Introduction
Alternative splicing (AS) of pre-mRNA allows many
gene products with different functions to be produced from a single
coding sequence. This is a mechanism by which higher-order
diversity is generated in compact human genomes
(>9×104 different proteins vs. <25×103
genes) (1–2). However, errors in pre-mRNA splicing
leading to altered gene expression patterns can result in cell
transformation and cancer. Affected proteins can include
transcription factors, cell signal transducers and components of
the extracellular matrix (2). One
of these transducers is insulin-like growth factor-I (IGF-I), a key
player in the somatotropic axis (3). It is a 70 amino acid long peptide
regulating processes such as cell growth, proliferation,
differentiation in almost all cell types (4). It has been considered for many years
that for acquisition of its biological activity IGF-I needs to be
fully processed at the transcript and protein level in order to
become mature and active hormone, although in the last two decades,
activities to pro-IGF and resulting E-peptides were assigned
(5–8). The Igf-1 gene structure is very
complex and the number of alternative splicing products is
impressive; in humans, six exons can be spliced to two IGF classes
(I and II depending on which promoter is used) and three isoforms
are present in each class, A, B and C depending on exons 4, 5 and 6
combination fused to exon 3 and 4 coding for mature peptide
(9). The combination of the last
three Igf-1 exons is called C-terminal extension or
E-peptide (10,11). These E-peptides are either cleaved
by proteases to release mature IGF or stay attached and together
with ‘mature IGF sequence’ to form pro-IGF-I (A, B or C). It has
been recently demonstrated that pro-IGF-1A form is as potent as
mature IGF-1 to activate IGF-1R and is a predominant form present
in muscle (12). Another level of
complexity in the IGF-1 activity is glycosylation of IGF-1A
isoform. A gly-pro-IGF-1A can be generated since only C-terminal
extension of an A form can be glycosylated in rodents and humans.
This particular aspect has not yet been studied extensively.
The longest pro-IGF-1 isoform is human pro-IGF-1B
composed of 147 amino acids as a product of Igf-1 gene
splicing pattern exon 1/2-exon 3-exon 4-exon 5 (13). It can be cleaved to mature IGF-I and
E-peptide of 70 and 77 amino acids, respectively. It is of note
that in case of IGF-1B isoform, the C-terminal extension is even
bigger than the mature IGF product. There have been a very limited
number of studies concerning human Eb-peptide, which may be due, in
part, to the lack of an appropriate and specific antibody. Previous
studies used only hybrid proteins and immunodetection of human Eb
peptide was based on either anti-GFP or anti-RFP antibodies
(13,14), which is a less precise approach as
compared to the one specifically targeting an antigen of interest.
Afforded detection of endogenous IGF-I is always better than
relying on transfection models and overexpression.
The aim of the present study was to analyze human
IGF-I isoforms at the protein and transcript level, taking
advantage of oligonucleotides specific for each form, as well as
newly generated antibodies for the A isoform (15) and B isoform produced specifically
for this study. We compared IGF-I levels in 4 cancer cell lines:
HepG2, K562, HeLa and U2OS. There are multiple advantages of these
cell lines from our study perspective. First, they are all
immortalized human cells that can grow and divide indefinitely
under optimal culture conditions. Second, they exhibit different
levels of IGF-I production. HepG2 and K562 cells are known to have
high IGF-1 expression level; the former originated from liver being
the main source of IGF-1 in the circulation and the latter have one
of the highest levels of total IGF-1 among all cell lines
(www.proteinatlas.org) (16,17).
Both cell lines were expected to show detectable levels of
endogenous IGF-1B at the protein level in western blotting
experiments. Third, U2OS cells produce low levels of IGF-1 and can
be considered as a cell line very poor in IGF-1 (‘IGF negative cell
line’), whereas the HeLa line is of considerable interest since it
is transformed with human papillomavirus 18 (HPV-18). Fourth,
influence of viral oncogenes on splicing patterns and crucial
cellular proteins has already been described and we sought to
verify if such interference would be possible for IGF-1.
Materials and methods
Cell culture, cell lines and cell
fractionation
The following cell lines were used: homo
sapiens cervix adenocarcinoma transformed with human
papillomavirus HPV18 (HeLa), homo sapiens osteosarcoma
(U2OS), hepatocellular carcinoma (HepG2) and human immortalized
myelogenous leukemia (K562). In addition, U2OS stable monoclonal
cell lines were used expressing RFP-hEb fusion protein as
previously described (14). All
cell lines with the exception of K562 were cultured in DMEM
containing 10% fetal bovine serum (FBS) supplemented with 100 U/ml
ampicillin and 100 U/ml streptomycin. Stable cell lines were
supplemented with G-418 at 200 μg/ml instead of
ampicillin/streptomycin, whereas K562 cells, known to produce high
level of overall IGF-1 (www.proteinatlas.org) (16,17),
were cultured in a special medium (RPMI, 10% FBS, penicillin and
streptomycin at 100 U/ml, non-essential amino acid, 200 mM
L-glutamine, 10 mM HEPES and 1 mM sodium puryvate). Before
harvesting, cells were washed with PBS and trypsinized using
standard procedures (K562 cells do not require trypsinization). All
culture reagents were purchased from Life Technologies (Grand
Island, NY, USA).
Protease and phosphatase inhibitor cocktails were
added to all buffers according to the manufacturer’s suggestions
(P8340 and P5726, Sigma Chemical Co., St. Louis, MO, USA). Briefly,
cells were transferred from culture dishes to Eppendorf tubes and
an estimated packed cell volume (PCV) of 200 μl was mixed with 5×
PCV of hypotonic lysis buffer (10 mM Hepes pH=7.9, 1.5 mM
MgCl2 and 10 mM KCl) supplemented with 0.5 mM DTT and
protease inhibitors. Cells were incubated on ice for 15 min
allowing them to swell. They were centrifuged for 5 min at 420 × g
and resuspended in 400 μl of isotonic lysis buffer (10 mM Tris-HCl,
pH 7.5, 2 mM MgCl2, 3 mM CaCl2 and 0.3 M
sucrose with the same supplements described above). Cells were
transferred into a glass tissue grind tube and ground on ice slowly
with 15–20 up and down strokes using a type B pestle. Trypan blue
solution was added to a small portion of cells and cell lysis was
confirmed under light microscope. Disrupted cells were centrifuged
for 20 min at 10,000 × g. The supernatant was transferred to a
fresh tube as a cytoplasmic fraction. For protocol details,
http://www.sigmaaldrich.com/technical-documents/protocols/biology/nuclear-protein-extraction.html.
The crude nuclei pellet and total cell extracts were resuspended in
RIPA buffer [20 mM Tris-HCl (pH 7.5), 150 mM NaCl, 1 mM
Na2EDTA, 1 mM EGTA, 1% NP-40, 1% sodium deoxycholate,
2.5 mM sodium pyrophosphate, 1 mM β-glycerophosphate, 1 mM
Na3VO4 and 1 μg/ml leupeptin) containing also
0.5 mM DTT and other protease inhibitors (P8340). The protein
contents of all preparations were measured using the Bradford
procedure (Bio-Rad protein assay; Bio-Rad Laboratories, Hercules,
CA, USA).
RNA extraction, cDNA synthesis and
qRT-PCR
Total RNA was isolated from cells using TRIzol
reagent (Invitrogen, Life Technologies, Grand Island, NY, USA). RNA
concentration was evaluated by reading optical density in 260 nm
(SpectraMax M5 plate reader; Molecular Devices, Sunnyvale, CA,
USA).
Equal amounts (1 μg) of total RNA from each sample
were subjected to single-strand reverse transcription using GenAmp
RNA PCR kit with both random hexamers and oligo d(T)16 (Applied
Biosystems, Foster City, CA, USA). The resultant cDNA was utilized
for quantitative RT-PCR (qRT-PCR) using the Applied Biosystems 7300
real-time PCR System and reagents (Power SYBR Green PCR Master
Mix). Oligonucleotides listed in Table
I specific to Igf-1 isoforms were first checked for
accuracy by PCR (PuReTaq Ready-To-Go PCR Beads; GE Healthcare,
Buckinghamshire, UK) and resolved in 1.5% agarose gel stained with
ethidium bromide. Before qRT-PCR was run on test samples, serial
dilutions of cDNA obtained from K562 cells were prepared (10-fold
dilutions over 7 logs) and the amplification efficiency for all
pairs of oligonucleotides designed according to sequence no.
NT_019546.15 was calculated from the slope(s) of the standard curve
according to the following formula: E = 10(−1/slope) −1 (Table I). For an efficiency of 100%, the
slope is −3.32. A good reaction should have an efficiency between
90 and 110%, which corresponds to a slope between −3.58 and −3.10.
R2 values were ≥0.99 and only 1 pair of nucleotides
showed a lower value of 0.98, which was still acceptable. All
oligonucleotides used in this study met these gold standards for
qRT-PCR parameters. Details are shown in Table I. Regrading qRT-PCR, all samples
were loaded in duplicate in 96-well plates. Expression of
β-actin was used to control for cDNA content. Fold change
(relative copy number) was calculated by comparing ΔCt values for
each Igf-1 splice variant, total Igf-1 and
β-actin.
 | Table ISequences of oligonucleotides used in
real-time PCR for amplification of each of the IGF isoforms using
cDNA from human cancer cell lines. |
Table I
Sequences of oligonucleotides used in
real-time PCR for amplification of each of the IGF isoforms using
cDNA from human cancer cell lines.
| IGF isoform | Sequences
(5′-3′) | Product size (bp)
(ref) | Slope | qPCR efficiency
(E-value %) | R2 |
|---|
| IGF total |
GAGCTGGTGGATGCTCTTCA
CATCCACGATGCCTGTCTGA | 112 (present
study) | −3.4167 | 96.19 | 0.9969 |
| IGF-1A |
TCGTGGATGAGTGCTGCTTCCG
TCAAATGTACTCCCTTCTGGGTCTTG | 144 (26) | −3.5875 | 90.00 | 0.9987 |
| IGF-1B |
GGAGGGGACAGAAGCAAGTC
ATCATCTAGCTCCAGCAGGC | 224 (present
study) | −3.5512 | 91.25 | 0.9986 |
| IGF-1C |
ACCAACAAGAACACGAAGTC
CATGTCACTCTTCACTCCTC | 126 (26) | −3.5538 | 91.15 | 0.9873 |
| β-actin |
TCTGGCACCACACCTTCTAC
GATAGCACAGCCTGGATAGC | 169 (26) | −3.5827 | 90.20 | 0.9896 |
E-peptide synthesis, immunization and
antibody purification
A 45 amino acid sequence of human Eb peptide was
synthesized to generate an Eb specific antibody. The sequence
encompassed the last portion of C-terminus of human IGF-1B isoform
(GWPKTHPGGEQKEGTEASLQIRGKKKEQRRE IGSRNAECRGKKGK) and it is not
present in any other IGF isoform. Human Eb (based from GenBank:
X56774.1) was synthesized using a Thuramed TETRAS 106 peptide
synthesizer at a 10 mg scale and purified via HPLC >95%
(Shimadzu LC-10AT pump). The final product was confirmed by MALDI
mass spectrometry (Waters ZQ 2000 mass spectrometer and Shimadzu
LCMS-2010EV). Furthermore, the peptide was coupled with keyhole
limpet haemocyanin (KLH) through an additional cysteine. The hapten
carrier was used in order to elicit a better immune response in
immunized rabbits as KLH is a very strong immunogen. A very similar
approach was used for production of anti-MGF antibody (18,19).
Two New Zealand white male rabbits (NZW), aged 3 months, ~2.5 kg
mass, were immunized by four injections of 1 mg of synthetic
Eb-peptide-KLH conjugate in 0.5 ml phosphate-buffered saline (PBS),
administered subcutaneously at week 0, 4, 8 and 12. The first
immunization was performed with the addition of 0.5 ml of complete
Freund’s adjuvant. Further immunizations to boost the rabbits were
performed with incomplete Freund’s adjuvant (1, 2 and 3 months
after first immunization). Blood samples from rabbits were
collected every week immediately before (pre-immune) and 8 times
after first immunization. Serum was immediately separated from
blood. The animals were sacrificed on day 101 and blood was
collected by heart puncture. Serum from the last blood collection
was used for antibody purification using magnetic beads containing
protein A (Novazym, Poznań, Poland) and antibody was eluted and
concentration was measured, which was 7.41 mg/ml and 1.52 mg/ml for
animal 1 and 2, respectively. The antibody concentration was
standardized to 1 mg/ml before further use. The immunization
process was carried out according to the guidelines for the use of
laboratory animals and the Polish Ethics Committee for animal
research.
Immunoblotting
Equal amounts of protein were subjected to SDS-PAGE
using 16.5% Tris/Tricine (Bio-Rad, USA) and transferred to
polyvinylidene fluoride membranes (Immobilon-P; Millipore
Corporation, Billerica, MA, USA). Membranes were blocked in
Tris-buffered saline (TBS) plus 0.1% Tween-20 (TTBS) and 5% non-fat
dry milk. Membranes were incubated in primary antibody diluted in
5% milk-TTBS overnight at 4°C. The following primary antibodies
were used in the present study: rabbit polyclonal anti-hEb peptide
described above (dilution, 1:30,000), rabbit polyclonal
anti-IGF-1Ea (dilution, 1:30,000) (15), mouse monoclonal anti-lamin A/C
(dilution, 1:1,000) (Cell Signaling Technology, Danvers, MA, USA),
rabbit polyclonal anti-DsRed (dilution, 1:1,000) (Clontech
Laboratories, Mountain View, CA, USA) and mouse monoclonal
anti-α-tubulin (1:4,000) (T5168, Sigma).
For detection of primary antibodies, secondary
antibodies were used: HRP-conjugated anti-rabbit and HRP-conjugated
anti-mouse (dilution, 1:2,000) (Cell Signaling Technology). Between
each incubation with antibodies, membranes were washed 5 times with
TTBS and before applying ECL they were washed 2 times with TBS.
Specific bands were visualized by X-ray film and by ImageQuant LAS
4000 (GE Healthcare Biosciences, Pittsburg, PA, USA), after
incubation with an enhanced chemiluminescent (ECL) substrate
(Western Lightning-ECL, PerkinElmer, Waltham, MA, USA).
Prior to using the anti-hEb peptide antibody for the
cell lysates, a series of dilutions and blotting conditions were
tested on lysates of U2OS stable monoclonal cell lines expressing
RFP-hEb fusion protein as previously described, to optimize the
conditions. Antibody was diluted 1:1,000–1:50,000, nitrocellulose
vs. PVDF membranes were compared and blocking solutions containing
5% milk, 2% bovine serum albumin and/or 2% horse serum were also
compared.
Results
In order to compare Igf-1 isoform content at
the transcript level in cancer cells, qRT-PCR was performed. Total
RNA was extracted from HeLa, U2OS, HepG2 and K562 cells and reverse
transcribed. Resultant cDNA was analyzed for total Igf-1 and
all isoforms. The specificity of all oligonucleotides used was
verified with cDNA derived from mRNA of K562 cells and PCR products
were resolved in 1.5% agarose gels. This cell line was used as it
is known to have strong levels of Igf-1 expression
(www.proteinatlas.org). Sets of Igf-1
oligonucleotide pairs previously described in the literature turned
out to be below expected specificity within the range of annealing
temperatures tested (55–60°C) (data not shown) and for this reason
new oligonucleotides for Igf-1b and total Igf-1 were
designed to meet expected parameters. Annealing temperature for all
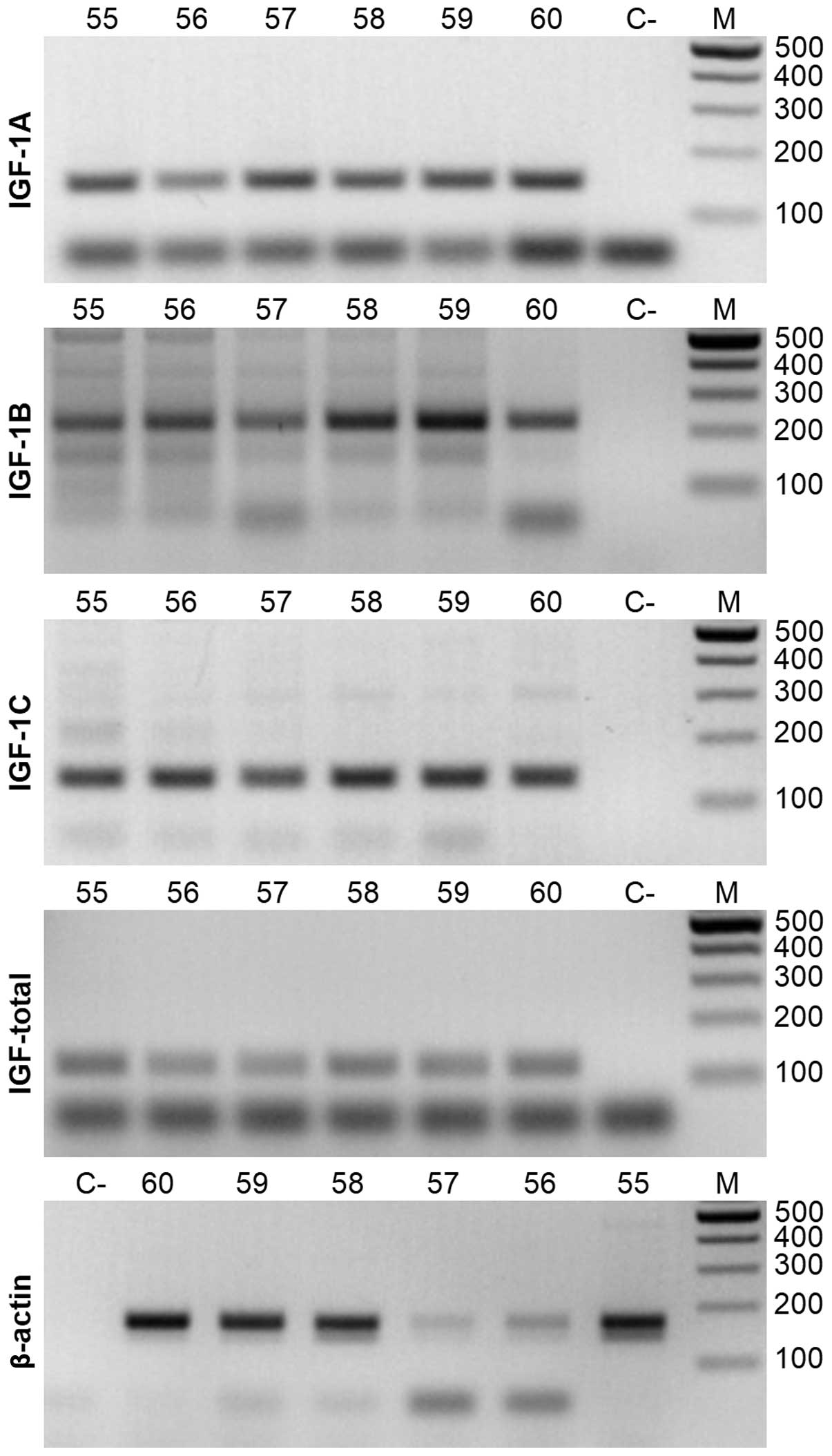
subsequent qRT-PCR reactions was set at 60°C (Fig. 1, all panels).
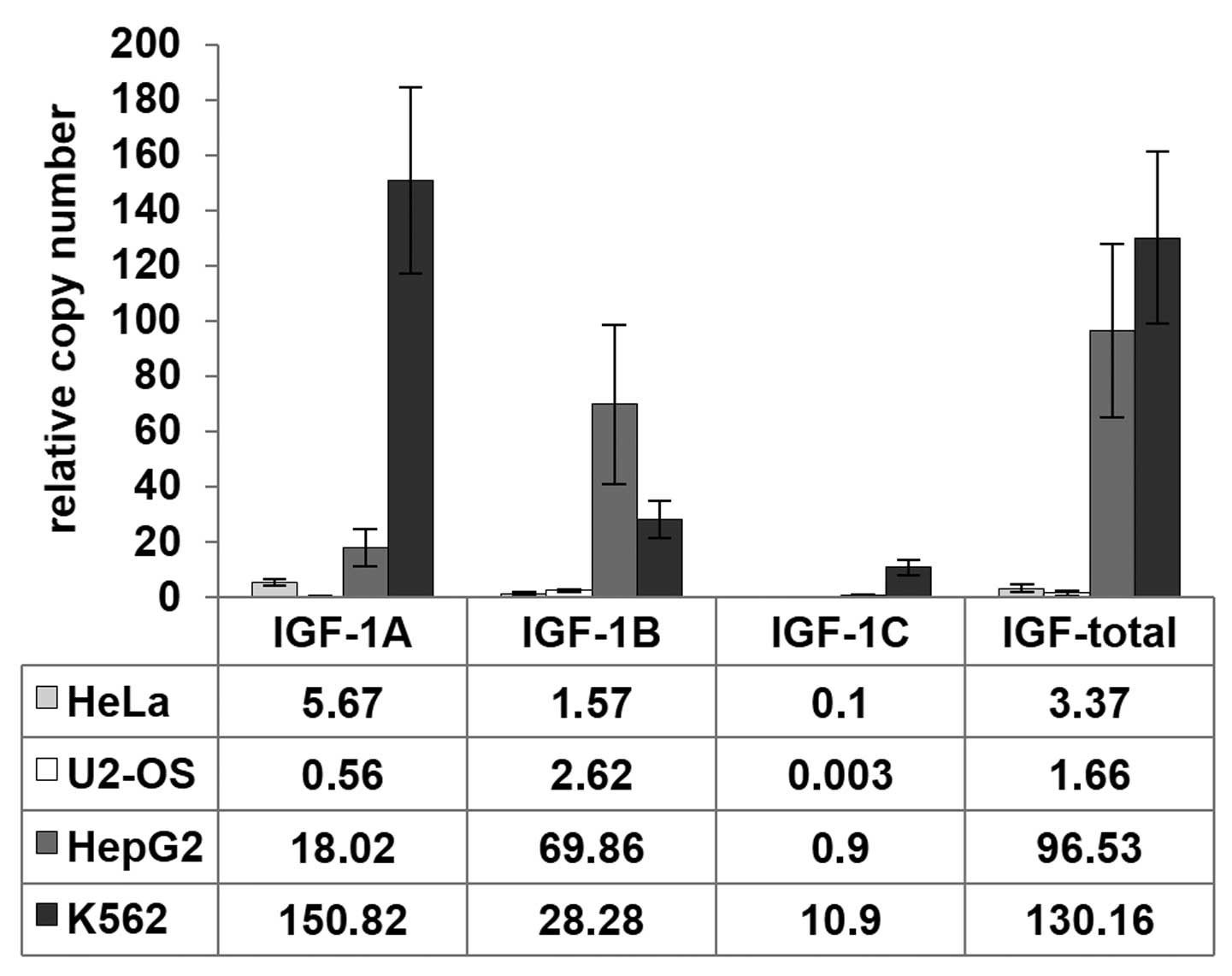
The qRT-PCR presented notable findings. First, the
highest overall relative Igf-1 expression level was observed
in K562 cells when compared to β-actin (~130±31.2), slightly
lower in HepG2 (~96±31.2) and the lowest in HeLa and U2OS cells
(3.4±1.3 and 1.7±1.1, respectively) (Fig. 2). The high expression of
Igf-1 in K562 cells was consistent with database
measurements (http://www.proteinatlas.org). Igf-1a is the
predominant isoform expressed in most tissues (20,21).
Similar to the total Igf-1 results, K562 cells exhibited the
highest expression for the Igf-1a isoform, where it was
10–300-fold higher than in the 3 other cell lines. Igf-1b
expression was most pronounced in HepG2 cells, followed by K562
cells. Igf-1c levels were the highest in K562 cells, with
minimal expression in the three other cells lines with respect to
the β-actin housekeeping gene. Collectively, we estimated
that the predominant isoform in K562 cells was Igf-1a,
whereas Igf-1b appeared to be predominant in HepG2 and U2OS
cells. Second, a switch between Igf-1a and Igf-1b
transcript copy number was observed in HepG2 and U2OS cells. In
both cell lines, this difference was several fold; for example, in
HepG2 cells, a relative copy number of Igf-1b transcripts
was 69.9±28.6 and of Igf-1a transcripts 28.3±6.7 (Fig. 2).
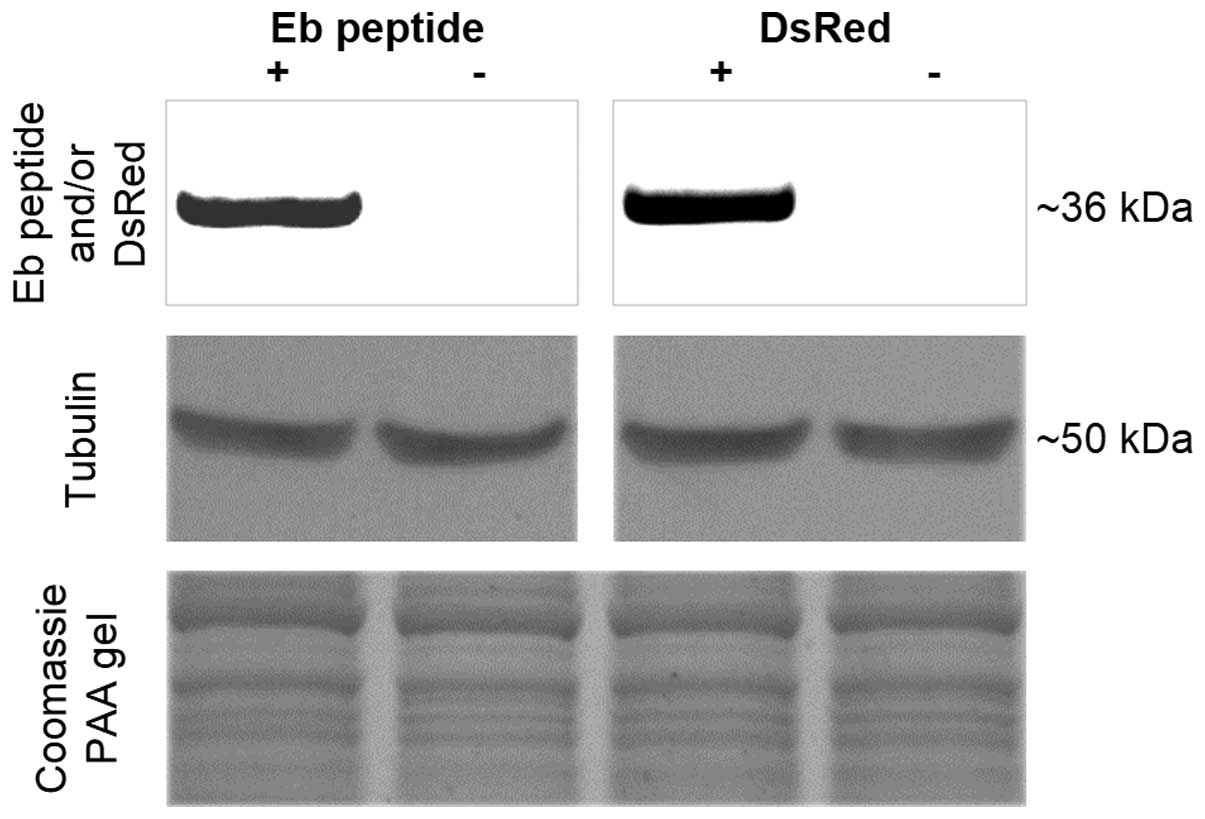
To assess IGF-1B protein expression level and
compare it with Igf-1b transcript level, a purified rabbit
polyclonal anti-Eb peptide antibody was generated. It was first
verified using U2OS stable cell lines expressing human Eb-peptide
fused with dsRed protein (dsRed-hEb) or naïve U2OS cells. These
stable cell lines were described previously (14). They express the fusion protein at a
high level as its mRNA is transcribed under control of a strong
promoter (PCMV). For validation, two separate
immunoblots were performed; one with a rabbit anti-Eb antibody and
a second with a rabbit anti-dsRed to confirm the presence of the
same hybrid protein using a different epitope and the same stable
cell line preparation. The results are presented in Fig. 3. It can be concluded that both
antibodies revealed a presence of a band of the same molecular mass
(36 kD) and anti-Eb peptide antibody specifically recognizes hEb
peptide. Thus, the new antibody was specific for hEb.
Since our previous studies were all performed using
hybrid dsRed-hEb fusion, we extended our analysis to measure
endogenous human Eb peptide and human Ea peptide levels and
cellular localization without the aid of epitope tags or fused
sequences (14,22). Based on the expression results, we
anticipated that the HeLa, U2OS, HepG2, K562 cancer cells would
also display a range of IGF-1 protein levels. Immunoblotting with
anti-Eb antibody revealed a higher protein content of Eb peptide in
K562 cell lysates compared with other cell lines. Only with long
exposure times could hEb be detected in the HepG2, U2OS, or HeLa
cell lysates. The ~9 kD band size was consistent with hEb cleaved
from mature IGF-I. However, in the HeLa cells, a ~16 kD band was
also evident, which was the predicted size for pro-IGF-IB. No
signal corresponding to human Eb peptide or pro-IGF-IB was observed
in the cytoplasmic fraction from any cell line (Fig. 4, panels 1 and 2). However, all cell
lines had hEb present in the nuclear fractions, where it was most
prominent in the K562 cells, with no evidence of pro-IGF-IB in any
nuclear fraction. The 20 kD bands detected by anti-hEb peptide
antibody were presumed to be non-specific as there was no
Eb-related protein predicted to be that size and the bands were
present in all lanes regardless of the cellular fraction.
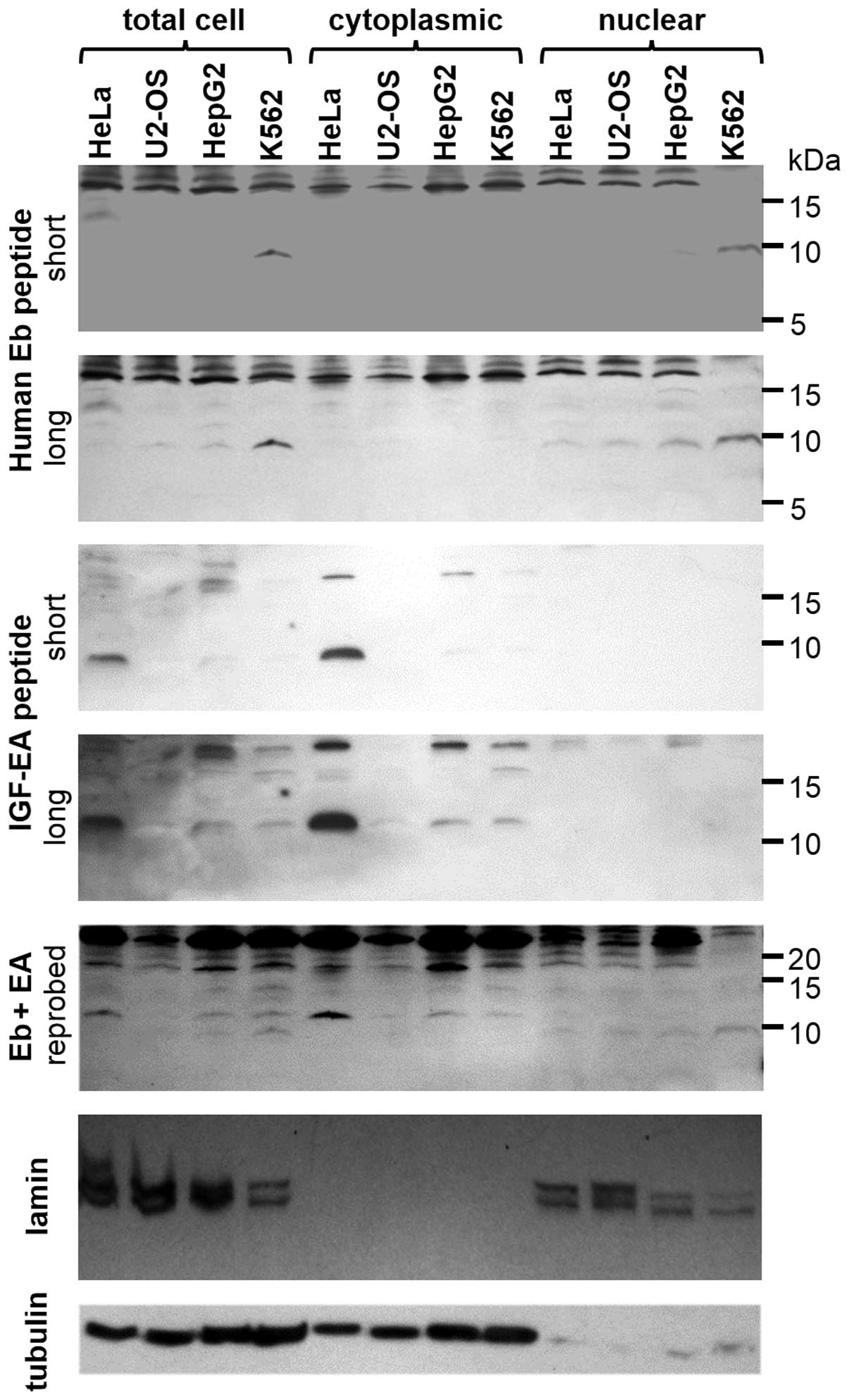 | Figure 4Immunoblotting showing endogenous IGF
isoforms in human cancer cells. Sixty micrograms of protein were
resolved in each lane. A 9 kD band is present in total cell extract
and nuclear fraction from K562 cells and, to a smaller extent, in
HepG2 when probed with anti-Eb peptide antibody on the first
membrane (short exposure to X-ray film). After longer exposure,
this band was also slightly visible in the other two cell lines
used in this study (HeLa, U2OS), second panel. It never appeared in
cytoplasmic fraction of these cells. A 11–12 kD band was detected
when the same membrane was probed with anti-EA antibody,
corresponding to human pro-IGF-1A (panel 3). However, this band was
evident only in total cell extract and cytoplasmic fraction and
never present in the nuclear one (panel 4). Also, another blotting
is shown (panel 5), the same membrane already probed with anti-Eb
antibody was stripped and reprobed with anti EA-peptide antibody. A
merge of both anti-Eb and anti-EA pattern can be observed and a
molecular mass difference of 2–3 kD between human Eb-peptide (9 kD)
and pro-IGF-1A (11–12 kD). For confirmation of cellular
fractionation, 2 different antigens are shown: lamin (nuclear
marker), tubulin (cytoplasmic marker) (panels 6 and 7). IGF,
insulin-like growth factor. |
In parallel, the same preparations (total cell
extracts, cytoplasmic and nuclear fractions) were probed with
rabbit anti-Ea antibody. A distinct 11–12 kD band corresponding to
pro-IGF1A was observed in most samples, in addition to multiple
higher molecular weight bands most likely representing glycosylated
pro-IGF-IA. However, no hEa peptide (predicted to be ~4 kD) was
detected. In contrast to the expression results, pro-IGF-1A levels
were the greatest in HeLa cell extracts, with moderate levels of
this protein in HepG2 and K562 lysates and virtually no detectable
pro-IGF-IA in U2OS cell extracts. All pro-IGF-1A was present in the
cytoplasmic fraction and no detection was observed in the nuclear
fraction (Fig. 4, panels 1 and 2
vs. panels 3 and 4). In order to demonstrate the presence of both
IGF forms, the immunoblots were reprobed with anti-Eb and anti-Ea
antibodies simultaneously to ensure that the bands were distinct
from each other with respect to size. Although the signals are
slightly weaker when compared to immunoblots presented in other
panels, both bands corresponding to pro-IGF-1A and hEb peptide can
be observed at the expected 2–3 kD difference in size (Fig. 4, panel 5). To demonstrate efficient
fractionation of cells, two markers were used for immunoblotting:
lamin for nuclear fraction and tubulin for cytoplasmic fraction
(Fig. 4, panels 6 and 7). These
results confirmed that the cytoplasmic and nuclear fractions were
separated.
Upon further analysis, the size of hEb was
consistent with the protein being exclusively the E peptide,
whereas the size of hEa was only detected as a pro-IGF-I form
(attached to mature IGF-I). Thus, the IGF-IA and IGF-IB isoforms
not only display different localization and processing in the range
of cell lines studied, but there are also clear differences in the
relationship between transcription and protein accumulation for the
isoforms. Comparisons of IGF-IA and IGF-IB in each cell line showed
discordance between RNA and protein levels. In comparison with the
expression levels measured by qRT-PCR, it appeared that the K562
cell line exhibited greater post-transcriptional stabilization of
hEb than HepG2 cells, for the ~2-fold greater expression of
Igf-1b in HepG2 vs. K562 cells was reversed in the
immunoblotting results, where, at most, hEb levels in HepG2 cells
were half of those in K562. For Igf-1a, HeLa cells had the
greatest intensity of pro-IGF-IA although expression of this
isoform was >3–20-fold higher in HepG2 and K562 cells,
respectively. Thus, the level of IGF-I production and retention
cannot be predicted from measurements of transcription.
Additional blots for total IGF-I and IGF-IC may have
helped to clarify the levels of all isoforms in the cell lines.
Initial experiments to detect total IGF-I and IGF-IC were
unsuccessful, presumably since the endogenous levels of these
proteins were below the level of detection by the available
antibodies. For instance, several antibodies specific for human
IGF-I were tried, but none of them could resolve a clean signal in
lanes containing <2 ng human recombinant IGF-I. Thus, we did not
pursue the measurements of total IGF-I by immunoblotting. In
addition, we did not pursue human Ec as the levels of expression
were very low in all cell lines (Fig.
2).
Discussion
Alternative splicing of the Igf-1 gene
produces several different transcripts, including Igf-1a,
Igf-1b and Igf-1c (23). However, understanding the biological
significance of these splicing events has been the subject of wide
debate for more than 2 decades. It is clear from our study that for
Igf-1a, -1b and -1c splice variants, the
transcript content does not necessarily reflect the protein
content. It is particularly true for pro-IGF-1A expression in HeLa
and K562 cells (lower transcript level vs. higher protein content
for HeLa cells and vice versa for K562 cells). Several explanations
are plausible. First, depending on the cellular context, the
post-transcriptional efficiency of one isoform processing into
mature IGF-1 protein may vary. Second, in case of IGF-1A protein,
processing is even more complex as glycosylated forms can be
produced in the cell. We have previously described that both
N-sites present in the rodent E peptide extension could carry sugar
residues and, depending on cell type, different glycosylation
patterns are possible. The proportion of glycosylated pro-IGF1A can
be overwhelming as demonstrated with rabbit anti-mouse mature IGF
antibody (12, unpublished observations). Closer observation of
HepG2 and K562 total cell extract lanes shows bands of molecular
mass in the range of 12–15 kD; they could correspond to human
glycosylated pro-IGF-1A, which contains one N-glycosylation site.
However, due to antigen-antibody affinity reduction by sugar
residues, the real protein content of gly-pro-IGF-1A is likely to
be largely underestimated. One more explanation to be considered is
that pro-IGF-1A protein in HeLa cells is stabilized in direct or
indirect interactions with E6 and E7 HPV proteins. It would be
consistent with anti-apoptotic properties of these two viral
oncogenes that could enhance pro-IGF-1A concentration and activity.
These reasons reflect why we do not observe concordant results for
IGF-1A between transcript copy number and protein level by western
blotting.
It is difficult to compare transcript copy numbers
of each IGF isoform, as different cells or tissues were used
between studies. Another difficulty is to compare transcript copy
numbers of each IGF isoform as different calculations were used
between researchers (absolute copy number vs. relative copy
number). However, proportions and shifts of IGF splice variants can
be compared between studies and within cell lines used in the
present study. Igf-1 expression between normal and cancerous
tissues and cells was evaluated by Kasprzak and co-workers and a
downshift of the Igf-1b content in the favor of
Igf-1a isoform was reported when colorectal cancer cells and
non-tumor tissue were analyzed (24). It is interesting to note that in a
previous study, the same group also observed a 10-fold decrease in
Igf-1b transcript level between cancerous and non-tumor
colorectal tissue (25). In our
study, a similar Igf-1 splicing pattern was observed in HeLa
and K562 cells where Igf-1a transcripts prevailed; however,
Igf-1b dominated over other isoforms in HepG2 cells. It is
not surprising since the fold increase in Igf-1b mRNA levels
described elsewhere was approximately 3 times greater than the fold
increase in Igf-1a mRNA levels for liver cells compared with
other tissues (20). Furthermore,
in a different study, an increase of Igf-1b and decrease of
Igf-1a expression was found when cervical cancer and control
cells were compared (26). Thus, it
appears that the alterations in Igf-1 expression and
splicing are cell-specific.
There are multiple possible mechanisms by which
there is higher human Eb peptide content in K562 compared to HepG2
cells. As there was little pro-IGF-IB found in any cell lines, the
pro-peptide could have been cleaved during processing, leading to
secretion of mature IGF-I and retention of Eb peptide that becomes
sequestered in the nucleus. This would require K562 cells to hold
on to significantly more Eb peptide per mole produced, compared to
the HepG2 cells. Since the HepG2 cells are derived from liver,
which is the primary endocrine organ for IGF-I secretion, it is
entirely possible that these cells secrete IGF-I, pro-IGF-I and the
E peptides more efficiently that K562. We did not determine how
much IGF-I was secreted by any of the cell lines and so this has
not been tested directly.
An alternative mechanism for heightened Eb present
in the K562 cells, is more efficient Eb peptide uptake from the
media, which then accumulates in the nucleus. We know from our
previous study that Ea and Ec peptides produced by transfection can
be secreted and internalized by neighboring cells (22). One possibility is that pro-IGF-1B is
cleaved quickly after translation and there is little pro-IGF-1B
present within the cell. Pre-IGF-1 (mature IGF-1 with a signal
peptide) would be directed for secretion and Eb-petide containing a
nuclear localization signal (NoLS) would be directed to nuclei. Or,
human Eb peptide produced in cells from same culture would be
secreted as pre-pro-IGF-1 form, cleaved in extracellular milieu and
trafficked back to nuclei of target cells. Results presented herein
show that K562 and HepG2 cells may be a suitable tool for more
extensive studies of IGF-IA and IGF-1B in terms of protein
processing and nuclear interactions at endogenous levels. It would
be interesting to find out why the IGF-1B isoform and, by
extension, human Eb peptide are modulated in different types of
cancer cells.
IGF-IC is also known as mechano growth factor (MGF),
which has been the focus of much study in skeletal muscle for its
potential growth-promoting properties (15,27–29).
However, Igf-1c levels have also been evaluated in human
prostate cancer (PCa) tissues and in human PC-3 and LNCaP cells.
IGF-IC/MGF expression was markedly higher in PCa and prostatic
intraepithelial neoplasia (PIN) than in normal prostate tissues,
while the normal prostate epithelial cells (HPrEC) did not express
MGF (30). In our study, IGF-1C
transcript copy number was very low in HeLa, U2OS and HepG2 cells,
except for K562 cells, in which the proportion of Igf-1c
transcript vs. total Igf-1 transcripts was also bigger than
in other cells. We did not include the PC-3 cells in the present
study, but it is worth noting that PCa growth is regulated by
IGF-IEc transcript via Ec peptide-specific and
IGF-IR/IR-independent signaling (31). However, it is not known if the
levels of Igf-1c are predominant in human prostate cancer
cells and tissues, or if other isoforms are also high.
The differences between the transcript and resultant
protein level among different cell types may arise from the
efficiency of transcript-protein translation and a number of
studies showed that depending on the cellular context, this
efficiency may vary (32–34). A good example of how discrepant mRNA
and protein levels can be is NOXA expression. NOXA belongs to the
Bcl-2 family, promotes activation of caspases and apoptosis. In
mantle cell lymphoma (MCL) cells NOXA mRNA was found to be highly
expressed, whereas NOXA protein levels were low (35). It has been previously reported that
RNA splicing factors were regulated by HPV16 during cervical tumour
progression (36); in this context,
HPV18 proteins could influence IGF-I splicing pattern in HeLa cells
observed in our study. Since viruses use cellular RNA binding
proteins for viral RNA processing, it is possible that the splicing
of cellular pre-mRNAs is affected by viral infection. For example,
the expression of promyelocytic leukemia (PML) isoforms differs in
various cell lines or tissues, where PML-I is the dominant form
expressed in the brain but not in the liver (37). However, the regulation mechanisms of
the cell type- or tissue-specific alternative splicing of PML
pre-mRNA have not yet been clarified. In one study, it was
demonstrated that the viral protein ICP27 regulates the switching
of the PML isoform from PML-II to PML-V by promoting the retention
of PML intron 7a (38). Further
studies are required to elucidate cells expressing viral proteins,
which can interfere not only in splicing events as described
earlier, but can also stabilize cellular protein products involved
in many important processes: hepatitis B virus × protein (HBx) was
reported to stabilize amplified in breast cancer 1 protein (AIB1)
and cooperate with it to promote human hepatocellular carcinoma
cell invasiveness (39) and it was
also reported that cellular proteins were actively stabilized by
HSV-1 viral gene products (40). In
HeLa cells, such stabilizing activity towards pro-IGF-1A protein
could be linked to HPV18 oncoproteins. This hypothesis merits
further investigation.
To conclude, it should be emphasized that the
presence of human Eb peptide was clearly demonstrated for the first
time in nuclear fractions of cancer cells in the present study.
Similarly, a novel and sensitive antibody to human Eb peptide
proved to be sensitive and specific. Moreover, mouse Ea peptide
antibody proved to be suitable for detection of its human
counterpart despite four amino acids difference between the two
species. A larger picture emerges from our study as well as those
of others cited in this manuscript; in different types of cancer, a
different IGF isoform and a resultant IGF E-peptide may be down- or
upregulated and further involved in growth, proliferation, cell
transformation, cancer progression. Viral proteins are also
involved in these processes, modifying gene splicing and
stabilizing cellular proteins of important actions.
Acknowledgements
This study was financed by the National Science
Center, Poland (NN 401 555 440 to Dr Julia Durzyńska) and a
Research Grant from National Institutes of Health (R01 AR057363 to
Professor Elisabeth Barton). The authors thank Adam Burzyński and
his employees from Novazym, Poznań, Poland (http://www.novazym.com) for the assistance with the
anti-Eb antibody production and purification.
References
|
1
|
Brett D, Pospisil H, Valcárcel J, Reich J
and Bork P: Alternative splicing and genome complexity. Nat Genet.
30:29–30. 2002. View
Article : Google Scholar : PubMed/NCBI
|
|
2
|
Roy B, Haupt LM and Griffiths LR: Review:
alternative splicing (AS) of genes as an approach for generating
protein complexity. Curr Genomics. 14:182–194. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Renaville R, Hammadi M and Portetelle D:
Role of the somatotropic axis in the mammalian metabolism. Domest
Anim Endocrinol. 23:351–360. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Langdahl BL, Kassem M, Møller MK and
Eriksen EF: The effects of IGF-I and IGF-II on proliferation and
differentiation of human osteoblasts and interactions with growth
hormone. Eur J Clin Invest. 28:176–183. 1998. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Siegfried JM, Kasprzyk PG, Treston AM,
Mulshine JL, Quinn KA and Cuttitta F: A mitogenic peptide amide
encoded within the E peptide domain of the insulin-like growth
factor IB prohormone. Proc Natl Acad Sci USA. 89:8107–8111. 1992.
View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Kuo YH and Chen TT: Novel activities of
pro-IGF-I E peptides: regulation of morphological differentiation
and anchorage-independent growth in human neuroblastoma cells. Exp
Cell Res. 280:75–89. 2002. View Article : Google Scholar
|
|
7
|
Matheny RW Jr and Nindl BC: Loss of
IGF-IEa or IGF-IEb impairs myogenic differentiation. Endocrinology.
152:1923–1934. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Hede MS, Salimova E, Piszczek A, et al:
E-peptides control bioavailability of IGF-1. PLoS One.
7:e511522012. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Barton ER: The ABCs of IGF-I isoforms:
impact on muscle hypertrophy and implications for repair. Appl
Physiol Nutr Metab. 31:791–797. 2006. View
Article : Google Scholar : PubMed/NCBI
|
|
10
|
Tian XC, Chen MJ, Pantschenko AG, Yang TJ
and Chen TT: Recombinant E-peptides of pro-IGF-I have mitogenic
activity. Endocrinology. 140:3387–3390. 1999. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Brisson BK and Barton ER: New modulators
for IGF-I activity within IGF-I processing products. Front
Endocrinol (Lausanne). 4:422013.PubMed/NCBI
|
|
12
|
Durzyńska J, Philippou A, Brisson BK,
Nguyen-McCarty M and Barton ER: The pro-forms of insulin-like
growth factor I (IGF-I) are predominant in skeletal muscle and
alter IGF-I receptor activation. Endocrinology. 154:1215–1224.
2013.PubMed/NCBI
|
|
13
|
Tan DS, Cook A and Chew SL: Nucleolar
localization of an isoform of the IGF-I precursor. BMC Cell Biol.
3:172002. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Durzyńska J, Wardziński A, Koczorowska M,
Goździcka-Józefiak A and Barton ER: Human Eb peptide: not just a
by-product of pre-pro-IGF1b processing? Horm Metab Res. 45:415–422.
2013.PubMed/NCBI
|
|
15
|
Brisson BK and Barton ER: Insulin-like
growth factor-I E-peptide activity is dependent on the IGF-I
receptor. PLoS One. 7:e455882012. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Aro AL, Savikko J, Pulkkinen V and von
Willebrand E: Expression of insulin-like growth factors IGF-I and
IGF-II, and their receptors during the growth and megakaryocytic
differentiation of K562 cells. Leuk Res. 26:831–837. 2002.
View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Wahner Hendrickson AE, Haluska P,
Schneider PA, et al: Expression of insulin receptor isoform A and
insulin-like growth factor-1 receptor in human acute myelogenous
leukemia: effect of the dual-receptor inhibitor BMS-536924 in
vitro. Cancer Res. 69:7635–7643. 2009.PubMed/NCBI
|
|
18
|
Philippou A, Stavropoulou A, Sourla A, et
al: Characterization of a rabbit antihuman mechano growth factor
(MGF) polyclonal antibody against the last 24 amino acids of the E
domain. In Vivo; 22:27–35. 2008.PubMed/NCBI
|
|
19
|
Harris JR and Markl J: Keyhole limpet
hemocyanin (KLH): a biomedical review. Micron. 30:597–623. 1999.
View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Lowe WL Jr, Lasky SR, LeRoith D and
Roberts CT Jr: Distribution and regulation of rat insulin-like
growth factor I messenger ribonucleic acids encoding alternative
carboxyterminal E-peptides: evidence for differential processing
and regulation in liver. Mol Endocrinol. 2:528–535. 1988.
View Article : Google Scholar
|
|
21
|
Wallis M: New insulin-like growth factor
(IGF)-precursor sequences from mammalian genomes: the molecular
evolution of IGFs and associated peptides in primates. Growth Horm
IGF Res. 19:12–23. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Pfeffer LA, Brisson BK, Lei H and Barton
ER: The insulin-like growth factor (IGF)-I E-peptides modulate cell
entry of the mature IGF-I protein. Mol Biol Cell. 20:3810–3817.
2009. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Adamo ML, Neuenschwander S, LeRoith D and
Roberts CT Jr: Structure, expression, and regulation of the IGF-I
gene. Adv Exp Med Biol. 343:1–11. 1993. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Kasprzak A, Szaflarski W, Szmeja J, et al:
Differential expression of IGF-1 mRNA isoforms in colorectal
carcinoma and normal colon tissue. Int J Oncol. 42:305–316.
2013.PubMed/NCBI
|
|
25
|
Kasprzak A, Szaflarski W, Szmeja J, et al:
Expression of various insulin-like growth factor-1 mRNA isoforms in
colorectal cancer. Contemp Oncol (Pozn). 16:147–153.
2012.PubMed/NCBI
|
|
26
|
Koczorowska MM, Kwasniewska A and
Gozdzicka-Jozefiak A: IGF1 mRNA isoform expression in the cervix of
HPV-positive women with pre-cancerous and cancer lesions. Exp Ther
Med. 2:149–156. 2011.PubMed/NCBI
|
|
27
|
Cui H, Yi Q, Feng J, Yang L and Tang L:
Mechano growth factor E peptide regulates migration and
differentiation of bone marrow mesenchymal stem cells. J Mol
Endocrinol. 52:111–120. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Wu J, Wu K, Lin F, et al: Mechano-growth
factor induces migration of rat mesenchymal stem cells by altering
its mechanical properties and activating ERK pathway. Biochem
Biophys Res Commun. 441:202–207. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Fornaro M, Hinken AC, Needle S, et al:
Mechano-growth factor peptide, the COOH terminus of unprocessed
insulin-like growth factor 1, has no apparent effect on myoblasts
or primary muscle stem cells. Am J Physiol Endocrinol Metab.
306:E150–E156. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Armakolas A, Philippou A, Panteleakou Z,
et al: Preferential expression of IGF-1Ec (MGF) transcript in
cancerous tissues of human prostate: evidence for a novel and
autonomous growth factor activity of MGF E peptide in human
prostate cancer cells. Prostate. 70:1233–1242. 2010. View Article : Google Scholar
|
|
31
|
Philippou A, Armakolas A and Koutsilieris
M: Evidence for the possible biological significance of the IGF-1
gene alternative splicing in prostate cancer. Front Endocrinol
(Lausanne). 4:312013.PubMed/NCBI
|
|
32
|
Straub L: Beyond the transcripts: what
controls protein variation? PLoS Biol. 9:e10011462011. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Vogel C and Marcotte EM: Insights into the
regulation of protein abundance from proteomic and transcriptomic
analyses. Nat Rev Genet. 13:227–232. 2012.PubMed/NCBI
|
|
34
|
Klevebring D, Fagerberg L, Lundberg E,
Emanuelsson O, Uhlén M and Lundeberg J: Analysis of transcript and
protein overlap in a human osteosarcoma cell line. BMC Genomics.
11:6842010. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Dengler MA, Weilbacher A, Gutekunst M, et
al: Discrepant NOXA (PMAIP1) transcript and NOXA protein levels: a
potential Achilles’ heel in mantle cell lymphoma. Cell Death Dis.
5:e10132014.PubMed/NCBI
|
|
36
|
Mole S, McFarlane M, Chuen-Im T, Milligan
SG, Millan D and Graham SV: RNA splicing factors regulated by HPV16
during cervical tumour progression. J Pathol. 219:383–391. 2009.
View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Condemine W, Takahashi Y, Zhu J, et al:
Characterization of endogenous human promyelocytic leukemia
isoforms. Cancer Res. 66:6192–6198. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Nojima T, Oshiro-Ideue T, Nakanoya H, et
al: Herpesvirus protein ICP27 switches PML isoform by altering mRNA
splicing. Nucleic Acids Res. 37:6515–6527. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Liu Y, Tong Z, Li T, et al: Hepatitis B
virus × protein stabilizes amplified in breast cancer 1 protein and
cooperates with it to promote human hepatocellular carcinoma cell
invasiveness. Hepatology. 56:1015–1024. 2012.
|
|
40
|
Kalamvoki M and Roizman B: HSV-1 degrades,
stabilizes, requires, or is stung by STING depending on ICP0, the
US3 protein kinase, and cell derivation. Proc Natl Acad Sci USA.
111:E611–E617. 2014. View Article : Google Scholar : PubMed/NCBI
|