Introduction
Acute myeloid leukemia (AML) is the most common
hematological malignancy in adults, with a worldwide incidence of
~3.6 cases in 100,000 individuals (National Cancer Institute. SEER
Stat Fact Sheets: Acute Myeloid Leukemia; http://seer.cancer.gov/statfacts/html/amyl.html).
A number of AML- related chromosomal abnormalities and gene
mutations have been identified, and their involvement in the
regulation of DNA methylation, signal transduction, energy
synthesis, and cellular structure has been reported (1).
The identification of new AML-related genes may
provide insight into disease prognosis, treatment response, and new
therapeutic regimens for patients with AML. Mutations in CCAAT
enhancer binding factor α (CEBPA) have been associated with
prolonged recurrence-free survival in AML patients (2). Mutations in nucleophosmin-1
(NPM1) and the internal tandem duplications in FLT3
predict positive AML prognosis (3),
and mutations in several genes such as MLL-PTD, FLT3/ITD,
WT1, and C-KIT predict negative AML prognosis (4–7). Thus,
screening for mutations in CEBPA, NPM1, FLT3,
and KIT is part of the routine workup for AML (8,9).
A number of drugs targeting AML-related genes have
been tested for their efficacy in treating AML, including
TNF-related apoptosis inducing ligand (TRAIL) and tipifarnib, an
inhibitor of farnesyltransferase (FTase), alone or in combination
with other anti-leukemia drugs (10,11).
Inhibitors of histone deacetylase, proteasome and Bcl-2 antisense
oligonucleotides have also been used as targeted drugs in the
clinical treatment of AML (12).
Furthermore, inhibitors of Fms-like tyrosine kinase 2 (FLT-2), such
as lestaurtinib (CEP701), tandutinib (MLN518) and PKC412 have shown
benefit in treating FLT3-associated AML (13).
Previously, we identified a large family with a high
incidence of AML in Fujian China (14). A total of 11 family members in 4
generations were diagnosed with AML; two patients developed acute
erythroleukemia (M6), one suffered from minimally differentiated
acute myeloblastic leukemia (M1), one had AML, one had acute
monocytic leukemia (M5), and the remaining 6 were diagnosed with
acute myeloid leukemia with partial differentiation (M2).
Using subtractive hybridization and
rapid-amplification of cDNA ends (RACE) techniques, the bone marrow
biopsy of an 11-year-old boy with M2 from the above family and the
expressed sequence tag (EST) zywb4 were used to clone the
full-length cDNA of a novel familial acute myelogenous
leukemia-associated gene (15). A
sequence was cloned, mapped to human chromosome 4, and contained
one open reading frame of 2257 bp coding for a sequence of 144
amino acids and a poly(A) tail. The newly predicted gene was
designated as ELF2C (GenBank accession no. DQ359746).
In addition, another EST sequence zywb87 was also
used to clone a familial acute myelogenous leukemia-associated gene
(16). A full-length cDNA of 2313
bp with a complete open reading frame (ORF) of 249 bp was obtained
from the bone marrow specimen of the above-mentioned familial AML
male patient. Localized on human chromosome 1q31.3, the gene coded
an 82-amino acid polypeptide with a leucine-rich repeat and was
named familial acute myelogenous leukemia-related factor (FAMLF;
GenBank accession no. EF413001; protein accession no. ABN58747).
The FAMLF gene expression level was significantly higher in
AML patients outside this family than in normal healthy persons
outside this family (16).
In the present study, the polyclonal antibody to
FAMLF was generated and used to assess the protein expression of
FAMLF in bone marrow cells collected from an AML patient and a
chronic myeloid leukemia (CML) patient, as well as in several human
cell lines. In addition, cell lines were transfected with the
FAMLF/GFP fusion protein expression vector to investigate the
subcellular localization of the FAMLF protein.
Materials and methods
The present study was approved by the Ethics and
Institutional Animal Care and Use Committees of the Fujian Medical
University Union Hospital.
Preparation of a polyclonal antibody
against FAMLF
The hydrophilicity and antigenicity of the 14-amino
acid N-terminal polypeptide of the FAMLF protein (amino acids 3–16)
were predicted using MacVector (IBI Ltd., Cambridge, UK), and it
was synthesized by Shanghai Chaobao Biotech (Shanghai, China). The
polypeptide was subsequently purified by high pressure liquid
chromatography and mass spectrometry; the purity of the resulting
polypeptide was as high as 92%. The purified polypeptide was
conjugated to keyhole limpet hemocyanin (KLH) protein (Shanghai
Chaobao Biotech). The resulting polypeptide-KLH was used for
immunization.
Preimmunization venous blood (1–2 ml) collected from
the ear vein of 2 healthy male New Zealand white rabbits 7 days
before immunization was used as a negative control. Then, equal
volumes of 200 μg of polypeptide-KLH complex and complete Freund's
adjuvant (CFA) were mixed and emulsified. This emulsion was
subcutaneously injected at multiple sites along the back. The
animals were immunized once per week with CFA being replaced with
incomplete Freund’s adjuvant (IFA) on alternate weeks.
At week 7, blood samples were collected from the ear
vein daily, and the IgG titer was then quantified by enzyme-linked
immunosorbent assay (ELISA) using horseradish peroxidase
(HRP)-conjugated goat anti-rabbit IgG (1:10,000; Pierce Thermo
Scientific, Rockford, IL USA) according to the manufacturer's
instructions. When the IgG titer was higher than 10,000, the
anti-FAMLF antibody in the collected serum was purified using an
IgG affinity column (Pierce Thermo Scientific) according to the
manufacturer's instructions.
Western blot assay
Antibody specificity was examined using western blot
analysis. Total protein lysates were prepared from bone marrow
cells derived from an AML patient and a CML patient, NIH3T3 cells
expressing the FAMLF-GFP fusion protein (described below), and
several human cell lines, including the COLO205 human colon
adenocarcinoma cell line; the NB4 human acute promyelocytic
leukemia cell line; the U937 human macrophage-like cell line; the
k562 human myeloid leukemia cell line; the U266 human myeloma cell
line; the HL60 human promyelocytic cell line; the CA46 human
lymphoma cell line; and the CEM human T cell lymphoblast-like cell
line (Institute of Hematologic Diseases, Fujian, China). Total
protein lysates (40 μg/lane) were separated by Tricine SDS-PAGE.
Blots were incubated with the primary anti-FAMLF antibody (1:250)
or the anti-GFP antibody (1:1,000), and subsequently with
HRP-conjugated goat anti-rabbit IgG (1:4,000; Pierce Thermo
Scientific). Protein bands were visualized using an enhanced
chemiluminescence detection kit (Amersham, Little Chalfont,
UK).
PCR amplification of FAMLF DNA
Total RNA was isolated from U937 cells, a human
macrophage-like cell line, using TRIzol (Invitrogen, Carlsbad, CA,
USA), and RNA samples were reverse transcribed by MultiScribe™
reverse transcriptase (Invitrogen), according to the manufacturer's
instructions. The sequences of the FAMLF forward and reverse
primers (which were designed to include the BglII and
SalI restriction sites but not the stop codon) were:
forward, 5′-GGAAGATCTATGAAAACAGGGATTC-3′ and reverse,
5′-ACGCGTCGACTTGATTGTTGGTAAT-3′. The above cDNA samples along with
FAMLF PCR primers were amplified by Taq DNA polymerase
(Invitrogen). The PCR conditions for FAMLF amplification were as
follows: 94°C for 5 min, followed by 35 cycles of 94°C for 45 sec,
54°C for 30 sec, and 72°C for 30 sec and a final extension at 72°C
for 10 min. The products were purified using agarose gel
electrophoresis and subsequently sequenced.
Construction of the eukaryotic expression
vector expressing GFP/FAMLF
The purified PCR product was digested with
BglII and SalI, and then was ligated by T4 DNA ligase
(Invitrogen) with the vector pAcGFP-N1 (Clontech, Mountain View,
CA, USA) that was digested with the same enzymes. Consequently, the
FAMLF coding region was inserted upstream of GFP. The
sequence of the FAMLF/GFP was verified using DNA sequencing.
The resulting FAMLF-containing pAcGFP-N1 vector was used to
transform the competent TOP-10 cells (Invitrogen), which were then
plated on kanamycin agar medium. The positive colonies were
selected, and the recombinant plasmid DNA was subsequently
extracted and sequenced to confirm the existence of the
FAMLF gene sequence.
FAMLF subcellular localization as
determined using the FAMLF/GFP fusion protein
NIH3T3 cells were maintained in RPMI-1640 medium
containing 10% newborn calf serum and incubated with 5%
CO2 at 37°C for 24 h. The FAMLF/GFP plasmid was
transfected into NIH3T3 cells using Lipofectamine 2000 (Invitrogen)
according to the manufacturer's instructions, cultured for an
additional 24 h, fixed with 4% formaldehyde and viewed under a
laser scanning confocal microscope (LSCM). Since the expression of
FAMLF was driven by the GFP promoter in the
FAMLF/GFP fusion gene, green fluorescence was able to be
used as a marker for the expression and localization of the FAMLF
protein.
Results
Antibodies were constructed against a 14-amino acid
sequence located at the N-terminus of the predicted open reading
frame of the FAMLF gene. ELISA detected specific antibody in
the blood collected after but not before immunization (data not
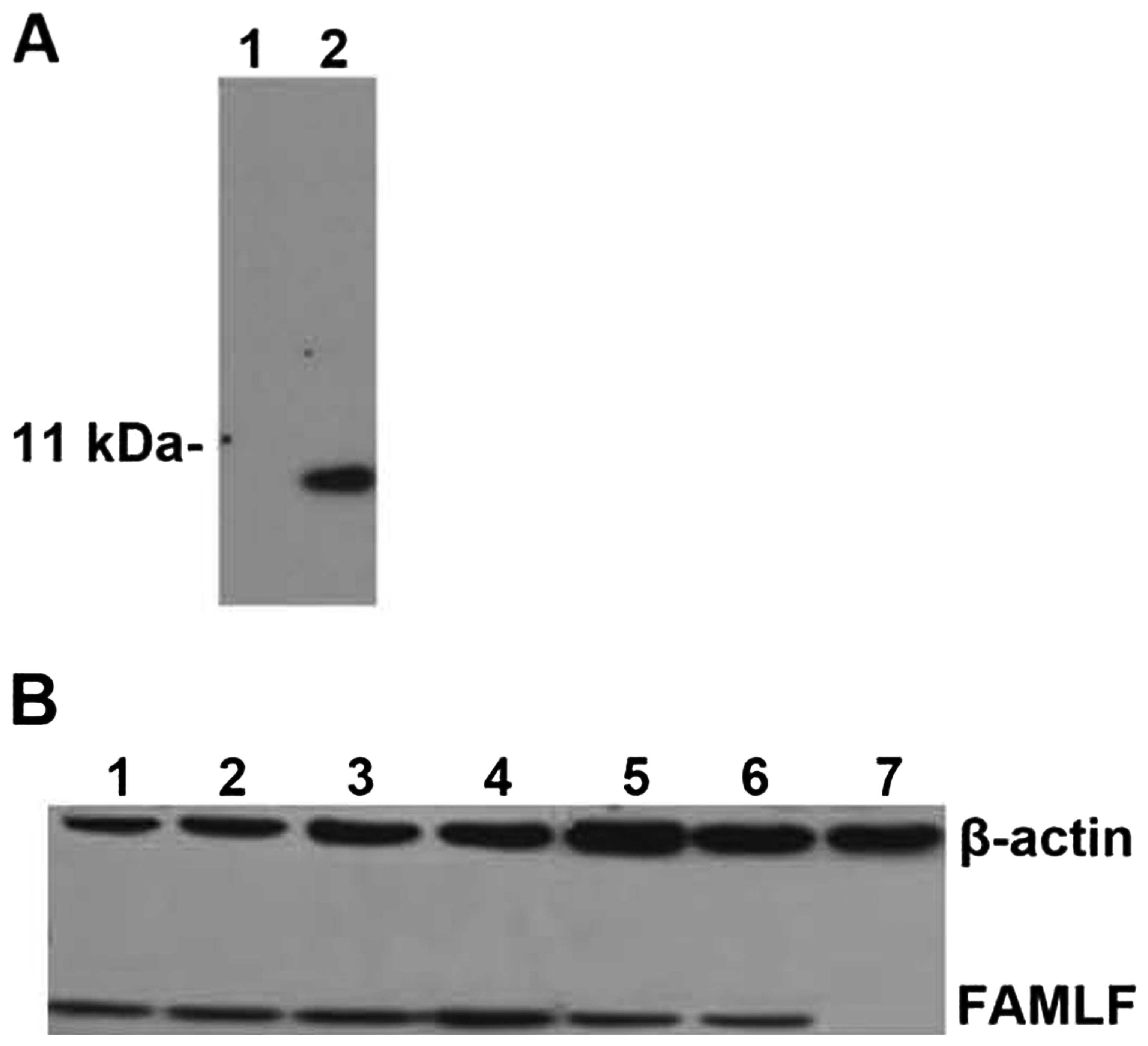
shown). Western blot analysis using the purified anti-FAMLF
polyclonal antibody identified an 8-kDa protein (consistent with
the predicted size of the FAMLF protein) in the COLO205 colon
adenocarcinoma cells but not in the bone marrow cells of a healthy
control (Fig. 1A) and detected the
FAMLF protein in NB4 acute promyelocytic leukemia cells, U937
macrophage-like cells, k562 myeloid leukemia cells, U266 myeloma
cells, HL60 promyelocytic cells, CA46 lymphoma cells, but not in
CCRF CEM (T-cell lymphoblast-like) cells (Fig. 1B).
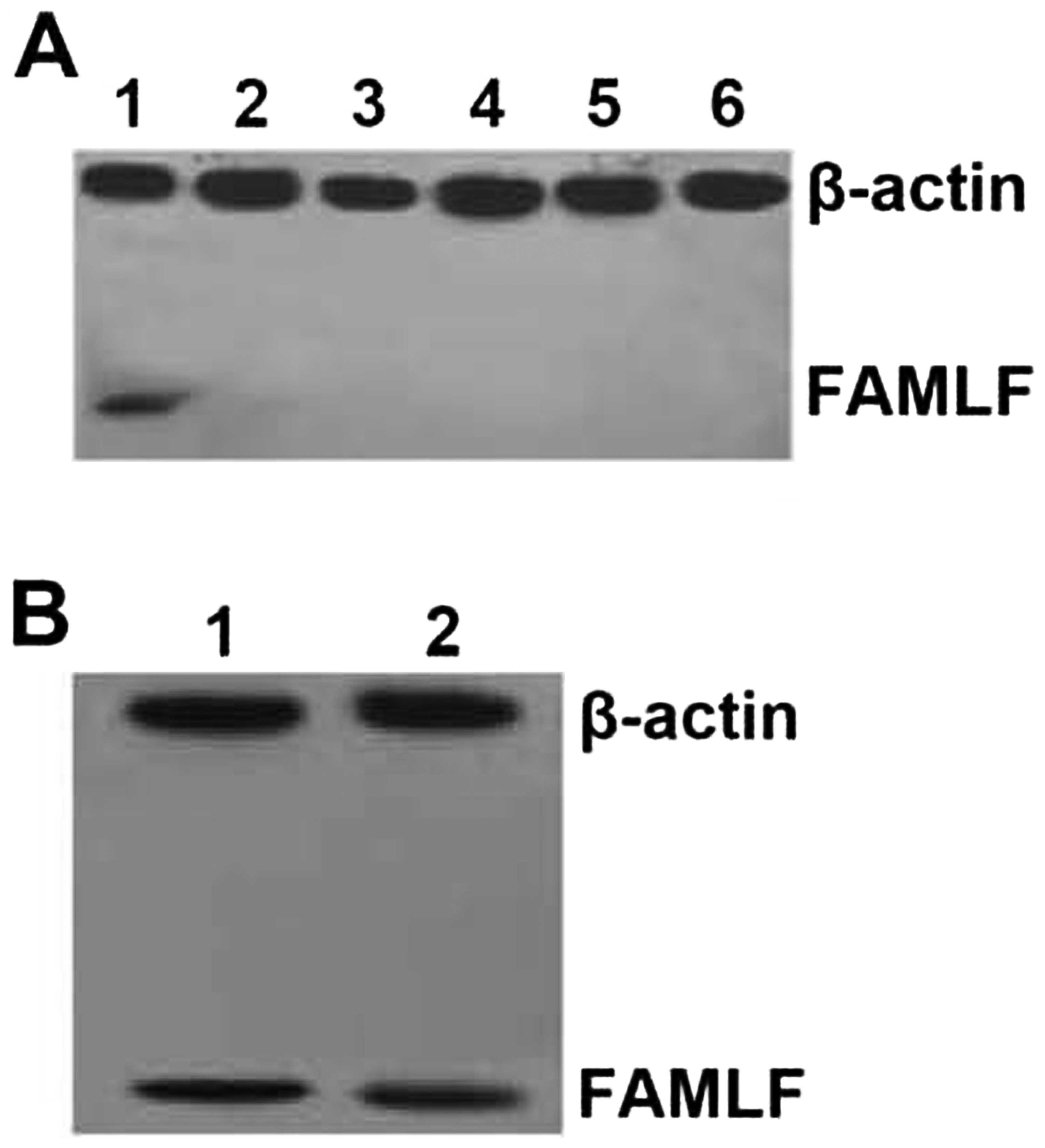
Western blot analysis further detected FAMLF protein
expression in the bone marrow cells from one AML patient (Fig. 2A, lane 1) and one CML patient
(Fig. 2B, lane 2) but not in the
healthy controls (Fig. 2A, lanes
2–6).
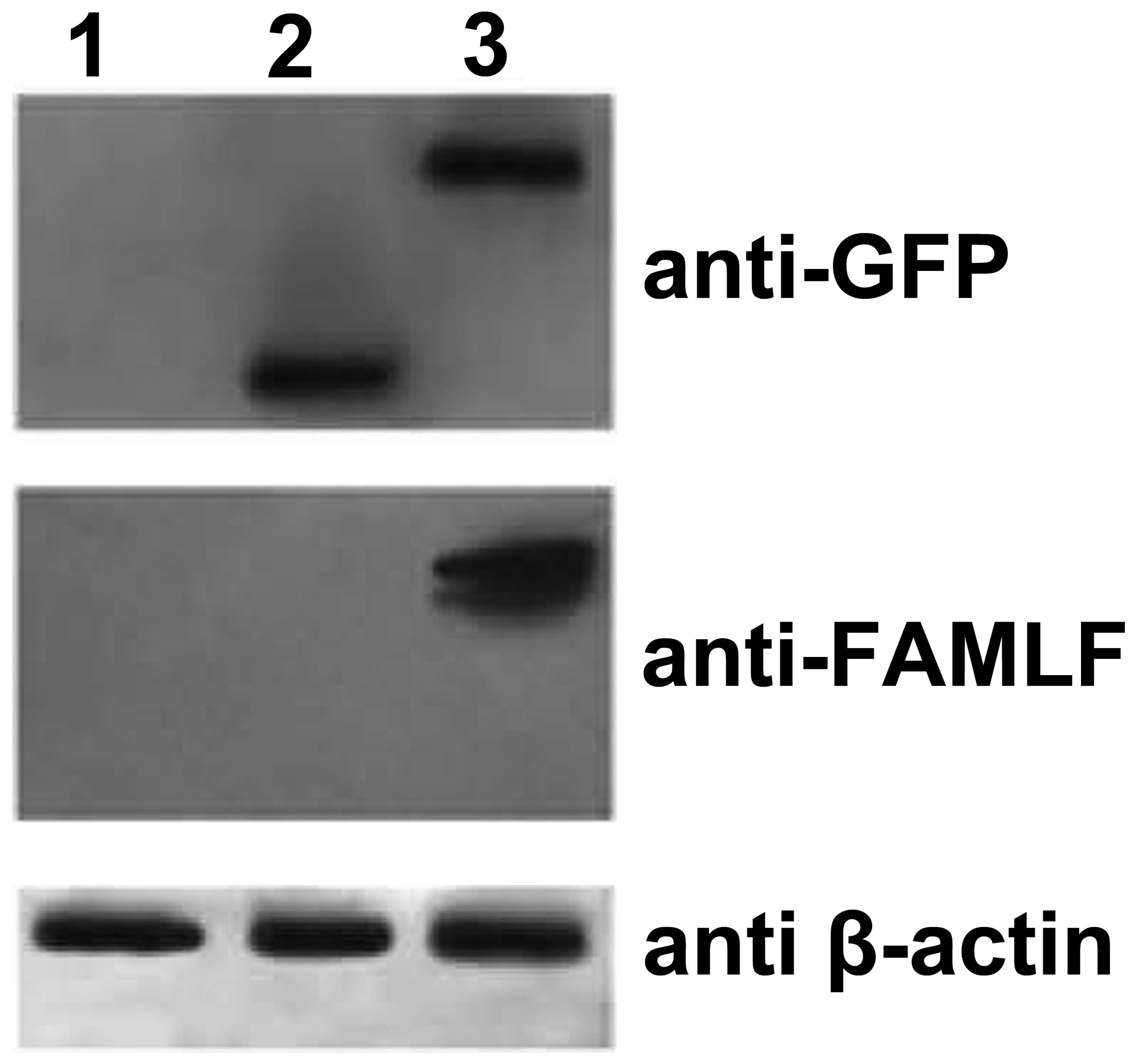
Western blot assay of FAMLF/GFP fusion protein
expression also cross-validated the specificity of the FAMLF
antibody. The anti-GFP antibody identified a 27- and 35-kDa protein
in the NIH3T3 cells transfected with vectors expressing GFP or the
FAMLF-GFP fusion protein, respectively (Fig. 3; top panel). The anti-FAMLF antibody
also detected a 35-kDa protein from the same protein extracts
(Fig. 3; middle panel), suggesting
that both anti-GFP and anti-FAMLF antibodies specifically
recognized the FAMLF/GFP fusion protein.
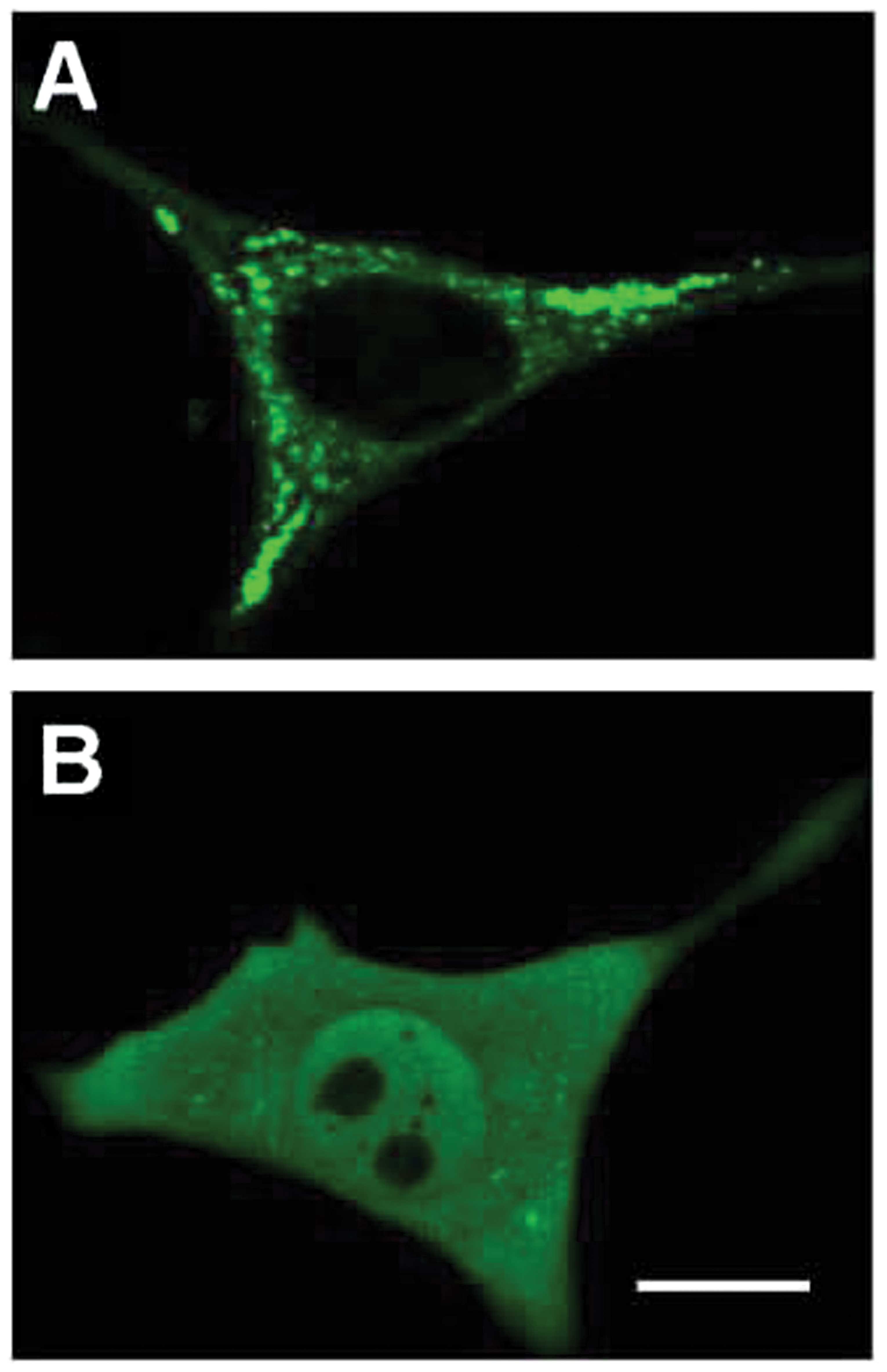
Furthermore, confocal microscopy of NIH3T3 cells
revealed localization of GFP alone only in the cytoplasm (Fig. 4A) and the FAMLF/GFP fusion protein
in both the nucleus and the cytoplasm (Fig. 4B).
Discussion
The FAMLF antibody detected a single protein in the
extract of bone marrow cells from one AML patient and one CML
patient, but not from the healthy controls. The size of the
detected protein (8 kDa) was consistent with the predicted
molecular weight of the FAMLF protein. FAMLF/GFP fusion protein was
found in the nucleus and cytoplasm. Our results indicate for the
first time that the FAMLF protein is associated with AML and may
participate in activities within the nucleus.
Prior to the present study, no antibody against
FAMLF had been developed. In the present study, we used a
previously described strategy for generating antibodies against
particular proteins (3,4,6). This
approach requires the selection of a specific peptide epitope. In
general, to enhance antigen immunoreactivity, the peptide should
have strong hydrophilicity and a stable conformation, and either
end of the peptide should be linear to maximize its availability
for antibody binding (5,7). Using this criterion, we selected a
14-amino acid sequence (CALWVSGIFVDEVI) at the N-terminus of the
FAMLF protein that was predicted to have high hydrophilicity, be
readily accessible to antibody, and be specific to FAMLF, thereby
reducing the likelihood of non-specific binding.
We also maximized the likelihood of producing high
quality antibodies by careful selection of animals, preparation of
the adjuvant, selection of the antigen dose injected and
maximization of immunogenicity. In regards to maximizing the
immunogenicity of the antigen, the synthesized peptide was purified
to ~92% purity, conjugated to KLH, and emulsified in adjuvant (CFA
and IFA). The IgG titer was monitored using ELISA until the titer
was higher than 1:10,000, and the specificity of the FAMLF antibody
was confirmed using western blot analysis.
Families with familial AML are powerful resources
for the identification of possible underlying genetic determinants
of AML. Many families who carry mutations in the GATA2,
TERT, TERC, RUNX1 and CEBPA genes have
been repeatedly identified (17–22).
In contrast, the FAMLF gene identified in the Fujian AML
family (14) was expressed only in
affected individuals but not in healthy controls, suggesting that
this gene has a distinct pathophysiological role.
A relatively large number of genes with diverse
functions have been associated with AML (1–7),
suggesting distinct underlying mechanisms of AML. The biological
function of FAMLF and its pathophysiological role in AML are
still unclear. In the present study, the expression of FAMLF was
evaluated in only one Fujian family member who had AML. It will be
of interest to determine how penetrant this expression phenotype is
in family members of the Fujian family and other familial and
sporadic AML patients and whether other mutations within the
FAMLF gene, including regulatory regions, are associated
with AML in the Fujian family.
Our findings suggest that the FAMLF protein is a
relatively small protein (8 kDa), may translocate to the nucleus or
be associated with the nuclear membrane, and may act as a
transcription factor to modulate gene expression. The FAMLF/GFP
protein was also found in the cytoplasm, suggesting that the FAMLF
protein may be shuttled between the nucleus and cytoplasm similar
to a transcription factor. Additional research is warranted to
investigate the subcellular localization and biological function of
the FAMLF protein.
In conclusion, we generated an antibody to the
predicted protein of the FAMLF gene and showed, for the
first time, that the FAMLF protein was expressed in several
leukemia cell lines and in the bone marrow cells of an AML patient
and a CML patient. This protein was present in both the nucleus and
the cytosol.
Acknowledgements
The present study was supported by the Science and
Technology Project of the Fujian Provincial Department of Science
and Technology (grant nos. 2003F003 and 2012Y4012, the National
Natural Science Foundation of China (grant nos. 81270609 and
30770909), and major scientific research project funds of Fujian
Medical University (grant no. 09ZD008). We would like to thank all
the family members and volunteers for their participation in the
present study. We also greatly appreciate the guidance and help
from Professor Xu Lin from the Basic Medical School of Fujian
Medical University.
Abbreviations:
|
AML
|
acute myeloid leukemia
|
|
FAMLF
|
familial acute myelogenous
leukemia-related factor
|
References
|
1
|
Riva L, Luzi L and Pelicci PG: Genomics of
acute myeloid leukemia: the next generation. Front Oncol. 2:402012.
View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Marcucci G, Maharry K, Radmacher MD,
Mrozek K, Vukosavljevic T, Paschka P, Whitman SP, Langer C, Baldus
CD, Liu CG, Ruppert AS, Powell BL, Carroll AJ, Caligiuri MA, Kolitz
JE, Larson RA and Bloomfield CD: Prognostic significance of, and
gene and microRNA expression signatures associated with, CEBPA
mutations in cytogenetically normal acute myeloid leukemia with
high-risk molecular features: a Cancer and Leukemia Group B Study.
J Clin Oncol. 26:5078–5087. 2008. View Article : Google Scholar
|
|
3
|
Schlenk RF, Döhner K, Krauter J, Frohling
S, Corbacioglu A, Bullinger L, Habdank M, Spath D, Morgan M, Benner
A, Schlegelberger B, Heil G, Ganser A and Döhner H; German-Austrian
Acute Myeloid Leukemia Study Group. Mutations and treatment outcome
in cytogenetically normal acute myeloid leukemia. New Engl J Med.
358:1909–1918. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Birg F, Courcoul M, Rosnet O, Bardin F,
Pebusque MJ, Marchetto S, Tabilio A, Mannoni P and Birnbaum D:
Expression of the FMS/KIT-like gene FLT3 in human acute leukemias
of the myeloid and lymphoid lineages. Blood. 80:2584–2593.
1992.PubMed/NCBI
|
|
5
|
Cairoli R, Beghini A, Grillo G, Nadali G,
Elice F, Ripamonti CB, Colapietro P, Nichelatti M, Pezzetti L,
Lunghi M, Cuneo A, Viola A, Ferrara F, Lazzarino M, Rodeghiero F,
Pizzolo G, Larizza L and Morra E: Prognostic impact of c-KIT
mutations in core binding factor leukemias: an Italian
retrospective study. Blood. 107:3463–3468. 2006.PubMed/NCBI
|
|
6
|
Gale RE, Hills R, Kottaridis PD, Srirangan
S, Wheatley K, Burnett AK and Linch DC: No evidence that FLT3
status should be considered as an indicator for transplantation in
acute myeloid leukemia (AML): an analysis of 1135 patients,
excluding acute promyelocytic leukemia, from the UK MRC AML10 and
12 trials. Blood. 106:3658–3665. 2005. View Article : Google Scholar
|
|
7
|
Ommen HB, Nyvold CG, Braendstrup K,
Andersen BL, Ommen IB, Hasle H, Hokland P and Ostergaard M: Relapse
prediction in acute myeloid leukaemia patients in complete
remission using WT1 as a molecular marker: development of a
mathematical model to predict time from molecular to clinical
relapse and define optimal sampling intervals. Br J Haematol.
141:782–791. 2008. View Article : Google Scholar
|
|
8
|
Dohner H, Estey EH, Amadori S, Appelbaum
FR, Büchner T, Burnett AK, Dombret H, Fenaux P, Grimwade D, Larson
RA, Lo-Coco F, Naoe T, Niederwieser D, Ossenkoppele GJ, Sanz MA,
Sierra J, Tallman MS, Löwenberg B and Bloomfield CD; European
LeukemiaNet. Diagnosis and management of acute myeloid leukemia in
adults: recommendations from an international expert panel, on
behalf of the European LeukemiaNet. Blood. 115:453–474. 2010.
View Article : Google Scholar : PubMed/NCBI
|
|
9
|
O'Donnell MR, Abboud CN, Altman J,
Appelbaum FR, Arber DA, Attar E, Borate U, Coutre SE, Damon LE,
Goorha S, Lancet J, Maness LJ, Marcucci G, Millenson MM, Moore JO,
Ravandi F, Shami PJ, Smith BD, Stone RM, Strickland SA, Tallman MS,
Wang ES, Naganuma M and Gregory KM: Acute myeloid leukemia. J Natl
Compr Cancer Netw. 10:984–1021. 2012.
|
|
10
|
Karp JE, Flatten K, Feldman EJ, Greer JM,
Loegering DA, Ricklis RM, Morris LE, Ritchie E, Smith BD, Ironside
V, Talbott T, Roboz G, Le SB, Meng XW, Schneider PA, Dai NT, Adjei
AA, Gore SD, Levis MJ, Wright JJ, Garrett-Mayer E and Kaufmann SH:
Active oral regimen for elderly adults with newly diagnosed acute
myelogenous leukemia: a preclinical and phase 1 trial of the
farnesyltransferase inhibitor tipifarnib (R115777, Zarnestra)
combined with etoposide. Blood. 11:4841–4852. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Tazzari PL, Tabellini G, Ricci F, Papa V,
Bortul R, Chiarini F, Evangelisti C, Martinelli G, Bontadini A,
Cocco L, McCubrey JA and Martelli AM: Synergistic proapoptotic
activity of recombinant TRAIL plus the Akt inhibitor perifosine in
acute myelogenous leukemia cells. Cancer Res. 68:9394–9403. 2008.
View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Moore J, Seiter K, Kolitz J, Stock W,
Giles F, Kalaycio M, Zenk D and Marcucci G: A Phase II study of
Bcl-2 antisense (oblimersen sodium) combined with gemtuzumab
ozogamicin in older patients with acute myeloid leukemia in first
relapse. Leuk Res. 30:777–783. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Furukawa Y, Vu HA, Akutsu M, Odgerel T,
Izumi T, Tsunoda S, Matsuo Y, Kirito K, Sato Y, Mano H and Kano Y:
Divergent cytotoxic effects of PKC412 in combination with
conventional antileukemic agents in FLT3 mutation-positive versus
-negative leukemia cell lines. Leukemia. 21:1005–1014. 2007.
|
|
14
|
Zhang YW, Wang SY, Lin X and Wang CY:
Identification of differentially expressed genes in familial acute
myelogenous leukemia by suppression subtractive hybridization.
Zhonghua Yi Xue Za Zhi. 87:533–537. 2007.(In Chinese).
|
|
15
|
Wang CY, Wang SY, Lin X, Zhang YW and Li
JG: Full-length cloning of a novel gene, ELF2C, related to familial
acute myelo-genous leukemia. Zhonghua Yi Xue Za Zhi. 87:2245–2248.
2007.(In Chinese).
|
|
16
|
Li JG, Wang SY, Huang YM and Wang CY:
Full-length cDNA cloning and biological function analysis of a
novel gene FAMLF related to familial acute myelogenous leukemia.
Zhonghua Yi Xue Za Zhi. 88:2667–2671. 2008.(In Chinese).
|
|
17
|
Hahn CN, Chong CE, Carmichael CL, Wilkins
EJ, Brautigan PJ, Li XC, Babic M, Lin M, Carmagnac A, Lee YK, Kok
CH, Gagliardi L, Friend KL, Ekert PG, Butcher CM, Brown AL, Lewis
ID, To LB, Timms AE, Storek J, Moore S, Altree M, Escher R, Bardy
PG, Suthers GK, D'Andrea RJ, Horwitz MS and Scott HS: Heritable
GATA2 mutations associated with familial myelodysplastic syndrome
and acute myeloid leukemia. Nat Genet. 43:1012–1017. 2011.
View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Holme H, Hossain U, Kirwan M, Walne A,
Vulliamy T and Dokal I: Marked genetic heterogeneity in familial
myelodysplasia/acute myeloid leukaemia. Br J Haematol. 158:242–248.
2012. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Preudhomme C, Renneville A, Bourdon V,
Philippe N, Roche-Lestienne C, Boissel N, Dhedin N, André JM,
Cornillet-Lefebvre P, Baruchel A, Mozziconacci MJ and Sobol H: High
frequency of RUNX1 biallelic alteration in acute myeloid leukemia
secondary to familial platelet disorder. Blood. 113:5583–5587.
2009. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Renneville A, Mialou V, Philippe N,
Kagialis-Girard S, Biggio V, Zabot MT, Thomas X, Bertrand Y and
Preudhomme C: Another pedigree with familial acute myeloid leukemia
and germline CEBPA mutation. Leukemia. 23:804–806. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Ripperger T, Steinemann D, Göhring G,
Finke J, Niemeyer CM, Strahm B and Schlegelberger B: A novel
pedigree with heterozygous germline RUNX1 mutation causing familial
MDS-related AML: can these families serve as a multistep model for
leukemic transformation? Leukemia. 23:1364–1366. 2009. View Article : Google Scholar
|
|
22
|
Stelljes M, Corbacioglu A, Schlenk RF,
Döhner K, Frühwald MC, Rossig C, Ehlert K, Silling G, Müller-Tidow
C, Juergens H, Döhner H, Berdel WE, Kienast J and Koschmieder S:
Allogeneic stem cell transplant to eliminate germline mutations in
the gene for CCAAT-enhancer-binding protein α from hematopoietic
cells in a family with AML. Leukemia. 25:1209–1210. 2011.PubMed/NCBI
|