Introduction
Pleomorphic leiomyosarcoma is a rare but distinct
variant of leiomyosarcoma, accounting for only 8.6% of all
leiomyosarcomas (1). It occurs
predominantly in the retroperitoneum and extremities of middle-aged
to elderly adults. In general, pleomorphic leiomyosarcoma has an
aggressive behavior with a 48–89% metastatic rate and an overall
tumor-associated mortality of 50–65% (1,2).
Retroperitoneal leiomyosarcomas tend to correlate with a higher
frequency of local recurrence and distant metastasis, probably due
to the difficulty in achieving wide or even clear margins.
Histologically, pleomorphic leiomyosarcoma is
defined as a tumor composed of typical leiomyosarcoma with
pleomorphic areas occupying more than two-thirds of the lesion
(1). The typical component consists
of spindle-shaped cells with brightly eosinophilic cytoplasm and
cigar-shaped nuclei arranged in a fascicular pattern. In contrast,
the pleomorphic component is composed of polygonal cells,
spindle-shaped cells, and/or epithelioid cells, simulating the
morphology of so-called malignant fibrous histiocytoma (MFH).
Multinucleated bizarre cells and rhabdoid cells may be found in the
pleomorphic component (1,2). Immunohistochemically, the tumor cells
are positive for at least one of the smooth muscle markers (smooth
muscle actin, desmin, muscle-specific actin, calponin, and
caldesmon) in both the typical and pleomorphic components.
Pleomorphic soft tissue leiomyosarcomas are
generally associated with highly complex karyotypes lacking
specific structural or numerical aberrations (3–5).
Metaphase and array-based comparative genomic hybridization (CGH)
analyses have demonstrated gains of 1q, 5p, 6q, 8q, 17p, and Xp and
losses of 2p, 10q, 11q, 13q, and 16q. Amplifications of 1q21,
5p14-pter, 12q13-q15, 17p11-p12, 19p13, and 20q13 have also been
observed (5–13).
In this study, we report a case of pleomorphic
leiomyosarcoma with giant marker and ring chromosomes occurring in
the deep soft tissue of the thigh. We also review the cytogenetic
and molecular cytogenetic features of pleomorphic soft tissue
leiomyosarcoma as well as its clinicopathological
characteristics.
Materials and methods
Case presentation
A 60-year-old man was referred to our hospital with
a history of a non-painful left thigh mass first noticed one month
previously. Physical examination revealed an 8×7 cm, firm,
non-tender mass in the medial aspect of the proximal left thigh.
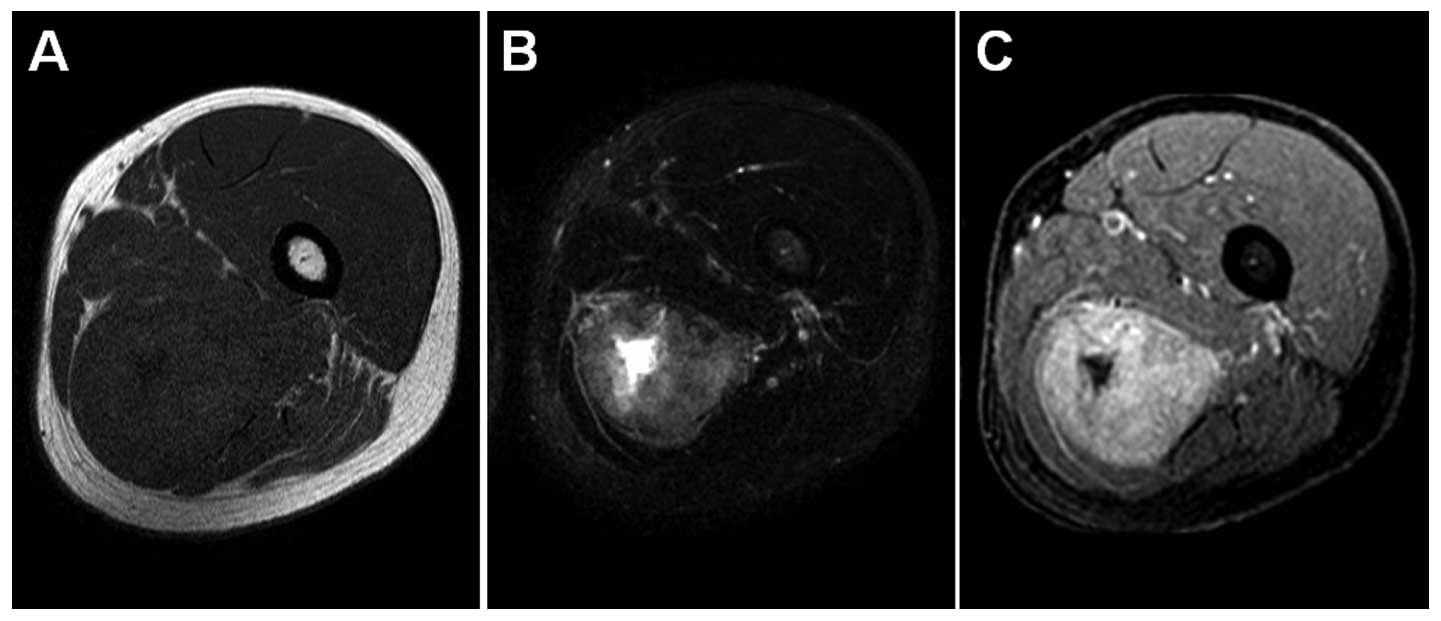
Magnetic resonance imaging revealed an ill-defined intramuscular
mass in the left adductor magnus. The mass was low to intermediate
signal intensity on T1-weighted sequences (Fig. 1A) and heterogeneously high signal
intensity on T2-weighted spectral presaturation with inversion
recovery sequences (Fig. 1B).
T1-weighted contrast-enhanced fat-suppressed sequences showed
heterogeneous enhancement of the mass (Fig. 1C). Positron emission tomography
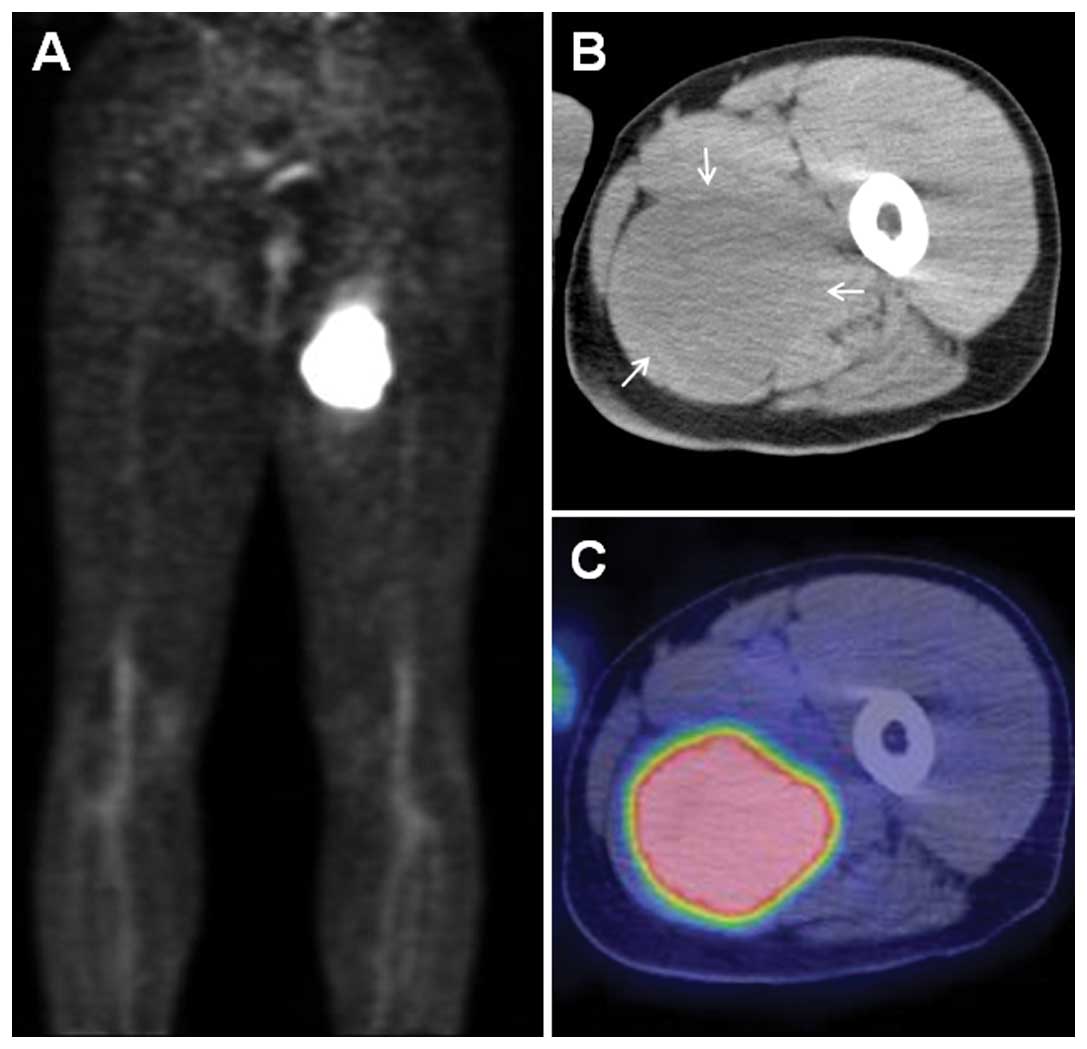
imaging demonstrated a markedly increased uptake of
fluorine-18-deoxyglucose in the proximal left thigh (Fig. 2A). The maximum standardized uptake
value was 20.9. Computed tomography (CT) images showed a hypodense
intramuscular mass, with corresponding tracer uptake (Fig. 2B and C). No evidence of distant
metastasis was found on chest and abdominal CT scans.
The patient underwent a core needle biopsy and the
pathologic diagnosis was pleomorphic leiomyosarcoma. A wide
resection of the tumor was performed. Macroscopically, the resected
tumor, measuring 10×8×7 cm, had a solid, grayish-white cut surface.
Microscopically, the tumor was composed of a mixture of atypical
spindle cells, polygonal cells, and bizarre giant cells forming
interlacing bundles and storiform pattern (Fig. 3A and B). Mitotic figures including
abnormal forms were frequently seen. Immunohistochemically, the
tumor cells were positive for vimentin, smooth muscle actin
(Fig. 3C), and desmin, but negative
for muscle-specific actin, h-caldesmon, MyoD1, or myogenin. The
MIB-1 labeling index was 19.7% in the highest spot (Fig. 3D). These findings confirmed the
diagnosis of pleomorphic leiomyosarcoma. The patient received
postoperative adjuvant radiotherapy with a total dose of 60 Gy and
chemotherapy with five cycles of doxorubicin. At 19 months of
follow-up, the patient is well without any evidence of local
recurrence or distant metastasis.
Cytogenetic analysis
A representative fresh tissue sample was obtained
from the surgical resection. Culturing, harvesting, and preparation
of slides were performed as described previously (14). Karyotypic descriptions were
expressed according to the International System for Human
Cytogenetic Nomenclature 2009 (15).
Molecular cytogenetic analysis
Spectral karyotyping (SKY) analysis was performed on
unstained cytogenetic preparations according to the manufacturer’s
instructions (Applied Spectral Imaging, Carlsbad, CA, USA) and as
described previously (16).
Spectral images were acquired with an SD200 spectral bio-imaging
system (ASI) and analyzed using SkyView software (ASI). Five
metaphase cells were analyzed by SKY.
High molecular weight DNA was extracted from the
fresh frozen tissue. CGH was performed as described previously
(17). Tumor DNA and reference DNA
were labelled by nick translation and hybridized karyotypically
normal male slides. The location of aberrant CGH signals was
analyzed using an image analysis system (Isis, Carl Zeiss Vision,
Oberkochen, Germany) based on an integrated high-sensitivity
monochrome charge-coupled device camera and automated CGH analysis
software (MetaSystems GmbH, Altlussheim, Germany). Based on the
control experiments, 1.2 and 0.8 were used as cutoff levels for
gains and losses, respectively. Gains exceeding the 1.5 threshold
were termed high-level amplifications. The heterochromatic regions
in chromosomes 1, 9, and 16, the p-arms of the acrocentric
chromosomes, and Y chromosome were excluded from the analysis
because of suppression of hybridization with Cot-1 DNA in these
regions. Three-color images, green (fluorescein-12-dUTP) for the
tumor DNA, red (Spectrum Red) for the reference DNA, and blue
(DAPI) for the DNA counterstain, were acquired from at least 10
metaphases.
Results
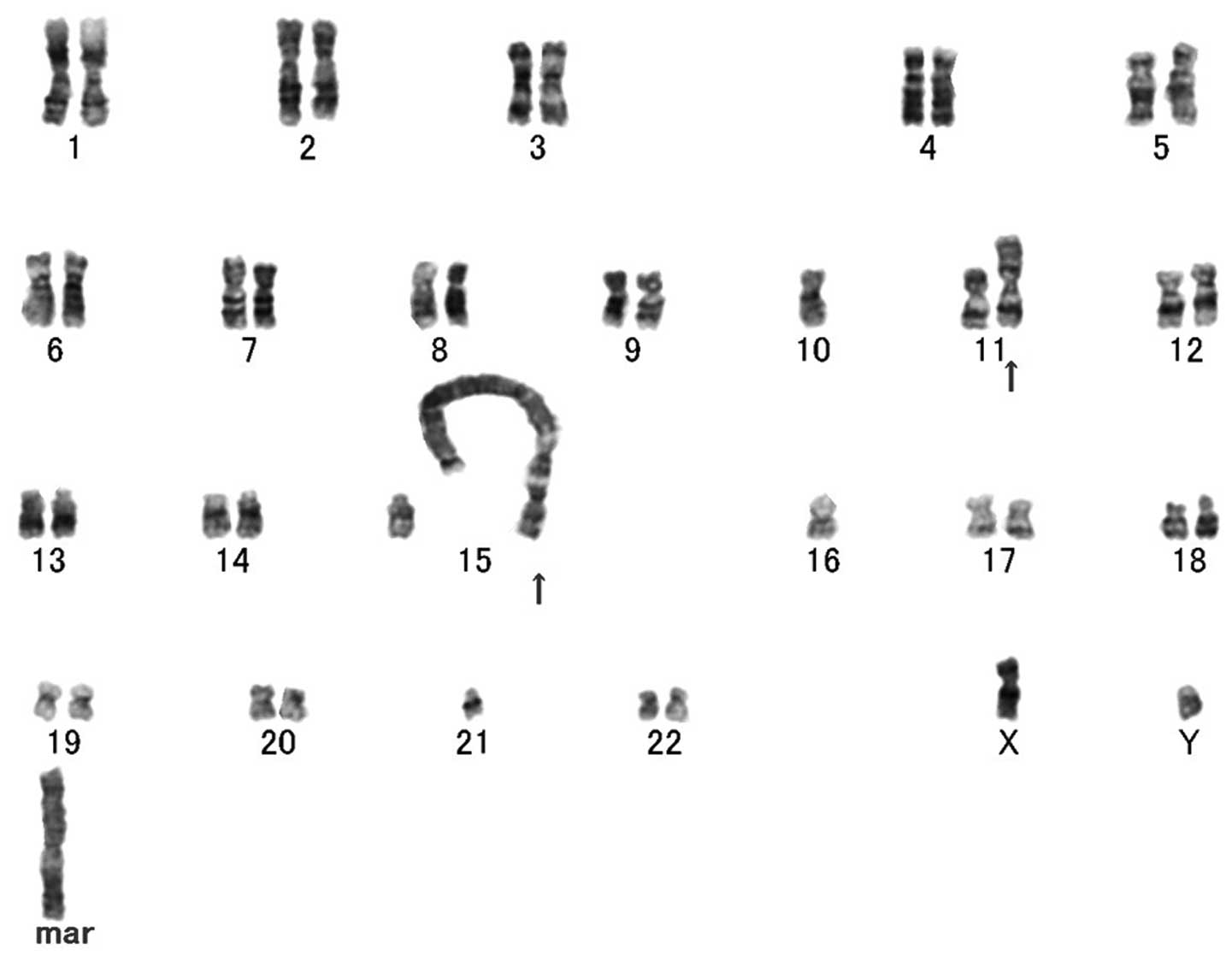
Cytogenetic analysis exhibited a complex karyotype
with several numerical and structural alterations, including giant
marker and ring chromosomes (Fig.
4). The composite karyotype was as follows:
44~48,XY,der(7;21)t(7;?)(p11.2;?)t(21;?)(p11.2;?),-8,-10,
add(11)(p13),
der(15;21)t(15;?)(p11.2;?)t(21;?)(p11.2;?),-16,-21,dic(21;?)(p11.2;?),-22,+1~2r,+1~2mar
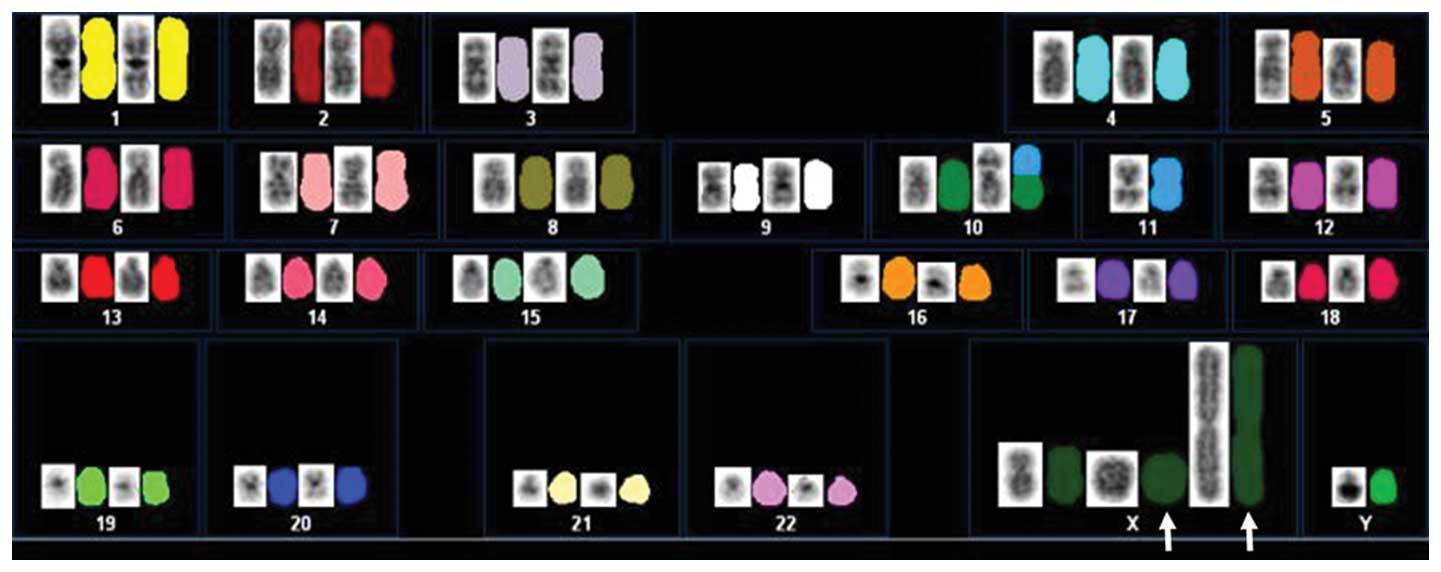
[cp20]. SKY analysis revealed that giant marker and ring
chromosomes were composed of material from X chromosome (Fig. 5).
Metaphase-based CGH analysis showed high-level
amplifications of 1q21-q25 and 12q13-q21 and gains of 1p31-p32,
10p11-p13, 17p11, and 19p13 (Fig.
6). Significant loss of DNA sequences was not detected.
Discussion
Pleomorphic leiomyosarcoma is a recently described
morphological variant of leiomyosarcoma with an aggressive clinical
course (18). On the other hand,
the term ‘dedifferentiated leiomyosarcoma’ has also been used to
describe leiomyosarcoma with a poorly differentiated pleomorphic
component (2,9,19). The
pleomorphic component of dedifferentiated leiomyosarcoma is
immunohistochemically negative for all muscle markers. Based on
staining for muscle markers in the pleomorphic component, we
diagnosed the present case as pleomorphic leiomyosarcoma.
Giant marker and/or ring chromosomes have been found
in a variety of human neoplasms, but they are particularly common
in a subgroup of low-grade or borderline malignant mesenchymal
tumors, such as well-differentiated liposarcoma/atypical lipomatous
tumor (20). The origin of these
chromosomes cannot be disclosed by chromosome banding techniques
due to the suboptimal banding quality and morphological
variability. In the present study, we used SKY technique to
identify the origin of giant marker and ring chromosomes in a
pleomorphic soft tissue leiomyosarcoma. SKY analysis showed that
giant marker and ring chromosomes were derived from X chromosome.
Interestingly, high-level amplification of Xp has been described in
soft tissue and non-soft tissue leiomyosarcomas (6,21–23).
The amplified material is frequently carried in mitotically
unstable dicentric, multicentric, or ring-shaped chromosomes
(24,25). Based on these findings, we
speculated that ring and/or giant marker chromosomes may contain
amplified material from X chromosome. However, no amplification or
gain of X chromosome was detected by CGH. In general, telomeres
play a key role in the maintenance of chromosomal stability. Many
human neoplasms exhibit significantly shortened telomeric repeat
sequences, which may trigger the formation of telomeric fusions
between chromosome arms (26). We
suggest that such fusions may lead to ring chromosomes in the
present case.
High-level amplifications of 1q21-q25 and 12q13-q21
were found in the present case. Coamplification of these chromosome
regions has been observed in several malignant mesenchymal tumors
including leiomyosarcoma (14,27,28).
Amplification of COAS and PRUNE genes, located at
1q21, has been identified in a subset of leiomyosarcomas (28,29).
El-Rifai et al(6) reported
that 1q gain was seen in large and very large tumors, but not in
any of the small tumors. On the other hand, Ragazzini et
al(30) demonstrated that
MDM2, CDK4, and TSPAN31 (formerly known as
SAS) genes, located at 12q13-q15, are amplified or
overrepresented in at least a subset of pleomorphic soft tissue
leiomyosarcomas. These gene alterations seem to play an important
role in pleomorphic leiomyosarcoma development, but no clear
relationship with clinical outcome has yet been found.
In addition to the amplifications of 1q and 12q,
gains/amplifications of 17p and 19p have been observed in soft
tissue and non-soft tissue leiomyosarcomas, as in our case
(6,11,12,21–23,31).
Pérot et al(32)
demonstrated that MYOCD gene, located at 17p11.2, is highly
amplified and overexpressed in retroperitoneal leiomyosarcomas.
MYOCD encodes a smooth muscle-specific transcriptional
co-activator of the serum response factor (33–35).
Moreover, they showed that MYOCD expression in sarcoma cell lines
not only induces smooth muscle differentiation but also promotes
cell migration. These data suggest that MYOCD might constitute a
promising therapeutic target. On the other hand, there are several
candidate targets on 19p, including LPSA, MAP2K2,
CDKN2D, and NOTCH3(36–38).
Choong et al(39) found that
19p13 rearrangements were associated with an increased risk of
local recurrence and/or metastasis in MFH. However, the biological
significance of 19p aberrations is still unclear in pleomorphic
leiomyosarcomas.
Losses of 10q and 13q have also been reported to be
recurrent alterations in pleomorphic soft tissue leiomyosarcomas
(10,11,40).
The tumor suppressor genes PTEN and RB1 are located
at 10q23 and 13q14, respectively. Functional inactivation of these
genes may play a critical role in the development of leiomyosarcoma
(41,42). Of note, Hu et al(11) reported that loss of 10q was
associated with an aggressive behavior of soft tissue
leiomyosarcoma. Loss of 13q14-q21 was also found to be associated
with a shortened survival (10).
In summary, our study demonstrates the constitution
of giant marker and ring chromosomes in a pleomorphic
leiomyosarcoma of soft tissue, and suggests that X chromosome
rearrangements may play a role in the development of this
tumor.
Acknowledgements
This study was supported in part by the Fukuoka
Cancer Society and Medical Care Education Research Foundation.
References
|
1
|
Oda Y, Miyajima K, Kawaguchi K, Tamiya S,
Oshiro Y, Hachitanda Y, Oya M, Iwamoto Y and Tsuneyoshi M:
Pleomorphic leiomyosarcoma: clinicopathologic and
immunohistochemical study with special emphasis on its distinction
from ordinary leiomyosarcoma and malignant fibrous histiocytoma. Am
J Surg Pathol. 25:1030–1038. 2001. View Article : Google Scholar
|
|
2
|
Nicolas MM, Tamboli P, Gomez JA and
Czerniak BA: Pleomorphic and dedifferentiated leiomyosarcoma:
clinicopathologic and immunohistochemical study of 41 cases. Hum
Pathol. 41:663–671. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Mertens F, Fletcher CD, Dal Cin P, De
Wever I, Mandahl N, Mitelman F, Rosai J, Rydholm A, Sciot R,
Tallini G, et al: Cytogenetic analysis of 46 pleomorphic soft
tissue sarcomas and correlation with morphologic and clinical
features: a report of the CHAMP Study Group. Genes Chromosomes
Cancer. 22:16–25. 1998. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Mandahl N, Fletcher CD, Dal Cin P, De
Wever I, Mertens F, Mitelman F, Rosai J, Rydholm A, Sciot R,
Tallini G, et al: Comparative cytogenetic study of spindle cell and
pleomorphic leiomyosarcomas of soft tissues: a report from the
CHAMP Study Group. Cancer Genet Cytogenet. 116:66–73. 2000.
View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Sandberg AA: Updates on the cytogenetics
and molecular genetics of bone and soft tissue tumors:
leiomyosarcoma. Cancer Genet Cytogenet. 161:1–19. 2005. View Article : Google Scholar
|
|
6
|
El-Rifai W, Sarlomo-Rikala M, Knuutila S
and Miettinen M: DNA copy number changes in development and
progression in leiomyosarcomas of soft tissues. Am J Pathol.
153:985–990. 1998. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Wang R, Lu YJ, Fisher C, Bridge JA and
Shipley J: Characterization of chromosome aberrations associated
with soft-tissue leiomyosarcomas by twenty-four-color karyotyping
and comparative genomic hybridization analysis. Genes Chromosomes
Cancer. 31:54–64. 2001. View
Article : Google Scholar
|
|
8
|
Derré J, Lagacé R, Nicolas A, Mairal A,
Chibon F, Coindre JM, Terrier P, Sastre X and Aurias A:
Leiomyosarcomas and most malignant fibrous histiocytomas share very
similar comparative genomic hybridization imbalances: an analysis
of a series of 27 leiomyosarcomas. Lab Invest. 81:211–215.
2001.
|
|
9
|
Evans HL and Shipley J: Leiomyosarcoma.
World Health Organization Classification of Tumours: Pathology and
Genetics of Tumours of Soft Tissue and Bone. Fletcher CDM, Unni KK
and Mertens F: IARC Press; Lyon: pp. 131–134. 2002
|
|
10
|
Wang R, Titley JC, Lu YJ, Summersgill BM,
Bridge JA, Fisher C and Shipley J: Loss of 13q14-q21 and gain of
5p14-pter in the progression of leiomyosarcoma. Mod Pathol.
16:778–785. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Hu J, Rao UN, Jasani S, Khanna V, Yaw K
and Surti U: Loss of DNA copy number of 10q is associated with
aggressive behavior of leiomyosarcomas: a comparative genomic
hybridization study. Cancer Genet Cytogenet. 161:20–27. 2005.
View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Larramendy ML, Kaur S, Svarvar C, Böhling
T and Knuutila S: Gene copy number profiling of soft-tissue
leiomyosarcomas by array-comparative genomic hybridization. Cancer
Genet Cytogenet. 169:94–101. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Yang J, Du X, Chen K, Ylipää A, Lazar AJ,
Trent J, Lev D, Pollock R, Hao X, Hunt K and Zhang W: Genetic
aberrations in soft tissue leiomyosarcoma. Cancer Lett. 275:1–8.
2009. View Article : Google Scholar
|
|
14
|
Nishio J, Aoki M, Nabeshima K, Iwasaki H
and Naito M: Cytogenetic and molecular cytogenetic findings in
giant dedifferentiated liposarcoma of the thigh. Oncol Rep.
27:764–768. 2012.PubMed/NCBI
|
|
15
|
Shaffer LG, Slovak ML and Campbell LJ:
ISCN (2009): An International System for Human Cytogenetic
Nomenclature. Karger; Basel: 2009
|
|
16
|
Nishio J, Iwasaki H, Althof PA, Naumann S,
Ishiguro M, Haraoka S, Iwashita A, Iwasaki A, Kaku Y, Kaneko Y, et
al: Identification of a ring chromosome with spectral karyotyping
in a pleural synovial sarcoma. Cancer Genet Cytogenet. 160:174–178.
2005. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Nishio J, Iwasaki H, Ohjimi Y, Ishiguro M,
Koga T, Isayama T, Naito M and Kikuchi M: Gain of Xq detected by
comparative genomic hybridization in elastofibroma. Int J Mol Med.
10:277–280. 2002.PubMed/NCBI
|
|
18
|
Fletcher CDM: Pleomorphic malignant
fibrous histiocytoma: fact or fiction? A critical reappraisal based
on 159 tumors diagnosed as pleomorphic sarcoma. Am J Surg Pathol.
16:213–218. 1992. View Article : Google Scholar
|
|
19
|
Chen E, O’Connell F and Fletcher CDM:
Dedifferentiated leiomyosarcoma: clinicopathological analysis of 18
cases. Histopathology. 59:1135–1143. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Nishio J: Contributions of cytogenetics
and molecular cytogenetics to the diagnosis of adipocytic tumors. J
Biomed Biotechnol. 2011:5240672011. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Otaño-Joos M, Mechtersheimer G, Ohl S,
Wilgenbus KK, Scheurlen W, Lehnert T, Willeke F, Otto HF, Lichter P
and Joos S: Detection of chromosomal imbalances in leiomyosarcoma
by comparative genomic hybridization and interphase cytogenetics.
Cytogenet Cell Genet. 90:86–92. 2000.PubMed/NCBI
|
|
22
|
Levy B, Mukherjee T and Hirschhorn K:
Molecular cytogenetic analysis of uterine leiomyoma and
leiomyosarcoma by comparative genomic hybridization. Cancer Genet
Cytogenet. 121:1–8. 2000. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Hu J, Khanna V, Jones M and Surti U:
Genomic alterations in uterine leiomyosarcomas: potential markers
for clinical diagnosis and prognosis. Genes Chromosomes Cancer.
31:117–124. 2001. View
Article : Google Scholar : PubMed/NCBI
|
|
24
|
Gisselsson D, Björk J, Höglund M, Mertens
F, Dal Cin P, Akerman M and Mandahl N: Abnormal nuclear shape in
solid tumors reflects mitotic instability. Am J Pathol.
158:199–206. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Iwasaki H, Nabeshima K, Nishio J, Jimi S,
Aoki M, Koga K, Hamasaki M, Hayashi H and Mogi A: Pathology of
soft-tissue tumors: daily diagnosis, molecular cytogenetics and
experimental approach. Pathol Int. 59:501–521. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Gisselsson D, Jonson T, Petersén A,
Strömbeck B, Dal Cin P, Höglund M, Mitelman F, Mertens F and
Mandahl N: Telomere dysfunction triggers extensive DNA
fragmentation and evolution of complex chromosome abnormalities in
human malignant tumors. Proc Natl Acad Sci USA. 98:12683–12688.
2001. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Forus A, Weghuis DO, Smeets D, Fodstad O,
Myklebost O and van Kessel AG: Comparative genomic hybridization
analysis of human sarcomas: I. Occurrence of genomic imbalances and
identification of a novel major amplicon at 1q21-q22 in soft tissue
sarcomas. Genes Chromosomes Cancer. 14:8–14. 1995. View Article : Google Scholar
|
|
28
|
Nilsson M, Meza-Zepeda LA, Mertens F,
Forus A, Myklebost O and Mandahl N: Amplification of chromosome 1
sequences in lipomatous tumors and other sarcomas. Int J Cancer.
109:363–369. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Forus A, D’Angelo A, Henriksen J, Merla G,
Maelandsmo GM, Flørenes VA, Olivieri S, Bjerkehagen B, Meza-Zepeda
LA, del Vecchio Blanco F, et al: Amplification and overexpression
of PRUNE in human sarcomas and breast carcinomas - a possible
mechanism for altering the nm23-H1 activity. Oncogene.
20:6881–6890. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Ragazzini P, Gamberi G, Pazzaglia L, Serra
M, Magagnoli G, Ponticelli F, Ferrari C, Ghinelli C, Alberghini M,
Bertoni F, et al: Amplification of CDK4, MDM2, SAS and GLI genes in
leiomyosarcoma, alveolar and embryonal rhabdomyosarcoma. Histol
Histopathol. 19:401–411. 2004.PubMed/NCBI
|
|
31
|
Meza-Zepeda LA, Kresse SH,
Barragan-Polania AH, Bjerkehagen B, Ohnstad HO, Namløs HM, Wang J,
Kristiansen BE and Myklebost O: Array comparative genomic
hybridization reveals distinct DNA copy number differences between
gastrointestinal stromal tumors and leiomyosarcomas. Cancer Res.
66:8984–8993. 2006. View Article : Google Scholar
|
|
32
|
Pérot G, Derré J, Coindre JM, Tirode F,
Lucchesi C, Mariani O, Gibault L, Guillou L, Terrier P and Aurias
A: Strong smooth muscle differentiation is dependent on myocardin
gene amplification in most human retroperitoneal leiomyosarcomas.
Cancer Res. 69:2269–2278. 2009.PubMed/NCBI
|
|
33
|
Du KL, Ip HS, Li J, Chen M, Dandre F, Yu
W, Lu MM, Owens GK and Parmacek MS: Myocardin is a critical serum
response factor cofactor in the transcriptional program regulating
smooth muscle cell differentiation. Mol Cell Biol. 23:2425–2437.
2003. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Wang Z, Wang DZ, Pipes GC and Olson EN:
Myocardin is a master regulator of smooth muscle gene expression.
Proc Natl Acad Sci USA. 100:7129–7134. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Pipes GC, Creemers EE and Olson EN: The
myocardin family of transcriptional coactivators: versatile
regulators of cell growth, migration, and myogenesis. Genes Dev.
20:1545–1556. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Gill S, Stratton MR, Patterson H, Spurr
NK, Fisher C, Gusterson BA and Cooper CS: Detection of transforming
genes by transfection of DNA from primary soft-tissue tumours.
Oncogene. 6:1651–1656. 1991.PubMed/NCBI
|
|
37
|
Pryor JG, Brown-Kipphut BA, Iqbal A and
Scott GA: Microarray comparative genomic hybridization detection of
copy number changes in desmoplastic melanoma and malignant
peripheral nerve sheath tumor. Am J Dermatopathol. 33:780–785.
2011. View Article : Google Scholar
|
|
38
|
Bellavia D, Checquolo S, Campese AF, Felli
MP, Gulino A and Screpanti I: Notch3: from subtle structural
differences to functional diversity. Oncogene. 27:5092–5098. 2008.
View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Choong PF, Mandahl N, Mertens F, Willén H,
Alvegård T, Kreicbergs A, Mitelman F and Rydholm A: 19p+ marker
chromosome correlates with relapse in malignant fibrous
histiocytoma. Genes Chromosomes Cancer. 16:88–93. 1996.
|
|
40
|
Larramendy ML, Gentile M, Soloneski S,
Knuutila S and Böhling T: Does comparative genomic hybridization
reveal distinct differences in DNA copy number sequence patterns
between leiomyosarcoma and malignant fibrous histiocytoma? Cancer
Genet Cytogenet. 187:1–11. 2008. View Article : Google Scholar
|
|
41
|
Dei Tos AP, Maestro R, Doglioni C,
Piccinin S, Libera DD, Boiocchi M and Fletcher CDM: Tumor
suppressor genes and related molecules in leiomyosarcoma. Am J
Pathol. 148:1037–1045. 1996.PubMed/NCBI
|
|
42
|
Hernando E, Charytonowicz E, Dudas ME,
Menendez S, Matushansky I, Mills J, Socci ND, Behrendt N, Ma L,
Maki RG, et al: The AKT-mTOR pathway plays a critical role in the
development of leiomyosarcomas. Nat Med. 13:748–753. 2007.
View Article : Google Scholar : PubMed/NCBI
|