Introduction
Osteosarcoma (OS), the most common malignant tumor
of the bone, occurs mainly in adolescents and young adults with a
morbidity of ~5 cases per million (1). It originates from common mesenchymal
stem cell (MSC) progenitors that undergo disruption to normal
osteoblast differentiation (2).
Although the 5-year survival rate has risen to ~60–70%, a
substantial percentage of patients still respond poorly to
chemotherapy and have a high risk of relapse or metastasis even
after curative resection (3).
microRNAs (miRNAs), a class of short (~22 nt)
endogenous non-coding RNAs, post-transcriptionally regulate the
expression of target genes involved in many types of cancers
including osteosarcoma (4) and act
as oncogenes or tumor-suppressor genes. A single miRNA can silence
a large number of genes, allowing these molecules extensive control
of many cellular functions (5).
Emerging evidence of individual miRNAs affecting developmental
biology, cellular differentiation programs and oncogenesis
continues to grow (6).
In osteosarcoma, expression levels of miRNAs as
tumor suppressors, such as miR-34 and miR-142, were recently
demonstrated to be downregulated. It was suggested that miR-142 may
play an important role in maintaining the self-renewal capacity of
bronchioalveolar stem cells (7). It
was also found that esophageal squamous cell carcinoma patients
with high expression of miR-142 had poorer survival rates than
those with low expression of miR-142, suggesting that miR-142 may
act as a tumor suppressor (8).
miR-142 was found to be downregulated in osteosarcoma cell lines
(9), however, the role of miR-142
in osteosarcoma remains unknown.
Ras-related C3 botulinum toxin substrate 1 (Rac1) is
a member of the Ras homologue (Rho) family that plays a vital role
in multiple cell functions, including cell migration, invasion and
cell cycle arrest (10). Rac1 has
been found to be upregulated in many cancers, including glioma
(11), breast cancer (12) and skin tumors (13). Rac1 as a novel target of miR-142 has
been demonstrated in hepatocellular carcinoma cells (14). However, the underlying mechanism of
miR-142 and Rac1 in osteosarcoma remains to be elucidated.
In the present study, we found that the expression
of miR-142 was significantly reduced in OS tissues and OS cell
lines, while Rac1 expression was increased in OS tissues and OS
cell lines compared with expression levels in the controls. We then
demonstrated that miR-142 regulated Rac1 expression at the
transcriptional and translational levels by directly targeting its
3′-untranslated region (3′UTR). In addition, by loss- and
gain-of-function experiments, we investigated the role of miR-142
in OS cell lines, and found that miR-142 acted as a tumor
suppressor in the OS cell lines, inhibiting cell proliferation and
cell invasion and arresting the cell cycle in the S phase.
Furthermore, miR-142 inhibited osteosarcoma cell invasion by
inducing E-cadherin expression and reducing expression of matrix
metalloproteinase 2 (MMP2) and MMP9. Thus, overexpression of
miR-142 and/or knockdown of Rac1 may be a novel target for OS
therapy.
Materials and methods
Cell culture and cancer tissue
collection
hFOB1.19, Saos-2, MG63, U2OS and HOS cells were
obtained from the American Type Culture Collection (ATCC). All the
cells were cultivated in RPMI-1640 (Gibco) basic medium
supplemented with 10% FBS. All of the cells were cultured in
conditions of 95% air and 5% carbon dioxide (CO2) at
37°C. A total of 6 OS tissue and 3 adjacent tissue samples were
obtained from The Second Xiangya Hospital of Central South
University according to the legislation and the Ethics Board of The
Second Xiangya Hospital. Informed consent was acquired from all
subjects or from their caregivers. All samples were collected and
classified according to the World Health Organization (WHO)
criteria. All of the samples were stored at −80°C until used.
Cell treatment
To investigate the role of miR-142 in OS cells,
ectopic expression of miR-142 was achieved by transfecting
Lv-pre-miR-142 or Lv-anti-miR-142 using Lipofectamine®
2000 (Invitrogen, Carlsbad, CA, USA) according to the
manufacturer’s instructions. Rac1 siRNA oligonucleotides were
purchased from Thermo Fisher Scientific (Waltham, MA, USA). Rac1
siRNA was transfected into the Saos-2 and MG63 cells
(1×105 cells/ml) at concentrations of 50 nM for 48 h
using Lipofectamine® 2000 Transfection Reagent (Life
Technologies, Bedford, MA, USA). The group design consisted of a
blank control group (con), an empty vector group (vector) and
transfection groups (si-Rac1).
Quantitative polymerase chain reaction
(qPCR) analysis Real-time PCR for miRNA
Total RNA was extracted from the indicated cells
using TRIzol reagent (Invitrogen) following the manufacturer’s
instructions. The specific primers for miRNA-142 and U6 were
purchased from GeneCopoeia. The relative expression of miR-142 was
measured using the miScript SYBR®-Green PCR kit
(Qiagen). Expression of U6 was used as an endogenous control. Data
were processed using the 2−ΔΔCT method.
Real-time PCR for mRNA
Total RNA (0.5 μg) was reverse transcribed using the
RevertAid First Strand cDNA synthesis kit (Fermentas). The
following PCR amplification was carried out in a Thermal Cycler
Dice Real-Time System using SYBR-Green qRCR Mix (Toyobo). For each
sample, the relative mRNA level was normalized to β-actin. The
following primer pairs were used: Rac1 sense,
5′-GAGAAACTGAAGGAGAAGAAG-3′ and antisense,
5′-AAGGGACAGGACCAAGAACGA-3′; β-actin sense,
5′-AGGGGCCGGACTCGTCATACT3′ and antisense,
5′-GGCGGCACCACCATGTACCCT-3′.
Western blotting
Total protein was extracted from the indicated cells
using RIPA buffer (Auragene, Changsha, China). Protein
concentration was determined with the BCA protein assay kit
(Beyotime, Hangzhou, China). Fifty micrograms of proteins mixed in
loading buffer was separated by SDS-PAGE and transferred onto a
polyvinylidene fluoride (PVDF) membrane (Corning Inc., Corning, NY,
USA). After incubation with the appropriate primary antibody
overnight at 4°C, the membrane was washed and the antigen-antibody
complex was incubated using a horseradish peroxidase-conjugated
secondary antibody. The signal was visualized by the Chemi-Lumi One
system (Nacalai Tesque, Kyoto, Japan). Data were analyzed by
densitometry using Image-Pro Plus software 6.0 and normalized to
β-actin expression. Antibodies were obtained from the following
sources: mouse monoclonal anti-Rac1 antibody from Cell Signaling
Technology, rabbit polyclonal anti-E-cadherin, anti-MMP2 and
anti-MMP9 antibodies from Immunoway (Newark, DE, USA); mouse
monoclonal anti-β-actin antibody from Boster (Wuhan, China); and
horseradish peroxidase-conjugated secondary anti-rabbit/mouse IgG
from Cell Signaling Technology.
Dual luciferase reporter assay
A wild-type 3′UTR of Rac1 (wt-Rac1) or a mutant
3′UTR of Rac1 (mut-Rac1) was constructed into the dual luciferase
reporter vector. For luciferase assay, 105 cells were
seeded in 6-well plates for 12 h. Then, the cells were
co-transfected with wt-Rac1 or mut-Rac1 dual luciferase reporter
vector and miR-142 mimic (pre-miR-142), or miR-142 inhibitor
(anti-miR-142), respectively. Following a 5-h incubation with
transfection reagent, the medium was refreshed with fresh complete
medium. After transfection for 48 h, the luciferase activities in
each group were measured by using the dual luciferase reporter gene
assay kit and detected on an LD400 luminometer (both from Promega).
Renilla luciferase activity was normalized to firefly
luciferase activity.
CCK-8 cell proliferation assay
CCK-8 was used to evaluate cell proliferation. Cells
(5×103) were seeded in each 96-well plate for 24 h,
treated with the indicated drugs, and further incubated for 0, 24,
48 and 72 h, respectively. One hour before the end of the
incubation, 10 μl CCK-8 reagents was added to each well. Optical
density (OD) 570 nm value in each well was determined by an enzyme
immunoassay analyzer.
Transwell assay
The cells treated with the indicated drugs for 72 h
were starved in serum-free medium for 24 h and were then
resuspended in serum-free medium. The cells were added to the upper
chamber, while the lower chamber was filled with base medium
containing 10% FBS. After incubation for 24 h, the cells that
attached to the bottom were fixed and stained with crystal violet
for 20 min. The redundant crystal violet was washed by 0.1 M PBS,
and dried in air. The OD of the crystal violet dissolved by 10%
acetic acid at 570 nm was detected by an enzyme immunoassay
analyzer.
Flow cytometric analysis of the cell
cycle
Cells were treated with the indicated drugs for 72
h. After trypsinization and washing with ice-cold PBS, the cell
suspensions were stained using BD CycletestTM Plus (BD
Biosciences) according to the manufacturer’s instructions, and then
the cell cycle was analyzed by flow cytometry (Beckman Coulter,
Fullerton, CA, USA). The experiments were performed in
triplicate.
Statistical analysis
A t-test or one-way ANOVA were used to analyze the
statistical data by GraphPad Prism 5 software, depending on the
experimental conditions. All data are presented as mean ± SD.
Compared with the respective controls, P-values of <0.05 were
considered to indicate statistically significant differences.
Results
miR-142 is downregulated in OS tissues
and OS cell lines, while Rac1 is upregulated in OS tissues and OS
cell lines
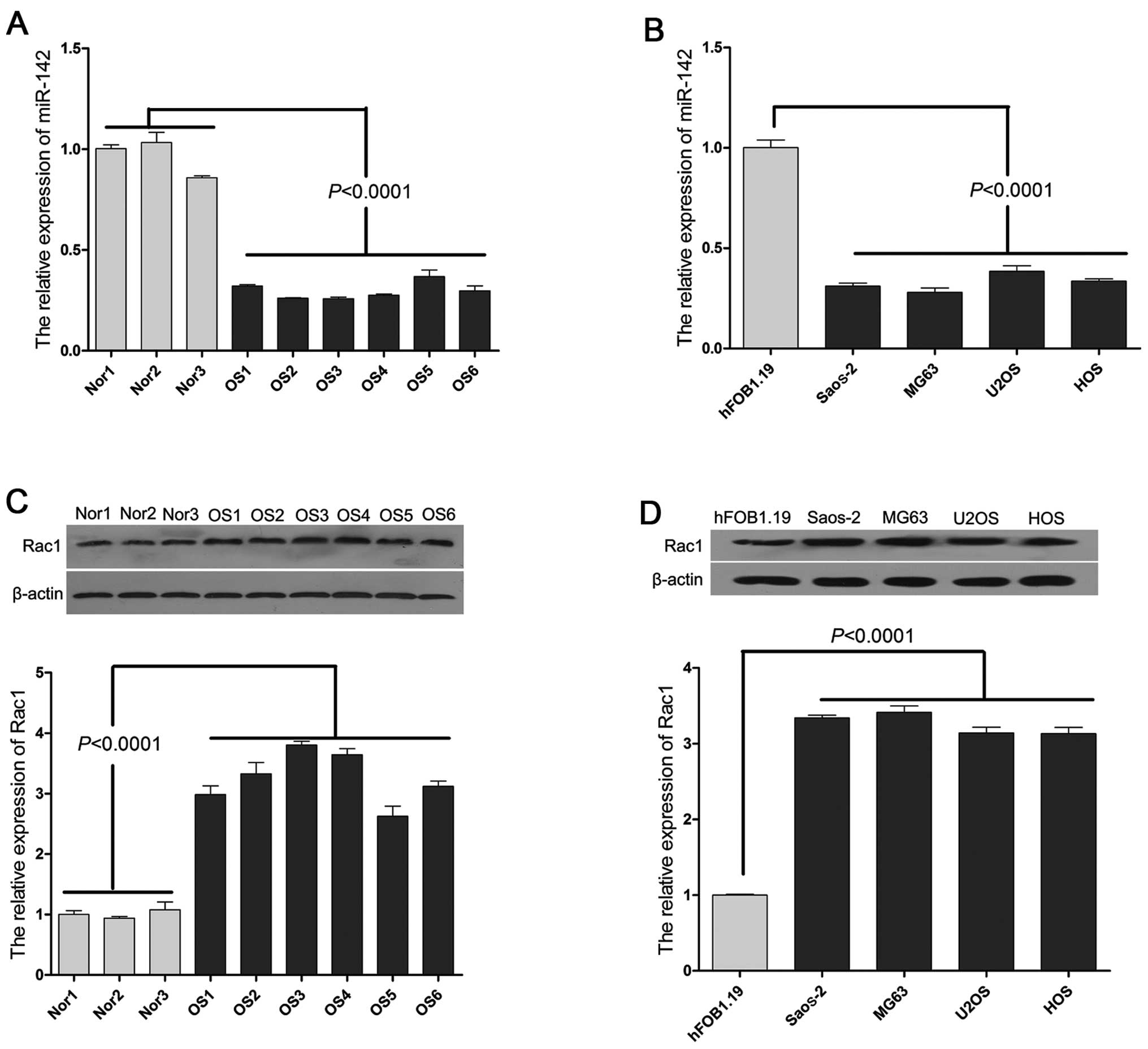
The average expression level of miR-142 was
significantly down-regulated (P<0.001) in the OS tissue samples
from the 6 OS patients compared with the 3 normal controls, as
indicated by qPCR (Fig. 1A).
Similar results were observed in the OS cell lines, particularly in
the Saos-2 and MG63 cells (Fig.
1B). In addition, we also detected the expression of Rac1 in
the OS tissue samples and OS cell lines. As shown in Fig. 1C and D, we found that Rac1 was
upregulated in the OS tissues and OS cell lines, which was
inversely correlated with the expression of miR-142.
miR-142 regulates Rac1 expression at the
transcriptional and translational levels by directly targeting its
3′UTR
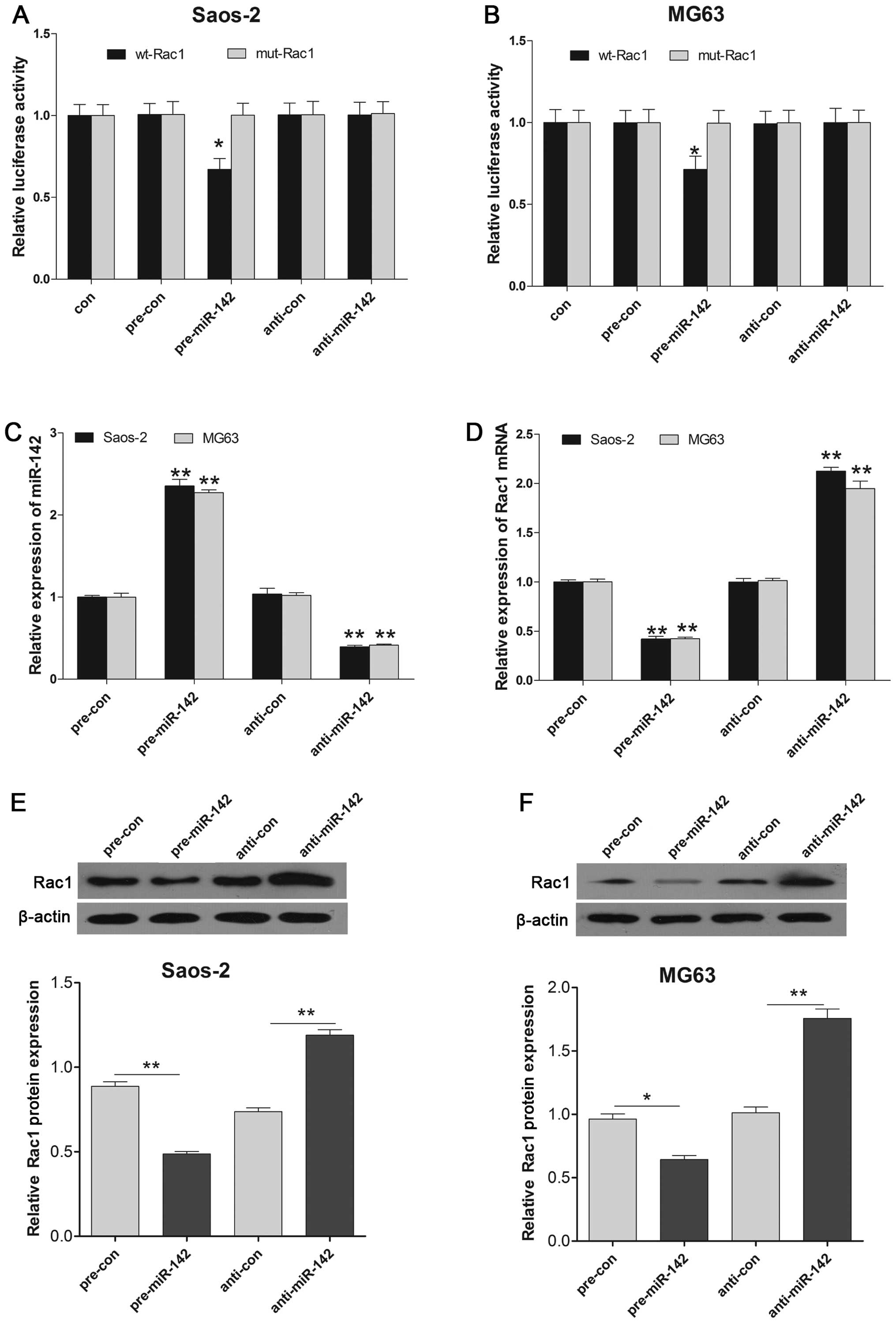
Considering the inverse correlation between miR-142
and Rac1, we then aimed to ascertain whether the 3′UTR of Rac1
contains a direct target site for miR-142. A dual luciferase
reporter assay was performed using a vector encoding the wild-type
(Wt) and mutant (Mut) 3′UTR of Rac1 mRNA. We found that the
luminescence activity was significantly suppressed in the miR-142
transfectants compared to the negative control transfectant.
Moreover, miR-142-mediated repression of luciferase activity was
abolished by the mutant type 3′UTR of Rac1 (Fig. 2A and B). Furthermore, Rac1 mRNA and
protein expression levels were markedly down-regulated by
pre-miR-142 transfection, compared with the control in the Saos-2
and MG63 cell lines (Fig. 2C–F).
These results determined that miR-142 directly targets Rac1 and
regulates its expression at the transcriptional and translational
levels.
Effects of miR-142 on cell proliferation
and the cell cycle in the OS cell lines
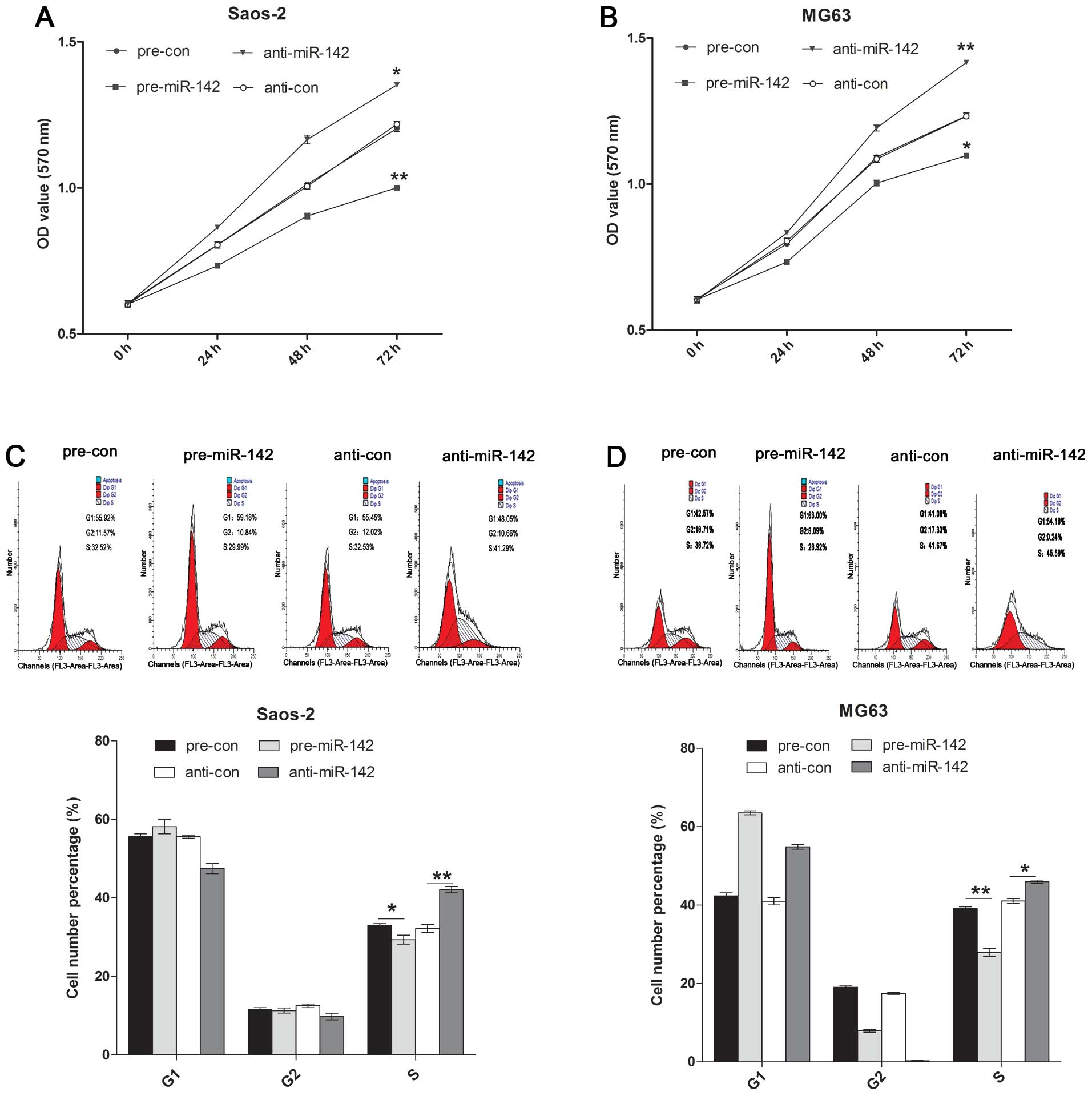
CCK-8 was used to detect the Saos-2 and MG63 cell
proliferation affected by miR-142. It was found that overexpression
of miR-142 induced inhibition of proliferation, while
downregulation of miR-142 promoted cell proliferation in the Saos-2
and MG63 cell lines (Fig. 3A and
B). Flow cytometric analysis was used to analyze cell cycle
alterations after pre-miR-142 or anti-miR-142 treatment.
Upregulation of miR-142 induced cell cycle arrest at the S phase
and decreased the percentage of cells in the S phase in the Saos-2
and MG63 cell lines. Treatment of Saos-2 and MG63 cells with
anti-miR-142 significantly increased the percentage of cells in the
S phase compared with the percentage in the control group (Fig. 3C and D).
Overexpression of miR-142 suppresses cell
invasion and regulates invasion-related genes
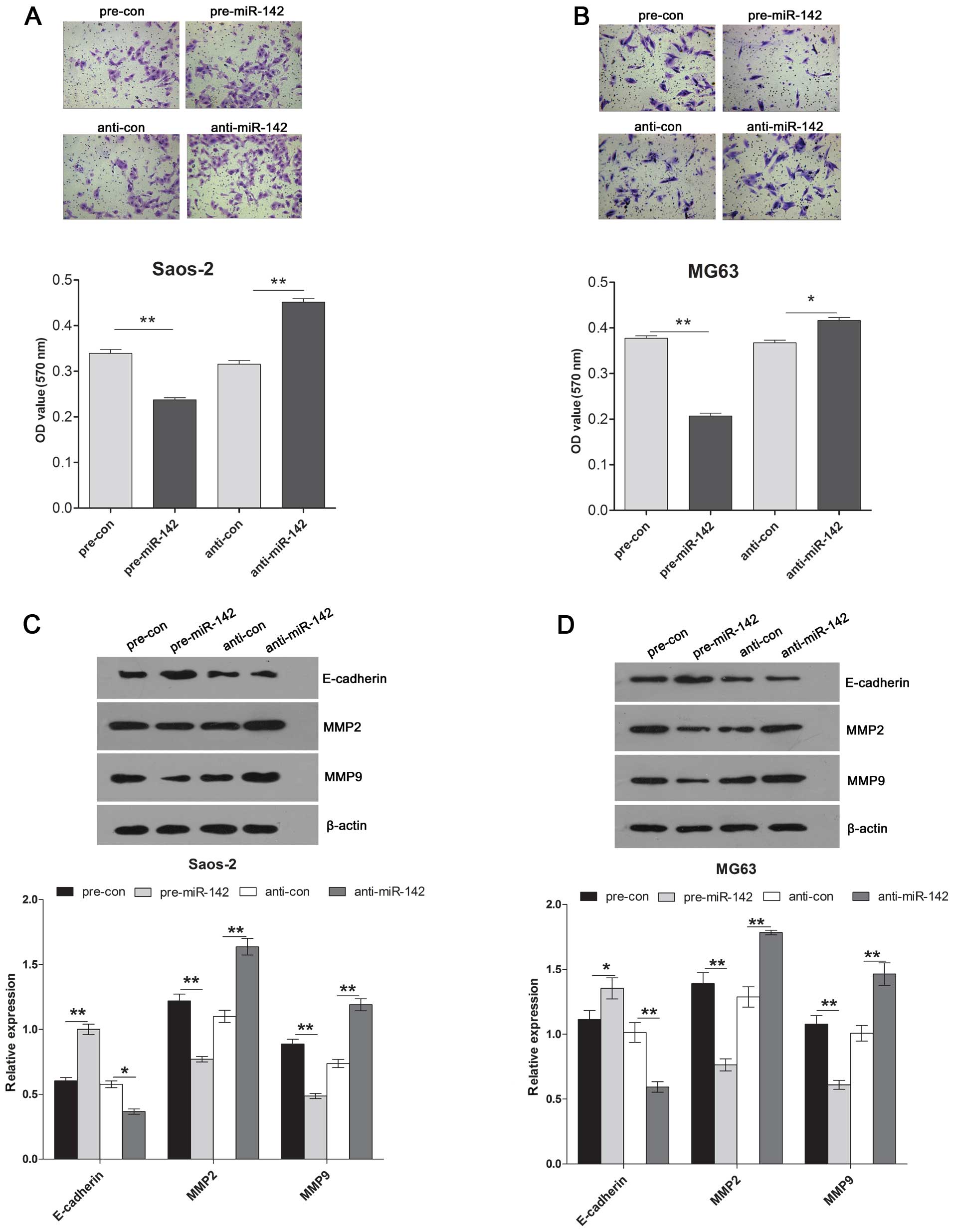
A Transwell assay was used to measure the invasive
ability of the Saos-2 and MG63 cells following pre-miR-142 or
anti-miR-142 transfection. The results showed that overexpression
of miR-142 significantly decreased the invasive ability, while
downregulation of miR-142 induced the invasive ability of the
Saos-2 and MG63 cells (Fig. 4A and
B). In addition, to explore the potential downstream molecular
pathway underlying miR-142 targeting to Rac1, we tested the
expression of invasion-related genes including E-cadherin, MMP2 and
MMP9 by western blotting in the Saos-2 and MG63 cells after
pre-miR-142 or anti-miR-142 transfection. We observed a significant
decrease in expression of MMP2 and MMP9 proteins and a significant
increase in expression of E-cadherin protein in the cells treated
with pre-miR-142. Inversely, downregulation of miR-142 induced the
expression of MMP2 and MMP9 proteins and significantly decreased
the expression of E-cadherin in the Saos-2 and MG63 cells compared
with that of control (Fig. 4C and
D).
Knockdown of Rac1 suppresses cell
proliferation and invasion, and induces cell cycle arrest at the S
phases
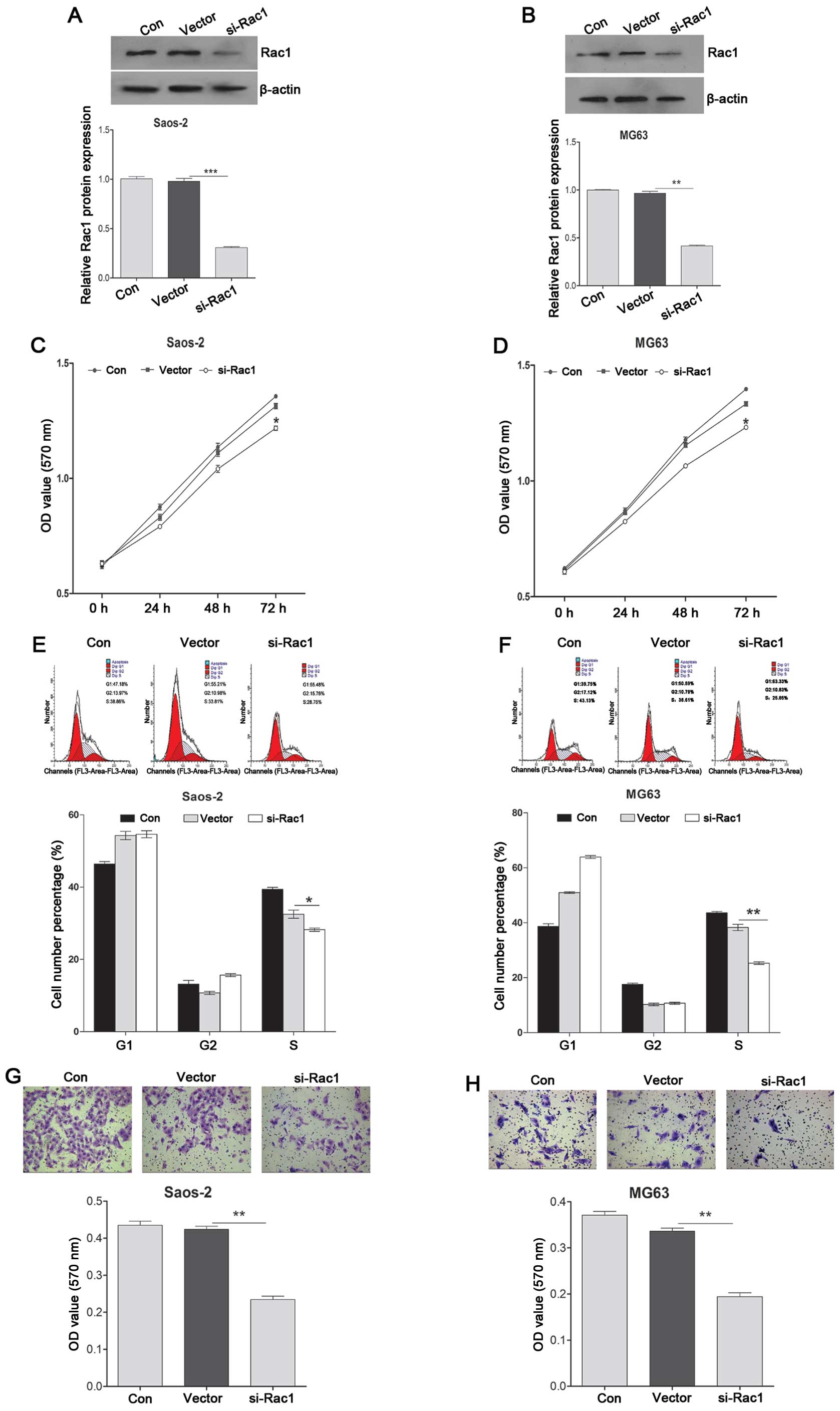
To investigate the role of Rac1 in OS cells, a
loss-of-function experiment was performed. As shown in Fig. 5A and B, siRac1 treatment
significantly decreased the expression of Rac1 in the Saos-2 and
MG63 cells, indicating that the efficiency was satisfied for
further analysis. CCK-8 was used to detect the Saos-2 and MG63 cell
proliferation affected by Rac1. It was found that knockdown of Rac1
promoted cell proliferation in the Saos-2 and MG63 cell lines
(Fig. 5C and D). Flow cytometric
analysis was used to analyze cell cycle alterations following
knockdown of Rac1. Knockdown of Rac1 induced cell cycle arrest at
the S phase and markedly decreased the percentage of cells in the S
phase in the Saos-2 and MG63 cell lines compared with the control
group (Fig. 5E and F). Transwell
assay was used to measure the invasive ability of the Saos-2 and
MG63 cells after knockdown of Rac1. The results showed that
knockdown of Rac1 significantly decreased the invasive ability
(Fig. 5G and H).
Discussion
miRNAs have been implicated in cancer growth and
metastasis (15). Moreover, miRNAs
function as tumor suppressors or oncogenes by targeting oncogenes
or suppressor genes (16). In this
study, we found that miR-142 expression was downregulated in the
osteosarcoma tissues and cell lines compared with expression in the
paired normal bone tissues and osteoblastic hFOB1.19 cell line,
whereas upregulated Rac1 expression was observed in the
osteosarcoma tissues and cell lines compared with expression in the
paired normal bone tissues and osteoblastic cell lines. In
addition, we identified Rac1 as a direct target of miR-142, which
regulates Rac1 expression at the transcriptional and translational
levels. Furthermore, we also found that overexpression of miR-142
suppressed osteosarcoma cell proliferation and invasion and
arrested the cell cycle in the S phase in the osteosarcoma Saos-2
and MG-63 cells. Moreover, our findings showed that the ability of
miR-142 to inhibit osteosarcoma cell invasion involved E-cadherin,
MMP2 and MMP9 expression. Similarly, knockdown of Rac1 by si-Rac1
transfection suppressed the ability of cell growth and invasion,
and induced cell cycle arrest in the S phase. Taken together, our
findings suggest that miR-142 plays a fundamental role in
tumorigenesis and cancer cell invasion by targeting Rac1.
miR-142 has been found to be downregulated in
several types of cancers, including hepatic cancer (17), squamous cell lung cancer (18) and human acute lymphoblastic leukemia
(19), and acts as a tumor
suppressor. It was reported that lower miR-142 expression in
hepatocellular carcinoma was significantly associated with poor
survival (17). Rac1 expression is
frequently upregulated in several types of cancers, including
hepatic cancer (14) and breast
cancer (20). In line with previous
studies, in the present study, we also found that miR-142 was
downregulated, while Rac1 was upregulated in 6 osteosarcoma tissue
samples compared with the paired normal bone tissues. We also found
similar results in the osteosarcoma Saos-2 and MG63 cells.
To further verify the role of miR-142 in the
development of osteosarcoma, loss- and gain-of-function experiments
were performed. Upregulation of miR-142 significantly inhibited
cell proliferation and invasion and arrested the cell cycle in the
S phase in the osteosarcoma cell lines, indicating that
upregulation of miR-142 may inhibit tumor progression in
osteosarcoma carcinogenesis. Meanwhile, downregulation of miR-142
significantly induced cell proliferation and invasion and increased
the percentage of cells in the S phase in the osteosarcoma cell
lines, indicating that downregulation of miR-142 may promote tumor
progression in osteosarcoma carcinogenesis. These results suggest
that miR-142 acts as a tumor-suppressor whose downregulation may
contribute to the progression and metastasis of osteosarcoma.
Rac1 as a target of miR-142 has been demonstrated in
diffuse large B-cell lymphoma cells (21) and hepatocellular carcinoma cell
lines (14). Consistent with
previous studies, we also identified Rac1 as a direct target of
miR-142 in the osteosarcoma Saos-2 and MG63 cells. miR-142
overexpression suppressed Rac1 3′UTR luciferase report activity,
which was abolished by mutation of the Rac1 3′UTR binding site.
Overexpression of miR-142 induced a significant decrease in Rac1 at
both the mRNA and protein levels. These results indicate that
miR-142 may function as a tumor suppressor, at least partly,
mediated by suppressing Rac1 expression in osteosarcoma cells.
Rac1 activation is correlated with metastatic
progression in many types of cancers such as medulloblastoma
(22) and breast cancer (20). Increasing evidence reveals that
Rac1-dependent cell signaling activation can promote cell adhesion,
invasion and metastasis in a variety of cancers, including
osteosarcoma (23), suggesting that
Rac1 plays an important role in cancer invasion (24). In the present study, we confirmed
that Rac1 is upregulated in osteosarcoma. Furthermore, our data
showed that knockdown of Rac1 by si-Rac1 suppressed cell
proliferation and invasion, and induced cell cycle arrest at the S
phase. In addition, we found that the ability of miR-142 to act as
a tumor suppressor not only suppressed Rac1 expression, but also
induced E-cadherin expression and decreased MMP2 and MMP9
expression. E-cadherin is a member of the type I classical cadherin
family. Studies, including those in osteosarcoma, have demonstrated
that E-cadherin plays a tumor-suppressor role (25). E-cadherin is frequently
downregulated during carcinoma metastasis (26). Loss of E-cadherin facilitates the
initial invasive behavior of cancer (27). MMPs promote cell invasion and
migration in several types of cancers, such as osteosarcoma,
prostate, lung, colon and pancreas cancer (28). Compared with normal tissues, a
higher expression of MMPs is often observed in malignant tumor
tissues (29). Taken together, our
study demonstrated that the ability of miR-142 to inhibit
osteosarcoma cell invasion was achieved by inducing E-cadherin
expression and reducing expression of MMP2 and MMP9.
In conclusion, the present study provides novel
evidence that miR-142 functions as a tumor-suppressor miRNA in
osteosarcoma via targeting the 3′UTR of Rac1. Furthermore, miR-142
inhibits osteosarcoma cell invasion by inducing E-cadherin
expression and reducing expression of MMP2 and MMP9. Our findings
revealed that upregulation of miR-142 could be a potential target
for the treatment of osteosarcoma in the future.
References
|
1
|
Kobayashi E, Hornicek FJ and Duan Z:
MicroRNA involvement in osteosarcoma. Sarcoma. 2012:3597392012.
View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Bennani-Baiti IM: Epigenetic and
epigenomic mechanisms shape sarcoma and other mesenchymal tumor
pathogenesis. Epigenomics. 3:715–732. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Geller DS and Gorlick R: Osteosarcoma: a
review of diagnosis, management, and treatment strategies. Clin Adv
Hematol Oncol. 8:705–718. 2010.
|
|
4
|
Maire G, Martin JW, Yoshimoto M,
Chilton-MacNeill S, Zielenska M and Squire JA: Analysis of
miRNA-gene expression-genomic profiles reveals complex mechanisms
of microRNA deregulation in osteosarcoma. Cancer Genet.
204:138–146. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Lewis BP, Burge CB and Bartel DP:
Conserved seed pairing, often flanked by adenosines, indicates that
thousands of human genes are microRNA targets. Cell. 120:15–20.
2005. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Croce CM: Causes and consequences of
microRNA dysregulation in cancer. Nat Rev Genet. 10:704–714. 2009.
View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Qian S, Ding JY, Xie R, et al: MicroRNA
expression profile of bronchioalveolar stem cells from mouse lung.
Biochem Biophys Res Commun. 377:668–673. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Lin RJ, Xiao DW, Liao LD, et al:
MiR-142–3p as a potential prognostic biomarker for esophageal
squamous cell carcinoma. J Surg Oncol. 105:175–182. 2012.
View Article : Google Scholar
|
|
9
|
Namløs HM, Meza-Zepeda LA, Barøy T, et al:
Modulation of the osteosarcoma expression phenotype by microRNAs.
PLoS One. 7:e480862012. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Bid HK, Roberts RD, Manchanda PK and
Houghton PJ: RAC1: an emerging therapeutic option for targeting
cancer angiogenesis and metastasis. Mol Cancer Ther. 12:1925–1934.
2013. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Yukinaga H, Shionyu C, Hirata E, et al:
Fluctuation of Rac1 activity is associated with the phenotypic and
transcriptional heterogeneity of glioma cells. J Cell Sci.
127:1805–1815. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Zhang H, An F, Tang L and Qiu R: Multiple
effects of a novel epothilone analog on cellular processes and
signaling pathways regulated by Rac1 GTPase in the human breast
cancer cells. Korean J Physiol Pharmacol. 18:109–120. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Chen R, Fu M, Zhang G, et al: Rac1
regulates skin tumors by regulation of keratin 17 through
recruitment and interaction with CD11b+Gr1+ cells. Oncotarget.
5:4406–4417. 2014.PubMed/NCBI
|
|
14
|
Wu L, Cai C, Wang X, Liu M, Li X and Tang
H: MicroRNA-142-3p, a new regulator of RAC1, suppresses the
migration and invasion of hepatocellular carcinoma cells. FEBS
Lett. 585:1322–1330. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Nugent M: MicroRNA function and
dysregulation in bone tumors: the evidence to date. Cancer Manag
Res. 6:15–25. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Miao J, Wu S, Peng Z, Tania M and Zhang C:
MicroRNAs in osteosarcoma: diagnostic and therapeutic aspects.
Tumour Biol. 34:2093–2098. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Chai S, Tong M, Ng KY, et al: Regulatory
role of miR-142-3p on the functional hepatic cancer stem cell
marker CD133. Oncotarget. 5:5725–5735. 2014.PubMed/NCBI
|
|
18
|
Su YH, Zhou Z, Yang KP, Wang XG, Zhu Y and
Fa XE: MIR-142-5p and miR-9 may be involved in squamous lung cancer
by regulating cell cycle related genes. Eur Rev Med Pharmacol Sci.
17:3213–3220. 2013.PubMed/NCBI
|
|
19
|
Dou L, Li J, Zheng D, et al:
MicroRNA-142-3p inhibits cell proliferation in human acute
lymphoblastic leukemia by targeting the MLL-AF4 oncogene. Mol Biol
Rep. 40:6811–6819. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Lin CW, Sun MS, Liao MY, et al:
Podocalyxin-like 1 promotes invadopodia formation and metastasis
through activation of Rac1/Cdc42/cortactin signaling in breast
cancer cells. Carcinogenesis. 35:2425–2435. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Kwanhian W, Lenze D, Alles J, et al:
MicroRNA-142 is mutated in ~20% of diffuse large B-cell lymphoma.
Cancer Med. 1:141–155. 2012. View
Article : Google Scholar
|
|
22
|
Chen HH, Yu HI, Cho WC and Tarn WY: DDX3
modulates cell adhesion and motility and cancer cell metastasis via
Rac1-mediated signaling pathway. Oncogene. Jul 21–2014.(Epub ahead
of print). View Article : Google Scholar
|
|
23
|
Geng S, Zhang X, Chen J, et al: The tumor
suppressor role of miR-124 in osteosarcoma. PLoS One. 9:e915662014.
View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Lane J, Martin T, Weeks HP and Jiang WG:
Structure and role of WASP and WAVE in Rho GTPase signalling in
cancer. Cancer Genomics Proteomics. 11:155–165. 2014.PubMed/NCBI
|
|
25
|
Yang H, Zhang Y, Zhou Z, Jiang X and Shen
A: Transcription factor Snai1-1 induces osteosarcoma invasion and
metastasis by inhibiting E-cadherin expression. Oncol Lett.
8:193–197. 2014.PubMed/NCBI
|
|
26
|
Dong S, Zhao J, Wei J, et al: F-box
protein complex FBXL19 regulates TGFβ1-induced E-cadherin
downregulation by mediating Rac3 ubiquitination and degradation.
Mol Cancer. 13:762014. View Article : Google Scholar
|
|
27
|
Lee SJ, Jung YH, Oh SY, Yong MS, Ryu JM
and Han HJ: Netrin-1 induces MMP-12-dependent E-cadherin
degradation via the distinct activation of PKCα and FAK/Fyn in
promoting mesenchymal stem cell motility. Stem Cells Dev.
23:1870–1882. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Wen X, Liu H, Yu K and Liu Y: Matrix
metalloproteinase 2 expression and survival of patients with
osteosarcoma: a meta-analysis. Tumour Biol. 35:845–848. 2014.
View Article : Google Scholar
|
|
29
|
Gyurkó DM, Veres DV, Módos D, Lenti K,
Korcsmáros T and Csermely P: Adaptation and learning of molecular
networks as a description of cancer development at the
systems-level: potential use in anticancer therapies. Semin Cancer
Biol. 23:262–269. 2013. View Article : Google Scholar
|