Introduction
Cervical cancer is a common cancer type in women
that severely affects women's health worldwide (1). Although cancer treatments have been
improved in recent decades, the treatment outcomes of cervical
cancer patients have not been significantly impaired and cervical
cancer remains the second leading cause of cancer-related death in
women (2). Although much research
has focused on cervical cancer, the precise molecular mechanisms
underlying the pathogenesis of this disease remain poorly
understood. Thus, finding new targets for the development of
effective therapeutics for cervical cancer is important.
Forkhead box (FOX) proteins are a family of
transcriptional regulators that are associated with regulating
various cellular processes, including cell proliferation and
survival, apoptosis, differentiation and metabolism (3). Thus, FOX proteins contribute to the
development and progression of cancers (4). Among the FOX family members, forkhead
box Q1 (FOXQ1) is reportedly associated with various types of
cancers. FOXQ1 regulates the proliferation, migration, invasion,
metastasis, epithelial-mesenchymal transition (EMT), and
chemosensitivity of cancer cells by regulating gene networks
(5–9). The increased EMT of cancer cells
accounts for cancer metastasis by promoting the migratory and
invasive abilities of cancer cells (10,11).
FOXQ1 has been suggested as an oncogene in various cancers,
including colorectal (12), lung
(13,14), gastric (15), ovarian (16) and breast cancers (17). However, whether or not FOXQ1 plays
an important role in cervical cancer remains poorly understood.
The role of microRNAs (miRNAs) in tumorigenesis has
been widely studied in recent decades (18). miRNAs negatively regulate gene
expression by directly targeting the 3′-untranslated region (UTR)
of mRNAs, leading to translation inhibition (19). miRNAs modulate numerous cancer cell
processes, including proliferation, apoptosis, migration and
invasion, by regulating target genes (20). Thus, miRNAs are promising targets
for developing anticancer drugs. A previous study suggested that
miRNAs can be used as biomarkers or therapeutic targets for the
diagnosis or treatment of cervical cancer (21). Various studies have demonstrated
that miRNAs such as miR-21 (22)
and miR-1246 (23) are dysregulated
in cervical cancer. Some miRNAs could inhibit cervical cancer cell
growth by suppressing target genes (24,25).
However, the precise role of miRNAs in cervical cancer pathogenesis
remains unclear.
In this study, we investigated the role of FOXQ1 in
cervical cancer and tested the potential of specific miRNAs to
inhibit FOXQ1 expression. FOXQ1 was overexpressed in cervical
cancer cell lines, and knockdown of FOXQ1 by small interfering RNA
(siRNA) significantly inhibited the proliferation and EMT of the
cervical cancer cells. Notably, FOXQ1 was indicated as the target
gene of miR-506. The overexpression of miR-506 significantly
suppressed FOXQ1 expression and inhibited the proliferation and EMT
of cervical cancer cells. Furthermore, restoration of FOXQ1
expression partially reversed the suppression of miR-506. Overall,
our study demonstrated that miR-506 targets FOXQ1 to inhibit the
proliferation and EMT of cervical cancer cells.
Materials and methods
Cell lines and clinical specimens
The human normal skin keratinocyte line HaCaT and
the human cervical cancer cell lines CaSki and SiHa were purchased
from the American Type Culture Collection (ATCC; Manassas, VA, USA)
and were then cultured in Dulbecco's modified Eagle's medium (DMEM)
containing 10% fetal bovine serum (FBS) (both from Gibco,
Rockville, MD, USA) and 1% penicillin/streptomycin (Sigma, St.
Louis, MO, USA) in accordance with the recommended protocols. The
cells were maintained at 37°C in a humidified incubator containing
5% CO2. Twenty-two clinical specimens of cervical cancer
were collected from the Women and Infants Hospital of Zhengzhou.
Frozen cancer tissues in liquid nitrogen were stored at -80°C and
prepared for RNA isolation. The use of human clinical samples was
approved by the Institutional Human Experiment and Ethics Committee
of the Women and Infants Hospital of Zhengzhou.
Quantitative real-time PCR (qRT-PCR)
analysis
The level of mRNA expression was determined by
qRT-PCR analysis. In brief, total RNA from cells or clinical
specimens was isolated using the miRNeasy Mini kit (Qiagen,
Shanghai, China). The total RNA was reverse-transcribed into cDNA
by using M-MLV reverse transcriptase (Takara, Dalian, China) or
miScript Reverse Transcription kit (Qiagen). qRT-PCR analysis was
performed using SYBR Premix Ex Taq™ II (Takara) with the following
primers: FOXQ1 forward, 5′-CGCGGACTTTGCACTTTGAA-3′ and reverse,
5′-AGCTTTAAGGCACGTTTGATGGAG-3′; E-cadherin forward,
5′-TGCCCAGAAAATGAAAAAGG-3′ and reverse, 5′-GTGTATGTGGCAATGCGTTC-3′;
vimentin forward, 5′-GAGAACTTTGCCGTTGAAGC-3′ and reverse,
5′-TCCAGCAGCTTCCTGTAGGT-3′; GAPDH forward,
5′-AATGGGCAGCCGTTAGGAAA-3′ and reverse, 5′-TGAAGGGGTCATTGATGGCA-3′;
miR-506 forward, 5′-GGGTATTGAGGAAGGTGTT-3′ and reverse,
5′-CAGTGCGTGTCGTGGAGT-3′ and U6 forward,
5′-CTCGCTTCGGCAGCACATATACT-3′ and reverse,
5′-ACGCTTCACGAATTTGCGTGTC-3′. Gene quantification data were
normalized to GAPDH for mRNA expression analysis or normalized to
U6 for miRNA expression analysis using the 2−ΔΔCt
method.
Western blot analysis
Proteins were extracted from cells using RIPA lysis
buffer containing protease inhibitor cocktail (Beyotime, Haimen,
China), and the concentration was measured and quantified using the
Bradford method. A total of 25 µg proteins from each cell
group were separated via 12.5% sodium dodecyl
sulfate-polyacrylamide gel electrophoresis. The proteins on the gel
were transferred onto a nitrocellulose membrane (Millipore, Boston,
MA, USA), which was then blocked by blocking solution (3%
nonfat-dried milk) for 1 h at 37°C. After blocking, the membrane
was incubated with primary antibodies, including anti-FOXQ1 (1:400)
and anti-GAPDH (1:10,000) (both from Abcam, Cambridge, UK), which
were diluted in blocking buffer at 4°C overnight. The membrane was
incubated with horseradish peroxidase-conjugated secondary
antibodies (1:2,000; Beyotime) for 1 h at 37°C. The protein bands
were detected using chemiluminescence (Amersham, Little Chalfont,
UK). Densitometric analysis of the protein bands was performed
using Image-Pro Plus 6.0 software (Media Cybernetics, Inc.,
Rockville, MD, USA).
Cell treatment
The expression of FOXQ1 was suppressed by FOXQ siRNA
or miR-506 mimic transfection. FOXQ1 siRNA
(5′-CGCGGACUUUGCACUUUGA-3′) and negative control siRNA (NC-siRNA;
5′-UUCUCCGAACGUGUCACGU-3′), miR-506 mimics
(5′-UAAGGCACCCUUCUGAGUAGA-3′) and negative control miRNA mimics
(miR-NC; 5′-UUCUCCGAACGUGUCACGUTT-3′) were synthesized by Shanghai
GenePharma Co., Ltd. (Shanghai, China). The miR-506 mimics were
transfected into cells at final concentrations of 50 nM using
Lipofectamine 2000 (Invitrogen) in accordance with the
manufacturer's instructions. For knockdown of FOXQ1, ~1 µg
of FOXQ1 siRNA was diluted into 100 µl of DMEM containing 6
µl of Lipofectamine 2000 and incubated for 45 min at room
temperature. The siRNA transfection reagent mixture was added to
each well and incubated for 7 h. Then, the medium was replaced with
fresh medium and cultured for 48 h. The open reading frame of FOXQ1
without 3′-UTR was inserted into pcDNA3.0 plasmids. For the rescue
experiment, miR-506 mimics (50 nm) and pcDNA3/FOXQ1 overexpression
vectors (3 µg) were co-transfected into cervical cancer
cells for 48 h. Empty vectors were used as control.
Cell proliferation assay
Cells were grown into 96-well plates
(1×104 cells/well), transfected with FOXQ1 siRNA or
miR-506 mimics, and then incubated for 24, 48, and 72 h. Cell
proliferation was measured using the
3-(4,5-dimethyl-2-thiazolyl)-2,5-diphenyl-2-H-tetrazolium bromide
(MTT) method. In brief, 20 µl of MTT solution (5 mg/ml;
Sigma) was added to each well after replacement with fresh medium.
After 4 h, the formazan crystals were melted by adding 200
µl of dimethyl sulfoxide (DMSO) to each well. After the
crystals were dissolved, the optical density of each well was
measured at 490 nm using an ELISA reader.
Luciferase reporter assay
The cDNA fragments of FOXQ1 3′-UTR containing the
predicted binding site for miR-506 were inserted into pmirGLO
luciferase promoter vectors (Promega, Madison, WI, USA). The cells
were seeded into 24-well plates (5×104 cells/well) and
were then transfected with 500 ng of pmirGLO-FOXQ1 3′-UTR and 50 nM
of miR-506 mimics for 48 h using Lipofectamine 2000. The cells were
lysed, and the activity of firefly or Renilla luciferase was
detected using a dual-luciferase reporter assay kit (Promega) in
accordance with the manufacturer's instructions.
Statistical analysis
Data are shown as means ± standard deviation. SPSS
11.5 for Windows (SPSS Inc., Chicago, IL, USA) was used for the
statistical analyses. Differences between two groups were analyzed
using the Student's t-test. One-way ANOVA was conducted in multiple
groups for statistical analyses. The correlation between FOXQ1 and
miR-506 was evaluated using Spearman's correlation analysis.
Statistical significance was considered at P<0.05.
Results
High expression of FOXQ1 is detected in
cervical cancer cells
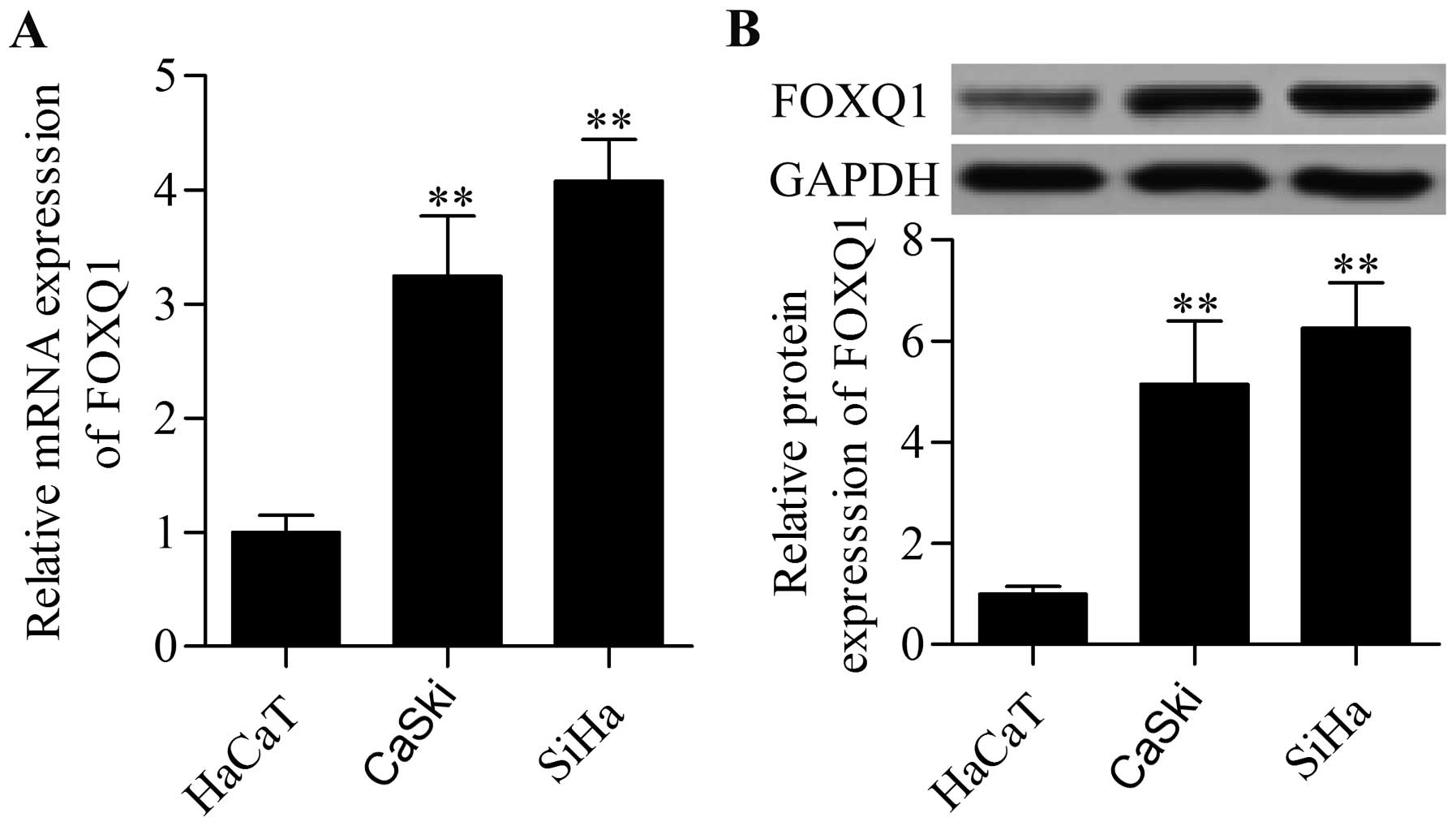
To explore the potential role of FOXQ1 in cervical
cancer, we detected FOXQ1 expression in the cervical cancer cell
lines CaSki and SiHa through qRT-PCR and Western blot analyses.
qRT-PCR data showed that the mRNA expression of FOXQ1 was
significantly higher in the CaSki and SiHa cells than that in the
HaCaT cells (Fig. 1A). Western blot
analysis also revealed a high protein expression level of FOXQ1 in
the CaSki and SiHa cells (Fig. 1B).
The data indicated that FOXQ1 may function as an oncogene in
cervical cancer.
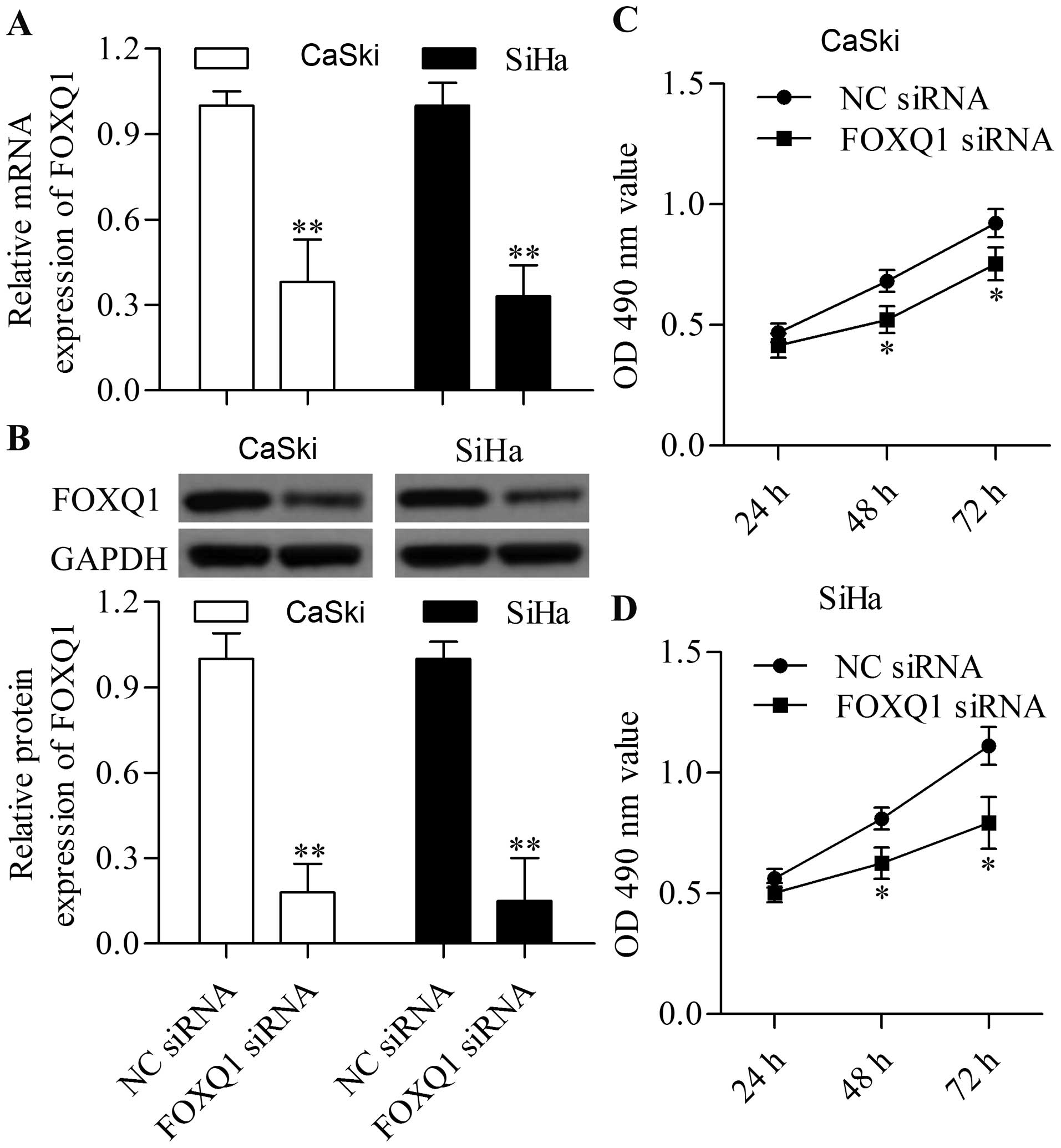
Knockdown of FOXQ1 by siRNA suppresses
cervical cancer cell proliferation
To understand the biological role of FOXQ1 in
cervical cancer, we performed loss-of-function experiments in the
CaSki and SiHa cells by using siRNA targeting FOXQ1. Cells were
transfected with FOXQ1 siRNA, and the expression of FOXQ1 was
detected by qRT-PCR and western blot analyses. The mRNA and protein
expression levels of FOXQ1 were significantly decreased by FOXQ1
siRNA transfection (Fig. 2A and B).
Furthermore, the effect of FOXQ1 silencing on cell proliferation
was detected by MTT assay. Transfection of FOXQ1 siRNA markedly
impeded the proliferation of CaSki (Fig. 2C) and SiHa (Fig. 2D) cells. These results imply that
FOXQ1 is a potential molecular target for inhibiting the
proliferation of cervical cancer cells.
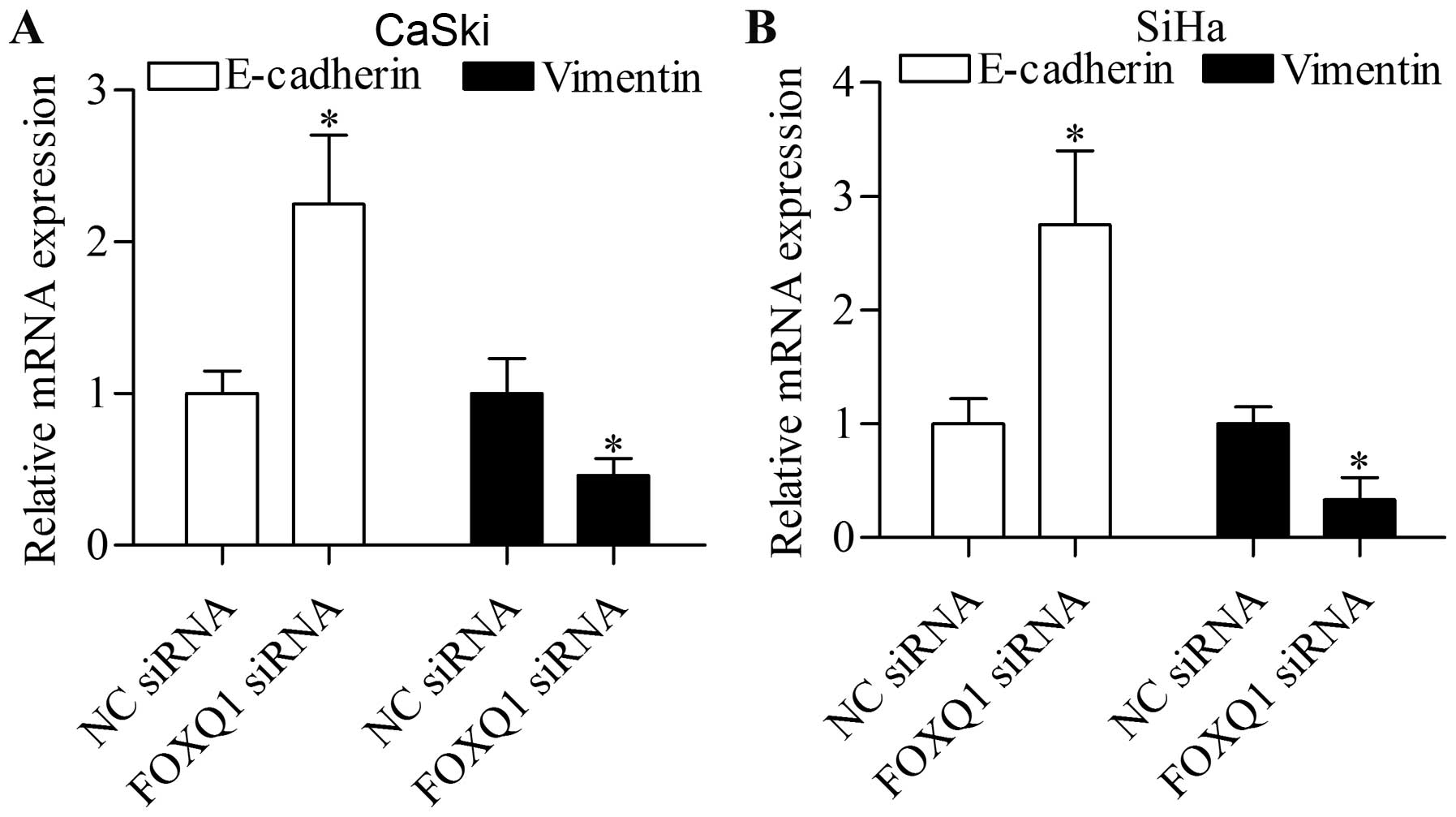
Knockdown of FOXQ1 by siRNA inhibits the
EMT of cervical cancer cells
To further investigate the function of FOXQ1, we
determined the effect of FOXQ1 silencing on the EMT of cervical
cancer cells. Results showed that FOXQ1 knockdown significantly
increased the expression of the epithelial marker E-cadherin and
decreased the expression of the mesenchymal marker vimentin in the
CaSki (Fig. 3A) and SiHa (Fig. 3B) cells transfected with FOXQ1
siRNA. The data suggest that FOXQ1 may be a potential target for
inhibiting cervical cancer metastasis.
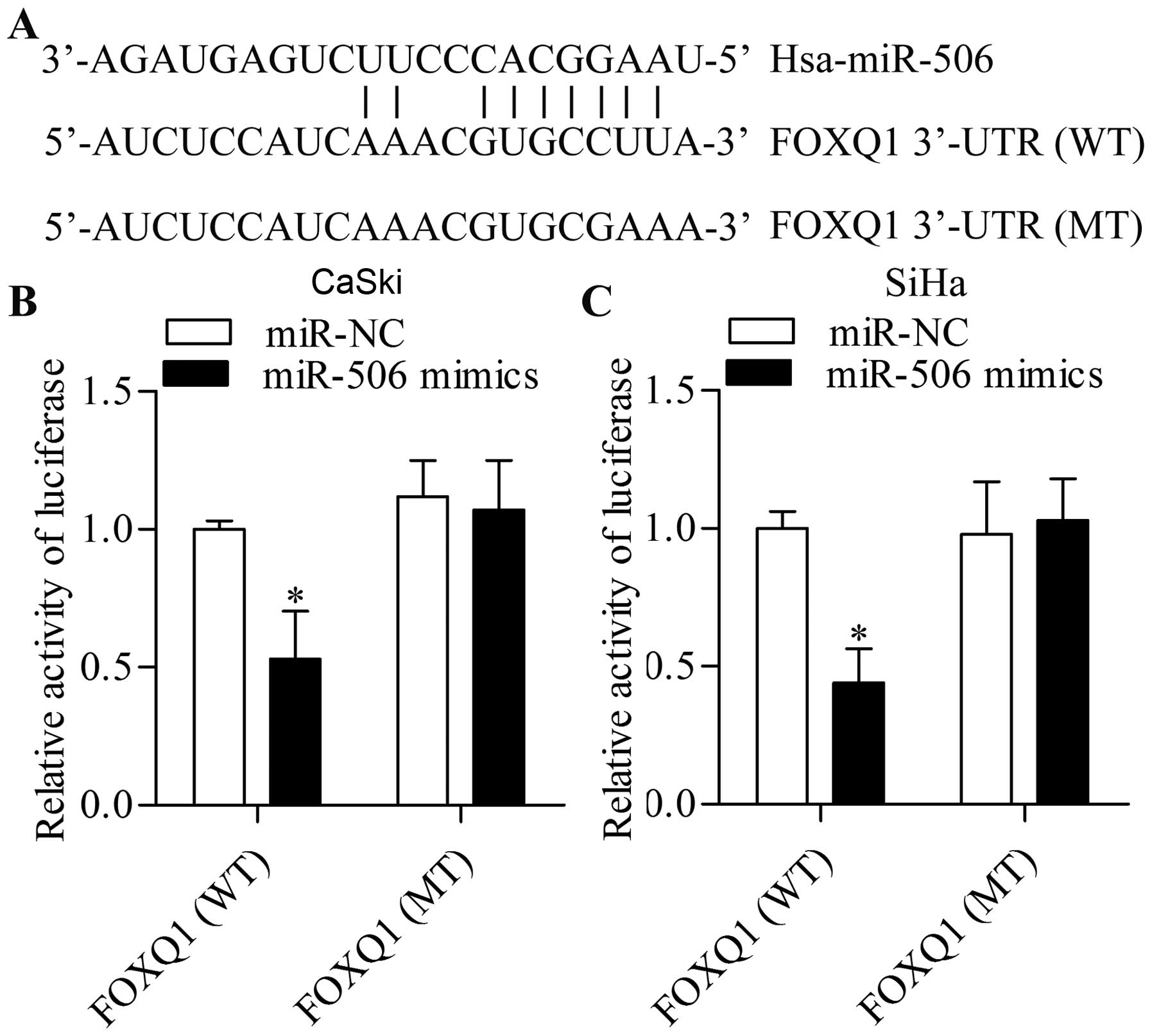
FOXQ1 is a potential target of
miR-506
Previous studies have reported that FOXQ1 expression
can be regulated by specific miRNAs in cancers (26,27).
Bioinformatic research revealed that FOXQ1 has a putative binding
site for miR-506 (Fig. 4A), a
well-recognized tumor-suppressor miRNA (28,29). A
dual-luciferase reporter assay was performed to verify whether or
not FOXQ1 is a target gene of miR-506. The results of the
luciferase assay showed that miR-506 overexpression significantly
inhibited the luciferase activity of the reporter gene with the
wild-type FOXQ1 3′-UTR but not with the mutant FOXQ1 3′-UTR in the
CaSki (Fig. 4B) and SiHa cells
(Fig. 4C). These results indicate
that FOXQ1 is a direct target gene of miR-506.
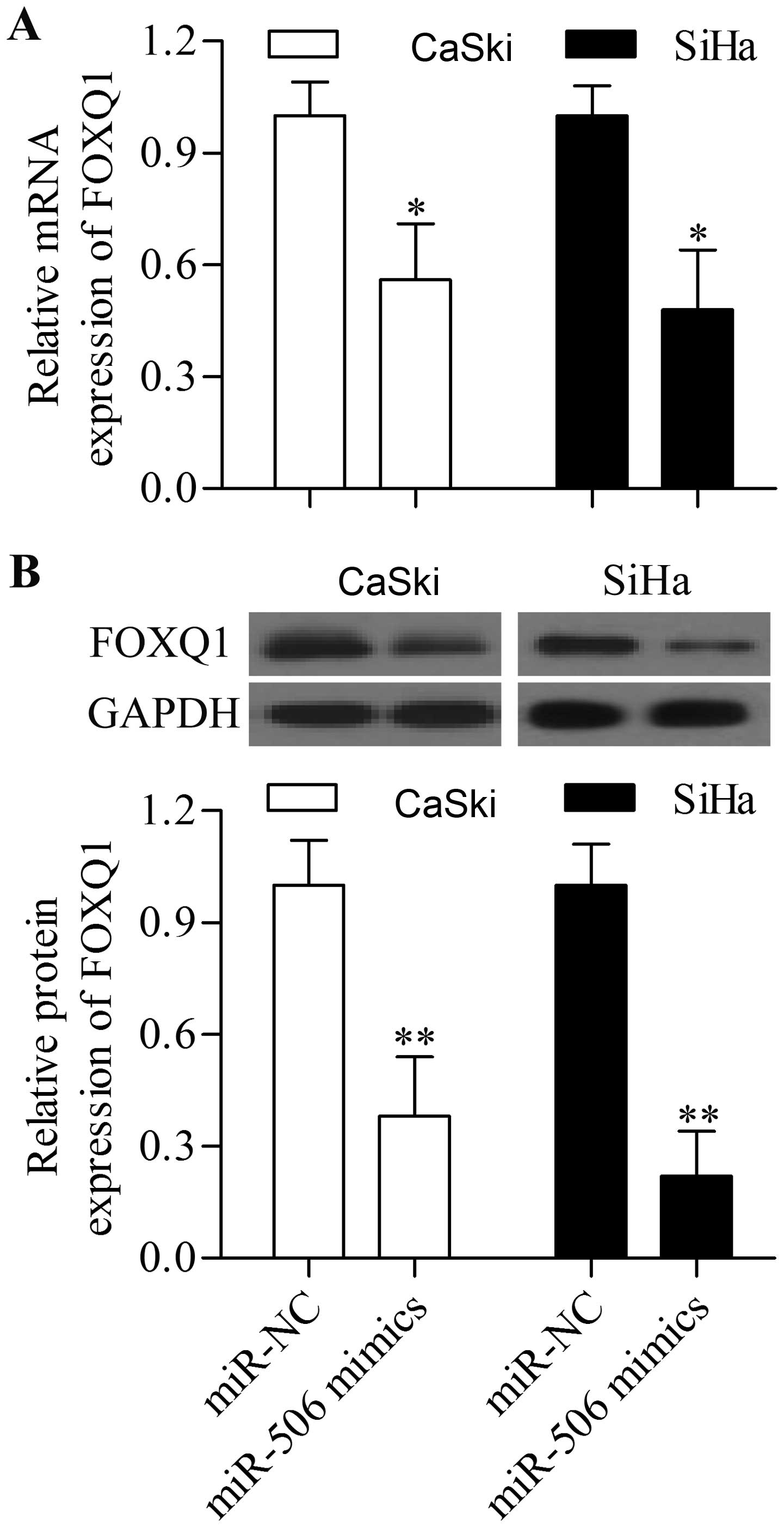
miR-506 negatively regulates FOXQ1
expression
To further confirm that FOXQ1 is the target gene of
miR-506, we detected the expression change of FOXQ1 in response to
miR-506 mimic transfection. The overexpression of miR-506 through
the transfection of miR-506 mimics significantly suppressed the
mRNA expression of FOXQ1 in the cervical cancer cell lines
(Fig. 5A). Furthermore, the protein
expression of FOXQ1 was also significantly decreased by miR-506
overexpression (Fig. 5B).
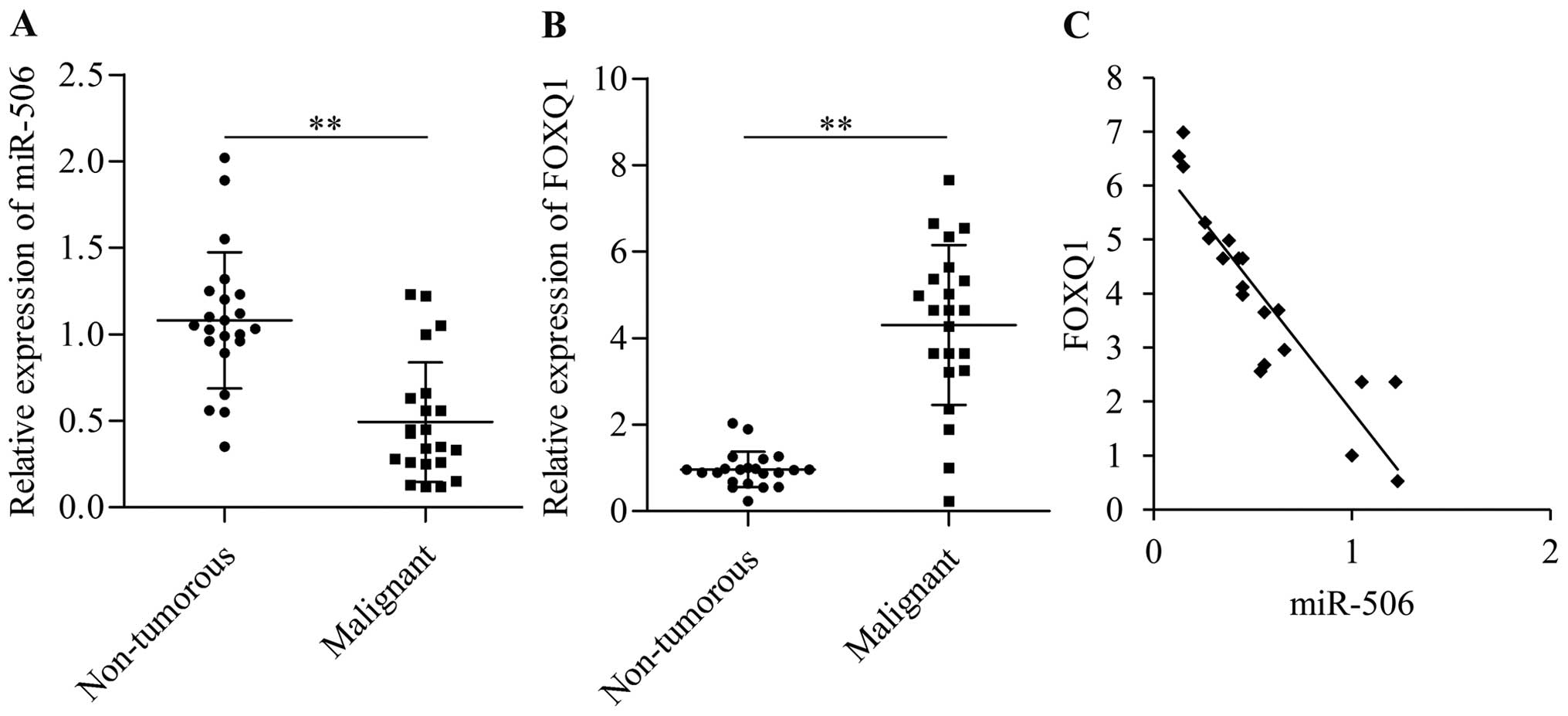
To further investigate the relationship between
miR-506 and FOXQ1, we examined the expression levels of miR-506 and
FOXQ1 in clinical specimens and analyzed their correlation. The
expression of miR-506 (Fig. 6A) was
significantly downregulated in the cervical cancer tissues, whereas
that of FOXQ1 (Fig. 6B) was
significantly upregulated in the cervical cancer tissues when
compared with the adjacent non-tumor tissues. Furthermore,
correlation analysis revealed a marked inverse correlation between
FOXQ1 and miR-506 expression in the cervical cancer tissues
(Fig. 6C). Overall, these results
indicate that miR-506 negatively regulates the expression of FOXQ1,
and the high expression of FOXQ1 in cervical cancer may be caused
by the dysregulation of miR-506.
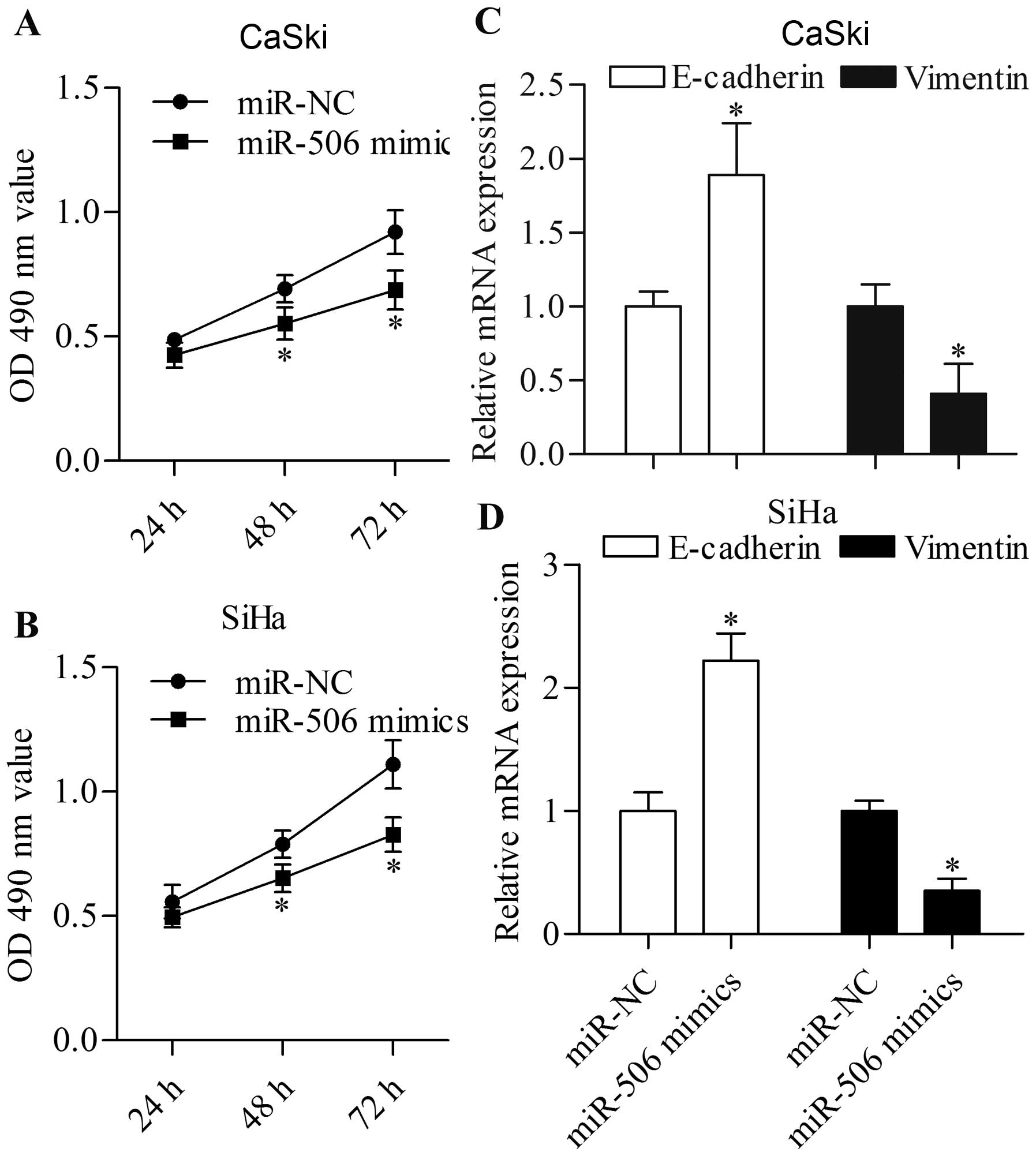
Overexpression of miR-506 inhibits the
proliferation and EMT of cervical cancer cells
Considering the regulatory function of miR-506 on
FOXQ1 expression as described above, we speculated that miR-506
plays an important role in cervical cancer. To test this
hypothesis, we detected the effect of miR-506 overexpression on
cervical cancer cell proliferation. MTT assay showed that the
proliferation of the cervical cancer CaSki (Fig. 7A) and SiHa (Fig. 7B) cells transfected with the miR-506
mimics was significantly decreased. Furthermore, the transfection
of miR-506 mimics significantly increased the expression of
E-cadherin and decreased the expression of vimentin in the CaSki
(Fig. 7C) and SiHa (Fig. 7D). These results revealed that
miR-506 inhibited the proliferation and EMT of cervical cancer
cells.
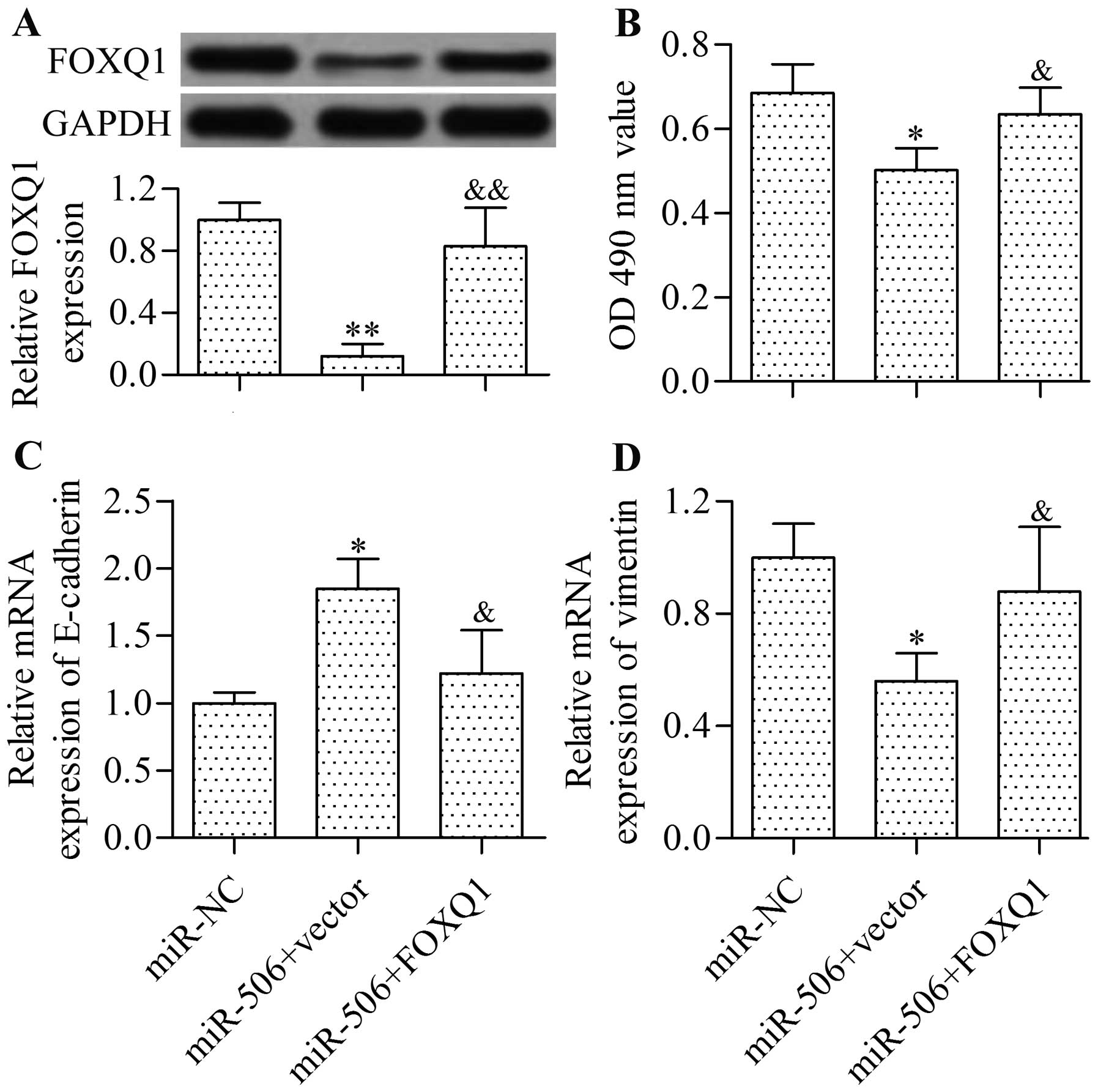
miR-506 suppresses the proliferation and
EMT of cervical cancer cells through FOXQ1
To validate whether miR-506 exerts its
tumor-suppressive function by suppressing FOXQ1, we performed
rescue experiments. Results showed that the restoration of FOXQ1
expression (Fig. 8A) by
transfecting pcDNA3/FOXQ1 overexpression vectors partially rescued
the inhibitory effect of miR-506 overexpression on the
proliferation (Fig. 8B) and EMT
(Fig. 8C and D) in CaSki cells.
Similar results were obtained in the SiHa cells (data not shown).
Thus, these results revealed that miR-506 functions through FOXQ1
in cervical cancer.
Discussion
The present study was the first to report that FOXQ1
regulates the proliferation and EMT of cervical cancer cells.
Moreover, miR-506 directly targeted and regulated FOXQ1 expression,
representing a novel tool for the treatment of cervical cancer by
targeting FOXQ1.
The FOXQ1 gene, which encodes a protein of 403 amino
acids, is predominantly expressed in the bladder, salivary gland,
stomach and trachea (13). FOXQ1 is
overexpressed in colorectal and lung cancer cell lines (13). FOXQ1 was later found to be an
oncogene in various types of cancer. Kaneda et al (12) reported that FOXQ1 expression is
increased in colorectal cancer tissues and promotes tumorigenicity
and tumor growth in vitro and in vivo. High FOXQ1
expression is tightly linked to the aggressive malignant phenotype
of hepatocellular carcinoma (30).
FOXQ1 expression is significantly upregulated in gastric cancer
tissues and is positively related to tumor size, histological
grade, lymph node metastasis, and poor prognosis of gastric cancer
(15). Furthermore, high expression
of FOXQ1 promotes the proliferation and metastasis of glioma
(7) and esophageal cancer cells
(5). However, the role of FOXQ1 in
cervical cancer is poorly understood. In the present study, we
found that the mRNA and protein levels of FOXQ1 were overexpressed
in cervical cancer cell lines. This study is the first to report
the role of FOXQ1 in cervical cancer. Knockdown of FOXQ1 inhibited
the proliferation of cervical cancer cells. These results further
indicate that FOXQ1 is an oncogene, which may be a promising
molecular target for cervical cancer.
EMT is a critical step in cancer metastasis
(10,11). Increasing studies have demonstrated
that FOXQ1 plays an important role in regulating EMT. FOXQ1
promotes the EMT of mammary epithelial cells and breast cancer
metastasis (17). FOXQ1 induces EMT
by inhibiting the epithelial regulator E-cadherin in human cancers
(8). High FOXQ1 expression in lung
cancer tissues correlates with the loss of E-cadherin (14). Fan et al (31) reported that FOXQ1 protein interacts
with the promoter region of E-cadherin and promotes the EMT of
mammary epithelial cells. Knockdown of FOXQ1 suppresses invasion
and metastasis by the reversal of EMT in bladder cancer (32). Consistent with these findings, the
present results demonstrated that FOXQ1 regulated the EMT of
cervical cancer cells. We found that FOXQ1 knockdown increased the
expression of the epithelial marker E-cadherin and decreased the
expression of the mesenchymal marker vimentin, implying the EMT
suppression of cervical cancer. These reports and our findings
support the notion that FOXQ1 is a potential molecular target for
inhibiting EMT and the metastasis of cervical cancer.
Inhibiting oncogene expression by specific miRNAs
could be a novel strategy for the treatment of cancer. To date, the
miRNAs that specifically target and inhibit FOXQ1 have not been
well recognized. In the present study, miR-506 directly targeted
the 3′-UTR of FOXQ1 and inhibited the mRNA and protein expression
of FOXQ1. Further analysis showed that miR-506 expression was
decreased in the cervical cancer tissues and was inversely
correlated with the increased expression of FOXQ1. Thus, the
overexpression of miR-506 inhibited the proliferation and EMT of
cervical cancer cells. Moreover, the restoration of FOXQ1
expression significantly reversed the inhibitory effect of miR-506.
These data indicate that miR-506 suppresses the proliferation and
EMT of cervical cancer cells by targeting FOXQ1. Our findings agree
with a recent study that reported that miR-506 suppresses the
proliferation and invasion of nasopharyngeal cancer cells by
targeting and inhibiting FOXQ1 (33). Overall, miR-506 may be a promising
miRNA for targeting FOXQ1. In addition, miR-124 is reportedly
capable of targeting and inhibiting FOXQ1 to suppress tumor growth
and metastasis in nasopharyngeal cancer (27). Zhang et al (26) reported that FOXQ1 is a target gene
of miR-422a and that the overexpression of miR-422a inhibited the
proliferation and migration of hepatocellular carcinoma. miR-1271
targets FOXQ1 to suppress the proliferation and EMT of gastric
cancer cells (34). These findings
further support that targeting FOXQ1 by using specific miRNAs could
be potential strategy for cancer therapy.
In this study, miR-506 inhibited the proliferation
and EMT of cervical cancer cells by targeting FOXQ1. We further
demonstrated that miR-506 expression was downregulated in cervical
cancer tissues. miR-506 functioned as a tumor-suppressive miRNA. In
fact, increasing evidence supports the tumor-suppressive role of
miR-506 in various cancer types. The overexpression of miR-506
inhibited the proliferation, metastasis, and EMT of breast cancer
cells (35,36). Furthermore, miR-506 suppressed the
metastasis of breast cancer cells by targeting the IQ motif
containing GTPase-activating protein 1 (29). In gastric cancer, miR-506 inhibited
EMT by targeting ETS1 (37) or
SNAI2 (38). miR-506 also
suppressed the proliferation and invasion of gastric cancer cells
by targeting Yap1 (39).
Furthermore, miR-506 served a tumor-suppressive function in
hepatocellular carcinoma (40),
colon cancer (28), and oral
squamous cell carcinoma (41) by
targeting various oncogenes. In the present study, we demonstrated
the critical role of miR-506 in cervical cancer. Our results were
consistent with those of a recent study that reported that miR-506
is significantly downregulated in cervical cancer tissues (42). These authors further demonstrated
that miR-506 overexpression suppressed the growth and enhanced
apoptosis and chemosensitivity of cervical cancer cells by
targeting Gli3 (42). Overall,
decreased miR-506 expression may result in the overexpression of
oncogenes, such as FOXQ1, which contributes to the development and
progression of cervical cancer.
In conclusion, our study elucidated that FOXQ1
functions as an oncogene and is related to the proliferation and
EMT of cervical cancer cells. FOXQ1 could be targeted and inhibited
by miR-506, leading to the suppression of the proliferation and EMT
of cervical cancer cells. These results suggest that the
miR-506/FOXQ1 axis plays an important role in the pathogenesis of
cervical cancer, representing potential molecular targets for the
development of anticancer agents.
Abbreviations:
|
miRNAs
|
microRNAs
|
|
FOXQ1
|
forkhead box Q1
|
|
FOX
|
forkhead box
|
|
siRNA
|
small interfering RNA
|
|
EMT
|
epithelial-mesenchymal transition
|
References
|
1
|
Ojesina AI, Lichtenstein L, Freeman SS,
Pedamallu CS, Imaz-Rosshandler I, Pugh TJ, Cherniack AD, Ambrogio
L, Cibulskis K, Bertelsen B, et al: Landscape of genomic
alterations in cervical carcinomas. Nature. 506:371–375. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Rob L, Halaska M and Robova H:
Nerve-sparing and individually tailored surgery for cervical
cancer. Lancet Oncol. 11:292–301. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Lam EW, Brosens JJ, Gomes AR and Koo CY:
Forkhead box proteins: Tuning forks for transcriptional harmony.
Nat Rev Cancer. 13:482–495. 2013. View
Article : Google Scholar : PubMed/NCBI
|
|
4
|
Myatt SS and Lam EW: The emerging roles of
forkhead box (Fox) proteins in cancer. Nat Rev Cancer. 7:847–859.
2007. View
Article : Google Scholar : PubMed/NCBI
|
|
5
|
Pei Y, Wang P, Liu H, He F and Ming L:
FOXQ1 promotes esophageal cancer proliferation and metastasis by
negatively modulating CDH1. Biomed Pharmacother. 74:89–94. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Xia L, Huang W, Tian D, Zhang L, Qi X,
Chen Z, Shang X, Nie Y and Wu K: Forkhead box Q1 promotes
hepatocellular carcinoma metastasis by transactivating ZEB2 and
VersicanV1 expression. Hepatology. 59:958–973. 2014. View Article : Google Scholar
|
|
7
|
Sun HT, Cheng SX, Tu Y, Li XH and Zhang S:
FoxQ1 promotes glioma cells proliferation and migration by
regulating NRXN3 expression. PLoS One. 8:e556932013. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Qiao Y, Jiang X, Lee ST, Karuturi RK, Hooi
SC and Yu Q: FOXQ1 regulates epithelial-mesenchymal transition in
human cancers. Cancer Res. 71:3076–3086. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Feng J, Xu L, Ni S, Gu J, Zhu H, Wang H,
Zhang S, Zhang W and Huang J: Involvement of FoxQ1 in NSCLC through
regulating EMT and increasing chemosensitivity. Oncotarget.
5:9689–9702. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Scheel C, Onder T, Karnoub A and Weinberg
RA: Adaptation versus selection: The origins of metastatic
behavior. Cancer Res. 67:11476–11480. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Thiery JP: Epithelial-mesenchymal
transitions in development and pathologies. Curr Opin Cell Biol.
15:740–746. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Kaneda H, Arao T, Tanaka K, Tamura D,
Aomatsu K, Kudo K, Sakai K, De Velasco MA, Matsumoto K, Fujita Y,
et al: FOXQ1 is overexpressed in colorectal cancer and enhances
tumorigenicity and tumor growth. Cancer Res. 70:2053–2063. 2010.
View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Bieller A, Pasche B, Frank S, Gläser B,
Kunz J, Witt K and Zoll B: Isolation and characterization of the
human forkhead gene FOXQ1. DNA Cell Biol. 20:555–561. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Feng J, Zhang X, Zhu H, Wang X, Ni S and
Huang J: FoxQ1 overexpression influences poor prognosis in
non-small cell lung cancer, associates with the phenomenon of EMT.
PLoS One. 7:e399372012. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Liang SH, Yan XZ, Wang BL, Jin HF, Yao LP,
Li YN, Chen M, Nie YZ, Wang X, Guo XG, et al: Increased expression
of FOXQ1 is a prognostic marker for patients with gastric cancer.
Tumour Biol. 34:2605–2609. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Gao M, Shih IEM and Wang TL: The role of
forkhead box Q1 transcription factor in ovarian epithelial
carcinomas. Int J Mol Sci. 13:13881–13893. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Zhang H, Meng F, Liu G, Zhang B, Zhu J, Wu
F, Ethier SP, Miller F and Wu G: Forkhead transcription factor
foxq1 promotes epithelial-mesenchymal transition and breast cancer
metastasis. Cancer Res. 71:1292–1301. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Ling H, Fabbri M and Calin GA: MicroRNAs
and other non-coding RNAs as targets for anticancer drug
development. Nat Rev Drug Discov. 12:847–865. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Bartel DP: MicroRNAs: Genomics,
biogenesis, mechanism, and function. Cell. 116:281–297. 2004.
View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Shi XB, Tepper CG and deVere White RW:
Cancerous miRNAs and their regulation. Cell Cycle. 7:1529–1538.
2008. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Wang F, Li B and Xie X: The roles and
clinical significance of microRNAs in cervical cancer. Histol
Histopathol. 31:131–139. 2016.
|
|
22
|
Han Y, Xu GX, Lu H, Yu DH, Ren Y, Wang L,
Huang XH, Hou WJ, Wei ZH, Chen YP, et al: Dysregulation of miRNA-21
and their potential as biomarkers for the diagnosis of cervical
cancer. Int J Clin Exp Pathol. 8:7131–7139. 2015.PubMed/NCBI
|
|
23
|
Yang Y, Xie YJ, Xu Q, Chen JX, Shan NC and
Zhang Y: Down-regulation of miR-1246 in cervical cancer tissues and
its clinical significance. Gynecol Oncol. 138:683–688. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Zhao Q, Zhai YX, Liu HQ, Shi YA and Li XB:
MicroRNA-491-5p suppresses cervical cancer cell growth by targeting
hTERT. Oncol Rep. 34:979–986. 2015.PubMed/NCBI
|
|
25
|
Song X, Shi B, Huang K and Zhang W:
miR-133a inhibits cervical cancer growth by targeting EGFR. Oncol
Rep. 34:1573–1580. 2015.PubMed/NCBI
|
|
26
|
Zhang J, Yang Y, Yang T, Yuan S, Wang R,
Pan Z, Yang Y, Huang G, Gu F, Jiang B, et al: Double-negative
feedback loop between microRNA-422a and forkhead box (FOX)G1/Q1/E1
regulates hepatocellular carcinoma tumor growth and metastasis.
Hepatology. 61:561–573. 2015. View Article : Google Scholar
|
|
27
|
Peng XH, Huang HR, Lu J, Liu X, Zhao FP,
Zhang B, Lin SX, Wang L, Chen HH, Xu X, et al: miR-124 suppresses
tumor growth and metastasis by targeting Foxq1 in nasopharyngeal
carcinoma. Mol Cancer. 13:1862014. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Zhang Y, Lin C, Liao G, Liu S, Ding J,
Tang F, Wang Z, Liang X, Li B, Wei Y, et al: MicroRNA-506
suppresses tumor proliferation and metastasis in colon cancer by
directly targeting the oncogene EZH2. Oncotarget. 6:32586–32601.
2015.PubMed/NCBI
|
|
29
|
Sun G, Liu Y, Wang K and Xu Z: miR-506
regulates breast cancer cell metastasis by targeting IQGAP1. Int J
Oncol. 47:1963–1970. 2015.PubMed/NCBI
|
|
30
|
Wang W, He S, Ji J, Huang J, Zhang S and
Zhang Y: The prognostic significance of FOXQ1 oncogene
overexpression in human hepatocellular carcinoma. Pathol Res Pract.
209:353–358. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Fan DM, Feng XS, Qi PW and Chen YW:
Forkhead factor FOXQ1 promotes TGF-β1 expression and induces
epithelial-mesenchymal transition. Mol Cell Biochem. 397:179–186.
2014. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Zhu Z, Zhu Z, Pang Z, Xing Y, Wan F, Lan D
and Wang H: Short hairpin RNA targeting FOXQ1 inhibits invasion and
metastasis via the reversal of epithelial-mesenchymal transition in
bladder cancer. Int J Oncol. 42:1271–1278. 2013.PubMed/NCBI
|
|
33
|
Zhang Z, Ma J, Luan G, Kang L, Su Y, He Y
and Luan F: miR-506 suppresses tumor proliferation and invasion by
targeting FOXQ1 in nasopharyngeal carcinoma. PLoS One.
10:e01228512015. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Xiang XJ, Deng J, Liu YW, Wan LY, Feng M,
Chen J and Xiong JP: miR-1271 inhibits cell proliferation, invasion
and EMT in gastric cancer by targeting FOXQ1. Cell Physiol Biochem.
36:1382–1394. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Arora H, Qureshi R and Park WY: miR-506
regulates epithelial mesenchymal transition in breast cancer cell
lines. PLoS One. 8:e642732013. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Yu F, Lv M, Li D, Cai H, Ma L, Luo Q, Yuan
X and Lv Z: miR-506 over-expression inhibits proliferation and
metastasis of breast cancer cells. Med Sci Monit. 21:1687–1692.
2015. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Li Z, Liu Z, Dong S, Zhang J, Tan J, Wang
Y, Ge C, Li R, Xue Y, Li M, et al: miR-506 inhibits
epithelial-to-mesenchymal transition and angiogenesis in gastric
cancer. Am J Pathol. 185:2412–2420. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Sakimura S, Sugimachi K, Kurashige J, Ueda
M, Hirata H, Nambara S, Komatsu H, Saito T, Takano Y, Uchi R, et
al: The miR-506-induced epithelial-mesenchymal transition is
involved in poor prognosis for patients with gastric cancer. Ann
Surg Oncol. 22(Suppl 3): 1436–1443. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Deng J, Lei W, Xiang X, Zhang L, Yu F,
Chen J, Feng M and Xiong J: MicroRNA-506 inhibits gastric cancer
proliferation and invasion by directly targeting Yap1. Tumour Biol.
36:6823–6831. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Deng Q, Xie L and Li H: miR-506 suppresses
cell proliferation and tumor growth by targeting Rho-associated
protein kinase 1 in hepatocellular carcinoma. Biochem Biophys Res
Commun. 467:921–927. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Deng L and Liu H: MicroRNA-506 suppresses
growth and metastasis of oral squamous cell carcinoma via targeting
GATA6. Int J Clin Exp Med. 8:1862–1870. 2015.PubMed/NCBI
|
|
42
|
Wen SY, Lin Y, Yu YQ, Cao SJ, Zhang R,
Yang XM, Li J, Zhang YL, Wang YH, Ma MZ, et al: miR-506 acts as a
tumor suppressor by directly targeting the hedgehog pathway
transcription factor Gli3 in human cervical cancer. Oncogene.
34:717–725. 2015. View Article : Google Scholar
|