Introduction
Oesophageal cancer is the eighth most common cancer
worldwide and the sixth most common cause of death from cancer
(1). It is three to four times more
common among males than females (2). Survival rates are poor, as patients
with oesophageal cancer usually present with disease that is
advanced and has already metastasised at the time of initial
diagnosis. Oesophageal squamous cell carcinoma (OSCC) is the
predominant histological subtype, comprising approximately 70% of
cases. This cancer arises in the middle and lower third of the
oesophagus. OSCC is particularly prevalent in the developing world,
where highest incidence rates are in Southern and Eastern Africa
and Eastern Asia (2). Risk factors
include poor nutritional status, drinking beverages at high
temperatures, smoking and alcohol consumption (2). However, the molecular basis of OSCC is
still largely unknown. The identification of new molecular
biomarkers would be of great importance for the early detection of
the disease. Moreover, identification of molecular targets for
therapeutic drugs would result in better treatment options for OSCC
patients.
Crm1 (the chromosome region maintenance 1 protein or
exportin 1) is a protein that has recently been identified as being
highly expressed and playing a functional role in several types of
cancers. Crm1 is a member of the karyopherin β protein family and
the major nuclear export receptor in the cell (3). It mediates the nuclear export of cargo
proteins and certain RNAs from the nucleus into the cytoplasm,
across the nuclear pore complex (NPC), thus facilitating the
appropriate subcellular localisation of target proteins and RNAs.
Crm1 has a broad substrate range, recognising cargo proteins that
carry a leucine-rich nuclear export signal (NES) (4). Since cargo proteins include various
transcription factors, cell cycle proteins and signalling proteins,
for example, p53 (5) and Akt1
(6), it would appear that many of
the integral processes in the cell depend on appropriate Crm1
expression and function.
Recent studies have reported that the expression of
Crm1 is altered in cancer, with elevated Crm1 levels reported in
cervical cancer (7), ovarian cancer
(8), osteosarcoma (9), glioma (10), pancreatic cancer (11), gastric cancer (12) and multiple myeloma (13) compared to normal tissue. We
previously identified increased Crm1 expression in transformed
fibroblasts compared to normal fibroblasts, suggesting that the
increased expression of Crm1 is a feature of the transformed
phenotype (7). Furthermore,
elevated Crm1 expression appears to be necessary for the survival
of cancer cells, as we showed that its inhibition using siRNA
resulted in cervical cancer cell death via apoptosis (7). Moreover, Crm1 inhibition using
leptomycin B (LMB) has been well documented to have an anticancer
effect both in vitro and in vivo (14). Although LMB failed clinical trials,
studies are currently underway in an attempt to develop more
effective inhibitors of Crm1 for cancer therapy (15,16).
In the present study, we examined the expression and
functional relevance of Crm1 expression in oesophageal squamous
cell carcinoma. We found that Crm1 levels are elevated in
oesophageal tumours compared to normal epithelium and that its
cellular localisation is also altered. We also found that Crm1 is
required for the proliferation and survival of oesophageal cancer
cells, supporting its use as a potential target for anticancer
drugs.
Materials and methods
Tissue sections
Immunohistochemical examination of Crm1 was
performed on archived paraffin-embedded tissue sections of matched
normal and cancer tissues obtained from 56 patients with
oesophageal squamous cell carcinoma (Groote Schuur Hospital, Cape
Town, South Africa). Immunohistochemistry was performed using
standard procedures. Briefly, slides were heat-fixed for 10 min,
deparaffinised and rehydrated, followed by antigen retrieval by
incubation in 10 mM EDTA in a pressure cooker for 2 min. Sections
were blocked for endogenous peroxidase activity by submerging the
slides in hydrogen peroxide for 20 min and were then incubated at
room temperature with a 1:20 dilution of goat serum (Dako,
Glostrup, Denmark) for 30 min. Slides were next incubated with a
1:500 dilution of Crm1 primary antibody (sc-5595; Santa Cruz
Biotechnology, Santa Cruz, CA, USA) for 1 h at room temperature. A
negative control was included where no primary antibody was used.
The Dako REAL™ EnVision™/HRP rabbit/mouse detection system was
subsequently used to detect Crm1 expression, according to the
manufacturer’s instructions. The intensity and localisation of Crm1
staining were evaluated and scored independently by a pathologist.
Crm1 expression was noted in nuclear and cytoplasmic cellular
compartments and thus a separate scoring for Crm1 expression was
performed in each case. For nuclear staining, the Allred system for
oestrogen receptor was used (17),
where the staining intensity was scored as 0 (negative), 1 (weak),
2 (moderate), or 3 (strong). The percentage of positive cells was
not included in the scoring criteria as in all cases the proportion
of positive cells was >75%. For cytoplasmic staining the
following criteria were used: 0 (negative), 1 (weak), 2 (moderate),
or 3 (strong) (18).
Quantitative real-time RT-PCR
For the analysis of Crm1 mRNA expression, RNA was
isolated from matched normal and tumour tissue biopsies obtained
from 22 patients with oesophageal squamous cell carcinoma (Groote
Schuur Hospital and Tygerberg Hospital, Cape Town, South Africa).
For cDNA synthesis, 1 μg RNA was reverse transcribed using
ImProm-II™ Reverse Transcriptase (Promega, Madison, WI, USA).
Quantitative real-time PCR was performed using the KAPA SYBR qPCR
kit (KAPA Biosystems, Cape Town, South Africa) using the following
primers: Crm1 (F 5′-GCA CCT CTT GGA CTG AAT CG-3′ and R 5′-AAG CGA
CAG CAC ACA CAC AC-3′), β-glucuronidase (F 5′-CTC ATT TGG AAT TTT
GCC GAT T-3′ and R 5′-CCG AGT GAA GAT CCC CTT TTT A-3′) and
cyclophilin D (F 5′-TGA GAC AGC AGA TAG AGC CAA GC-3′ and R 5′-TCC
CTG CCA ATT TGA CAT CTT C-3′), where β-glucuronidase and
cyclophilin D were used to normalise for Crm1 expression. The
StepOne Real-time PCR System (Applied Biosystems, USA) was used.
The comparative threshold cycle (CT) method (19) was used for the calculation of
expression fold change between samples.
Cell culture
Oesophageal carcinoma cell lines, WHCO1, WHCO5 and
WHCO6, were originally established from South African patients with
oesophageal squamous cell carcinoma and were provided by Dr R.
Veale (20). Oesophageal carcinoma
cell lines, KYSE30, KYSE70, KYSE150, KYSE180, KYSE420 and KYSE450,
were either obtained from DSMZ (Berlin, Germany) or were a gift
from Professor Y. Shimada (21).
Cells were maintained in Dulbecco’s modified Eagle’s medium (DMEM),
supplemented with penicillin (100 U/ml), streptomycin (100 μg/ml)
and 10% fetal calf serum (Gibco, Paisley, Scotland). Normal
hTERT-immortalised human oesophageal keratinocytes, EPC2-hTERT,
were a gift from Professor A.K. Rustgi (University of Pennsylvania,
Philadelphia, PA, USA). The cells were maintained in keratinocyte
growth medium supplemented with 1 ng/ml EGF and 50 μg/ml pituitary
extract. All cells were cultured at 37°C in a humidified atmosphere
of 5% CO2.
Materials
For the inhibition of gene expression, Crm1 siRNA
was used (sc-35116; Santa Cruz Biotechnology). Control siRNA-A
consisting of a scrambled sequence (sc-37007; Santa Cruz
Biotechnology) was used as a non-silencing control. Leptomycin B
(LMB), an inhibitor of Crm1 activity, was obtained from Sigma (St.
Louis, MO, USA) and stored as a 10.2 μM stock in methanol.
Protein harvest and western blot
analysis
Cells in culture were grown to 80% confluency and
lysed on ice in RIPA buffer (10 mM Tris-Cl, pH 7.4, 150 mM NaCl, 1%
deoxycholate, 0.1% SDS, 1% Triton X-100 and 1X Complete Protease
Inhibitor Cocktail (Roche, Basel, Switzerland). Western blot
analyses were performed using the rabbit anti-Crm1 (H-300)
(sc-5595) and rabbit anti-β-tubulin (H-235 (sc-9104) antibodies
(Santa Cruz Biotechnology).
Immunofluorescence
For immunofluorescence analysis of Crm1, cells were
plated on coverslips and fixed with 4% paraformaldehyde. After
fixing, cells were permeabilised in 0.1% Triton X-100 in PBS,
followed by quenching in 50 mM NH4Cl in PBS. Cells were
blocked in 0.2% gelatin for 30 min and subsequently incubated with
an α-Crm1 primary antibody (1:100 dilution, sc-5595; Santa Cruz
Biotechnology) for 45 min in a humidified chamber. After washing in
PBS, Cy3-conjugated goat anti-rabbit antibody (1:300; Jackson
ImmunoResearch Laboratories, West Grove, PA, USA) was applied for a
further 45 min. Cell nuclei were stained with DAPI (100 ng/ml) and
coverslips were mounted in Mowiol. Fluorescence was visualised
using standard fluorescence microscopy.
Cell proliferation assay
To examine the effect of Crm1 siRNA or LMB on cell
viability, the MTT assay was used. For the analysis of the effect
of Crm1 siRNA on cell viability, 2.0×103 cells were
seeded into each well of a 96-well plate and transfected with 20 nM
siRNA on the following day, whereas for treatment with LMB,
1.0×104 cells were seeded into each well of a 96-well
plate and treated with varying concentrations of LMB on the
following day. Cell proliferation was measured five days after
siRNA transfection and two days after LMB treatment by the addition
of MTT reagent (Sigma), and crystals were subsequently solubilized
using solubilization buffer. Absorbance was measured at an OD of
595 nM using a microplate reader.
Cell cycle analysis
For analysis of the effect of Crm1 inhibition on the
cell cycle, 1.8×105 KYSE30 and WHCO5 cells were plated
on 60-mm dishes and transfected with 20 nM control or Crm1 siRNA
for 72 h. Subsequently, cells were harvested and fixed in 95%
ethanol, after which the cells were stained with propidium iodide
and the cell cycle profiles were analyzed using the BD FACSVerse™
flow cytometer (BD Biosciences, Franklin Lakes, NJ, USA).
Quantification of the percentage of cells at different stages of
the cell cycle was performed using Modfit 3.3 software.
Caspase-3/7 assay
To assay for caspase-3/7 activity, the Caspase-Glo™
3/7 assay (Promega) was performed, according to the manufacturer’s
instructions. Briefly, 2.0×103 cells were plated/well in
96-well plates and transfected with 20 nM control or Crm1 siRNA.
Caspase-3/7 activity was measured 48 h after siRNA transfection,
and luminescence was monitored using the Veritas™ microplate
luminometer (Promega). MTT assays were performed concurrently and
caspase-3/7 luminescence values were normalised to OD595 values to
control for any differences in cell number.
Statistical analysis
The statistical significance of the means was
calculated using the Student’s t-test. Fisher’s exact tests were
used to compare the expression of Crm1 in the different groups.
Survival analysis was carried out using the Kaplan-Meier method. A
p-value of <0.05 was required for statistical significance.
Results
Crm1 immunostaining in normal and tumour
tissues of the oesophagus
Altered Crm1 expression has been described in
certain cancer types. In the present study we investigated Crm1
expression in oesophageal squamous cell carcinoma (OSCC). Crm1
expression was investigated in formalin-fixed tissue sections of
matched tumour and normal stratified squamous epithelium, obtained
from 56 South African patients with oesophageal cancer. The basic
patient demographics and pathological characteristics are
documented in Table I. Among our
patient cohort, 4% had stage I disease, 55% had stage II disease
and 41% had stage III disease, based on the TNM
(tumour-node-metastasis) staging system (Table I). Due to the late stage at which
oesophageal carcinoma is diagnosed, this distribution was as
expected.
 | Table IPatient demographics. |
Table I
Patient demographics.
| Characteristics | No. of patients
(%) |
|---|
| Gender |
| Male | 39 (70) |
| Female | 17 (30) |
| Age (years) |
| Average | 53.4 |
| Range | 32–80 |
| Race |
| Black | 18 (32) |
| Mixed ancestry | 32 (57) |
| Caucasian | 6 (11) |
| Smoker |
| Yes | 48 (86) |
| No | 8 (14) |
| Alcohol intake |
| Heavy | 17 (30) |
| Light | 15 (27) |
| No | 18 (32) |
| Unknown | 6 (11) |
| Stage |
| I | 2 (4) |
| II | 31 (55) |
| III | 23 (41) |
| Differentiation
status |
| Well | 1 (2) |
| Moderate | 40 (71) |
| Poor | 13 (23) |
| Unknown | 2 (4) |
| Presence of
keratinisation |
| Keratinisation | 32 (57) |
| No
keratinisation | 24 (43) |
Immunohistochemical analysis, followed by
independent scoring by a pathologist, revealed that most normal
sections had weak nuclear staining for Crm1, with no observable
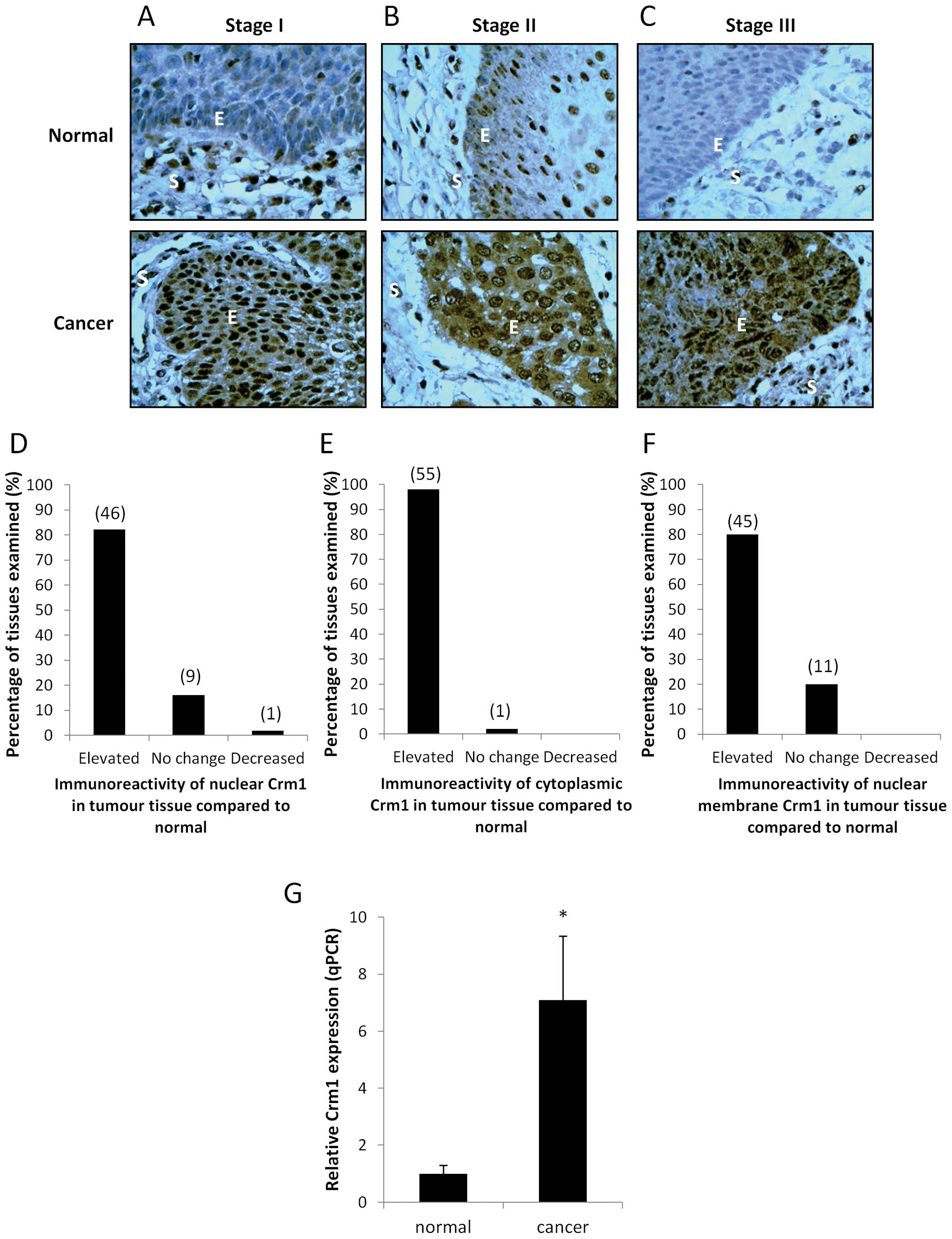
cytoplasmic staining (Fig. 1A–C).
The corresponding cancer sections, however, displayed much more
intense nuclear staining, as represented in Fig. 1D, with 82% of the patient specimens
displaying elevated nuclear staining in the cancer tissues compared
to the paired normal tissues. It was also noted that the cancer
specimens displayed a much more intense cytoplasmic and nuclear
membrane staining compared to the paired normal sections (Fig. 1E and F). Fig. 1A–C shows normal and cancer sections
from representative patients with stage I, II and III cancer,
showing intense nuclear and cytoplasmic staining of Crm1 in all
three cancer stages, compared to their corresponding normal
tissues. A statistical analysis revealed that the difference in
staining pattern between normal and cancer specimens was
statistically significant for all staining categories (nuclear,
cytoplasmic and nuclear membrane staining) (Table II).
 | Table IIExpression of Crm1 in normal and
cancer tissue of the oesophagus. |
Table II
Expression of Crm1 in normal and
cancer tissue of the oesophagus.
| No. of patients
(%) | |
|---|
|
| |
|---|
| Crm1
expression | Normal | Cancer | P-valuea |
|---|
| Nuclear
staining |
| Negative | 8 (14) | 3 (5) | |
| Weak | 48 (86) | 8 (14) | <0.0001 |
| Strong | 0 (0) | 45 (80) | |
| Cytoplasmic
staining |
| Negative | 55 (98) | 2 (4) | |
| Weak | 1 (2) | 26 (46) | <0.0001 |
| Strong | 0 (0) | 28 (50) | |
| Nuclear membrane
staining |
| Negative | 55 (98) | 10 (18) | |
| Weak | 1 (2) | 41 (73) | <0.0001 |
| Strong | 0 (0) | 5 (9) | |
It was next determined whether any correlation
exists between the expression of Crm1 and tumour stage. Correlation
analyses were performed and demonstrated that the expression of
Crm1 (where nuclear, cytoplasmic and nuclear membrane staining
patterns were taken into account) did not significantly associate
with tumour stage (Table III).
Since a similar proportion of stage II and stage III patients
displayed strong Crm1 expression, it was suggested that the
increase in expression of Crm1 is a relatively early event in
oesophageal carcinogenesis (occurring by the time the cancer has
progressed to stage II), rather than a late stage event.
 | Table IIIRelationship between Crm1 expression
and tumour stage. |
Table III
Relationship between Crm1 expression
and tumour stage.
| No. of patients
(%) | |
|---|
|
| |
|---|
| Crm1
expression | Stage I | Stage II | Stage III | P-valuea |
|---|
| Nuclear
expression |
| Negative | 0 (0) | 3 (10) | 0 (0) | |
| Weak | 1 (50) | 4 (13) | 4 (17) | 0.200 |
| Strong | 1 (50) | 24 (77) | 19 (83) | |
| Cytoplasmic
expression |
| Negative | 0 (0) | 2 (6) | 0 (0) | |
| Weak | 2 (100) | 13 (42) | 11 (48) | 0.437 |
| Strong | 0 (0) | 16 (52) | 12 (52) | |
| Nuclear membrane
expression |
| Negative | 0 (0) | 7 (23) | 3 (13) | |
| Weak | 2 (100) | 22 (71) | 18 (78) | 0.679 |
| Strong | 0 (0) | 2 (6) | 2 (9) | |
| Overall
expression |
| Weak | 1 (50) | 6 (19) | 2 (9) | |
| Moderate | 1 (50) | 15 (48) | 11 (48) | 0.184 |
| Strong | 0 (0) | 10 (32) | 10 (43) | |
Since Crm1 levels were found to be significantly
elevated in oesophageal tumour tissues in both the cytoplasmic and
nuclear cellular components, a Kaplan-Meier survival analysis was
performed to determine whether Crm1 expression predicts patient
survival. A trend, where high cytoplasmic Crm1 expression appeared
to be associated with poor overall survival, was observed (data not
shown). There was no correlation between nuclear Crm1 expression
and overall survival. These findings were likely influenced by the
limited data and tissue specimens available for early stage
oesophageal cancer patients. Access to early stage patient material
may have a significant effect on the interpretation of overall
survival analyses.
Crm1 mRNA expression in patient
tissues
As an increase in Crm1 protein expression was
observed in oesophageal cancer patient specimens, it was next
investigated whether Crm1 was similarly upregulated at the mRNA
level. Crm1 mRNA levels were thus examined by quantitative
real-time RT-PCR using RNA obtained from matched normal and cancer
tissue biopsies obtained from 22 patients with OSCC. A significant
increase in Crm1 mRNA expression was observed in the oesophageal
tumour specimens compared to the normal tissues (Fig. 1G), suggesting that the increase in
Crm1 protein expression observed by IHC was derived from elevated
Crm1 mRNA expression.
Expression of Crm1 in normal and cancer
oesophageal cell lines
As elevated Crm1 expression was observed in
oesophageal cancer tissues compared to that in the normal tissues,
Crm1 expresssion was next evaluated in 9 oesophageal cancer cell
lines (from South African and Japanese origin) and compared to the
expression in hTERT-immortalised EPC2 cells derived from normal
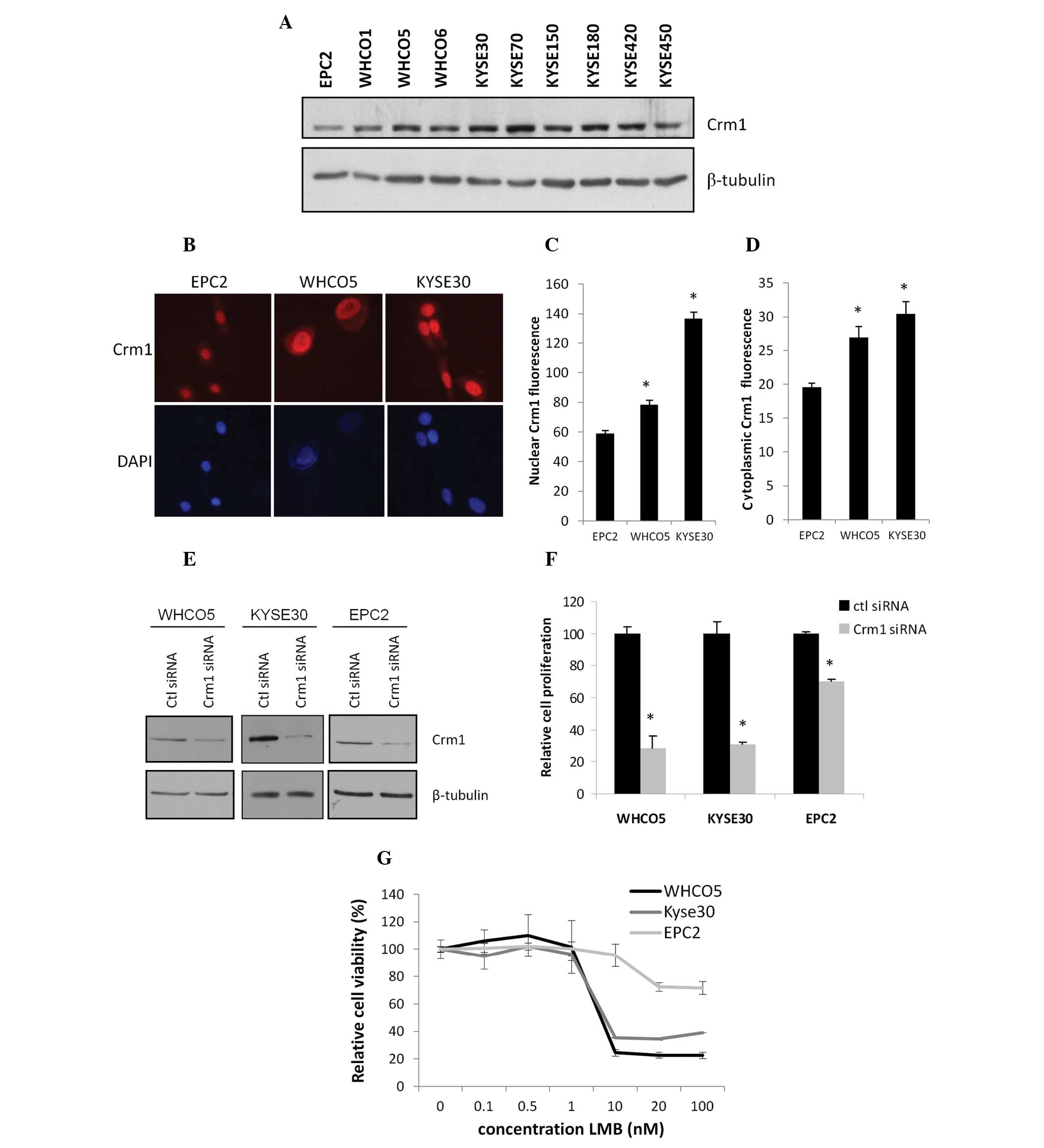
oesophageal epithelium. Western blot analysis revealed that Crm1
protein expression was high in all the oesophageal cancer cell
lines (WHCO1, WHCO5, WHCO6, KYSE30, KYSE70, KYSE150, KYSE180,
KYSE420, KYSE450), while lowest expression was observed in the
normal epithelial cell line, EPC2 (Fig.
2A). Moreover, Crm1 localisation in the normal and
representative cancer (WHCO5 and KYSE30) cell lines was determined
by immunofluorescence, and quantitation revealed that there was a
significant increase in Crm1 fluorescence in the cancer cell lines,
in both the nuclear and cytoplasmic compartments (Fig. 2B–D). These results are in agreement
with the patient data showing elevated nuclear and cytoplasmic Crm1
expression in oesophageal tumour tissues compared to normal
tissues.
Effect of Crm1 inhibition on cell
survival
To evaluate the functional significance of elevated
Crm1 expression in oesophageal cancer, Crm1 siRNA was used to
inhibit its expression. A control siRNA with no known silencing
effect was used to control for the non-specific effects of siRNA
transfection. WHCO5 and KYSE30 oesophageal cancer cells and EPC2
normal oesophageal cells were transfected with 20 nM siRNA, and
protein knockdown was confirmed by western blot analysis (Fig. 2D). The effect of Crm1 siRNA on cell
proliferation was next determined. As shown in Fig. 2E, a significant reduction in
oesophageal cancer cell proliferation was noted five days after
siRNA transfection. The effect of Crm1 knockdown on EPC2 cell
proliferation, on the other hand, was much less pronounced
(Fig. 2E).
To corroborate the effect of Crm1 siRNA on
oesophageal cancer cell proliferation, the commercially available
Crm1 inhibitor, leptomycin B (LMB), was used. Cells were treated
with varying concentrations of LMB, and cell proliferation was
determined 48 h later. In line with the siRNA data, WHCO5 and
KYSE30 cancer cells were significantly more sensitive to LMB
treatment (70–80% decrease in cell number at 100 nM LMB) when
compared to the normal EPC2 cells, which showed little
cytotoxicity, even at high LMB concentrations (30% decrease in cell
number at 100 nM LMB) (Fig.
2F).
These results suggest that Crm1 expression is
upregulated in oesophageal cancer cell lines and that these cells
are highly dependent on its expression for cell survival and
proliferation, whereas non-cancer oesophageal cells appear less
reliant on its expression.
Effect of Crm1 inhibition on the cell
cycle and apoptosis
Since Crm1 inhibition reduced oesophageal cancer
cell proliferation, we next investigated whether Crm1 inhibition
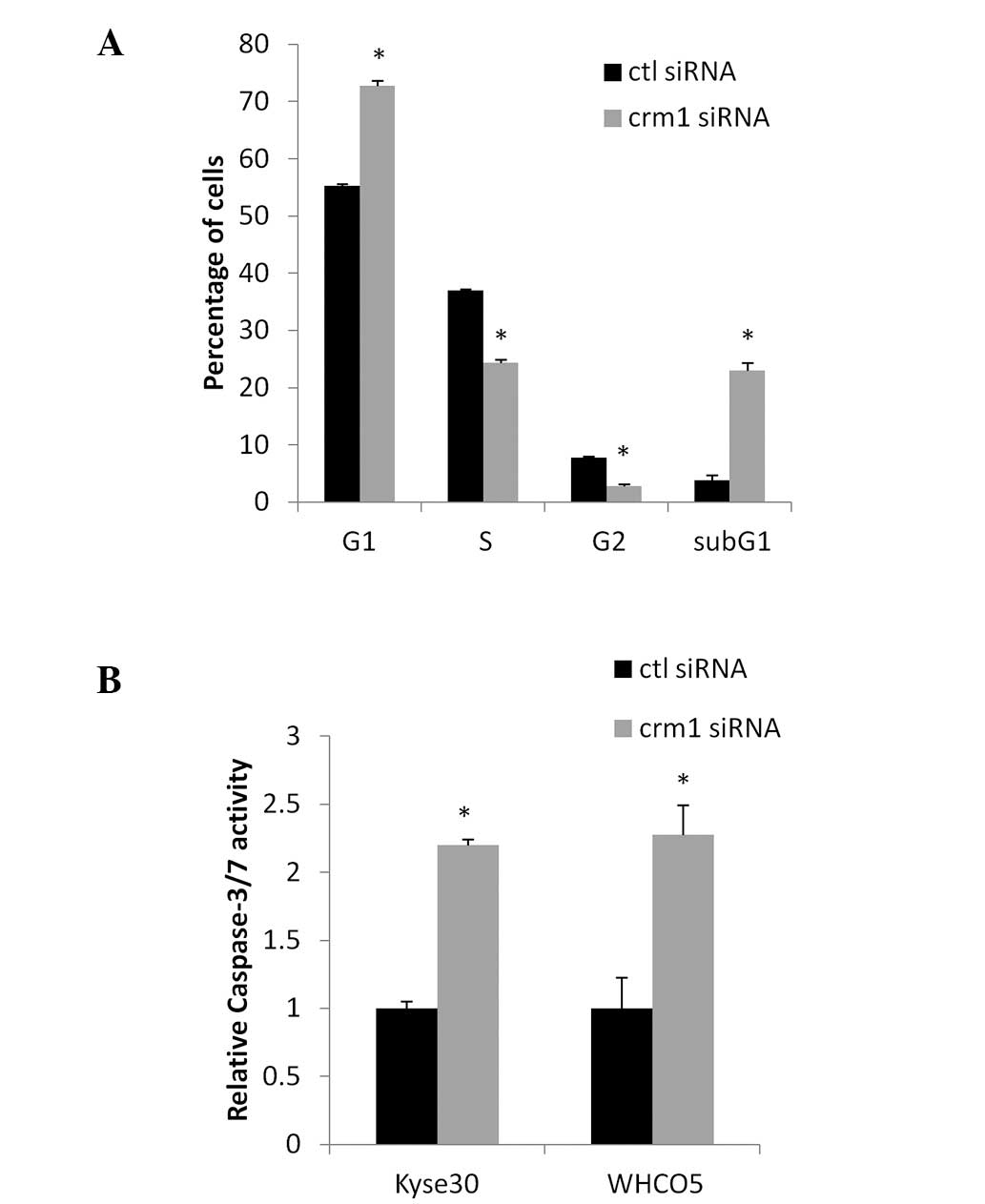
associated with alterations in cell cycle distribution. Crm1
expression was inhibited in KYSE30 oesophageal cancer cells
transfected with siRNA for 72 h. FACS analysis revealed that the
inhibition of Crm1 resulted in a significant increase in cell in
G1, associated with a significant decrease in the number of cells
in the S and G2/M phases of the cell cycle (Fig. 3A). In addition, Crm1 inhibition
resulted in the induction of a subG1 population, indicative of
apoptosis. Similar results were obtained using the WHCO5
oesophageal cancer cell line (data not shown). To confirm that Crm1
inhibition induces apoptosis in oesophageal cancer cells, a
caspase-3/7 assay was performed where caspase-3/7 activity was
measured in KYSE30 and WHCO5 cells after transfection with control
or Crm1 siRNA. Fig. 3B shows a
significant increase in caspase-37 activity in cells transfected
with Crm1 siRNA compared to control siRNA-transfected cells,
confirming the induction of apoptosis after Crm1 inhibition.
Discussion
This study investigated the expression of the
nuclear transport receptor, Crm1, in oesophageal cancer. Crm1
showed weak nuclear expression patterns in normal oesophageal
epithelium, while its expression was much stronger and localised to
the nucleus, cytoplasm and nuclear envelope in oesophageal tumour
tissues. Real-time RT-PCR analysis revealed that Crm1 was also
elevated at the mRNA level. Western blot analysis confirmed the
elevated expression of Crm1 in oesophageal cancer cells compared to
normal oesophageal epithelial cells. Crm1 has recently been
reported to be expressed at elevated levels in cervical squamous
cell carcinoma, ovarian carinoma, osteosarcoma, glioma, pancreatic
cancer, gastric cancer and multiple myeloma (7–13). To
the best of our knowledge, this is the first study reporting
elevated expression of Crm1 in oesophageal cancer.
It is likely that the elevated levels of Crm1 in
tumour tissues are derived from its cell cycle-dependent
regulation. Evidence suggests that Crm1 is a cell cycle-regulated
gene, as its mRNA levels have been reported to oscillate with the
cell cycle, where Crm1 expression is initiated in late G1 and
reaches a peak at G2/M (22). We
recently reported that the Crm1 promoter is regulated by NFY and
Sp1, both transcription factors that function in concert with the
cell cycle and are found at elevated levels in transformed and
cancer cells (23). Furthermore, we
found that p53 represses the Crm1 promoter in response to DNA
damage (23), also likely
contributing to elevated Crm1 in cancer cells, as most cancer cells
either contain low levels of wild-type p53 protein (through
enhanced p53 degradation) or harbor p53 mutations, including
oesophageal cancer (24,25). The regulation of Crm1 expression by
NFY, Sp1 and p53 transcription factors in cancer cells likely
results in an increased expression of Crm1 compared to that in
normal cells.
We report here that the nuclear, cytoplasmic and
nuclear membrane expression of Crm1 is increased in oesophageal
tumours. This localisation pattern is in line with Noske et
al, who observed an increase in both nuclear and cytoplasmic
Crm1 staining in ovarian carcinomas, when compared to normal
tissues (8). Notably, however, Shen
et al showed that Crm1 expression in gliomas was
predominantly nuclear, with only weak cytoplasmic immunoreaction
(10), suggesting that the
localization pattern of Crm1 does vary with tumour type. Both
studies made use of the same antibody to stain for Crm1 expression
as in the present study. Furthermore, we report that there is a
trend where high Crm1 expression associates with poor patient
survival, supporting studies showing a prognostic role for Crm1 in
other cancer types (8–11). However, due to the limited sample
size of early stage disease, prospective large-scale studies are
required to investigate the true prognostic role of high Crm1
expression in human oesophageal squamous cell carcinoma.
While the prognostic value of elevated Crm1 in
oesophageal cancer has not yet been determined conclusively, our
findings suggest that it may have value as an oesophageal cancer
therapeutic target. Our results demonstrate a role for Crm1 in the
biology of oesophageal cancer as its inhibition resulted in G1 cell
cycle arrest and the induction of apoptosis. We similarly found
that treatment with the Crm1 inhibitor, leptomycin B, resulted in
oesophageal cancer cell death. Interestingly, normal oesophageal
epithelial cells were less sensitive to Crm1 inhibition using siRNA
and LMB, suggesting that targeting Crm1 could be an attractive
therapy for oesophageal cancer. Other studies have similarly
reported an increased sensitivity of cancer cells to Crm1
inhibition compared to their normal counterparts (7,26,27),
thus highlighting the need for the development of further Crm1
inhibitors. It is interesting to note that Crm1 overexpression
appears to be necessary for the therapeutic efficacy of drugs
targeting Crm1 as cells with low Crm1 expression, for example
normal cells, are less sensitive to Crm1 inhibition. The increased
reliance of cancer cells on Crm1 compared to normal cells is likely
due to the increased nuclear transport rates in cancer cells and
thus an increased reliance on the nuclear transport machinery. This
is supported by a recent study by Kuusisto et al who
reported that nuclear import and export activity is increased in
transformed cells compared to non-transformed cells due to
increased expression of the importer and exporter proteins,
respectively (28).
Overall, we demonstrated that oesophageal cancer
cells contain elevated levels of Crm1 protein and are highly
reliant on its expression, as its inhibition results in cancer cell
death, while in normal cells its inhibition has only a minor
effect. Based on this, we propose that Crm1 has potential as a
diagnostic marker and/or therapeutic target for patients with
oesophageal squamous cell carcinoma. Further study however is
required in order to clarify its true clinical significance.
Acknowledgements
We thank Ms. Antionette Olivier for providing
patient information. This study was supported by grants from the
Cancer Association of South Africa (CANSA), the University of Cape
Town and the Carnegie Corporation of New York.
References
|
1
|
Parkin DM, Bray F, Ferlay J and Pisani P:
Global cancer statistics, 2002. CA Cancer J Clin. 55:74–108. 2005.
View Article : Google Scholar
|
|
2
|
Jemal A, Bray F, Center MM, Ferlay J, Ward
E and Forman D: Global cancer statistics. CA Cancer J Clin.
61:69–90. 2011. View Article : Google Scholar
|
|
3
|
Ossareh-Nazari B, Bachelerie F and
Dargemont C: Evidence for a role of CRM1 in signal-mediated nuclear
protein export. Science. 278:141–144. 1997. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Fukuda M, Asano S, Nakamura T, Adachi M,
Yoshida M, Yanagida M and Nishida E: CRM1 is responsible for
intracellular transport mediated by the nuclear export signal.
Nature. 390:308–311. 1997. View
Article : Google Scholar : PubMed/NCBI
|
|
5
|
Stommel JM, Marchenko ND, Jimenez GS, Moll
UM, Hope TJ and Wahl GM: A leucine-rich nuclear export signal in
the p53 tetramerization domain: regulation of subcellular
localization and p53 activity by NES masking. EMBO J. 18:1660–1672.
1999. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Saji M, Vasko V, Kada F, Allbritton EH,
Burman KD and Ringel MD: Akt1 contains a functional leucine-rich
nuclear export sequence. Biochem Biophys Res Commun. 332:167–173.
2005. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
van der Watt PJ, Maske CP, Hendricks DT,
et al: The Karyopherin proteins, Crm1 and Karyopherin β1, are
overexpressed in cervical cancer and are critical for cancer cell
survival and proliferation. Int J Cancer. 124:1829–1840. 2009.
|
|
8
|
Noske A, Weichert W, Niesporek S, et al:
Expression of the nuclear export protein chromosomal region
maintenance/exportin 1/Xpo1 is a prognostic factor in human ovarian
cancer. Cancer. 112:1733–1743. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Yao Y, Dong Y, Lin F, et al: The
expression of CRM1 is associated with prognosis in human
osteosarcoma. Oncol Rep. 21:229–235. 2009.PubMed/NCBI
|
|
10
|
Shen A, Wang Y, Zhao Y, Zou L, Sun L and
Cheng C: Expression of CRM1 in human gliomas and its significance
in p27 expression and clinical prognosis. Neurosurgery. 65:153–159.
2009. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Huang WY, Yue L, Qiu WS, Wang LW, Zhou XH
and Sun YJ: Prognostic value of CRM1 in pancreas cancer. Clin
Invest Med. 32:E3152009.PubMed/NCBI
|
|
12
|
Zhou F, Qiu W, Yao R, et al: CRM1 is a
novel independent prognostic factor for the poor prognosis of
gastric carcinomas. Med Oncol. 30:7262013. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Tai YT, Landesman Y, Acharya C, et al:
CRM1 inhibition induces tumor cell cytotoxicity and impairs
osteoclastogenesis in multiple myeloma: molecular mechanisms and
therapeutic implications. Leukemia. 28:155–165. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Roberts BJ, Hamelehle KL, Sebolt JS and
Leopold WR: In vivo and in vitro anticancer activity of the
structurally novel and highly potent antibiotic CI-940 and its
hydroxy analog (PD 114,721). Cancer Chemother Pharmacol. 16:95–101.
1986. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Mutka SC, Yang WQ, Dong SD, Ward SL, Craig
DA, Timmermans PB and Murli S: Identification of nuclear export
inhibitors with potent anticancer activity in vivo. Cancer Res.
69:510–517. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Van Neck T, Pannecouque C, Vanstreels E,
Stevens M, Dehaen W and Daelemans D: Inhibition of the
CRM1-mediated nucleocytoplasmic transport by N-azolylacrylates:
structure-activity relationship and mechanism of action. Bioorg Med
Chem. 16:9487–9497. 2008.PubMed/NCBI
|
|
17
|
Allred DC, Brown P and Medina D: The
origins of estrogen receptor α-positive and estrogen receptor
α-negative human breast cancer. Breast Cancer Res. 6:240–245.
2004.
|
|
18
|
Maae E, Nielsen M, Steffensen KD, Jakobsen
EH, Jakobsen A and Sorensen FB: Estimation of immunohistochemical
expression of VEGF in ductal carcinomas of the breast. J Histochem
Cytochem. 59:750–760. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2−ΔΔCT
method. Methods. 25:402–408. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Jones GJ, Heiss NS, Veale RB and Thornley
AL: Amplification and expression of the TGF-α, EGF receptor and
c-myc genes in four human oesophageal squamous cell carcinoma
lines. Biosci Rep. 13:303–312. 1993.
|
|
21
|
Shimada Y, Imamura M, Wagata T, Yamaguchi
N and Tobe T: Characterization of 21 newly established esophageal
cancer cell lines. Cancer. 69:277–284. 1992. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Kudo N, Khochbin S, Nishi K, Kitano K,
Yanagida M, Yoshida M and Horinouchi S: Molecular cloning and cell
cycle-dependent expression of mammalian CRM1, a protein involved in
nuclear export of proteins. J Biol Chem. 272:29742–29751. 1997.
View Article : Google Scholar : PubMed/NCBI
|
|
23
|
van der Watt PJ and Leaner VD: The nuclear
exporter, Crm1, is regulated by NFY and Sp1 in cancer cells and
repressed by p53 in response to DNA damage. Biochim Biophys Acta.
1809:316–326. 2011.PubMed/NCBI
|
|
24
|
Casson AG, Tammemagi M, Eskandarian S,
Redston M, McLaughlin J and Ozcelik H: p53 alterations in
oesophageal cancer: association with clinicopathological features,
risk factors, and survival. Mol Pathol. 51:71–79. 1998. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Uchino S, Saito T, Inomata M, Osawa N,
Chikuba K, Etoh K and Kobayashi M: Prognostic significance of the
p53 mutation in esophageal cancer. Jpn J Clin Oncol. 26:287–292.
1996. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Lecane PS, Kiviharju TM, Sellers RG and
Peehl DM: Leptomycin B stabilizes and activates p53 in primary
prostatic epithelial cells and induces apoptosis in the LNCaP cell
line. Prostate. 54:258–267. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Smart P, Lane EB, Lane DP, Midgley C,
Vojtesek B and Lain S: Effects on normal fibroblasts and
neuroblastoma cells of the activation of the p53 response by the
nuclear export inhibitor leptomycin B. Oncogene. 18:7378–7386.
1999. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Kuusisto HV, Wagstaff KM, Alvisi G, Roth
DM and Jans DA: Global enhancement of nuclear
localization-dependent nuclear transport in transformed cells.
FASEB J. 26:1181–1193. 2011. View Article : Google Scholar : PubMed/NCBI
|