Introduction
Female breast cancer is the most commonly diagnosed
cancer with more than one million new cases every year and breast
cancer is one of the leading causes of cancer-related death in
women (1). The
phosphatidylinositol-3-kinase (PI3K) pathway is a major signaling
pathway in cells and is involved in essential cell processes such
as metabolism, survival, proliferation, growth and motility
(2). Dysregulation of the PI3K
pathway occurs in a large variety of human cancers (3) and has been proven to be implicated in
breast cancer development and progression (4). PI3K converts
phosphatidylinositol-4,5-bisphosphate (PIP2) to
phosphatidylinositol-3,4,5-triphosphate (PIP3). The phosphatase and
tensin homolog (PTEN) antagonizes and negatively regulates PI3K by
converting PIP3 to PIP2 (5).
The PI3K/AKT/mTOR pathway appears to have a major
role in the response to treatment and in the development of
resistance to anticancer drugs. Overactivation of the PI3K pathway
downstream of human epidermal growth factor receptors (HER) can be
driven by mutations of PI3K, an enzyme from the lipid kinase family
involved in cell signaling. These activating mutations occur mainly
on p110α, the catalytic subunit of PI3K encoded by the
PIK3CA (phosphoinositide-3-kinase, catalytic, α polypeptide)
gene located on chromosome 3. PIK3CA mutations are present
in 25% of breast carcinomas, and the most common activating
mutations occur on exons 9 and 20 according to the COSMIC database
(Catalogue Of Somatic Mutations in Cancer Database, Wellcome Trust
Genome Campus, Hinxton, Cambridge; accessed June 2012; http://www.sanger.ac.uk). More precisely, E542K
(c.1624G→A, p.Glu542Lys), E545K (c.1633G→A, p.Glu545Lys) in exon 9
and H1047R (c.3140A→G, p.His1047Arg), H1047L (c.3140A→T,
p.His1047Leu), represent more than 90% of the mutations encountered
in breast carcinomas. These four mutations have been shown to have
an oncogenic role in breast cancers (6–9).
Recent studies have shown that PI3K may be
implicated in the resistance of breast cancers to anti-estrogen
therapy agents (10,11), anti-HER2 tyrosine kinase inhibitor
(lapatinib) (12) and anti-HER2
monoclonal antibody (trastuzumab) (5,11,13).
Mutations of PIK3CA and loss of the PTEN protein are keys
factors in the development of resistance to these drugs (14–16).
Moreover, lapatinib and trastuzumab resistance can occur in
HER2-amplified breast cancers bearing a PIK3CA mutation
(5,14,15).
HER2 overexpression in breast cancers is present in 15–25% of
tumors (17,18). Approximately 75% of the breast
cancers express estrogen receptors and/or progesterone receptors
and a relationship between anti-estrogen resistance and activation
of the PI3K pathway has recently been found (11). These new findings imply that the
PI3K pathway may be an important target for novel targeted
therapies.
New anti-PI3K and anti-mTOR drugs are currently
under development (19,20) for breast cancer treatment.
Activation of the PI3K/AKT pathway and overexpression of PI3K may
play a major role in the use of new therapeutic schemes, and
identification of PIK3CA mutations could be a major
biomarker for predicting the response to these new therapies.
In light of these issues, there is a huge interest
in developing rapid, reliable and sensitive methods that can be
used for clinical routine detection of PIK3CA mutations in
breast tumors. In the present study, we used a polymerase chain
reaction (PCR)-high resolution melting assay (HRM) and a
PCR-amplification refractory mutation system (ARMS) to analyze
alcohol-formalin-acetic acid (AFA)-fixed paraffin-embedded breast
tumor specimens. PCR-HRM is a cost-effective post-PCR method that
enables the identification of alterations in single nucleotides,
i.e., mutations through the analysis of thermal denaturation of
double-stranded DNA. PCR-ARMS is a powerful mutation-specific
real-time PCR-based technique combining ARMS and a bi-functional
fluorescent probe/primer molecule (Scorpion).
The present study evaluated the relationship between
PIK3CA exon 9 and 20 mutations and conventional
clinicopathological criteria and compared the sensitivity of the
two techniques by examining the correlation of the results achieved
using both methods. The final goal was to validate a double
technique approach, ensuring a cost-effective, rapid process
yielding a high quality level of analysis, according to the
recommendations of the French National Cancer Institute (INCa) for
clinical routine analysis of mutations in tumors in a
treatment-choosing process. In such a context, PCR-HRM could be
proposed to determine the PIK3CA mutational status yielding
binary results (mutated or wild-type) and PCR-ARMS-Scorpion to
accurately identify the four main hotspot mutations of
PIK3CA.
Patients and methods
Population
One hundred and forty-nine invasive breast carcinoma
tumor specimens, from patients diagnosed between 2008 and 2009,
were retrospectively included in this study. All specimens were
collected as AFA-fixed paraffin-embedded tissues from our
institutional Biobank. The tumor characteristics of this population
were consistent with literature data regarding mean age at
diagnosis, histological type (ductal and lobular),
Scarff-Bloom-Richardson (SBR) grade as well as hormone receptor and
HER2 status (Tables I and II). According to the Biobank procedure of
our institute, all women involved in this study were informed that
their tumor samples might be used for research purposes and had the
opportunity to decline. No opposition was expressed.
 | Table IRelationship between the
PIK3CA mutation status analyzed using PCR-HRM and main
standard clinicopathological and biological characteristics of the
breast cancer cases. |
Table I
Relationship between the
PIK3CA mutation status analyzed using PCR-HRM and main
standard clinicopathological and biological characteristics of the
breast cancer cases.
| | No. of patients
(%) | |
|---|
| |
| |
|---|
| Total/class | PIK3CA
wild-type | PIK3CA
mutated | P-valuea |
|---|
| Total | 118 (100.0) | 88 (72.5) | 30 (27.5) | |
| Age at diagnosis
(years) |
| ≤50 | 36 (30.5)f | 26 (72.2)g | 10 (27.8) | 0.873 |
| >50 | 82 (69.5) | 62 (75.6) | 20 (24.4) | |
| Tumor
histology |
| Ductal | 102 (86.4) | 73 (71.6) | 29 (28.4) | 0.137 |
| Lobular | 15 (12.7) | 14 (93.3) | 1 (6.7) | |
| Others | 1 (0.8) | 1 (100.0) | 0 (0.0) | |
| Estrogen receptor
(ER) α statusb |
| Positive | 94 (81.7) | 71 (75.5) | 23 (24.5) | 0.910 |
| Negative | 21 (19.3) | 15 (71.4) | 6 (28.6) | |
| Progesterone
receptor (PR) statusb |
| Positive | 74 (64.3) | 57 (77.0) | 17 (23.0) | 0.603 |
| Negative | 41 (35.7) | 29 (70.7) | 12 (29.3) | |
| HER2 statusb |
| Positivec | 12 (10.4) | 9 (75.0) | 3 (25.0) | 1.000 |
| Negative | 103 (89.6) | 77 (74.8) | 26 (25.2) | |
| Hormone receptor
(HR) statusb,d |
| Positive | 95 (82.3) | 72 (75.8) | 23 (24.2) | 0.796 |
| Negative | 20 (27.7) | 14 (70.0) | 6 (30.0) | |
| Combined HER and HR
statusb |
|
HER2+/HR+ | 8 (7.0) | 7 (87.5) | 1 (12.5) | 0.575 |
|
HER2−/HR+ | 87 (7.6) | 65 (74.7) | 22 (25.3) | |
|
HER2+/HR− | 4 (3.5) | 2 (50.0) | 2 (50.0) | |
|
HER2−/HR− | 16 (13.9) | 12 (75.0) | 4 (25.0) | |
| SBR gradee |
| I | 14 (11.9) | 8 (57.1) | 6 (42.9) | 0.050 |
| II | 56 (47.5) | 39 (69.6) | 17 (30.4) | |
| III | 48 (40.7) | 41 (85.4) | 7 (14.6) | |
 | Table IIRelationship between PIK3CA
mutation status analyzed using PCR-ARMS and the main standard
clinicopathological and biological characteristics of the breast
cancer cases. |
Table II
Relationship between PIK3CA
mutation status analyzed using PCR-ARMS and the main standard
clinicopathological and biological characteristics of the breast
cancer cases.
| | No. of patients
(%) | |
|---|
| |
| |
|---|
| Total/class | PIK3CA
wild-type | PIK3CA
mutated | P-valuea |
|---|
| Total | 149 (100.0) | 122 (81.9) | 27 (18.1) | |
| Age at diagnosis
(years) |
| ≤50 | 48 (30.5)f | 36 (72.2)g | 12 (27.8) | 0.202 |
| >50 | 101 (69.5) | 86 (75.6) | 15 (24.4) | |
| Tumor
histology |
| Ductal | 127 (85.2) | 102 (71.6) | 25 (28.4) | 0.521 |
| Lobular | 19 (12.8) | 17 (93.3) | 2 (6.7) | |
| Others | 3 (2.0) | 2 (66.7) | 1 (33.3) | |
| Estrogen receptor
(ER) α statusb |
| Positive | 113 (81.7) | 93 (75.5) | 20 (24.5) | 0.981 |
| Negative | 30 (19.3) | 24 (71.4) | 6 (28.6) | |
| Progesterone
receptor (PR) statusb |
| Positive | 88 (64.3) | 71 (77.0) | 17 (23.0) | 0.823 |
| Negative | 55 (35.7) | 46 (70.7) | 9 (29.3) | |
| HER2 statusb |
| Positivec | 19 (10.4) | 16 (75.0) | 3 (25.0) | 1.000 |
| Negative | 124 (89.6) | 101 (74.8) | 23 (25.2) | |
| Hormone receptor
(HR) statusb,d |
| Positive | 115 (82.3) | 95 (75.8) | 20 (24.2) | 0.823 |
| Negative | 28 (27.7) | 22 (70.0) | 6 (30.0) | |
| Combined HER and HR
statusb |
|
HER2+/HR+ | 13 (7.0) | 12 (87.5) | 1 (12.5) | 0.593 |
|
HER2−/HR+ | 102 (7.6) | 83 (74.7) | 19 (25.3) | |
|
HER2+/HR− | 6 (3.5) | 4 (50.0) | 2 (50.0) | |
|
HER2−/HR− | 22 (13.9) | 18 (75.0) | 4 (25.0) | |
| SBR gradee |
| I | 14 (11.9) | 7 (57.1) | 7 (42.9) | 0.004 |
| II | 67 (47.5) | 55 (69.6) | 12 (30.4) | |
| III | 64 (40.7) | 56 (85.4) | 8 (14.6) | |
DNA extraction
For each tumor specimen, hematoxylin and eosin slide
analysis was conducted by a pathologist to ensure a minimum of 20%
tumor tissue content as recommended in previous studies (21). Selected areas were macrodissected
and 5 10-μm serial sections were cut from each paraffin block and
collected in RNAase DNAase-free vials (SafeSeal Microcentrifuge
Tubes, Sorenson Biosciences, Salt Lake City, UT, USA). Paraffin was
removed by extraction with toluene (VWR BDH Prolabo, Fontenay Sous
Bois, France) and centrifuged. DNA isolation was performed using
the QIAamp DNA FFPE tissue kit (Qiagen, Courtaboeuf, France)
protocol. The pellet was washed with ethanol, centrifuged and
resuspended with 180 μl of tissue lysis buffer (ATL buffer; Qiagen)
and 20 μl of proteinase K (Qiagen). The sample was then gently
mixed, incubated at 56°C for 1 h and at 90°C for 1 h under
agitation. DNA was extracted with MinElute Columns (Qiagen) as
recommended by the manufacturer. The nucleic acids were eluted in a
volume of 100 μl. Final concentration of eluates, ranging from 33.6
to 729.0 ng/ml, were suitable for PCR-HRM and PCR-ARMS analyses
(Table III). DNA extracts from
cell lines bearing E542K (Cal51), E545K (MCF7), H1047R (HCT116) and
H1047R (SUM159PT) PI3-kinase mutations were used as positive
controls. DNA extracted from the MDA231 cell line was used as a
wild-type negative control. DNA quality was controlled using
agarose gel electrophoresis with GAPDH
(glyceraldehyde-3-phosphate dehydrogenase) as the control
housekeeping gene. Only DNA with no degradation was used. DNA
concentrations were determined using a Bio-photometer (Eppendorf,
Hamburg, Germany).
 | Table IIIContingency table of the 102 samples
analyzed by combined PCR-HRM and PCR-ARMS assays. |
Table III
Contingency table of the 102 samples
analyzed by combined PCR-HRM and PCR-ARMS assays.
| Sample no. | Sample DNA
concentration (ng/μl) | PCR-HRM | PCR-ARMS |
|---|
|
|
|---|
| Test result | Mutation
result | Test result | Mutation
result |
|---|
| 1 | 33.6 | Mutation not
detected | N/A | Mutation not
detected | N/A |
| 2 | 61.2 | Mutation not
detected | N/A | Mutation not
detected | N/A |
| 3 | 95.9 | Mutation
detected | Exon 20 | Mutation
detected | c.3140A→G |
| 4 | 64.3 | Mutation not
detected | N/A | Mutation not
detected | N/A |
| 5 | 105.2 | Mutation not
detected | N/A | Mutation not
detected | N/A |
| 6 | 164.4 | Mutation not
detected | N/A | Mutation not
detected | N/A |
| 7 | 71.1 | Mutation
detected | Exon 9 | Mutation
detected | c.1624G→A |
| 8 | 69.8 | Mutation not
detected | N/A | Mutation not
detected | N/A |
| 9 | 95.6 | Mutation not
detected | N/A | Mutation not
detected | N/A |
| 10 | 39.9 | Mutation not
detected | N/A | Mutation not
detected | N/A |
| 11 | 68.7 | Mutation not
detected | N/A | Mutation not
detected | N/A |
| 12 | 60.7 | Mutation
detected | Exon 9 | Mutation not
detected | N/A |
| 13 | 62.0 | Mutation not
detected | N/A | Mutation not
detected | N/A |
| 14 | 108.0 | Mutation not
detected | N/A | Mutation not
detected | N/A |
| 15 | 162.7 | Mutation
detected | Exon 9 | Mutation not
detected | N/A |
| 16 | 319.9 | Mutation not
detected | N/A | Mutation not
detected | N/A |
| 17 | 221.4 | Mutation
detected | Exon 20 | Mutation not
detected | N/A |
| 18 | 109.7 | Mutation not
detected | N/A | Mutation not
detected | N/A |
| 19 | 146.3 | Mutation not
detected | N/A | Mutation not
detected | N/A |
| 20 | 201.3 | Mutation not
detected | N/A | Mutation not
detected | N/A |
| 21 | 199.3 | Mutation not
detected | N/A | Mutation not
detected | N/A |
| 22 | 224.7 | Mutation not
detected | N/A | Mutation not
detected | N/A |
| 23 | 35.5 | Mutation not
detected | N/A | Mutation not
detected | N/A |
| 24 | 201.2 | Mutation not
detected | N/A | Mutation not
detected | N/A |
| 25 | 175.8 | Mutation
detected | Exon 20 | Mutation
detected | c.31401→T |
| 26 | 104.6 | Mutation not
detected | N/A | Mutation not
detected | N/A |
| 27 | 211.4 | Mutation not
detected | N/A | Mutation not
detected | N/A |
| 28 | 662.0 | Mutation
detected | Exon 9 | Mutation
detected | c.1633G→A |
| 29 | 457.8 | Mutation not
detected | N/A | Mutation not
detected | N/A |
| 30 | 223.3 | Mutation not
detected | N/A | Mutation not
detected | N/A |
| 31 | 199.8 | Mutation not
detected | N/A | Mutation not
detected | N/A |
| 32 | 68.0 | Mutation not
detected | N/A | Mutation not
detected | N/A |
| 33 | 122.1 | Mutation not
detected | N/A | Mutation not
detected | N/A |
| 34 | 232.4 | Mutation not
detected | N/A | Mutation not
detected | N/A |
| 35 | 252.4 | Mutation
detected | Exon 20 | Mutation
detected | c.31401→T |
| 36 | 155.3 | Mutation
detected | Exon 20 | Mutation
detected | c.3140A→G |
| 37 | 75.0 | Mutation not
detected | N/A | Mutation not
detected | N/A |
| 38 | 486.5 | Mutation not
detected | N/A | Mutation not
detected | N/A |
| 39 | 164.2 | Mutation not
detected | N/A | Mutation not
detected | N/A |
| 40 | 204.1 | Mutation not
detected | N/A | Mutation not
detected | N/A |
| 41 | 124.4 | Mutation
detected | Exon 9 | Mutation not
detected | N/A |
| 42 | 291.3 | Mutation not
detected | N/A | Mutation not
detected | N/A |
| 43 | 206.5 | Mutation not
detected | N/A | Mutation not
detected | N/A |
| 44 | 67.6 | Mutation
detected | Exon 9 | Mutation
detected | c.1633G→A |
| 45 | 96.4 | Mutation
detected | Exon 20 | Mutation
detected | c.3140A→G |
| 46 | 26.9 | Mutation
detected | Exon 20 | Mutation
detected | c.31401→T |
| 47 | 99.5 | Mutation not
detected | N/A | Mutation not
detected | N/A |
| 48 | 41.4 | Mutation
detected | Exon 9 | Mutation not
detected | N/A |
| 49 | 127.8 | Mutation not
detected | N/A | Mutation not
detected | N/A |
| 50 | 184.0 | Mutation not
detected | N/A | Mutation not
detected | N/A |
| 51 | 86.9 | Mutation not
detected | N/A | Mutation not
detected | N/A |
| 52 | 499.3 | Mutation not
detected | N/A | Mutation not
detected | N/A |
| 53 | 188.5 | Mutation not
detected | N/A | Mutation not
detected | N/A |
| 54 | 140.1 | Mutation not
detected | N/A | Mutation not
detected | N/A |
| 55 | 357.4 | Mutation not
detected | N/A | Mutation not
detected | N/A |
| 56 | 252.5 | Mutation
detected | Exon 20 | Mutation
detected | c.3140A→G |
| 57 | 211.7 | Mutation not
detected | N/A | Mutation not
detected | N/A |
| 58 | 213.5 | Mutation not
detected | N/A | Mutation not
detected | N/A |
| 59 | 230.3 | Mutation
detected | Exon 20 | Mutation
detected | c.3140A→G |
| 60 | 227.3 | Mutation
detected | Exon 9 | Mutation not
detected | N/A |
| 61 | 729.0 | Mutation not
detected | N/A | Mutation not
detected | N/A |
| 62 | 144.4 | Mutation not
detected | N/A | Mutation not
detected | N/A |
| 63 | 202.3 | Mutation not
detected | N/A | Mutation not
detected | N/A |
| 64 | 45.6 | Mutation not
detected | N/A | Mutation not
detected | N/A |
| 65 | 154.0 | Mutation not
detected | N/A | Mutation not
detected | N/A |
| 66 | 301.8 | Mutation
detected | Exon 20 | Mutation
detected | c.31401→T |
| 67 | 178.7 | Mutation not
detected | N/A | Mutation not
detected | N/A |
| 68 | 151.5 | Mutation not
detected | N/A | Mutation not
detected | N/A |
| 69 | 543.7 | Mutation not
detected | N/A | Mutation not
detected | N/A |
| 70 | 70.3 | Mutation
detected | Exon 9 | Mutation
detected | c.1624G→A |
| 71 | 179.8 | Mutation not
detected | N/A | Mutation not
detected | N/A |
| 72 | 202.1 | Mutation not
detected | N/A | Mutation not
detected | N/A |
| 73 | 244.9 | Mutation
detected | Exon 9 | Mutation
detected | c.1633G→A |
| 74 | 130.5 | Mutation not
detected | N/A | Mutation not
detected | N/A |
| 75 | 256.0 | Mutation not
detected | N/A | Mutation not
detected | N/A |
| 76 | 90.2 | Mutation
notdetected | N/A | Mutation
detected |
c.1624G→A |
| 77 | 302.6 | Mutation not
detected | N/A | Mutation not
detected | N/A |
| 78 | 138.8 | Mutation not
detected | N/A | Mutation not
detected | N/A |
| 79 | 117.0 | Mutation not
detected | N/A | Mutation not
detected | N/A |
| 80 | 130.9 | Mutation not
detected | N/A | Mutation not
detected | N/A |
| 81 | 120.6 | Mutation not
detected | N/A | Mutation not
detected | N/A |
| 82 | 149.4 | Mutation not
detected | N/A | Mutation not
detected | N/A |
| 83 | 221.2 | Mutation
detected | Exon 20 | Mutation
detected | c.3140A→G |
| 84 | 107.8 | Mutation
detected | Exon 9 | Mutation
detected | c.1624G→A |
| 85 | 166.1 | Mutation not
detected | N/A | Mutation not
detected | N/A |
| 86 | 277.2 | Mutation not
detected | N/A | Mutation not
detected | N/A |
| 87 | 111.1 | Mutation not
detected | N/A | Mutation not
detected | N/A |
| 88 | 81.1 | Mutation not
detected | N/A | Mutation not
detected | N/A |
| 89 | 170.2 | Mutation not
detected | N/A | Mutation not
detected | N/A |
| 90 | 252.6 | Mutation not
detected | N/A | Mutation not
detected | N/A |
| 91 | 127.9 | Mutation
detected | Exon 20 | Mutation
detected | c.3140A→G |
| 92 | 102.3 | Mutation not
detected | N/A | Mutation not
detected | N/A |
| 93 | 111.9 | Mutation not
detected | N/A | Mutation not
detected | N/A |
| 94 | 306.6 | Mutation not
detected | N/A | Mutation not
detected | N/A |
| 95 | 96.3 | Mutation
detected | Exon 20 | Mutation
detected | c.3140A→G |
| 96 | 88.7 | Mutation not
detected | N/A | Mutation not
detected | N/A |
| 97 | 122.6 | Mutation
detected | Exon 20 | Mutation
detected | c.3140A→G |
| 98 | 222.9 | Mutation not
detected | N/A | Mutation not
detected | N/A |
| 99 | 194.4 | Mutation
detected | Exon 9 | Mutation
detected | c.1624G→A |
| 100 | 44.5 | Mutation not
detected | N/A | Mutation not
detected | N/A |
| 101 | 185.3 | Mutation
detected | Exon 20 | Mutation
netected | c.3140A→G |
| 102 | 127.3 | Mutation
detected | Exon 9 | Mutation
detected | c.1624G→A |
PCR-HRM
HRM analysis was performed using the LightCycler
480® Real-Time PCR system (Roche Diagnostics, Meylan,
France) and the LightCycler 480 HRM Master kit (Roche Diagnostics)
in 384-well plates (Roche Diagnostics). Twenty micrograms of DNA
was amplified in a final volume of 20 μl. All data and melting
curves were analyzed using LightCycler SW v. 1.5.0.39 software
(Roche Diagnostics). One mix was prepared for each exon. For each
sample, 10 μl of Master Mix (Roche Diagnostics), 2.8 μl of
MgCl2 25 mM, PCR-quality grade water and 1 μl of the
primers (forward and reverse) were added. Eighteen microliters of
mix was added to each well, and 2 μl of the sample was used for the
analysis. One set of primers were used for each of the
PIK3CA exons 9 and 20. All primers were designed as
previously described (22).
PCR-HRM was divided into different phases: a phase
of pre-incubation (10 min at 95°C) was followed by 45 cycles of
classic PCR (10 sec at 95°C, a temperature decrease from 60 to 54°C
by 0.5°C/cycle in 15 sec and, finally, 10 sec at 72°C). The PCR
phase was followed by the high resolution melting phase which
consisted of 1 min at 95°C, 1 min at 40°C and a temperature
increase by 0.2°C/sec from 65 to 95°C. A cooling phase of 1 min at
40°C was finally performed.
PCR-ARMS
ARMS analysis was performed using the LightCycler
480 Real-Time PCR system (Roche Diagnostics) in 384-well plates.
Eighty micrograms of DNA was amplified in a final volume of 20 μl.
Data and fluorescence curves were analy’ed using LightCycler SW v.
1.5.0.39 software. All primers were designed as previously
described (23). One mix was made
for each tested mutation and all samples were proceeded as simplex.
For each sample, 0.06 μl of Hot Diamond Taq polymerase (Eurogentec,
Angers, France) was added together with 2 μl of reaction buffer 10X
(Eurogentec), 3.2 μl of MgCl2 25 mM (Eurogentec), 0.4 μl
of dNTP 10 mM (Eurogentec), PCR-quality water, 0.8 μl of ARMS
primers 6.25 μM (Eurogentec) and 0.8 μl of Scorpion primers 6.25 μM
(ATD Bio, Southampton, UK). Eighteen microliters of mix was added
to each well, and 2 μl of the sample was used for the analysis. The
mix for exon 9 mutations contained ARMS control primers for exon
15, specific ARMS primers, respectively, of E542K and E545K
mutations and exon 9 and 15 specific Scorpion primers. The mix for
exon 20 mutations contained ARMS control primers for exon 15,
specific ARMS primers, respectively, of H1047R and H1047L mutations
and exon 20 and 15 specific Scorpion primers.
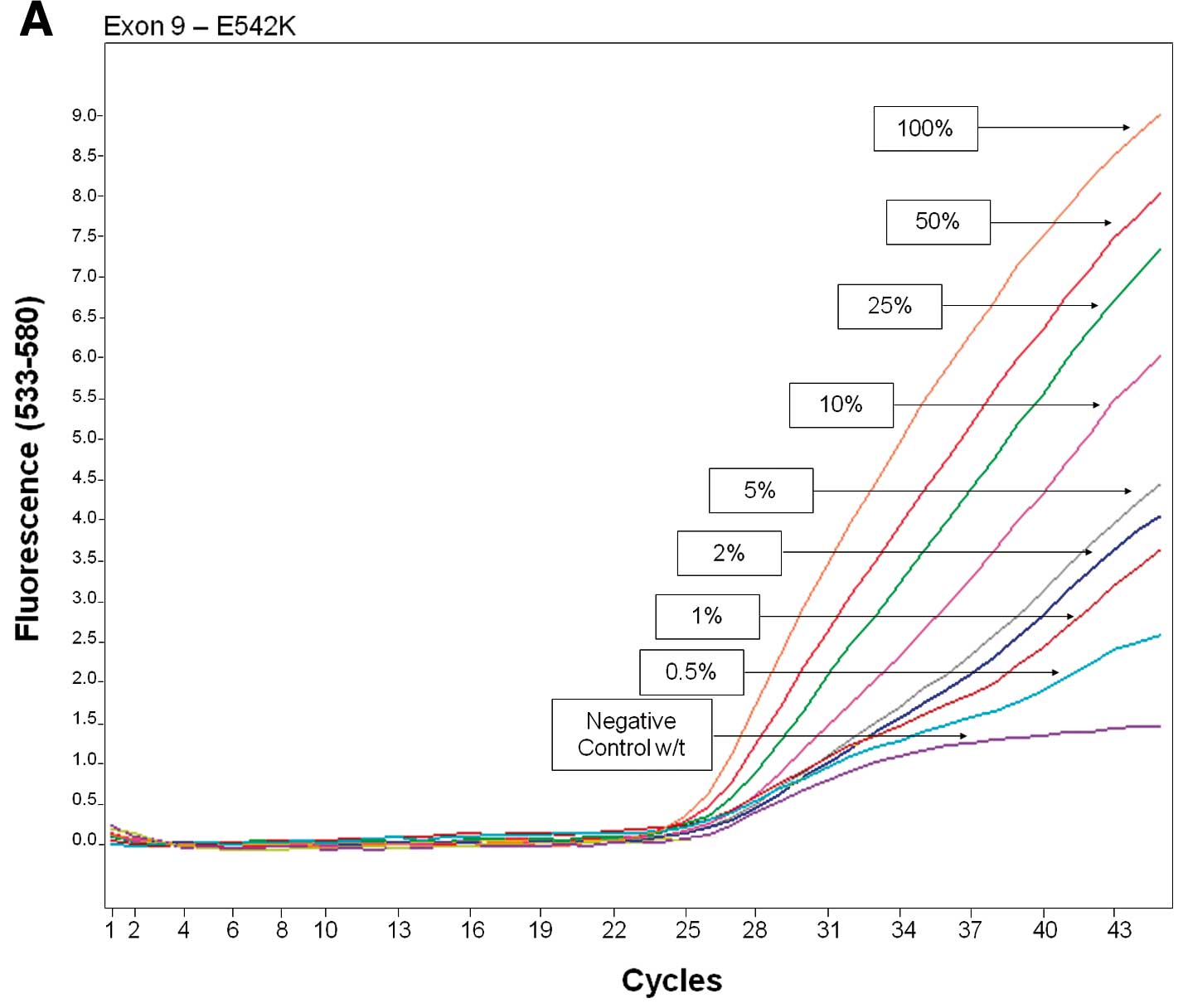
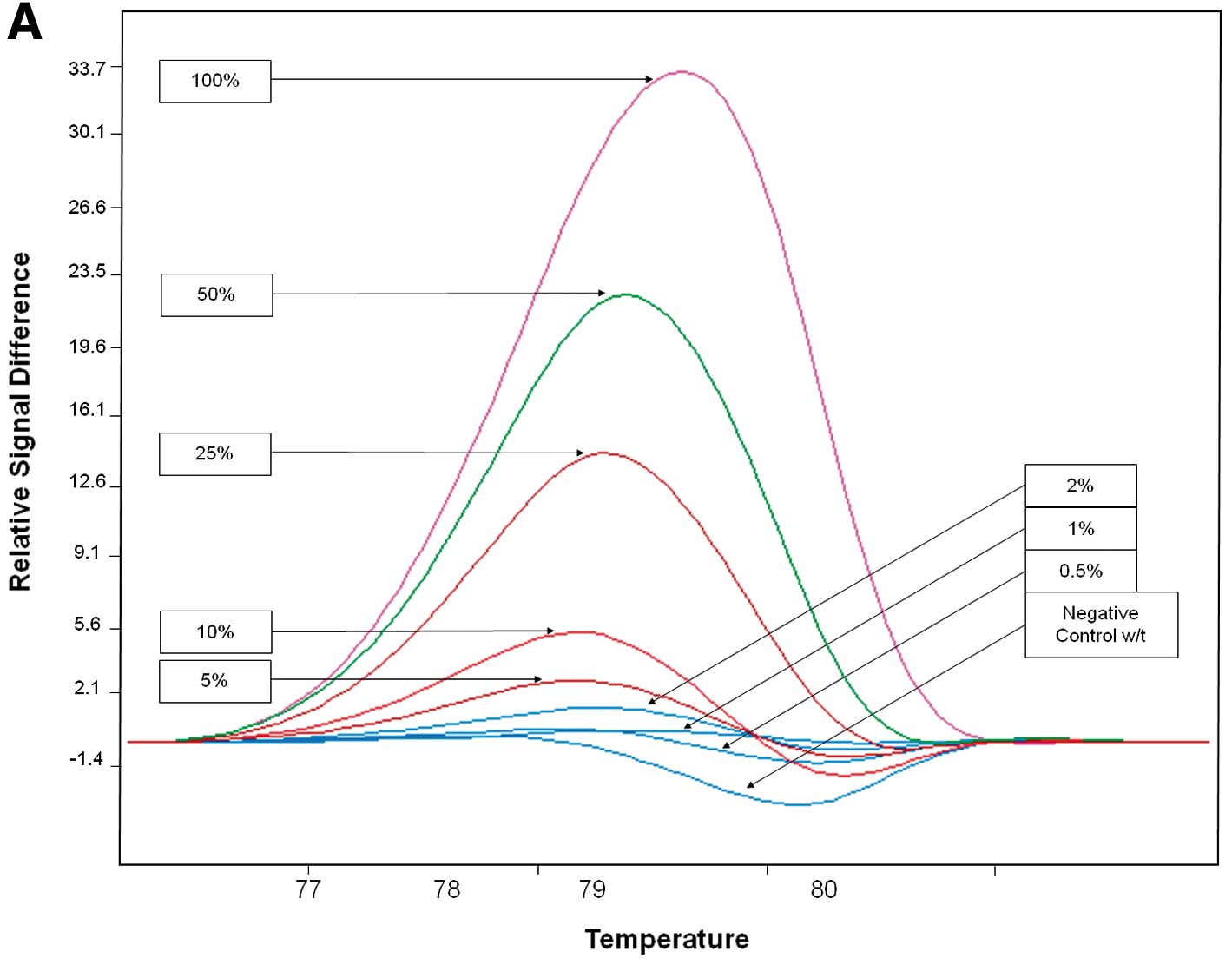
Sensitivity
The sensitivity of PCR-HRM and PCR-ARMS was
evaluated by mixing mutated and wild-type DNA from the cell lines
at 50, 25, 10, 5, 2, 1 and 0.5% ratios.
Statistical analysis
The significance of the concordance of mutation
detection using the two methods was assessed using κ statistics.
κ>0.8 was considered as indicative of significance to conclude
that both methods provide similar results. The χ2 test
was also used to compare mutation frequencies with those obtained
from the literature. χ2 and Fisher’s exact tests were
used to test for differences between classes of patients and tumors
based on clinical, pathological and biological characteristics.
Limit of statistical significance was set at P<0.05.
Results
Mutation analysis using PCR-HRM
One hundred and eighteen specimens were analyzed
using PCR-HRM (Table I).
PIK3CA mutations (exons 9 and 20) were detected in 30
(27.5%) of the specimens. No correlation was found regarding
patient age (≤50 or >50 years), ductal or luminal type, estrogen
and progesterone status (alone or combined as hormonal receptor
status), HER2 status, and for the four subtypes identified as
HER2+/HR+, HER2+/HR−,
HER2−/HR+ and HER2−/HR−
(triple negative).
PIK3CA mutations were found to be correlated
with SBR grade (P=0.050) with a lower mutation rate noted in the
highest grades. Regarding the frequency of exon 9 and 20 mutations,
a low rate of exon 9 mutations was observed in SBR grade III tumors
(P=0.025) while no difference was observed for the exon 20 mutation
rate. Exon 9 mutations were found to be more frequent in SBR grade
I tumors while exon 20 mutations were predominantly observed in SBR
grade II and III tumors.
Mutation analysis using PCR-ARMS
One hundred and forty-nine specimens were analyzed
using PCR-ARMS (Table II).
PIK3CA mutations (exons 9 and 20) were detected in 27
(18.1%) specimens. No difference was found regarding patient age
(≤50 or >50 years), ductal or luminal type, estrogen and
progesterone status (alone or combined as hormonal receptor
status), HER2 status, and for the four subtypes
(HER2+/HR+, HER2+/HR−,
HER2−/HR+ and
HER2−/HR−, i.e., triple negative). As with
PCR-HRM, PIK3CA mutations were found to be related with SBR
grade (P=0.004) with a lower mutation rate in the highest grades.
Regarding the frequency of exon 9 and 20 mutations, a lower rate of
exon 9 and exon 20 mutations was observed in SBR grade III tumors
(P=0.009). Again, exon 9 mutations were found to be predominant in
SBR grade I tumors (5/7) while exon 20 mutations were predominantly
observed in SBR grade II and III tumors (13/20).
Comparative analysis of PCR-HRM and
PCR-ARMS
One hundred and two breast tumor samples (Table III) were analyzed using both
PCR-HRM and PCR-ARMS. PIK3CA mutations were detected
(Table IV) in 28 tumors (27.5%)
when PCR-HRM was used and 23 (22.5%) when PCR-ARMS was used.
 | Table IVFrequencies of mutations detected
with combined PCR-ARMS and PCR-HRM assays. |
Table IV
Frequencies of mutations detected
with combined PCR-ARMS and PCR-HRM assays.
| Nucleotide
change | Protein change | No. of samples | Relative (%) | Total (%) |
|---|
| PCR-ARMS |
| Exon 9 |
| c.1624G→A | E542K | 6 | 26.1 | 5.9 |
| c.1633G→A | E545K | 3 | 13.0 | 2.9 |
| Exon 20 |
| c.3140A→G | H1047R | 10 | 43.5 | 9.8 |
| c.31401→T | H1047L | 4 | 17.4 | 3.9 |
| Total | | 23 | 100.0 | 22.5 |
| PCR-HRM |
| Exon 9 | Not available | 13 | 46.4 | 12.8 |
| Exon 20 | Not available | 15 | 53.6 | 14.7 |
| Total | | 28 | 100.0 | 27.5 |
Among the 28 mutated tumors (Table IV), PCR-HRM results showed that 13
(46.4%) carried a mutation on exons 9 and 15 (53.6%) on exon
20.
Among the 23 samples in which a mutation was
detected using PCR-ARMS assay (Table
IV), 9 (39.1%) carried a mutation in exon 9, identified as
c.1624G→A in 6 cases (26.1%) and as c.1633G→A in the 3 others
(13.0%). Among the remaining 14 (60.9%) specimens identified as
mutated in exon 20, 10 (43.5%) carried the c.3140A→G and 4 (17.4%)
the c.3140A→T mutation. No sample was found with mutations in both
exons 9 and 20 with any of the assays.
Sensitivity
The analytical sensitivity of each assay was
evaluated from dilution of DNA extracted from PIK3CA-mutated
cell lines into DNA from PIK3CA wild-type cell lines (MDA
231) from 0.5 to 50%. PCR-HRM was able to discriminate a dilution
corresponding to 5% of exon 9 mutated DNA and 10% of exon 20
mutated DNA (Fig. 1, Table VI). PCR-ARMS was able to
discriminate a DNA dilution corresponding to 0.5% of c.1624G→A,
0.5% of c.1633G→A, 0.5% of c.3140A→G and 0.5% of c.3140A→T mutated
DNA (Fig. 2, Table VI).
 | Table VILimits of sensitivity of PCR-HRM and
PCR-ARMS assays. |
Table VI
Limits of sensitivity of PCR-HRM and
PCR-ARMS assays.
| Nucleotide
change | Protein change | Sensitivity
(%) | DNA quantity
(ng) |
|---|
| PCR-HRM |
| Exon 9 | Not available | 5 | 1.0 |
| Exon 20 | Not available | 10 | 2.0 |
| PCR-ARMS |
| Exon 9 |
| c.1624G→A | E542K | 0.5 | 0.8 |
| c.1633G→A | E545K | 0.5 | 0.8 |
| Exon 20 |
| c.3140A→G | H1047R | 0.5 | 0.8 |
| c.31401→T | H1047L | 0.5 | 0.8 |
Results obtained using PCR-ARMS and PCR-HRM
(Table V), were found to be
statistically comparable (κ=0.845, P<0.001). Contingency table
is shown in Table III.
 | Table VSummary of the results achieved with
combined PCR-HRM and PCR-ARMS assays. |
Table V
Summary of the results achieved with
combined PCR-HRM and PCR-ARMS assays.
| PCR-HRM |
|---|
|
|
|---|
| Mutated | Wild-type | Total |
|---|
| PCR-ARMS |
| Mutated | 22 | 1 | 23 |
| Wild-type | 6 | 73 | 79 |
| Total | 28 | 74 | 102 |
No statistical difference in the frequencies of
PIK3CA mutations was found between results achieved with any
of the two assays and data from Sanger database.
Discussion
Recent studies have shown that PIK3CA
mutations play a major role in resistance to trastuzumab or
lapatinib (5) or to hormonal
therapy (11,13) of breast carcinomas and could
represent a potent response predictive marker for PI3-kinase- and
mTOR-targeted therapies (24) and
be used as a treatment-choosing parameter. PIK3CA mutations
are mostly located within exons 9 and 20 and four hotspots
(c.1624G→A, c.1633G→A, c.3140A→G and c.3140A→T) represent more than
90% of all mutations. Although the prognostic value of
PIK3CA mutations in breast cancer remains controversial, in
a recent evaluation of 2587 breast cancers cases from 12
independent studies, Dumont et al(24) reported a more favorable clinical
outcome in patients with PIK3CA mutated tumors and that
improved prognosis may pertain only to patients with a mutation in
the kinase domain of p110α and to post-menopausal women with
estrogen-positive cancers.
The relative prognostic value of exon 9 vs. 20
mutations also remains controversial. Barbareschi et
al(25), found that exon 9
mutations have a negative prognostic value while exon 20 mutations
were associated with favorable outcome while Lai et
al(26) reported exon 20
mutations as associated with poor prognosis. Furthermore, Lerma
et al(27) reported a
decrease in survival in PIK3CA exon 20-mutated and
HER2-positive patients. This study was retrospective and was based
on a small number of cases (only 6 patients in the exon 20-mutated
PIK3CA and HER2-positive group) and was never confirmed
prospectively. Finally, in a larger series of patients, Cizkova
et al(28) did not find any
difference between exon 9- and exon 20-mutated tumors regarding
metastasis-free survival. Using immunohistochemistry, Aleskandarany
et al(29) reported that
PIK3CA protein expression in invasive breast cancer was associated
with poor prognosis.
The frequencies of PIK3CA mutations reported
here, with PCR-HRM and PCR-ARMS, respectively, are fully consistent
with COSMIC database values and previously published results
reporting 18–40% of PIK3CA mutations in breast tumors
(7,26,30–33).
In the present series, with a relatively limited
number of cases, no significant correlation was observed between
PIK3CA mutations and several clinical and histopathological
criteria. Concerning the age at diagnosis, a higher PIK3CA
mutation rate was noted in older patients (34) that was not observed in other series
(25,26,28).
The same is true for the histological type of breast cancer, when
comparing ductal and lobular types. Some studies reported
PIK3CA mutations to be more frequently observed in ductal
than in lobular carcinomas (25,28,34)
while others did not (35).
Although it has been reported that PIK3CA
mutations are related to hormone receptor and HER2 status (28,34),
this issue still remains controversial since no correlation between
PIK3CA mutations and hormonal status has been observed
(25,36,37)
similarly to the results of the present study, or limited to
estrogen receptor status, but not to progesterone receptor status
(35). Similar findings have been
found concerning the correlation between PIK3CA mutations
and HER2 status (28,34) but again are controversial in other
series (25,35) or observed when HER2 is determined
using immunohistochemistry but not FISH (37). A higher frequency of PIK3CA
mutation has also been noted in low SBR grade tumors similarly to
the observation in the present series and consistently with other
studies (28,34) but contrary with others (35).
All these discrepancies among the different studies
could probably be explained by the relatively limited number of
cases that were analyzed and the different techniques that were
used for PIK3CA mutation analysis with different sensitivity
limits. This also probably explains why a large range of mutation
rates were reported. This point clearly justifies that
consideration should be given to the validation and especially the
endpoints and sensitivity limits of the assays that are routinely
used.
Combining PCR-HRM and PCR-ARMS assays, several
discrepancies were observed between the two techniques in 6 (5.9%)
samples. All were identified as PIK3CA wild-type using
PCR-ARMS and mutated using PCR-HRM; 5 samples with mutations on
exon 9 and 1 sample with mutation on exon 20. Because our PCR-ARMS
assay was designed to specifically, but only, identify the four
main PIK3CA mutations (c.1624G→A, c.1633G→A, c.3140A→G and
c.3140A→T), it is obvious that the 6 discordant cases should bear
other mutations on exon 9 or 20, that were only detected using
PCR-HRM which is an exon-specific method, able to detect all the
somatic mutations on the entire exon 9 and 20. Exon 9 and 20
mutations, not located in the four main hotspots, were reported to
represent approximately 10% of all exon 9 and 20 mutations in
breast cancers (28). Our data were
consistent with these values. In the present series, this rate of
PIK3CA mutations, differing from the four main mutations
identified by PCR-ARMS was found to be consistent with data
recently reported in breast cancer (28).
Direct Sanger sequencing of these samples could
confirm the hypothesis that they may carry another mutation but the
lack of sensitivity of Sanger sequencing (approximately 20% mutant
DNA) could be not discriminatory between a false-positive with HRM
or a false-negative with sequencing (38).
The PCR-HRM and PCR-ARMS assays used here are highly
sensitive, able to detect as low as 5% of mutated DNA for exon 9
(2.0 ng) and 10% for exon 20 (4.0 ng) and 0.5% (0.8 ng) of mutated
DNA. The PCR-ARMS assay is more sensitive than pyrosequencing-based
assay as previously described (38)
and which was able to detect 5% of mutated DNA and even higher than
another PCR-ARMS assay as previously reported (5 ng) (23). This high sensitivity could be
explained by the high specificity of the primers used for this
technique.
High sensitivity assays are required for routine
analysis of mutations in clinical specimens. Recent results
(39) achieved in colon cancer
using KRAS mutation analysis, reported that sequencing was
not sensitive enough to provide clinically relevant results
yielding two many false-negative results. The authors recommended
the use of more sensitive techniques with a detection limit
approximating 1–2% to overcome this problem.
In conclusion, the present study highlighted the
potential of PCR-HRM- and PCR-ARMS-based assays for the evaluation
of the PIK3CA mutation status in breast cancers. No
correlation was observed with patient age at diagnosis,
histological type, hormone receptor and HER2 status. PIK3CA
exon 9 and 20 mutations were found to be related to
Scarff-Bloom-Richardson (SBR) grade with a lower rate of mutations
and a higher frequency of exon 9 mutations in SBRI and exon 20
mutations in SBRII/III tumors. No difference was observed in the
frequency of the two different mutations screened for each exon in
any subcategory. Thus, we propose to use a combination of both
assays, with a screening of full exon 9 and 20 using PCR-HRM and
further identification of the four main mutations using PCR-ARMS.
Following analysis of our data, using such a procedure would have
led to only one false-negative result (<1%) which is quite
satisfactory. Combining these two assays should represent a
cost-effective rapid procedure that provides highly reliable
results that are fully consistent with the attempts of
practitioners in view of personalizing therapy for invasive breast
cancers.
Acknowledgements
The authors are grateful to the French ‘Ligue contre
le Cancer, Comités Lorrains’ for supporting this study.
References
|
1
|
Jemal A, Bray F, Center MM, Ferlay J, Ward
E and Forman D: Global cancer statistics. CA Cancer J Clin.
61:69–90. 2011. View Article : Google Scholar
|
|
2
|
Engelman JA, Luo J and Cantley LC: The
evolution of phosphatidylinositol 3-kinases as regulators of growth
and metabolism. Nat Rev Genet. 7:606–619. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Tokunaga E, Oki E, Egashira A, et al:
Deregulation of the Akt pathway in human cancer. Curr Cancer Drug
Targets. 8:27–36. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
McAuliffe PF, Meric-Bernstam F, Mills GB
and Gonzalez-Angulo AM: Deciphering the role of PI3K/Akt/mTOR
pathway in breast cancer biology and pathogenesis. Clin Breast
Cancer. 10:S59–S65. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Wang L, Zhang Q, Zhang J, et al: PI3K
pathway activation results in low efficacy of both trastuzumab and
lapatinib. BMC Cancer. 11:2482011. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Jiang BH and Liu LZ: PI3K/PTEN signaling
in angiogenesis and tumorigenesis. Adv Cancer Res. 102:19–65. 2009.
View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Bachman KE, Argani P, Samuels Y, et al:
The PIK3CA gene is mutated with high frequency in human breast
cancers. Cancer Biol Ther. 3:772–775. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Karakas B, Bachman KE and Park BH:
Mutation of the PIK3CA oncogene in human cancers. Br J Cancer.
94:455–459. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Dunlap J, Le C, Shukla A, et al:
Phosphatidylinositol-3-kinase and AKT1 mutations occur early in
breast carcinoma. Breast Cancer Res Treat. 120:409–418. 2009.
View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Orlando L, Schiavone P, Fedele P, et al:
Molecularly targeted endocrine therapies for breast cancer. Cancer
Treat Rev. 36:S67–S71. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Miller TW, Balko JM and Arteaga CL:
Phosphatidylinositol 3-kinase and antiestrogen resistance in breast
cancer. J Clin Oncol. 29:4452–4461. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Abramson V and Arteaga CL: New strategies
in HER2-overexpressing breast cancer: many combinations of targeted
drugs available. Clin Cancer Res. 17:952–958. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Ma CX, Crowder RJ and Ellis MJ: Importance
of PI3-kinase pathway in response/resistance to aromatase
inhibitors. Steroids. 76:750–752. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Razis E, Bobos M, Kotoula V, et al:
Evaluation of the association of PIK3CA mutations and PTEN loss
with efficacy of trastuzumab therapy in metastatic breast cancer.
Breast. 128:447–456. 2011.PubMed/NCBI
|
|
15
|
Dave B, Migliaccio I, Gutierrez MC, et al:
Loss of phosphatase and tensin homolog or phosphoinositol-3 kinase
activation and response to trastuzumab or lapatinib in human
epidermal growth factor receptor 2-overexpressing locally advanced
breast cancers. J Clin Oncol. 29:166–173. 2011. View Article : Google Scholar
|
|
16
|
Esteva FJ, Guo H, Zhang S, et al: PTEN,
PIK3CA, p-AKT, and p-p70S6K status: association with trastuzumab
response and survival in patients with HER2-positive metastatic
breast cancer. Am J Pathol. 177:1647–1656. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Baselga J: Treatment of
HER2-overexpressing breast cancer. Ann Oncol. 21:vii36–40.
2011.
|
|
18
|
Garrett JT and Arteaga CL: Resistance to
HER2-directed antibodies and tyrosine kinase inhibitors: mechanisms
and clinical implications. Cancer Biol Ther. 11:793–800. 2011.
View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Tanaka H, Yoshida M, Tanimura H, et al:
The selective class I PI3K inhibitor CH5132799 targets human
cancers harboring oncogenic PIK3CA mutations. Clin Cancer Res.
17:3272–3281. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Dan S, Okamura M, Mukai Y, et al: ZSTK474,
a specific phosphatidylinositol 3-kinase inhibitor, induces G1
arrest of the cell cycle in vivo. Eur J Cancer. 48:936–943. 2012.
View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Bibeau F, Frugier H, Denouel A, Sabourin
JC and Boissiere-Michot F: Technical considerations for KRAS
testing in colorectal cancer. The pathologist’s point of view. Bull
Cancer. 96:S15–S22. 2009.(In French).
|
|
22
|
Vorkas PA, Poumpouridou N, Agelaki S,
Kroupis C, Georgoulias V and Lianidou ES: PIK3CA hotspot mutation
scanning by a novel and highly sensitive high-resolution small
amplicon melting analysis method. J Mol Diagn. 12:697–704. 2010.
View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Board RE, Thelwell NJ, Ravetto PF, et al:
Multiplexed assays for detection of mutations in PIK3CA. Clin Chem.
54:757–760. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Dumont AG, Dumont SN and Trent JC: The
favorable impact of PIK3CA mutations on survival: an analysis of
2587 patients with breast cancer. Chin J Cancer. 31:327–334. 2012.
View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Barbareschi M, Buttitta F, Felicioni L, et
al: Different prognostic roles of mutations in the helical and
kinase domains of the PIK3CA gene in breast carcinomas. Clin Cancer
Res. 13:6064–6069. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Lai YL, Mau BL, Cheng WH, Chen HM, Chiu HH
and Tzen CY: PIK3CA exon 20 mutation is independently associated
with a poor prognosis in breast cancer patients. Ann Surg Oncol.
15:1064–1069. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Lerma E, Catasus L, Gallardo A, et al:
Exon 20 PIK3CA mutations decrease survival in aggressive (HER-2
positive) breast carcinomas. Virchows Arch. 453:133–139. 2008.
View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Cizkova M, Susini A, Vacher S, et al:
PIK3CA mutation impact on survival in breast cancer patients and in
ERalpha, PR and ERBB2-based subgroups. Breast Cancer Res.
14:R282012. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Aleskandarany MA, Rakha EA, Ahmed MA, et
al: PIK3CA expression in invasive breast cancer: a biomarker of
poor prognosis. Breast Cancer Res Treat. 122:45–53. 2009.
View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Campbell IG, Russell SE, Choong DY, et al:
Mutation of the PIK3CA gene in ovarian and breast cancer. Cancer
Res. 64:7678–7681. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Levine DA, Bogomolniy F, Yee CJ, et al:
Frequent mutation of the PIK3CA gene in ovarian and breast cancers.
Clin Cancer Res. 11:2875–2878. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Miron A, Varadi M, Carrasco D, et al:
PIK3CA mutations in in situ and invasive breast carcinomas. Cancer
Res. 70:5674–5678. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Wu G, Xing M, Mambo E, et al: Somatic
mutation and gain of copy number of PIK3CA in human breast cancer.
Breast Cancer Res. 7:R609–R616. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Kalinsky K, Jacks LM, Heguy A, et al:
PIK3CA mutation associates with improved outcome in breast cancer.
Clin Cancer Res. 15:5049–5059. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Maruyama N, Miyoshi Y, Taguchi T, Tamaki
Y, Monden M and Noguchi S: Clinicopathologic analysis of breast
cancers with PIK3CA mutations in Japanese women. Clin Cancer Res.
13:408–414. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Ellis MJ, Lin L, Crowder R, et al:
Phosphatidyl-inositol-3-kinase alpha catalytic subunit mutation and
response to neoadjuvant endocrine therapy for estrogen receptor
positive breast cancer. Breast Cancer Res Treat. 119:379–390. 2010.
View Article : Google Scholar
|
|
37
|
Perez-Tenorio G, Alkhori L, Olsson B, et
al: PIK3CA mutations and PTEN loss correlate with similar
prognostic factors and are not mutually exclusive in breast cancer.
Clin Cancer Res. 13:3577–3584. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Baker CL, Vaughn CP and Samowitz WS: A
PIK3CA pyrosequencing-based assay that excludes pseudogene
interference. J Mol Diagn. 14:56–60. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Tougeron D, Lecomte T, Pages JC, et al:
Effect of low-frequency KRAS mutations on the response to anti-EGFR
therapy in metastatic colorectal cancer. J Clin Oncol.
30:35202012.PubMed/NCBI
|