Introduction
Gastric cancer is a common disease worldwide and the
second most frequent cause of cancer-associated mortality,
affecting approximately one million individuals annually (1). Of 880,000 people diagnosed with
gastric cancer in 2000, approximately 650,000 (74%) succumbed to
the disease (2). The genesis and
progression of human gastric cancer is thought to be crucially
influenced by genetic and epigenetic alterations, including the
activation of oncogenes and the inactivation of tumor-suppressor
genes (3). Oncogenes and
tumor-suppressor genes have always been regulated by microRNAs
(miRNAs).
miRNAs are non-coding RNA molecules ~21–23 nucleo
tides long that regulate gene expression at the
post-transcriptional level (4–6). miRNA
expression profiling analyses have revealed a global dysregulation
of mature miRNA levels in primary human tumors compared to normal
tissues (7,8). miRNAs act as novel oncogenes or
tumor-suppressor genes (9,10) and it has been shown that alterations
in microRNA expression correlate highly with the progression and
prognosis of human tumors (11,12).
Furthermore, a number of differentially expressed miRNAs derived
from circulation are used as potential biomarkers in various types
of cancer, including liver, prostate, lymphoma and ovarian cancer
(13–16). Thus, focusing on miRNAs in gastric
cancer resulted in insight into the diagnosis and treatment of this
disease.
An increasing body of evidence indicates miR-130a is
differentially expressed in various tumors. miR-130a was
overexpressed in adult T-cell leukemia (ATL), basal cell carcinoma
and esophageal cancer tissue, but underexpressed in bladder and
ovarian cancer, and glioblastoma (17–22).
However, few studies have focused on miR-130a expression and its
function in gastric cancer. Xu et al found that miR-130a
directly inhibited the expression of tumor-suppressor gene
runt-related transcription factor 3 (RUNX3) (23). RUNX3, a member of the family of
transcription factors that contain the runt domain, is located at
human chromosome 1p36 and was identified as a tumor-suppressor in
breast, bladder and lung cancer (24–26).
Previous findings showed that, RUNX3 was identified as a pivotal
tumor-suppressor in gastric cancer (27), and a loss or substantial decrease in
RUNX3 expression may be causally associated with gastric cancer, as
it correlates with differentiation, lymph node metastasis and poor
prognosis of this disease (28).
Thus, miR-130a may act as an oncogene in gastric cancer by
targeting RUNX3. Therefore, further systemic delineation of
miR-130a expression and function in gastric cancer is needed.
Materials and methods
Human tissue specimens and cell
lines
The present study utilized fresh tissues, including
41 human gastric cancer samples and 41 samples of adjacent normal
mucosal tissues derived from 41 patients who underwent surgery at
the Second Affiliated Hospital of Chongqing Medical University
(Chongqing, China) between 2010 and 2011. The present study was
conducted according to the 'Biomedical Research Involving Human
Ethics Review (Tentative)' regulation of the Ministry of Health and
the Declaration of Helsinki on Ethical Principles for Medical
Research Involving Human Subjects. All the samples were obtained
with the informed consent of the patients, and the experiments were
approved by the Institutional Review Board of the Second Affiliated
Hospital of Chongqing Medical University (Chongqing, China). All
the participants provided written informed consent to participate
in the present study.
The SGC-7901, HGC-27, AGS, MKN45 and N87 cell lines
were obtained from the American Type Culture Collection (ATCC;
Manassas, VA, USA), and the GES-1 cell line was purchased from the
Type Culture Collection of the Chinese Academy of Sciences
(Shanghai, China). The cell lines were cultured in RPMI-1640
(HyClone, Logan, UT, USA) supplemented with 10% fetal bovine serum
(FBS) and were incubated at 37°C with 5% CO2.
Serum collection
Whole blood (2 ml) from the gastric cancer patients
and healthy controls was collected in regular tubes and immediately
processed to prevent contamination by cellular nucleic acids. Blood
samples were centrifuged at 2,000 rpm for 10 min at room
temperature, and then the upper supernatant, which was the serum
sample, was transferred to new RNase-free collection tubes,
respectively, and stored at −80°C for further processing.
Detection of CEA and CA-199
Values for carcinoembryonic antigen (CEA) and
carbohydrate antigen 199 (CA-199) levels in the serum of the
gastric cancer patients and healthy controls were determined at the
Clinical Laboratory of the Second Affiliated Hospital of Chongqing
Medical University (Chongqing, China).
Primers, RNA isolation and miRNA
detection
The primers for miR-338-3p and U6 were produced
using the miScript Primer Assay kit (Qiagen, Dusseldorf, Germany).
The sequences of the miRNAs used in the present study were as
follows: miR-130a, CAGUGCAAUGUUAAAAGGGCAU; and U6,
CGCAAGGAUGACACGCAAAUUCGUGAAGCGUUCCAUAUUUUU. The reverse primers
were also used in the reverse transcription step. Total miRNA was
extracted from the cultured cells, human tissue specimens and serum
sample using RNAiso for small RNA (Takara Bio, Otsu, Japan)
according to the manufacturer's instructions. Poly(A) tails were
added to miR-338 and U6 with the miRNA Reaction Buffer Mix, and
then cDNA was produced from 5 ng of total RNA using the miRNA
PrimeScript RT Enzyme Mix (both from Takara Bio). RT-qPCR was
performed in a CFX96™ Real-Time PCR Detection System (Bio-Rad,
Hercules, CA, USA) with SYBR® Premix Ex Taq™ II
(Takara Bio). The PCR conditions used were 95°C for 30 sec,
followed by 40 cycles of 95°C for 5 sec, and 60°C for 30 sec. The
data were normalized against the U6 snRNA. After amplification, a
melting curve analysis was performed to confirm the specificity of
the products.
Expression levels of the miRNAs were calculated by
cycle threshold (Ct) values with SDS 2.0 software (Applied
Biosystems, Foster City, CA, USA). The concentrations from serum,
tissues or cell lines samples were normalized using the
2−ΔΔCt method relative to U6 small nuclear RNA (RNU6B).
The value of ΔCt was calculated by subtracting the Ct values of
RNU6B from the Ct values of the miRNAs of interest in the present
study. The values of ΔΔCt were then calculated by subtracting the
ΔCt of the control samples from the ΔCt of the cancer samples. The
change in gene expression was calculated using the equation
2−ΔΔCt.
Oligonucleotide transfection
miR-130a mimics, inhibitor and cont-miR were
produced by Sangon Biotechnology (Sangon, Shanghai, China), and
co-transfections were performed with Lipofectamine 2000
(Invitrogen-Life Technologies, Carlsbad, CA, USA). Twenty-four
hours after transfection, the cells were plated for hte
proliferation, migration and invasion assays. The cells were
collected for RNA and protein analyses 48 h after transfection.
pcDNA expression plasmids and plasmid
transfection
The ORF sequences of RUNX3 were amplified from
genomic DNA isolated from the AGS cell line and were then subcloned
into the GV230 vector (GeneChem Corporation, Shanghai, China). The
plasmid was transfected into AGS cells using Lipofectamine 2000.
Twenty-four hours after transfection, the cells were used for a
rescue experiment.
Luciferase reporter assay
A Psicheck™-2 Dual-Luciferase miRNA target
expression vector was used for the 3′UTR luciferase assays (Sangon
Biotechnology). The target gene of miRNA-130a was selected based on
the online microRNA target database, http://www.microrna.org/microrna/home.do. The primer
sequences used for the wild-type 3′UTR of RUNX3 were: forward,
5′-CCGCCCTGGTGGACTCCT-3′ and reverse,
5′-CCTTCCACACATCTCAGAGTTATAT-3′. Since there was one binding site
in the RUNX3 3′UTR, primer sequences were designed for the mutant
3′UTR as follows: forward,
5′-GTGGAAACTGTGGCGCGCCATCGTTTGCTTGGTGTTTG-3′ and reverse,
5′-GCAAACGATAGTGCA AAGCAGTTTCCACCCAGCTCCAT-3′. For the luciferase
assay, Lipofectamine 2000 was used to co-transfect MKN45 cells with
the miR-130a mimics and Psicheck™-2 Dual-Luciferase miRNA target
expression vectors containing wild-type or mutant target sequences.
The Dual-Luciferase Assay (Promega, Madison, WI, USA) was used to
measure the firefly luciferase activity 18 h after transfection,
and the results were normalized against the Renilla
luciferase. Each reporter plasmid was transfected at least three
times (on different days), and each sample was assayed in
triplicate.
Cell viability assays
The transfected cells were seeded in 96-well plates
at a density of 1×104 cells/well. A
3-(4,5-dimethyl-thazol-2-yl)-2,5-diphenyltetrazolium bromide (MTT)
solution (20 ml of 5 mg/ml MTT) was added to the cultures (for a
total volume of 250 µl) and incubated for 4 h at 37°C.
Following the removal of the culture medium, the remaining crystals
were dissolved in dimethylsulfoxide (DMSO), and the absorbance at
570 nm was measured.
Migration and invasion assays
For the Transwell migration assays, 1×104
cells were plated in the top chamber with a non-coated membrane
(24-well insert; 8-mm pore size; BD Biosciences, Franklin Lakes,
NJ, USA). For the invasion assays, 2×105 cells were
plated in the top chamber with a Matrigel-coated membrane (24-well
insert; 8 mm pore size; BD Biosciences). For the two assays, the
cells were plated in a serum-free medium, and medium supplemented
with 10% serum was used as a chemoattractant in the lower chamber.
The cells were incubated for 16 h at 37°C and 5% CO2 in
a tissue culture incubator. After 16 h, the
non-migrated/non-invading cells were removed from the upper sides
of the Transwell membrane filter inserts using cotton-tipped swabs.
The migrated/invaded cells on the lower sides of the inserts were
stained with Giemsa, and the cells were counted.
Antibodies and immunoblotting
Antibodies against RUNX3 and Smad4 were purchased
from Abcam (Cambridge, UK). Antibody against GAPDH was purchased
from Santa Cruz Biotechnology, Inc. (Santa Cruz, CA, USA).
HRP-conjugated goat anti-rabbit IgG was purchased from Santa Cruz
Biotechnology, Inc.. The total protein was extracted from the
transfected cells and gastric cancer tissues using RIPA lysis
buffer (Beyotime, China) according to the manufacturer's
instructions. After the whole-cell protein extracts were quantified
using the BCA protein assay, equivalent amounts of cell lysates
were resolved by 10% SDS polyacrylamide gel electrophoresis, and
were transferred onto a polyvinylidene fluoride membrane, which was
then blocked in 5% non-fat milk in TBST for 1 h at 4°C. The blots
were then incubated with primary antibodies. After incubation with
HRP-conjugated secondary antibodies, the protein bands were
visualized using an enhanced chemiluminescence reagent (Millipore,
Billerica, MA, USA). The following antibody dilutions were used:
anti-RUNX and anti-Smad4, 1:1,500; and HRP-conjugated IgG,
1:7,000.
Statistical analysis
SPSS 17.0 software was used for the statistical
analysis. The data are presented as the means ± standard deviation
(SD). Group comparisons were performed using the Student's t-test.
The relationships between miR-130a expression in the serum of
gastric cancer patients and healthy control were analyzed using the
non-parametric Mann-Whitney U test. Receiver-operating
characteristic (ROC) curves and the area under the ROC curve (AUC)
were used to assess the feasibility of serum and tissue miRNA as a
diagnostic tool for detecting gastric cancer. For disease
progression, the Kaplan-Meier (log-rank test) analysis was
performed. The Spearman's rank test was used to evaluate the
relationships among the relative expression levels of miR-130a and
RUNX3 in gastric cancer tissues. The relationships, in the AGS cell
line with the forced expression of miR-130a or cont-miR and with or
without RUNX3 restoration, were analyzed using the one-way ANOVA
Dunnett's test. Differences were considered to indicate a
statistically significant result when P<0.05.
Results
miR-130a is upregulated in gastric cancer
tissues and cell lines
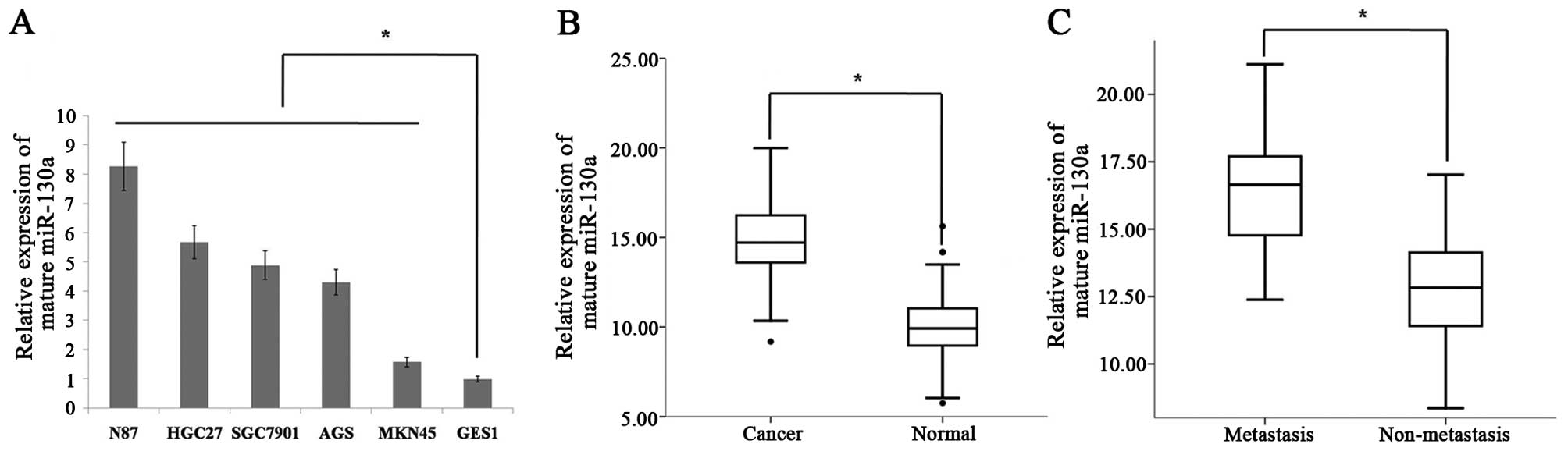
To investigate the miR-130a expression levels in
human gastric cancer cell lines, we monitored its expression in
several cancer cell lines (SGC7901, HGC27, AGS, MKN45 and N87) and
a normal gastric mucosal cell line (GES1). We found that miR-130a
expression was increased in gastric cancer cell lines compared to
the normal gastric mucosal cells (Fig.
1A). To further examine the role of miR-130a in human gastric
cancer development, we detected the levels of its expression in 41
cases of human gastric cancer and 41 cases adjacent normal mucosa
tissues. The RT-qPCR analysis revealed that, the levels of miR-130a
expression were significantly increased in tumor tissues compared
to that of the adjacent normal mucosal tissues (Fig. 1B). To determine whether miR-130a
expression was associated with gastric cancer metastasis, we
examined the miR-130a expression levels in 41 archived primary
gastric tumors. These tumors were divided into two groups: in one
group tumors were resected from 25 patients with lymph node
metastasis, while in the second group the tumors were resected from
16 patients without metastasis. The RT-qPCR analysis revealed that,
the miR-130a expression levels were significantly higher in the
patients with metastasis than in those without metastasis.
Diagnostic and prognostic significance of
miR-130a in gastric cancer
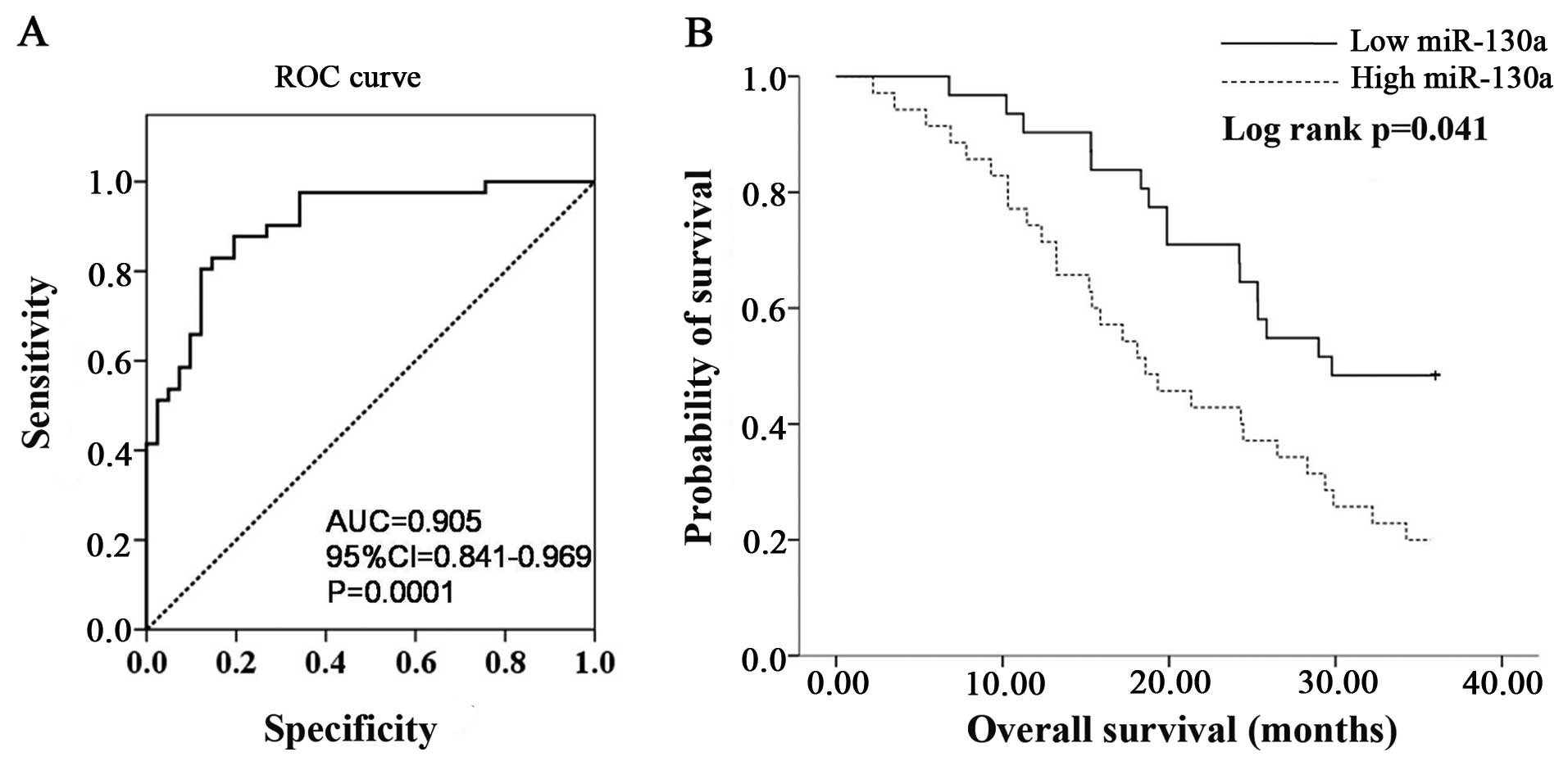
Clinical information for the 41 patients is shown in
Table I. Receiver-operating
characteristic (ROC) curve analyses were performed to evaluate the
ability of miR-130a expression to discriminate between normal and
tumor cases using tissue samples. According to ROC curve, miR-130a
had the best sensitivity and specificity when the miR-130a Ct value
was 11.13 (YI=0.634), and an AUC of 0.905 (P=0.0001; 95% CI,
0.841–0.969) (Fig. 2A) was
obtained, suggesting that miR-130a expression discriminated between
malignant and non-malignant samples and may therefore be used as a
diagnostic marker for gastric cancer. To determine whether the
levels of miR-130a in tumor tissues correlate with the survival of
the gastric cancer patients, the patients were divided into
low-miR-130a (expression Ct value <11.13) and high-miR-130a
(expression Ct value ≥11.13) groups and a Kaplan-Meier survival
analysis was performed. The results of the Kaplan-Meier analysis
revealed that the low-miR-130a group had a significantly improved
overall survival compared to the high-miR-130a group (Fig. 2B).
 | Table IClinicopathological characteristics
of the patient cohort. |
Table I
Clinicopathological characteristics
of the patient cohort.
|
Characteristics | Total cases
(n=41) |
|---|
| Age (years) |
| Range | 33–88 |
| Mean | 54 |
| Median | 57 |
| Pathological T
(%) |
|
PT0-Tis | 12 (29.2) |
|
PT1 | 9 (22.0) |
|
PT2 | 6 (14.6) |
|
PT3 | 8 (19.5) |
|
PT4 | 6 (14.7) |
| Lymph node
metastases (%) |
|
PN0 | 16 (39.0) |
|
PN1–3 | 25 (61.0) |
| Dead/alive (%) |
| Dead | 25 (61.0) |
| Alive | 16 (39.0) |
miR130a expression and diagnostic
significance in the serum of gastric cancer patients and healthy
controls
Clinicopathological characteristics for the gastric
cancer patients are shown in Table
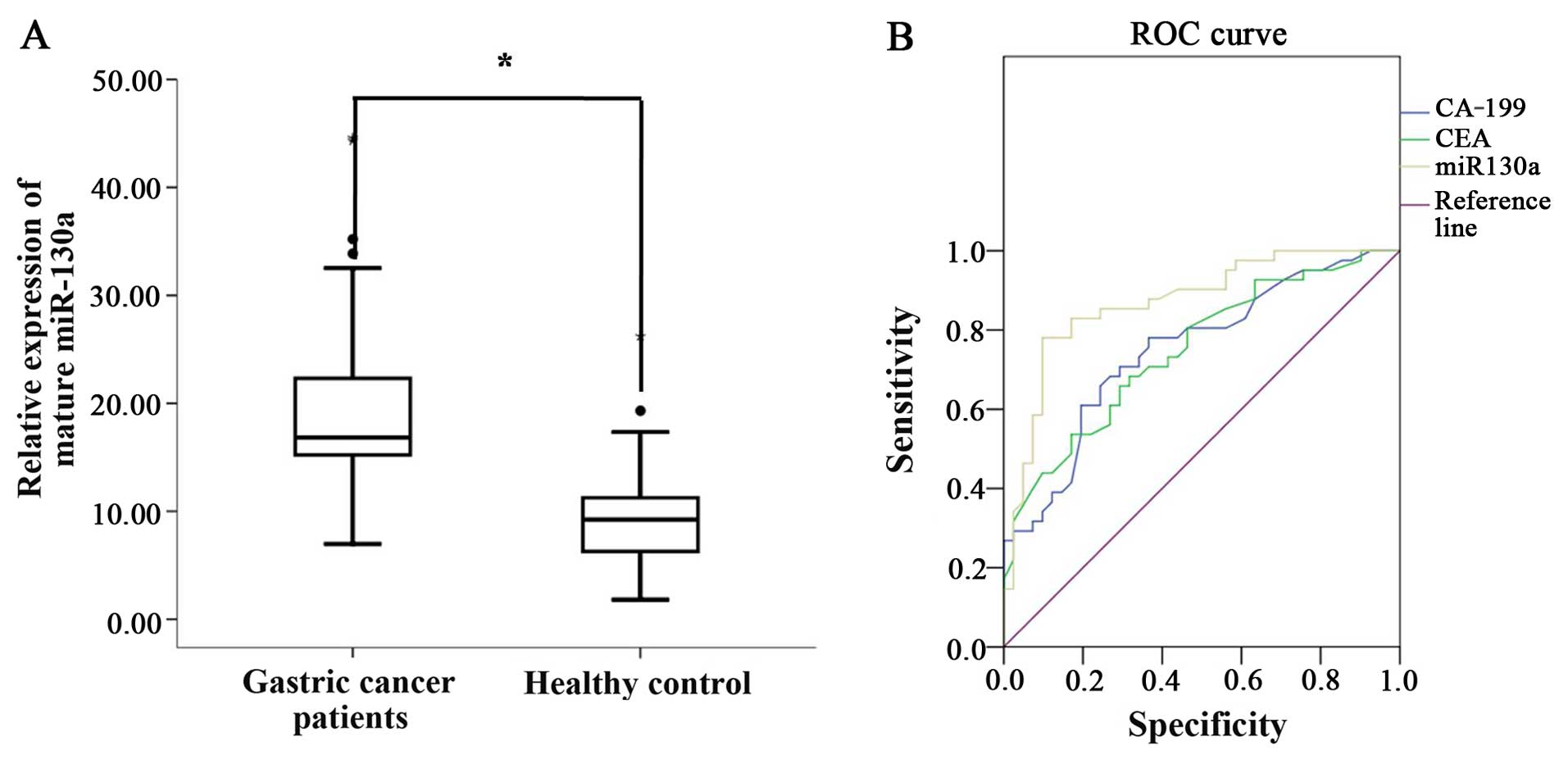
I. miR-130a expression was detected in the serum of gastric
cancer patients and healthy controls. We also found that the
expression of miR-130a in plasma in 41 gastric cancer patients was
significantly higher than that from the healthy controls (Fig. 3A). To further understand the
significance of the diagnostic value between miR-130a and
traditional tumor markers CEA and CA-199, a ROC analysis of CEA,
CA-199 and miR-130a was performed. Regarding the area under the ROC
curve, miR-130a had the highest value at 0.870, while values for
CEA and CA-199 were at 0.749 and 0.750, respectively (Fig. 3B and Table II). Our findings showed that the
diagnostic value of miR-130a was more effective than that for tumor
markers CEA and CA-199 in the serum of gastric cancer patients and
healthy controls.
 | Table IIArea under the ROC curve analysis of
CEA, CA-199 and miR-130a expression in the serum samples of the
gastric cancer patients and controls. |
Table II
Area under the ROC curve analysis of
CEA, CA-199 and miR-130a expression in the serum samples of the
gastric cancer patients and controls.
| Variables | Area | SEa | Asymptotic
significanceb | Asymptotic 95% CI
|
|---|
| Lower bound | Upper bound |
|---|
| CA-199 | 0.750 | 0.054 | 0.000 | 0.644 | 0.855 |
| CEA | 0.749 | 0.053 | 0.000 | 0.644 | 0.853 |
| miR-130a | 0.870 | 0.040 | 0.000 | 0.792 | 0.948 |
miR-130a directly targets RUNX3
We predicted 26 targets for miR-130a using the
following prediction tools: miRanda (http://www.microrna.org), TargetScan (http://www.targetscan.org/) and miRDB (mirdb.org/miRDB/). In those 26 targets, we found some
genes associated with tumors, such as RUNX3, MET, Smad4 and KLF4
(Table III). Recent findings
showed that, RUNX3 and Smad4, which function as tumor-suppressor
genes, are associated with gastric cancer (29–31).
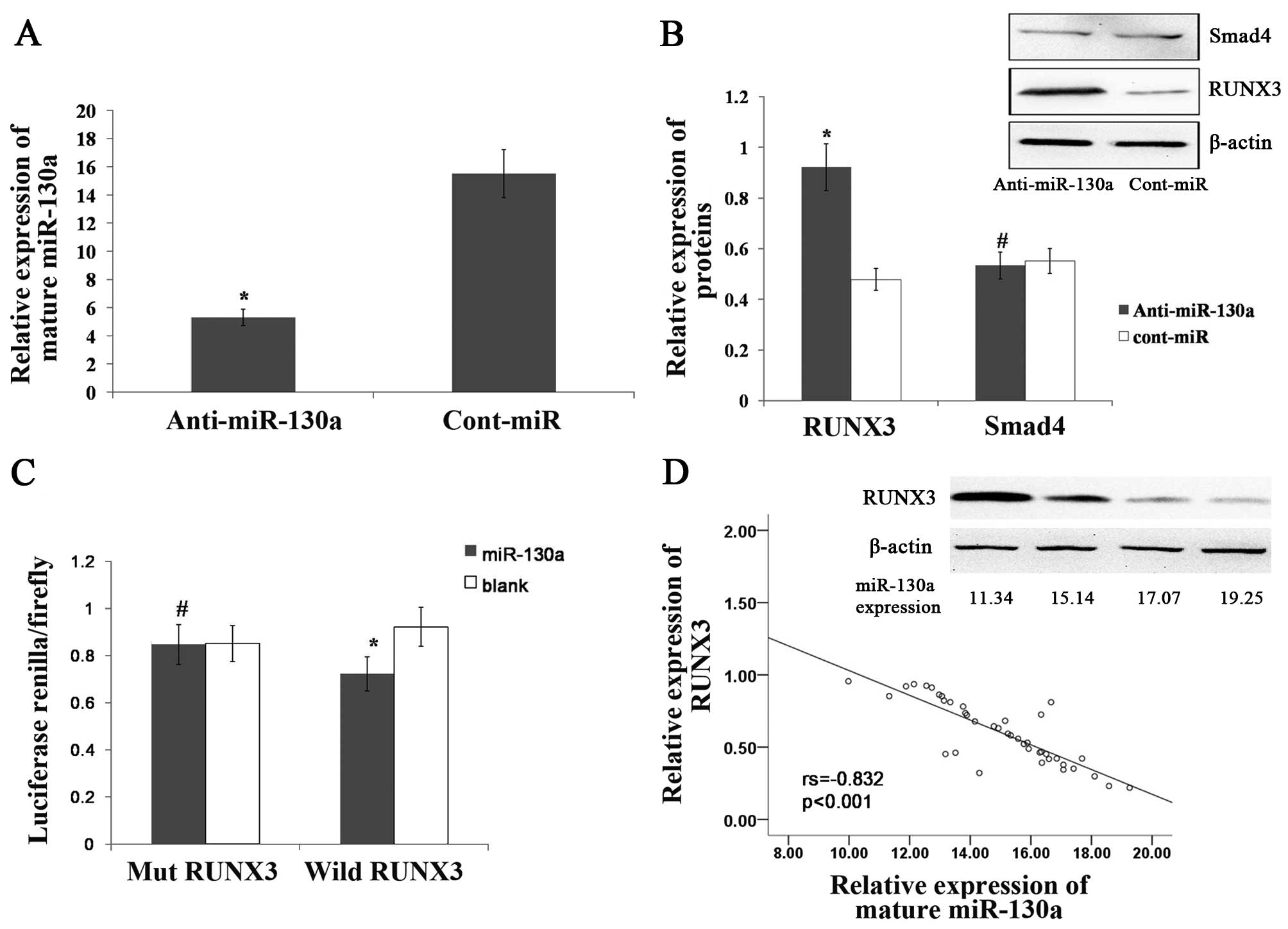
To determine whether miR-130a targets RUNX3 and Smad4, we examined
RUNX3 and Smad4 expression in the AGS gastric cell line following
transfection with miR-130a inhibitor. Compared with the expression
of cont-miR, miR-130a was significantly downregulated in the AGS
cell line following transfection with the miR-130a inhibitor
(Fig. 4A), and we found that RUNX3
expression was significantly increased in miR-130a-low-expressing
gastric cancer cell lines, although Smad4 expression was not
increased (Fig. 4B). Our findings
showed that RUNX3 may be the target gene for miR-130a. To confirm
that miR-130a directly targeted RUNX3, we performed luciferase
reporter assays to examine whether miR-130a interacts directly with
RUNX3. We found that the co-transfection of miR-130a and the
wild-type RUNX3 3′UTR caused a significant decrease in luciferase
expression when compared with the controls. However, the
co-transfection of miR-130a and the mutant RUNX3 3′UTR did not
cause a decrease in luciferase expression (Fig. 4C). The RUNX3 expression was also
detected in 41 gastric cancer tissues by western blotting, and we
found that the expression of miR-130a was inversely correlated with
that of RUNX3 (Fig. 4D). These
results suggested that miR-130a directly targets RUNX3.
 | Table IIImiR-130a predicted targets. |
Table III
miR-130a predicted targets.
| Total target
genes | Target genes
associated with tumors |
|---|
| CTSA,
STK38L, ZFPM2, BMPR2, ACVR1,
LDLR, DNM2, ATG2B, DICER1,
PPARG, KLF4, HOXA10, CSF1,
MEOX2, APP, ATXN1, TAC1, HoxA5,
MAFB, ESR1, IFITM1, RUNX3, MET,
RAB5A, Smad4, Smad5 | HoxA5,
MAFB, ESR1, IFITM1, RUNX3, MET,
RAB5A, Smad4, Smad5, ATG2B,
KLF4, HOXA10 |
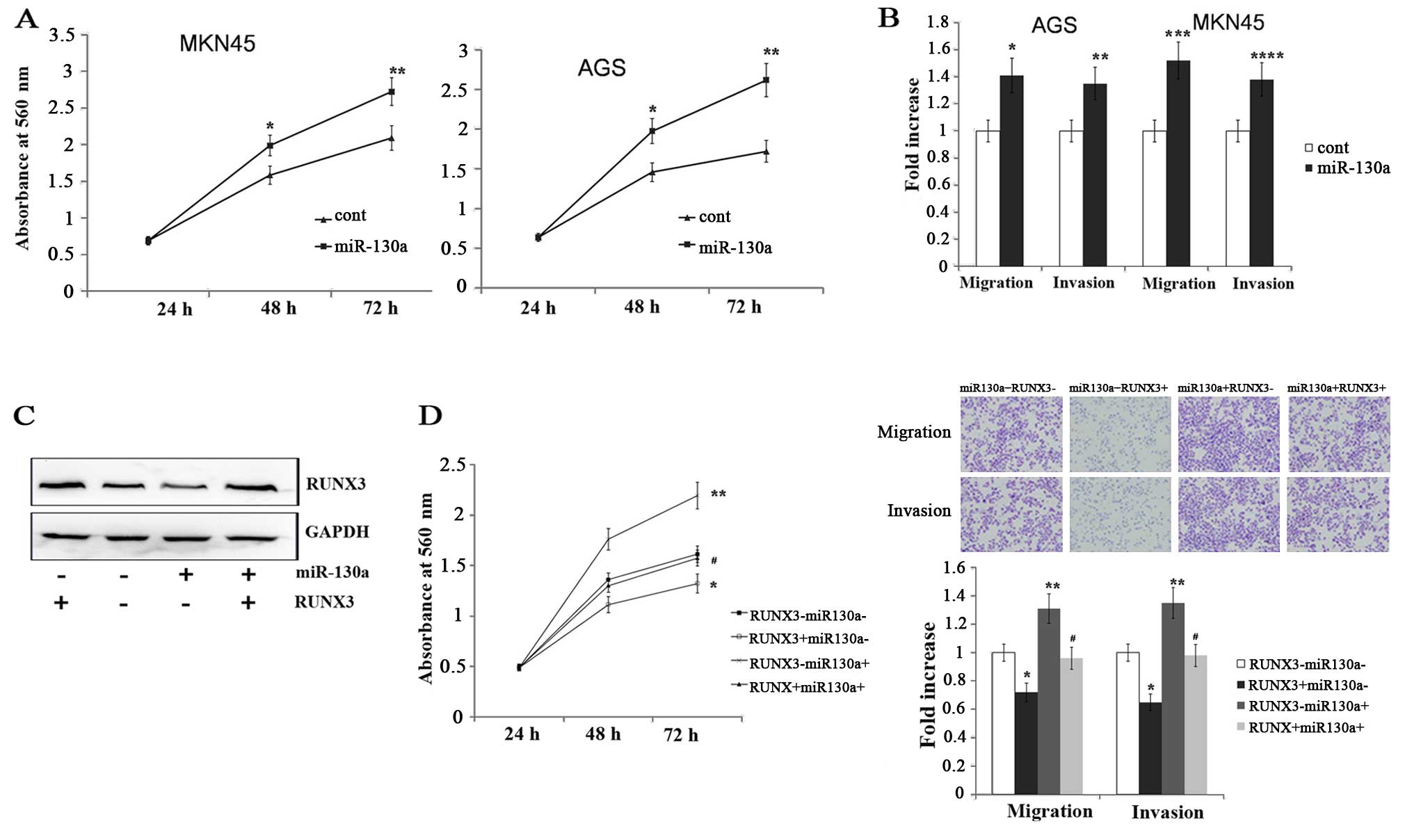
miR-130a promotes gastric cancer cell
migration, invasion and proliferation by targeting RUNX3
To determine the functional significance of miR-130a
in gastric cancer, we transfected the AGS and MKN45 gastric cancer
cell lines with miR-130a mimics. We found that the cells with a
forced expression of miR-130a significantly increased proliferation
compared with the cells with a forced expression of cont-miR
(Fig. 5A). Transwell migration and
Matrigel invasion assays demonstrated that miR-130a significantly
increased the migration and invasion of AGS and MKN45 cells
(Fig. 5B). To confirm whether
miR-130a promotes gastric cancer migration, invasion and
proliferation by targeting RUNX3, we forced the expression of
miR-130a in AGS and MKN45 cell lines along with a construct
containing the RUNX3 coding sequence but lacking the 3′UTR of the
RUNX3 mRNA. As a result, this construct yielded a RUNX3 mRNA that
was resistant to miR-130a. The restoration of RUNX3 expression was
confirmed through an immunoblot analysis (Fig. 5C). We found that the gastric cancer
cell migration, invasion and proliferation were completely restored
in the AGS cell line with a forced miR-130a expression and RUNX3
restoration (Fig. 5D). Therefore,
miR-130a regulated gastric cancer cell migration, invasion and
proliferation by targeting RUNX3.
Discussion
In the present study, we did examined miR-130a
expression in gastric cancer cell lines and detected miR-130a
expression in gastric cancer tissues, and found that miR-130a
expression was significantly upregulated in gastric cancer cell
lines and tissues. Furthermore, miR-130a expression was
significantly higher in the patients with metastasis than in the
patients without metastasis. The results show that miR-130a as an
oncogene was overexpressed in gastric cancer and was associated
with metastasis. miRNAs possess several features that make them
attractive candidates as new prognostic biomarkers and powerful
tools for the early diagnosis of cancer (32,33).
ROC and Kaplan-Meier analysis were than performed to determine the
miR-130a diagnostic and prognostic potential in gastric cancer. Our
findings show that miR-130a can be used as a diagnostic marker for
gastric cancer and low levels of miR-130a were significantly
associated with an extended overall survival of gastric cancer
patients. Thus, miR-130a serves as a molecular diagnostic and
prognostic marker for gastric cancer patients.
Screening for early gastric cancer potentially
reduces mortality of the disease (34–36).
Since the currently known tumor markers have the limitation of low
sensitivity and specificity (37),
diagnosis of gastric cancer is performed using a gastroscopic
biopsy sample and histology specified by WHO criteria. Although
gastroscopic screening for gastric cancer is currently the most
reliable screening tool, it is expensive and unsuitable as a
first-line examination due to the invasive nature. Thus,
identification of novel non-invasive biomarkers for tumor detection
is imperative. Previous findings have shown that some miRNAs have
potential diagnostic value in different types of cancer (19,38,39).
In the present study, we found that miR-130a expression in serum
was overexpressed in gastric cancer patients than healthy controls.
Compared with the diagnostic value of CEA, CA-199 and miR-130a
individually, miR-130a had the highest value at 0.870 on the area
under the ROC curve. miR-130a is sufficiently reliable for the
diagnosis of gastric cancer. Thus, miR-130a acts as a potential
circulating diagnostic biomarker for gastric cancer.
Some miRNAs inhibit the expression of
tumor-suppressor genes in normal tissues, elucidating the reason
for miRNAs acting as oncogenes. Chu et al found that
miR-590-5p acts as an oncogene by targeting the CHL1 gene
and promotes cervical cancer proliferation (40). Ma et al also found that
miR-34a targets GAS1 to promote cell proliferation and inhibit
apoptosis in papillary thyroid carcinoma (41). To predict target genes of miR-130a,
we screened 26 targets for miR-130a by prediction tools, and found
that 12 target genes were associated with tumors. We focused on
RUNX3 and Smad4 since it has been previously shown that RUNX3 and
Smad4 as tumor suppressor genes are associated with gastric cancer.
RUNX3 has been thought to be the target gene of miR-130a in other
tissues. It has been shown that miR-130a directly inhibited the
expression of the tumor-suppressor gene RUNX3, which
resulted in the activation of Wnt/β-catenin signaling and increased
drug resistance (23). miR-130a
also contributes to endothelial progenitor cell dysfunction by
targeting RUNX3 (42).
In the present study, Smad4 was excluded since the
inhibited expression of miR-130a did not increase Smad4 expression,
although RUNX3 expression was significantly increased following
transfection with anti-miR-130a. miR-130a expression was inversely
correlated with that of RUNX3 in gastric cancer tissues. The result
shows that miR-130a attenuates RUNX3 expression in gastric cancer
tissues. Thus, RUNX3 may be a target gene of miR-130a. Luciferase
reporter assays were performed to confirm miR-130a directly targets
RUNX3, and we found that miR-130a directly interacts with RUNX3 by
binding to RUNX3 3′UTR. Thus, miR-130a can directly target
tumor-suppressor gene RUNX3. Furthermore we found that
miR-130a significantly increases gastric cancer cell migration,
invasion and proliferation, which was completely restored after
RUNX3 restoration. The results show that miR-130 promotes gastric
cancer tumorigenic abilities by targeting RUNX3, and it elucidates
the reason for miR-130a acting as an oncogene in gastric
cancer.
In conclusion, the results show that miR-130a
expression was increased in gastric cancer and was associated with
the overall survival of gastric cancer. miR-130a can be a potential
circulating diagnostic biomarker for gastric cancer. Thus, miR-130a
may be used as a biomarker for gastric cancer diagnosis and
prognosis. Furthermore, miR-130a acts as an oncogene that promotes
gastric cancer tumorigenesis by targeting the tumor-suppressor gene
RUNX3. Thus, future studies on the anticancer mechanisms of
miR-130a may contribute to the development of new therapeutic
strategies for gastric cancer.
Acknowledgments
We thank Dr Xiao-Qiu Xiao for his assistance with
the RT-qPCR analysis.
References
|
1
|
Hartgrink HH, Jansen EP, van Grieken NC
and van de Velde CJ: Gastric cancer. Lancet. 374:477–490. 2009.
View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Kamangar F, Dores GM and Anderson WF:
Patterns of cancer incidence, mortality, and prevalence across five
continents: Defining priorities to reduce cancer disparities in
different geographic regions of the world. J Clin Oncol.
24:2137–2150. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Wu WK, Lee CW, Cho CH, Fan D, Wu K, Yu J
and Sung JJ: MicroRNA dysregulation in gastric cancer: A new player
enters the game. Oncogene. 29:5761–5771. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Rana TM: Illuminating the silence:
Understanding the structure and function of small RNAs. Nat Rev Mol
Cell Biol. 8:23–36. 2007. View
Article : Google Scholar
|
|
5
|
Valencia-Sanchez MA, Liu J, Hannon GJ and
Parker R: Control of translation and mRNA degradation by miRNAs and
siRNAs. Genes Dev. 20:515–524. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Pillai RS, Bhattacharyya SN and Filipowicz
W: Repression of protein synthesis by miRNAs: How many mechanisms?
Trends Cell Biol. 17:118–126. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Lu J, Getz G, Miska EA, Alvarez-Saavedra
E, Lamb J, Peck D, Sweet-Cordero A, Ebert BL, Mak RH, Ferrando AA,
et al: MicroRNA expression profiles classify human cancers. Nature.
435:834–838. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Thomson JM, Newman M, Parker JS,
Morin-Kensicki EM, Wright T and Hammond SM: Extensive
post-transcriptional regulation of microRNAs and its implications
for cancer. Genes Dev. 20:2202–2207. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Esquela-Kerscher A and Slack FJ: Oncomirs
- microRNAs with a role in cancer. Nat Rev Cancer. 6:259–269. 2006.
View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Calin GA and Croce CM: MicroRNA signatures
in human cancers. Nat Rev Cancer. 6:857–866. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Schetter AJ, Leung SY, Sohn JJ, Zanetti
KA, Bowman ED, Yanaihara N, Yuen ST, Chan TL, Kwong DL, Au GK, et
al: MicroRNA expression profiles associated with prognosis and
therapeutic outcome in colon adenocarcinoma. JAMA. 299:425–436.
2008. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Garzon R, Volinia S, Liu CG,
Fernandez-Cymering C, Palumbo T, Pichiorri F, Fabbri M, Coombes K,
Alder H, Nakamura T, et al: MicroRNA signatures associated with
cytogenetics and prognosis in acute myeloid leukemia. Blood.
111:3183–3189. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Xu J, Wu C, Che X, Wang L, Yu D, Zhang T,
Huang L, Li H, Tan W, Wang C, et al: Circulating microRNAs, miR-21,
miR-122, and miR-223, in patients with hepatocellular carcinoma or
chronic hepatitis. Mol Carcinog. 50:136–142. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Yaman Agaoglu F, Kovancilar M, Dizdar Y,
Darendeliler E, Holdenrieder S, Dalay N and Gezer U: Investigation
of miR-21, miR-141, and miR-221 in blood circulation of patients
with prostate cancer. Tumour Biol. 32:583–588. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Lawrie CH, Gal S, Dunlop HM, Pushkaran B,
Liggins AP, Pulford K, Banham AH, Pezzella F, Boultwood J,
Wainscoat JS, et al: Detection of elevated levels of
tumour-associated microRNAs in serum of patients with diffuse large
B-cell lymphoma. Br J Haematol. 141:672–675. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Resnick KE, Alder H, Hagan JP, Richardson
DL, Croce CM and Cohn DE: The detection of differentially expressed
microRNAs from the serum of ovarian cancer patients using a novel
real-time PCR platform. Gynecol Oncol. 112:55–59. 2009. View Article : Google Scholar
|
|
17
|
Qiu S, Lin S, Hu D, Feng Y, Tan Y and Peng
Y: Interactions of miR-323/miR-326/miR-329 and
miR-130a/miR-155/miR-210 as prognostic indicators for clinical
outcome of glioblastoma patients. J Transl Med. 11(10)2013.
View Article : Google Scholar
|
|
18
|
Ratert N, Meyer HA, Jung M, Lioudmer P,
Mollenkopf HJ, Wagner I, Miller K, Kilic E, Erbersdobler A, Weikert
S, et al: miRNA profiling identifies candidate miRNAs for bladder
cancer diagnosis and clinical outcome. J Mol Diagn. 15:695–705.
2013. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Ishihara K, Sasaki D, Tsuruda K, Inokuchi
N, Nagai K, Hasegawa H, Yanagihara K and Kamihira S: Impact of
miR-155 and miR-126 as novel biomarkers on the assessment of
disease progression and prognosis in adult T-cell leukemia. Cancer
Epidemiol. 36:560–565. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Sand M, Skrygan M, Sand D, Georgas D, Hahn
SA, Gambichler T, Altmeyer P and Bechara FG: Expression of
microRNAs in basal cell carcinoma. Br J Dermatol. 167:847–855.
2012. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Liu SG, Qin XG, Zhao BS, Qi B, Yao WJ,
Wang TY, Li HC and Wu XN: Differential expression of miRNAs in
esophageal cancer tissue. Oncol Lett. 5:1639–1642. 2013.PubMed/NCBI
|
|
22
|
Zhang X, Huang L, Zhao Y and Tan W:
Downregulation of miR-130a contributes to cisplatin resistance in
ovarian cancer cells by targeting X-linked inhibitor of apoptosis
(XIAP) directly. Acta Biochim Biophys Sin. 45:995–1001. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Xu N, Shen C, Luo Y, Xia L, Xue F, Xia Q
and Zhang J: Upregulated miR-130a increases drug resistance by
regulating RUNX3 and Wnt signaling in cisplatin-treated HCC cell.
Biochem Biophys Res Commun. 425:468–472. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Huang B, Qu Z, Ong CW, Tsang YH, Xiao G,
Shapiro D, Salto-Tellez M, Ito K, Ito Y and Chen LF: RUNX3 acts as
a tumor suppressor in breast cancer by targeting estrogen receptor
α. Oncogene. 31:527–534. 2012. View Article : Google Scholar
|
|
25
|
Kim WJ, Kim EJ, Jeong P, Quan C, Kim J, Li
QL, Yang JO, Ito Y and Bae SC: RUNX3 inactivation by point
mutations and aberrant DNA methylation in bladder tumors. Cancer
Res. 65:9347–9354. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Araki K, Osaki M, Nagahama Y, Hiramatsu T,
Nakamura H, Ohgi S and Ito H: Expression of RUNX3 protein in human
lung adenocarcinoma: Implications for tumor progression and
prognosis. Cancer Sci. 96:227–231. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Li QL, Ito K, Sakakura C, Fukamachi H,
Inoue K, Chi XZ, Lee KY, Nomura S, Lee CW, Han SB, et al: Causal
relationship between the loss of RUNX3 expression and gastric
cancer. Cell. 109:113–124. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Hsu PI, Hsieh HL, Lee J, Lin LF, Chen HC,
Lu PJ and Hsiao M: Loss of RUNX3 expression correlates with
differentiation, nodal metastasis, and poor prognosis of gastric
cancer. Ann Surg Oncol. 16:1686–1694. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Kundu J, Wahab SM, Kundu JK, Choi YL,
Erkin OC, Lee HS, Park SG and Shin YK: Tob1 induces apoptosis and
inhibits proliferation, migration and invasion of gastric cancer
cells by activating Smad4 and inhibiting β-catenin signaling. Int J
Oncol. 41:839–848. 2012.PubMed/NCBI
|
|
30
|
Fukamachi H: Runx3 controls growth and
differentiation of gastric epithelial cells in mammals. Dev Growth
Differ. 48:1–13. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Cheng HC, Liu YP, Shan YS, Huang CY, Lin
FC, Lin LC, Lee L, Tsai CH, Hsiao M and Lu PJ: Loss of RUNX3
increases osteo-pontin expression and promotes cell migration in
gastric cancer. Carcinogenesis. 34:2452–2459. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Schaefer A, Jung M, Mollenkopf HJ, Wagner
I, Stephan C, Jentzmik F, Miller K, Lein M, Kristiansen G and Jung
K: Diagnostic and prognostic implications of microRNA profiling in
prostate carcinoma. Int J Cancer. 126:1166–1176. 2010.
|
|
33
|
Majid S, Dar AA, Saini S, Shahryari V,
Arora S, Zaman MS, Chang I, Yamamura S, Chiyomaru T, Fukuhara S, et
al: MicroRNA-1280 inhibits invasion and metastasis by targeting
ROCK1 in bladder cancer. PLoS One. 7:e467432012. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Gotoda T, Yamamoto H and Soetikno RM:
Endoscopic submu-cosal dissection of early gastric cancer. J
Gastroenterol. 41:929–942. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Kosuke N, Oguma H and Yamamoto M: Early
gastric cancer with lymph node metastasis. Ann Surg. 253:840–841.
2011. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Kim BS, Cho SW, Min SK and Lee BH:
Differences in prognostic factors between early and advanced
gastric cancer. Hepatogastroenterology. 58:1032–1040.
2011.PubMed/NCBI
|
|
37
|
Lai IR, Lee WJ, Huang MT and Lin HH:
Comparison of serum CA72-4, CEA, TPA, CA19-9 and CA125 levels in
gastric cancer patients and correlation with recurrence.
Hepatogastroenterology. 49:1157–1160. 2002.PubMed/NCBI
|
|
38
|
Gao F, Chang J, Wang H and Zhang G:
Potential diagnostic value of miR-155 in serum from lung
adenocarcinoma patients. Oncol Rep. 31:351–357. 2014.
|
|
39
|
Sun Y, Wang M, Lin G, Sun S, Li X, Qi J
and Li J: Serum microRNA-155 as a potential biomarker to track
disease in breast cancer. PLoS One. 7:e470032012. View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Chu Y, Ouyang Y, Wang F, Zheng A, Bai L,
Han L, Chen Y and Wang H: MicroRNA-590 promotes cervical cancer
cell growth and invasion by targeting CHL1. J Cell Biochem.
115:847–853. 2014. View Article : Google Scholar
|
|
41
|
Ma Y, Qin H and Cui Y: miR-34a targets
GAS1 to promote cell proliferation and inhibit apoptosis in
papillary thyroid carcinoma via PI3K/Akt/Bad pathway. Biochem
Biophys Res Commun. 441:958–963. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Meng S, Cao J, Zhang X, Fan Y, Fang L,
Wang C, Lv Z, Fu D and Li Y: Downregulation of microRNA-130a
contributes to endothelial progenitor cell dysfunction in diabetic
patients via its target Runx3. PLoS One. 8:e686112013. View Article : Google Scholar : PubMed/NCBI
|