Introduction
Breast cancer is the most common cause of
cancer-related mortality among women in developed and developing
countries (1). In 2013 in the
United States, 232,340 women were diagnosed with invasive breast
cancer, and 39,620 succumbed to the disease (2). The exploration of the underlying
mechanisms involved in breast cancer have been the subject of
extensive research over the past decades. However, the mechanisms
involved in breast cancer tumorigenesis and progression remain
poorly understood.
MicroRNAs (miRNAs) are small, conserved, non-coding
RNA sequences that suppress mRNA translation and/or degrade mRNA
molecules by binding to their 3′-untranslated regions. Differential
expression of miRNAs has been widely described in breast cancer
tissue compared to normal tissue suggesting that miRNAs may
function as oncogenes or tumorsuppressors (3–5).
The miR-129 family is one of the typical
cancer-associated miRNAs. It has two precursors, miR-129-1 and
miR-129-2. Two mature miRNAs are processed from the 5′-prime and
3′-prime of its pre-miRNA precursor. For miR-129-1 and miR-129-2,
their 5′-prime product, miR-129-5p, is the same. However, their
3′-prime products are different, which are named miR-129-1-3p and
miR-129-2-3p, respectively (6,7).
Dysregulation of miR-129-2 is frequently detected in solid tumors
(8–13). However, the role of miR-129-2 in
breast cancer remains unknown.
The methylation of CpG islands in gene promoters has
been strongly linked to the silencing of the expression of
tumor-suppressor genes in different types of cancers (14–17).
The hypermethylation of promoter CpG islands has been found to
affect not only tumor-suppressor mRNAs, but also tumor-suppressor
miRNAs. Several tumor-associated miRNAs have been reported to be
silenced by the aberrant hypermethylation of their promoter regions
in breast cancer, including miR-124, miR-34c, miR-148a, miR-155,
miR-203 and miR-129 (3,18–22).
In the present study, we found that the miR-129-2
promoter was hypermethylated in breast cancer cells. Furthermore,
we found that miR-129-2-3p functions as a tumor suppressor by
inducing the apoptosis of breast cancer cells by targeting
BCL2L2.
Materials and methods
Cell lines and culture
Human breast cancer cell lines (MCF-7, T47D, HMLER,
and MDA-MB-231) were cultured in Dulbecco's modified Eagle's medium
(DMEM; Life Technologies) supplemented with 10% fetal bovine serum
[Biological Industries (BI)]. Immortalized breast epithelial cell
lines (MCF-10A and 76N) were cultured in DMEM/F-12 1:1 mix
supplemented with human insulin (10 µg/ml), epidermal growth
factor (20 ng/ml), cholera toxin (50 µl), hydrocortisone
(0.5 µg/ml), and horse serum (5%). All cell lines were
incubated in a humidified chamber with 5% CO2 at
37°C.
Fifteen pairs of fresh primary breast cancer tissues
and non-cancerous tissues were obtained at the time of diagnosis
prior to any therapy from the Cancer Hospital of Jiangxi Province
(Nanchang, China). The clinical processes were approved by the
Ethics Committees of Nanchang University and informed consent was
collected from each patient.
Oligonucleotide transfection
The miR-129-2-5p and miR-129-2-3p mimics and a
non-specific miRNA control (NC) were synthesized by GenePharma
(Shanghai, China). miRNAs were transfected at a working
concentration of 50 nmol/l using RNAiMAX reagent (Invitrogen, USA)
according to the manufacturer's instructions. The transfected cells
were incubated at 37°C for 24 h in complete medium and harvested at
the indicated time-points.
RNA extraction, reverse transcription,
and quantitative real-time PCR
Total RNA was extracted using TRIzol (Invitrogen)
according to the manufacturer's instructions. cDNA was synthesized
with the MLV transcriptase kit (Invitrogen). The quantitative
analysis of miR-129-2-3p expression was assayed using a Bulge-Loop™
miRNA qRT-PCR primer (RiboBio, Guangzhou, China) and
Platinum® SYBR® Green qPCR SuperMix-UDG with
ROX (Invitrogen) on an ABI 7900HT instrument (Applied Biosystems,
Foster, CA, USA) according to the manufacturer's instructions, and
U6 small nuclear RNA (U6-snRNA) purchased from RiboBio was used as
an internal control. The fold changes were calculated through
relative quantification with 2−ΔΔCt. All of the reac
tions were performed in a 20-µl reaction vol ume in
triplicate.
Western blot analysis
This procedure was detailed previously (23). Briefly, protein lysates were
resolved through 10% SDS-PAGE and electrophoretically transferred
to a PVDF (polyvinylidene difluoride) membrane (Millipore, USA).
Then, the membrane was probed with an antibody against human BCL2L2
(1:1,000 dilution; Cell Signaling Technology) or β-actin (Bioworld,
USA) as a protein loading control, followed by
peroxidase-conjugated goat anti-mouse IgG (H+L; Proteintech, China)
as the secondary antibody. The intensity of the protein fragments
was visualized with an X-ray image film processing machine (Kodak,
Japan).
MTT and colony formation assays
For cell proliferation assays, cells transfected
with miRNAs or co-transfected with miRNAs and plasmids for 24 h
were reseeded in 96-well plates at 1.5×103 cells/well in
a final volume of 150 µl and incubated overnight. The effect
of miR-129-2-3p/BCL2L2 on cell growth and proliferation was
determined with an MTT assay as described previously (24).
For the colony formation assays, after 24 h of
transfection, the cells were reseeded in 6-well plates at 1,000
cells/well, and the medium was replaced every 3 days. After
incubation at 37°C for 2 weeks, the cells were washed twice with
PBS, fixed and stained with 0.5% crystal violet. The number of
colonies was counted under a microscope.
Cell cycle profile assay
A cell cycle profile assay was performed as
described previously (25).
Briefly, the cells were harvested by trypsinization and collected
by centrifugation. The cells were washed twice with
phosphate-buffered saline (PBS) and fixed in 1 ml of 70% ethanol at
4°C for 1 h to overnight. The cells were washed twice with PBS/1%
bovine serum albumin (BSA) and then incubated with 1 ml of PBS/1%
BSA containing 30 mg/ml propidium iodide (PI) and 0.25 mg/ml RNase
A for 30 min at room temperature. The cells were then analyzed for
DNA content by flow cytometry using a cytomics FACSCalibur (BD
Biosciences, USA). The data were analyzed using Modifit.
Apoptosis assay
Apoptosis was tested using Annexin V and PI double
staining kits according to the manufactory's instructions. Briefly,
MDA-MB-231 and MCF-7 cells were transfected with miR-129-2-3p or
the control for 48 h. The cells were then harvested and washed
twice with PBS. Then the cells were incubated with Annexin V and PI
for 30 min. Cells were analyzed by flow cytometry using a cytomics
FACSCalibur (BD Biosciences).
Luciferase reporter assay
The 3′UTR sequence of BCL2L2 containing the putative
miR-129-2-3p binding site was amplified with PCR and cloned into
the pGL3-control vector (Promega, Madison, WI, USA) downstream of
the firefly luciferase gene, which was designated as wild-type
3′UTR (wt 3′UTR). Mutagenesis was performed using a Quik-Change
Site-Directed Mutagenesis kit (Stratagene, USA) according to the
manufacturer's instructions, resulting in mutated 3′UTR (mut
3′UTR). The primers used for the construc tion of luciferase
reporters were BCL2L2 wt (forward, AATCTAGAACCCTGCCTGTGGTCCTGA and
reverse, AACTCTAGAAGAGAGTCCCTAGT). The Wt or mut 3′UTR vector and
the control vector pRL-TK (Promega) coding for Renilla
luciferase were co-transfected with miR-129-2-3p mimics or NC into
the MDA-MB-231 cells using Lipofectamine 2000 (Invitrogen). The
luciferase activity was measured 48 h later using the
Dual-Luciferase Reporter Assay system (Promega). The firefly
luciferase values were normalized to Renilla, and the ratio
of firefly/Renilla values is presented. The experiments were
performed independently in triplicate.
Rescue experiments
The open reading frame (ORF) encoding BCL2L2 protein
was amplified and cloned into pcDNA3.1 with the Myc tag located in
the N-terminal of BCL2L2. The primers used for amplified BCL2L2
were forward, AAGGTACCACCATGGCGACCCCAGCCTCG and reverse,
AACTCGAGTCACTTGCTAGCAAAAAAGGC. Breast cancer cells co-transfected
with miR-129-2-3p mimics and the BCL2L2-expressing vector or empty
plasmid for 24 h were trypsinized and subjected to apoptosis
assay.
Genomic DNA isolation and bisulfite DNA
sequencing PCR (BSP) analysis
Genomic DNAs were isolated from the cells using the
DNeasy Tissue kit (Qiagen, USA). DNA samples were treated with
sodium bisulfite to convert cytosine to uracil using the Methyl
Detector™ Bisulfite Modification kit (Active Motif, North America)
according to the manufacturer's instructions. For BSP, a
1-µl aliquot of sodium bisulfite-treated DNA was amplified
by PCR with commonly used primers for methylated and unmethylated
DNA sequences. The PCR products were cloned into the pGEMT Easy
vector (Promega), and 10 clones from each sample were sequenced to
determine the methylation status of each CpG site. BSP primers for
miR-129-2 were forward, TAGGGATTTGAAGATAGTGTTTTTAT and reverse,
AAAAAAACCTCACCCAAAATAAATTA, and were designed according to
previously validated oligonucleotides and synthesized commercially
(Invitrogen).
Statistical analysis
Statistical analyses were performed using SPSS 16.0
software (SPSS Inc., Chicago, IL, USA). The values are presented as
the mean ± standard deviation (SD) of three independent exper
iments. Differences between two groups were evaluated by a
two-tailed Student's t-test. The relationship between BCL2L2 and
miR-129-2-3p expression was assessed with two-tailed Pearson's
correlation. Differences were con sidered to be statistically
significant at P<0.05 and P<0.01 as indicated in the figure
legends.
Results
miR-129-2 is downregulated in breast
cancer cells as well as in clinical samples
To explore the potential role of miR-129-2 in breast
cancer progression, we examined the miR-129-2 level in breast
cancer cells and tissues. Since the sequence of miR-129-1-5p and
miR-129-2-5p are the same, we evaluated the miR-129-2 level using
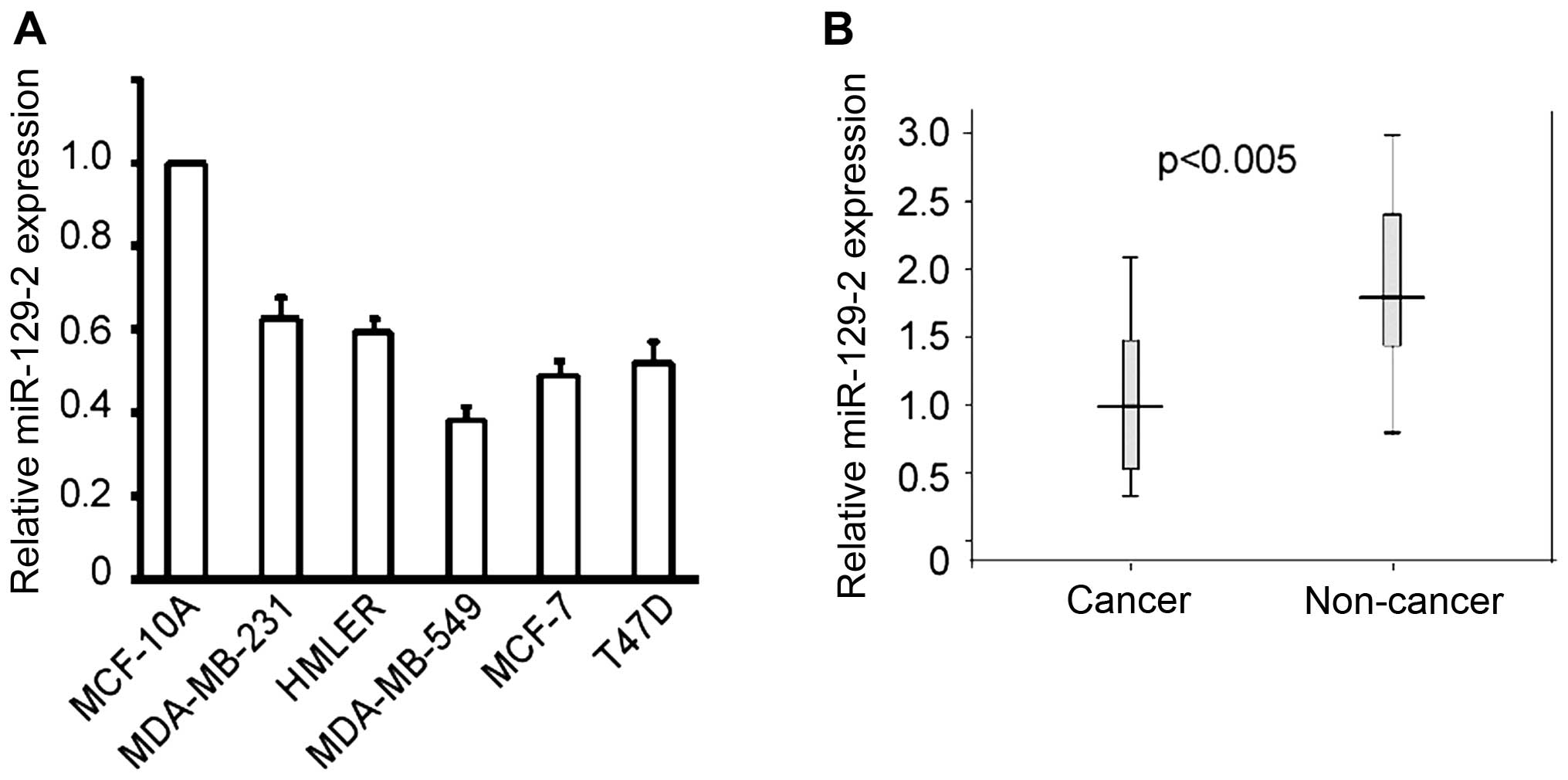
primers specific to miR-129-2-3p. As shown in Fig. 1A, the level of miR-129-2-3p detected
in the breast cancer cells was significantly lower than that in
immortalized breast cells. In addition, the level of miR-129-2-3p
was also downregulated in the breast cancer tissues compared with
normal breast tissues (Fig. 1B).
These results suggest that miR-129-2-3p may exert a
tumor-suppressor function in breast cancer.
miR-129-2-3p suppresses the cell growth
and colony formation of breast cancer cells
The above results led to us to investigate the
functional impact of miR-129-2 on breast cancer cells. We firstly
transfected miR-129-5p into MDA-MB-231 cells and found that it did
not affect the proliferation of MDA-MB-231 cells (data not shown).
We next investigated the effect of miR-129-2-3p on the breast
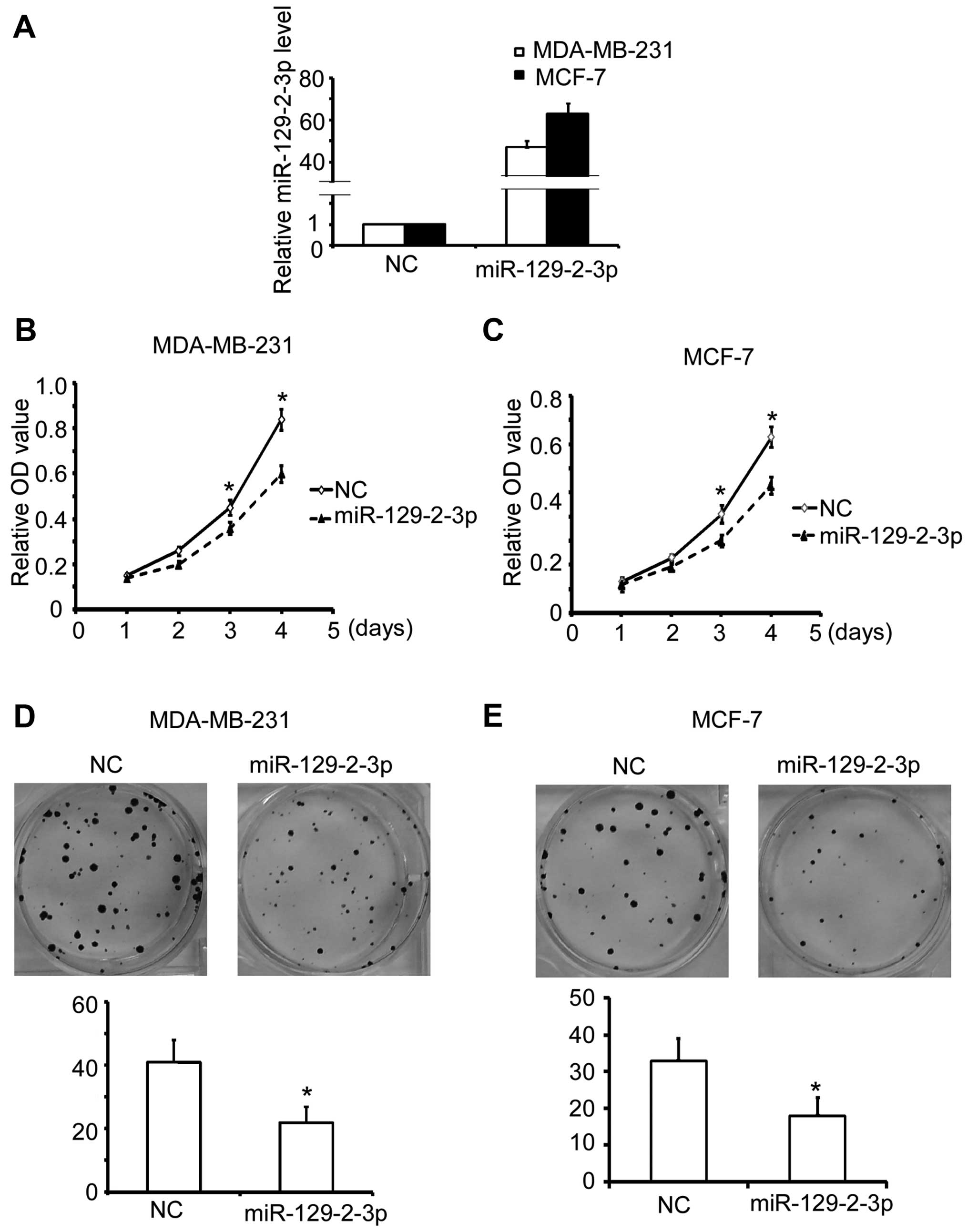
cancer cells. The efficient transfection of miR-129-2-3p mimics
into the breast cancer cells was validated by qRT-PCR (Fig. 2A). Notably, we observed a major
reduction in the cell growth of the MDA-MB-231 cells upon
miR-129-2-3p transfection compared with the cell growth noted in
the control (Fig. 2B). Accordingly,
similar results were also observed in the MCF-7 cells (Fig. 2C). Furthermore, consistent with the
proliferation assay, transfection of miR-129-2-3p mimics markedly
suppressed the colony formation of both the MDA-MB-231 (Fig. 2D) and MCF-7 cells (Fig. 2E).
miR-129-2-3p induces the apoptosis of
breast cancer cells
Give that the miR-129-2-3p mimic transfection
suppressed the cell growth of breast cancer cells, we sought to
explore its underlying mechanisms. We first detected the effect of
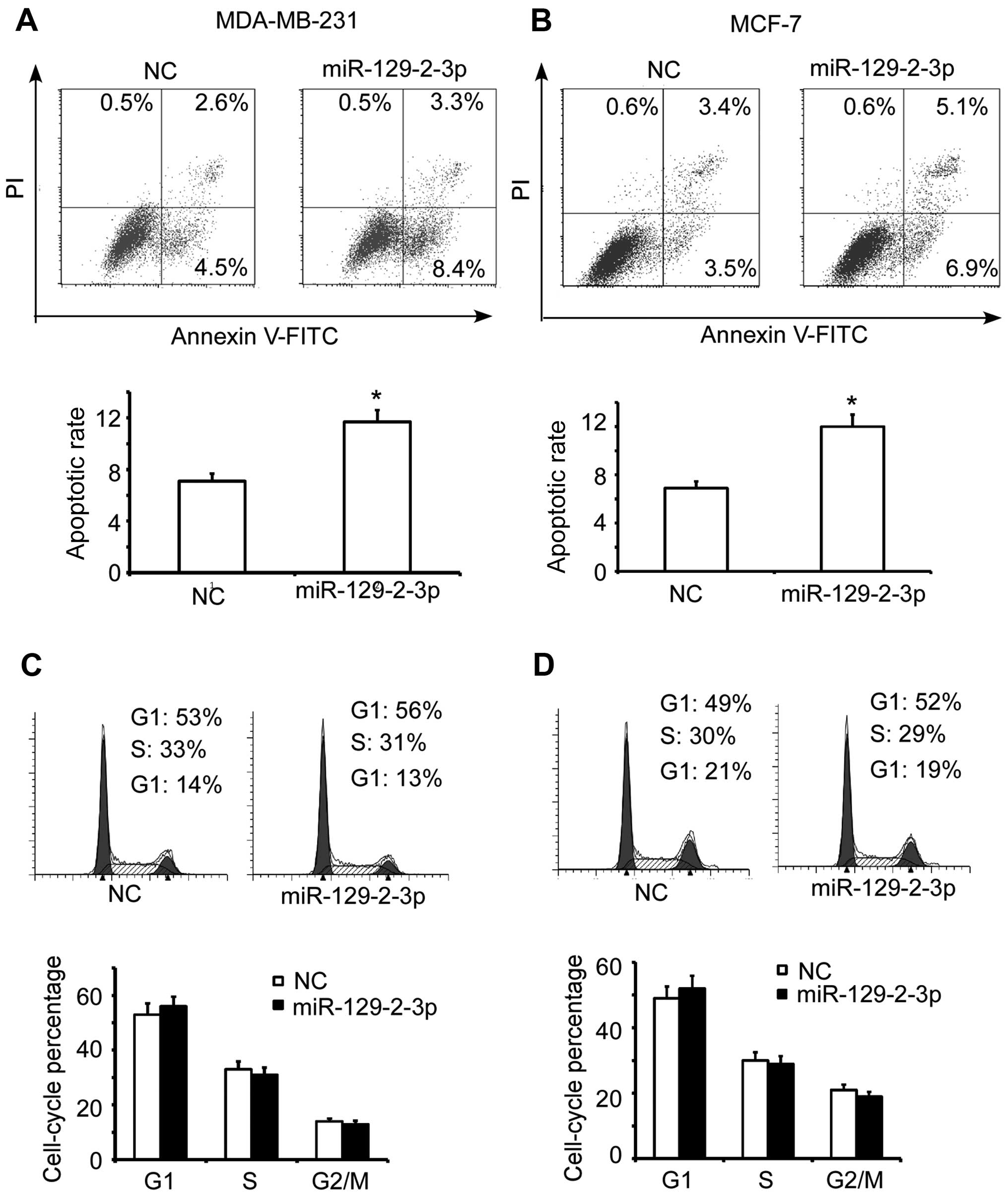
miR-129-2-3p mimics on the apoptosis of breast cancer cells. As
shown in Fig. 3A, miR-129-2-3p
mimic transfection led to a notably elevated apoptotic rate in the
MDA-MB-231 cells compared to the apoptotic rate noted in the NC
control. Consistently, this similar result was also observed in the
MCF-7 cells (Fig. 3B). However,
transfection of the miR-129-2-3p mimics only had a slight influence
on the cell cycle profile of both the MDA-MB-231 (Fig. 3C) and MCF-7 cells (Fig. 3D). Taken together, these results
indicate that miR-129-2-3p functions as a tumor suppressor of
breast cancer mainly through induction of apoptosis.
BCL2L2 is a direct target of miR-129-2-3p
in breast cancer cells
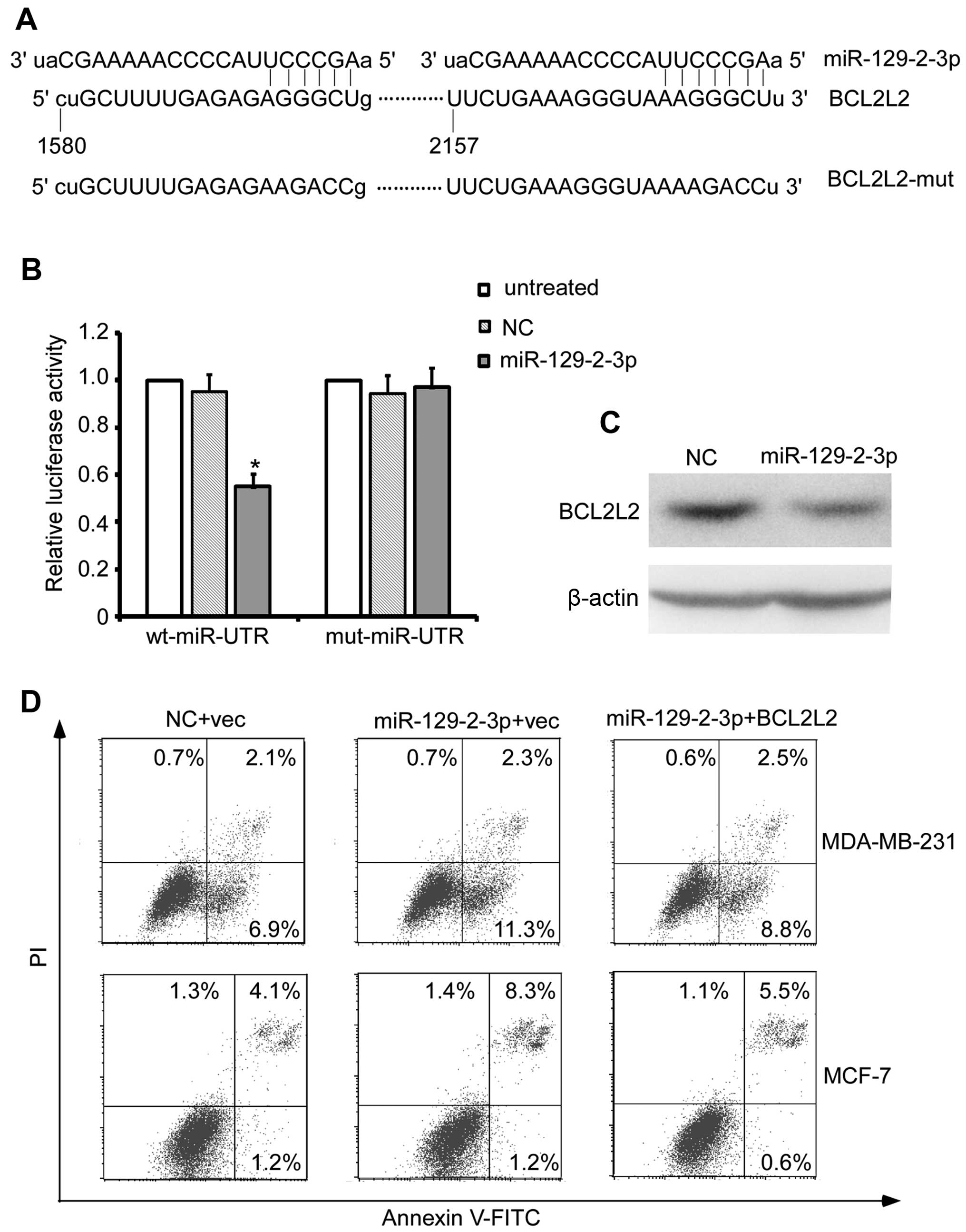
We used the online bioinformatic prediction
algorithm (http://ferrolab.dmi.unict.it/miro/) to analyze the
direct mRNA targets of miR-129-2-3p. Of all the hypothetical
targets of miR-129-2-3p, BCL2L2, an oncogene with a 3′UTR
containing the miR-129-2-3p targeting site provoked our interest
(Fig. 4A). The dual-luciferase
reporter assay showed that miR-129-2-3p mimics reduced the
fluorescence activity of wt 3′UTR by 50% compared with the negative
control (Fig. 4B, P<0.01),
whereas mut 3′UTR showed an abrogated response to miR-129-2-3p
(Fig. 4B). In addition,
miR-129-2-3p mimic transfection reduced the protein level of BCL2L2
in the MDA-MB-231 cells (Fig. 4C).
More importantly, reintroduction of BCL2L2 reversed the
miR-129-2-3p mimic transfection-induced apoptosis of the breast
cancer cells (Fig. 4D). Taken
together, these results indicate that BCL2L2 is a direct target of
miR-129-2-3p and is responsible for the miR-129-2-3p-induced
apoptosis of breast cancer cells.
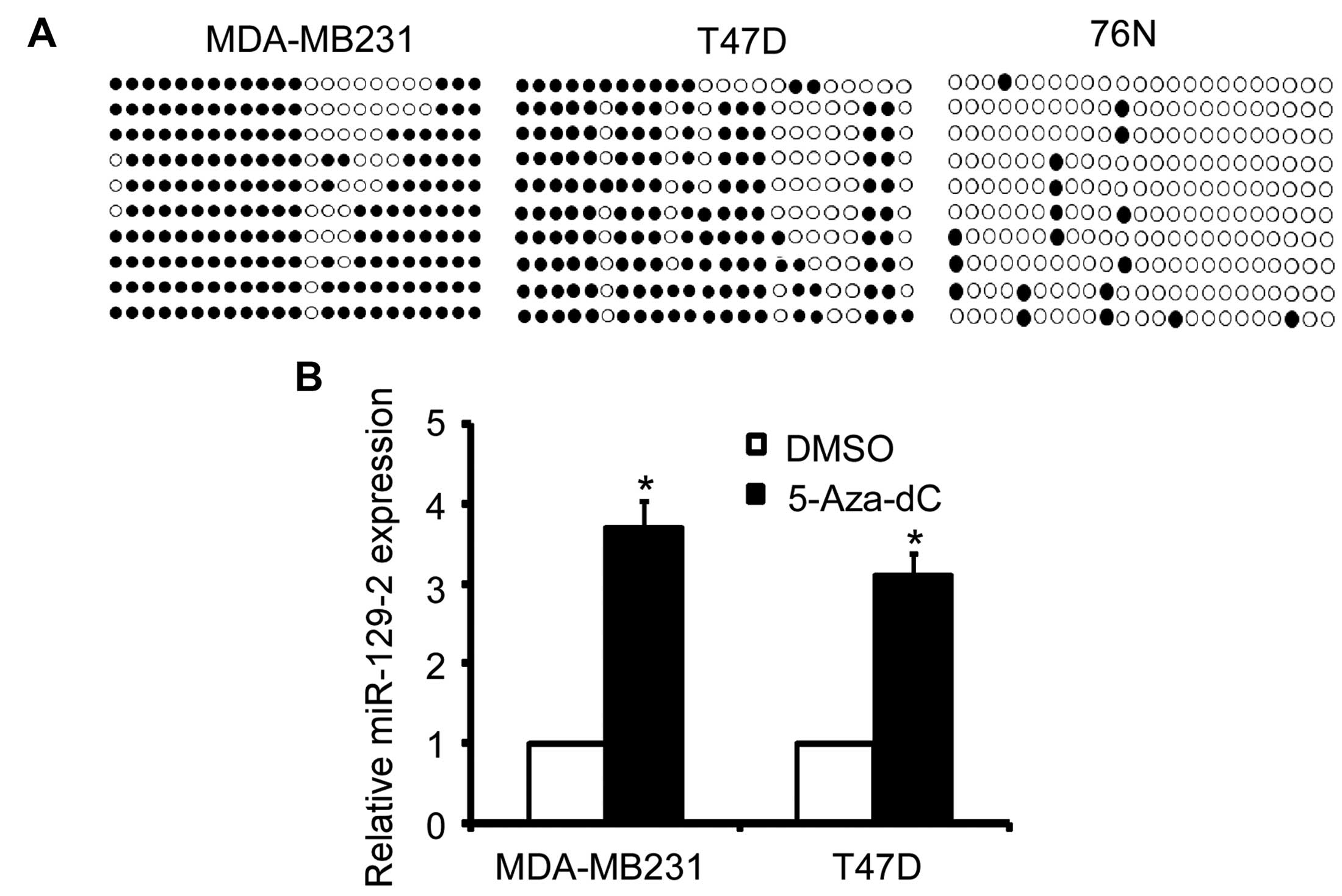
Transcriptional downregulation of
miR-129-2 in breast cancer is attributed in part to promoter DNA
hypermethylation
To explore the potential mechanisms involved in the
down-regulation of miR-129-2 in breast cancer cells, we analyzed
the methylation status of the CpG island of the miR-129-2 promoter
using BSP sequencing. As shown in Fig.
5A, the promoter of miR-129-2 was heavily methylated in the
MDA-MB-231 and T47D breast cancer cells but slightly methylated in
the immortalized breast 76N cells (Fig.
5A). More importantly, treatment of MDA-MB-231 and T47D cells
with de-methylation reagent 5-Aza-dC increased the miR-129-2 level
(Fig. 5B). Taken together, these
results indicate that miR-129-2 expression is regulated by promoter
hypermethylation.
Discussion
In recent years, growing evidence has revealed that
miRNAs play important roles in tumor development and progression.
While in different tumor types tissue-specific differences exist,
diverse molecular pathways in miRNA signaling and regulation are
also important factors to consider when approaching the
discrepancies in mRNA expression in cancer. Therefore, it is
necessary to profile the molecular function and the underlying
mechanisms of cancer-related miRNAs in each tumor type. Few studies
have investigated the roles of the miR-129 family members in cell
proliferation and apoptosis in breast cancer. Here, we demonstrated
that miR-129-2 was downregulated in human breast cancer cells and
clinical samples. Functional studies demonstrated that
overexpression of miR-129-2-3p suppressed the cell growth, colony
formation and induced apoptosis of the breast cancer cells. In
addition, prediction and a luciferase reporter assay revealed that
miR-129-2-3p directly targets BCL2L2 and inhibits its expression.
Furthermore, BSP sequence results showed that miR-129-2 was heavily
methylated in the breast cancer cells and tissues. 5′-Aza-dC
treatment increased the expression of miR-129-2-3p in the
MDA-MB-231 and T47D cells, indicating that promoter
hypermethylation is an important factor contributing to the
downregulation of miR-129-2-3p in breast cancer.
The miR-129 family includes two precursors miR-129-1
and miR-129-2 which are processed to three mature miRNAs,
miR-129-5p, miR-129-1-3p and miR-129-2 (7). Dual functions for miR-129, as a tumor
suppressor and oncogene, have been observed in different types of
cancers. It has been reported that miR-129-5p is downregulated in
neuroendocrine tumors, gastric cancer, ovarian cancer and medullary
thyroid carcinoma and plays a tumor-suppressor role in these tumors
(22,26–28).
Epigenetic regulation of miR-129-2 was reported in glioma and lung
cancer (8,9). In contrast, however, miR-129 is also
upregulated in several solid tumors and non-cancerous tissues from
cancer patients with lymph node metastases (29,30).
Our results revealed the tumor-suppressor functions of
miR-129-2.miRNAs degrade or repress translation of target mRNAs.
Therefore, to validate the oncogenic function of miR-129-2, an
oncogenic target was required to be identified. We predicted
several potential target candidates for miR-129-2-3p. Among them,
BCL2L2 provoked our interesting due to its oncogenic role in many
solid tumors (31–33). Luciferase report assay confirmed
that miR-129-2-3p directly targeted BCL2L2 3′UTR for degradation.
More importantly, rescue assay results indicated that BCL2L2 is
crucial for miR-129-2-3p-induced breast cancer apoptosis.
DNA hypermethylation is closely associated with gene
inactivation and is a common and widespread phenomenon in human
tumors. Many previous studies have reported that promoter
hypermethylation-induced downregulation of tumor-suppressor miRNAs
correlates with carcinogenesis. Our data revealed that the
hypermethylation of the miR-129-2 promoter led to the
downregulation of the miR-129-2 level in breast cancer patients. In
addition, demethylation agent 5-Aza-dC treatment increased
miR-129-2 expression in the breast cancer cells. These results
indicate that promoter hypermethylation leads to the downregulation
of miR-129-2-3p in breast cancer, at least in part.
In conclusion, our study suggests that miR-129-2 is
down-regulated by DNA methylation and plays a tumor-suppressor role
in breast cancer by targeting BCL2L2 for degradation. Transfection
of miR-129-2-3p was found to suppress cell growth and promote the
cell apoptosis of breast cancer cells, which may provide a novel
therapeutic strategy for the treatment of breast cancer.
Acknowledgments
The present study was supported by grants from the
Natural Science Foundation of China (81560452 and 81102023 to
X.B.L.; 81560403 to J.T.).
References
|
1
|
Torre LA, Bray F, Siegel RL, Ferlay J,
Lortet-Tieulent J and Jemal A: Global cancer statistics, 2012. CA
Cancer J Clin. 65:87–108. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
DeSantis C, Ma J, Bryan L and Jemal A:
Breast cancer statistics, 2013. CA Cancer J Clin. 64:52–62. 2014.
View Article : Google Scholar
|
|
3
|
Diao Y, Guo X, Jiang L, Wang G, Zhang C,
Wan J, Jin Y and Wu Z: miR-203, a tumor suppressor frequently
downregulated by promoter hypermethylation in rhabdomyosarcoma. J
Biol Chem. 289:529–539. 2014. View Article : Google Scholar :
|
|
4
|
Gong C, Qu S, Lv XB, Liu B, Tan W, Nie Y,
Su F, Liu Q, Yao H and Song E: BRMS1L suppresses breast cancer
metastasis by inducing epigenetic silence of FZD10. Nat Commun.
5:54062014. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Lu K, Wang J, Song Y, Zhao S, Liu H, Tang
D, Pan B, Zhao H and Zhang Q: miRNA-24-3p promotes cell
proliferation and inhibits apoptosis in human breast cancer by
targeting p27Kip1. Oncol Rep. 34:995–1002.
2015.PubMed/NCBI
|
|
6
|
Cui L, Zhang X, Ye G, Zheng T, Song H,
Deng H, Xiao B, Xia T, Yu X, Le Y, et al: Gastric juice microRNAs
as potential biomarkers for the screening of gastric cancer.
Cancer. 119:1618–1626. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Katada T, Ishiguro H, Kuwabara Y, Kimura
M, Mitui A, Mori Y, Ogawa R, Harata K and Fujii Y: microRNA
expression profile in undifferentiated gastric cancer. Int J Oncol.
34:537–542. 2009.PubMed/NCBI
|
|
8
|
Tian XY, Zhang L, Sun LG and Li M:
Epigenetic regulation of miR-129-2 leads to overexpression of
PDGFRa and FoxP1 in glioma cells. Asian Pac J Cancer Prev.
16:6129–6133. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Xiao Y, Li X, Wang H, Wen R, He J and Tang
J: Epigenetic regulation of miR-129-2 and its effects on the
proliferation and invasion in lung cancer cells. J Cell Mol Med.
19:2172–2180. 2015.PubMed/NCBI
|
|
10
|
Yang Y, Huang JQ, Zhang X and Shen LF:
miR-129-2 functions as a tumor suppressor in glioma cells by
targeting HMGB1 and is down-regulated by DNA methylation. Mol Cell
Biochem. 404:229–239. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
He Y, Huang C, Zhang L and Li J:
Epigenetic repression of miR-129-2 in cancer. Liver Int.
34:6462014. View Article : Google Scholar
|
|
12
|
Kang M, Li Y, Liu W, Wang R, Tang A, Hao
H, Liu Z and Ou H: miR-129-2 suppresses proliferation and migration
of esophageal carcinoma cells through downregulation of SOX4
expression. Int J Mol Med. 32:51–58. 2013.PubMed/NCBI
|
|
13
|
Lu CY, Lin KY, Tien MT, Wu CT, Uen YH and
Tseng TL: Frequent DNA methylation of miR-129-2 and its potential
clinical implication in hepatocellular carcinoma. Genes Chromosomes
Cancer. 52:636–643. 2013.PubMed/NCBI
|
|
14
|
Bedi U, Mishra VK, Wasilewski D, Scheel C
and Johnsen SA: Epigenetic plasticity: A central regulator of
epithelial-to-mesenchymal transition in cancer. Oncotarget.
5:2016–2029. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Chen QW, Zhu XY, Li YY and Meng ZQ:
Epigenetic regulation and cancer (Review). Oncol Rep. 31:523–532.
2014.
|
|
16
|
Sang Y, Cheng C, Tang XF, Zhang MF and Lv
XB: Hypermethylation of TET1 promoter is a new diagnosic marker for
breast cancer metastasis. Asian Pac J Cancer Prev. 16:1197–1200.
2015. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Dong Y and Wang A: Aberrant DNA
methylation in hepatocellular carcinoma tumor suppression (Review).
Oncol Lett. 8:963–968. 2014.PubMed/NCBI
|
|
18
|
Lv XB, Jiao Y, Qing Y, Hu H, Cui X, Lin T,
Song E and Yu F: miR-124 suppresses multiple steps of breast cancer
metastasis by targeting a cohort of pro-metastatic genes in vitro.
Chin J Cancer. 30:821–830. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Yu F, Jiao Y, Zhu Y, Wang Y, Zhu J, Cui X,
Liu Y, He Y, Park EY, Zhang H, et al: MicroRNA 34c gene
downregulation via DNA methylation promotes self-renewal and
epithelial-mesenchymal transition in breast tumor-initiating cells.
J Biol Chem. 287:465–473. 2012. View Article : Google Scholar :
|
|
20
|
Li HP, Huang HY, Lai YR, Huang JX, Chang
KP, Hsueh C and Chang YS: Silencing of miRNA-148a by
hypermethylation activates the integrin-mediated signaling pathway
in nasopharyngeal carcinoma. Oncotarget. 5:7610–7624. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Yim RL, Wong KY, Kwong YL, Loong F, Leung
CY, Chu R, Lam WW, Hui PK, Lai R and Chim CS: Methylation of
miR-155-3p in mantle cell lymphoma and other non-Hodgkin's
lymphomas. Oncotarget. 5:9770–9782. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Wu Q, Yang Z, Xia L, Nie Y, Wu K, Shi Y
and Fan D: Methylation of miR-129-5p CpG island modulates
multi-drug resistance in gastric cancer by targeting ABC
transporters. Oncotarget. 5:11552–11563. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Lv XB, Wu W, Tang X, Wu Y, Zhu Y, Liu Y,
Cui X, Chu J, Hu P, Li J, et al: Regulation of SOX10 stability via
ubiquitination-mediated degradation by Fbxw7α modulates melanoma
cell migration. Oncotarget. 6:36370–36382. 2015.PubMed/NCBI
|
|
24
|
Lv XB, Zhang X, Deng L, Jiang L, Meng W,
Lu Z and Wang X: miR-92a mediates AZD6244 induced apoptosis and
G1-phase arrest of lymphoma cells by targeting Bim. Cell Biol Int.
38:435–443. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Salilew-Wondim D, Hölker M, Rings F,
Phatsara C, Mohammadi-Sangcheshmeh A, Tholen E, Schellander K and
Tesfaye D: Depletion of BIRC6 leads to retarded bovine early
embryonic development and blastocyst formation in vitro. Reprod
Fertil Dev. 22:564–579. 2010. View
Article : Google Scholar : PubMed/NCBI
|
|
26
|
Døssing KB, Binderup T, Kaczkowski B,
Jacobsen A, Rossing M, Winther O, Federspiel B, Knigge U, Kjær A
and Friis-Hansen L: Down-regulation of miR-129-5p and the let-7
family in neuro-endocrine tumors and metastases leads to
up-regulation of their targets Egr1, G3bp1, Hmga2 and Bach1. Genes
(Basel). 6:1–21. 2014.
|
|
27
|
Tan G, Cao X, Dai Q, Zhang B, Huang J,
Xiong S, Zhang Y, Chen W, Yang J and Li H: A novel role for
microRNA-129-5p in inhibiting ovarian cancer cell proliferation and
survival via direct suppression of transcriptional co-activators
YAP and TAZ. Oncotarget. 6:8676–8686. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Duan L, Hao X, Liu Z, Zhang Y and Zhang G:
miR-129-5p is down-regulated and involved in the growth, apoptosis
and migration of medullary thyroid carcinoma cells through
targeting RET. FEBS Lett. 588:1644–1651. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Li M, Tian L, Wang L, Yao H, Zhang J, Lu
J, Sun Y, Gao X, Xiao H and Liu M: Down-regulation of miR-129-5p
inhibits growth and induces apoptosis in laryngeal squamous cell
carcinoma by targeting APC. PLoS One. 8:e778292013. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Huang ZM, Yang J, Shen XY, Zhang XY, Meng
FS, Xu JT, Zhang BF and Gao HJ: MicroRNA expression profile in
non-cancerous colonic tissue associated with lymph node metastasis
of colon cancer. J Dig Dis. 10:188–194. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Hu T, Weng S, Tang W, Xue R, Chen S, Cai
G, Cai Y, Shen X, Zhang S and Dong L: Overexpression of BIRC6 is a
predictor of prognosis for colorectal cancer. PLoS One.
10:e01252812015. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Luk SU, Xue H, Cheng H, Lin D, Gout PW,
Fazli L, Collins CC, Gleave ME and Wang Y: The BIRC6 gene as a
novel target for therapy of prostate cancer: Dual targeting of
inhibitors of apoptosis. Oncotarget. 5:6896–6908. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Tang W, Xue R, Weng S, Wu J, Fang Y, Wang
Y, Ji L, Hu T, Liu T, Huang X, et al: BIRC6 promotes hepatocellular
carcinogenesis: Interaction of BIRC6 with p53 facilitating p53
degradation. Int J Cancer. 136:E475–E487. 2015. View Article : Google Scholar
|