Introduction
DNA topoisomerase I (Top1) is an essential enzyme in
higher eukaryotes as well as the prime intracellular target of
various classes of anticancer drugs, such as camptothecins,
indenoisoquinolines and indolocarbazoles (1–3). Top1
catalyzes the relaxation of DNA supercoiling to allow the processes
of replication, transcription, and recombination to occur by
reversibly nicking one strand and forming transient DNA cleavage
complexes (4). Under physiological
conditions, cleavage complexes are transient. Top1-targeting drugs,
which act as ‘interfacial inhibitors’, stabilize covalent Top1-DNA
complexes and cause DNA strand breaks that lead to the apoptosis of
drug-treated cells (5).
The underlying mechanisms of the resistance to
Top1-targeting drugs may involve the inappropriate accumulation of
drug in the tumor cells, mutations in Top1, or changes in the
cellular response to DNA strand breaks. Mutations of Top1 that give
culture cells resistance to Top1-targeting drugs have been
identified (6). We previously
established a camptothecin-resistant colon cancer cell line, which
was designated DLDSNR6, and identified a missense mutation of the
Top1 gene that resulted in a glycine to serine substitution at
codon 365. In these resistant cells, Top1 shows lower catalytic
activity and camptothecin traps fewer Top1-DNA complexes than
parent DLD-1 cells (7).
Camptothecin is a plant alkaloid produced by the
Chinese tree Camptotheca acuminata. Camptothecin and its
derivatives are potent poisons to most eukaryotic cells, including
those of higher plants, but camptothecin-producing trees are
insensitive to these self-producing toxic metabolites.
Sirikantaramas et al(8)
demonstrated that camptothecin-producing plants have point
mutations in the Top1 gene at Asn421, Leu530 and Asn722, which
confer resistance to camptothecins. Although Top1 mutations at
codon 722 have been identified in several camptothecin-resistant
human cancer cell lines, the other mutations have yet to be found
(9).
Materials and methods
Materials
SN-38 was kindly provided by Yakult Co., Ltd.
(Tokyo, Japan), and J-107088 was kindly supplied by MSD K.K.
(Tokyo, Japan, formerly Banyu Pharmaceutical Co., Ltd). Other
chemicals were purchased from Sigma-Aldrich Japan K.K. (Tokyo,
Japan). SN-38, J-107088, camptothecin and Ko143 were resuspended
with Me2SO as stock solutions and stored at −20ºC.
Verapamil was resuspended with water and stored at −20ºC. Rabbit
anti-Top1 antibody was purchased from TopoGEN, Inc. (Columbus, OH,
USA), and mouse anti-DNA topoisomerase II α (Top2α) antibody was
purchased from Medical & Biological Laboratories Co., Ltd.
(Nagoya, Japan).
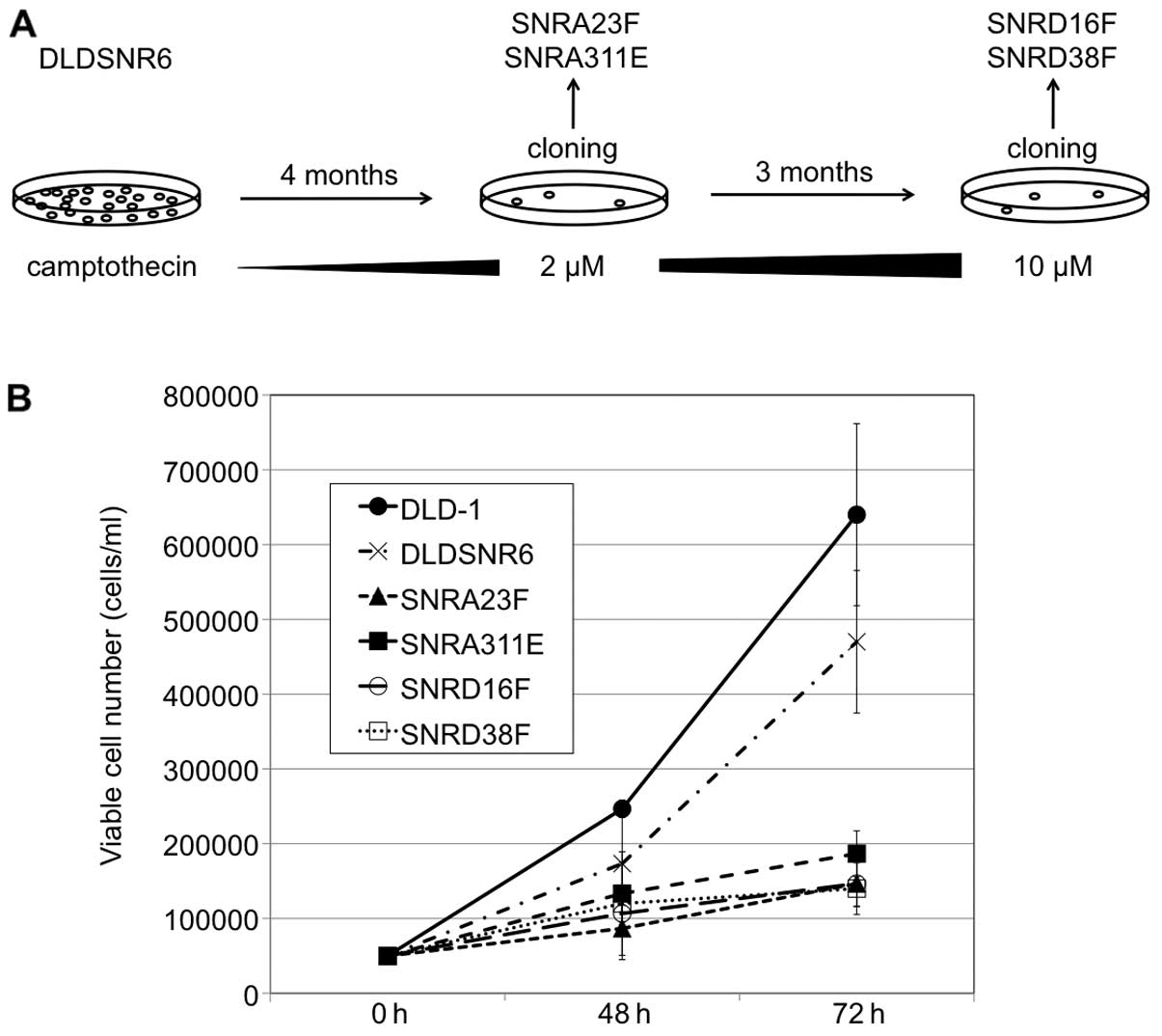
Establishment of highly
camptothecin-resistant colon cancer cell sublines
The DLD-1 human colon cancer cell line was provided
by the Cell Resource Center for Biochemical Research of Tohoku
University (Sendai, Japan). We previously established the DLDSNR6
cell line from parental DLD-1 cells through the continuous exposure
to stepwise increases in SN-38 concentrations (7). In this study, DLDSNR6 cells were
exposed to stepwise increases in camptothecin concentrations (up to
2 μM) over a period of 4 months and then SNRA23F and SNRA311E
sublines were established by the limiting dilution technique. The
camptothecin-resistant cell pool was again exposed to camptothecin
with concentrations up to 10 μM for 3 months, and SNRD16F and
SNRD38F sublines were obtained (Fig.
1A). These cell lines were cultured at 37ºC in RPMI-1640 medium
(Life Technologies Japan, Tokyo, Japan) that was supplemented with
10% fetal bovine serum (Thermo Fisher Scientific K.K., Yokohama,
Japan) and Antibiotic-Antimycotic Mixed Solution (Nacalai Tesque,
Inc., Kyoto, Japan) under a humidified atmosphere containing 5%
CO2.
Cell growth, viability and cytotoxicity
assays
DLD-1, DLDSNR6, SNRA23F, SNRA311E, SNRD16F and
SNRD38F cells (5.0×105/ml) were cultured in 6-cm culture
dishes for 48–72 h. The number of viable cells was counted by the
trypan blue dye exclusion method with a hemocytometer. The
cytotoxicity of Top1-targeting drugs was measured by an MTS assay
(Cell Titer 96 Aqueous One Solution Cell Proliferation Assay;
Promega, San Luis Obispo, CA, USA) with minor technical
modifications. Three to ten thousand cells were incubated in a
96-well tissue culture plate for 96 h in the presence of the
indicated concentrations of camptothecin, SN-38, and J-107088,
after which the assay was performed according to the manufacturer's
instructions. All the experiments were performed in triplicate.
Detection of Top1 mutations
Total RNA was extracted from each cell line and
reverse transcription was performed. The full-length Top1 cDNA was
amplified by polymerase chain reaction (PCR), and the resulting
fragments were inserted into the cloning vector. The entire Top1
open reading frame was sequenced with the BigDye Terminator Version
3.1 Cycle Sequencing kit (Life Technologies Japan) and the ABI 3700
DNA Analyzer (7,10).
Preparation of protein samples and
immunoblotting analysis
Crude cell extracts were prepared by suspending the
cells in radioimmunoprecipitation assay lysis buffer containing 1
μM phenylmethylsulfonyl fluoride, and the protein concentrations of
each sample were measured by the Bradford method. Protein samples
were separated by 7.5% sodium dodecyl sulfate-polyacrylamide gel
electrophoresis and immunoblotting was performed with antibodies
for Top1 and Top2α.
Top1-mediated DNA relaxation assay
The Top1 catalytic activities of the nuclear
extracts from each cell line were determined by measuring the
relaxation of the supercoiled pHOT1 plasmid (TopoGEN, Inc.), which
contained a Top1-cleavage site that was derived from the
tetrahymena ribosomal gene repeat (11). The supercoiled pHOT1 plasmid (0.25
μg) was incubated with the indicated amounts of nuclear extracts in
10 mM Tris-HCl (pH 7.5), 150 mM NaCl, and 1 mM
ethylenediaminetetraacetic acid (EDTA) at 37ºC for 60 min in a
final volume of 20 μl (7). The
reaction was terminated by the addition of 5 μl of 0.05% sodium
dodecyl sulfate and the samples were loaded onto 1% agarose gels.
After electrophoresis, the gels were stained with Tris-borate EDTA
buffer (89 mM Tris-borate, 2 mM EDTA, pH 8.0) containing 0.5 μg/ml
ethidium bromide and visualized by transillumination with UV light.
Relaxation activity was identified by the disappearance of the
supercoiled DNA.
Quantitative reverse transcription
(qRT)-PCR array analysis of ATP-binding cassette transporters
To assess the relative expression of ATP-binding
cassette transporters in each cell line, we used the TaqMan™ Array
Gene Signature 96-well plates (human ABC transporters; Life
Technologies Japan). After 2 months of passage in drug-free medium,
the cells were harvested, and the total RNA was extracted with
ISOGEN reagent (Nippon Gene, Tokyo, Japan). The analysis was
performed according to the manufacturer's directions with the
Applied Biosystems 7500 Real-Time PCR system (Life Technologies
Japan). As a measure of the relative levels of expression between
the parental DLD-1 and the resistant cell lines, ΔΔCt values were
calculated and converted to fold-change values
(2−ΔΔCt).
Flow cytometry
The effects of J-107088 on cell cycle distribution
in each cell line were determined with propidium iodide staining
and analyzed with flow cytometry. Each cell line was either treated
with vehicle alone [Me2SO (0.2%)], J-107088 (5 μM),
J-107088 (5 μM) plus verapamil (10 μM), or J-107088 (5 μM) plus
Ko143 (0.3 μM) for 48 h. Cells were harvested, fixed in 70%
precooled ethanol, and incubated in phosphate-buffered saline
containing 10 μg/ml propidium iodide and 10 μg/ml RNase for 30 min
at room temperature. The fluorescence (excitation at 488 nm and
emission at 585/42 nm) of 2×104 cells from each sample
was analyzed with FACSCalibur (Becton-Dickinson, San Jose, CA, USA)
flow cytometry, and the cell population at each cell cycle phase
was determined with ModiFit software (Becton-Dickinson).
Results
Establishment of highly
camptothecin-resistant colon cancer cells
In this study, we established the SNRA23F, SNRA311E,
SNRD16F and SNRD38F sublines, which were highly
camptothecin-resistant cell lines (Fig.
1A). These cells exhibited markedly retarded growth compared
with that of parental DLD-1 and DLDSNR6 cells (Fig. 1B). Observations of the cells stained
with 4′-6-diamidino-phenylindole under a fluorescence microscope
revealed no signs of apoptotic cell death in the resistant cell
lines (data not shown). The newly established sublines grew in the
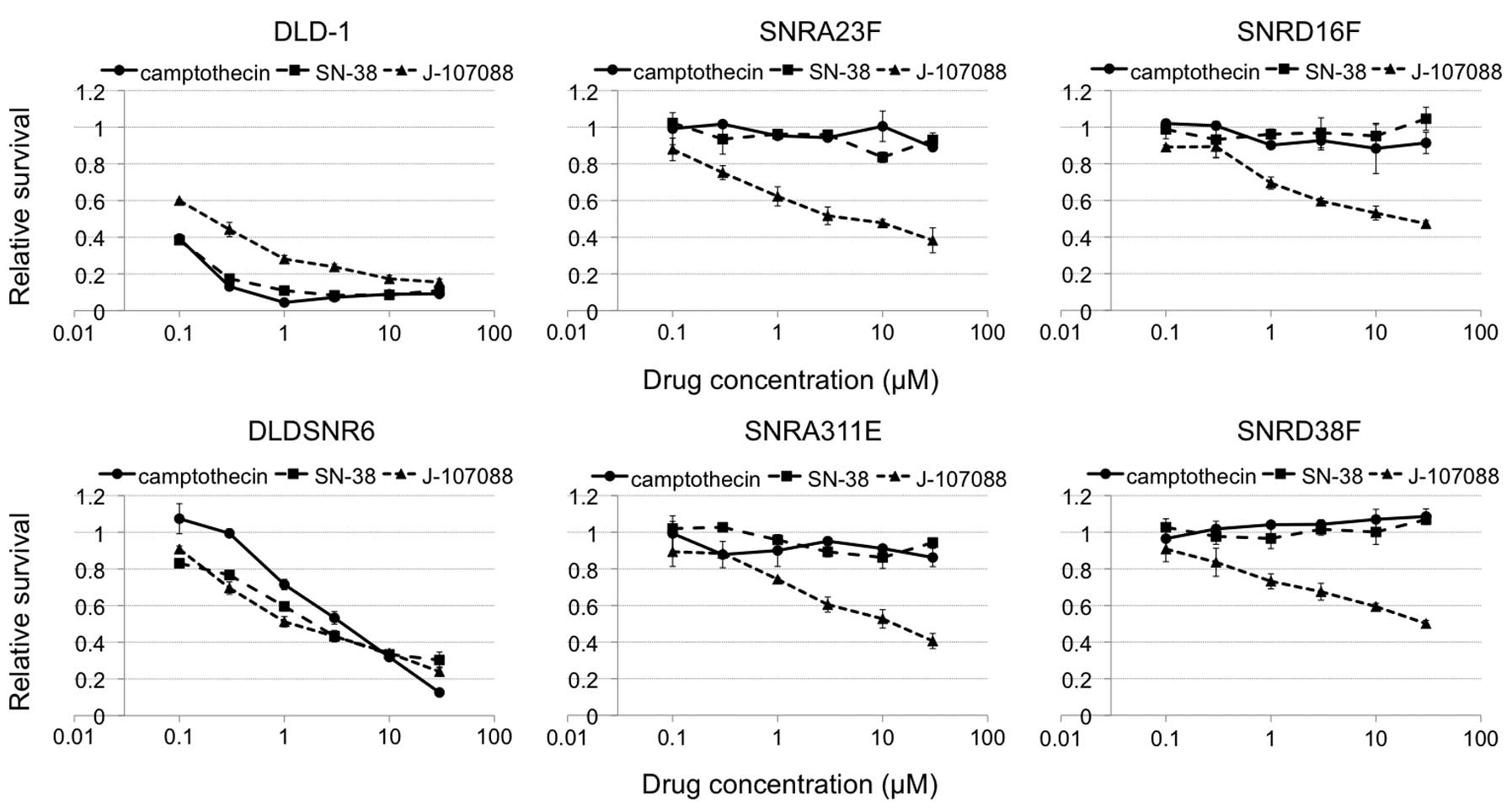
presence of 30 μM of camptothecin. In addition, these cells were
resistant to the indolocarbazole derivative, J-107088 (Fig. 2).
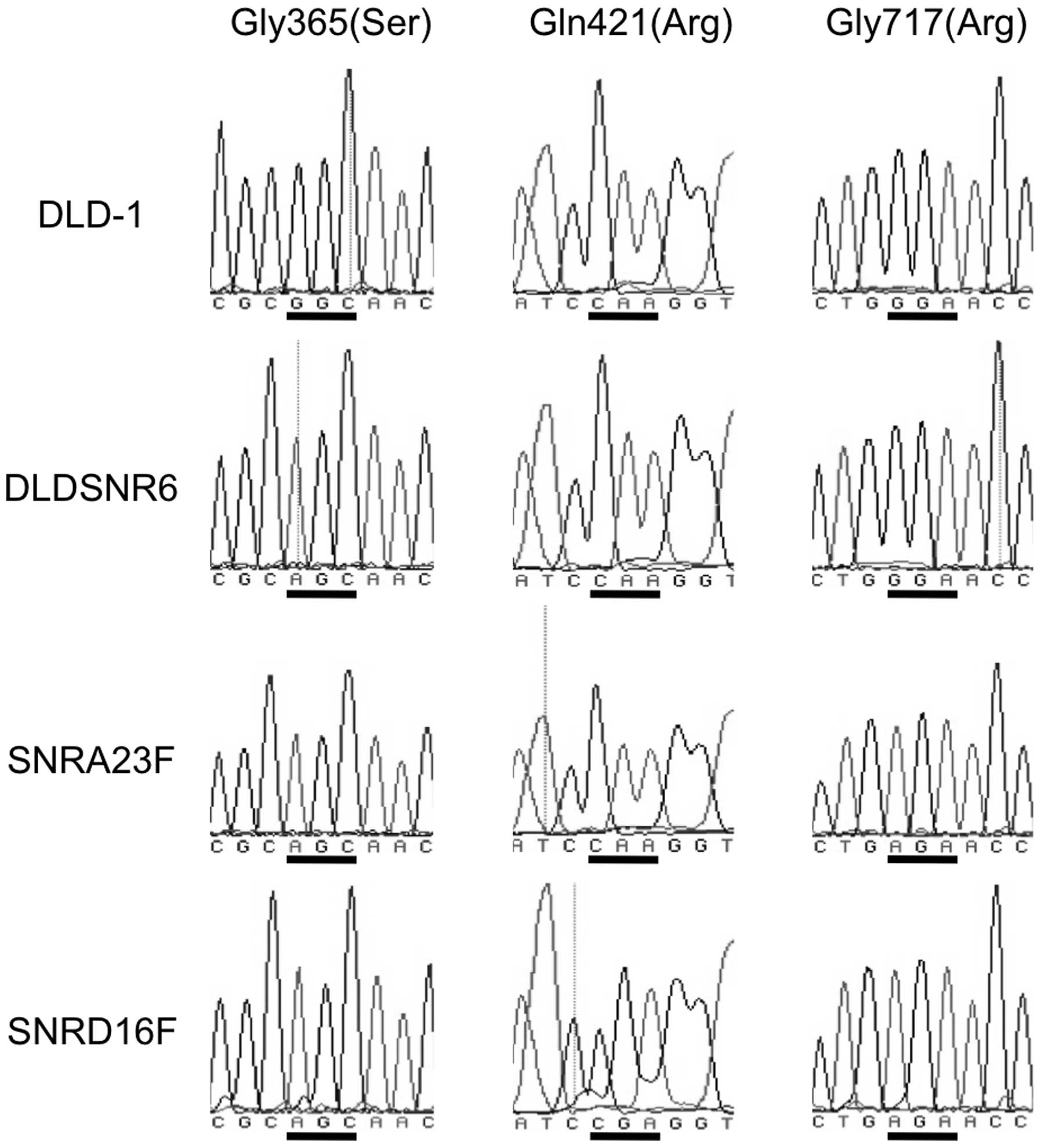
Three missense mutations of the Top1 gene
in resistant cell lines
We previously identified a Top1 missense mutation in
DLDSNR6 cells at codon 365, which resulted in the amino acid
alteration of glycine (GGC) to serine (AGC) (7). In addition, the parental DLD-1 cells
had a heterozygous Top1 missense mutation resulting in a Met675Ile
alteration (12,13). The highly camptothecin-resistant
cells (SNRA23F, SNRA311E, SNRD16F and SNRD38F) harbored a Top1
missense mutation at codon 717, which resulted in a glycine (GGA)
to arginine (AGA) substitution. Furthermore, the SNRD16F and
SNRD38F cell lines carried a codon 421 mutation that resulted in
the substitution from glutamine (CAA) to arginine (CGA) (Fig. 3).
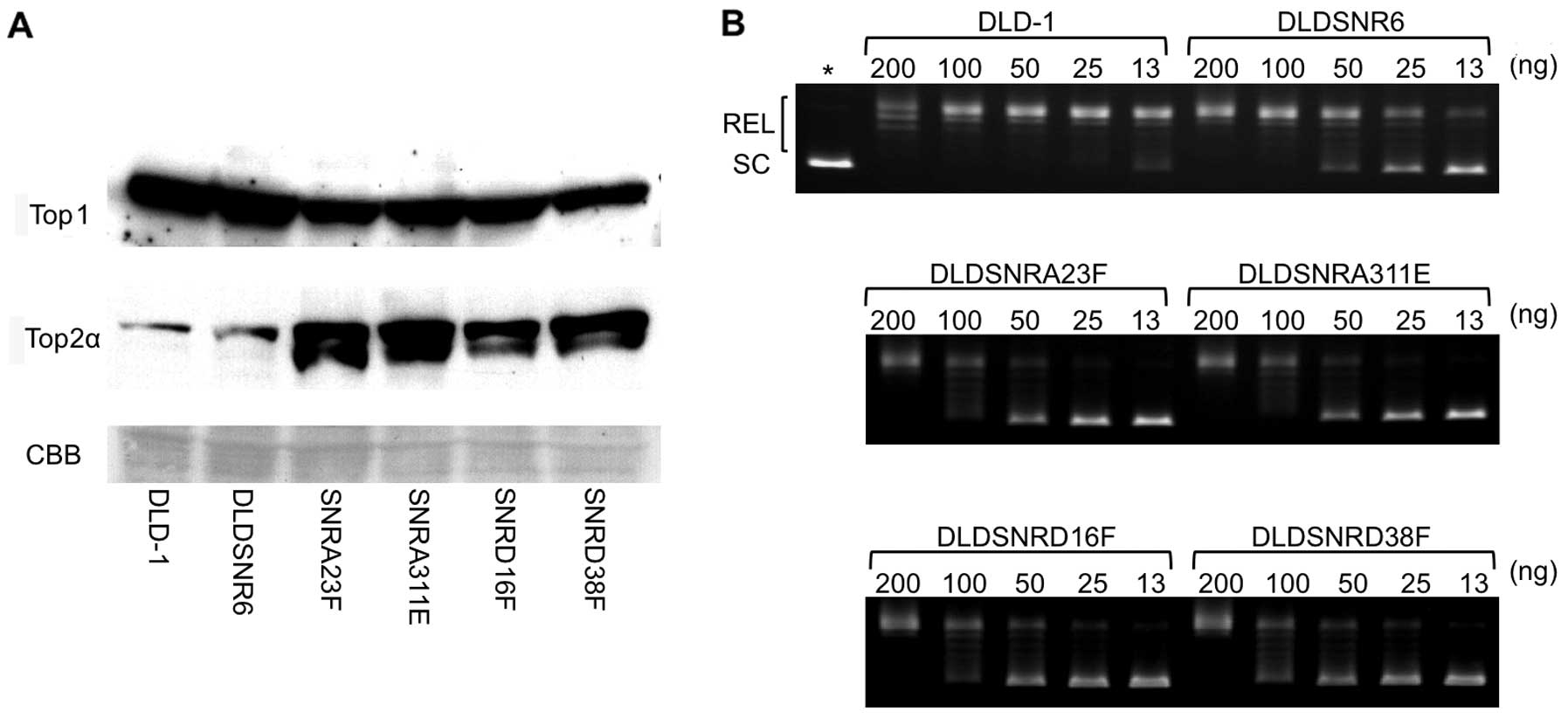
Top1 protein expression and enzymatic
function in resistant cell lines
There were slightly lower levels of Top1 protein
expression in highly camptothecin-resistant cell lines compared
with those of the parental DLD-1 and DLDSNR6 cells, whereas the
levels of Top2 protein expression were increased in these sublines
(Fig. 4A). The DNA relaxation assay
revealed that Top1 activity was markedly reduced to approximately
one-eighth in highly camptothecin-resistant cell lines compared
with that in DLD-1 cells (Fig.
4B).
Expression of ATP-binding cassette
transporters
The levels of mRNA expression of the ATP-binding
cassette transporters ABCB1 [multidrug resistance protein 1 (MDR1)]
and ABCG2 [breast cancer resistance protein (BCRP)] were
significantly increased in camptothecin-resistant cell lines,
including DLDSNR6. However, the levels of mRNA expression of ABCC2
(multidrug resistance-associated protein 2) were reduced in SNRA23F
and SNRA311E cells (Table I).
 | Table IqRT-PCR array analysis of ATP-binding
cassette transporters. |
Table I
qRT-PCR array analysis of ATP-binding
cassette transporters.
| Gene | DLD-1 | DLDSNR6 | SNRA23F | SNRA311E | SNRD16F | SNRD38F |
|---|
| ABCA2 | 1.000 | 0.871 | 0.871 | 0.824 | 1.602 | 0.807 |
| ABCB1 | 1.000 | 6.774 | 11.314 | 17.268 | 12.295 | 7.945 |
| ABCB4 | 1.000 | 2.028 | 2.990 | 2.621 | 1.505 | 1.117 |
| ABCC1 | 1.000 | 1.141 | 1.087 | 0.914 | 0.927 | 0.883 |
| ABCC2 | 1.000 | 0.486 | 0.156 | 0.071 | 0.620 | 0.374 |
| ABCC3 | 1.000 | 0.753 | 0.901 | 0.883 | 0.835 | 0.901 |
| ABCC4 | 1.000 | 1.050 | 0.801 | 0.889 | 0.914 | 1.007 |
| ABCC5 | 1.000 | 1.035 | 1.173 | 1.240 | 1.000 | 0.841 |
| ABCC6 | 1.000 | 1.338 | 2.868 | 3.364 | 3.227 | 2.445 |
| ABCC10 | 1.000 | 1.548 | 2.085 | 2.532 | 1.753 | 2.129 |
| ABCC11 | 1.000 | 0.940 | 0.274 | 0.406 | 0.339 | 0.412 |
| ABCG2 | 1.000 | 3.031 | 6.105 | 2.497 | 9.580 | 4.199 |
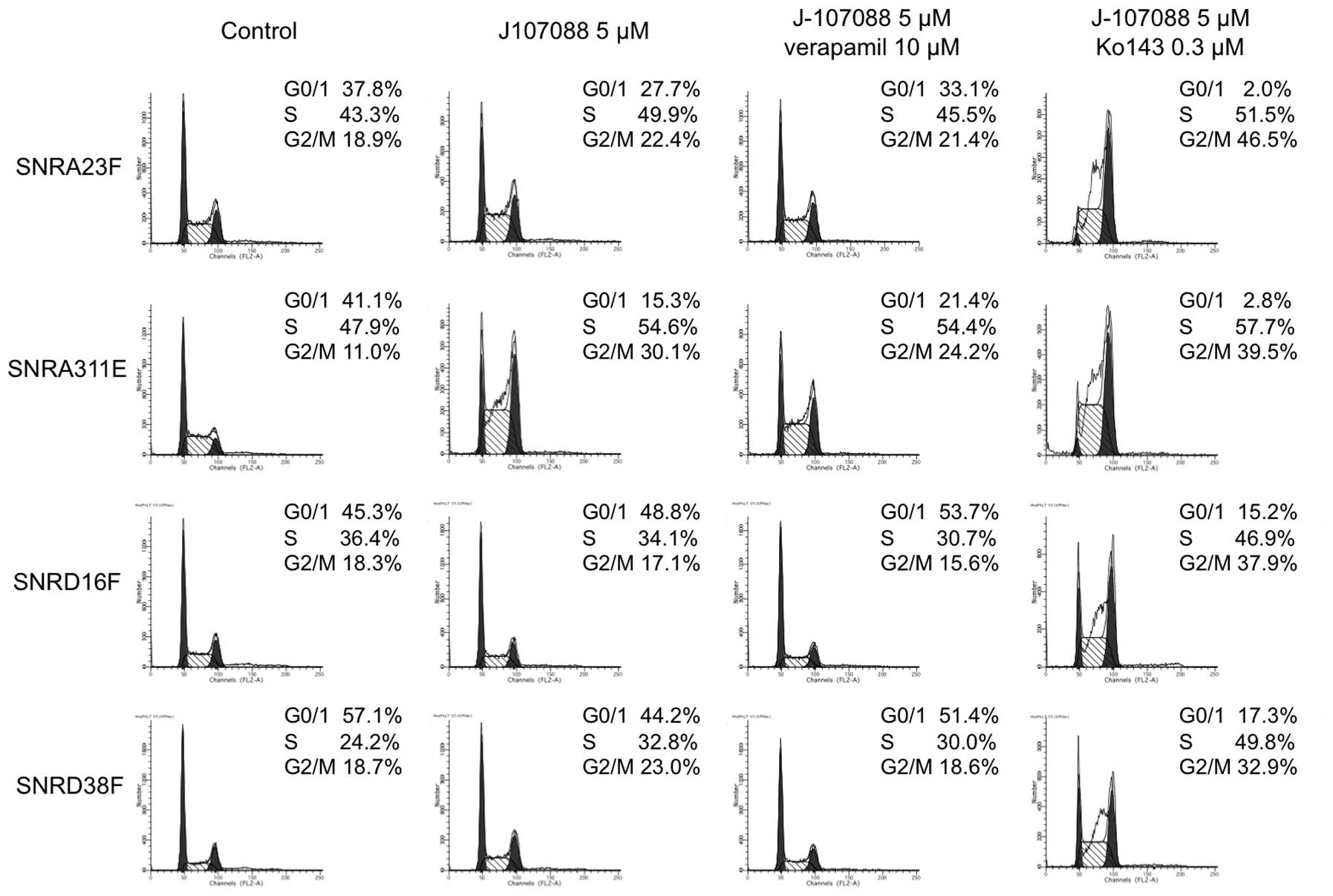
Cell cycle analysis of J-107088-treated
cells
The flow cytometry analysis of untreated cells
revealed that the S-phase fraction was not reduced in highly
camptothecin-resistant cells (Fig.
5) compared with parental cells (~24% in DLD-1 cells, ~30% in
DLDSNR6 cells, data not shown). When cells were treated with 5 μM
of J-107088 for 48 h, accumulation was observed in the late
S-G2/M phase in SNRA23F and SNRA311E cells, while the
cell cycle distribution was not clearly affected in SNRD16F and
SNRD38F cells (Fig. 5). Treatment
with camptothecin at the higher concentration of 10 μM caused
marginal changes in the cell cycle distribution in highly
camptothecin-resistant cells (data not shown). The addition of the
MDR1 inhibitor, verapamil, to 5 μM of J-107088 did not evidently
affect the cell cycle distribution. When cells were coincubated
with the BCRP inhibitor, Ko143, and 5 μM of J-107088 for 48 h, a
marked accumulation in late S to G2/M phase was observed
in all highly camptothecin-resistant cells, while the
G0/1 population was still observed in SNRD16F
and SNRD38F cells (Fig. 5).
Discussion
In the present study, we established highly
camptothecin-resistant colon cancer cell lines and characterized
these cells. First, the highly resistant clones (SNRA23F, SNRA311E,
SNRD16F, and SNRD38F) were retarded in growth and showed slightly
lower levels of Top1 protein expression (Fig. 1B and 4A). The previously established DLDSNR6
cells exhibited a loss of heterozygosity in the Top1 gene, but
these cells did not exhibit significant growth retardation. Toyoda
et al(14) demonstrated the
heterozygous disruption of the Top1 gene in a human pre-B cell
line, Nalm-6. The TOP1-heterozygous Nalm-6 cells exhibited ~70%
protein expression levels of Top1, but they showed no significant
differences in the growth rate compared with that of the parental
cells. In our highly camptothecin-resistant cells, Top1 enzymatic
activity levels were reduced to approximately one-eighth mainly due
to the new missense mutations (Fig.
4B). The TOP1 wild-type allele was not expressed in our cells.
Top1 knockdown that was conducted with small interfering RNA to the
level of ~10–20% in cancer cell lines caused genomic instability
and replication defects (15).
Minor deficits in Top1 activity may not affect the cell growth
rate. Human Top2α, which can relax the positively supercoiled DNA,
has been shown to partially compensate for Top1 activity (16). Our newly established resistant cells
showed high levels of expression of Top2α (Fig. 4A). The increased levels of Top2α
expression in these cells but not in DLDSNR6 cells may suggest that
a substantial reduction of Top1 activity levels occurred only in
highly resistant cells.
Our highly camptothecin-resistant clones grew in the
presence of 30 μM of camptothecin (Fig.
2). These clones harbored two or three missense mutations in
the Top1 gene. The X-ray structure of human Top1 has been
determined (17,18), and it shows that the cap region
(residues 215–433) is sterically close to the catalytic Tyr723.
When Top1 clamps double-stranded DNA, some loop regions focus on
one side of the DNA (17,19). It has been proposed that amino acids
360–370 of Top1 form a loop region, which contacts other loop
regions (residues 417–423, 496–505 and 529–538) to create a salt
bridge and two non-covalent bonds between the cap region and the
bottom lobe of the enzyme (19).
Several structural models have demonstrated that camptothecin
derivatives mimic a DNA pair and inhibit the DNA religation
activity of Top1 by stabilizing the covalent Top1-DNA complexes
(1,20). Moreover, structural models have
indicated that camptothecin derivatives interact with Arg364,
Asp533 and Asn722 of Top1 (17,20).
The newly identified Top1 mutations in this study were positioned
in or near the residues that have been shown to be important for
enzyme-DNA interactions or enzyme-drug interactions.
The camptothecin-resistant cell lines overexpressed
MDR1 and BCRP (Table I). The
overexpression of ATP-binding cassette transporters is often
responsible for the cellular resistance to anticancer drugs.
Camptothecin derivatives and indolocarbazole Top1 inhibitors have
been demonstrated to be effectively effluxed by BCRP, while
camptothecin is a relatively poor substrate for MDR1 (21–23).
Verapamil and Ko143 could not enhance the effects of camptothecin
on the highly resistant cells in terms of the growth rate or cell
cycle progression (data not shown). The higher concentration of
camptothecin plus verapamil or Ko143 could not trap covalent
enzyme-DNA complexes by a band depletion assay in these cells (data
not shown). The established cells in this study were ~15 to
150-fold resistant to the indolocarbazole derivative, J-107088
(Fig. 2). The exposure of cells to
5 μM of J-107088 plus 0.3 μM Ko143 caused the accumulation of
SNRA23F and SNRA311E cells in the late S to G2/M phase.
In the SNRD16F and SNRD38 cells, this combined exposure caused
accumulation in the late S-G2/M phase, but the G0/1
population remained in these cells (Fig. 5). These data suggested that J-107088
effectively caused DNA damage in SNRA23F and SNRA311E cells
compared with SNRD16F and SNRD38F cells, although the SNRA23 cells
expressed more BCRP mRNA than SNRD38F. A Top1Gln412Arg mutation may
confer further resistance to this indolocarbazole derivative. In
the cytotoxicity assay, the SNRD16F and SNRD38F cells were slightly
more resistant to J-107088 at higher concentrations compared with
SNRA23F and SNRA311E cells (Fig.
2).
The DLDSNR6 cells have shown a loss of
heterozygosity in the TOP1 gene and exhibited genomic instability
due to homozygous mutations in the hMSH6 gene (7). This background enabled us to establish
the highly camptothecin-resistant cell lines that had three
mutations in one allele of the TOP1 gene. To the best of our
knowledge, such cell lines have not previously been reported. The
Top1Gly717 mutation has been reported in camptothecin-resistant
ovarian cancer cells (24). A
mutation corresponding to human Top1Glu421 was previously
identified in camptothecin-producing plants, but it has not been
identified in mammalian cells (8).
The camptothecin producing plants have three mutations in the TOP1
gene and mutations in residues sterically near the catalytic
tyrosine in addition to the Glu421 mutation. Our newly established
cell lines may be useful for understanding enzyme-drug interactions
and the molecular evolution of drug resistance.
Acknowledgements
This study was supported by Japan Society for the
Promotion of Science KAKENHI grant no. 24701011 (Y.A.) and was also
supported by a grant from the Vehicle Racing Commemorative
Foundation (Y.A.).
References
|
1
|
Staker BL, Hjerrild K, Feese MD, Behnke
CA, Burgin AB and Stewart L: The mechanism of topoisomerase I
poisoning by a camptothecin analog. Proc Natl Acad Sci USA.
99:15387–15392. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Pommier Y and Cushman M: The
indenoisoquinoline noncamptothecin topoisomerase I inhibitors:
update and perspectives. Mol Cancer Ther. 8:1008–1014. 2009.
View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Sáchez C, Médez C and Salas JA:
Indolocarbazole natural products: occurrence, biosynthesis, and
biological activity. Nat Prod Rep. 23:1007–1045. 2006.PubMed/NCBI
|
|
4
|
Wang JC: Cellular roles of DNA
topoisomerases: a molecular perspective. Nat Rev Mol Cell Biol.
3:430–440. 2002. View
Article : Google Scholar : PubMed/NCBI
|
|
5
|
Marchand C, Antony S, Kohn KW, et al: A
novel norindenoisoquinoline structure reveals a common interfacial
inhibitor paradigm for ternary trapping of the topoisomerase I-DNA
covalent complex. Mol Cancer Ther. 5:287–295. 2006. View Article : Google Scholar
|
|
6
|
Urasaki Y, Laco G, Takebayashi Y, Bailly
C, Kohlhagen G and Pommier Y: Use of camptothecin-resistant
mammalian cell lines to evaluate the role of topoisomerase I in the
antiproliferative activity of the indolocarbazole, NB-506, and its
topoisomerase I binding site. Cancer Res. 61:504–508.
2001.PubMed/NCBI
|
|
7
|
Arakawa Y, Suzuki H, Saito S and Yamada H:
Novel missense mutation of the DNA topoisomerase I gene in
SN-38-resistant DLD-1 cells. Mol Cancer Ther. 5:502–508. 2006.
View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Sirikantaramas S, Yamazaki M and Saito K:
Mutations in topoisomerase I as a self-resistance mechanism
coevolved with the production of the anticancer alkaloid
camptothecin in plants. Proc Natl Acad Sci USA. 105:6782–6786.
2008. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Rasheed ZA and Rubin EH: Mechanisms of
resistance to topoisomerase I-targeting drugs. Oncogene.
22:7296–7304. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Urasaki Y, Laco GS, Pourquier P, et al:
Characterization of a novel topoisomerase I mutation from a
camptothecin-resistant human prostate cancer cell line. Cancer Res.
61:1964–1969. 2001.PubMed/NCBI
|
|
11
|
Bonven BJ, Gocke E and Westergaard O: A
high affinity topoisomerase I binding sequence is clustered at
DNAase I hypersensitive sites in Tetrahymena R-chromatin. Cell.
41:541–551. 1985. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Chen TR, Dorotinsky CS, McGuire LJ, Macy
ML and Hay RJ: DLD-1 and HCT-15 cell lines derived separately from
colorectal carcinomas have totally different chromosome changes but
the same genetic origin. Cancer Genet Cytogenet. 81:103–108. 1995.
View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Moisan F, Longy M, Robert J and Le Morvan
V: Identification of gene polymorphisms of human DNA topoisomerase
I in the National Cancer Institute panel of human tumour cell
lines. Br J Cancer. 95:906–913. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Toyoda E, Kurosawa A, Fujii M and Adachi
N: Heterozygous disruption of the DNA topoisomerase I gene confers
cellular resistance to camptothecin in human cells. Biol Pharm
Bull. 32:724–727. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Miao ZH, Player A, Shankavaram U, et al:
Nonclassic functions of human topoisomerase I: genome-wide and
pharmacologic analyses. Cancer Res. 67:8752–8761. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
McClendon AK, Rodriguez AC and Osheroff N:
Human topoisomerase IIα rapidly relaxes positively supercoiled DNA:
implications for enzyme action ahead of replication forks. J Biol
Chem. 280:39337–39345. 2005.
|
|
17
|
Redinbo MR, Stewart L, Kuhn P, Champoux JJ
and Hol WG: Crystal structures of human topoisomerase I in covalent
and noncovalent complexes with DNA. Science. 279:1504–1513. 1998.
View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Stewart L, Redinbo MR, Qiu X, Hol WG and
Champoux JJ: A model for the mechanism of human topoisomerase I.
Science. 279:1534–1541. 1998. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Chrencik JE, Staker BL, Burgin AB, et al:
Mechanisms of camptothecin resistance by human topoisomerase I
mutations. J Mol Biol. 339:773–784. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Laco GS, Collins JR, Luke BT, et al: Human
topoisomerase I inhibition: docking camptothecin and derivatives
into a structure-based active site model. Biochemistry.
41:1428–1435. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Nakagawa H, Saito H, Ikegami Y,
Aida-Hyugaji S, Sawada S and Ishikawa T: Molecular modeling of new
camptothecin analogues to circumvent ABCG2-mediated drug resistance
in cancer. Cancer Lett. 234:81–89. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Komatani H, Kotani H, Hara Y, et al:
Identification of breast cancer resistant protein/mitoxantrone
resistance/placenta-specific, ATP-binding cassette transporter as a
transporter of NB-506 and J-107088, topoisomerase I inhibitors with
an indolocarbazole structure. Cancer Res. 61:2827–2832. 2001.
|
|
23
|
Pommier Y: Topoisomerase I inhibitors:
camptothecins and beyond. Nat Rev Cancer. 6:789–802. 2006.
View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Wang LF, Ting CY, Lo CK, et al:
Identification of mutations at DNA topoisomerase I responsible for
camptothecin resistance. Cancer Res. 57:1516–1522. 1997.PubMed/NCBI
|