Introduction
Prostate cancer is the most frequently diagnosed
cancer and the second leading cause of cancer-related death among
American men. In 2012, an estimated 241,740 men were diagnosed with
prostate cancer, and 28,170 died of the disease (1). Radical prostatectomy, radiation
therapy or hormonal therapy is often effective for newly diagnosed
prostate cancer patients. However, a number of patients will
progress after two to three years, and the growth of cancer will
resume despite hormone therapy (2,3).
Unfortunately, the clinical treatment of advanced prostate cancer
still remains a challenge.
Gemcitabine is a nucleoside analog that inhibits DNA
biosynthesis, and is active against a broad spectrum of solid
tumors (4), such as non-small cell
lung cancer, pancreatic, bladder and breast cancer. Although
gemcitabine significantly inhibits the growth of prostate cancer
cell lines (5,6), clinical data suggest only modest
activity when gemcitabine is used as a single agent for metastatic
androgen-independent prostate cancer (AIPC), and the efficacy of
gemcitabine for AIPC varies widely between patients (7). Therefore, it is imperative to
investigate the mechanism of gemcitabine inhibition of prostate
cancer, and define the gemcitabine-sensitive patient subgroups.
HMGN5, also known as nucleosome binding protein 1
(NSBP1), is a new member of the high mobility group N (HMGN)
protein family (8). HMGN5 is
localized to the nucleus by a nucleosome-binding domain, and binds
to histone proteins by its negatively charged C-terminus, which
unfolds chromatin and counteracts linker histone-mediated chromatin
compaction and affects transcription and DNA repair (9–11).
HMGN5 plays an important role during development and tumorigenesis
(8,12,13),
and HMGN5 expression is upregulated in prostate (14), bladder (15), clear cell renal cell carcinoma
(16), breast (17), cutaneous squamous (18) and ovarian cancer (19), suggesting an association between
high HMGN5 expression and tumorigenesis. In our previous studies,
we found that HMGN5 was overexpressed in prostate cancer tissues
and prostate cancer cell lines, and downregulation of HMGN5 with
shRNA caused cell cycle arrest, growth inhibition and apoptosis
in vitro and in vivo (20,21).
We also found that gemcitabine downregulated the expression of
HMGN5 (22). This finding motivated
us to investigate whether gemcitabine exerts antitumor effects
through inhibition of the HMGN5 pathway in prostate cancer.
In the present study, we demonstrated that HMGN5
promotes prostate cancer development through activation of the MAPK
signaling pathway, and we also found that gemcitabine suppressed
growth of prostate cancer cells by inhibiting HMGN5, and that
prostate cancer cells expressing a high level of HMGN5 are more
sensitive to gemcitabine. These findings indicate that HMGN5 is a
potential treatment marker for prostate cancer, and that patients
expressing a high level of HMGN5 will benefit from gemcitabine
treatment.
Materials and methods
Cell culture
The human prostate cancer cell lines, LNCaP, DU145,
22RV1 and PC-3, were purchased from the American Type Culture
Collection (ATCC; Manassas, VA, USA). The PC-3M cell line was
purchased from the National Platform of Experimental Cell Resources
for Sci-Tech (Beijing, China). Cells were cultured in RPMI-1640
(HyClone, Logan, UT, USA) containing 10% fetal bovine serum (FBS;
Gibco, Carlsbad, CA, USA) with 1% antibiotics. The human prostate
epithelial cell line RWPE-1 was purchased from ATCC, and was
maintained in keratinocyte serum-free medium supplemented with 0.05
mg/ml bovine pituitary extract and 5 ng/ml epidermal growth factor
(Invitrogen, Carlsbad, CA, USA). All cells were cultured at 37°C in
a humidified incubator with 5% CO2 and 95%
O2.
shRNA lentiviral vector and
HMGN5-expressing lentiviral vector infection
HMGN5 shRNA sequences and construction of the
lentivirus were identical to those used in a previous study
(23). PC-3 cells were seeded into
6-well plates and grown to 70% confluency. On the day of infection,
the purified lentivirus (shCtrl or shHMGN5) was added to the cells
at a multiplicity of infection (MOI) of 20 for 6 h, and cells were
washed twice with medium. Successful knockdown of HMGN5 was
analyzed by western blotting and real-time quantitative PCR (qPCR)
after 72 h.
Human HMGN5 cDNA was obtained from the German
Science Centre for Genome Research (RZPD), and the fragments
containing the HMGN5 coding sequence were subcloned between the
AgeI and NheI sites of the GV205 lentiviral vector
(synthesized by GeneChem Corporation, Shanghai, China). The
lentiviral construct was verified by standard DNA sequencing.
qPCR
Total RNA from the cultured cells was isolated using
TRIzol reagent (Invitrogen) following the manufacturer’s protocol.
We reverse-transcribed total RNA (3 μg) using the reverse
transcription system (Promega, Madison, WI, USA). qPCR was
performed using SYBR-Green PCR Mix (Roche, Indianapolis, IN, USA)
in an Applied Biosystems 7300 Fast Real-Time PCR system. The primer
sequences for real-time qPCR were as follows: HMGN5,
GCAGTCAGGCAGTGACTGCCTTCG (forward) and CCCTTTTCTGTGGCATCTTC
(reverse); GAPDH, CAGTCAGCCGCATCTTCTTTT (forward) and
GTGACCAGGCGCCCAATAC (reverse). Gene expression analysis was
performed using the comparative ΔΔCt method; expression was
normalized to GAPDH.
Western blot analysis
Protein lysate was prepared by homogenization in
RIPA lysis buffer containing phosphatase and protease inhibitors.
Western blot assay was performed according to previously described
protocols (24). Anti-HMGN5 (Sigma,
St. Louis, MO, USA), -ERK1/2, -pERK1/2, -GAPDH and -PARP (Cell
Signaling Technology, Inc., Beverly, MA, USA) were used as primary
antibodies, and either goat anti-mouse IgG or goat anti-rabbit IgG
(Sigma) was used as secondary antibody. The membrane was visualized
using the ECL detection system (GE Healthcare Biosciences,
Piscataway, NJ, USA).
Cell proliferation assay
Cell proliferation was assessed using the
CellTiter-Blue reagent (Promega) according to the manufacturer’s
instructions. Cells (2,000–5,000/well) were seeded into a 96-well
plate and cultured for 12 h before new medium containing different
doses of gemcitabine was added; cells were cultured for an
additional 48 h, then 20 μl of CellTiter-Blue reagent was added to
each well. Plates were incubated for 4 h at 37°C, then fluorescence
was recorded at an excitation wavelength of 560 nm and an emission
wavelength range of 590 nm using a Labsystems Fluoroskan Ascent
plate reader (Thermo Fisher Scientific, Boston, MA, USA).
Cell invasion assay
The cell invasion assay was performed with 8-μm cell
culture inserts (Millipore Corporation, Billerica, MA, USA) coated
with 60 μl ECM gel (BD Biosciences, Bedford, MA, USA) mixed with
RPMI-1640 serum-free medium in 1:4 dilution for 5 h at 37°C.
Infected PC-3 cells (5×104) were resuspended in 100 μl
serum-free RPMI-1640 and placed into Matrigel, and the lower
chamber of the Transwell was filled with 600 μl of RPMI-1640
containing 10% FBS before incubating cells at 37°C for 24 h. The
Transwells were removed from the 24-well plates and fixed with 100%
methanol for 30 min, and then stained with 0.1% crystal violet for
30 min. Non-invaded cells on the top of the Transwell were removed
with a cotton swab. Five visual fields were chosen randomly under a
light microscope at ×200 magnification, and invaded cells were
counted. The data are expressed as means ± SD (standard
deviation).
Colony formation assay
After infection for 24 h, cells (1,000/well) were
seeded into a 6-well plate, and culture medium was changed every 3
days for 2 weeks. Colonies (50 or more cells/colony) were counted
after staining with gentian violet.
Apoptosis assay
The apoptosis assay was performed using the
Apo-ONE® Homogeneous Caspase-3/7 kit (Promega) according
to the manufacturer’s instructions. Following infection for 24 h,
the PC-3 cells (5,000/well) were seeded into a 96-well plate and
cultured for 48 h, and then 100 μl of Apo-ONE Caspase-3/7 reagent
was added to each well, and the plate was incubated at room
temperature for 2 h before measuring the fluorescence of each well
at an excitation wavelength of 480 nm and an emission wavelength
range of 530 nm.
Statistical analysis
All data groups were analyzed by one-way ANOVA using
SPSS 13.0 (SPSS, Inc., Chicago, IL, USA), and the graphs were
generated in MS Excel. Data are expressed as means ± SD. A
probability (P) value of <0.05 was assigned to indicate a
statistically significant difference when compared with the control
and is indicated by an asterisk in the figures.
Results
HMGN5 expression is high in prostate
cancer cells
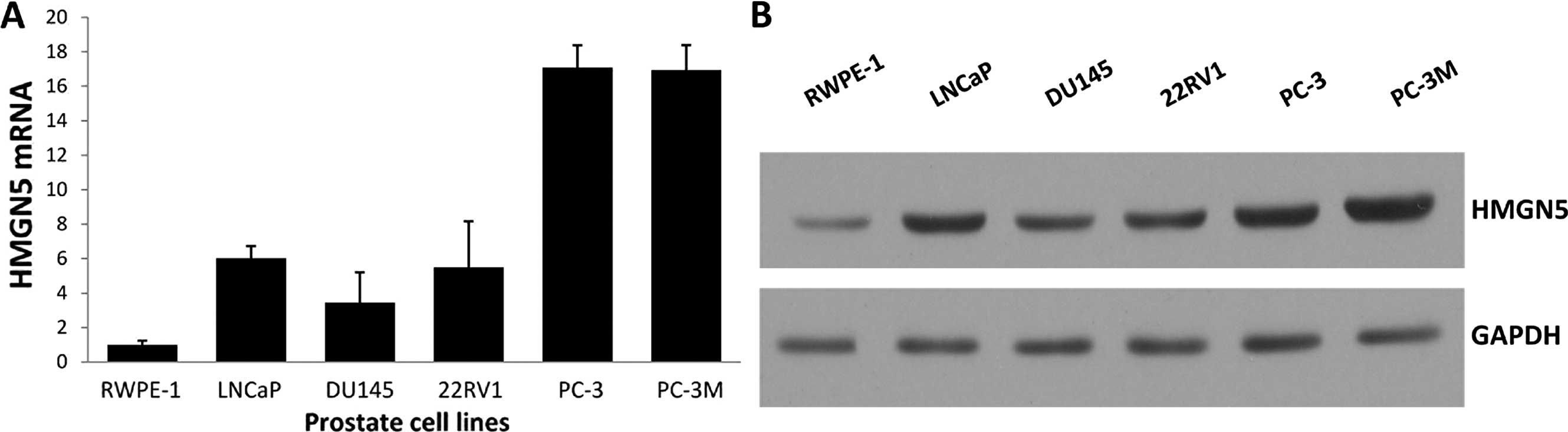
HMGN5 expression was determined using real-time qPCR
and western blot analysis in the prostate cancer cell lines,
including LNCaP, DU145, 22RV1, PC-3 and PC-3M, as well as human
prostate epithelial cell line RWPE-1. Both the mRNA and protein
levels of HMGN5 were higher in the prostate cancer cell lines than
the levels in the control human prostate epithelial cell line, and
the HMGN5 protein level was correlated with its mRNA level
(Fig. 1A and B).
HMGN5 knockdown decreases prostate cancer
cell growth and invasion, and promotes prostate cancer cell
apoptosis
To explore the function of HMGN5 in prostate cancer,
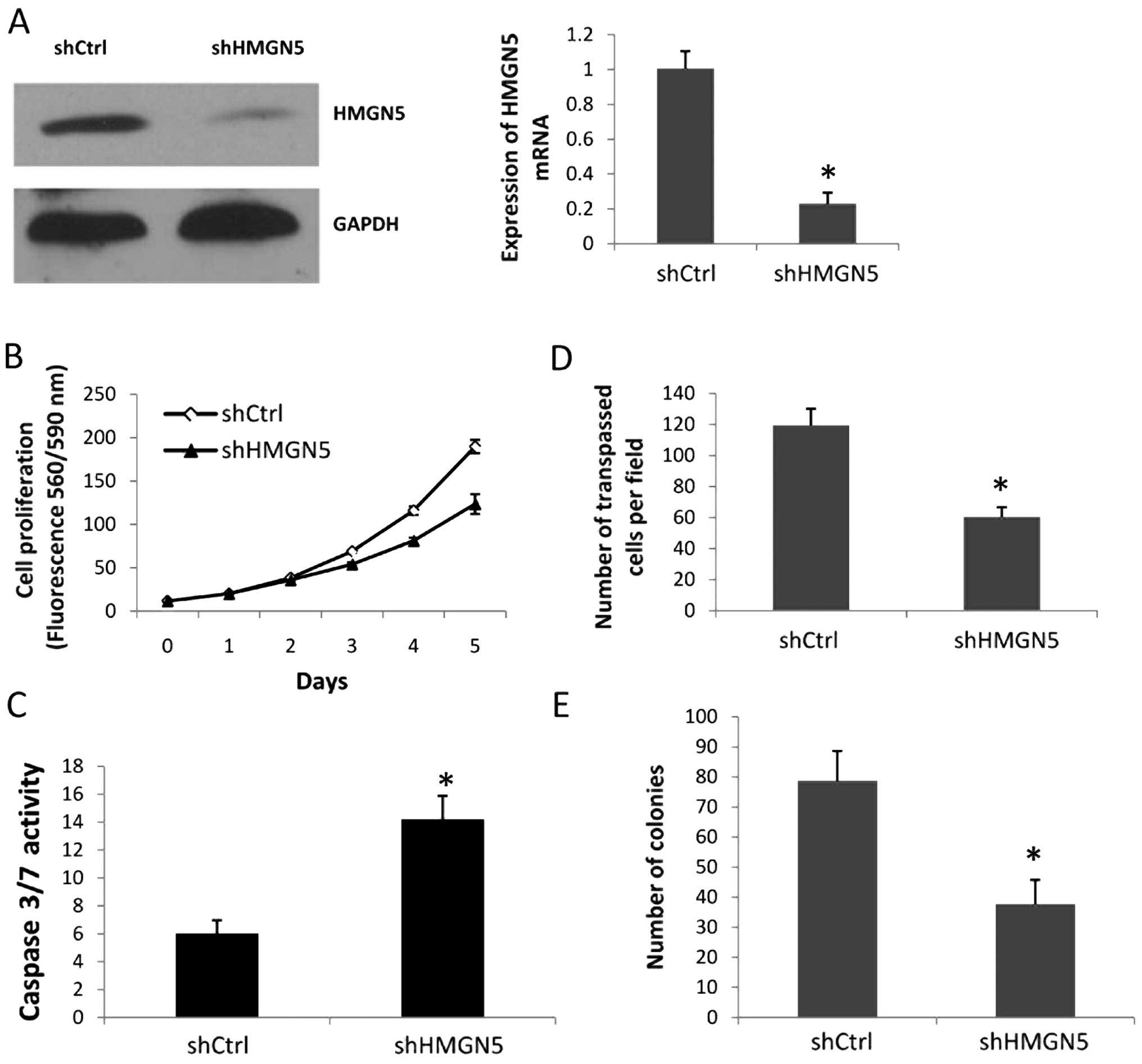
we used a loss-of-function approach. PC-3 cells were infected with
the shHMGN5 or shCtrl lentiviral vectors for 72 h; the shHMGN5
reduced HMGN5 expression at both the mRNA and protein levels
(Fig. 2A). Cell proliferation assay
results showed that the PC-3 cells with shHMGN5 exhibited a
consistent decrease over the time course examined, with a decrease
in proliferation of 30.03% at day 4 and 34.91% at day 5, compared
with the PC-3 cells with shCtrl (Fig.
2B). To determine whether the downregulation of HMGN5 promoted
PC-3 cell apoptosis, we used a homogeneous caspase-3/7 analysis, in
which caspase-3/7 activity denoted the apoptosis level. The results
showed that shHMGN5 induced the activity of caspase-3/7 compared
with shCtrl in the PC-3 cells (Fig.
2C). Next, we assessed the role of HMGN5 in PC-3 cell invasion.
The cell invasion assay indicated that there were fewer PC-3 cells
in the shHMGN5-infected group when compared with the shCtrl group;
the number of cells crossing the Matrigel was 60.3±6.4 in the
shHMGN5 group vs. 119.4±10.8 in the shCtrl group (Fig. 2D). To further assess the tumor
promoter functions of HMGN5 in prostate cancer cells, infected PC-3
cells were assessed with anchorage-dependent colony formation
assays. After 2 weeks of culture, the number of colonies in the
PC-3 cells infected with shHMGN5 was significantly less than that
in the PC-3 with the shCtrl (Fig.
2E). These data provide evidence that HMGN5 promotes cell
proliferation, invasion and clonogenicity, and antagonizes cell
apoptosis.
HMGN5 overexpression induces colony
formation and invasion
To further investigate the function of HMGN5 in
prostate cancer tumorigenesis, we constructed an HMGN5-expressing
lentiviral vector, and the human prostate epithelial cell line
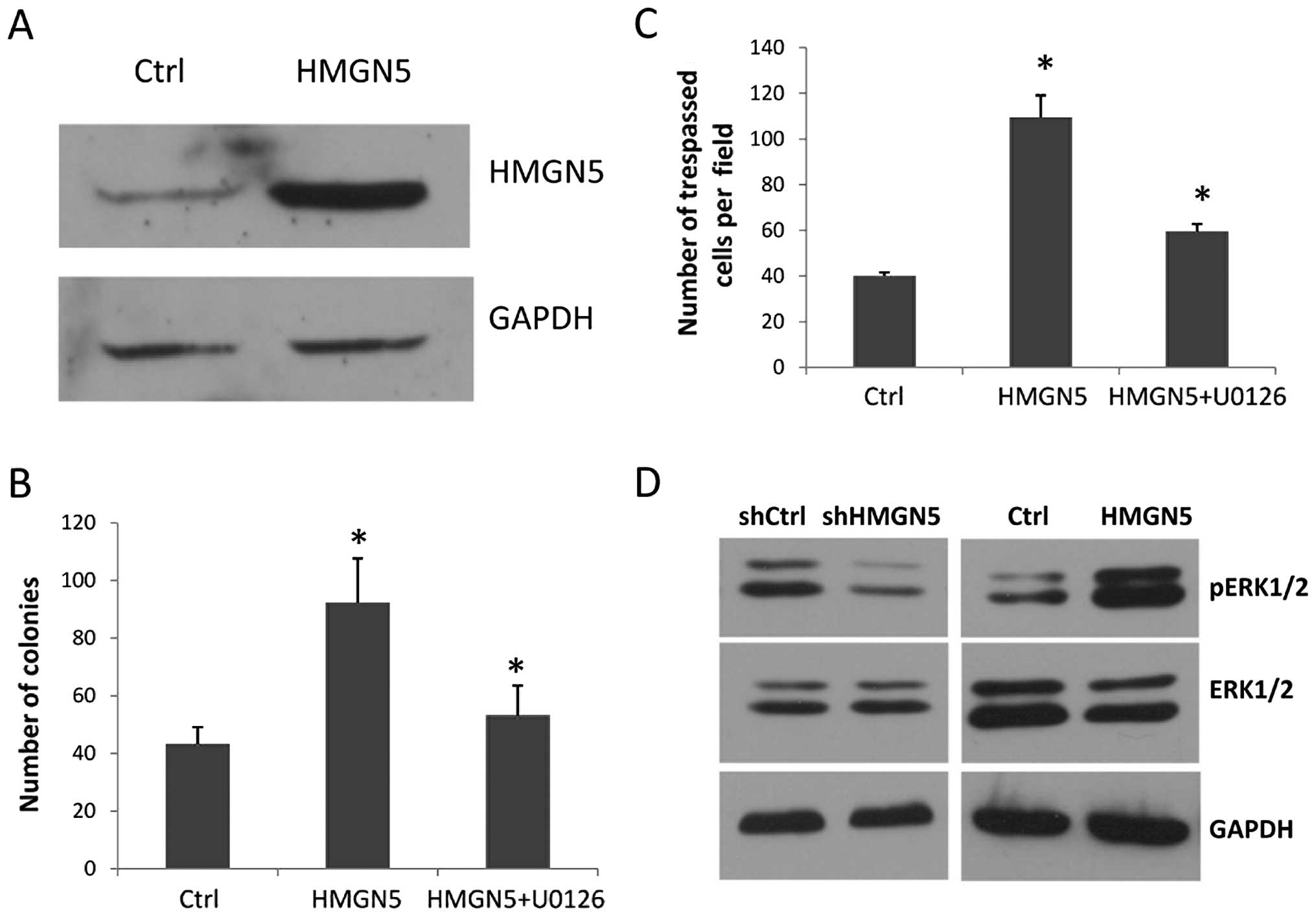
RWPE-1 was engineered to stably express HMGN5. Western blot
analysis showed that the HMGN5-transfected clones (RWPE-1-HMGN5)
had a high level of HMGN5 protein compared with the GV205 (empty
vector)-transfected controls (RWPE-1-Ctrl) (Fig. 3A). Then, the colony formation assay
and cell invasion assay were performed, which mimic crucial events
in tumorigenesis and cancer progression. We found that the
RWPE-1-HMGN5 cells formed more, larger colonies compared with the
RWPE-1-Ctrl cells in monolayer culture for 14 days (Fig. 3B). Furthermore, ectopic HMGN5
expression significantly increased the migration of the RWPE-1
cells in the cell invasion assay (Fig.
3C).
HMGN5 activates the MAPK signaling
pathway
Abnormal activation of the MAPK signaling pathway
plays an important role in prostate cancer tumorigenesis and
progress; thus, we studied whether HMGN5 contributes to prostate
cancer development by activating the MAPK signaling pathway.
Western blot results showed that the level of pERK1/2 protein,
which indicates the activity of the MAPK signaling pathway, was
significantly increased in the RWPE-1 cells ectopically expressing
HMGN5, and was strongly decreased in the PC-3 cells with HMGN5
knockdown when compared with the control (Fig. 3D). However, for ERK1/2, there was no
difference in expression between the RWPE-1-HMGN5 and RWPE-1-Ctrl
cells or the PC-3 with shHMGN5 and PC-3 with shCtrl cells (Fig. 3D). We observed the reverse effect
following treatment with the ERK inhibitor U0126. As shown in
Fig. 3B and C, the invasion and
clonogenicity mediated by ectopic HMGN5 expression in the RWPE-1
cells was attenuated by U0126. These results demonstrated that
HMGN5 contributes to prostate cancer tumorigenesis and progression
by activating the MAPK signaling pathway.
HMGN5 expression determines the response
of prostate cancer cells to gemcitabine
Our previous study showed that gemcitabine
downregulates HMGN5 mRNA levels, thus this next study focused on
the relationship between the HMGN5 expression level and the
response to gemcitabine in prostate cancer. First, we assessed the
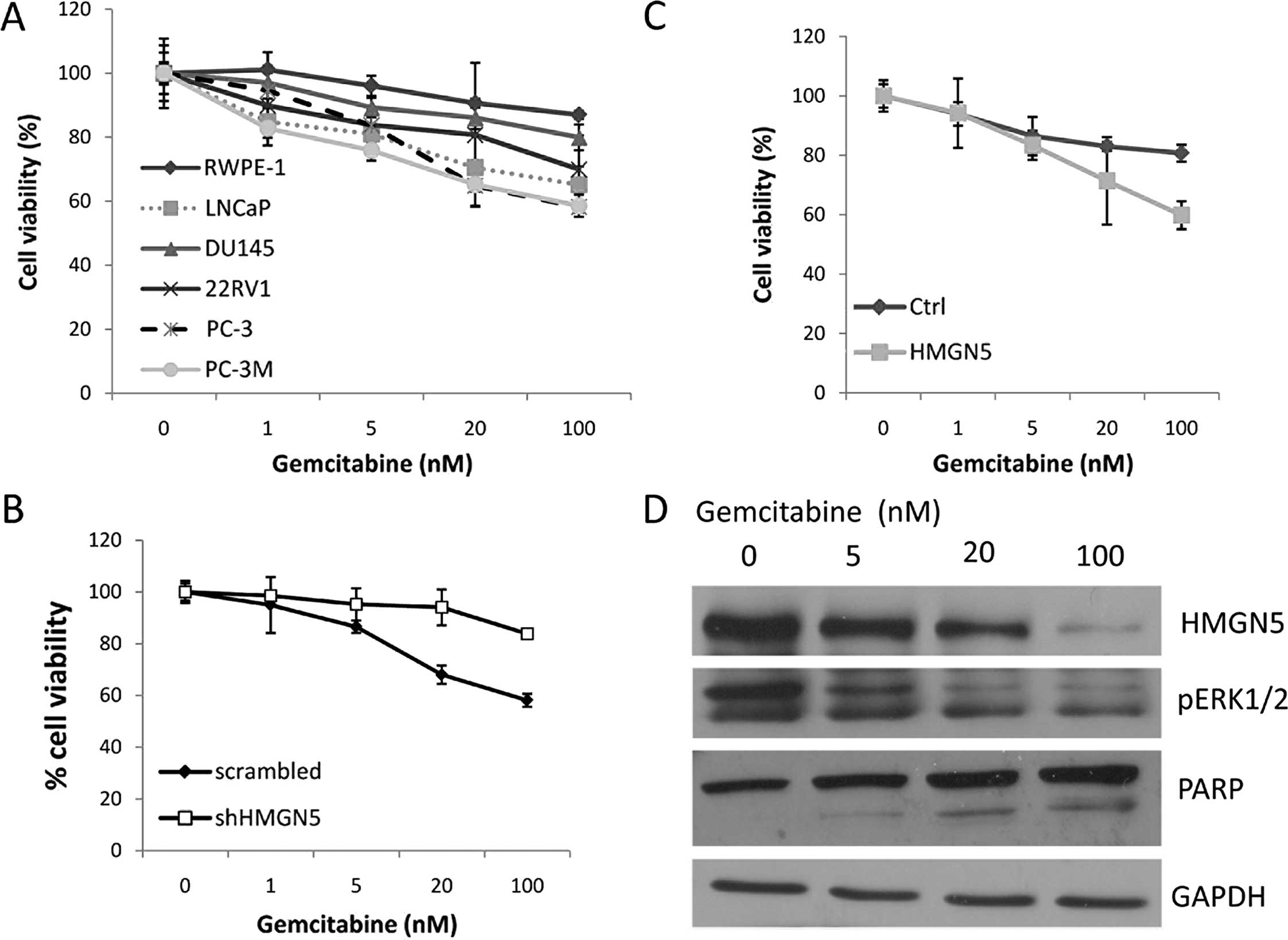
effect of gemcitabine on prostate cell lines with various levels of
HMGN5 (Fig. 4A). Cell viability was
detected by the CellTiter-Blue assay after 48 h of treatment with
different doses of gemcitabine. Notably, cells with a higher level
of HMGN5 expression (PC-3, PC-3M and LNCaP) showed more sensitivity
to gemcitabine, and cells with a lower level of HMGN5 expression
(DU145, 22RV1) showed less sensitivity to gemcitabine; cells with
the lowest level of HMGN5 expression (RWPE-1) showed resistance to
gemcitabine (Fig. 4A). Furthermore,
downregulation of HMGN5 in the PC-3 cells with shHMGN5 resulted in
the resistance of PC-3 cells to gemcitabine. PC-3 cells with shCtrl
were sensitive to gemcitabine (Fig.
4B). Upregulation of HMGN5 in DU145 cells induced their
sensitivity to gemcitabine (Fig.
4C). Next, we explored the mechanism of HMGN5-mediated
sensitivity to gemcitabine. We first tested whether gemcitabine
treatment downregulated the HMGN5 protein level in PC-3 cells.
After 48 h of treatment with different doses of gemcitabine,
western blotting showed that gemcitabine downregulated HMGN5
expression in a dose-dependent manner (Fig. 4D), meanwhile, consistent with HMGN5
expression, the MAPK signaling pathway marker, pERK1/2, was also
downregulated by gemcitabine. Cleavage of PARP was upregulated by
gemcitabine (Fig. 4D).
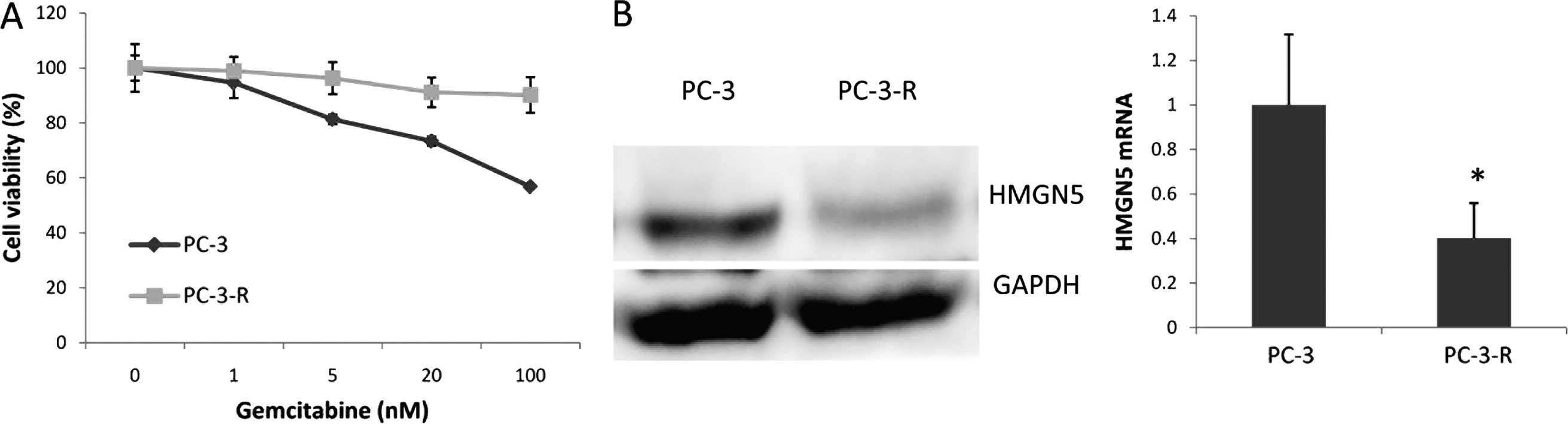
We established a gemcitabine-resistant PC-3 cell
line (PC-3-R) by culturing PC-3 cells with a low level of
gemcitabine for 3 months. Notably, the PC-3-R cell line expressed
less HMGN5 protein and mRNA compared with its parental cells
(Fig. 5). Taken together, these
results indicate that HMGN5 may be a potential biomarker for the
treatment of prostate cancer patients with gemcitabine.
Discussion
In the present study, we found new evidence to prove
that HMGN5 plays an oncogenic role in prostate cancer, and we
demonstrated that knockdown of HMGN5 suppressed cell proliferation,
reduced cell invasion and induced apoptosis in PC-3 cells, which
express the highest level of HMGN5 among prostate cancer cell
lines. Conversely, ectopic expression of HMGN5 in human prostate
epithelial cell line RWPE-1 promoted cell colony formation and
induced cell invasion.
We demonstrated that HMGN5 exerted its function by
activating the MAPK signaling pathway, which plays a vital role in
prostate cancer tumorigenesis and progression (25). Upregulation of HMGN5 activated the
MAPK signaling pathway in RWPE-1 cells, and PC-3 cells with HMGN5
knockdown showed less pERK1/2 expression, indicating that the MAPK
signaling pathway was less active. In addition, the HMGN5-mediated
malignant phenotype was partially reversed by U0126, which is an
ERK inhibitor. The mechanism by which HMGN5 activates the MAPK
signaling pathway is unclear. HMGN5 is a nucleosome-binding
protein. By binding to histone H1, HMGN5 can unfold chromatin and
regulate transcription; therefore, HMGN5 may activate the MAPK
signaling pathway by elevating the transcription of genes upstream
of the MAPK signaling pathway.
Although gemcitabine has been clinically proven to
act against a broad spectrum of solid tumors and inhibit prostate
cancer cell lines effectively, the clinical effect of gemcitabine
on AIPC is limited. Gemcitabine is usually used to treat AIPC in
combination with other chemotherapy drugs, such as docetaxel or
prednisone (26–28). In the present study, we found that
there was a variation in the response of prostate cancer cell lines
to gemcitabine; prostate cancer cells with higher HMGN5 expression
showed more sensitivity to gemcitabine, and cells with lower
expression of HMGN5 showed less sensitivity to gemcitabine.
Knockdown of HMGN5 with shHMGN5 reduced the sensitivity of PC-3
cells to gemcitabine while ectopic expression of HMGN5 in DU145
cells promoted sensitivity to gemcitabine. Furthermore, gemcitabine
was found to downregulate HMGN5 and pERK1/2 expression and
upregulated cleaved-PARP. The mechanism of this result is unclear;
gemcitabine may have killed the subpopulation of PC-3 cells with a
high level HMGN5, and the remaining cells expressed a low level of
HMGN5. Another explanation for this may be that gemcitabine causes
downregulation directly in prostate cancer cells.
Moreover, we also found a decrease in HMGN5 in
secondary gemcitabine-resistant PC-3 cells, indicating that
prostate cancer cells may gain gemcitabine resistance by
downregulating HMGN5 expression. Collectively, we found that HMGN5
may be a biomarker which predicts the response of prostate cancer
to gemcitabine.
HMGN5 is a nucleosome-binding protein. It can bind
to histone protein H1 by its negatively charged C-terminus, and
unfold chromatin and counteract linker histone-mediated chromatin
compaction, as gemcitabine exerts its antitumor activity by
inhibiting DNA biosynthesis. HMGN5 may promote the effect of
gemcitabine by loosening chromatin and facilitating gemcitabine to
combine and react with DNA.
Based on our data, we conclude that HMGN5 is an
oncogene and plays an important role in prostate cancer
tumorigenesis and progression. HMGN5 has the potential to be a
therapeutic target for prostate cancer treatment. Moreover, the
level of HMGN5 may be used as a biomarker to predict the patients
that would benefit from gemcitabine treatment.
Acknowledgements
The present study was supported by grants from the
National Natural Science Foundation of China (no. 30672099), and
the Beijing National Science Foundation of China (no. 7122183).
References
|
1
|
Siegel R, Naishadham D and Jemal A: Cancer
statistics, 2012. CA Cancer J Clin. 62:10–29. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Harris WP, Mostaghel EA, Nelson PS and
Montgomery B: Androgen deprivation therapy: progress in
understanding mechanisms of resistance and optimizing androgen
depletion. Nat Clin Pract Urol. 6:76–85. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Watson PA, Chen YF, Balbas MD, et al:
Constitutively active androgen receptor splice variants expressed
in castration-resistant prostate cancer require full-length
androgen receptor. Proc Natl Acad Sci USA. 107:16759–16765. 2010.
View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Abbruzzese JL: Phase I studies with the
novel nucleoside analog gemcitabine. Semin Oncol. 23(Suppl 10):
S25–S31. 1996.
|
|
5
|
Muenchen HJ, Quigley MM, Pilat MJ, et al:
The study of gemcitabine in combination with other chemotherapeutic
agents as an effective treatment for prostate cancer. Anticancer
Res. 20:735–740. 2000.PubMed/NCBI
|
|
6
|
Cronauer MV, Klocker H, Talasz H, et al:
Inhibitory effects of the nucleoside analogue gemcitabine on
prostatic carcinoma cells. Prostate. 28:172–181. 1996. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Morant R, Bernhard J, Maibach R, et al:
Response and palliation in a phase II trial of gemcitabine in
hormone-refractory metastatic prostatic carcinoma. Swiss Group for
Clinical Cancer Research (SAKK). Ann Oncol. 11:183–188. 2000.
View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Rochman M, Malicet C and Bustin M:
HMGN5/NSBP1: a new member of the HMGN protein family that affects
chromatin structure and function. Biochim Biophys Acta. 1799:86–92.
2010. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Rochman M, Postnikov Y, Correll S, et al:
The interaction of NSBP1/HMGN5 with nucleosomes in euchromatin
counteracts linker histone-mediated chromatin compaction and
modulates transcription. Mol Cell. 35:642–656. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Rochman M, Taher L, Kurahashi T, et al:
Effects of HMGN variants on the cellular transcription profile.
Nucleic Acids Res. 39:4076–4087. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Hock R, Furusawa T, Ueda T and Bustin M:
HMG chromosomal proteins in development and disease. Trends Cell
Biol. 17:72–79. 2007. View Article : Google Scholar
|
|
12
|
Shirakawa H, Rochman M, Furusawa T, et al:
The nucleosomal binding protein NSBP1 is highly expressed in the
placenta and modulates the expression of differentiation markers in
placental Rcho-1 cells. J Cell Biochem. 106:651–658. 2009.
View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Shirakawa H, Landsman D, Postnikov YV and
Bustin M: NBP-45, a novel nucleosomal binding protein with a
tissue-specific and developmentally regulated expression. J Biol
Chem. 275:6368–6374. 2000. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Wang JW, Zhou LQ, Yang XZ, et al:
Up-regulated expression of a prostate cancer-related gene, NSBP1,
in prostate cancer cells. Basic Med Sci Clin. 24:393–397. 2004.
|
|
15
|
Wahafu W, He ZS, Zhang XY, et al: The
nucleosome binding protein NSBP1 is highly expressed in human
bladder cancer and promotes the proliferation and invasion of
bladder cancer cells. Tumour Biol. 32:931–939. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Ji SQ, Yao L, Zhang XY, Li XS and Zhou LQ:
Knockdown of the nucleosome binding protein 1 inhibits the growth
and invasion of clear cell renal cell carcinoma cells in vitro and
in vivo. J Exp Clin Cancer Res. 31:222012. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Han HJ, Russo J, Kohwi Y and
Kohwi-Shigematsu T: SATB1 reprogrammes gene expression to promote
breast tumour growth and metastasis. Nature. 452:187–193. 2008.
View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Green J, Ikram M, Vyas J, et al:
Overexpression of the Axl tyrosine kinase receptor in cutaneous
SCC-derived cell lines and tumours. Br J Cancer. 94:1446–1451.
2006. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Rizzolio F, Bione S, Sala C, et al:
Chromosomal rearrangements in Xq and premature ovarian failure:
mapping of 25 new cases and review of the literature. Hum Reprod.
21:1477–1483. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Jiang N, Zhou LQ and Zhang XY:
Downregulation of the nucleosome-binding protein 1 (NSBP1) gene can
inhibit the in vitro and in vivo proliferation of prostate cancer
cells. Asian J Androl. 12:709–717. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Zhang XY, Guo ZQ, Ji SQ, et al: Small
interfering RNA targeting HMGN5 induces apoptosis via modulation of
a mitochondrial pathway and Bcl-2 family proteins in prostate
cancer cells. Asian J Androl. 14:487–492. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Zhang CJ, Li XS, Wahafu W, et al: Effect
of paclitaxel and gemcitabine on the expression of nucleosomal
binding protein 1 in bladder cancer cell line T24. Chin J Urol.
31:536–540. 2010.
|
|
23
|
Jiang N, Zhou LQ, Yao K and Huang C: The
experimental investigation of androgen independent prostate cancer
cell line inhibited growth in vivo by recombinant lentivirus of
small interfering RNA (siRNA) targeting NSBP1. Chin J Clinicians.
4:154–157. 2010.
|
|
24
|
Wang Y, Guo Z, Ge Q, Wang E, Zhang X and
Jin G: Preparation and characterization of RGD
tumour-homing-peptide-modified plasminogen K5. Biotechnol Appl
Biochem. 57:17–24. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Rodriguez-Berriguete G, Fraile B,
Martinez-Onsurbe P, Olmedilla G, Paniagua R and Royuela M: MAP
kinases and prostate cancer. J Signal Transduct. 2012:1691702012.
View Article : Google Scholar
|
|
26
|
Buch-Hansen TZ, Bentzen L, Hansen S, et
al: Phase I/II study on docetaxel, gemcitabine and prednisone in
castrate refractory metastatic prostate cancer. Cancer Chemother
Pharmacol. 66:295–301. 2010. View Article : Google Scholar
|
|
27
|
Di Lorenzo G, Autorino R, Giuliano M, et
al: Phase II trial of gemcitabine, prednisone, and zoledronic acid
in pretreated patients with hormone refractory prostate cancer.
Urology. 69:347–351. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Garcia JA, Hutson TE, Shepard D, Elson P
and Dreicer R: Gemcitabine and docetaxel in metastatic,
castrate-resistant prostate cancer: results from a phase 2 trial.
Cancer. 117:752–757. 2011. View Article : Google Scholar
|