Introduction
Ionizing radiation, used alone or in combination
with surgery and chemotherapy, plays an essential role in the
management of breast cancer from the early to advanced stages. More
than half of all patients with breast cancer are treated with
radiotherapy (1). Ionizing
radiation damages cells by using free radicals from the radiolysis
of water that cause DNA double-strand breaks. Extensive biological
effects are induced in the cells after radiation exposure,
including apoptosis and DNA repair (2,3). A
complex cell response is elicited to repair radiation damage,
including an alteration in gene expression, particularly in genes
involved in stress response, cell cycle control and DNA repair
(4). Radiation sensitivity varies
widely among people, and much of the difference in the effects of
radiation exposure is observed at the gene expression level
(5). A more comprehensive
understanding of the tumor radiation-related genes can be
particularly useful for predicting tumor response to radiotherapy
and potentially modulating the treatment outcome of breast cancer
patients.
Clinically, fractionation is widely used in
radiotherapy with multiple 2-Gy fractions over the course of
several weeks with total radiation doses of 50–60 Gy. This process
allows for sufficient time between dose fractions, it provides
treatment benefits involving the further sparing of normal tissue
by repairing the sublethal damage, and it increases the damage to
the tumor through the reoxygenation and reassortment of cells into
radiosensitive phases of the cycle between dose fractions. However,
in recent years, radiotherapy involving large fraction size
[including stereotactic body radiation therapy (SBRT)] and doses
from 5 Gy to 25 Gy per fraction has become a mainstay for treating
lung cancer or metastatic tumors to achieve more effective tumor
control. Tsai et al (6)
determined that, at the molecular level, the patterns of gene
expression vary substantially between single-dose (SD) and
fractionated radiation in 3 cancer cell lines, which results in the
differential expression of 463 genes, of which 13 are commonly
upregulated in MCF7, SF539 and DU145 cells. Compared with SD
radiation, another study revealed that a more robust induction of
genes occurs during fractionated radiation (7).
MicroRNAs (miRNAs) are small RNAs of ~22 nucleotides
in length that act as crucial negative regulators of protein
expression at the post-transcriptional level (8). These molecules function as inhibitors
of target mRNA for either the degradation or inhibition of gene
function. Researchers have suggested that miRNAs are responsible
for controlling ~50% of all protein-coding genes (9). Studies have indicated that the
expression of miRNAs has been clearly involved in cancer
development, and the alternation of miRNAs has been observed in
various types of cancer, including breast cancer. Iorio et
al (10) identified miRNA
aberrant expression in human breast cancer by using systematic
profiling, and revealed that it is specifically correlated with
pathological features of breast cancer, such as estrogen and
progesterone receptor expression, tumor stage, vascular invasion
and cell proliferation. miRNAs have also been determined to play
vital roles in a majority of biological processes in breast cancer,
including tumor cell growth, apoptosis, invasion and metastasis.
miR-125a suppresses cell growth and induces apoptosis in breast
cancer cells by targeting the mRNA encoding the RNA-stabilizing
protein HuR (11). In a murine
xenograft model study, invasion and metastasis were induced by the
overexpression of miR-10b in a breast cancer tumor (12). Among these dysfunctional miRNAs,
several have been demonstrated to play a critical role in breast
cancer radiosensitivity, including let-7 family miR-7, miR-21,
miR-31, miR-200c, miR-199a and miR-302a (13–18).
However, these radioresponse miRNA expressions may differ depending
on whether cells are exposed to an SD or multifractionated (MF)
radiation dose. In the present study, miRNA expression profiles
were compared in human MDA-MB-361 cells exposed to an SD or MF
radiation dose by using the next-generation sequence approach. The
data revealed that differential expression patterns of miRNAs
occurred when using 2 distinct radiation protocols. Among these
patterns, the response of miR-17-92 cluster expression clearly
differed between the SD and MF radiation dose in MDA-MB-361 cells.
We further investigated the role of the miR-17-92 cluster in breast
cancer by using in silico analysis. We concluded that these
radiation-response miRNAs can be used as therapeutic targets for
improving the efficacy of radiation treatment in future breast
cancer therapy.
Materials and methods
Cell culture and radiation treatment
Breast cancer cells, MDA-MB-361, were obtained from
the American Type Culture Collection and were maintained in
Dulbecco’s modified Eagle’s medium, supplemented with 10%
inactivated fetal bovine serum (FBS; Invitrogen, Carlsbad, CA,
USA). The cells were exposed to various radiation dosages (0, 2, 6,
10, 14 and 18 Gy), and were subsequently cultured in fresh medium.
The total RNA was obtained at 15 h following radiation treatment,
using TRIzol (Invitrogen) according to the manufacturer’s
instructions. The concentration, purity and amount of total RNA
were determined using a NanoDrop 1000 spectrophotometer (NanoDrop
Technologies Inc., USA).
Clinical breast cancer samples
Breast cancer samples (including 44 breast tumors
and 29 adjacent normal tissues) were collected from 44 breast
cancer patients who received a surgical operation at the Department
of Surgery, Kaohsiung Veterans General Hospital. Informed consent
was obtained from all the patients. The total RNA of the tissue was
extracted using a TRIzol reagent, according to the
instructions.
Collection and preprocessing of sequence
reads
The MDA-MB-361 cells were exposed to 10 Gy of
radiation, administered either as SD or MF radiation. To administer
MF radiation, the cells were exposed to 2 Gy a day for 5 days.
After the final dose of radiation treatment was administered, the
cells were lysed at 15 h for RNA extraction. The RNA samples were
prepared using an Illumina small RNA preparation kit, and were
subsequently sequenced using the Illumina HiSeq platform. The
generated sequence reads were first subjected to quality control to
remove low-quality reads. The sequence reads were then subjected to
3′ adaptor trimming to generate clean reads, as previously
described (19,20). To attain a high confidence level,
only the clean reads with a read count ≥2 and with a length ranging
from 15 to 27 nt were included in further analyses.
Mapping clean reads to pre-miRNAs
To investigate the miRNA expression profiles in
various libraries (control, SD and MF radiation), the qualified
clean reads were mapped back to human pre-miRNAs (miRBase 19). To
eliminate ambiguous multiple hits during the mapping procedure, no
mismatch was allowed. Previous studies have reported that, when
mapped back to pre-miRNAs, sequence reads typically carry
mismatches preferentially located at the terminal 3′ ends (21–24).
This mismatch was termed the 3′ end modification. Based on the read
count of all the isomiRs belonging to the same mature miRNAs, the
miRNA expression levels were evaluated and presented in transcripts
per million (TPM).
Stem-loop reverse transcription and
real-time polymerase chain reaction
Reverse transcription (RT) primers were specifically
designed for examining miRNAs according to the methods used by Chen
et al (25). One microgram
of total RNA was reverse transcribed in a stem-loop RT reaction,
using RT primers and a SuperScript III Reverse Transcriptase
according to the user manual (Invitrogen). The reaction was
performed under the following incubation conditions: 30 min at
16°C, followed by 50 cycles of 20°C for 30 sec, 42°C for 30 sec and
50°C for 1 sec. The enzyme was subsequently inactivated by
incubating it at 85°C for 5 min. Real-time polymerase chain
reactions (PCRs) were performed using a miRNA-specific forward
primer and a universal reverse primer combined with incubation at
94°C for 10 min, followed by 40 cycles of 94°C for 15 sec and 60°C
for 32 sec. The gene expression levels were detected using the
SYBR-Green I assay (Applied Biosystems, Foster City, CA, USA), and
the miRNA expression levels were normalized to that of U6. The
primer sequences for the examined miRNAs are listed as follows:
miR-17-RT, CTCAACTG GTGTCGTGGAGTCGGCAATTCAGTTGAGCTACCTGC and
miR-17-GSF, CGGCGGCAAAGTGCTTACAGTG; miR-18a-RT,
CTCAACTGGTGTCGTGGAGTCGGCAATTCAGTTGAGCTATCTGC and miR-18a-GSF,
CGGCGGTAAGGTGCATCTAGTG; miR-19a-RT,
CTCAACTGGTGTCGTGGAGTCGGCAATTCAGTTGAGTCAGTTTT and miR19a-GSF,
CGGCGGTGTGCAAATCCATGCA; miR-19b-RT,
CTCAACTGGTGTCGTGGAGTCGGCAATTCAGTTGAGTCAGTTTT and miR-19b-GSF,
CGGCGGTGTGCAAATCCATGCA; miR-20a-RT,
CTCAACTGGTGTCGTGGAGTCGGCAATTCAGTTGAGCTACCTGC and miR-20a-GSF,
CGGCGGTAAAGTGCTTATAGTG; miR-92a-RT,
CTCAACTGGTGTCGTGGAGTCGGCAATTCAGTTGAGACAGGCCG and miR-92a-GSF,
CGGCGGTATTGCACTTGTCCCG.
miRNA expression level according to The
Cancer Genome Atlas data
Members of The Cancer Genome Atlas (TCGA) project
collect both cancer and corresponding normal tissues from hundreds
of breast cancer patients. All the level-3 miRNA expression data
for breast cancer were downloaded from the TCGA data portal
(https://tcga-data.nci.nih.gov/tcga/dataAccessMatrix.htm).
These level-3 data included calculated expressions for each miRNA
derived from the next-generation sequencing results. In addition,
the expression profiles of miRNAs observed in the tumor and
corresponding normal samples of 102 patients were downloaded. The
normalized quantification expression levels for these 102
participants were further examined for each investigated miRNA.
Pathway enrichment analysis
The target genes of miR-17, miR-18a, miR-19a,
miR-19b, miR-20a and miR-92a were downloaded from TargetScan 6.0,
and were then mapped onto the Kyoto Encyclopedia of Genes and
Genomes (KEGG) pathways based on the Enzyme Commission (EC) numbers
by using the R package SubPathwayMiner v.3.1 software (26). Subsequently, a hypergeometric test
was performed to identify significantly enriched pathways and to
calculate the false positive discovery rate in the FDR-corrected
q-value.
Results
miRNA profiling of radiation-treated
breast cancer cells
By using the next-generation sequencing approach, we
comprehensively analyzed the distribution of miRNAs in MDA-MB-361
cells after administering SD and MF radiation at 10 Gy. As shown in
Table I, we obtained >8 million
clean reads in 3 libraries. By summarizing the read count of all
the isomiRs belonging to the same mature miRNAs, we quantified the
miRNA expression abundances, presented in TPM. After mapping the
clean reads to the genome, >500 miRNAs expressed in the
MDA-MB-361 cells (TPM >1) were detected. Most of these miRNAs
were consistently expressed between the control group and the group
that received radiation treatment with SD or MF radiation
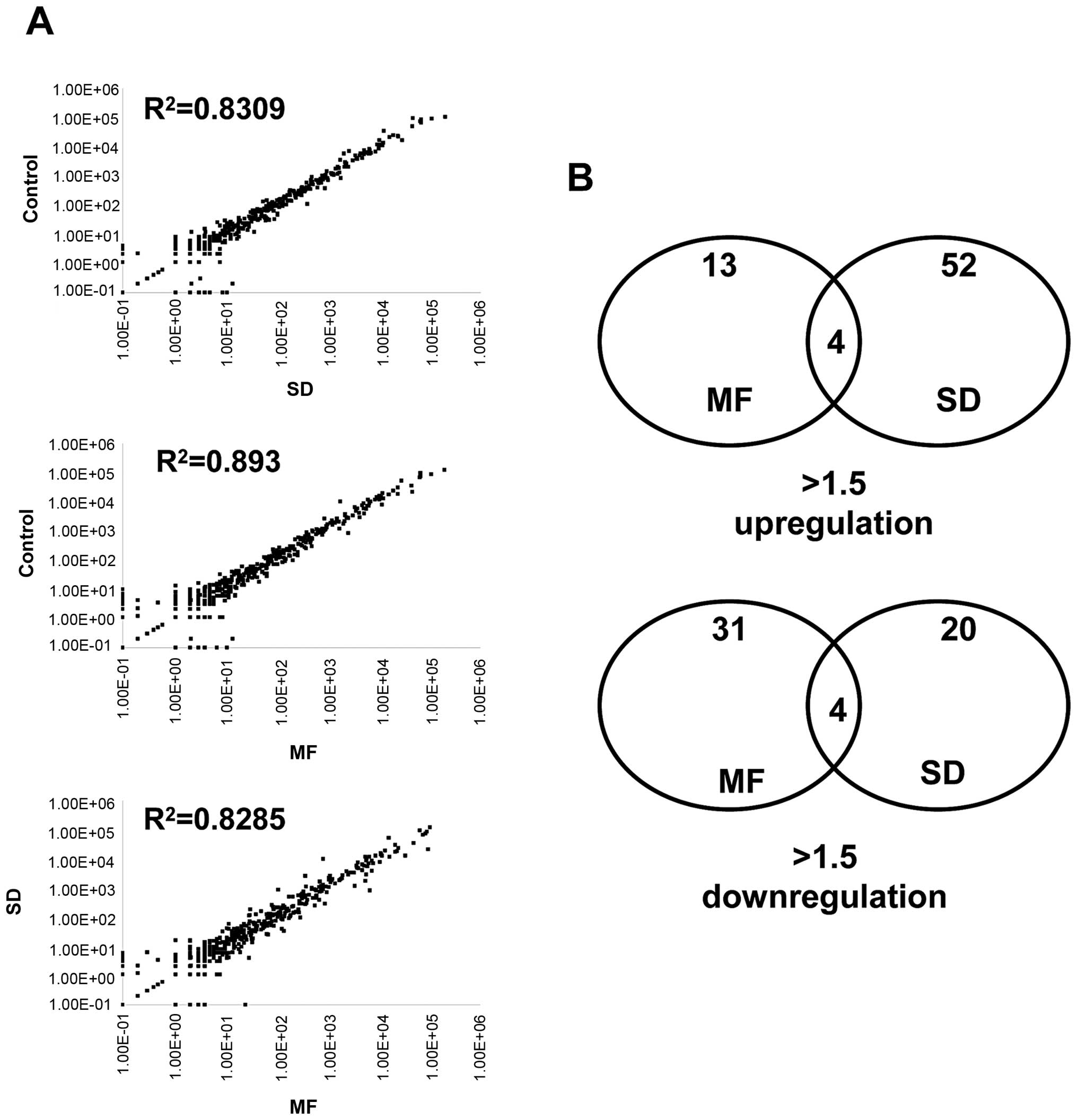
(R2 >0.8; Fig. 1A).
Only a small fraction of the miRNAs were identified as being
differentially expressed (>1.5-fold) after the MDA-MB-361 cells
were exposed to a 10-Gy SD and 2-Gy × 5 fractionated radiation.
Comparing the control cells revealed that 13 and 31 miRNAs were
upregulated and downregulated, respectively, after undergoing 2-Gy
× 5 fractionated radiations. In addition, 52 and 20 miRNAs were
upregulated and downregulated, respectively, after undergoing 10-Gy
SD radiation (Fig. 1B). Tables II and III list the top 20 upregulated and top
20 downregulated miRNAs after undergoing MF or SD radiation.
 | Table ISummary of sequences reads
information in three libraries. |
Table I
Summary of sequences reads
information in three libraries.
| Sample name | Total Illumina
reads | Clean read, R
>=2 | Percentage (%) | Detected miRs |
|---|
| Control | 11,229,160 | 8,741,817 | 77.85 | 510 |
| MF | 11,086,472 | 8,780,482 | 79.20 | 637 |
| SD | 11,685,185 | 8,982,545 | 76.87 | 626 |
 | Table IIMicroRNAs differentially expressed
between control and multi-fractionated radiation. |
Table II
MicroRNAs differentially expressed
between control and multi-fractionated radiation.
| Upregulation | Control (RPM) | MF (RPM) | MF/control |
|---|
| hsa-let-7e-5p | 2113.1 | 5788.1 | 2.739151 |
| hsa-let-7a-5p | 12845.3 | 34454.3 | 2.68225 |
| hsa-miR-423-5p | 2644.1 | 7037.1 | 2.661435 |
| hsa-let-7f-5p | 47084.2 | 95634.2 | 2.031131 |
| hsa-let-7d-5p | 1791.1 | 3516.1 | 1.963095 |
| hsa-miR-32-5p | 35.1 | 66.1 | 1.883191 |
| hsa-miR-744-5p | 500.1 | 933.1 | 1.865827 |
|
hsa-miR-181c-5p | 191.1 | 319.1 | 1.669806 |
| hsa-miR-16-5p | 10650.2 | 16872.2 | 1.584214 |
| hsa-miR-424-3p | 182.1 | 285.1 | 1.565623 |
| hsa-miR-126-3p | 71.1 | 109.1 | 1.534459 |
| hsa-miR-128 | 984.2 | 1473.2 | 1.49685 |
| hsa-miR-361-3p | 85.1 | 127.1 | 1.493537 |
| hsa-miR-320b | 484.2 | 720.2 | 1.487402 |
| hsa-miR-877-5p | 393.1 | 584.1 | 1.485881 |
| hsa-miR-574-5p | 85.1 | 125.1 | 1.470035 |
| hsa-miR-151b | 64.1 | 92.1 | 1.436817 |
| hsa-miR-218-5p | 103.2 | 148.2 | 1.436047 |
| hsa-miR-10a-5p | 3512.1 | 5020.1 | 1.429373 |
| hsa-miR-92b-3p | 5372.1 | 7672.1 | 1.428138 |
| Downregulation |
| hsa-miR-1296 | 406.1 | 110.1 | 0.271115 |
|
hsa-let-7f-1-3p | 37.1 | 12.1 | 0.326146 |
| hsa-let-7a-3p | 92.2 | 32.2 | 0.349241 |
| hsa-miR-4521 | 74.1 | 31.1 | 0.419703 |
| hsa-miR-19a-3p | 901.1 | 382.1 | 0.424037 |
| hsa-miR-1246 | 108.1 | 47.1 | 0.435708 |
| hsa-miR-19b-3p | 1829.2 | 855.2 | 0.467527 |
| hsa-miR-4286 | 63.1 | 30.1 | 0.477021 |
| hsa-miR-149-5p | 107.1 | 51.1 | 0.477124 |
| hsa-miR-30a-5p | 201261.1 | 105788.1 | 0.525626 |
| hsa-miR-221-3p | 28915.1 | 16665.1 | 0.576346 |
| hsa-miR-4454 | 64.1 | 37.1 | 0.578783 |
| hsa-miR-143-3p | 61.1 | 36.1 | 0.590835 |
| hsa-miR-20a-5p | 644.1 | 388.1 | 0.602546 |
| hsa-miR-339-5p | 158.1 | 105.1 | 0.664769 |
| hsa-miR-598 | 135.1 | 91.1 | 0.674315 |
| hsa-miR-660-5p | 320.1 | 218.1 | 0.68135 |
|
hsa-miR-181b-3p | 91.1 | 62.1 | 0.681668 |
|
hsa-miR-374b-5p | 60.1 | 41.1 | 0.68386 |
| hsa-miR-561-5p | 203.1 | 140.1 | 0.689808 |
 | Table IIIMicroRNAs differentially expressed
between control and single-dose radiation. |
Table III
MicroRNAs differentially expressed
between control and single-dose radiation.
| Upregulation | Control (RPM) | SD (RPM) | SD/control |
|---|
| hsa-miR-19b-3p | 1829.2 | 10280.2 | 5.620052 |
| hsa-miR-19a-3p | 901.1 | 2663.1 | 2.955388 |
|
hsa-miR-374a-5p | 88.1 | 225.1 | 2.555051 |
|
hsa-miR-1285-3p | 118.2 | 267.2 | 2.260575 |
| hsa-miR-1296 | 406.1 | 867.1 | 2.135188 |
|
hsa-miR-374b-5p | 60.1 | 128.1 | 2.131448 |
| hsa-miR-454-3p | 242.1 | 511.1 | 2.111111 |
| hsa-miR-210 | 281.1 | 577.1 | 2.053006 |
| hsa-miR-424-5p | 104.1 | 213.1 | 2.04707 |
| hsa-let-7a-3p | 92.2 | 186.2 | 2.019523 |
| hsa-miR-769-5p | 527.1 | 1029.1 | 1.952381 |
| hsa-miR-30c-5p | 6496.2 | 12008.2 | 1.848496 |
| hsa-miR-101-3p | 1258.2 | 2322.2 | 1.845653 |
| hsa-miR-10a-5p | 3512.1 | 6375.1 | 1.815182 |
| hsa-miR-96-5p | 69.1 | 125.1 | 1.81042 |
| hsa-miR-4286 | 63.1 | 114.1 | 1.808241 |
| hsa-miR-221-3p | 28915.1 | 51925.1 | 1.795778 |
| hsa-miR-625-3p | 33.1 | 59.1 | 1.785498 |
| hsa-miR-20a-5p | 644.1 | 1144.1 | 1.776277 |
| hsa-miR-126-5p | 316.1 | 561.1 | 1.775071 |
| Downregulation |
| hsa-miR-423-5p | 2644.1 | 847.1 | 0.320374 |
| hsa-let-7f-5p | 47084.2 | 21041.2 | 0.446885 |
|
hsa-miR-138-1-3p | 218.1 | 100.1 | 0.458964 |
| hsa-miR-222-5p | 273.1 | 147.1 | 0.538631 |
| hsa-miR-143-3p | 61.1 | 34.1 | 0.558101 |
| hsa-miR-744-5p | 500.1 | 288.1 | 0.576085 |
|
hsa-miR-92a-1-5p | 154.1 | 94.1 | 0.610642 |
| hsa-miR-18a-3p | 138.1 | 85.1 | 0.61622 |
| hsa-miR-30a-5p | 201261.1 | 124649.1 | 0.61934 |
| hsa-miR-877-5p | 393.1 | 253.1 | 0.643857 |
| hsa-miR-423-3p | 4484.1 | 2966.1 | 0.661471 |
| hsa-miR-339-3p | 234.1 | 156.1 | 0.666809 |
| hsa-let-7d-5p | 1791.1 | 1195.1 | 0.667244 |
| hsa-miR-30d-5p | 12115.1 | 8090.1 | 0.66777 |
| hsa-miR-1246 | 108.1 | 73.1 | 0.676226 |
| hsa-miR-25-5p | 271.1 | 185.1 | 0.682774 |
| hsa-miR-1275 | 157.1 | 109.1 | 0.694462 |
| hsa-miR-92b-3p | 5372.1 | 3880.1 | 0.722269 |
| hsa-miR-224-5p | 190.1 | 141.1 | 0.742241 |
| hsa-miR-484 | 374.1 | 279.1 | 0.746057 |
SD and MF radiation induces dissimilar
miRNA responses in MDA-MB-361
Although most of the miRNA expression was consistent
after MF and SD radiation treatment (R2=0.8285; Fig. 1A), we observed that several
radiation-induced miRNA responses were dissimilar between those
treated with either SD or MF radiation. In the present study, we
revealed that using various radiation exposure methods caused 8
common miRNAs in MDA-MB-361 cells to change >1.5-fold under MF
and SD radiation, including the upregulation of 4 miRNAs:
miR-32-3p, miR-126-5p, miR-181c-5p and miR-424-5p, and the
downregulation of 4 miRNAs: miR-30a-5p, miR-143-3p, miR-339-3p and
miR-1246. In addition, the converse response of these miRNAs was
frequently observed in the cells that underwent either MF or SD
radiation treatment in the present study. The levels of 4 miRNA
expressions increased after MF radiation, but after SD radiation,
these miRNA expression levels decreased (let-7f-5p, let-7d-5p,
miR-423-5p and miR-744-5p). In addition, 16 miRNA expressions were
inhibited in 2-Gy MF radiation-treated MDA-MB-361 cells, but were
activated following 10-Gy SD radiation treatment.
Radiation-induced miR-17-92 cluster
expression
According to the NGS data, we identified several
radiation-induced miRNAs and observed that several miRNAs exhibited
differential responses to various radiation treatment methods.
Among these miRNAs, we observed that the expression levels of
miR-19a, miR-20a and miR-19b clearly increased in the MDA-MB-361
cells after undergoing 10-Gy SD radiation treatment (Table II). These miRNAs were located at
chromosome 13 and belonged to the miR-17-92 cluster, which has been
determined to play an oncogenic role in tumorigenesis by regulating
cell survival, proliferation, differentiation and cell cycles
(27–29). The miR-17-92 cluster was embedded
with 6 miRNAs: miR-17, miR-18a, miR-19a, miR-20a, miR-19b and
miR-92a (Fig. 2A). As shown in
Fig. 2B, the expression levels of
most of the miRNAs in the miR-17-92 cluster increased after
undergoing SD radiation treatment. Conversely, the miR-19a-3p,
miR-20a-5p and miR-19b-3p expressions were inhibited >1.5-fold
in the MDA-MB-361 cells following MF treatment (Fig. 2B). We further examined the
expression levels of the miR-17-92 cluster (only the major arm of
the miRNAs was selected) in the MDA-MB-361 cells following
treatment with various doses of radiation for 15 h, using the
real-time PCR approach. As shown in Fig. 2C, the expression levels of the
miRNAs in the miR-17-92 cluster gradually increased in a
dose-dependent manner (0, 2, 6, 10, 14 and 18 Gy). These results
indicated that, except for miR-92a, miR-17-92 cluster expression
can be activated by radiation treatment in MDA-MB-361 cells.
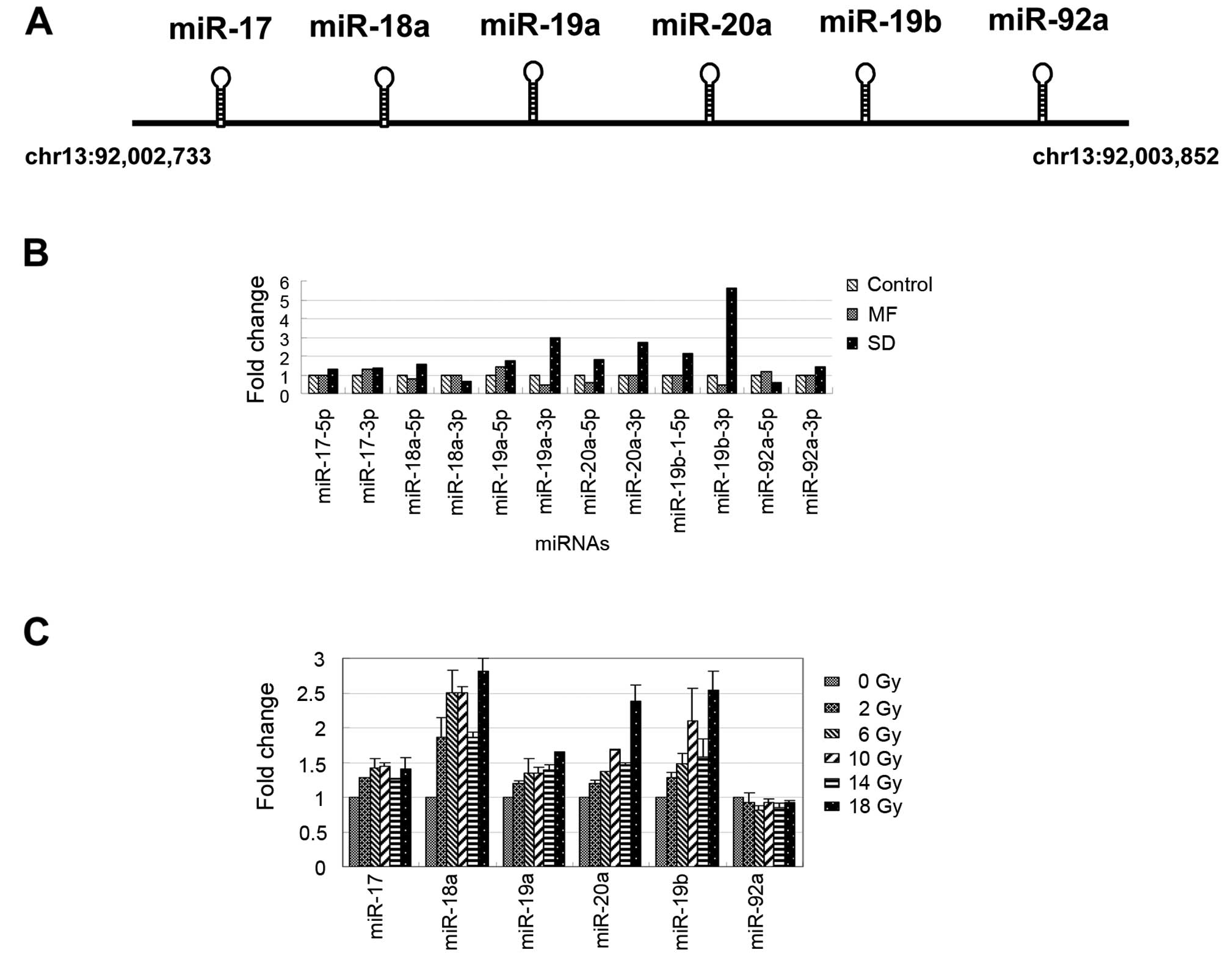 | Figure 2Radiation-induced expression levels
of miR-17-92 in breast cancer cells, MDA-MB-361. (A) Schematic
displays of the structure and location of the miR-17-92 cluster.
(B) Fold changes of the 5p/3p arm of the miR-17-92 cluster after SD
and MF radiation treatment were observed using the NGS data. (C)
The expression pattern of the miR-17-92 cluster was induced at
various radiation treatment doses (0, 2, 4, 6, 8, 10, 12, 14, 16
and 18 Gy) in the MDA-MB-361 cells. SD, single-dose; MF,
multifractionated. |
Analysis of the role of the miR-17-92
cluster in breast cancer by using an in silico approach
Generally, one miRNA typically contains hundreds of
target genes and slightly suppresses its own target gene;
therefore, conducting a biological function by using a group of
miRNAs comodulated with the same signaling pathway may be more
efficient. According to this theory, we further examined the
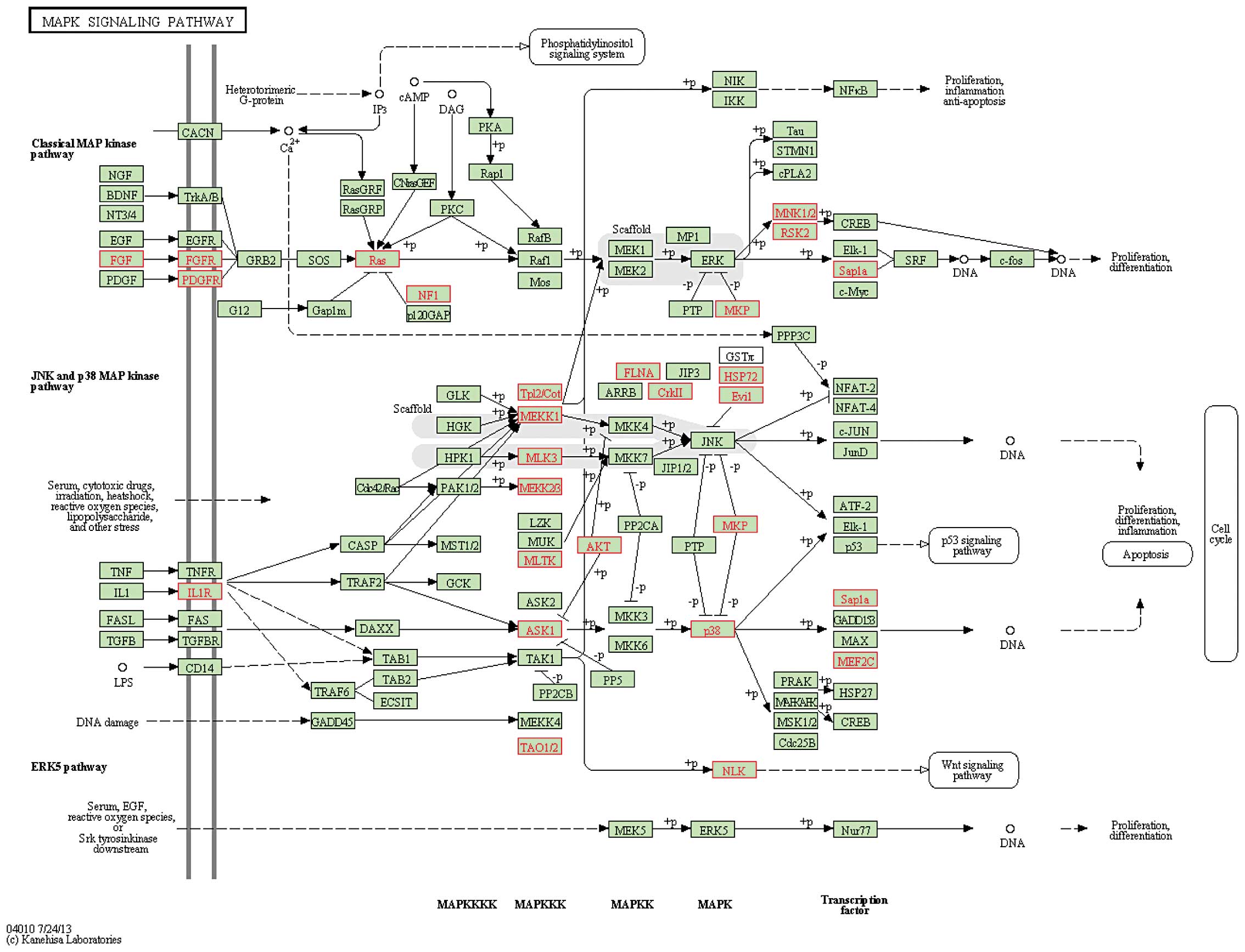
miR-17-92 cluster function by using pathway enrichment analysis. We
first obtained the putative target genes of a miR-17-92 cluster
from TargetScan 6.0; subsequently, these target genes were mapped
onto KEGG pathways. The data indicated that the target genes of the
miR-17-92 cluster were frequently and significantly enriched in
several cancer-related and radiation-related pathways, including
the mitogen-activated protein kinase (MAPK), ErbB, p53, Wnt,
transforming growth factor-β (TGF-β) and mTOR signaling pathways,
and cell cycle with an FDR <0.05 (Table IV; Fig.
3). These data indicated that the miR-17-92 cluster
participated in cancer cell progression and played a crucial role
in radiation therapy by regulating radiation-response pathways.
 | Table IVPredicted targets of miR-17-92
cluster involved in radiation-relative pathways. |
Table IV
Predicted targets of miR-17-92
cluster involved in radiation-relative pathways.
| Pathway | AnnMolecule
ratio | AnnBg ratio | FDR |
|---|
| Endocytosis | 67/2785 | 203/21796 | 1.21E-11 |
| MAPK signaling
pathwaya | 73/2785 | 266/21796 | 1.10E-08 |
| Axon guidance | 42/2785 | 126/21796 | 1.38E-07 |
| Pathways in
cancera | 78/2785 | 322/21796 | 6.70E-07 |
| mTOR signaling
pathwaya | 22/2785 | 52/21796 | 5.06E-06 |
| Neurotrophin
signaling pathway | 37/2785 | 126/21796 | 2.37E-05 |
| Regulation of actin
cytoskeleton | 51/2785 | 211/21796 | 0.000122 |
| Melanogenesis | 30/2785 | 100/21796 | 0.000122 |
| Focal adhesion | 48/2785 | 199/21796 | 0.000188 |
| GnRH signaling
pathway | 29/2785 | 98/21796 | 0.000188 |
| Glioma | 22/2785 | 65/21796 | 0.000196 |
| Long-term
potentiation | 23/2785 | 70/21796 | 0.000199 |
| Dilated
cardiomyopathy | 26/2785 | 85/21796 | 0.000217 |
| Melanoma | 23/2785 | 71/21796 | 0.000222 |
| Chronic myeloid
leukemia | 23/2785 | 72/21796 | 0.000268 |
| Wnt signaling
pathwaya | 38/2785 | 150/21796 | 0.000285 |
| Renal cell
carcinoma | 22/2785 | 68/21796 | 0.000285 |
|
Progesterone-mediated oocyte
maturation | 26/2785 | 88/21796 | 0.000309 |
| Pancreatic
cancer | 22/2785 | 70/21796 | 0.000423 |
| Calcium signaling
pathway | 42/2785 | 177/21796 | 0.000487 |
|
Phosphatidylinositol signaling
systema | 23/2785 | 78/21796 | 0.000787 |
| Oocyte meiosis | 29/2785 | 111/21796 | 0.001083 |
| Salivary
secretion | 24/2785 | 86/21796 | 0.00132 |
| TGF-β signaling
pathwaya | 23/2785 | 82/21796 | 0.001591 |
| Prostate
cancer | 24/2785 | 88/21796 | 0.001777 |
| Small cell lung
cancer | 23/2785 | 83/21796 | 0.001777 |
| Hypertrophic
cardiomyopathy (HCM) | 22/2785 | 78/21796 | 0.001777 |
| Bladder cancer | 14/2785 | 40/21796 | 0.002097 |
| Adipocytokine
signaling pathway | 19/2785 | 65/21796 | 0.002643 |
| ErbB signaling
pathwaya | 23/2785 | 86/21796 | 0.002728 |
| Insulin signaling
pathway | 32/2785 | 136/21796 | 0.002821 |
| Ubiquitin mediated
proteolysis | 32/2785 | 137/21796 | 0.003136 |
| Non-small cell lung
cancer | 16/2785 | 54/21796 | 0.005639 |
| Circadian
rhythm-mammal | 9/2785 | 22/21796 | 0.005986 |
| Gastric acid
secretion | 19/2785 | 72/21796 | 0.00868 |
| p53 signaling
pathwaya | 18/2785 | 67/21796 | 0.008943 |
| Acute myeloid
leukemia | 16/2785 | 57/21796 | 0.009507 |
| Colorectal
cancer | 17/2785 | 63/21796 | 0.010764 |
| Cell cyclea | 28/2785 | 127/21796 | 0.014135 |
|
Vasopressin-regulated water
reabsorption | 13/2785 | 44/21796 | 0.014273 |
| Dorso-ventral axis
formation | 9/2785 | 26/21796 | 0.019367 |
| Inositol phosphate
metabolism | 15/2785 | 57/21796 | 0.022744 |
| Vascular smooth
muscle contraction | 25/2785 | 114/21796 | 0.022744 |
|
Aldosterone-regulated sodium
reabsorption | 12/2785 | 42/21796 | 0.025281 |
| Fc ɛ RI signaling
pathway | 18/2785 | 75/21796 | 0.026074 |
| Hepatitis C | 28/2785 | 134/21796 | 0.026074 |
| Amyotrophic lateral
sclerosis (ALS) | 14/2785 | 53/21796 | 0.026074 |
| Fc γ R-mediated
phagocytosis | 20/2785 | 87/21796 | 0.027332 |
| Arrhythmogenic
right ventricular cardiomyopathy (ARVC) | 17/2785 | 71/21796 | 0.031624 |
| Chemokine signaling
pathway | 36/2785 | 187/21796 | 0.032834 |
| Glycosphingolipid
biosynthesis-ganglio series | 6/2785 | 15/21796 | 0.03323 |
| ECM-receptor
interaction | 19/2785 | 84/21796 | 0.036636 |
| Hedgehog signaling
pathway | 14/2785 | 56/21796 | 0.038697 |
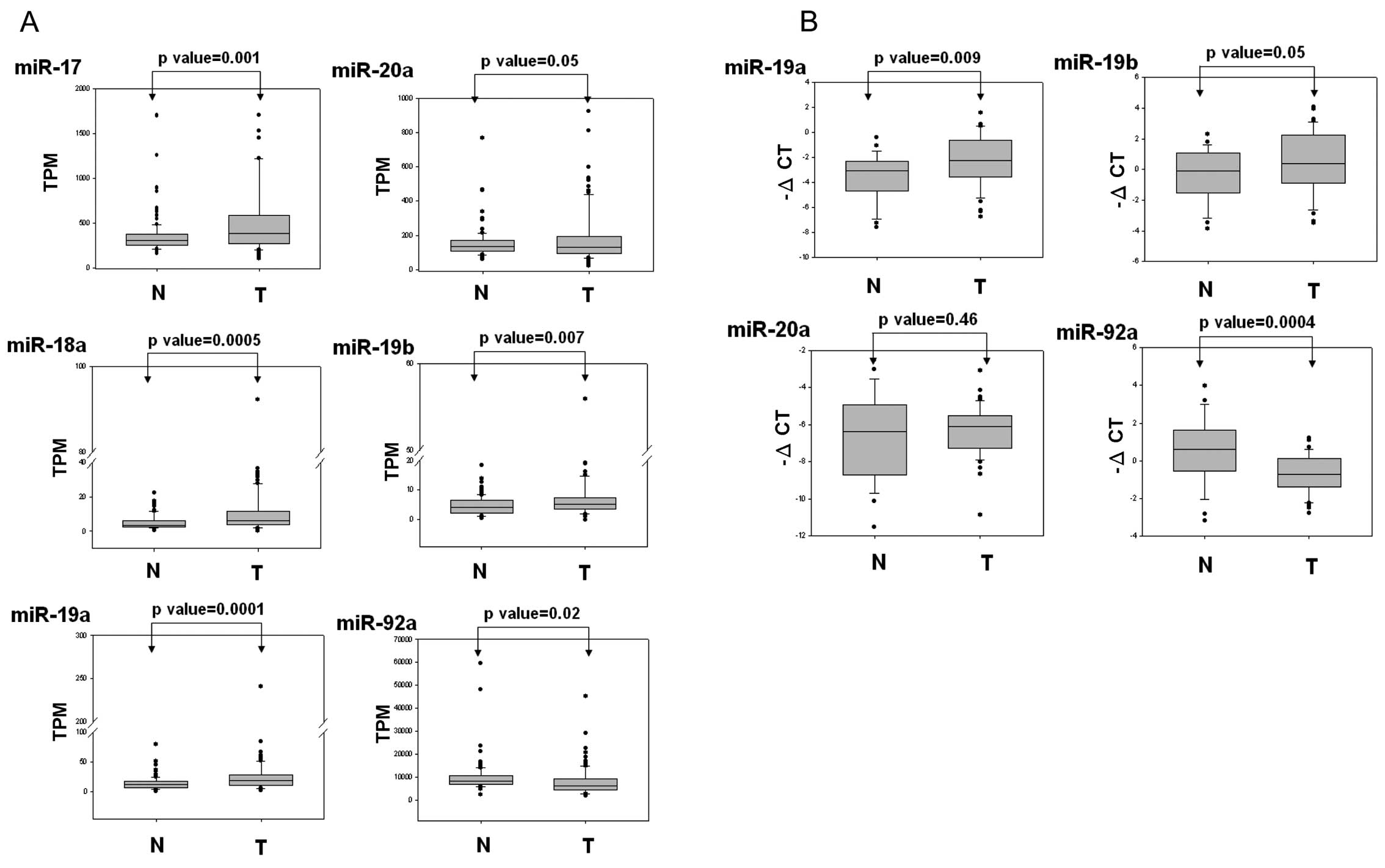
To further elucidate the role of the miR-17-92
cluster in breast cancer, we analyzed the expression levels of a
miR-17-92 cluster in breast cancer, which were provided by the TCGA
dataset, using in silico analysis. We downloaded 204 miRNA
expression profiles characterizing 102 breast cancer patients.
These profiles included breast cancer lesions and the corresponding
normal tissues. As shown in Fig.
4A, the expression levels of miR-17, miR-18a, miR-19a, miR-20a
and miR-19b were significantly upregulated in the breast cancer
cells compared with those of the corresponding normal tissue cells.
miR-92a expression was significantly decreased in the breast cancer
cells compared with that of the corresponding adjacent cells. We
also further examined the expression levels of miR-19a, miR-19b,
miR-20a and miR-92a by using real-time PCR in the breast cancer
cells. These results were consistent with the TCGA data, indicating
that miR-19a and miR-19b expression is significantly upregulated
and miR-92a is downregulated in breast cancer cells compared with
that of adjacent normal tissues (Fig.
4B). Collectively, the data indicated that expression levels of
the miR-17-92 cluster were deregulated in breast cancer cells,
suggesting that the miR-17-92 cluster plays a vital role in breast
cancer progression.
Discussion
miRNAs modulate gene expression by degrading mRNA
and inhibiting protein translation, and thereby contribute to the
regulation of numerous cancer-relevant cellular phenotypes,
including cell proliferation, apoptosis, cell cycle, cell motility
and stress response (30,31). Moreover, dysfunctional miRNAs are
involved in breast cancer carcinogenesis through the impairment of
cancer-related pathways (32).
Understanding the functions of miRNAs may contribute to cancer
diagnosis, prognosis and therapy (33,34).
Previous studies have indicated that several miRNAs can serve as
biomarkers for the diagnosis and prognosis of breast cancer
(35). In a previous study, the
expression of miR-210 was determined to be associated with the
clinical prognosis of breast cancer. The upregulation of miR-210
was reported to contribute to decreased rates of disease-free and
overall survival (36). Kovalchuk
et al (37) revealed that
miR-451 is correlated with the expression of multidrug resistant
genes and that drug sensitivity to doxorubicin-resistant breast
cancer could be enhanced by restoring miR-451 into the cell. Thus,
correcting the altered expression of miRNA by restoring miR-451 can
also be used as a strategy to improve responsiveness to
chemotherapy. In addition, miRNAs were observed to exhibit high
stability in human serum and plasma (38). One recent study demonstrated that a
decrease in the levels of miR-92a and an increase in the levels of
miR-21 were consistent and specific in the serum of breast cancer
patients, and suggested that miRNA can be used as biomarkers for
early breast cancer detection in the future (39).
Radiation-induced DNA damage initiates cellular
responses, such as cell cycle arrest, DNA damage repair and
apoptosis (40). Previous studies
have revealed that miRNAs also act as potential agents for
predicting radiation responses or are used to modulate tumor
radiation response to cancer cells further, thus enhancing the
efficacy of radiation therapy against resistant cancers (34,35,40).
In breast cancer, several miRNAs have been identified as critical
in influencing the radiosensitivity of breast cancer during
radiation therapy, including let-7 family miR-7, miR-21, miR-31,
miR-200c, miR-199a and miR-302a (13–18).
Among the miRNAs identified as potential agents that contribute to
radiation responsiveness, let-7 family was one of the first miRNAs
to be investigated. A previous study revealed that let-7a
overexpression can suppress the expression of K-Ras and
radiosensitize lung cancer cells (41). Another study determined that the
ectopic overexpression of miR-7 enhanced EGFR and Akt expression
and radiosensitized laryngeal, breast and lung cancer cell
(14). In addition, a recent study
demonstrated that miR-21 expression in breast cancer cells
contributes to radiation resistance by inhibiting the G2/M check
point (16). The upregulation of
miR-302a sensitizes radioresistant breast cancer cells and reduces
the expression of AKT1 and RAD52 (42). Furthermore, miR-199a-5p suppresses
radiation-induced autophagy, and inhibits DRAM1 and Beclin1
expression in breast cancer cells (17).
The miR-17-92 cluster is an oncogenic miRNA that
regulates cell survival, proliferation, differentiation and
angiogenesis in most human types of cancer, including breast cancer
(43–45). Previous studies revealed that the
overexpression of miR-17-92 in human cancer cells markedly
decreases the radiosensitivity of these cells through the
repression of PTEN and PHLPP2, which causes the activation of the
PI3K/AKT pathway to become enhanced (46). In the present study, the data
revealed that the numerous miRNA responses differed between the
cells that underwent either SD or MF radiation. Among the miRNAs,
we observed that the expressions of the miR-17-92 cluster were
upregulated under SD radiation treatment. Conversely, these
expressions were frequently inhibited following MF radiation
treatment (Fig. 2B). Tsai et
al (6) reported that the
survival rate of breast cancer cells following MF (5 × 2 Gy) was
higher than that following SD, which was expected since sublethal
damage was repaired between fractions. Exposing the breast cancer
cells to SD at 10 Gy led to the predominant entry of these cells
into the cell death pathway. The observation that the cells were
affected differently following SD and MF radiation treatment could
explain why miRNA expressions frequently vary based on the
treatment method used. John-Aryankalayil et al (20) also demonstrated the differential
expression patterns of several miRNAs, including miR-17-92 cluster,
miR-34a, let-7 family miRNAs and has-miR-146a, in 3 prostate cancer
cell lines after undergoing SD and fractionated radiation. Their
results revealed that the miR-17-92 cluster is markedly
downregulated by radiation treatment in p53-positive prostate
cells. Following radiation treatment, increasing wild-type p53
expression causes cells to accumulate in the G1 phase (47). The status of p53 was demonstrated to
significantly influence the expression profile of genes following
fractionated irradiation (48).
Since a mutated p53 gene cell line, MDA-MB-361, was used in the
present study, p53-regulated genes should not be induced in
MDA-MB-361 cells following radiation treatment. The elevated
expression levels of a miR-17-92 cluster in MDA-MB-361 cells
undergoing radiation treatment should be p53-independent pathway.
Yan et al (49) reported
that the miR-17-92 cluster is a novel target for p53-mediated
transcriptional repression. The transcription factors, c-Myc and
E2F family, have been observed to directly bind to the promoter of
the miR-17-92 cluster and increase the transcriptional activity of
the cluster (50–52). Previous studies have reported that
expression levels of c-Myc and E2F can be induced in breast cancer
cells following radiation treatment (53–55).
According to these studies, cells with wild-type p53 cause the
expression levels of the miR-17-92 cluster to decrease through
radiation-induced p53 activity. Conversely, an increase in
miR-17-92 transcriptional activity results from c-Myc or E2F
expression levels in mutated p53 cells undergoing radiation
treatment.
In the present study, we identified several
radiation-associated miRNAs that were clearly altered under either
SD or MF radiation treatment, including let-7 family, miR-138 and
the miR-17-92 cluster (>2-fold changes; Tables II and III). Wang et al (13) demonstrated that overexpression of
Let-7 enhanced the sensitivity of breast cancer cells to radiation.
In addition, miR-138 expression increased under SD treatment.
However, overexpression of miR-138 inhibited homologous
recombination and enhanced the sensitivity of osteosarcoma cells to
multiple DNA-damaging agents including radiation (56). Niemoeller et al (57) identified the expression levels of
numerous miRNAs known to be involved in the regulation of cellular
processes such as apoptosis, proliferation, invasion, local immune
response and radioresistance. These miRNAs, including let-7,
miR-138 and miR-1285, displayed 2–3-fold changes after undergoing
irradiation. Moreover, several miRNAs previously not known to be
radiation responsive were determined in the present study,
including miR-149, miR-374a/b, miR-423, miR-424, miR-454, miR-1246
and miR-1296. These miRNA candidates can serve as effective targets
for improving the efficacy of radiation treatment in future breast
cancer therapy.
Acknowledgements
This study was supported by grants from the
Kaohsiung Veterans General Hospital (VGHKS 103-G01-3 and VGHKS
103-043). The authors would like to thank Genomics and Proteomics
Core Laboratory, Department of Medical Research, Kaohsiung Chang
Gung Memorial Hospital, for the help with NGS data analysis.
References
|
1
|
Delaney G, Jacob S, Featherstone C and
Barton M: The role of radiotherapy in cancer treatment: estimating
optimal utilization from a review of evidence-based clinical
guidelines. Cancer. 104:1129–1137. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Rzeszowska-Wolny J, Przybyszewski WM and
Widel M: Ionizing radiation-induced bystander effects, potential
targets for modulation of radiotherapy. Eur J Pharmacol.
625:156–164. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Runkle EA, Zhang H, Cai Z, et al:
Reversion of the ErbB malignant phenotype and the DNA damage
response. Exp Mol Pathol. 93:324–333. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Yin E, Nelson DO, Coleman MA, Peterson LE
and Wyrobek AJ: Gene expression changes in mouse brain after
exposure to low-dose ionizing radiation. Int J Radiat Biol.
79:759–775. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Correa CR and Cheung VG: Genetic variation
in radiation-induced expression phenotypes. Am J Hum Genet.
75:885–890. 2004. View
Article : Google Scholar : PubMed/NCBI
|
|
6
|
Tsai MH, Cook JA, Chandramouli GV, et al:
Gene expression profiling of breast, prostate, and glioma cells
following single versus fractionated doses of radiation. Cancer
Res. 67:3845–3852. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
John-Aryankalayil M, Palayoor ST, Cerna D,
et al: Fractionated radiation therapy can induce a molecular
profile for therapeutic targeting. Radiat Res. 174:446–458. 2010.
View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Bartel DP: MicroRNAs: genomics,
biogenesis, mechanism, and function. Cell. 116:281–297. 2004.
View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Krol J, Loedige I and Filipowicz W: The
widespread regulation of microRNA biogenesis, function and decay.
Nat Rev Genet. 11:597–610. 2010.PubMed/NCBI
|
|
10
|
Iorio MV, Ferracin M, Liu CG, et al:
MicroRNA gene expression deregulation in human breast cancer.
Cancer Res. 65:7065–7070. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Guo X, Wu Y and Hartley RS: MicroRNA-125a
represses cell growth by targeting HuR in breast cancer. RNA Biol.
6:575–583. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Ma L, Teruya-Feldstein J and Weinberg RA:
Tumour invasion and metastasis initiated by microRNA-10b in breast
cancer. Nature. 449:682–688. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Wang L, Yuan C, Lv K, et al: Lin28
mediates radiation resistance of breast cancer cells via regulation
of caspase, H2A.X and Let-7 signaling. PLoS One. 8:e673732013.
View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Lee KM, Choi EJ and Kim IA: microRNA-7
increases radiosensitivity of human cancer cells with activated
EGFR-associated signaling. Radiother Oncol. 101:171–176. 2011.
View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Lin J, Liu C, Gao F, et al: miR-200c
enhances radiosensitivity of human breast cancer cells. J Cell
Biochem. 114:606–615. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Anastasov N, Höfig I, Vasconcellos IG, et
al: Radiation resistance due to high expression of miR-21 and G2/M
checkpoint arrest in breast cancer cells. Radiat Oncol. 7:2062012.
View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Yi H, Liang B, Jia J, et al: Differential
roles of miR-199a-5p in radiation-induced autophagy in breast
cancer cells. FEBS Lett. 587:436–443. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Körner C, Keklikoglou I, Bender C, Wörner
A, Münstermann E and Wiemann S: MicroRNA-31 sensitizes human breast
cells to apoptosis by direct targeting of protein kinase C ɛ
(PKCɛ). J Biol Chem. 288:8750–8761. 2013.PubMed/NCBI
|
|
19
|
Cloonan N, Wani S, Xu Q, et al: MicroRNAs
and their isomiRs function cooperatively to target common
biological pathways. Genome Biol. 12:R1262011. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
John-Aryankalayil M, Palayoor ST, Makinde
AY, et al: Fractionated radiation alters oncomir and tumor
suppressor miRNAs in human prostate cancer cells. Radiat Res.
178:105–117. 2012. View
Article : Google Scholar : PubMed/NCBI
|
|
21
|
Ebhardt HA, Tsang HH, Dai DC, Liu Y,
Bostan B and Fahlman RP: Meta-analysis of small RNA-sequencing
errors reveals ubiquitous post-transcriptional RNA modifications.
Nucleic Acids Res. 37:2461–2470. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Landgraf P, Rusu M, Sheridan R, et al: A
mammalian microRNA expression atlas based on small RNA library
sequencing. Cell. 129:1401–1414. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Reid JG, Nagaraja AK, Lynn FC, et al:
Mouse let-7 miRNA populations exhibit RNA editing that is
constrained in the 5′-seed/ cleavage/anchor regions and stabilize
predicted mmu-let-7a:mRNA duplexes. Genome Res. 18:1571–1581.
2008.PubMed/NCBI
|
|
24
|
Morin RD, O’Connor MD, Griffith M, et al:
Application of massively parallel sequencing to microRNA profiling
and discovery in human embryonic stem cells. Genome Res.
18:610–621. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Chen C, Ridzon DA, Broomer AJ, et al:
Real-time quantification of microRNAs by stem-loop RT-PCR. Nucleic
Acids Res. 33:e1792005. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Li C, Li X, Miao Y, et al:
SubpathwayMiner: a software package for flexible identification of
pathways. Nucleic Acids Res. 37:e1312009. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
He L, Thomson JM, Hemann MT, et al: A
microRNA polycistron as a potential human oncogene. Nature.
435:828–833. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Hayashita Y, Osada H, Tatematsu Y, et al:
A polycistronic microRNA cluster, miR-17-92, is
overexpressed in human lung cancers and enhances cell
proliferation. Cancer Res. 65:9628–9632. 2005.PubMed/NCBI
|
|
29
|
Cho WC: OncomiRs: the discovery and
progress of microRNAs in cancers. Mol Cancer. 6:602007. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Pan HW, Li SC and Tsai KW: MicroRNA
dysregulation in gastric cancer. Curr Pharm Des. 19:1273–1284.
2013.PubMed/NCBI
|
|
31
|
Zhao L, Bode AM, Cao Y and Dong Z:
Regulatory mechanisms and clinical perspectives of miRNA in tumor
radiosensitivity. Carcinogenesis. 33:2220–2227. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Chang HT, Li SC, Ho MR, et al:
Comprehensive analysis of microRNAs in breast cancer. BMC Genomics.
13(Suppl 7): S182012.PubMed/NCBI
|
|
33
|
Ferracin M, Querzoli P, Calin GA and
Negrini M: MicroRNAs: toward the clinic for breast cancer patients.
Semin Oncol. 38:764–775. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Ng EK, Wong CL, Ma ES and Kwong A:
MicroRNAs as new players for diagnosis, prognosis, and therapeutic
targets in breast cancer. J Oncol. 2009:3054202009.PubMed/NCBI
|
|
35
|
Iorio MV, Casalini P, Piovan C, Braccioli
L and Tagliabue E: Breast cancer and microRNAs: therapeutic impact.
Breast. 20(Suppl 3): S63–S70. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Camps C, Buffa FM, Colella S, et al:
hsa-miR-210 is induced by hypoxia and is an independent prognostic
factor in breast cancer. Clin Cancer Res. 14:1340–1348. 2008.
View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Kovalchuk O, Filkowski J, Meservy J, et
al: Involvement of microRNA-451 in resistance of the MCF-7 breast
cancer cells to chemotherapeutic drug doxorubicin. Mol Cancer Ther.
7:2152–2159. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Mitchell PS, Parkin RK, Kroh EM, et al:
Circulating microRNAs as stable blood-based markers for cancer
detection. Proc Natl Acad Sci USA. 105:10513–10518. 2008.
View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Si H, Sun X, Chen Y, et al: Circulating
microRNA-92a and microRNA-21 as novel minimally invasive biomarkers
for primary breast cancer. J Cancer Res Clin Oncol. 139:223–229.
2013. View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Zhao L, Lu X and Cao Y: MicroRNA and
signal transduction pathways in tumor radiation response. Cell
Signal. 25:1625–1634. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Weidhaas JB, Babar I, Nallur SM, et al:
MicroRNAs as potential agents to alter resistance to cytotoxic
anticancer therapy. Cancer Res. 67:11111–11116. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Liang Z, Ahn J, Guo D, Votaw JR and Shim
H: MicroRNA-302 replacement therapy sensitizes breast cancer cells
to ionizing radiation. Pharm Res. 30:1008–1016. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
43
|
de Rinaldis E, Gazinska P, Mera A, et al:
Integrated genomic analysis of triple-negative breast cancers
reveals novel microRNAs associated with clinical and molecular
phenotypes and sheds light on the pathways they control. BMC
Genomics. 14:6432013.
|
|
44
|
Kim K, Chadalapaka G, Lee SO, et al:
Identification of oncogenic microRNA-17-92/ZBTB4/specificity
protein axis in breast cancer. Oncogene. 31:1034–1044. 2012.
View Article : Google Scholar : PubMed/NCBI
|
|
45
|
Li H, Bian C, Liao L, Li J and Zhao RC:
miR-17-5p promotes human breast cancer cell migration and invasion
through suppression of HBP1. Breast Cancer Res Treat. 126:565–575.
2011. View Article : Google Scholar : PubMed/NCBI
|
|
46
|
Jiang P, Rao EY, Meng N, Zhao Y and Wang
JJ: MicroRNA-17-92 significantly enhances radioresistance in human
mantle cell lymphoma cells. Radiat Oncol. 5:1002010. View Article : Google Scholar : PubMed/NCBI
|
|
47
|
Kuerbitz SJ, Plunkett BS, Walsh WV and
Kastan MB: Wild-type p53 is a cell cycle checkpoint determinant
following irradiation. Proc Natl Acad Sci USA. 89:7491–7495. 1992.
View Article : Google Scholar : PubMed/NCBI
|
|
48
|
Simone CB II, John-Aryankalayil M,
Palayoor ST, et al: mRNA expression profiles for prostate cancer
following fractionated irradiation are influenced by p53 status.
Transl Oncol. 6:573–585. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
49
|
Yan HL, Xue G, Mei Q, et al: Repression of
the miR-17-92 cluster by p53 has an important function in
hypoxia-induced apoptosis. EMBO J. 28:2719–2732. 2009.
|
|
50
|
Thomas M, Lange-Grunweller K, Hartmann D,
et al: Analysis of transcriptional regulation of the human
miR-17-92 cluster; evidence for involvement of Pim-1. Int J Mol
Sci. 14:12273–12296. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
51
|
Li Y, Li Y, Zhang H and Chen Y:
MicroRNA-mediated positive feedback loop and optimized bistable
switch in a cancer network involving miR-17-92. PLoS One.
6:e263022011. View Article : Google Scholar : PubMed/NCBI
|
|
52
|
Aguda BD, Kim Y, Piper-Hunter MG, Friedman
A and Marsh CB: MicroRNA regulation of a cancer network:
consequences of the feedback loops involving miR-17-92, E2F, and
Myc. Proc Natl Acad Sci USA. 105:19678–19683. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
53
|
Papanikolaou V, Iliopoulos D, Dimou I, et
al: Survivin regulation by HER2 through NF-κB and c-myc in
irradiated breast cancer cells. J Cell Mol Med. 15:1542–1550.
2011.
|
|
54
|
Orr MS, Watson NC, Sundaram S, Randolph
JK, Jain PT and Gewirtz DA: Ionizing radiation and teniposide
increase p21waf1/cip1 and promote Rb dephosphorylation
but fail to suppress E2F activity in MCF-7 breast tumor cells. Mol
Pharmacol. 52:373–379. 1997.PubMed/NCBI
|
|
55
|
Calaf GM and Hei TK: Ionizing radiation
induces alterations in cellular proliferation and c-myc, c-jun and
c-fos protein expression in breast epithelial cells. Int J Oncol.
25:1859–1866. 2004.PubMed/NCBI
|
|
56
|
Wang Y, Huang JW, Li M, et al:
MicroRNA-138 modulates DNA damage response by repressing histone
H2AX expression. Mol Cancer Res. 9:1100–1111. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
57
|
Niemoeller OM, Niyazi M, Corradini S, et
al: MicroRNA expression profiles in human cancer cells after
ionizing radiation. Radiat Oncol. 6:292011. View Article : Google Scholar : PubMed/NCBI
|