW, Chen CH, Hong KT, Tsai CW, Chang WS, Chen CC, Pei JS, Lee HT and Bau DT: Involvement of mitochondrial damage and oxidative stress in apoptosis induced by betulin plus arsenic trioxide in neuroblastoma cells. Anticancer Res. 43:2467–2476. 2023. View Article : Google Scholar
|
|
64
|
Yang K, Chen Z, Gao J, Shi W, Li L, Jiang S, Hu H, Liu Z, Xu D and Wu L: The key roles of GSK-3β in regulating mitochondrial activity. Cell Physiol Biochem. 44:1445–1459. 2017. View Article : Google Scholar
|
|
65
|
Chen K, Xue R, Geng Y and Zhang S: Galangin inhibited ferroptosis through activation of the PI3K/AKT pathway in vitro and in vivo. FASEB J. 36:e225692022. View Article : Google Scholar
|
|
66
|
Dong L, Du H, Zhang M, Xu H, Pu X, Chen Q, Luo R, Hu Y, Wang Y, Tu H, et al: Anti-inflammatory effect of Rhein on ulcerative colitis via inhibiting PI3K/Akt/mTOR signaling pathway and regulating gut microbiota. Phytother Res. 36:2081–2094. 2022. View Article : Google Scholar
|
|
67
|
Yang S, Xie Z, Pei T, Zeng Y, Xiong Q, Wei H, Wang Y and Cheng W: Salidroside attenuates neuronal ferroptosis by activating the Nrf2/HO1 signaling pathway in Aβ(1–42)-induced Alzheimer's disease mice and glutamate-injured HT22 cells. Chin Med. 17:822022. View Article : Google Scholar
|
|
68
|
Xiong Y, Ruan YT, Zhao J, Yang YW, Chen LP, Mai YR, Yu Q, Cao ZY, Liu FF, Liao W and Liu J: Magnesium-L-threonate exhibited a neuroprotective effect against oxidative stress damage in HT22 cells and Alzheimer's disease mouse model. World J Psychiatry. 12:410–424. 2022. View Article : Google Scholar
|






















