Introduction
Hepatocellular carcinoma (HCC) is one of the most
common and aggressive malignant tumors worldwide and the second
leading cause of cancer-related mortality (1). Approximately 10 million HCC patients
die every year in China. To date, surgical resection and liver
transplantation are the only curative treatment options (2). However, tumor metastasis remains the
leading cause of death in HCC patients after resection (3). The proliferation and migration of HCC
cells are markedly increased by various growth factors and
cytokines such as vascular endothelial growth factor (VEGF)
(4), tumor necrosis factor-α
(TNF-α) (5) and granulocyte
colony-stimulating factor (G-CSF) (6).
VEGF is one of the most potent stimulants of
progression in several tumor types, mainly because it modulates its
target transcription factors through multiple signaling pathways
(7,8). Indeed, expression of signaling
molecules in the VEGF pathway is elevated in several cardiovascular
disorders including acute myocardial infarction, coronary artery
disease and atherosclerosis (9–11).
However, the exact mechanism(s) involved in the process by which
VEGF modulates hepatoma cell proliferation and migration remain
unclear. Thus, identification of novel molecular mechanisms,
particularly novel inhibitors, in the VEGF-dependent hepatoma cell
proliferation and migration process has enormous therapeutic
potential.
MicroRNAs (miRNAs) are RNA fragments typically
composed of only 20–22 nucleotides. They regulate target genes
variously at the RNA and/or protein levels and as a result control
downstream cellular processes including proliferation,
differentiation, and survival (12). Many lines of evidence have
implicated miRNAs in modulating hepatoma cell function by targeting
the transcriptional factors or signaling molecules involved in
tumor cell proliferation and migration such as miR-25, miR-520e and
miR-21-3p (2,13,14).
Deregulation of miRNAs expression has also been reported in
numerous human cancer types, the miRNAs functioning as tumor
suppressors in these cases (15).
Recently, miR-199a-5p was reported to be an antagonist of tumor
cell proliferation. Shi et al revealed that overexpression
of miR-199a-5p in porcine preadipocytes significantly promoted cell
proliferation while attenuating lipid deposition in subsequent
adipocytes (16). Dai et al
also demonstrated repression of sustained endoplasmic reticulum
stress and hepatocyte apoptosis by endogenous miR-199a-5p by
blocking the IRE1α-related pathway (17). Underexpression of miR-199a-5p
contributes to the rise of cell invasion in HCC via the functional
deregulation of DDR1 activity (18). Hsu et al also showed that
upregulation of miR-199a-5p suppresses cell proliferation,
motility, and angiogenesis of these ectopic stem cells by targeting
the 3′-untranslated region of VEGFA (19).
Here, we found by miRNA microarray expression
analysis that miR-199a-5p, which is abundantly expressed in normal
human liver cells, was significantly downregulated in human
hepatoma cells. On this basis, we hypothesized that miR-199a-5p
regulates tumorigenesis through some unknown pathway. We then found
that miR-199a-5p inhibits human hepatoma cell proliferation and
migration through targeting the NOR1 gene, which was
previously cloned in our laboratory in the Third Xiang Ya Hospital
of Central South University (Hunan, China) and identified as a
novel tumor suppressor gene (20).
Materials and methods
Cell source and culture
The human normal liver (THLE-3) and human hepatoma
(HepG2) cell lines were obtained from ATCC. Both cell lines were
cultured in RPMI-1640 (Gibco, USA) supplemented with 10% fetal
bovine serum (FBS) (Gibco), 10 mg/ml streptomycin and 10,000 U/ml
penicillin in a humidified atmosphere of 5% CO2 at 37°C.
The sequence of miR-199a-5p is ccc agt gtt cag act acc tgt tc, and
the anti-sequence is ggg tca caa gtc tga tgg aca ag.
Microarray analysis
Total RNAs obtained from cells were subjected to the
Mammalian miRNA Array Service V4.0 (CapitalBio Corp., Beijing,
China) and analyzed.
PCR and quantitative RT-PCR analysis
Total RNAs were extracted using TRIzol reagent
(Invitrogen, Carlsbad, CA, USA) following the manufacturer's
instructions. PCR was performed for miR-199a-5p (F_Pr:
5′-TCCAGCTGGGCCCAGTGTTCAGACTAC-3′ ; R_Pr:
5′-GTGTCGTGGAGTCGGCAATTC-3′), human NOR1 (F_Pr:
5′-TGTTAGGCTAGCGATTGAGTTATTTGCTTACAC-3′ and R_Pr:
5′-CTGCACGAATTCGGTTAAGTATGGCCCGATCTA-3′). SYBR® Green
assays (Invitrogen) was used for quantitative RT-PCR. All samples
were run in triplicate in 96-well reaction plates using the Applied
BioSystems 7300 Sequence Detection system (Perkin-Elmer Applied
Biosystems, Foster City, CA, USA). The reactions started at 95°C
for 5 min followed by 38 cycles of 95°C for 15 sec, 60°C for 30 sec
and 72°C for 30 sec. All experiments were repeated three times.
Western blotting
The anti-NOR1 rabbit monoclonal antibody
(Epitomics, CA, USA) diluted at 1:1,000, and the anti-GAPDH rat
monoclonal antibody (Beyotime, China) at 1:1,000, were used for
western blotting. Band intensities were quantified by grayscale and
analyzed.
Cell proliferation and migration
Cell proliferation was measured by the MTT assay
(MTT cell proliferation assay kit, Invitrogen™). HepG2 cells
(2×104 per well) were seeded on cell culture inserts
(8-µm pore size) (Millipore Cell, USA) with serum-free
RPMI-1640 in triplicate. Those inserts were then put into a 24-well
culture plate. Twelve hours later the medium was removed and the
inserts were washed with PBS and stained after fixation. After
rinsing with water, images were photographed in three random fields
(×400). Cell migration was tested and quantified.
Luciferase reporter assay
The pMIR-NOR1-3′-(UTR) luciferase vector
containing the putative binding site for miR-199a-5p in multiple
cloning sites in the 3′-UTR of pMIR-REPORT™ microRNA Expression
Reporter Vector (Ambion) was constructed according to the
manufacturer's instructions. HepG2 cells were plated at
2×105 cells/well in 12-well plates in triplicate. The
pMIR-NOR1-3′-UTR vector (200 ng) together with the β-gal
expressing vector pMIR-REPORT β-gal (200 ng) (Ambion) were
co-transfected with either Pre-miR™ miRNA precursor molecules or
negative control miRNA precursors (Ambion). Luciferase assays and
β-gal enzyme assays were performed 24 h after the transfection
according to the manufacturer's protocol (Promega Corp., Madison,
WI, USA). Firefly luciferase activity was normalized to β-gal
expression for each sample.
Construction of lentiviral vector
The lentiviral miR-199a-5p overexpression system
(LV-miR-199a-5p) was amplified by PCR. The two ends of miR-199a-5p
were linked with restriction enzyme cutting sites. The PCR products
of the target gene and pSMPUW-U6-Puro lentiviral expression vector
(Cell Biolabs, Inc.) were respectively digested with BamHI
and SalI. After purification by electrophoretic separation,
the target fragments were induced into competent cells followed by
incubation in Luria-Bertani broth containing ampicillin to select
overnight. Single colonies were picked as putative products. The
products of PCR amplification were identified by restriction
endonuclease digestion and sequenced.
Lentiviral packaging and titer
determination
The lentiviral vector and pRSV-Rev lentivirus
package vector (Invitrogen) were co-transfected into HepG2 cells.
Supernatants were collected by centrifugation 24 and 48 h after
transfection, respectively, for concentration tests. The lentivirus
titers at each time-point were analyzed by the hole-by-dilution
titer assay.
Lentiviral transduction
HepG2 cells were infected with lentivirus
(LV-miR-199a-5p) when they were 50–70% confluent. In our
pre-experiment, the MOI (multiplicity of infection) value gradients
were 0, 10, 50 and 100. Corresponding doses of LV-miR-199a-5p were,
respectively, added. LV-GFP was transduced as control. The green
fluorescent protein (GFP) expression levels were measured 72 h
after transduction in order to determine the optimal MOI value,
which was used in subsequent experiments.
Statistical analysis
All results are presented as means ± SEM of at least
three independent experiments, unless otherwise indicated.
Student's t-test was used to assess differences between two groups.
P<0.05 was considered statistically significant.
Results
The expression levels of miRNAs in HepG2
and THLE-3
To identify miRNAs that contribute to the
proliferation of liver cells, we performed microarray analysis of
miRNA expression in a normal human liver cell line (THLE-3) and a
human hepatoma cell line (HepG2). There were significant
differences between the two cell lines in the expression of 149
miRNAs, 58 of which were upregulated and 91 downregulated.
Consistent with previous reports, miR-let-7e was significantly
downregulated in the cancer cells (21). miRNAs with significantly altered
expression levels are listed in Table
I; they include miR-199a-5p, which was significantly
downregulated in the hepatoma cells.
 | Table IThe expression levels of miRNAs in
HepG2 and THLE-3 cells. |
Table I
The expression levels of miRNAs in
HepG2 and THLE-3 cells.
| miRNA name | score(d) | Fold change |
|---|
| miR-199a-5p | −61.3854 | 0.16945 |
| miR-18b | −54.7323 | 0.11342 |
| miR-let-7e | −43.3242 | 0.12435 |
| miR-193a | −36.3287 | 0.08793 |
| miR-224 | −28.6543 | 0.15276 |
| miR-28 | −25.9657 | 0.25738 |
| miR-19a | −20.1985 | 0.09486 |
| miR-434 | −16.9874 | 0.28677 |
| miR-122a | 94.3249 | 28.43587 |
| miR-422b | 85.4535 | 25.45325 |
| miR-520e | 70.45278 | 23.97742 |
| miR-134 | 63.32455 | 14.554325 |
| miR-198 | 43.31445 | 8.43529 |
| miR-202 | 28.45289 | 3.23455 |
| miR-382 | 20.35185 | 2.55325 |
| miR-520b | 13.31457 | 1.98097 |
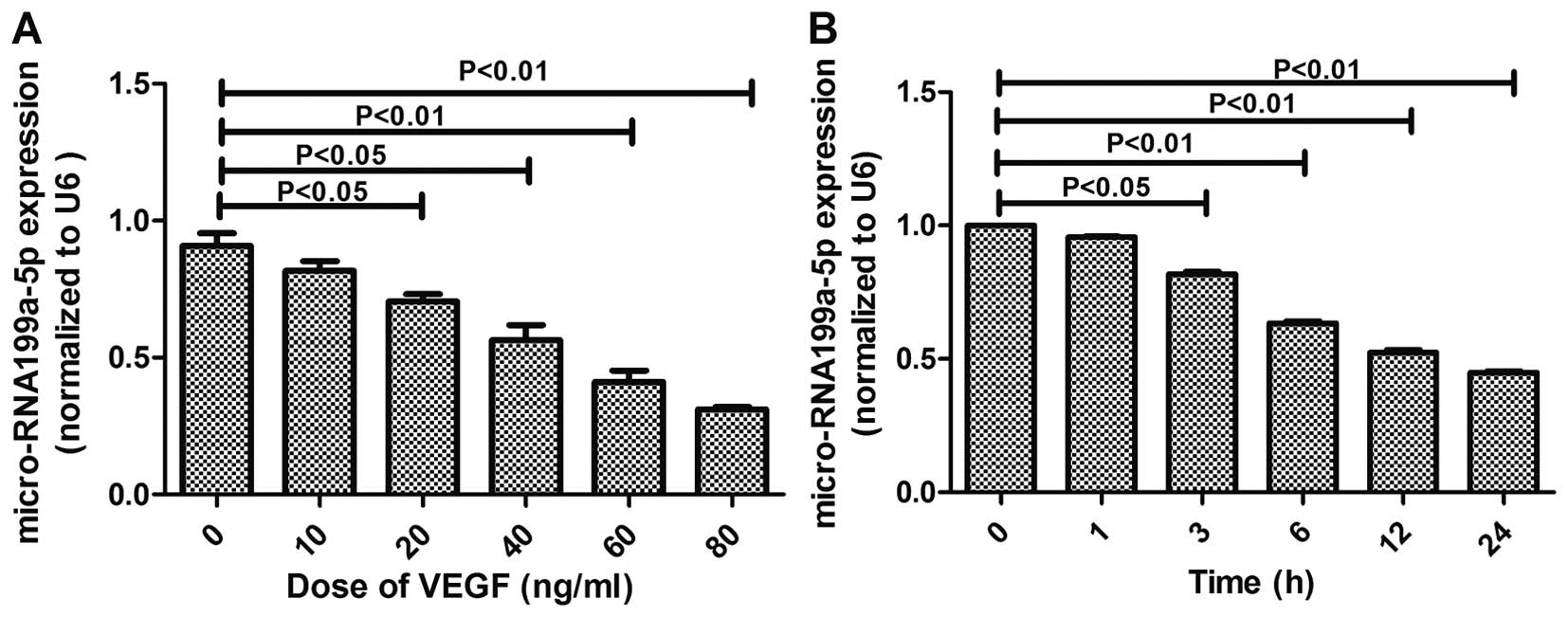
miR-199a-5p expression was attenuated in
VEGF-induced HepG2 cells
VEGF contributes to some key stages of
tumorigenesis, including the function of cancer stem cells and
tumor initiation (22). To confirm
the importance of miR-199a-5p in regulating tumorigenesis, we
performed miRNA microarray analysis in HepG2 cells at 0, 3, 6, and
24 h after VEGF stimulation with an IC50 of 60 ng/ml
(Fig. 1B).
The relative expression levels of miR-199a-5p were
normalized to U6, the endogenous control. The results showed that
VEGF stimulation on HepG2 cells caused a significant dose-dependent
reduction in the expression levels of miR-199a (Fig. 1A). These findings revealed that
miR-199a and VEGF expressions are negatively correlated.
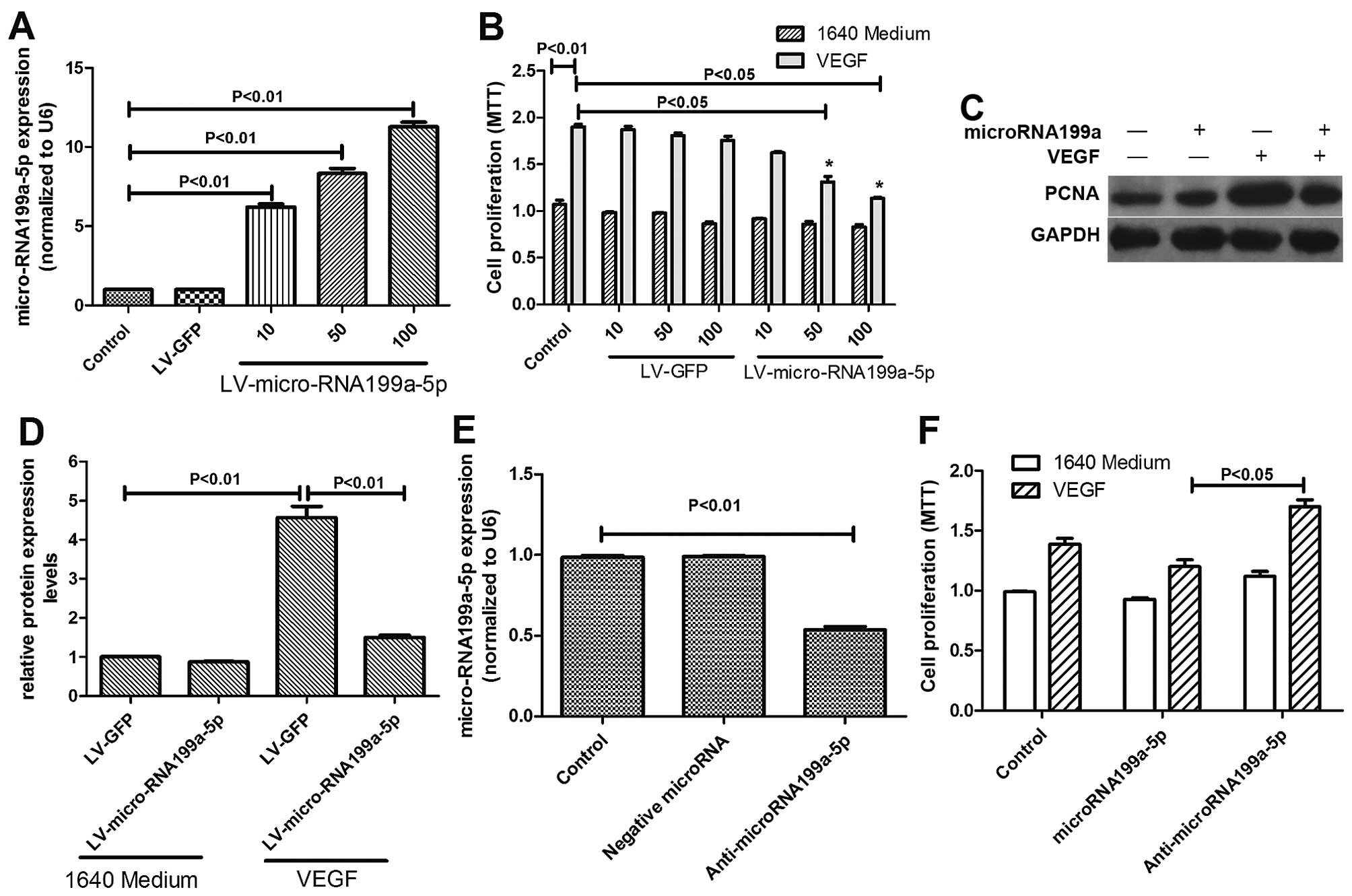
Effects of micro-RNA199a-5p and VEGF on
HepG2 proliferation
To examine the effects of microRNA199a-5p in HepG2
cells further, we examined whether its restoration would affect
VEGF-induced cell proliferation. We constructed a lentiviral vector
expressing miR-199a-5p (LV-miR-199a-5p) and showed that
transductions with increasing multiplicity of infections (MOI) of
LV-miR-199a-5p significantly elevated miR-199a-5p expression in
HepG2 cells (Fig. 2A). Forced
expression of miR-199a-5p by LV-miR-199a-5p strongly inhibited cell
proliferation over the range 10-100 MOI (Fig. 2B). The effects of miR-199a-5p on
HepG2 cell proliferation were further elucidated by measuring the
expression of PCNA, a marker of tumor cell proliferation. VEGF
significantly increased PCNA expression, but this was markedly
attenuated in miR-199a-5p-overexpressing cells (Fig. 2C and D), while miR-199a-5p
knock-down by anti-miR-199a-5p increased proliferation (Fig. 2E and F).
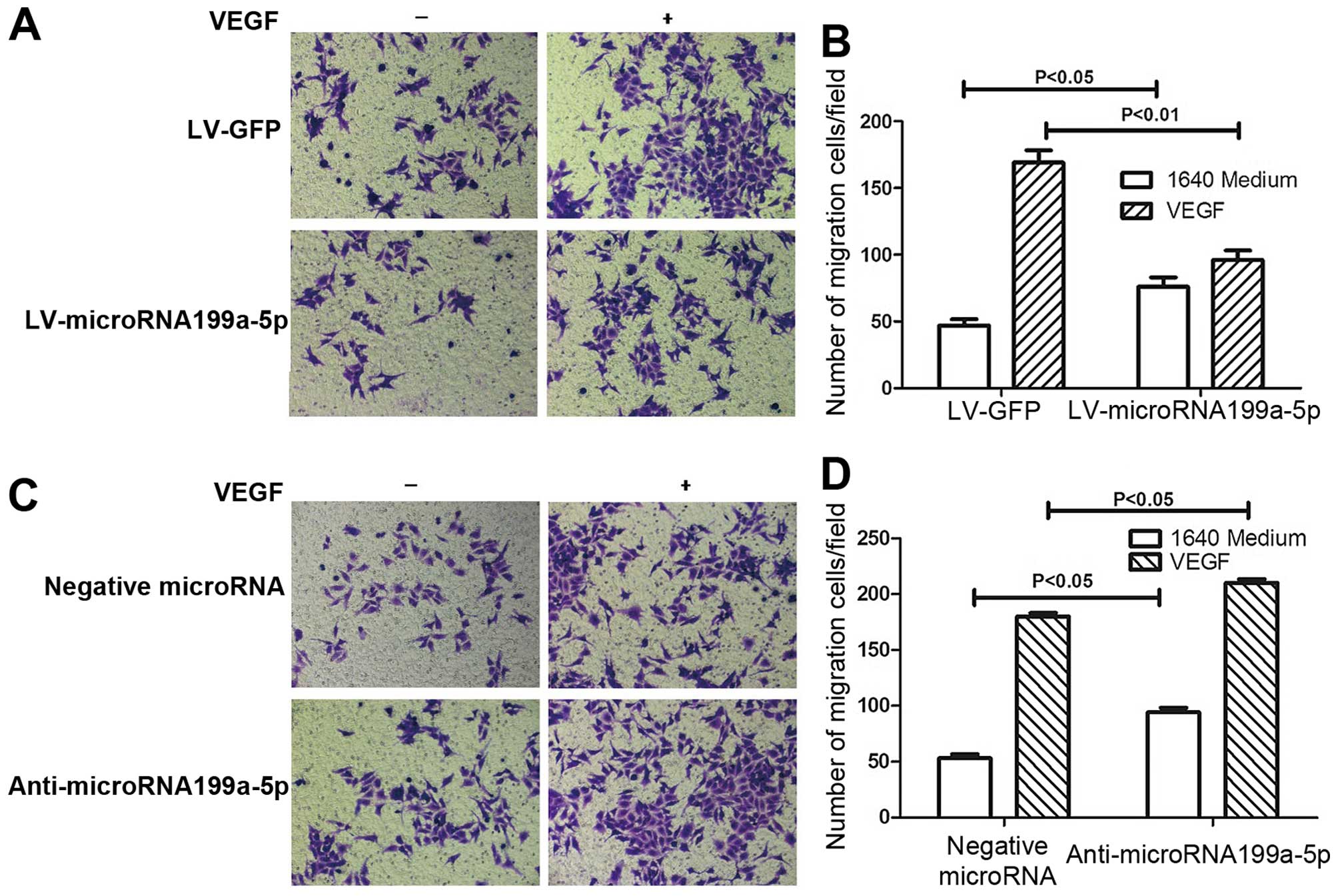
Effects of micro-RNA199a and VEGF on
hepatoma cell migration
Hepatoma cell migration is also thought to make an
important contribution to the progression of HCC. To determine
whether miR-199a-5p is also involved in this process, we performed
cell migration experiments, and the results verified that
miR-199a-5p suppresses tumorigenesis. miR-199a inhibited cell
migration induced by VEGF overexpression (Fig. 3A and B). Moreover, consistent with
our hypothesis, the inhibition of miR-199a expression by
anti-miR-199a restored the capacity of HepG2 cells for migration
(Fig. 3C and D). These results
suggest that miR-199a modulates tumorigenesis by changing key
factors in the tumor microenvironment.
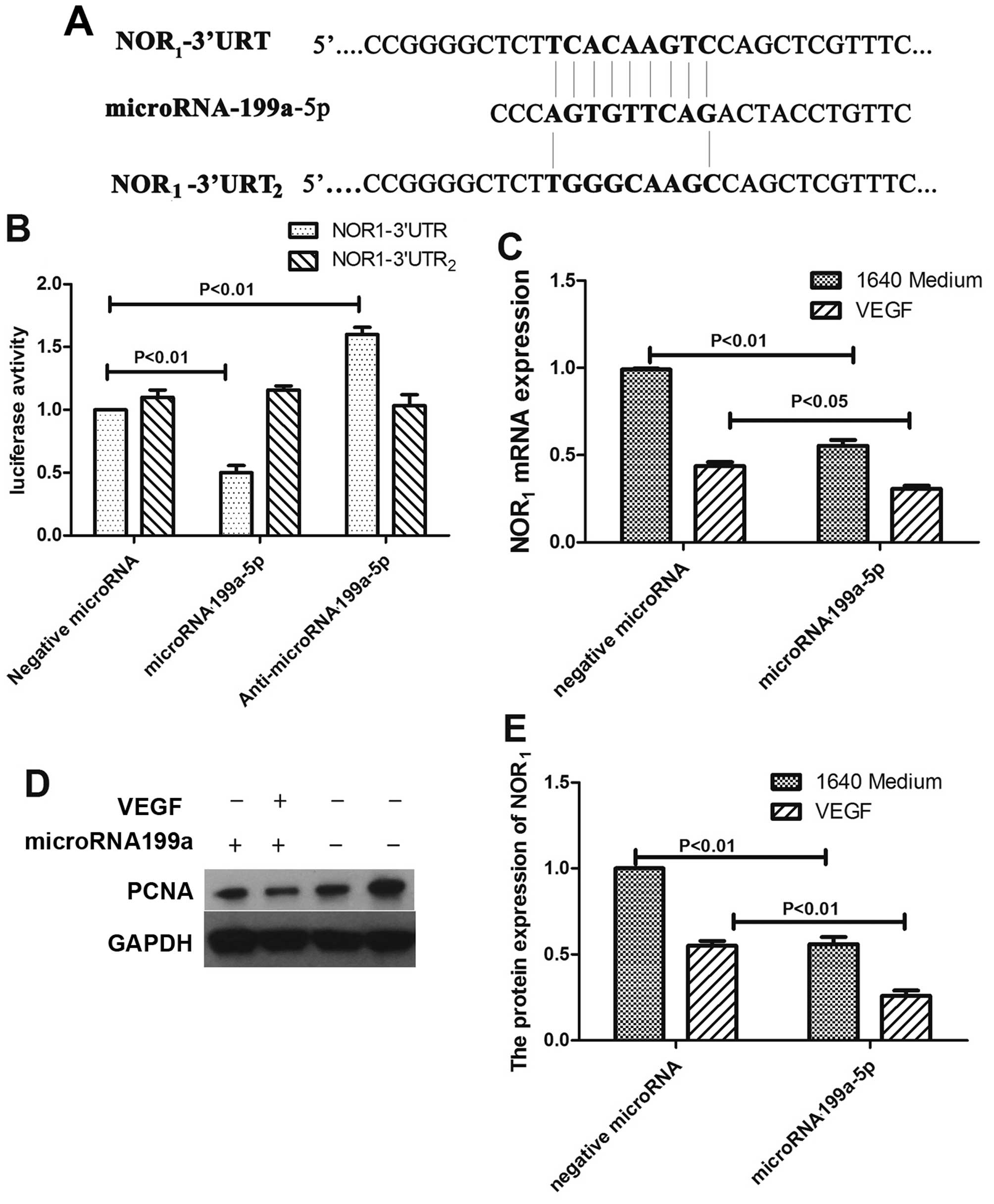
NOR1 is a direct target of
miR-199a-5p
Our previous study revealed NOR1 as a
candidate tumor suppressor that inhibits the development and/or
progression of tumors (23). In
this study, when we searched the target scan database, we found
that NOR1 is a potential target of miR-199a-5p. As shown
in Fig. 4A, human NOR1
mRNA has a potential miR-199a-5p binding site in its 3′-UTR. To
verify whether miR-199a-5p binds directly to the 3′-UTR sequence of
NOR1 mRNA, down-regulating its expression, the 3′-UTR
sequence containing the putative binding was cloned into an
assay-ready luc-UTR reporter vector. The constructed vector was
then co-transfected with either miR-199a-5p or control plasmid into
HepG2 cells. As shown in Fig. 4B,
the luciferase activity was inhibited in the miR-199a-5p-induced
cells but not in the control group. To verify that NOR1
is a functional target gene of miR-199a-5p, we induced miR-199a-5p
into HepG2 cells and found that NOR1 expression was
decreased at both the mRNA and protein levels despite VEGF
treatment (Fig. 4C–E). We therefore
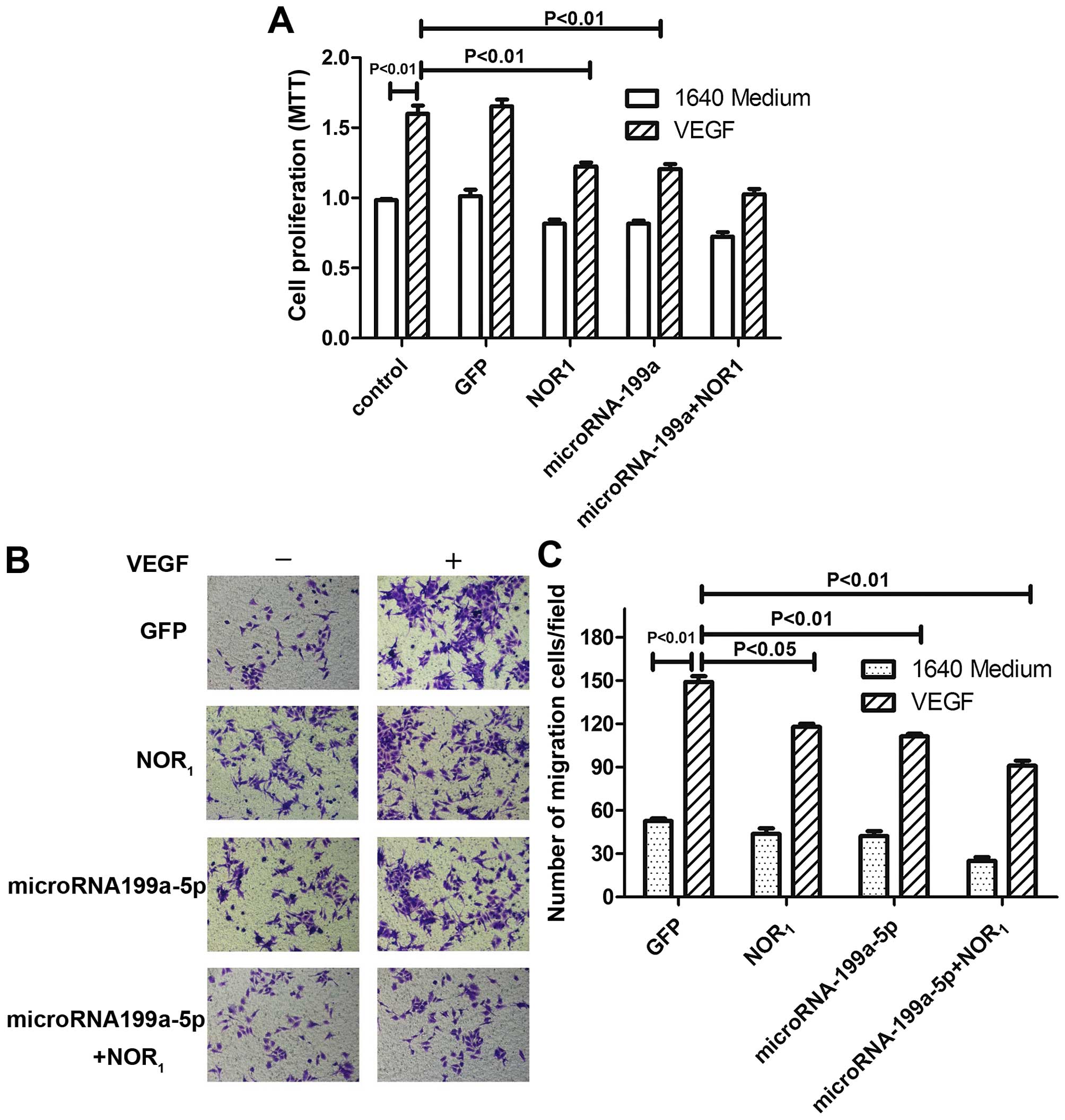
measured the proliferation and migration of HepG2 cells treated
with NOR1 to test its role in the inhibition of
tumorigenesis by miR-199a-5p. The results indicate that
overexpression of NOR1 significantly inhibits the
proliferation (Fig. 5A) and
migration (Fig. 5B and C) of HepG2
cells.
Discussion
miR-199a-5p has been shown to have a wide range of
functions and can behave differently in different systems and
diseases. For example, it is a major regulator of lung fibroblasts
and livers in injured tissues (24,25).
Rapid downregulation of miR-199a-5p is required for upregulation of
HIF1α to protect cells exposed to hypoxia (26). Its behavior in tumors could be more
complicated. In different contexts it can function either as an
oncogene (27,28) or as a tumor suppressor (29,30).
In our previous study, miR-199a-5p was confirmed as a tumor
suppressor in hepatocellular carcinoma. However, details of the
mechanism by which it regulates tumorigenesis remained unclear. In
our present study, we demonstrated that it inhibits HepG2 cell
proliferation and migration through targeting NOR1. We
also found that miR-199a-5p expression in hepatocellular carcinoma
was markedly lower than in normal liver cells because it was
downregulated both time- and dose-dependently by VEGF stimulation,
which induces the deterioration of hepatocellular carcinoma.
Restoration of miR-199a-5p markedly attenuated VEGF-induced HepG2
cell proliferation and migration, and knock-down of miR-199a-5p by
anti-miR-199a-5p had the opposite effect on tumorigenesis.
MicroRNAs are highly conserved small regulatory RNAs
that antagonize the expression of target genes by hybridizing to
specific binding sites in the 3′-untranslated regions (UTR) of many
mRNAs (31). Upon microRNA-guided
recruitment of a multi-protein complex, either the target mRNA is
degraded directly or its translation is blocked, depending on the
complementarity between the microRNA and its binding site (32). In our study, we confirmed that
miR-199a-5p inhibited cell proliferation and migration via
NOR1 by binding directly to the 3′-UTR of its mRNA.
NOR1, the gene for which is located in a 120 kb region
at 1p34.3, was first isolated from nasopharyngeal cells (NPCs)
(23). Zeng et al showed
that NOR1 mRNA expression was frequently downregulated
in NPCs (33). This epigenetic
silencing of NOR1 impaired the cellular protective
response to environmental stresses by normal NPCs and promoted
nasopharyngeal carcinogenesis (34). Our previous study confirmed
NOR1 as a tumor suppressor, the expression of which was
significantly downregulated in nasopharyngeal carcinoma,
hepatocellular carcinoma and gastric cancer (20). Previously, regulation of
NOR1 expression by microRNAs had not been demonstrated.
In this study, we showed that miR-199a-5p binds directly to the
3′-UTR of human NOR1 mRNA and downregulates
NOR1 expression at both the mRNA and protein levels with
or without VEGF stimulation. Overexpression of NOR1
strongly inhibited the proliferation and migration of HepG2 cells,
i.e., miR-199a-5p is a novel post-transcriptional regulator of
NOR1.
In conclusion, this study identified miR-199a-5p as
a potential regulator of human HepG2 cells that acts by targeting
the 3′-UTR of NOR1 mRNA. miR-199a-5p expression was
substantially downregulated in human hepatocellular carcinoma but
not in normal liver cells. Re-establishment of its expression
markedly inhibited both cell proliferation and migration in
response to VEGF stimulation. Our study provides novel insight into
the molecular mechanisms associated with tumorigenesis, and
suggests a potential therapeutic target for human HCC
treatment.
In conclusion, this study showed that miR-199a-5p
antagonizes tumorigenesis owing to its inhibitory role in cell
proliferation and migration. Through binding to the 3′-UTR of human
NOR1 mRNA, miR-199a-5p enhances NOR1
expression, and then NOR1 inhibits cell proliferation
and migration induced by VEGF.
Acknowledgments
This study was supported by National Natural Science
Foundation of China (NSFC) (grant nos. 81573091, 30300383, 81072270
and 81101828), and Fundamental Research Funds for the Central
Universities (no. 2011JQ030).
References
|
1
|
Jemal A, Bray F, Center MM, Ferlay J, Ward
E and Forman D: Global cancer statistics. CA Cancer J Clin.
61:69–90. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Su ZX, Zhao J, Rong ZH, Geng WM, Wu YG and
Qin CK: Upregulation of microRNA-25 associates with prognosis in
hepatocellular carcinoma. Diagn Pathol. 9:472014. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Li S, Dong P, Wang J, Zhang J, Gu J, Wu X,
Wu W, Fei X, Zhang Z, Wang Y, et al: Icariin, a natural flavonol
glycoside, induces apoptosis in human hepatoma SMMC-7721 cells via
a ROS/JNK-dependent mitochondrial pathway. Cancer Lett.
298:222–230. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Zhan P, Qian Q and Yu LK: Serum VEGF level
is associated with the outcome of patients with hepatocellular
carcinoma: A meta-analysis. Hepatobiliary Surg Nutr. 2:209–215.
2013.
|
|
5
|
Cao X, Zhang L, Shi Y, Sun Y, Dai S, Guo
C, Zhu F, Wang Q, Wang J, Wang X, et al: Human tumor necrosis
factor (TNF)-alpha-induced protein 8-like 2 suppresses
hepatocellular carcinoma metastasis through inhibiting Rac1. Mol
Cancer. 12:1492013. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Miyahara K, Nouso K, Morimoto Y, Takeuchi
Y, Hagihara H, Kuwaki K, Onishi H, Ikeda F, Miyake Y, Nakamura S,
et al Okayama Liver Cancer Group: Pro-angiogenic cytokines for
prediction of outcomes in patients with advanced hepatocellular
carcinoma. Br J Cancer. 109:2072–2078. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Duong T, Koltowska K, Pichol-Thievend C,
Le Guen L, Fontaine F, Smith KA, Truong V, Skoczylas R, Stacker SA,
Achen MG, et al: VEGFD regulates blood vascular development by
modulating SOX18 activity. Blood. 123:1102–1112. 2014. View Article : Google Scholar
|
|
8
|
Ryzhov S, Biktasova A, Goldstein AE, Zhang
Q, Biaggioni I, Dikov MM and Feoktistov I: Role of JunB in
adenosine A2B receptor-mediated vascular endothelial growth factor
production. Mol Pharmacol. 85:62–73. 2014. View Article : Google Scholar :
|
|
9
|
Yao HC, Liu T, Meng XY, Han QF, Zhang M
and Wang LX: Effect of basic fibroblast growth factor on the
myocardial expression of hypoxia-inducible factor-1α and vascular
endothelial growth factor following acute myocardial infarction.
Heart Lung Circ. 22:946–951. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Cui QT, Li Y, Duan CH, Zhang W and Guo XL:
Further evidence for the contribution of the vascular endothelial
growth factor gene in coronary artery disease susceptibility. Gene.
521:217–221. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Winnik S, Lohmann C, Siciliani G, von
Lukowicz T, Kuschnerus K, Kraenkel N, Brokopp CE, Enseleit F,
Michels S, Ruschitzka F, et al: Systemic VEGF inhibition
accelerates experimental atherosclerosis and disrupts endothelial
homeostasis - implications for cardiovascular safety. Int J
Cardiol. 168:2453–2461. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Ambros V: The functions of animal
microRNAs. Nature. 431:350–355. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Lo TF, Tsai WC and Chen ST:
MicroRNA-21-3p, a berberine-induced miRNA, directly down-regulates
human methionine adenosyltransferases 2A and 2B and inhibits
hepatoma cell growth. PLoS One. 8:e756282013. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Li BA: A novel tumor suppressor miRNA
miR-520e contributes to suppression of hepatoma. Acta Pharmacol
Sin. 33:3–4. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Iorio MV and Croce CM: microRNA
involvement in human cancer. Carcinogenesis. 33:1126–1133. 2012.
View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Shi XE, Li YF, Jia L, Ji HL, Song ZY,
Cheng J, Wu GF, Song CC, Zhang QL, Zhu JY, et al: MicroRNA-199a-5p
affects porcine preadipocyte proliferation and differentiation. Int
J Mol Sci. 15:8526–8538. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Dai BH, Geng L, Wang Y, Sui CJ, Xie F,
Shen RX, Shen WF and Yang JM: microRNA-199a-5p protects hepatocytes
from bile acid-induced sustained endoplasmic reticulum stress. Cell
Death Dis. 4:e6042013. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Shen Q, Cicinnati VR, Zhang X, Iacob S,
Weber F, Sotiropoulos GC, Radtke A, Lu M, Paul A, Gerken G, et al:
Role of microRNA-199a-5p and discoidin domain receptor 1 in human
hepatocellular carcinoma invasion. Mol Cancer. 9:2272010.
View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Hsu CY, Hsieh TH, Tsai CF, Tsai HP, Chen
HS, Chang Y, Chuang HY, Lee JN, Hsu YL and Tsai EM: miRNA-199a-5p
regulates VEGFA in endometrial mesenchymal stem cells and
contributes to the pathogenesis of endometriosis. J Pathol.
232:330–343. 2014. View Article : Google Scholar
|
|
20
|
Gui R, Li D, Qi G, Suhad A and Nie X:
Inhibition of Grb2-mediated activation of MAPK signal transduction
suppresses NOR1/CB1954-induced cytotoxicity in the HepG2 cell line.
Oncol Lett. 4:566–570. 2012.
|
|
21
|
Cai J, Yang C, Yang Q, Ding H, Jia J, Guo
J, Wang J and Wang Z: Deregulation of let-7e in epithelial ovarian
cancer promotes the development of resistance to cisplatin.
Oncogenesis. 2:e752013. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Goel HL and Mercurio AM: VEGF targets the
tumour cell. Nat Rev Cancer. 13:871–882. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Nie X, Zhang B, Li X, Xiang J, Xiao B, Ma
J, Zhou M, Zhu S, Lu H, Gui R, et al: Cloning, expression, and
mutation analysis of NOR1, a novel human gene down-regulated in
HNE1 naso-pharyngeal carcinoma cell line. J Cancer Res Clin Oncol.
129:410–414. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Lino Cardenas CL, Henaoui IS, Courcot E,
Roderburg C, Cauffiez C, Aubert S, Copin MC, Wallaert B, Glowacki
F, Dewaeles E, et al: miR-199a-5p Is upregulated during fibrogenic
response to tissue injury and mediates TGFbeta-induced lung
fibroblast activation by targeting caveolin-1. PLoS Genet.
9:e10032912013. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Ogawa T, Enomoto M, Fujii H, Sekiya Y,
Yoshizato K, Ikeda K and Kawada N: MicroRNA-221/222 upregulation
indicates the activation of stellate cells and the progression of
liver fibrosis. Gut. 61:1600–1609. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Sayed D and Abdellatif M: AKT-ing via
microRNA. Cell Cycle. 9:3213–3217. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Magrelli A, Azzalin G, Salvatore M,
Viganotti M, Tosto F, Colombo T, Devito R, Di Masi A, Antoccia A,
Lorenzetti S, et al: Altered microRNA expression patterns in
hepatoblastoma patients. Transl Oncol. 2:157–163. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Pencheva N, Tran H, Buss C, Huh D,
Drobnjak M, Busam K and Tavazoie SF: Convergent multi-miRNA
targeting of ApoE drives LRP1/LRP8-dependent melanoma metastasis
and angiogenesis. Cell. 151:1068–1082. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Cheung HH, Davis AJ, Lee TL, Pang AL,
Nagrani S, Rennert OM and Chan WY: Methylation of an intronic
region regulates miR-199a in testicular tumor malignancy. Oncogene.
30:3404–3415. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Wang F, Zheng Z, Guo J and Ding X:
Correlation and quantitation of microRNA aberrant expression in
tissues and sera from patients with breast tumor. Gynecol Oncol.
119:586–593. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Augustin R, Endres K, Reinhardt S, Kuhn
PH, Lichtenthaler SF, Hansen J, Wurst W and Trümbach D:
Computational identification and experimental validation of
microRNAs binding to the Alzheimer-related gene ADAM10. BMC Med
Genet. 13:352012. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Scherr M, Venturini L and Eder M:
Lentiviral vector-mediated expression of pre-miRNAs and antagomiRs.
Methods Mol Biol. 614:175–185. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Zeng Z, Zhou Y, Xiong W, Luo X, Zhang W,
Li X, Fan S, Cao L, Tang K, Wu M, et al: Analysis of gene
expression identifies candidate molecular markers in nasopharyngeal
carcinoma using microdissection and cDNA microarray. J Cancer Res
Clin Oncol. 133:71–81. 2007. View Article : Google Scholar
|
|
34
|
Li W, Li X, Wang W, Li X, Tan Y, Yi M,
Yang J, McCarthy JB, Xiong W, Wu M, et al: NOR1 is an HSF1- and
NRF1-regulated putative tumor suppressor inactivated by promoter
hyper-methylation in nasopharyngeal carcinoma. Carcinogenesis.
32:1305–1314. 2011. View Article : Google Scholar : PubMed/NCBI
|