Introduction
Colorectal cancer is a major medical issue
worldwide. It is the third most common cancer in both genders and
the second leading cause of cancer-related mortality in the United
States (1). In Thailand, although
the incidence is low, colorectal cancer is the second most common
cancer in men and the third most common cancer in women (2). Colorectal cancer is the most frequent
type of cancer among individuals aged 50 years and older,
suggesting that the majority of these cancers occur in elder
people. However, it has been reported that the incidence rate of
colorectal cancer in young individuals (ages 20–49 years) has
increased 1.5%/year in men and 1.6%/year in women (3). Therefore, early screening tests are of
great important to prevent colorectal cancer and deaths.
The development of colorectal cancer can take years
or decades and mainly progresses through the accumulation of many
genetic mutations (4). Elevated
glucose consumption is a necessary component of carcinogenesis
(5). Clinical imaging of primary
and metastatic cancers has clearly demonstrated that a common trait
of human malignancies including colorectal cancer is elevation in
glucose flux compared to normal tissues (6). Changes in the glucose metabolism of
tumors lead to the alteration of certain metabolic pathways,
including the hexosamine biosynthesis pathway (HBP), a relatively
minor branch of glycolysis. A small fraction of glucose enters the
HBP and produces uridine diphosphate N-acetylglucosamine
(UDP-GlcNAc) (7). UDP-GlcNAc is a
donor substrate used for classical glycosylation as well as
O-GlcNAcylation. Unlike classical glycosylation,
O-GlcNAcylation consists of the attachment of a single
N-acetylglucosamine (GlcNAc) molecule to serine and
threonine residues, which occurs mainly on cytoplasmic, nuclear and
mitochondrial proteins. This glycosylation is not static but
dynamically regulated by 2 key enzymes, O-GlcNAc transferase
(OGT) (8) and O-GlcNAcase
(OGA) (9) which add and remove a
sugar molecule from the proteins, respectively.
Abnormal O-GlcNAc modification is associated
with the pathological status of many diseases including diabetes,
neurodegenerative disease, cardiovascular diseases and cancers
(10). This modification is thought
to act as a nutrient sensor in highly proliferating cells including
cancer cells. The O-GlcNAcylation level was found to be
elevated in colorectal and lung cancer (11). Increasing O-GlcNAc
modification was found to enhance the anchorage-independent growth
of colorectal cells in vitro and vice versa for cells
observed to be reduced of this modification (11). This modification contributes to the
invasion of breast cancer cells in vitro and lung metastasis
in vivo(12). Recently, our
group demonstrated that O-GlcNAc modification was
upregulated in breast cancer tissues and decreased
O-GlcNAcylation was related to inhibition of
anchorage-independent growth in vitro(13). We also showed that a number of
proteins were selectively modified by O-GlcNAc and that
these may be novel potential tumor biomarkers in breast cancer
(13). However, there is little
information concerning O-GlcNAc-modified proteins and their
roles in other types of cancer. In this study, we explored the
expression of O-GlcNAcylation, OGT and OGA levels in primary
colorectal cancer tissues. Using the combination of proteomic
approaches, 2-D IEF/SDS-PAGE, O-GlcNAc immunoblotting,
followed by LC-MS/MS analysis, we identified a number of proteins
associated with aberrant O-GlcNAcylation. These results
provide the expandable spectrum of O-GlcNAcomic in cancer
and suggest that aberrant O-GlcNAc-modified proteins play a
vital role in colorectal cancer.
Materials and methods
Human colon specimens
Cancerous and adjacent normal parts of 7 colorectal
specimens were obtained from Pramongkutklao Hospital (Bangkok,
Thailand). Each sample was divided into 2 parts, one for
histopathological examination and the other was stored at −80°C
until processing. The clinicopathological data for each patient are
shown in Table I. The research
protocol was approved by the Institutional Review Board of the
Royal Thai Army Medical Department, Thailand.
 | Table IClinicopathological data of the
colorectal cancer patients. |
Table I
Clinicopathological data of the
colorectal cancer patients.
| Case no. | Age (years) | Gender | T | N | M | Stage | Grade | Histology |
|---|
| 1 | 65 | Female | 3 | 0 | 0 | IIA | II | Adenocarcinoma |
| 2 | 53 | Female | 2 | 0 | 0 | I | II | Adenocarcinoma |
| 3 | 61 | Female | 3 | 2A | 0 | IIIB | II | Adenocarcinoma |
| 4 | 78 | Male | 3 | 0 | 0 | IIA | II | Adenocarcinoma |
| 5 | 57 | Female | 2 | 1 | 0 | IIIA | II | Adenocarcinoma |
| 6 | 43 | Female | 3 | 0 | 0 | IIA | II | Adenocarcinoma |
| 7 | 53 | Female | 3 | 1 | 0 | IIIB | II | Adenocarcinoma |
Assessment of O-GlcNAc-modified protein,
O-GlcNAc transferase (OGT) and O-GlcNAcase (OGA) levels
Colorectal tissues (30–50 mg) were lysed with 1X
RIPA supplemented with 1% protease inhibitor cocktail and 100 μM
PUGNAc (both from Sigma), an O-GlcNAcase inhibitor,
homogenized and incubated on ice for 30 min. Soluble proteins (50
μg) were separated on 7.5% SDS-PAGE and transferred to PVDF
membranes (Millipore), which were probed with the
anti-O-GlcNAc antibody CTD110.6, OGT (both from Sigma), and
OGA (Abcam). Immunoblots were developed with enhanced
chemiluminescence (GE Healthcare), and the signal was recorded on
X-ray film and captured by a densitometer (Bio-Rad). Protein
loading was compared using gels stained with 0.1% Coomassie
brilliant blue R-250 (CCB) and captured by a densitometer.
Densitometric analysis of O-GlcNAc proteins was performed on
the entire lane of each sample using ImageJ 1.41v (NIH), and the
mean intensity was normalized to the normal groups.
Two-dimensional gel electrophoresis of
O-GlcNAc-modified proteins
Colorectal tissues (30–50 mg) were lysed in 2D lysis
buffer [7M urea, 2M thiourea, 4% CHAPS, 2% DTT, 2% ampholine
(3–10), 1% protease inhibitor cocktail
(Sigma), and 100 μM PUCNAc] homogenized and incubated on ice for 30
min. Due to limitations of sample amount and to reduce the
variation in samples, pooled protein samples were used for
O-GlcNAc identification. Pooled samples (1.5 mg) were
applied by overnight in-gel rehydration of 130-mm nonlinear pH 2.0,
IPG strips (GE Healthcare). The strips were then applied to the
second dimension and run on 10% SDS-PAGE, transferred to PVDF
membranes (Millipore) and immunoblotted with the anti
O-GlcNAc antibody. Immunoblots were developed with enhanced
chemiluminescence (GE Healthcare) for 1 min (short exposure time)
and 20 min (long exposure time), and the signal was recorded on
X-ray film and captured by a densitometer. Immunoblotted membranes
were then stripped with stripping buffer (2% SDS, 62.5 mM Tris-HCl,
pH 6.8 and 0.8% β-mercaptoethanol) and counter-stained using 0.1%
CBB. Duplicate gels were also stained with 0.1% CBB for protein
identification. O-GlcNAc signals of the X-ray film and total
protein expression spots stained on stripped membrane were captured
by a densitometer. Expression levels of individual
O-GlcNAc-modified protein spots of the cancer and normal
groups were measured by ImageJ 1.41v (NIH). Changes in intensity by
>1.5-fold were considered to indicate upregulation or
downregulation of expression of O-GlcNAc levels between
cancer and normal groups, respectively.
Protein identification by LC-MS/MS
O-GlcNAc signals obtained from X-ray film
were aligned with total protein spots on stripped membranes and CCB
gels using Master 2D Platinum 7.0 software (GE Healthcare).
Proteins spots from the CCB gels were excised, destained and
enzymatically digested by trypsin (Promega Corporation). For
protein spots from the PVDF membranes, Zwittergent 3–16 (Merck) at
0.5% was used for blocking prior to trypsin digestion as described
(14). The trypsinized peptides
were analyzed by a capillary LC system (Waters) coupled to a Q-TOF
mass spectrometer (Micromass) as described previously (13). Briefly, tryptic peptides were
injected into a C18 PepMap column (LC Packing) with the gradient of
0.1% formic acid in 3% ACN and 0.1% formic acid in 97% ACN.
Purified samples (6 μl) were injected into the nano-LC. For
ESI-Q-TOF analysis, the automatic scan rate was 1.0 sec with an
interscan delay of 0.1 sec. Parent mass peaks with a range of
400–1600 m/z were selected for MS/MS analysis. MS/MS data were
processed using MassLynx 4.0 software (Micromass) and converted to
PKL files by the ProteinLynx 2.2 software (Waters), which were
analyzed using the MASCOT search engine using the NCBInr database
(http://www.matrixscience.com). Peptide
and fragment mass tolerance were set at 1.2 and 0.6 Da,
respectively. Proteins with molecular weight and pI consistent with
the gel region, having at least one peptide exceeding the score
threshold (P<0.05) were considered as being positively
identified.
Validation of O-GlcNAc-modified
proteins
A subset of the potential O-GlcNAc-modified
proteins identified from mass spectrometry was confirmed using the
direct immunoprecipitation kit (Pierce Biotechnology, Inc.),
performed according to the manufacturer's recommendations. An
O-GlcNAc antibody (CTD110.6) was used for coupling reaction
with the slurry resin (agarose bead) in order to enrich the
O-GlcNAc proteins from all cancer and normal samples (7
cases). Briefly, proteins (1 mg) in 1X RIPA buffer were incubated
with the antibody-coupled resin (5 μl of CTD110.6) with gentle
shaking overnight at 4°C. After washing, 100 μl of 1X sampling
buffer supplemented with 100 mM DTT was added into the column tube
and heated at 95°C for 5 min. The eluted samples were centrifuged
and loaded in 10% SDS-PAGE and immunoblotted with the proteins of
interest including O-GlcNAc, annexin A2 (Abcam), cytokeratin
18 (Chemicon), heat shock cognate protein, and Hsc70 (Abcam).
Intensity of the O-GlcNAc-modified proteins of the cancer
and normal samples was measured by ImageJ 1.41v (NIH).
Data analysis
All data from the densitometric analysis are
presented as means ± standard error (SE). Comparisons were
performed using the Student's t-test, and statistically significant
differences between groups were defined as P<0.05 and
P<0.01.
Results
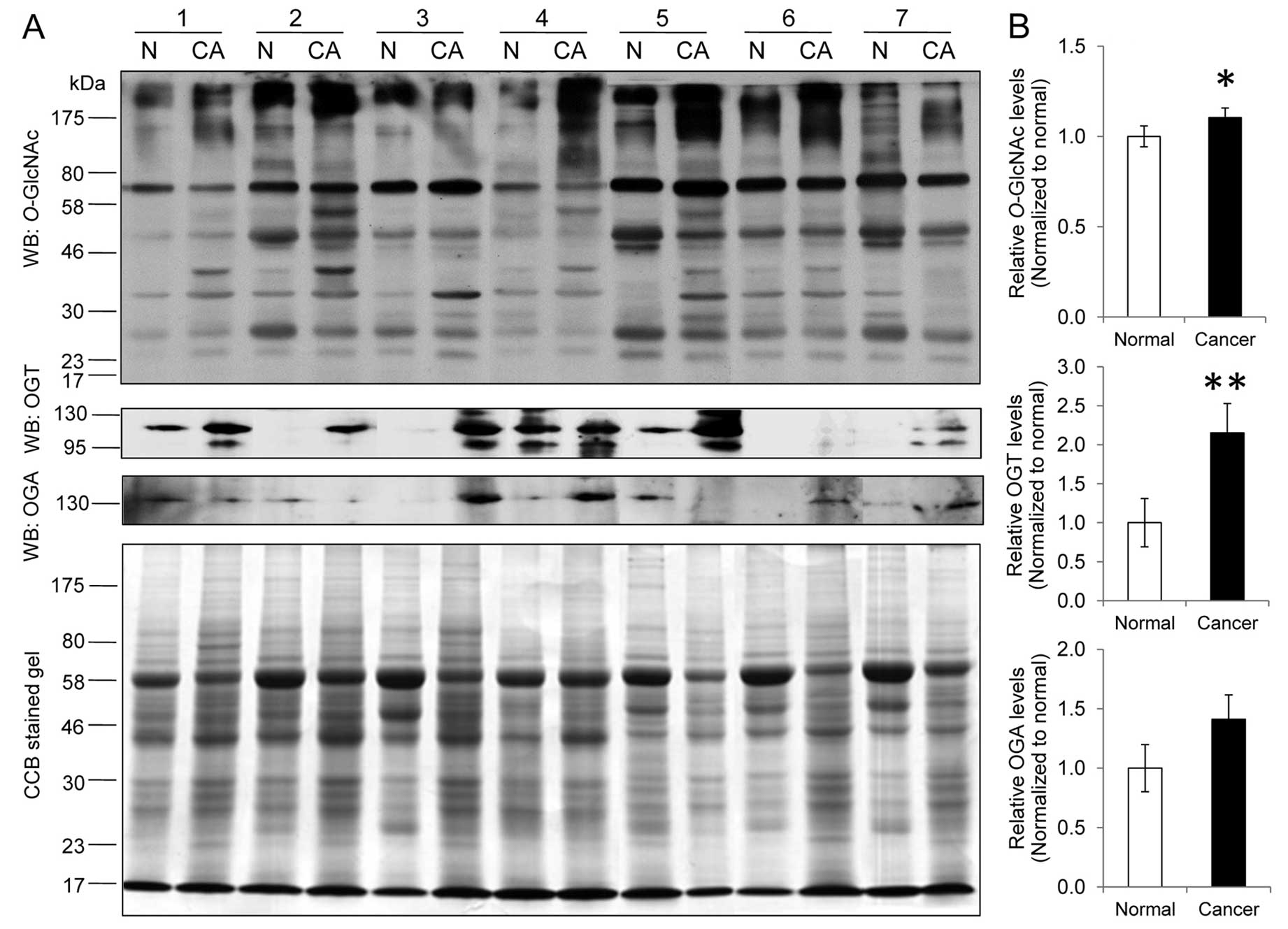
Alteration of O-GlcNAcylation in
colorectal cancer
The expression levels of O-GlcNAcylation and
O-GlcNAc controlling enzymes, OGT and OGA, were examined in
7 cases of colorectal cancer and their adjacent normal tissues
(Fig. 1A). O-GlcNAc
immunoblot analysis revealed a significant increase in the
O-GlcNAcylation expression level in the cancer group
(1.00±0.06 vs. 1.10±0.05, P<0.05) (Fig. 1B). Notably, several O-GlcNAc
bands between 17 and 58 kDa were more predominantly presented in
tumors compared to those in the normal group. The OGT level was
also significantly increased in cancer (1.00±0.31 vs. 2.15±0.37,
P<0.01) whereas the OGA level tended to increase but was not
statistically different (1.00±0.20 vs. 1.41±0.20, P=0.13) in
comparison to the normal groups, respectively (Fig. 1B).
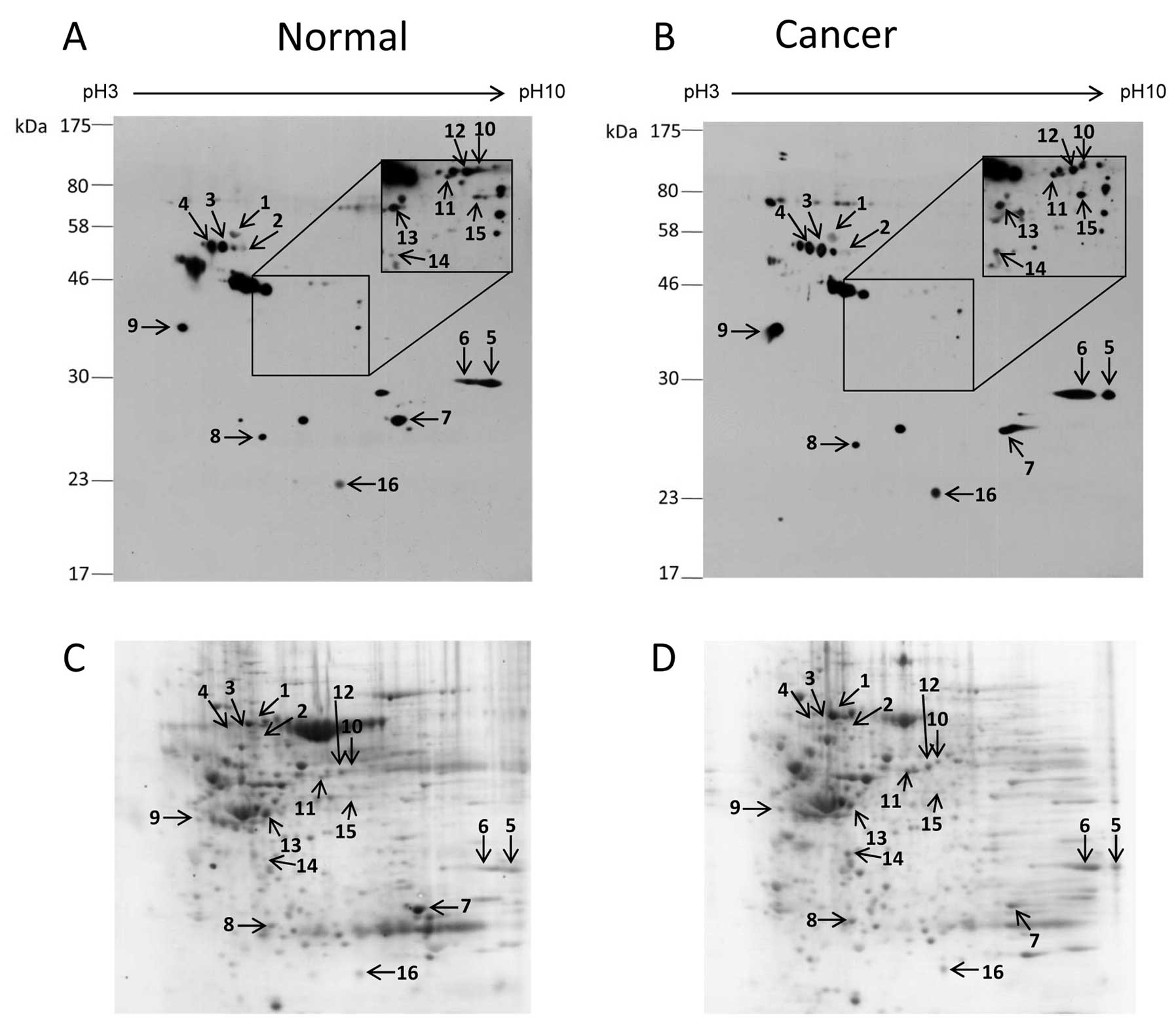
Comparison of O-GlcNAc-modified proteins
between colorectal cancer and normal tissues
Two dimensional gel electrophoresis and
O-GlcNAc immunoblot analysis were performed to identify the
different O-GlcNAc-modified proteins in colorectal cancer.
The pooled protein samples of cancerous and normal tissues were
used to reduce the complexity of the samples and other errors.
Numerous O-GlcNAc spots were present in both the cancer and
normal 2-D O-GlcNAc immunoblots (Fig. 2). However, only 16 O-GlcNAc
protein spots that matched with CCB protein spots were successfully
identified. A list of proteins identified and their O-GlcNAc
expression levels are summarized in Table II. Among the identified proteins, 8
showed increased O-GlcNAc expression in cancer (ratio
>1.5-fold) including cytokeratin 18, heterogeneous nuclear
ribonucleoproteins A2/B1 (hnRNP A2/B1), hnRNP H, annexins A2 and
A7, laminin binding protein, α-tubulin and protein DJ-1. In
contrast, 8 proteins did not differ in regards to the
O-GlcNAc expression level between the cancer and normal
groups (ratio <1.5-fold) including heat shock cognate protein
(Hsc70), hnRNP K, glyceraldehyde-3-phosphate dehydrogenase (GAPDH),
carbonic anhydrase 1, aldehyde dehydrogenase, NADH dehydrogenase,
selenium binding protein 1 and creatine kinase-B. All identified
proteins resulted from digestion of a single protein spot, except
for spot no. 6 which contained 2 proteins, hnRNP A2/B1 and annexin
A2.
 | Table IIList of identified
O-GlcNAc-modified proteins in colorectal tissues. |
Table II
List of identified
O-GlcNAc-modified proteins in colorectal tissues.
| Spot ID | Accession no. | Protein name | Mw/pI matches | Peptide | Score | O-GlcNAc
levela | O-GlcNAc
(refs.) |
|---|
| 1 | gi|62896815 | Heat shock cognate
proteins | 67938/5.32 | 11 | 192 | NC | (13,33,35,36) |
| 2 | gi|30311 | Cytokeratin 18 | 47305/5.27 | 8 | 125 | UP | (13,22–24) |
| 3 | gi|460789 | Heterogeneous
nuclear ribonucleoprotein K | 51040/5.13 | 3 | 27 | NC | (13,33) |
| 4 | gi|460789 | Heterogeneous
nuclear ribonucleoprotein K | 51040/5.13 | 3 | 48 | NC | (13,33) |
| 5 | gi|31645 |
Glyceraldehyde-3-phosphate
dehydrogenase | 36031/8.26 | 6 | 118 | NC | (13,33,35) |
| 6 | gi|4504447 | Heterogeneous
nuclear ribonucleoproteins A2/B1 | 35984/8.67 | 2 | 62 | UP | (13,33) |
| gi|18645167 | Annexin A2 | 38552/7.57 | 3 | 54 | UP | (13) |
| 7 | gi|4502517 | Carbonic anhydrase
1 | 28852/6.59 | 7 | 124 | NC | - |
| 8 | gi|4758788 | NADH
dehydrogenase | 30223/6.99 | 7 | 186 | NC | - |
| 9 | gi|34234 | Laminin binding
protein | 31774/4.84 | 6 | 175 | UP | - |
| 10 | gi|1263008 | Aldehyde
dehydrogenase | 57181/6.41 | 6 | 85 | NC | - |
| 11 | gi|5031753 | Heterogeneous
nuclear ribonucleoprotein H | 49198/5.89 | 3 | 67 | UP | - |
| 12 | gi|16306550 | Selenium-binding
protein 1 | 52358/5.93 | 14 | 203 | NC | - |
| 13 | gi|180555 | Creatine
kinase-B | 42460/5.34 | 6 | 172 | NC | - |
| 14 | gi|340021 | α-Tubulin | 50120/4.94 | 7 | 80 | UP | (25,26) |
| 15 | gi|4502111 | Annexin A7 | 50284/6.25 | 6 | 57 | UP | - |
| 16 | gi|50513593 | Protein DJ-1 | 19886/6.33 | 9 | 105 | UP | (13) |
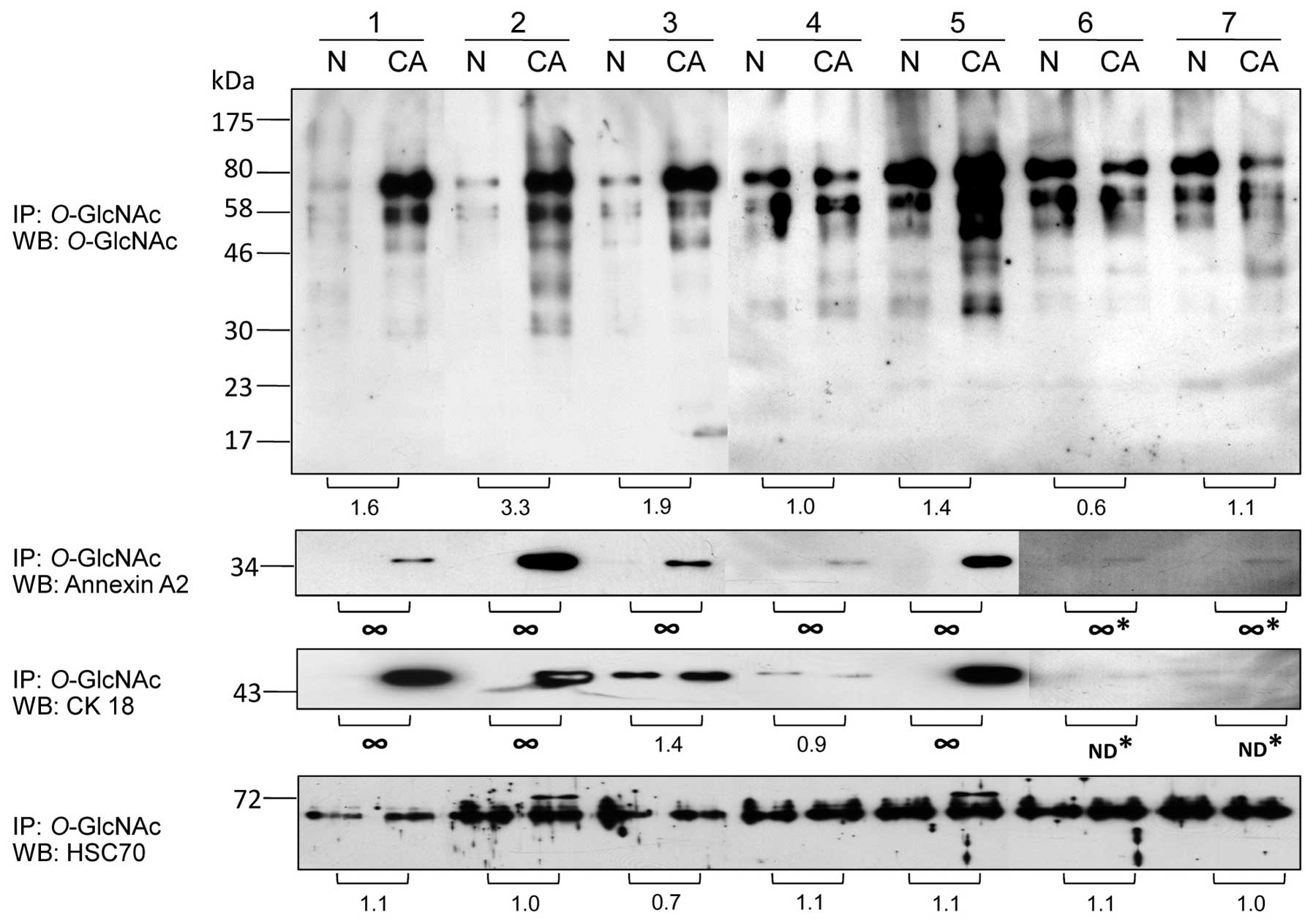
Confirmatory studies of O-GlcNAc-modified
proteins in colorectal cancer
A subset of potential O-GlcNAc-modified
proteins was confirmed using direct immunoprecipitation (IP) with
the O-GlcNAc antibody followed by immunoblotting with
antibodies against the proteins of interest. The results showed
that O-GlcNAc-modified proteins tended to be increased at
higher levels in all cancer samples except sample 6 which showed
lower O-GlcNAc levels in cancer compared to the normal
groups (Fig. 3). O-GlcNAc
association of annexin A2 was increased in all cancer samples
(7/7). O-GlcNAc association of cytokeratin 18 (CK18) was
increased in 4 cancer samples (4/7) while sample 4 had slighly
increased CK18-O-GlcNAc level in normal samples. However,
sample 6 and 7 did not show any O-GlcNAc-CK18 band in both
the normal and cancer samples. Hsc70 associated with
O-GlcNAc expression was slightly altered in all samples, but
sample 2 and 5 which had a high O-GlcNAc enrichment showed
an extra O-GlcNAc band of Hsc70 at 72 kDa which was
undetectable in all normal samples.
Discussion
Abnormal O-GlcNAcylation is implicated in
many metabolic diseases and cancers. In the present study,
O-GlcNAcylation in primary colorectal tumor tissues was
examined in terms of the expression levels of O-GlcNAc, OGT
and OGA, as well as the identification of O-GlcNAc-modified
proteins.
O-GlcNAc immunoblots showed a significantly
different O-GlcNAcylated protein pattern between the cancer
and normal groups, with several O-GlcNAc bands in the
molecular weight range from 17 to 58 kDa appearing to increase in
cancer (Fig. 1). This increase was
associated with an increased expression level of OGT while the OGA
level was not significantly different in cancer compared to the
normal groups (Fig. 1). This is
consistent with a report by Mi and co-workers who demonstrated by
immunohistochemistry that the global O-GlcNAcylation was
significantly elevated in colorectal cancer patients using a
different O-GlcNAc antibody (RL2) and this elevation was
related to an increased OGT level (11). Several lines of evidence have shown
that OGT and O-GlcNAc levels are increased in many malignant
tumors including breast, lung, liver and prostate cancer (11–13,15,16).
Recently, it was reported that a metastatic clone of colorectal
cancer, SW620, had increased O-GlcNAcylation when compared
with its primary clone, SW480, and this increase was related to the
level of OGA expression in the metastatic clone (17). OGT and OGA may act as key regulatory
proteins in cancer, and abnormal addition/removal of
O-GlcNAcylation in certain proteins, at least, affects
protein functions.
In this study, we successfully identified 16
proteins modified by O-GlcNAcylation using proteomic
approaches including 2-D IEF/SDS-PAGE followed by O-GlcNAc
immunoblotting and LC-MS/MS, and validated several
O-GlcNAc-modified proteins using O-GlcNAc
immunoprecipitation (Table II,
Figs. 2 and 3). 2-D proteomic profiles revealed
differences in various O-GlcNAc-modified proteins between
normal and malignant tumors. Eight proteins were associated with
increased O-GlcNAc in cancer, namely annexins A2 and A7,
CK18, α-tubulin, hnRNP A2/B1 and H, protein DJ-1 and laminin
binding protein.
Annexins are a group of phospholipid-binding
proteins in a calcium-dependent manner. They play important roles
in cellular signaling such as exocytosis and endoctyosis,
cytoskeletal formation, cell matrix interaction, cell division and
apoptosis (18,19). We found that
O-GlcNAc-annexins A2 and A7 were increased in colorectal
cancer cases (Fig. 2, Table II). We previously reported that
O-GlcNAc-annexin A2 was increased in breast cancer tissues
(13). It was reported that annexin
A2 was significantly increased in primary tumors compared to normal
colon and that increased expression of annexin A2 is related to
increasing tumor stage (20).
Annexin A7 has biological and genetic properties expected of a
tumor-suppressor gene. The loss of annexin A7 is associated with
distant metastasis in gastric cancer (21). At present, there is no information
concerning the glycosylation of annexins. Abnormal
O-GlcNAc-annexins may confer benefits to cancer in terms of
cellular networking, vesicle transportation, cell matrix formation
and metastasis.
Two proteins involved in cell structure, cytokeratin
18 (CK18) and α-tubulin, are O-GlcNAcylated. CK18 is a type
I cytokeratin which forms with its filament partner, cytokeratin 8
(CK8). We found that O-GlcNAc-CK18 was increased in
colorectal cancer samples similar to that found in our previous
study on breast cancer (13). CK18
was also reported to be modified by O-GlcNAc, at least in 3
sites (S30/S31/S49) (22).
Augmentation of O-GlcNAc-CK18 was associated with increased
solubility and decreased cellular levels whereas absence of
O-GlcNAc on CK18 made it more stable (23). O-GlcNAcylated CK18 has been
shown to play a protective role in epithelial injury and this
glycosylation is required for the recruitment and activation of AKT
and protein kinase Cθ (24).
Tubulin is a member of the microtubule family, which is involved in
many essential cellular processes, including cell division and
intracellular transport. We found that O-GlcNAc-α-tubulin
was increased in cancer. It was reported that α-tubulin was
identified as an O-GlcNAc-modified protein (25). Moreover, it was shown that an
increase in O-GlcNAc-α-tubulin led to reduced
heterodimerization and that O-GlcNAcylated tubulin did not
polymerize into microtubules (26).
Aberrant O-GlcNAc-CK18 and O-GlcNAc-tubulin,
therefore, at least may create appropriate structural organization
and signaling in the cytoskeletal network of cancer cells. However,
the role of this glycosylation on the protein cytoskeleton still
requires further study in cancer biology.
The hnRNPs are a group of RNA-binding proteins
implicated in a variety of processes in mRNA metabolism including
pre-mRNA splicing, mRNA transport and translation (27,28).
We found that hnRNP A2/B1 and H were increased in terms of
O-GlcNAc modification (Table
II). hnRNP A2/B1 interacts with at least 20 other different
hnRNPs and RNA in the nucleus which modulate pre-mRNA processing
and splicing (29). Fielding et
al(30) reported that hnRNP
A2/B1 is detected in patients with lung cancer or lung neoplasms
suggesting that it is a clinical marker for early lung cancer
detection. HnRNP H functions in the splicing of selected target
mRNAs such as Bcl-x, a member of the Bcl-2 family of proteins that
are key regulators of apoptosis (31). Although overexpression and
subcellular distribution of many hnRNPs have been described in
colorectal cancer, the pathogenesis of cancer linked with hnRNPs is
unclear (32). To date, the
regulation of hnRNPs is poorly understood; however, phosphorylation
is thought to be a regulator of the interaction between nucleic
acids and protein partners. Wang et al(33) reported that 9 members of the hnRNP
family including hnRNP K, U, R, U, A2/B1, A1, C1/C2, H3 and A/B,
are likely to be modified by O-GlcNAc in COS7 cells. We
previously reported that 5 hnRNPs including hnRNP U-like protein 2,
hnRNP K, hnRNP F, hnRNP M isoform a and hnRNP A2/B1, were all
increased in their O-GlcNAcylation in breast cancer
(13). At present, there is no
information concerning the functions of O-GlcNAc and the
association with the site(s) of this modification of hnRNPs. It is
possible that increasing O-GlcNAcylation of the hnRNP family
may interplay with phosphorylation that regulates alterative mRNA
splicing and processing, as well as gene expression of proteins and
promotes a phenotype of cancer.
Protein DJ-1 plays a role as an antioxidant and/or a
molecular chaperone. It is also a regulator of the tumor-suppressor
protein, PTEN, with stimulatory effects on PI3K-AKT/PKB signaling
in lung carcinoma. We previously reported that O-GlcNAc-DJ-1
was increased in breast cancer (13). Although DJ-1 was
O-GlcNAc-altered in colorectal cancer, the effect of this
modification in stress response remains unclear.
Laminin binding protein (ribosomal protein SA) is
involved in the assembly and/or stability of the 40S ribosomal
subunit and also functions as a cell surface receptor for laminin
which is implicated in a wide variety of biological processes
including cell adhesion, differentiation, migration, signaling and
metastasis (34). O-GlcNAc
modification of laminin binding protein was increased in colorectal
cancer which is similar to our results found in breast cancer
(13).
Another group of proteins associated with slightly
altered O-GlcNAc in colorectal cancer includes Hsc70, hnRNP
K, GAPDH, carbonic anhydrase 1, creatine kinase-B, selenium binding
protein 1, aldehyde dehydrogenase and NADH dehydrogenase. Hsc70, a
constitutively expressed protein, is a chaperone protein which
plays vital roles in protein folding and degradation. We found that
a major band of O-GlcNAc-Hsc70 (~70 kDa) was similar between
normal and cancer groups but a minor band at 72 kDa with high
O-GlcNAc enrichment was found in cancer samples. Our group
and others previously reported that Hsc70 was modified/associated
with O-GlcNAc (13,33,35,36).
GAPDH is a glycolytic enzyme, which breaks down glucose to provide
energy. Our and other groups have reported that GAPDH was modified
by O-GlcNAc (13,33,35).
Carbonic anhydrase (CA) catalyzes the hydration of CO2
to form HCO3− and H+ ions which
are subsequently contributed to an acidic condition in cells. CA1
is found in the cytoplasm of epithelial cells, specifically
expressed at high levels in the colon (37). Overexpression of CAs in cancer cells
was found to contribute to cancer invasiveness, and CA inhibitors
suppress the invasiveness of renal cancer cells in
vitro(38). Creatine kinase
(CK) is an enzyme catalyzing the reversible phosphorylation of
creatine by ATP, thus playing an important role in energy
metabolism. High concentrations of CK may appear in cells with
highly cellular activities such as cancer cells (39). Selenium binding protein 1 (SBP1) is
a 56-kDa protein which is found in various cell types. The function
of SBP1 is unknown. However, it was reported that levels of SBP1
are significantly decreased in various epithelial cancers including
colorectal cancer (40,41). Aldehyde dehydrogenase is an
important enzyme involved in acetaldehyde metabolism. NADH
dehydrogenase is an enzyme located in the inner mitochondrial
membrane which catalyzes the transfer of electrons from NADH to
coenzyme Q. It is still unknown how O-GlcNAc modification
occurs in mitochondria.
In conclusion, we demonstrated that
O-GlcNAcylation was increased in primary colorectal cancer
patients (grade II) and that this increase was associated with an
increased OGT expression level. Proteomic profiles revealed that
O-GlcNAc-modified proteins are involved in the cytoskeleton,
oxidative response and environment, and RNA metabolism. Some
proteins were increasingly modified by O-GlcNAc. Among these
modified proteins, aberrantly increased O-GlcNAc-modified
annexin A2 was overexpressed in all cancer samples. Taken together,
our findings on the alteration of O-GlcNAcylation provide
the expanding spectrum of O-GlcNAcomics in cancer and
suggest that abnormal O-GlcNAc-modified proteins,
particularly annexin A2, may be novel biomarkers of colorectal
cancer. Further studies are needed to clarify the biological roles
of O-GlcNAc-modified proteins in colorectal cancer in order
to understand their regulation and function in cancer.
Acknowledgements
This study was supported by the Chulabhorn Research
Institute and Chulabhorn Graduate Institute, Thailand.
References
|
1
|
Siegel R, Ward E, Brawley O and Jemal A:
Cancer statistics, 2011: the impact of eliminating socioeconomic
and racial disparities on premature cancer deaths. CA Cancer J
Clin. 61:212–236. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Attasara P: Hospital-Based Cancer Registry
2010. National Cancer Institute; Thailand, Bangkok: 2011
|
|
3
|
Siegel RL, Jemal A and Ward EM: Increase
in incidence of colorectal cancer among young men and women in the
United States. Cancer Epidemiol Biomarkers Prev. 18:1695–1698.
2009. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Gryfe R, Swallow C, Bapat B, Redston M,
Gallinger S and Couture J: Molecular biology of colorectal cancer.
Curr Probl Cancer. 21:233–300. 1997. View Article : Google Scholar
|
|
5
|
Gillies RJ, Robey I and Gatenby RA: Causes
and consequences of increased glucose metabolism of cancers. J Nucl
Med. 49(Suppl 2): S24–S42. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Chowdhury FU, Shah N, Scarsbrook AF and
Bradley KM: (18F)FDG PET/CT imaging of colorectal cancer: a
pictorial review. Postgrad Med J. 86:174–182. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Marshall S, Bacote V and Traxinger RR:
Discovery of a metabolic pathway mediating glucose-induced
desensitization of the glucose transport system. Role of hexosamine
biosynthesis in the induction of insulin resistance. J Biol Chem.
266:4706–4712. 1991.
|
|
8
|
Kreppel LK, Blomberg MA and Hart GW:
Dynamic glycosylation of nuclear and cytosolic proteins. Cloning
and characterization of a unique O-GlcNAc transferase with
multiple tetratricopeptide repeats. J Biol Chem. 272:9308–9315.
1997. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Gao Y, Wells L, Comer FI, Parker GJ and
Hart GW: Dynamic O-glycosylation of nuclear and cytosolic
proteins: cloning and characterization of a neutral, cytosolic
β-N-acetylglucosaminidase from human brain. J Biol Chem.
276:9838–9845. 2001.PubMed/NCBI
|
|
10
|
Hart GW, Slawson C, Ramirez-Correa G and
Lagerlof O: Cross talk between O-GlcNAcylation and phosphorylation:
roles in signaling, transcription, and chronic disease. Annu Rev
Biochem. 80:825–858. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Mi W, Gu Y, Han C, et al: O-GlcNAcylation
is a novel regulator of lung and colon cancer malignancy. Biochim
Biophys Acta. 1812:514–519. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Gu Y, Mi W, Ge Y, et al: GlcNAcylation
plays an essential role in breast cancer metastasis. Cancer Res.
70:6344–6351. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Champattanachai V, Netsirisawan P,
Chaiyawat P, et al: Proteomic analysis and abrogated expression of
O-GlcNAcylated proteins associated with primary breast
cancer. Proteomics. 13:2088–2099. 2013.PubMed/NCBI
|
|
14
|
Pham VC, Henzel WJ and Lill JR: Rapid
on-membrane proteolytic cleavage for Edman sequencing and mass
spectrometric identification of proteins. Electrophoresis.
26:4243–4251. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Zhu Q, Zhou L, Yang Z, et al:
O-GlcNAcylation plays a role in tumor recurrence of hepatocellular
carcinoma following liver transplantation. Med Oncol. 29:985–993.
2012. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Lynch TP, Ferrer CM, Jackson SR, Shahriari
KS, Vosseller K and Reginato MJ: Critical role of O-Linked
β-N-acetylglucosamine transferase in prostate cancer
invasion, angiogenesis, and metastasis. J Biol Chem.
287:11070–11081. 2012.
|
|
17
|
Yehezkel G, Cohen L, Kliger A, Manor E and
Khalaila I: O-linked β-N-acetylglucosaminylation
(O-GlcNAcylation) in primary and metastatic colorectal
cancer clones and effect of N-acetyl-β-D-glucosaminidase
silencing on cell phenotype and transcriptome. J Biol Chem.
287:28755–28769. 2012. View Article : Google Scholar
|
|
18
|
Gerke V and Moss SE: Annexins: from
structure to function. Physiol Rev. 82:331–371. 2002.PubMed/NCBI
|
|
19
|
Gerke V, Creutz CE and Moss SE: Annexins:
linking Ca2+ signalling to membrane dynamics. Nat Rev
Mol Cell Biol. 6:449–461. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Duncan R, Carpenter B, Main LC, Telfer C
and Murray GI: Characterisation and protein expression profiling of
annexins in colorectal cancer. Br J Cancer. 98:426–433. 2008.
View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Hsu PI, Huang MS, Chen HC, et al: The
significance of ANXA7 expression and its correlation with poor
cellular differentiation and enhanced metastatic potential of
gastric cancer. J Surg Oncol. 97:609–614. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Ku NO and Omary MB: Identification and
mutational analysis of the glycosylation sites of human keratin 18.
J Biol Chem. 270:11820–11827. 1995. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Srikanth B, Vaidya MM and Kalraiya RD:
O-GlcNAcylation determines the solubility, filament
organization, and stability of keratins 8 and 18. J Biol Chem.
285:34062–34071. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Ku NO, Toivola DM, Strnad P and Omary MB:
Cytoskeletal keratin glycosylation protects epithelial tissue from
injury. Nat Cell Biol. 12:876–885. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Walgren JL, Vincent TS, Schey KL and Buse
MG: High glucose and insulin promote O-GlcNAc modification
of proteins, including α-tubulin. Am J Physiol Endocrinol Metab.
284:E424–E434. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Ji S, Kang JG, Park SY, Lee J, Oh YJ and
Cho JW: O-GlcNAcylation of tubulin inhibits its
polymerization. Amino Acids. 40:809–818. 2011. View Article : Google Scholar
|
|
27
|
Krecic AM and Swanson MS: hnRNP complexes:
composition, structure, and function. Curr Opin Cell Biol.
11:363–371. 1999. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Martinez-Contreras R, Cloutier P, Shkreta
L, Fisette JF, Revil T and Chabot B: hnRNP proteins and splicing
control. Adv Exp Med Biol. 623:123–147. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Boukakis G, Patrinou-Georgoula M,
Lekarakou M, Valavanis C and Guialis A: Deregulated expression of
hnRNP A/B proteins in human non-small cell lung cancer: parallel
assessment of protein and mRNA levels in paired tumour/non-tumour
tissues. BMC Cancer. 10:4342010. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Fielding P, Turnbull L, Prime W, Walshaw M
and Field JK: Heterogeneous nuclear ribonucleoprotein A2/B1
up-regulation in bronchial lavage specimens: a clinical marker of
early lung cancer detection. Clin Cancer Res. 5:4048–4052.
1999.PubMed/NCBI
|
|
31
|
Garneau D, Revil T, Fisette JF and Chabot
B: Heterogeneous nuclear ribonucleoprotein F/H proteins modulate
the alternative splicing of the apoptotic mediator Bcl-x. J Biol
Chem. 280:22641–22650. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Hope NR and Murray GI: The expression
profile of RNA-binding proteins in primary and metastatic
colorectal cancer: relationship of heterogeneous nuclear
ribonucleoproteins with prognosis. Hum Pathol. 42:393–402. 2011.
View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Wang Z, Pandey A and Hart GW: Dynamic
interplay between O-linked N-acetylglucosaminylation
and glycogen synthase kinase-3-dependent phosphorylation. Mol Cell
Proteomics. 6:1365–1379. 2007.
|
|
34
|
Gauczynski S, Peyrin JM, Haïk S, et al:
The 37-kDa/67-kDa laminin receptor acts as the cell-surface
receptor for the cellular prion protein. EMBO J. 20:5863–5875.
2001. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Wells L, Vosseller K, Cole RN, Cronshaw
JM, Matunis MJ and Hart GW: Mapping sites of O-GlcNAc
modification using affinity tags for serine and threonine
post-translational modifications. Mol Cell Proteomics. 1:791–804.
2002.
|
|
36
|
Lefebvre T, Cieniewski C, Lemoine J, et
al: Identification of N-acetyl-D-glucosamine-specific
lectins from rat liver cytosolic and nuclear compartments as
heat-shock proteins. Biochem J. 360:179–188. 2001.
|
|
37
|
Drummond FJ, Sowden J, Morrison K and
Edwards YH: Colon carbonic anhydrase 1: transactivation of gene
expression by the homeodomain protein Cdx2. FEBS Lett. 423:218–222.
1998. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Parkkila S, Rajaniemi H, Parkkila AK, et
al: Carbonic anhydrase inhibitor suppresses invasion of renal
cancer cells in vitro. Proc Natl Acad Sci USA. 97:2220–2224. 2000.
View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Zarghami N, Giai M, Yu H, et al: Creatine
kinase BB isoenzyme levels in tumour cytosols and survival of
breast cancer patients. Br J Cancer. 73:386–390. 1996. View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Li T, Yang W, Li M, et al: Expression of
selenium-binding protein 1 characterizes intestinal cell maturation
and predicts survival for patients with colorectal cancer. Mol Nutr
Food Res. 52:1289–1299. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Kim H, Kang HJ, You KT, et al: Suppression
of human selenium-binding protein 1 is a late event in colorectal
carcinogenesis and is associated with poor survival. Proteomics.
6:3466–3476. 2006. View Article : Google Scholar : PubMed/NCBI
|