Introduction
Neuroblastoma (NB) is one of the most common
malignant tumors in children (1).
In the US, NB is the third most common cause of cancer-related
mortality (~6%) among children aged 1–14 years (2). The subtypes of NB show great
heterogeneity in regard to genetic abnormalities, which are closely
associated with clinical outcomes among NB patients (3). For example, in patients presenting
with the MYC-related oncogene (MYCN) (amplified
subtype of NB), the MYCN gene is aberrantly repeated at
chromosome 2p24, resulting in NB progression to advanced stages,
aggressive metastasis and poor patient prognosis (3–5).
Unfortunately, the underlying molecular mechanisms contributing to
NB pathogenesis, metastasis and apoptosis are large unknown. It,
thus, presents a great challenge to identifying novel biomarkers or
therapeutic targets for early detection or treatment for young
patients with NB.
MicroRNAs (miRNAs) are families of highly conserved
non-coding small RNAs that attach to the 3′-untranslated region
(3′-UTR) of downstream target genes to post-transcriptionally
suppress gene expression, thus regulating various cellular
processes in both animals and human diseases (6). miRNAs have been shown to play critical
roles in NB pathogenesis, metastasis and apoptosis (7,8). Among
many of the oncogenic or tumor-suppressor miRNAs, miR-141 is highly
expressed in the circulating plasma in patients with advanced
stages of metastatic colon cancer, suggesting an oncogenic role as
a colon cancer biomarker (9). In
contrast, miR-141 was shown to be downregulated in gastric and
prostate cancer, presumably acting as a tumor-suppressor of cancer
progression and metastasis (10,11).
In NB, although a previous study showed that miR-141 is
differentially expressed among NB subtypes (12), the exact expression profiles or
mechanisms of miR-141 in NB remain elusive.
The fused in sarcoma (FUS) gene, which encodes an
RNA binding protein, is associated with genetic disorders
particularly neurodegenerative diseases such as amyotrophic lateral
sclerosis (ALS) (13,14). In human prostate cancer, FUS is
shown to be a tumor-suppressor gene as its overexpression inhibited
cancer growth and promoted cancer apoptosis (15). In a recent study, FUS was reported
to be highly expressed in NB SK-N-AS cells (16). However, similar to miR-141, the
exact expression and function of FUS in human NB are largely
unknown.
In the present study, we firstly evaluated the
expression of miR-141 in both MYCN- and
non-MYCN-amplified NB cell lines. We then upregulated
miR-141 in two NB cell lines IMR-32 and SH-SY5Y in to evaluate the
possible tumor-suppressive role of miR-141 on NB proliferation,
cell cycle progression, metastasis and chemosensitivity. In
addition, we hypothesized that the FUS gene is the downstream
target of miR-141, and is thereby inversely associated with miR-141
regulation in NB. To test this hypothesis, we utilized
siRNA-mediated FUS downregulation to evaluate its effect on NB
proliferation, cell cycle progression, metastasis and
chemosensitivity.
Materials and methods
Cell culture
Three human NB cell lines used in the present study,
IMR-32, SH-SY5Y and S-K-NAS, were purchased from the American Type
Culture Collection (ATCC; Manassas, VA, USA). Another three human
NB cell lines, NB-1691, LAN-5 and LAN-6, were purchased from the
Cell Bank of the Type Culture Collection of the Chinese Academy of
Sciences (Shanghai, China). The control cell line, human
retinal-pigmented epithelial cells immortalized with telom-erase
reverse transcriptase (hTERT-RPE1) was purchased from Clontech
(USA). All cells were maintained in RPMI-1640 medium supplemented
with 10% fetal bovine serum and penicillin/streptomycin (pen/strep)
(100 U/ml pen and 100 mg/ml strep) (all from Invitrogen, USA) in a
humidified chamber with 5% CO2 at 37°C.
RNA extraction and quantitative real-time
PCR
Total RNA was isolated from the NB cells using an
RNeasy Mini kit (Qiagen, USA) according to the manufacturer's
protocol. From each sample, 1 µg RNA was used for reverse
transcriptions using a Transcriptor First Strand cDNA Synthesis kit
(Roche, USA) according to the manufacturer's protocol. Quantitative
real-time PCR (qRT-PCR) was carried out in two manners. For miR-141
detection, a TaqMan miRNA assay (Applied Biosystems, USA) was
carried out with U6 snRNA as endogenous control. For FUS detection,
a Brilliant II SYBR-Green qPCR Master Mix (Stratagene, USA) was
used with glyceraldehyde-3-phosphate dehydrogenase (GAPDH) as the
endogenous control. Relative gene expression levels were calculated
using the 2−∆∆Ct method.
miR-141 overexpression assay
The forced overexpression of miR-141 in NB cell
lines was achieved by lentiviral transduction. The miR-141 mimic
lentivirus (miR-141-mimics), and its corresponding control miRNA
lentivirus (C-miRNA) were purchased from SunBio Medical
Biotechnology (Shanghai, China). In the NB culture, IMR-32 and
SH-SY5Y cells were transduced with 250 pmol lentiviruses for 48 h
in the presence of 8 µg/ml Polybrene (Sigma-Aldrich, USA)
and a multiplicity of infection (MOI) of ~20. Subsequently, the
cells were briefly washed with phosphate-buffered saline (PBS) and
continuously maintained in fresh culture medium for 48 or 72 h to
stabilize the lentivirus transduction. The floating cells were then
removed. The healthy cells were then subject to qRT-PCR examination
to verify the efficacy of the overexpression assay.
Proliferation assay
After lentiviral transduction or siRNA transfection,
the IMR-32 and SH-SY5Y cells were seeded in 96-well plates
(3.5×103/well) and maintained for 5 days. The
proliferation of NB cells was characterized using a
3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide (MTT)
assay (Sigma-Aldrich) according to the manufacturer's protocol.
Briefly, every 24 h during the proliferation assay, each well of
96-well plates was added with 200 µl MTT for 2 h, then
treated with 150 µl dimethyl sulfoxide (DMSO) for another 3
h. The optical density at 570 nm was measured using an
ultraviolet-visible spectrophotometer (Puxi Scientific Instruments,
Beijing, China).
Cell cycle assay
To analyze cell cycle distribution, IMR-32 and
SH-SY5Y cells (1×106) were quickly fixed by methanol
(4°C) and treated with 50 µg/ml propidium iodide (PI)
(Thermo Fisher Scientific, USA) for 20 min. A FACSCalibur™ flow
cytometer (Thermo Fisher Scientific) was used to obtain the cell
cycle histograms. MultiCycle AV software (Phoenix Flow System, USA)
was then used to calculate the percentages of cells in the G0/G1, S
or G2/M phases.
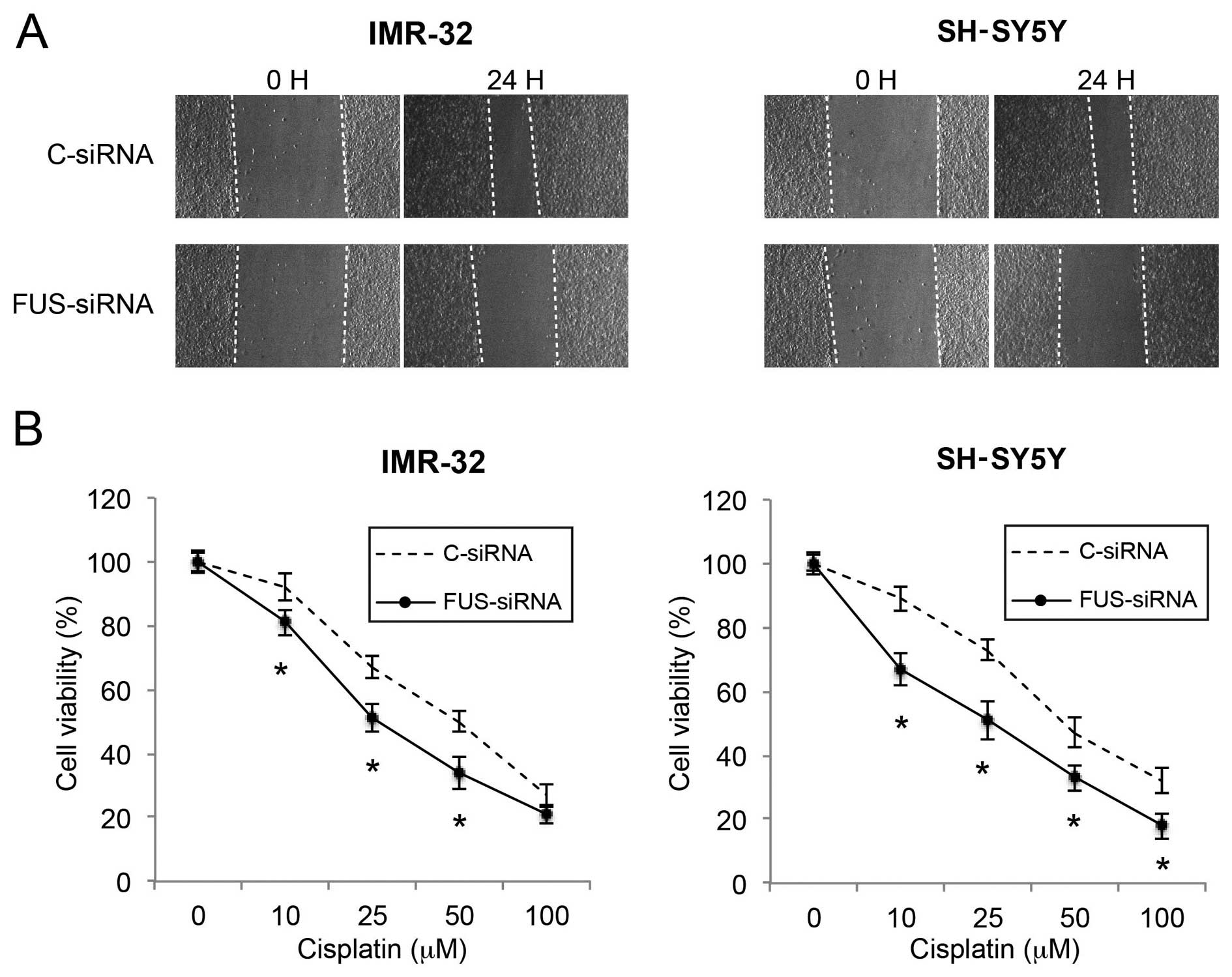
Wound healing assay
IMR-32 and SH-SY5Y cells were seeded into 12-well
plates (7.5×105/well). A sterile 1,000-µl pipette
tip was used to create the wound along the diameter of the well.
The detached cells were removed, and NB cultures were maintained
for 24 h. Phase contrast images aimed at the center of the wound
were captured at 0 and 24 h after wound creation.
Cisplatin sensitivity assay
IMR-32 and SH-SY5Y cells were seeded in 96-well
plates (7.5×103/well). Various concentrations of
cisplatin (0, 10, 25, 50 and 100 µM) were added into the NB
culture for 24 h. Cell survival was estimated through an MTT
assay.
In vivo tumor growth assay
To evaluate the in vivo growth of NB tumors,
2-month-old female nude mice were subcutaneously inoculated in the
right flank with either miR- 141-mimic- or C-miRNA-transduced
IMR-132 cells (1×106). The lengths and widths of the
subcutaneous tumors were measured weekly, and the tumor volume (V)
was calculated using the formula: V = length × width2/2.
Five weeks later, the mice were sacrificed. Subcutaneous tumors
were extracted and formalin-fixed and paraffin-embedded sections
were prepared. Immunohistochemistry was then performed using an
anti-Ki-67 antibody (Cell Signaling, USA).
Luciferase reporter assay
Wild-type (WT) human FUS 3′-UTR and mutated (MU) FUS
3′-UTR (with a MU sequence on the miR-141 binding site) were
amplified from a human brain cDNA library and inserted between the
XhoI/NotI restrictive sites of a
firefly/Renilla luciferase reporter pmiR-REPORT (RiboBio,
Guangzhou, China). Human HEK293T cells were co-transfected with 25
ng/ml of either the luciferase reporter with WT FUS 3′-UTR
(WT-3UTR), or the luciferase reporter with MU FUS 3′-UTR (MU-3UTR),
and 100 pmol of either miR-141-mimics or C-miRNA. Forty-eight hours
after co-transfection, a Dual-Luciferase Reporter Assay (Promega,
Madison, WI, USA) was carried out according to the manufacturer's
protocol. For each measurement, the relative firefly luciferase
activity was normalized to Renilla with transfection of
C-miRNA.
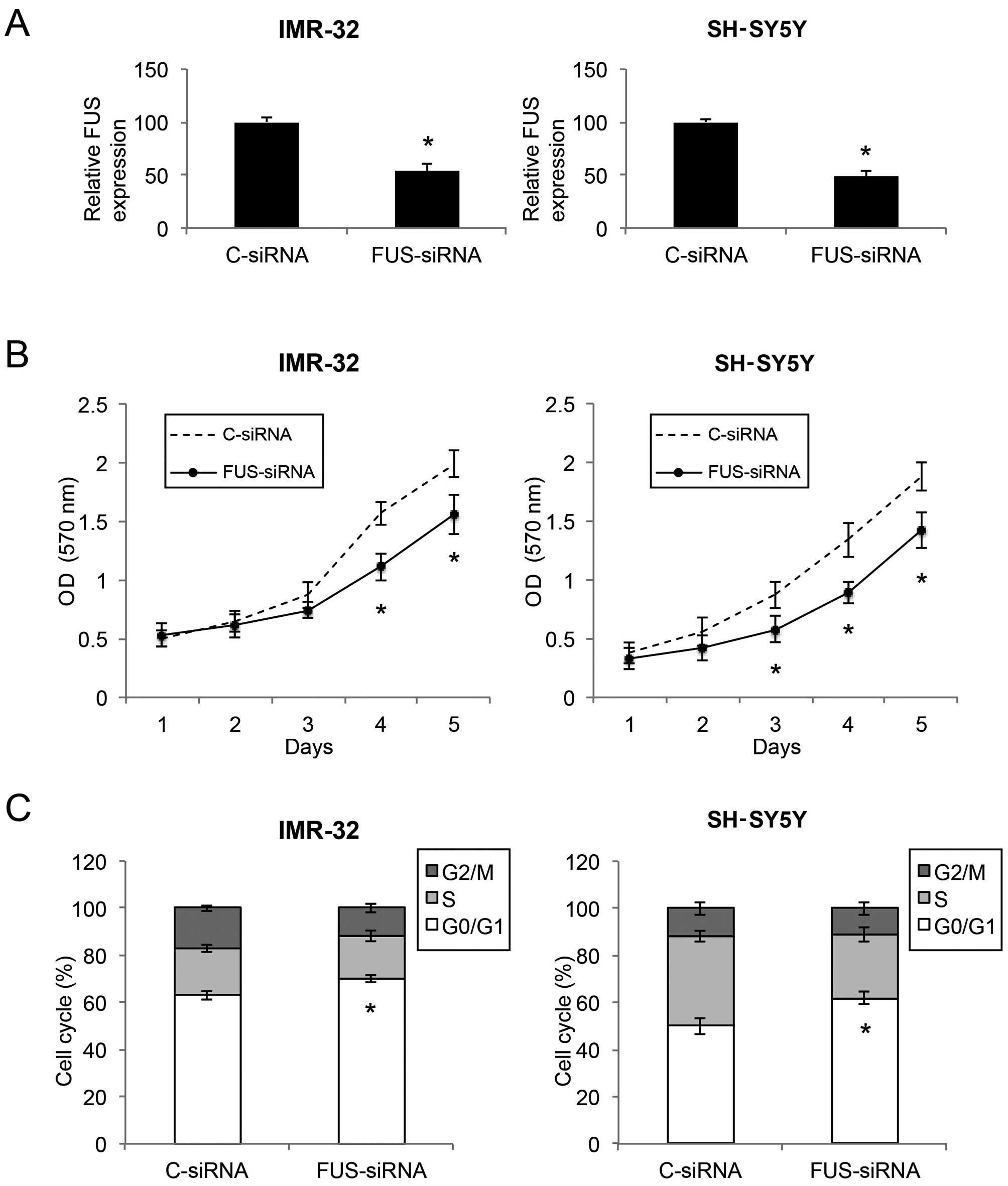
FUS downregulation assay
A small-interfering RNA (siRNA) against the human
FUS gene (FUS-siRNA), and its control siRNA (C-siRNA) were
purchased from RiboBio. In the NB culture, IMR-32 and SH-SY5Y cells
were transfected with 100 nM FUS-siRNA or C-siRNA. Forty-eight
hours after transfection, healthy cells were subject to qRT-PCR
examination to verify the efficacy of the downregulation.
Statistical analysis
All experiments were carried out in biological
triplicates. The results are presented as mean ± standard error.
SPSS 11.0 software (SPSS, Inc., Chicago, IL, USA) was applied for
statistical analysis, and the Student's t-test was performed to
compare the results. A statistically significant difference was
defined as P<0.05.
Results
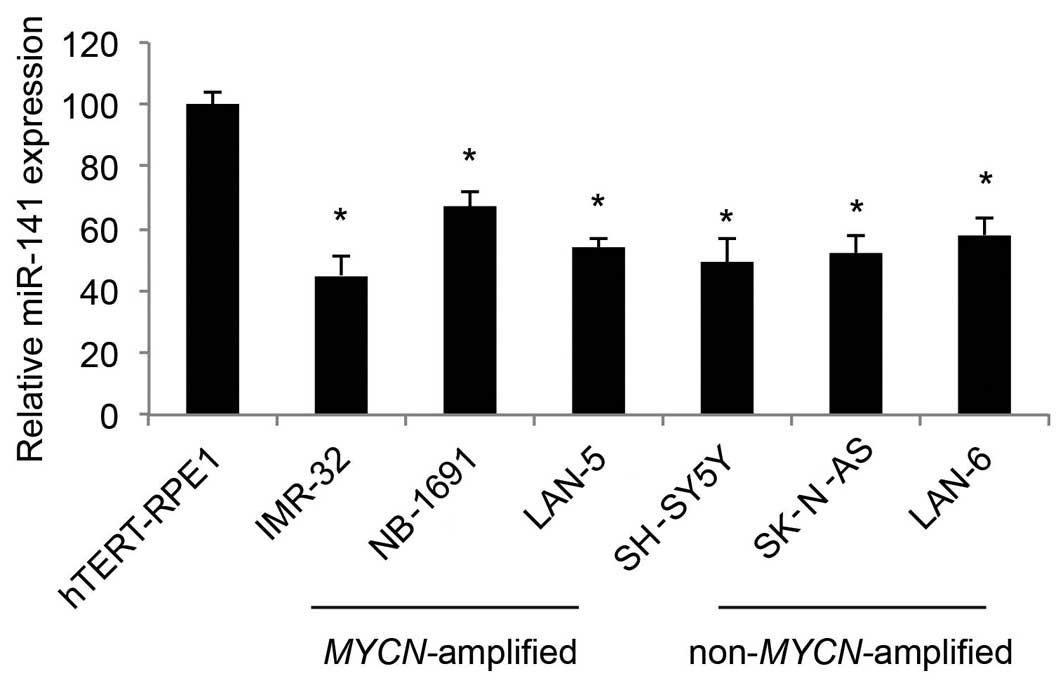
miR-141 is downregulated in human NB cell
lines
In the present study, we used qRT-PCR to determine
the expression profiles of miR-141 in human NB cell lines. As
compared to the expression level in the control cell line, human
retinal-pigmented epithelial cells immortalized with telomerase
reverse transcriptase (hTERT-RPE1), we found that miR-141 was
predominantly downregulated in both the MYCN-amplified NB
cell lines, IMR-32, NB-1691 and LAN-5, as well as the
non-MYCN-amplified NB cell lines, SH-SY5Y, SK-N-AS and LAN-6
(Fig. 1; P<0.05).
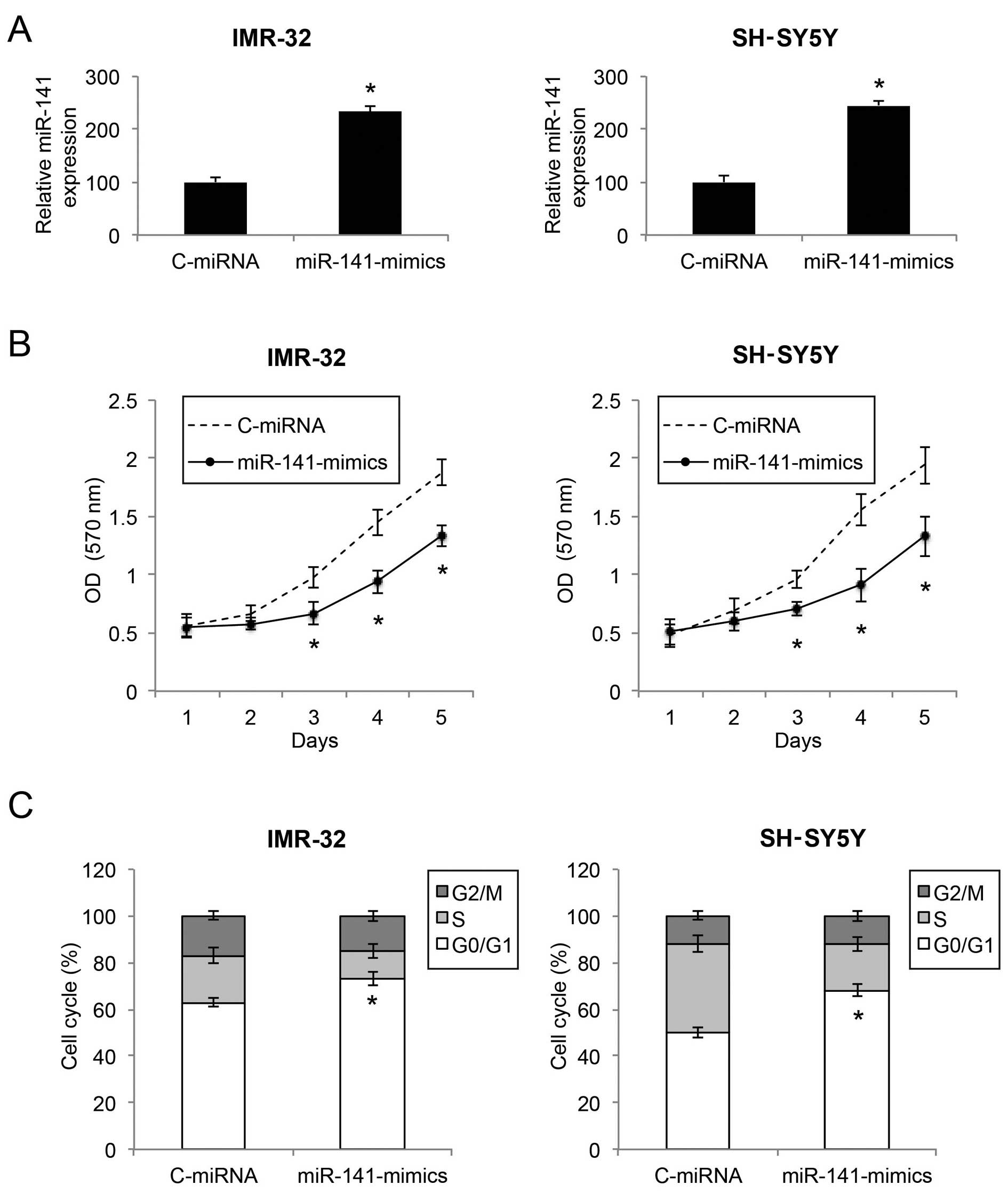
Upregulation of miR-141 inhibits the
proliferation and cell cycle transition in NB cells
We infected two NB cell lines, an
MYCN-amplified NB cell line IMR-32 and a non-
MYCN-amplified NB cell line SH-SY5Y, with either a
lentivirus of human mature miR-141 mimics (miR-141-mimics) or a
lentivirus of control miRNA mimics (C-miRNA). After lentiviral
transduction was allowed to stabilize for 48 or 72 h, qRT-PCR
demonstrated that miR-141 gene expression levels were markedly
upregulated in both the IMR-32 and SH-SY5Y cells by lentiviral
transduction of miR-141-mimics (Fig.
2A; P<0.05).
We then sought to ascertain the functional mechanism
of miR-141 upregulation in NB. Firstly, through a proliferation MTT
assay, we found that, in both IMR-32 and SH-SY5Y cells, cell
proliferation was significantly decreased by miR-141 upregulation
(Fig. 2B; P<0.05). Secondly,
through a cell cycle assay, we found that, in both the IMR-32 and
SH-SY5Y cells, miR-141 upregulation induced significant cell cycle
arrest at the G0/G1 phase (Fig. 2C;
P<0.05).
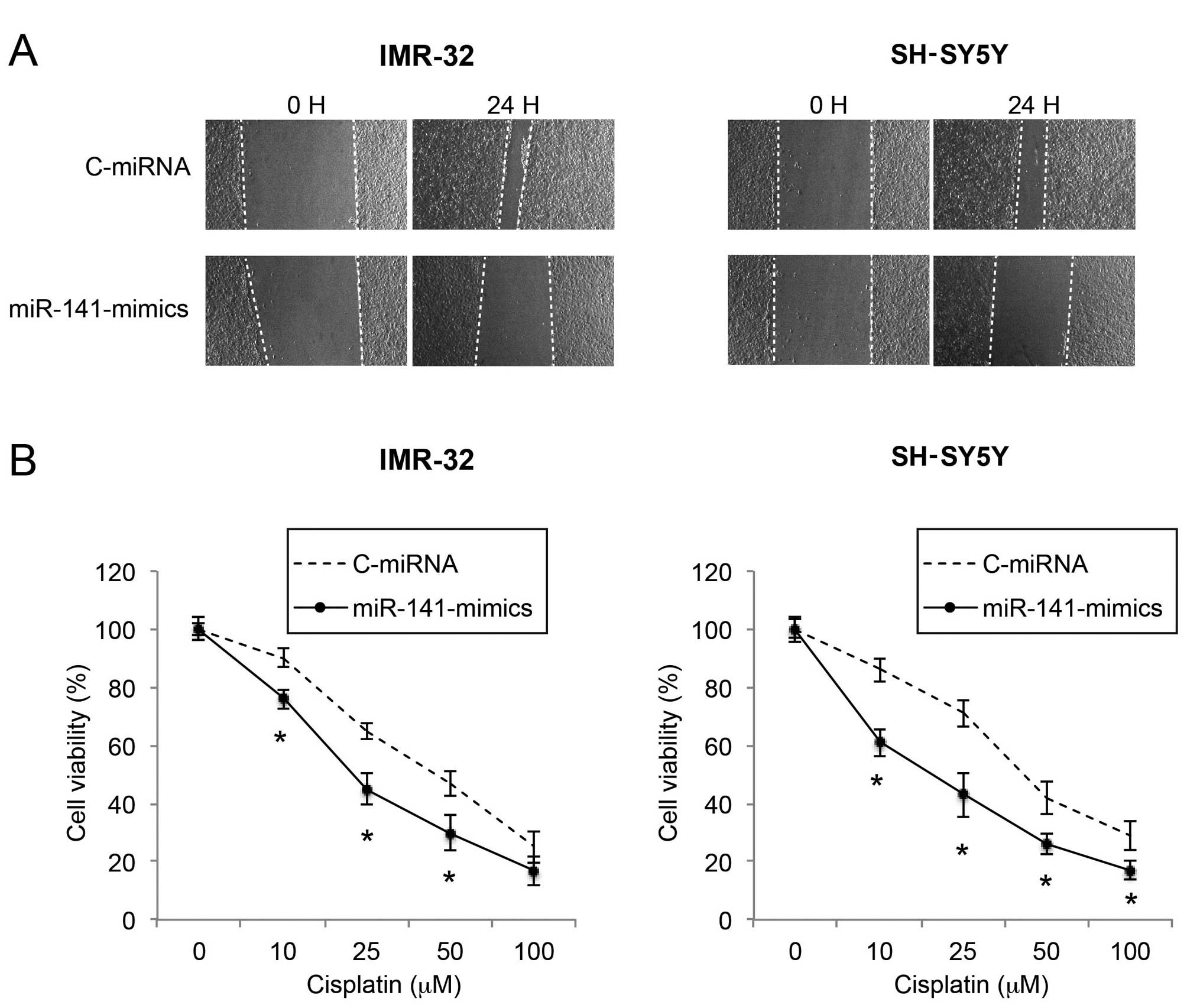
Upregulation of miR-141 reduces migration
and increases cisplatin sensitivity in NB cells
As metastatic potential and sensitivity to
chemotherapeutic drugs affect the prognosis of NB, we then sought
the functional mechanism of miR-141 upregulation on NB migration
and cisplatin sensitivity. Through a wound healing assay, we found
that the migration capability was significantly reduced by miR-141
upregulation in the IMR-32 and SH-SY5Y cells (Fig. 3A). In a cisplatin chemosensitivity
assay, we applied various concentrations of cisplatin to
lentiviral-infected IMR-32 and SH-SY5Y cells. We found that, in
response to moderate to high concentrations of cisplatin (10–100
µM), miR-141 upregulation significantly increased the
chemosensitivity of the IMR-32 and SH-SY5Y cell lines (Fig. 3B; P<0.05).
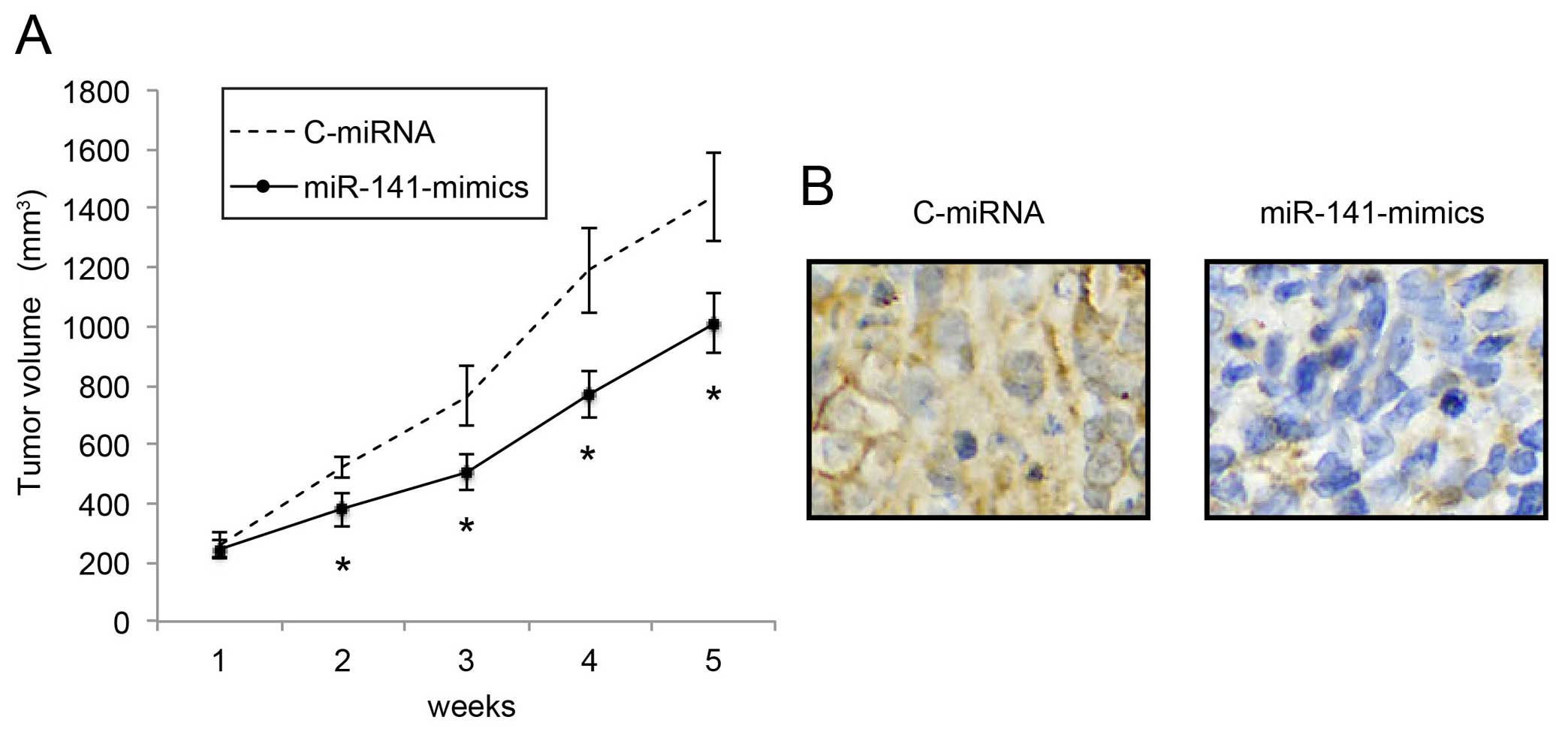
Upregulation of miR-141 inhibits in vivo
NB tumor growth
We also investigated the mechanism of miR-141
upregulation on the in vivo growth of NB tumors.
miR-141-mimic- or C-miRNA-infected IMR-132 cells (1×106
cells) were subcutaneously injected into 2-month-old female nude
mice. In vivo tumor volumes were compared between the
C-miRNA-infected IMR-132 transplantations and the
miR-141-mimic-infected IMR-132 transplantations every week for 5
weeks. The comparison demonstrated that, starting from 2 weeks
after transplantation, in vivo NB tumor growth was
significantly inhibited by miR-141 upregulation (Fig. 4A; P<0.05). Five weeks after
transplantation, the carrier mice were sacrificed. NB tumors were
extracted, formalin-fixed and prepared was paraffin-embedded
sections. Immunohistochemistry with the Ki-67 antibody confirmed
that the in vivo proliferation of IMR-32 cells was
significantly inhibited by miR-141 upregulation (Fig. 4B).
FUS is inversely associated with miR-141
in NB cells
We then sought the possible downstream signaling
pathway of miR-141 in NB. We searched some of the miRNA targeting
algorithms, including microRNA.org
(www.microrna.org) and TargetScanHuman (www.targetscan.org). It was noted that FUS is a
candidate gene with a complimentary miR-142 binding site on its
3′-UTR (Fig. 5A). A luciferase
activity assay confirmed this hypothesis by showing that WT FUS
3′-UTR was regulated by miR-141, whereas MU FUS 3′-UTR (without
miR-141 binding sequences) was not (Fig. 5B; P<0.05, P>0.05).
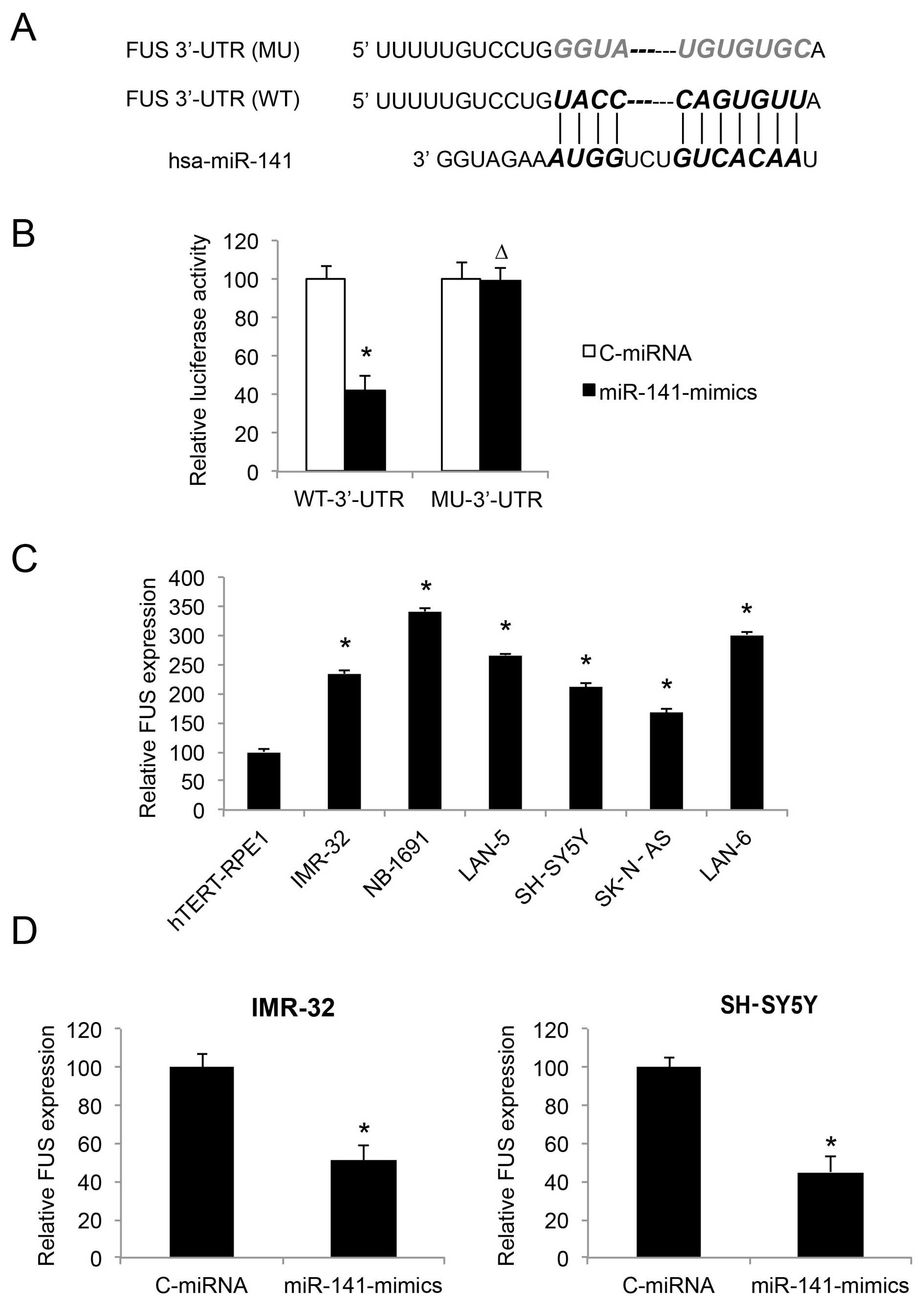 | Figure 5FUS is inversely regulated by miR-141
in neuroblastoma cell lines. (A) The complimentary binding of human
miR-141 on the 3′-UTR of the wild-type (WT) FUS gene is
highlighted. The binding sequence was also mutated (MU) to
deactivate the binding of miR-141. (B) Wild-type FUS 3′-UTR
(WT-3UTR) and MU FUS 3′-UTR (MU-3UTR) were inserted into
pmiR-REPORT luciferase vector, and co-transfected with
miR-141-mimics or C-miRNA into HEK293T cells, respectively.
Forty-eight fours after co-transfection, relative luciferase
activity was characterized by a dual-luciferase reporter assay
(*P<0.05; ∆P>0.05). (C) FUS expression
was compared by qRT-PCR between hTERT-RPE1, and both
MYCN-amplified NB cell lines, IMR-32, NB-1691 and LAN-5, as
well as non-MYCN-amplified NB cell lines, SH-SY5Y, SK-N-AS
and LAN-6 (*P<0.05, vs. hTERT-RPE1). (D) FUS
expression was compared by qRT-PCR between NB cells infected with
C-miRNA and NB cells infected with miR-141-mimics
(*P<0.05). |
We investigated the gene expression pattern of FUS
in NB cell lines. The results of qRT-PCR showed that FUS was
upregulated in both the MYCN- and non-MYCN-amplified
NB cells, as compared to the control cell line hTERT-RPE1 (Fig. 5C; P<0.05). Thus, the expression
levels of FUS and miR-141 are inversely correlated in NB cell
lines.
We then examined whether FUS expression was affected
by miR-141 upregulation, by comparing the qRT-PCR results between
NB cells transduced with C-miRNA and NB cells transduced with
miR-141-mimics. The results demonstrated that, in both the IMR-32
and SH-SY5Y cell lines, FUS was inversely regulated, or
downregulated by miR-141 upregulation (Fig. 5D; P<0.05).
Downregulation of FUS inhibits the
proliferation and cell cycle transition in NB cells
Since FUS was found to be inversely correlated with
miR-141 in NB, we aimed to ascertain whether downregulation of FUS
would have similar tumor-suppressive effects as miR-141
upregulation in NB. We transfected IMR-32 and SH-SY5Y cells with
either the FUS-targeted siRNA (FUS-siRNA) or a control siRNA
(C-siRNA). Results of qRT-PCR showed that the FUS level was
markedly downregulated by FUS-siRNA in both the IMR-32 and SH-SY5Y
cells (Fig. 6A; P<0.05). Based
on a proliferation MTT assay, we found that, in both IMR-32 and
SH-SY5Y cells, cell proliferation was significantly decreased by
FUS downregulation (Fig. 6B;
P<0.05). In addition, through a cell cycle assay, we found that
FUS downregulation induced significant cell cycle arrest at the
G0/G1 phase in the NB cells (Fig.
6C; P<0.05). Thus, FUS downregulation showed similar
inhibitory effects as miR-141 upregulation in regards to cell
proliferation and cell cycle arrest in the NB cell lines.
Downregulation of FUS reduces migration
and increases the cisplatin sensitivity in NB cells
Finally, we investigated the effects of FUS
downregulation on NB migration and cisplatin sensitivity. In a
wound healing assay, we found that the cell migratory capability
was significantly reduced by FUS downregulation in both the IMR-32
and SH-SY5Y cells (Fig. 7A). In the
cisplatin chemosensitivity assay, we also found that FUS
downregulation significantly increased cisplatin sensitivity in the
NB cells (Fig. 7B; P<0.05).
Thus, our data demonstrated that FUS downregulation also had
tumor-suppressive effects similar to those of miR-141 upregulation
on cancer metastasis and chemosensitivity in the NB cells.
Discussion
In human neuroblastoma (NB), miRNAs have been shown
to play important roles as both biomarkers for prediction of
prognosis and as genetic modulators for regulation of cancer
development (7,17). In the present study, we demonstrated
that miR-141, a cancer-associated miRNA that has never been
characterized in NB before the present study, was universally
downregulated in both MYCN- and non-MYCN-amplified NB
cell lines. The functional role of miR-141 in NB was investigated
by lentiviral transduction assay, in which miR-141 was stably
overexpressed in NB cell lines IMR-32 and SH-SY5Y. miR-141 was
found to negatively regulate NB development, including both in
vitro and in vivo proliferation, cell cycle progression,
migration and cisplatin chemosensitivity. These results strongly
suggest a tumor-suppressive role of miR-141 in NB. The
tumor-suppressive effect of miR-141 has been shown in other types
of cancers. In gastric cancer, miR-141 was found to be
significantly downregulated in both human tumors and gastric cancer
cell lines, and upregulation of miR-141 significantly inhibited the
tumor growth in gastric cancer cell lines (10). Notably, in other types of human
cancer, miR-141 may also exert an oncogenic effect. For example,
high expression of serum miR-141 was shown to be closely associated
with patients with advanced colon cancer (9). Thus, whether miR-141 acts as an
oncogene or a tumor-suppressor gene is largely dependent on the
host cancer type, and possibly the downstream signaling pathways
associated with miR-141.
We extended the present study to ascertain the
target gene of miR-141, and revealed that the FUS gene was
inversely correlated with miR-141. Luciferase reporter assay
demonstrated that 3′-UTR of FUS was attached by miR-141. qRT-PCR
assay showed that miR-141 upregulation downregulated FUS in the
IMR-32 and SH-SY5Y cells. Most importantly, in vitro
functional assays showed that siRNA-induced FUS downregulation had
similar tumor-suppressive effects as those noted by miR-141
upregulation on NB proliferation, cell cycle progression,
metastasis and chemosensitivity. Based on these data, miR-141 and
FUS are inversely associated, not only in terms of expression
pattern but also concerning the functional regulation on NB
development. Thus, miR-141 is a tumor suppressor, whereas FUS is an
oncogene in NB.
Notably, in other types of cancer, such as prostate
cancer, FUS acts as a tumor-suppressor gene and inhibits cancer
proliferation and induces apoptosis (15). The disparity in its oncogenic vs.
tumor-suppressive role in different cancer types may also be
attributed to the complex molecular pathways associated with FUS
regulation in different cancer types. Therefore, experiments to
explore the possible signaling pathways associated with FUS in NB
would be extremely helpful in deciphering the underlying mechanisms
of NB pathogenesis. Notably, the direct correlation of miR-141 and
FUS on their inverse effects on NB has yet to be established. Thus,
further experiments investigating FUS overexpression under the
circumstance of miR-141 upregulation are warranted, as it will shed
light on whether FUS may ameliorate the tumor suppressive functions
in NB induced by miR-141 upregulation.
In conclusion, we demonstrated a novel signaling
pathway of tumor-suppressor miR-141, inversely associated with
oncogene FUS, in human NB. The findings of the present study may
help identify new biomarkers or therapeutic targets for patient
with neuroblastoma.
References
|
1
|
Brodeur GM: Neuroblastoma: Biological
insights into a clinical enigma. Nat Rev Cancer. 3:203–216. 2003.
View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Siegel RL, Miller KD and Jemal A: Cancer
statistics, 2015. CA Cancer J Clin. 65:5–29. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Brodeur GM and Fong CT: Molecular biology
and genetics of human neuroblastoma. Cancer Genet Cytogenet.
41:153–174. 1989. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Corvi R, Amler LC, Savelyeva L, Gehring M
and Schwab M: MYCN is retained in single copy at chromosome 2 band
23–24 during amplification in human neuroblastoma cells. Proc Natl
Acad Sci USA. 91:5523–5527. 1994. View Article : Google Scholar
|
|
5
|
Tsuda T, Obara M, Hirano H, Gotoh S,
Kubomura S, Higashi K, Kuroiwa A, Nakagawara A, Nagahara N and
Shimizu K: Analysis of N-myc amplification in relation to disease
stage and histologic types in human neuroblastomas. Cancer.
60:820–826. 1987. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Alvarez-Garcia I and Miska EA: MicroRNA
functions in animal development and human disease. Development.
132:4653–4662. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Schulte JH, Horn S, Schlierf S, Schramm A,
Heukamp LC, Christiansen H, Buettner R, Berwanger B and Eggert A:
MicroRNAs in the pathogenesis of neuroblastoma. Cancer Lett.
274:10–15. 2009. View Article : Google Scholar
|
|
8
|
Das S, Bryan K, Buckley PG, Piskareva O,
Bray IM, Foley N, Ryan J, Lynch J, Creevey L, Fay J, et al:
Modulation of neuroblastoma disease pathogenesis by an extensive
network of epigenetically regulated microRNAs. Oncogene.
32:2927–2936. 2013. View Article : Google Scholar
|
|
9
|
Cheng H, Zhang L, Cogdell DE, Zheng H,
Schetter AJ, Nykter M, Harris CC, Chen K, Hamilton SR and Zhang W:
Circulating plasma miR-141 is a novel biomarker for metastatic
colon cancer and predicts poor prognosis. PLoS One. 6:e177452011.
View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Du Y, Xu Y, Ding L, Yao H, Yu H, Zhou T
and Si J: Down-regulation of miR-141 in gastric cancer and its
involvement in cell growth. J Gastroenterol. 44:556–561. 2009.
View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Yaman Agaoglu F, Kovancilar M, Dizdar Y,
Darendeliler E, Holdenrieder S, Dalay N and Gezer U: Investigation
of miR-21, miR-141, and miR-221 in blood circulation of patients
with prostate cancer. Tumour Biol. 32:583–588. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Chen Y and Stallings RL: Differential
patterns of microRNA expression in neuroblastoma are correlated
with prognosis, differentiation, and apoptosis. Cancer Res.
67:976–983. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Vance C, Rogelj B, Hortobágyi T, De Vos
KJ, Nishimura AL, Sreedharan J, Hu X, Smith B, Ruddy D, Wright P,
et al: Mutations in FUS, an RNA processing protein, cause familial
amyotrophic lateral sclerosis type 6. Science. 323:1208–1211. 2009.
View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Kwiatkowski TJ Jr, Bosco DA, Leclerc AL,
Tamrazian E, Vanderburg CR, Russ C, Davis A, Gilchrist J, Kasarskis
EJ, Munsat T, et al: Mutations in the FUS/TLS gene on chromosome 16
cause familial amyotrophic lateral sclerosis. Science.
323:1205–1208. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Brooke GN, Culley RL, Dart DA, Mann DJ,
Gaughan L, McCracken SR, Robson CN, Spencer-Dene B, Gamble SC,
Powell SM, et al: FUS/TLS is a novel mediator of androgen-dependent
cell-cycle progression and prostate cancer growth. Cancer Res.
71:914–924. 2011. View Article : Google Scholar
|
|
16
|
Khursheed K, Wilm TP, Cashman C, Quinn JP,
Bubb VJ and Moss DJ: Characterisation of multiple regulatory
domains spanning the major transcriptional start site of the FUS
gene, a candidate gene for motor neurone disease. Brain Res.
1595:1–9. 2015. View Article : Google Scholar
|
|
17
|
Lin RJ, Lin YC, Chen J, Kuo HH, Chen YY,
Diccianni MB, London WB, Chang CH and Yu AL: microRNA signature and
expression of Dicer and Drosha can predict prognosis and delineate
risk groups in neuroblastoma. Cancer Res. 70:7841–7850. 2010.
View Article : Google Scholar : PubMed/NCBI
|