Introduction
In mammals, many life activities vary in an
approximate 24 h periodic fluctuation, which is called the
circadian rhythm (1,2). The circadian rhythm, which is one of
the basically intrinsic characteristics of life activities, plays
an important role in maintaining complicated life activities in a
highly coordinated and orderly manner (3,4). The
clock genes, whose rhythmic expression is responsible for circadian
rhythms, exist in almost all cells in the body (5,6). To
date, at least 14 core clock genes have been described, including
Per1, period circadian clock 2 (Per2), Per3, Cry1, Cry2, Tim, Ck1ε,
Clock, Bmal1, Rors, Rev-Erbs, Npas2, Dec1 and Dec2 (7,8). In
mammals, circadian rhythms play an important role in physiological
activities, including cell proliferation, metabolism and hormone
secretion (9,10). Aberrant circadian rhythms lead to
cardiovascular diseases, gastrointestinal diseases, nervous and
mental diseases and cancers (11–13).
Per2 is an important clock gene that works as a
pacemaker of circadian rhythms in mammals (14). The absence of Per2 can lead to loss
of circadian rhythms (15–17). In recent years, studies have shown
that the aberrant expression of Per2 is responsible for not only
circadian rhythm alterations but also the occurrence and
development of cancers (6,18). Per2 is reduced in various types of
solid cancers, including breast and skin cancer, hepatocellular
carcinoma, colorectal cancer, renal carcinoma, gastric cancer and
head and neck squamous cell carcinoma (11,19–24).
Since many downstream cell cycle genes are regulated by Per2, the
aberrant expression of Per2 affects cell cycle progression and
leads to carcinogenesis (6,22,25).
Imbalance of cell proliferation and apoptosis caused
by cell cycle disorder is the main reason for carcinogenesis
(26,27). The cell cycle is under the control
of the cyclin/cyclin-dependent protein kinase
(CDK)/cyclin-dependent kinase inhibitor (CKI) cell cycle network
composed of cyclins, CDKs and CKIs (28,29).
To date, there have been only dispersed research studies on the
Per2 regulation of downstream cell cycle genes, and these are
cyclin A, B1, D1 and E, p53, c-myc, Rb, Mdm-2 and Gadd45α, which
are mainly focused on the Per2 regulation of cyclins (6,18,25,30).
However, the role Per2 plays in CDKs and CKIs, which are the two
other important aspects of the cyclin/CDK/CKI network, remains
unclear. We speculated that Per2 may regulate numerous molecules in
all the three aspects of the cyclin/CDK/CKI cell cycle network. In
the present study, we downregulated Per2 in oral squamous cell
carcinoma (OSCC) cell line Tca8113, and then detected the
alterations in the cell cycle, cell proliferation and apoptosis and
all the important genes in the cyclin/CDK/CKI network to further
illustrate the relationship of Per2 with the occurrence and
development of cancers.
Materials and methods
Cell culture
Normal oral mucosa was collected from the Department
of Maxillofacial Plastic Surgery at the Affiliated Hospital of
Stomatology, Chongqing Medical University following approval by the
local ethics committee. Patient samples were obtained after
informed consent following the tenets of the Declaration of
Helsinki, and written consent was obtained from all patients prior
to surgery. The mucosa was kept in 1.0 U/ml Dispase II (Roche,
Indianapolis, IN, USA) at 4°C overnight. Complete epithelial layer
was separated from the mucosa under a microscope (Leica, Wetzlar,
Germany). The epithelial layer was digested in 0.25% TrypLE Express
(Gibco, Grand Island, NY, USA) at 37°C, and then inoculated in a
6-well plate paved by rat tail collagen using OKM (ScienCell, San
Diego, CA, USA) at 37°C in a humidified atmosphere of 95% air and
5% CO2. The oral mucosal epithelial cells were
continuously passaged/4 days. The second generation of cells was
used for RNA and protein extraction, and the third generation of
cells was used for making cell climbing slices. Tca8113 cells
(Chongqing Key Laboratory of Oral Diseases and Biomedical Sciences,
China) were cultured in RPMI-1640 medium with 10% fetal bovine
serum (both from HyClone, Logan, UT, USA) at 37°C in an atmosphere
of 95% humidity and 5% CO2.
Purity identification of the oral mucosal
epithelial cells
Cell climbing slices were fixed by 4%
paraformaldehyde, and then incubated for 10 min with 3%
H2O2 at 37°C. After permeabilization in 0.1%
Triton X-100 for 15 min, the slices were blocked using goat serum
for 30 min at 37°C. The slices were incubated firstly with the
rabbit monoclonal anti-keratin antibody (1:100; ZA-0540) overnight
at 4°C, and secondly with the rabbit SP kit (SP-9001) (both from
ZSGB-BIo, Beijing, China) for 1 h at 37°C according to the
manufacturer's recommended protocol. Finally, slices were examined
under a microscope (Olympus, Tokyo, Japan). In negative controls
all reagents were used except the primary antibody.
Downregulation of Per2 in Tca8113 cells
by shRNA plasmids
The plasmids pGPU6-Per2-shRNA-I-III and
pGPU6-control-shRNA were obtained from Chengdu Biotechnology Co.,
Ltd. (Table I). The day before
transfection, the Tca8113 cells were incubated into a 6-well plate
to ensure that by the time of transfection the cells reached ~30%
confluency. Approximately 4 μg of the plasmids were
transfected using Lipofectamine 2000 (Invitrogen, Carlsbad, CA,
USA) according to the manufacturer's protocol. The effect of Per2
downregulation was examined 36–72 h later. There were five groups
in our experiment: Per2-shRNA-I, Per2-shRNA-II, Per2-shRNA-III,
control-shRNA and Tca8113 group. Per2-shRNA-I, Per2-shRNA-II,
Per2-shRNA-III and the control-shRNA group were transfected with
pGPU6-Per2-shRNA-I, pGPU6-Per2-shRNA-II, pGPU6-Per2-shRNA-III and
pGPU6-control-shRNA, respectively; the Tca8113 group did not accept
any reagents.
 | Table IThe RNA oligos of shRNA and negative
control plasmids. |
Table I
The RNA oligos of shRNA and negative
control plasmids.
| Plasmid | RNA oligos
(5′-3′) |
|---|
|
pGPU6-Per2-shRNA-I |
5′-CACCGAAGTACGCCCTCAGGAGCTTCAAGAGAGCTCCTGAGGGCGTACTTCTTTTTTG-3′ |
|
5′-GATCCAAAAAAGAAGTACGCCCTCAGGAGCTCTCTTGAAGCTCCTGAGGGCGTACTTC-3′ |
|
pGPU6-Per2-shRNA-II |
5′-CACCGTGAAGAATGCCGATATGTTTCAAGAGAACATATCGGCA
TTCTTCACTTTTTTG-3′ |
|
5′-GATCCAAAAAAGTGAAGAATGCCGATATGTTCTCTTGAAACATATCGGCATTCTTCAC-3′ |
|
pGPU6-Per2-shRNA-III |
5′-CACCGAAGTACGCCCTCAGGAGCTTCAAGAGAGCTCCTGAGGGCGTACTTCTTTTTTG-3′ |
|
5′-GATCCAAAAAAGAAGTACGCCCTCAGGAGCTCTCTTGAAGCTCCTGAGGGCGTACTTC-3′ |
|
pGPU6-control-shRNA |
5′-CCGGGCACTACGAGAGCTAACTCAGCTCGAGCTGAGTTAGCTCTCGTAGTGCTTTTTG-3′ |
|
5′-AATTCAAAAAGCACTACGAGAGCTAACTCAGCTCGAGCTGAGTTAGCTCTCGTAGTGC-3′ |
Western blotting
The cells were lysed using RIPA + PMSF (Beyotime,
Jiangsu, China) and centrifuged for 2 min, at 4°C and 167.7 × g.
The concentration of cell protein was detected using the enhanced
BCA protein assay kit (Beyotime). Proteins (50 μg) were
seperated by 8% SDS-PAGE using Mini-PROTEAN 3 system (Bio-Rad,
Hercules, CA, USA) and transferred to polyvinylidene difluoride
(PVDF) membranes (Pierce, Rockford, IL, USA) using Trans-Blot SD
Semi-Dry Transfer Cell (Bio-Rad). The membranes were incubated with
mouse monoclonal anti-Per2 antibody (1:500; 19-J6:sc-101105; Santa
Cruz Biotechnology, Inc., Santa Cruz, CA, USA) and mouse monoclonal
anti-β-actin antibody (1:1,000; 60008-1-lg), respectively,
overnight at 4°C, followed by goat monoclonal anti-mouse IgG
(1:1,000) (SA00001-1) (both from ProteinTech, Chicago, IL, USA) for
1 h at 37°C. Blots were detected using enhanced chemiluminescence
reagent (Pierce) under a fluorescent chemiluminescence imaging
system (ChemiDoc XRS+; Bio-Rad). The software Quality one (Bio-Rad)
was used to analyze the blots. To ensure accuracy the experiment
was performed in triplicate.
Flow cytometric analysis
For analysis of the cell cycle, after a 48-h
transient transfection, the cells were harvested and fixed using
70% ethanol overnight at 4°C. The cells were stained with propidium
iodide solution (Cell Cycle Detection kit; KGA, China) for 30 min
at 4°C in the dark, and subsequently detected by a flow cytometer
(FACSVantage; BD Biosciences, San Jose CA, USA). The following
formula was used to calculate the proliferation index (PI) of the
cells: PI = (S + G2/M)/(G0/G1 + S + G2/M) × 100% (G0, G1, S, G2 and
M represent the corresponding cell cycle phases). To ensure
accuracy, the experiment was performed in triplicate. For analysis
of apoptosis, after a 48-h transient transfection, the cells were
harvested and stained with the Annexin V-FITC cell apoptosis
analysis kit (with propidium iodide) (Sungene, Tianjin, China).
Apoptotic cells were quantified by a flow cytometer (FACSVantage).
The following formula was used to calculate apoptotic index (AI) of
the cells: AI = (number of apoptotic cells/number of total detected
cells) × 100%. To ensure accuracy, the experiment was performed in
triplicate.
Quantitative real-time PCR (RT-qPCR)
After a 36-h transient transfection, total RNA was
isolated from the cells using RNAiso Plus (9109; Takara, Japan).
The optical density and concentration of RNA were detected by an
ultraviolet spectrophotometer (NanoDrop, USA). RNA was
reverse-transcribed with 20 μl of the system with a Prime
Script RT reagent kit (RR047A; Takara) on a T100 thermal cycler
(Bio-Rad, Singapore) according to the manufacturer's instructions.
Oligo 17.0 software was used to design the specific primers of p53,
p16, p21, cyclin A2, B1, D1 and E, CDK1, CDK2, CDK4, CDK6, E2F1,
c-myc, cdc25, Wee1, Rb1 and GAPDH (endogenous reference) (Table II). According to the manufacturer's
instructions, cDNA was used as the template for amplification using
SYBR Premix Ex Taq™ II (RR820A; Takara) on a CFX96 Real-Time PCR
Detection system (Bio-Rad). The threshold cycle (Ct) value was
acquired. The relative expression levels of p53, p16, p21, cyclin
A2, B1, D1 and E, CDK1, CDK2, CDK4, CDK6, E2F1, c-myc, cdc25, Wee1
and Rb1 mRNA in cells were calculated using the 2−ΔΔCt
method. To ensure accuracy, the experiment was performed in
triplicate.
 | Table IIPrimers used for real-time PCR
amplification of gene expression |
Table II
Primers used for real-time PCR
amplification of gene expression
| Gene | Forward primer
sequence | Reverse primer
sequence |
|---|
| Per2 |
5′-CGTGTTCCACAGTTTCACCT-3′ |
5′-GGTAGCGGATTTCATTCTCG-3′ |
| cyclin A2 |
5′-ATGTCACCGTTCCTCCTTG-3′ |
5′-AGGGCATCTTCACGCTCTA-3′ |
| cyclin B1 |
5′-TGGTTGATACTGCCTCTCCA-3′ |
5′-TGACTGCTTGCTCTTCCTCA-3′ |
| cyclin D1 |
5′-GTGTATCGAGAGGCCAAAGG-3′ |
5′-CAACCAGAAATGCACAGACC-3′ |
| cyclin E |
5′-CTGGATGTTGACTGCCTTGA-3′ |
5′-ATGTCGCACCACTGATACCC-3′ |
| c-myc |
5′-ATCCTGTCCGTCCAAGCA-3′ |
5′-CGCACAAGAGTTCCGTAG-3′ |
| p53 |
5′-GTCCAACAACACCAGCTCCT-3′ |
5′-CTCTCGGAACATCTCGAAGC-3′ |
| CDK1 |
5′-GTCCGCAACAGGGAAGAAC-3′ |
5′-CGAAAGCCAAGATAAGCAACT-3′ |
| CDK2 |
5′-CAGGATGTGACCAAGCCAGT-3′ |
5′-TGAGTCCAAATAGCCCAAGG-3′ |
| CDK4 |
5′-CTGGACACTGAGAGGGCAAT-3′ |
5′-TGGGAAGGAGAAGGAGAAGC-3′ |
| CDK6 |
5′-TCTTCATTCACACCGAGTAGTGC-3′ |
5′-TGAGGTTAGAGCCATCTGGAAA-3′ |
| Rb1 |
5′-CACAAGCAACCTCAGCCTTC-3′ |
5′-GCGTTCACAAAGTGTATTTAGCC-3′ |
| E2F1 |
5′-CCAACTCCCTCTACCCTTGA-3′ |
5′-GTCTCCCTCCCTCACTTTCC-3′ |
| Wee1 |
5′-TGTGGTGGTGTGCTGCTTAT-3′ |
5′-TTCAAAGGGAGGGTATGTCTG-3′ |
| cdc25 |
5′-TACTCGGCCATGTCACCCTT-3′ |
5′-GGGTCGTATCGCCCTCATC-3′ |
| p16 |
5′-ACCAGAGGCAGTAACCATGC-3′ |
5′-TGATCTAAGTTTCCCGAGGTTT-3′ |
| p21 |
5′-TTAGCAGCGGAACAAGGAGT-3′ |
5′-CGTTAGTGCCAGGAAAGACA-3′ |
| GAPDH |
5′-ACAACTTTGGTATCGTGGAAGG-3′ |
5′-GCCATCACGCCACAGTTTC-3′ |
Statistical analysis
The SPSS 17.0 statistical software package was used
to analyze and calculate the mean ± SD of the data. The difference
in Per2 expression between epithelial and Tca8113 cells was
analyzed using group t-test. In addition, one-way ANOVA was used to
analyze the differences between the various groups transfected with
the different plasmids. A value of P<0.05 was considered
statistically significant.
Results
Culture and purity identification of the
oral mucosal epithelial cells
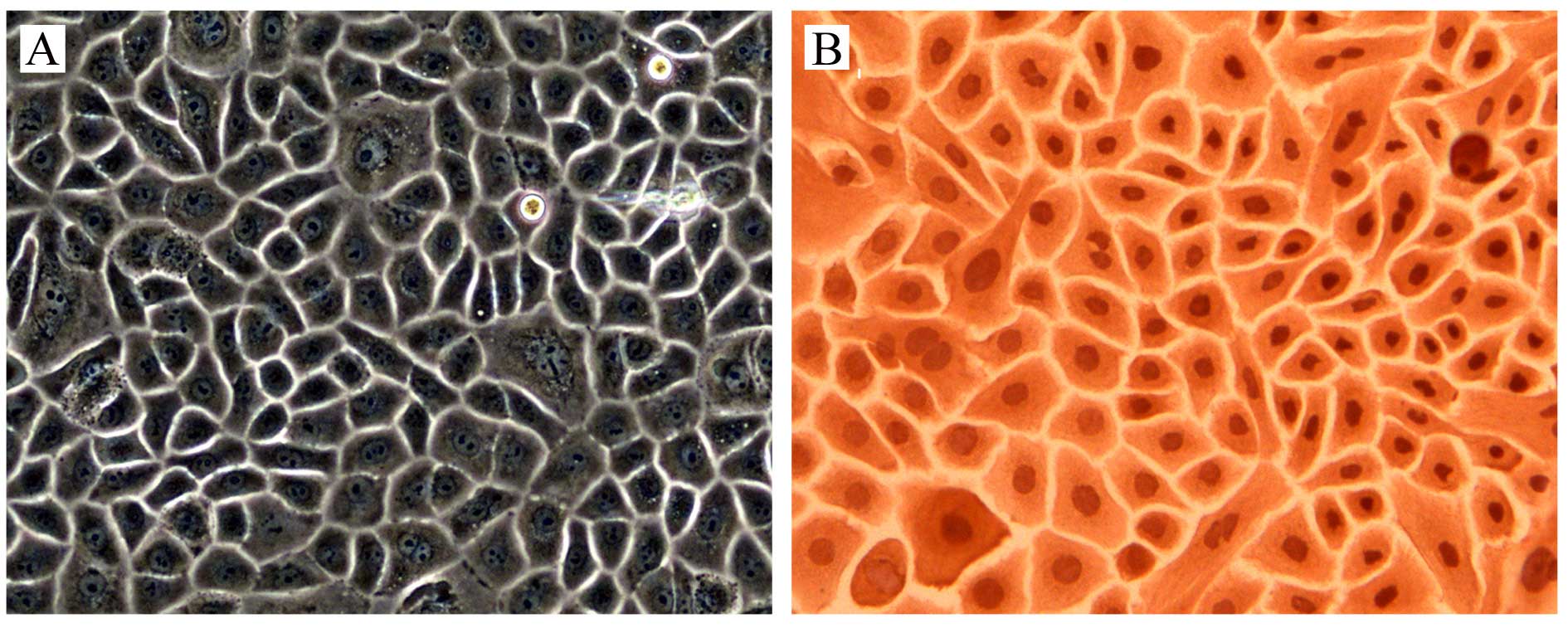
Under microscope, the oral mucosal epithelial cells
had a paving stone appearance (Fig.
1A), and keratin was expressed in 100% of the cells (Fig. 1B), which identified the purity of
the oral mucosal epithelial cells.
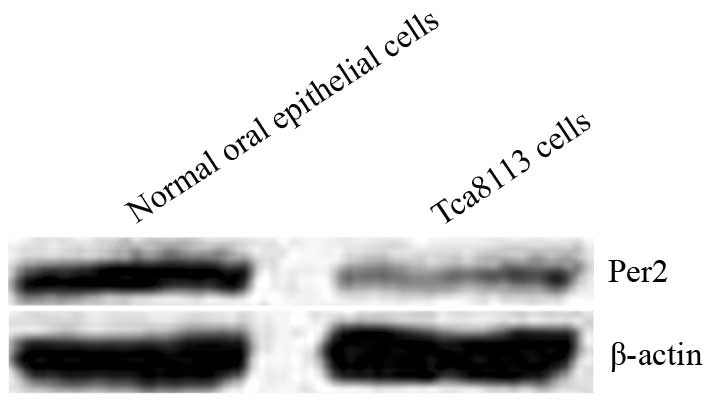
Expression of Per2 mRNA and protein in
the oral mucosal epithelial and Tca8113 cells
In the oral mucosal epithelial and Tca8113 cells,
the expression of Per2 mRNA was 2.41±0.21 and 1.00±0.12,
respectively; and the expression of Per2 protein was 2.87±0.26 and
1.11±0.13, respectively (Fig. 2).
The expression levels of Per2 mRNA and protein in the Tca8113 cells
were significantly lower than those of the oral mucosal epithelial
cells (P<0.05), indicating that Per2 expression is reduced in
OSCC.
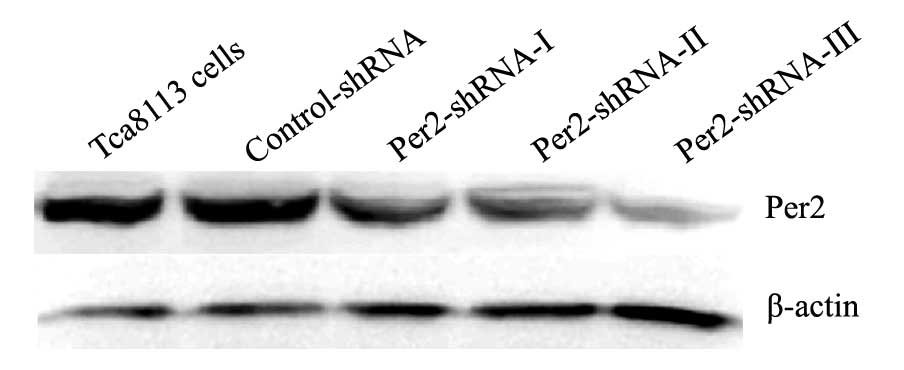
Alterations of Per2 mRNA and protein
expression after transfections in the Tca8113 cells
In the Tca8113, control-shRNA, Per2-shRNA-I,
Per2-shRNA-II and Per2-shRNA-III groups, the expression levels of
Per2 mRNA were 3.20±0.52, 3.01±0.11, 1.67±0.30, 1.45±0.34 and
1.00±0.13, respectively; and the expression levels of Per2 protein
were 3.21±0.42, 3.18±0.52, 1.52±0.11, 1.22±0.15 and 0.87±0.21,
respectively (Fig. 3). Per2 mRNA
and protein expression showed no significant difference between the
control-shRNA and Tca8113 groups (P>0.05). However, Per2 mRNA
and protein expression levels were significantly lower in the
Per2-shRNA-III group than those noted in the control-shRNA and
Tca8113 groups (P<0.05). Therefore, Per2 was effectively reduced
in the Per2-shRNA-III group, and the Per2-shRNA-III group was
adopted for subsequent experiment.
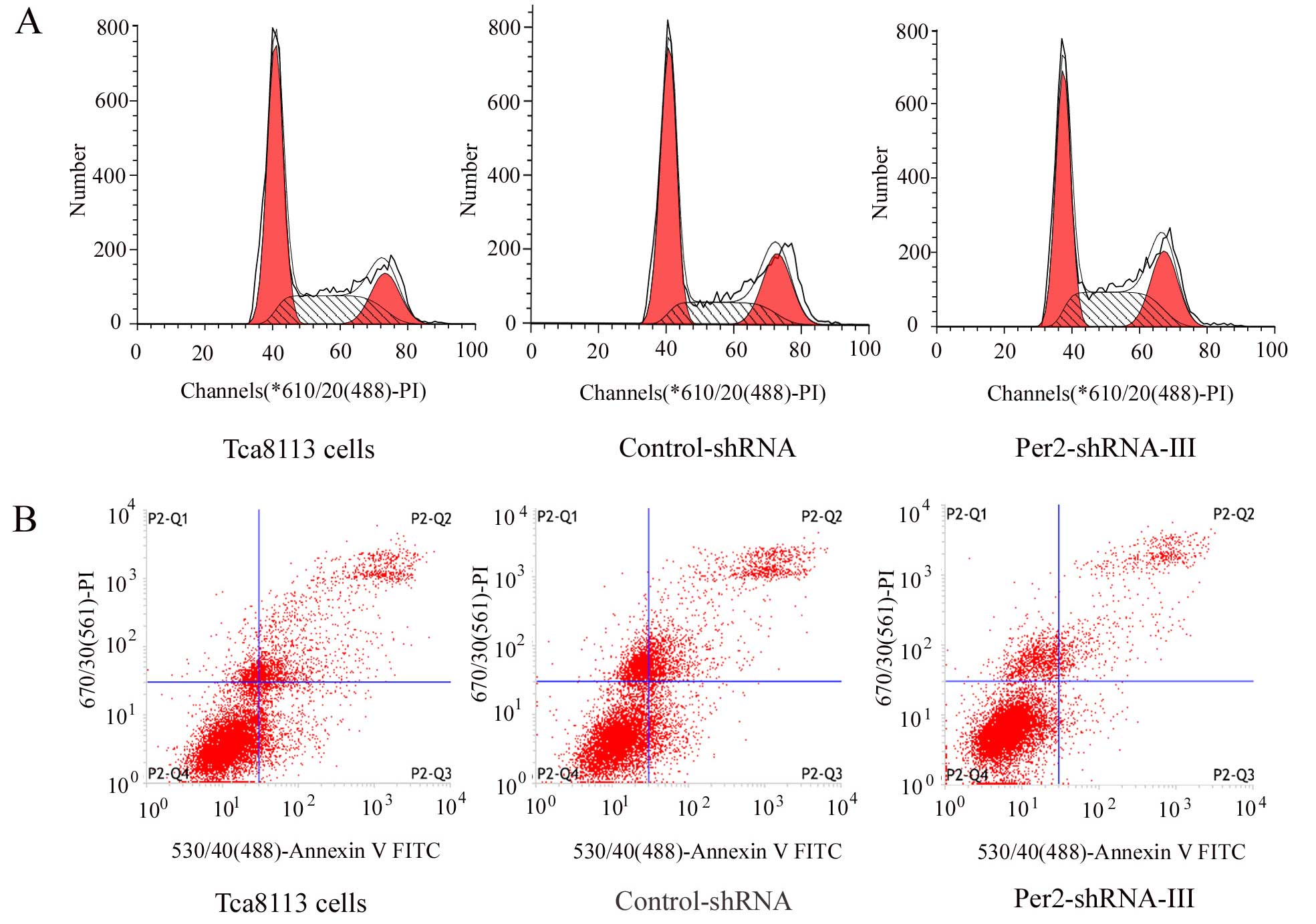
Effects of Per2 downregulation on cell
cycle distribution, PI and AI in the Tca8113 cells
The cells from the different groups were collected
and their cell cycle distribution (Fig.
4A; Table III) and apoptosis
(Fig. 4B; Table III) were analyzed, and then PI and
AI were calculated (Table III).
Compared with those of the Tca8113 and control-shRNA groups, the
Per2-shRNA-III group had a significantly decreased number of cells
in the G1/G0 phase (P<0.05), significantly increased PI
(P<0.05), and significantly decreased AI (P<0.05). The number
of cells in the G1/G0 phase, and PI and AI showed no significant
difference between the Tca8113 and control-shRNA group
(P>0.05).
 | Table IIIEffects of shRNA on cell cycle
distribution, PI and AI in Tca8113 cells (mean ± SD). |
Table III
Effects of shRNA on cell cycle
distribution, PI and AI in Tca8113 cells (mean ± SD).
| Index | Tca8113 | Control-shRNA | Per2-shRNA-III | P | P1 | P2 | P3 |
|---|
| G1/G0 (%) | 51.00±2.12 | 51.83±2.27 | 46.04±1.76 | 0.028 | 0.026 | 0.014 | 0.642 |
| S (%) | 29.23±1.10 | 27.41±4.19 | 30.93±1.94 | 0.355 | 0.476 | 0.167 | 0.447 |
| G2/M (%) | 19.77±2.75 | 20.76±3.17 | 23.03±0.71 | 0.319 | 0.155 | 0.301 | 0.638 |
| PI (%) | 49.00±2.12 | 48.17±2.27 | 53.96±1.77 | 0.028 | 0.026 | 0.014 | 0.642 |
| AI (%) | 14.66±1.93 | 14.13±0.94 | 10.26±1.02 | 0.015 | 0.008 | 0.014 | 0.651 |
Effects of Per2 downregulation on mRNA
expression of cell cycle-related genes in the Tca8113 cells
Results of the RT-qPCR are shown in Table IV. Compared with those of the
Tca8113 and control-shRNA groups, the Per2-shRNA-III group had a
significantly decreased mRNA expression of p53, p16 and p21
(P<0.05), significantly increased mRNA expression of cyclin A2,
B1 and D1, CDK4, CDK6, E2F1 (P<0.05), and a similar mRNA
expression of c-myc, cyclin E, CDK1, CDK2, cdc25, Wee1 and Rb1
(P>0.05). Between the Tca8113 and control-shRNA group, there was
no significant difference in mRNA expression of p53, p16, p21
cyclin A2, B1 and D1, CDK4, CDK6 and E2F1 (P>0.05).
 | Table IVEffects of shRNA on the mRNA
expression of cell cycle-related genes in the Tca8113 cells (mean ±
SD). |
Table IV
Effects of shRNA on the mRNA
expression of cell cycle-related genes in the Tca8113 cells (mean ±
SD).
| Gene | Tca8113 | Control-shRNA | Per2-shRNA-III | P | P1 | P2 | P3 |
|---|
| Cyclin A2 | 0.99±0.02 | 0.96±0.07 | 3.36±0.53 | 0.000 | 0.000 | 0.000 | 0.911 |
| Cyclin B1 | 0.97±0.35 | 1.02±0.22 | 2.77±0.83 | 0.01 | 0.006 | 0.007 | 0.926 |
| c-myc | 1.10±0.17 | 1.21±0.37 | 1.39±0.33 | 0.536 | 0.286 | 0.512 | 0.653 |
| Cyclin D1 | 0.86±0.13 | 1.01±0.47 | 3.11±0.55 | 0.001 | 0.001 | 0.001 | 0.672 |
| Cyclin E | 1.45±0.39 | 1.31±0.53 | 1.27±0.47 | 0.884 | 0.651 | 0.924 | 0.719 |
| p53 | 2.24±0.46 | 2.38±0.47 | 0.57±0.02 | 0.002 | 0.002 | 0.001 | 0.667 |
| CDK1 | 1.14±0.24 | 1.04±0.11 | 1.07±0.13 | 0.771 | 0.648 | 0.815 | 0.496 |
| CDK2 | 1.10±0.13 | 1.23±0.40 | 1.08±0.10 | 0.731 | 0.906 | 0.477 | 0.549 |
| CDK4 | 1.06±0.28 | 0.87±0.22 | 1.79±0.33 | 0.016 | 0.019 | 0.007 | 0.447 |
| CDK6 | 1.00±0.00 | 1.00±0.25 | 2.74±0.74 | 0.005 | 0.003 | 0.003 | 0.997 |
| p16 | 1.60±0.13 | 1.70±0.08 | 0.92±0.14 | 0.000 | 0.000 | 0.000 | 0.354 |
| p21 | 3.88±0.21 | 4.05±0.60 | 1.10±0.26 | 0.000 | 0.000 | 0.000 | 0.633 |
| cdc25 | 1.46±0.40 | 1.32±0.55 | 1.62±0.54 | 0.775 | 0.709 | 0.493 | 0.747 |
| Wee1 | 1.38±0.86 | 1.11±0.20 | 1.32±0.66 | 0.867 | 0.917 | 0.700 | 0.627 |
| Rb1 | 1.16±0.39 | 1.31±0.27 | 1.25±0.25 | 0.83 | 0.735 | 0.802 | 0.560 |
| E2F1 | 0.92±0.56 | 1.13±0.54 | 3.46±0.87 | 0.006 | 0.004 | 0.005 | 0.710 |
Discussion
Previous studies have shown that Per2 expression is
reduced in various types of solid cancers, including breast and
skin cancer, hepatocellular carcinoma, colorectal cancer, renal
carcinomas, gastric cancers and head and neck squamous cell
carcinomas (11,19–24).
The alteration in Per2 expression has a close relationship with the
occurrence and development of cancers (6,18). The
present study found that Per2 expression in OSCC Tca8113 cells was
significantly lower than that in the oral mucosal epithelial cells;
in Tca8113 cells, downregulation of Per2 significantly increased
PI, decreased AI and altered the cell cycle distribution by
significantly decreasing the number of cells in the G1/G0 phase,
which suggested that the clock gene Per2 has a tumor-suppressor
role in OSCC.
Cell cycle disorder is the main reason for
carcinogenesis (26,27). Normal cell cycle strictly and
chronologically progresses along the G1, S and G2 M phase under the
precise control of the cyclin/CDK/CKI cell cycle molecular network
(29). CDKs are the core of the
cell cycle, to which cyclins and CKIs are the positive and negative
regulators, respectively (27,29).
Cyclin A2, B1, D1 and E play an important role in cyclins; and
CDK1, CDK2, CDK4 and CDK6 play an important role in CDKs (27,29).
CKIs contain the Ink4 family and Cip/Kip family, in which p16 and
p21 play an important role, respectively (27,29).
In the different cell cycle phases, cyclins, CDKs and CKIs vary.
Cyclin/CDK complex formed by combinations of cyclins and CDKs can
promote orderly cell cycle progression by activating CDKs. While
CKIs inhibit CDKs by combining with the corresponding CDKs or the
cyclin/CDK complex, which may inhibit the transformation of the
cell cycle phase (27,29). To date, studies have confirmed that
Per2 can regulate cyclins and p53 which is a regulator of the cell
cycle checkpoint, and the mutation of Per2 expression is
responsible for the aberrant expression of cyclin A, B1, D1 and E,
and p53 (6,18,25,31).
Both changes in cell cycle progression and imbalance of cell
proliferation and apoptosis induce cancers. Previous studies have
mainly focused on the role of Per2 in cyclins (6,18,25,31),
but there is little research concerning the role of Per2 in the
other two important aspects of the cyclin/CDK/CKI network.
At the G1 phase, the p16/p21-cyclin
D1-CDK4/6-Rb1-E2F1 pathway which is an important transduction
pathway of molecular information is related to the occurrence and
development of tumors (32). E2F1
plays an important role in promoting the transition of cells from
the G1 to the S phase (33). At the
G0 and the early G1 phase, the transcriptional activity of E2F1 is
inhibited by combining with unphosphorylated Rb1 at specific
binding sites. At the late G1 phase, the cyclin D1/CDK4 and cyclin
D1/CDK6 complex, formed by combination of cyclin D1 and CDK4/6,
phosphorylate Rb1 from which E2F1 is consequently released to start
DNA biosynthesis and promote cells into the S phase (34). p16 and p21, as CDKIs, can inhibit
the activities of CDK4/6 by competing with cyclin D1 for CDK4/6
binding (35). The present study
found that downregulation of Per2 in Tca8113 cells significantly
reduced the expression of p16 and p21, significantly increased the
expression of cyclin D1, CDK4, CDK6 and E2F1 and significantly
reduced the number of cells at the G1 phase. These results
illustrate that in Tca8113 cells, Per2 downregulation decreases the
expression of p16 and p21, and consequently increases the binding
of cyclin D1 and CDK4/6, which can phosphorylate Rb1 to release
more E2F1 from the Rb1/E2F1 complex, resulting in promotion of the
cell transformation during the G1/S phase. Fu et al
(6) and Yang et al (36) both reported that the downregulation
of Per2 increased the expression of cyclin D1. In the present
study, there was no significant difference in the expression of Rb1
mRNA, and a difference in phosphorylated Rb1 and unphosphorylated
Rb1 was not detected.
p53 is an important regulator of the G1/S cell cycle
checkpoint in the cyclin/CDK/CKI network (37). In the G1/S checkpoint, p53 is
activated by damaged DNA to stagnate the progression of the cell
cycle, leading to either repair of the damaged DNA or apoptosis
(14,27). Meanwhile p53 in the cytoplasm can
directly react with the BCL-2 family to promote cell
permeabilization of mitochondria and apoptosis (18). The present study found that Per2
downregulation in the Tca8113 cells significantly reduced the
expression of p53, the number of cells at G1 and AI. This suggests
that in Tca8113 cells, Per2 downregulation reduces the expression
of p53, leading to a reduction in the repair of damaged DNA in the
G1/S phase checkpoint and a decreased ability to induce apoptosis,
resulting in the damaged DNA being translated into the S phase.
This can destroy the integrity and stability of the cell genome,
which promotes cell malignant transformation. Meanwhile, p53 can
reduce the expression of cyclin B1 (25). During the G2/M phase, the absence of
cyclin B1 could block cells at the G2 phase, resulting in the
inability to enter into the M phase (25). Thus, in the present study, Per2
downregulation significantly reduced the expression of p53, and
subsequently significantly increased the expression of cyclin B1,
which accelerated mitosis and significantly increased the PI. Gotoh
et al reported that Per2 is at the key site of the
transcription mediated by p53, and Per2 downregulation reduces p53
expression (31). Sun et al
reported that in leukemic K562 cells Per2 downregulation decreased
p53 expression, and Per2 overexpression increased p53 expression
(25). Hua et al reported
that Per2 overexpression increased p53 expression in Lewis lung
cancer cells (LLCs), decreased cell proliferation and accelerated
apoptosis in LLCs and breast cancer cells (EMT6) (18). The present study was in accordance
with the above reports. The present study also proved that Per2
down-regulation significantly increased and decreased the
expression of E2F1 and p21, respectively, which are the
dual-directional regulators of apoptosis (38,39),
resulting in a worsening of the imbalance of cell proliferation and
apoptosis.
The present study found that circadian clock gene
Per2 was reduced in OSCC. In the Tca8113 cells, Per2 downregulation
significantly increased the mRNA expression of cyclin A2, B1 and
D1, CDK4, CDK6 and E2F1, while significantly decreased the mRNA
expression of p53, p16 and p21. Cell proliferation was
significantly higher, apoptosis was significantly lower, and
progression of the cell cycle was altered. The present study
represents the first demonstration that in OSCC, the clock gene
Per2 plays an important role in the G1/S checkpoint and the three
aspects of the cyclin/CDK/CKI network at the transcriptional level.
On this basis, further research of Per2 at the protein level and
the modification level after protein translation may further define
the interaction of the circadian rhythm and the cell cycle, and
their relationship with carcinogenesis. This may provide effective
new molecular targets for the treatment of cancers.
Acknowledgments
We thank Wen-Ping Luo for her technical assistance.
The present study was supported by the Project Supported by the
Program for Innovation Team Building at Institutions of Higher
Education in Chongqing in 2013, and the Project Supported by
Chongqing Municipal Key Laboratory of Oral Biomedical Engineering
of Higher Education.
References
|
1
|
Eismann EA, Lush E and Sephton SE:
Circadian effects in cancer-relevant Psychoneuroendocrine and
immune pathways. Psychoneuroendocrinology. 35:963–976. 2010.
View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Zieker D, Jenne I, Koenigsrainer I,
Zdichavsky M, Nieselt K, Buck K, Zieker J, Beckert S, Glatzle J,
Spanagel R, et al: Circadian expression of clock- and tumor
suppressor genes in human oral mucosa. Cell Physiol Biochem.
26:155–166. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Hara Y, Onishi Y, Oishi K, Miyazaki K,
Fukamizu A and Ishida N: Molecular characterization of Mybbp1a as a
co-repressor on the Period2 promoter. Nucleic Acids Res.
37:1115–1126. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Reppert SM and Weaver DR: Coordination of
circadian timing in mammals. Nature. 418:935–941. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Schibler U, Ripperger J and Brown SA:
Peripheral circadian oscillators in mammals: Time and food. J Biol
Rhythms. 18:250–260. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Fu L, Pelicano H, Liu J, Huang P and Lee
C: The circadian gene Period2 plays an important role in tumor
suppression and DNA damage response in vivo. Cell. 111:41–50. 2002.
View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Rana S and Mahmood S: Circadian rhythm and
its role in malignancy. J Circadian Rhythms. 8:3–17. 2010.
View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Savvidis C and Koutsilieris M: Circadian
rhythm disruption in cancer biology. Mol Med. 18:1249–1260. 2012.
View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Mullenders J, Fabius AW, Madiredjo M,
Bernards R and Beijersbergen RL: A large scale shRNA barcode screen
identifies the circadian clock component ARNTL as putative
regulator of the p53 tumor suppressor pathway. PLoS one.
4:e47982009. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Kang TH, Reardon JT, Kemp M and Sancar A:
Circadian oscillation of nucleotide excision repair in mammalian
brain. Proc Natl Acad Sci USA. 106:2864–2867. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Chen ST, Choo KB, Hou MF, Yeh KT, Kuo SJ
and Chang JG: Deregulated expression of the PER1, PER2 and PER3
genes in breast cancers. Carcinogenesis. 26:1241–1246. 2005.
View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Viswanathan AN, Hankinson SE and
Schernhammer ES: Night shift work and the risk of endometrial
cancer. Cancer Res. 67:10618–10622. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Sack RL, Auckley D, Auger RR, Carskadon
MA, Wright KP Jr, Vitiello MV and Zhdanova IV; American Academy of
Sleep Medicine: Circadian rhythm sleep disorders: Part I, basic
principles, shift work and jet lag disorders. An American Academy
of Sleep Medicine Review. Sleep. 30:1460–1483. 2007.PubMed/NCBI
|
|
14
|
Rengarajan T, Nandakumar N, Rajendran P,
Haribabu L, Nishigaki I and Balasubramanian MP: D-pinitol promotes
apoptosis in MCF-7 cells via induction of p53 and Bax and
inhibition of Bcl-2 and NF-κB. Asian Pac J Cancer Prev.
15:1757–1762. 2014. View Article : Google Scholar
|
|
15
|
Zhu L, Yu J, Zhang W, Xie B and Zhu Y:
Research progress on the central mechanism underlying regulation of
visceral biological rhythm by per2 (Review). Mol Med Rep.
10:2241–2248. 2014.PubMed/NCBI
|
|
16
|
Qu X, Metz RP, Porter WW, Neuendorff N,
Earnest BJ and Earnest DJ: The clock genes period 1 and period 2
mediate diurnal rhythms in dioxin-induced Cyp1A1 expression in the
mouse mammary gland and liver. Toxicol Lett. 196:28–32. 2010.
View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Yang S, Liu A, Weidenhammer A, Cooksey RC,
McClain D, Kim MK, Aguilera G, Abel ED and Chung JH: The role of
mPer2 clock gene in glucocorticoid and feeding rhythms.
Endocrinology. 150:2153–2160. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Hua H, Wang Y, Wan C, Liu Y, Zhu B, Yang
C, Wang X, Wang Z, Cornelissen-Guillaume G and Halberg F: Circadian
gene mPer2 overexpression induces cancer cell apoptosis. Cancer
Sci. 97:589–596. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Okabe T, Kumagai M, Nakajima Y, Shirotake
S, Kodaira K, Oyama M, Ueno M and Ikeda M: The impact of HIF1α on
the Per2 circadian rhythm in renal cancer cell lines. PLoS One.
9:e1096932014. View Article : Google Scholar
|
|
20
|
Hsu CM, Lin SF, Lu CT, Lin PM and Yang MY:
Altered expression of circadian clock genes in head and neck
squamous cell carcinoma. Tumour Biol. 33:149–155. 2012. View Article : Google Scholar
|
|
21
|
Lengyel Z, Lovig C, Kommedal S, Keszthelyi
R, Szekeres G, Battyáni Z, Csernus V and Nagy AD: Altered
expression patterns of clock gene mRNAs and clock proteins in human
skin tumors. Tumour Biol. 34:811–819. 2013. View Article : Google Scholar
|
|
22
|
Lin YM, Chang JH, Yeh KT, Yang MY, Liu TC,
Lin SF, Su WW and Chang JG: Disturbance of circadian gene
expression in hepatocellular carcinoma. Mol Carcinog. 47:925–933.
2008. View
Article : Google Scholar : PubMed/NCBI
|
|
23
|
Soták M, Polidarová L, Ergang P, Sumová A
and Pácha J: An association between clock genes and
clock-controlled cell cycle genes in murine colorectal tumors. Int
J Cancer. 132:1032–1041. 2013. View Article : Google Scholar
|
|
24
|
Hu ML, Yeh KT, Lin PM, Hsu CM, Hsiao HH,
Liu YC, Lin HY, Lin SF and Yang MY: Deregulated expression of
circadian clock genes in gastric cancer. BMC Gastroenterol.
14:67–76. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Sun CM, Huang SF, Zeng JM, Liu DB, Xiao Q,
Tian WJ, Zhu XD, Huang ZG and Feng WL: Per2 inhibits k562 leukemia
cell growth in vitro and in vivo through cell cycle arrest and
apoptosis induction. Pathol Oncol Res. 16:403–411. 2010. View Article : Google Scholar
|
|
26
|
Murphy PJ and Campbell SS: Physiology of
the circadian system in animals and humans. J Clin Neurophysiol.
13:2–16. 1996. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Soták M, Sumová A and Pácha J: Cross-talk
between the circadian clock and the cell cycle in cancer. Ann Med.
46:221–232. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Borgs L, Beukelaers P, Vandenbosch R,
Belachew S, Nguyen L and Malgrange B: Cell 'circadian' cycle: New
role for mammalian core clock genes. Cell Cycle. 8:832–837. 2009.
View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Lim S and Kaldis P: Cdks, cyclins and
CKIs: Roles beyond cell cycle regulation. Development.
140:3079–3093. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Štorcelová M, Vicián M, Reis R, Zeman M
and Herichová I: Expression of cell cycle regulatory factors hus1,
gadd45a, rb1, cdkn2a and mre11a correlates with expression of clock
gene per2 in human colorectal carcinoma tissue. Mol Biol Rep.
40:6351–6361. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Gotoh T, Vila-Caballer M, Santos CS, Liu
J, Yang J and Finkielstein CV: The circadian factor Period 2
modulates p53 stability and transcriptional activity in unstressed
cells. Mol Biol Cell. 25:3081–3093. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Brennan P, Hainaut P and Boffetta P:
Genetics of lung-cancer susceptibility. Lancet Oncol. 12:399–408.
2011. View Article : Google Scholar
|
|
33
|
Bell LA and Ryan KM: Life and death
decisions by E2F-1. Cell Death Differ. 11:137–142. 2004. View Article : Google Scholar
|
|
34
|
Rivadeneira DB, Mayhew CN, Thangavel C,
Sotillo E, Reed CA, Graña X and Knudsen ES: Proliferative
suppression by CDK4/6 inhibition: Complex function of the
retinoblastoma pathway in liver tissue and hepatoma cells.
Gastroenterology. 138:1920–1930. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Herbst RS, Heymach JV and Lippman SM: Lung
cancer. N Engl J Med. 359:1367–1380. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Yang X, Wood PA, Oh EY, Du-Quiton J,
Ansell CM and Hrushesky WJ: Down regulation of circadian clock gene
Period 2 accelerates breast cancer growth by altering its daily
growth rhythm. Breast Cancer Res Treat. 117:423–431. 2009.
View Article : Google Scholar
|
|
37
|
Levine AJ: p53, the cellular gatekeeper
for growth and division. Cell. 88:323–331. 1997. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Hallstrom TC, Mori S and Nevins JR: An
E2F1-dependent gene expression program that determines the balance
between proliferation and cell death. Cancer Cell. 13:11–22. 2008.
View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Lin PY, Fosmire SP, Park SH, Park JY,
Baksh S, Modiano JF and Weiss RH: Attenuation of PTEN increases p21
stability and cytosolic localization in kidney cancer cells: A
potential mechanism of apoptosis resistance. Mol Cancer. 6:16–31.
2007. View Article : Google Scholar : PubMed/NCBI
|