Introduction
Gliomas are the most common primary brain tumors and
account for 40–50% of primary intracranial neoplasms (1). They can occur anywhere in the central
nervous system but primarily in the brain, and originate from
abnormally proliferating glial cells, which normally supply
nutrition and protection to neurons in the central nervous system
(2). The etiology and pathogenesis
of gliomas still remain obscure. Despite the advances in
therapeutic approaches, the treatment offers limited help to
prolong survival (3). In view of
the overall poor outcome with current therapies, a better
understanding of glioma etiology is crucial for future development
of more effective treatments to cure this rapidly progressing
disease.
Phosphoglycerate mutase 1 (PGAM1) catalyzes the
conversion of 3-phosphoglycerate (3-PG) to 2-phosphoglycerate
(2-PG) to release energy during glycolysis. Proteome reactivity
profiling was used to determine the drug target against breast
cancer, and PGAM1 was found a potential metabolic enzyme
participating in breast carcinogenesis (4). Several studies demonstrated that PGAM1
activity was increased in a variety of human cancers, including
lung (5), breast (4), prostate cancer (6), hepatocellular carcinoma (7), colorectal cancer (8), oral squamous cell carcinoma (9), and esophageal squamous cell carcinoma
(10) and also associated with
virus infection (11) and
spermatogenic dysfunction (12).
Previous reports have described that targeting PGAM1 by a
PGAM1-derived inhibitory peptide or PGAM inhibitor MJE3 could
attenuate breast cancer cell proliferation (4,13).
However, the association of PGAM1 with glioma grade and the role of
PGAM1 in glioma are poorly investigated.
In the present study, we aimed to evaluate the
expression patterns, the clinical roles and the functions of PGAM1
in human gliomas, as a possible diagnostic biomarker and
therapeutic target for glioma.
Materials and methods
Patients and tissue specimens
All 124 glioma specimens were obtained from patients
with primary glioma who underwent surgical treatment between July
2012 and May 2014 at Qilu Hospital; 20 normal brain tissue samples
were collected from patients undergoing surgery for epilepsy and
were reviewed to verify the absence of a tumor. For all patients,
the histological types and grade of gliomas were evaluated by two
experienced pathologists according to the 2007 WHO Classification
of Tumors of the Central Nervous System (14). All patients had a well-documented
clinical history. The study protocol was approved by the Ethics
Committee of Qilu Hospital (Shandong, China). Informed consent was
obtained from all patients included in the study.
RNA isolation and real-time PCR
Total RNA was extracted from frozen specimens by
using TRIzol reagent (Invitrogen/Life Technologies, Carlsbad, CA,
USA) according to the manufacturer's instructions. cDNA was
generated by reverse transcription of 1 µg total RNA with
primer oligonucleotides. qPCR reactions were conducted in a
10-µl reaction volume with SYBR-Green I, with a 1:25
dilution of the cDNA and 20 nM primers. β-actin was a reference
gene. The primers were designed by the use of Primer Premier 5.0
(Premier Biosoft International, Palo Alto, CA, USA) from mRNA
sequences in the GenBank database. Primer sequences were for PGAM1
(sense: 5′-GTGCAGAAGAGAGCGATCCG-3′ and antisense:
5′-CGGTTAGACCCCCATAGTGC-3′); β-actin (sense:
5′-CGTTGACATCCGTAAAGACC-3′ and antisense:
5′-TAGAGCCACCAATCCACACA-3′). Calculation of PGAM1 mRNA levels was
based on cycle threshold (Ct) values and determined by relative
quantification with β-actin as a normalizing gene according to the
following equation: 2−ΔCt [ΔCt = Ct (PGAM1) − Ct
(β-actin)]. All experiments were performed in triplicate.
Western blot analysis
Briefly, cultured cells and tissues were lysed in
RIPA buffer (Beyotime Institute of Biotechnology, Haimen, China)
containing 50 mM Tris (pH 7.4), 150 mM NaCl, 1% Triton X-100, 1%
sodium deoxycholate, 0.1% SDS and protease inhibitor cocktail with
1 mM phenylmethanesulfonyl fluoride (Beyotime Institute of
Biotechnology). Lysates were centrifuged at 12,000 × g for 15 min
and the protein concentration of the supernatant was quantified by
the BCA method. An amount of 20 µg protein from each sample
was separated by 10% SDS-PAGE and transferred to a polyvinylidene
difluoride membranes. After blocking with 5% non-fat milk in
phosphate-buffered saline (PBS) Tween-20 for 1 h at room
temperature, the membranes were incubated with primary antibody
overnight. The membrane was rinsed and washed 3 times with TBS
containing 0.1% Tween-20, then incubated with horseradish
peroxidase-conjugated secondary antibody for 2 h at room
temperature. Immunoreactive bands were visualized by using the
Immobilon Western Chemiluminescent HRP Substrate kit (Millipore).
Immunoreactive labeling was analyzed by the use of ImageJ 1.44
(National Institutes of Health, Bethesda, MD, USA) and standardized
against β-actin protein level.
Antibody for PGAM1 was from Santa Cruz Biotechnology
(Dallas, TX, USA); antibodies for Bax, Bcl-2, matrix
metalloproteinase 2 (MMP2)/9 and cleaved caspase-3 were from Cell
Signaling Technology (Danvers, MA, USA).
Immunohistochemistry
All samples were fixed with 10% buffered formalin
and embedded in paraffin. Slices of paraffin tissues were cut as
4-µm serial sections. Slides were stained according to the
manufacturer's protocol. In brief, slides were deparaffinized by
using xylene and rehydrated through an ethanol series to water. The
dewaxed slides underwent antigen retrieval in citrate buffer for 30
min by microwaving and then cooled to room temperature to expose
antigen epitopes. An amount of 3% hydrogen peroxide was added to
slides to inhibit endogenous peroxidase activity. The slides were
blocked with 10% normal goat serum for 1 h at 37°C, then incubated
with primary rabbit anti-human PGAM1 polyclonal antibody overnight
at 4°C, then anti-rabbit secondary antibody. Finally, DAB was used
as chromogen, and the slides were counterstained with hematoxylin.
Slides were dehydrated and mounted with neutral balsam according to
the laboratory protocol. Staining with PBS instead of the primary
antibody was a negative control.
Evaluation of immunohistochemistry
Each stained slide was simultaneously scored by two
independent pathologists who were blinded to the clinical
information. We selected 10 low-power fields randomly, and counted
cells at high-power field. The percentage of PGAM1-positive cells
was scored as 4 categories: 0, no staining; 1, <25% cells; 2,
25–75% cells; and 3, >75% cells. The intensity of positive
staining was scored as 4 grades: 0, negative; 1, weak; 2, moderate;
and 3, strong. The final staining score was then obtained (ranging
from 0 to 9) by the multiplication of the intensity and percentage
scores. The staining pattern of slides was scored as 0, −; 1–3, +;
4–6, ++; and 6–9, +++. In addition, samples with scores up to ++
were considered positive.
Cell culture
The human glioma cell line (U87) was obtained from
the Chinese Academy of Science and cultured in Dulbecco's modified
Eagle's medium (DMEM) supplemented with 10% fetal bovine serum
(FBS), 100 U/ml penicillin, and 100 µg/ml streptomycin at
37°C in a humidified atmosphere containing 5% CO2 in
60-mm flasks.
Transfection
When cultured U87 cells reached 90% confluence, they
were transfected with siRNA targeting PGAM1. The siRNA sequences
were as follows: PGAM1-siRNA: 5′-CGACUGGUAUUCCCAUUGUTT-3′ and
5′-ACAAUGGGAAUACCAGUCGTT-3′; Negative scramble control sequences:
5′-UUCUCCGAACGUGUCACGUTT-3′ and 5′-ACGUGACACGUUCGGAGAATT-3′. The
RNA duplexes were synthesized by Shanghai GenePharma Co., Ltd.
(Shanghai, China). Transfection of siRNA involved use of
Lipofectamine 2000 (Invitrogen/Life Technologies) according to the
manufacturer's instructions. The efficiency of gene silencing was
confirmed by assaying PGAM1 protein levels.
Cell proliferation assay
Proliferation of U87 cells was determined by
standard MTT assay (Beyotime Institute of Biotechnology). Briefly,
MTT solution (20 µl, 5 mg/ml) was added into each well for
incubation at 37°C for 4 h. Then the solution was removed by
aspiration, the insoluble formazan crystals were dissolved in 150
µl/well dimethyl sulfoxide (DMSO), and absorbance at 490 nm
was measured by the use of a Varioskan Flash spectral scanning
multimode reader (Thermo Electron Oy, Vantaa, Finland). The
spectrophotometer was calibrated to zero absorbance with culture
medium without cells. The percentage of cell survival was
determined by comparing the average absorbance of treated cells to
that of untreated cells. The experiment was repeated at least 3
times.
Cell apoptosis analysis
Cell apoptosis was evaluated by the use of an
Annexin V-FITC/PI kit (Invitrogen/Life Technologies). Briefly,
after treatment, attached cells were collected and washed with PBS
twice. An amount of 400 µl binding buffer, 5 µl
Annexin V FITC and 5 µl PI was successively added to the
cell suspension. After 15 min of incubation in the dark, cells
underwent flow cytometry (Becton-Dickinson, San Jose, CA, USA). At
least 3 independent experiments were carried out.
Cell cycle analysis
Cell cycle analysis was evaluated with a PI staining
kit (Beyotime Institute of Biotechnology). Briefly,
~12×105 U87 cells were seeded in a 6-well plate, allowed
to attach for 48 h, then cells were collected by the trypsin
method, washed with PBS, and fixed overnight at 4°C in 70% ethanol.
Fixed cells were washed with PBS; 20 µl RNase A was added
and incubated at 37°C for 30 min to hydrolyze RNA, then cells were
stained with PI in the dark for another 30 min, and the cell cycle
was evaluated by flow cytometry. The experiments were repeated at
least 3 times independently.
In vitro cell migration and invasion
Boyden chamber assay with some modifications
Briefly, 5×104 siRNA-transfected cells in
0.1 ml of serum-free DMEM were added to the wells of an 8-µm
pore membrane Boyden chamber (Corning, Inc., Corning, NY, USA)
uncoated (for the migration assay) or coated with Matrigel (BD
Biosciences; for the invasion assay). The bottom chamber contained
10% FBS in DMEM, which served as a chemoattractant. Cells were
allowed to invade for 48 h, and the cells that had not penetrated
the filters were removed from the filters with cotton swabs.
Chambers were fixed for 20 min at room temperature with 4%
formaldehyde in PBS, stained in 0.1% crystal violet for 30 min, and
rinsed in water. Cells that migrated to the bottom surface of the
filter were counted under a light microscope. Assays were performed
3 times with triplicate wells.
Statistical analysis
Data are expressed as mean ± SEM. SPSS 18.0 was used
for analysis. Data were analyzed by two-tailed Student's t-test and
one-way ANOVA. Chi-square test was used to analyze categorical
data. P<0.05 was considered statistically significant.
Results
PGAM1 is upregulated in human glioma
tissues
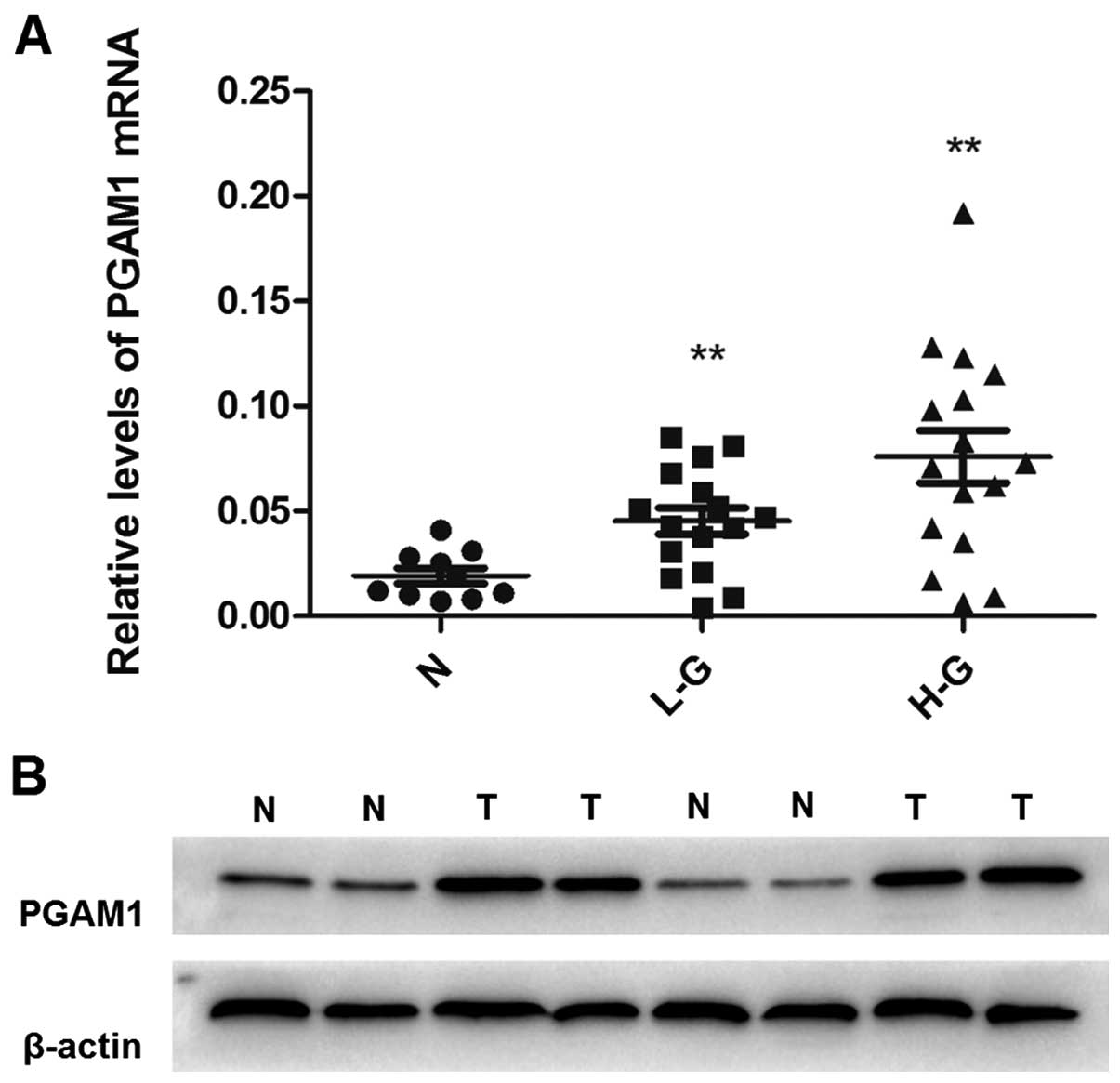
Expression of PGAM1 was compared at both
transcriptional and translational levels between glioma tissues and
normal brain tissues. We analyzed the mRNA expression of PGAM1 in
glioma tissues and normal brain tissues to assess the role of PGAM1
in the malignant progression of glioma. PGAM1 mRNA level was higher
in low-grade and high-grade glioma than normal brain tissue
(Fig. 1A) and PGAM1 protein
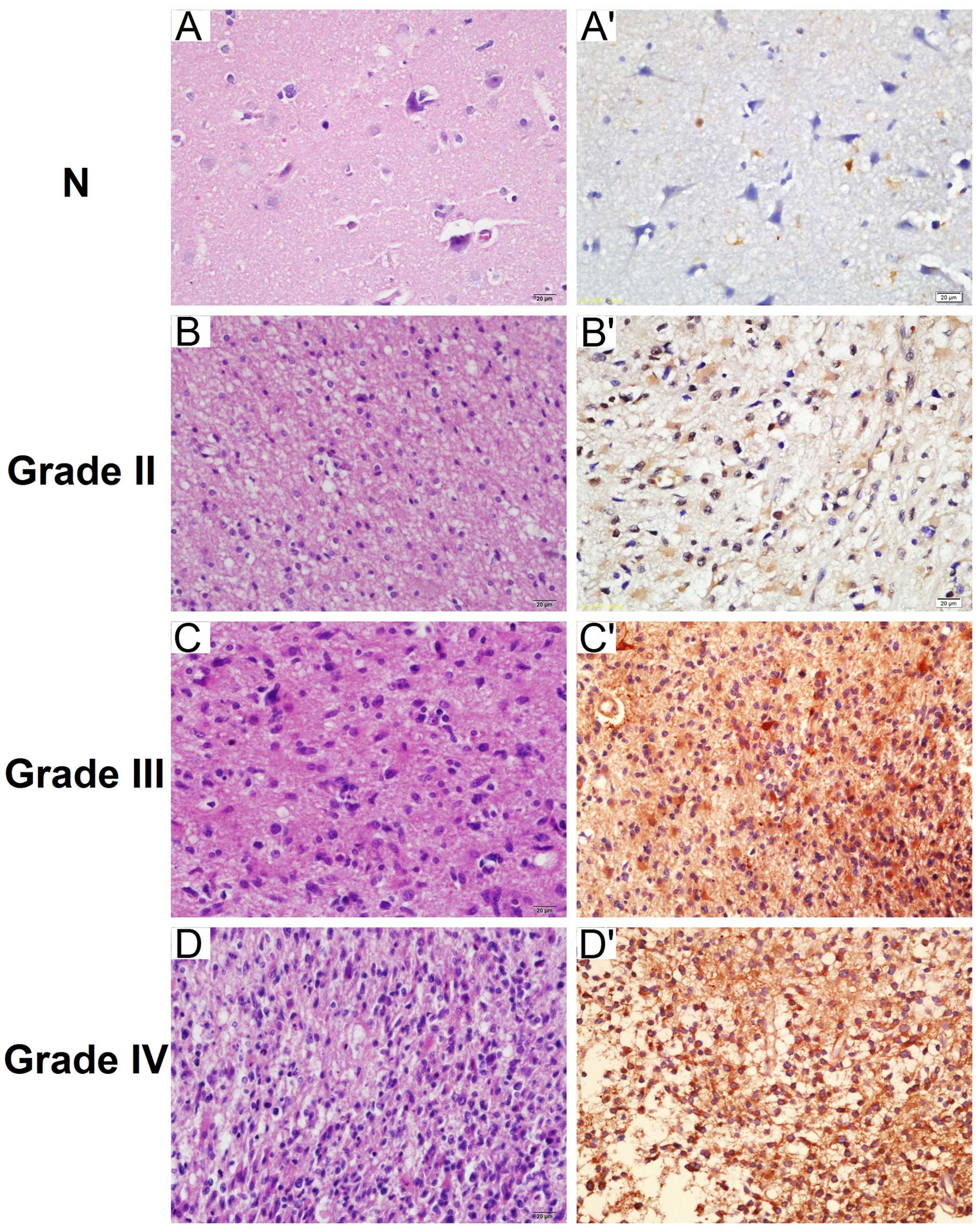
expression was higher in tumor tissue (Fig. 1B). Immunohistochemical staining was
used to detect the expression of PGAM1 in paraffin-embedded tissues
(Fig. 2). PGAM1 was predominantly
confined to the cytoplasm of tumor cells. PGAM1-positive expression
was present in 74.2% (92/124) of glioma tissues and in 25% (5/20)
of normal brain tissues (Table
I).
 | Table IPGAM1 expression in glioma tissues
and normal brain tissues. |
Table I
PGAM1 expression in glioma tissues
and normal brain tissues.
| Tissues | Total | −/+ (%) | ++/+++ (%) | P-value |
|---|
| Normal | 20 | 15 (75) | 5 (25) | |
| Glioma | 124 | 32 (25.8) | 92 (74.2) | <0.05 |
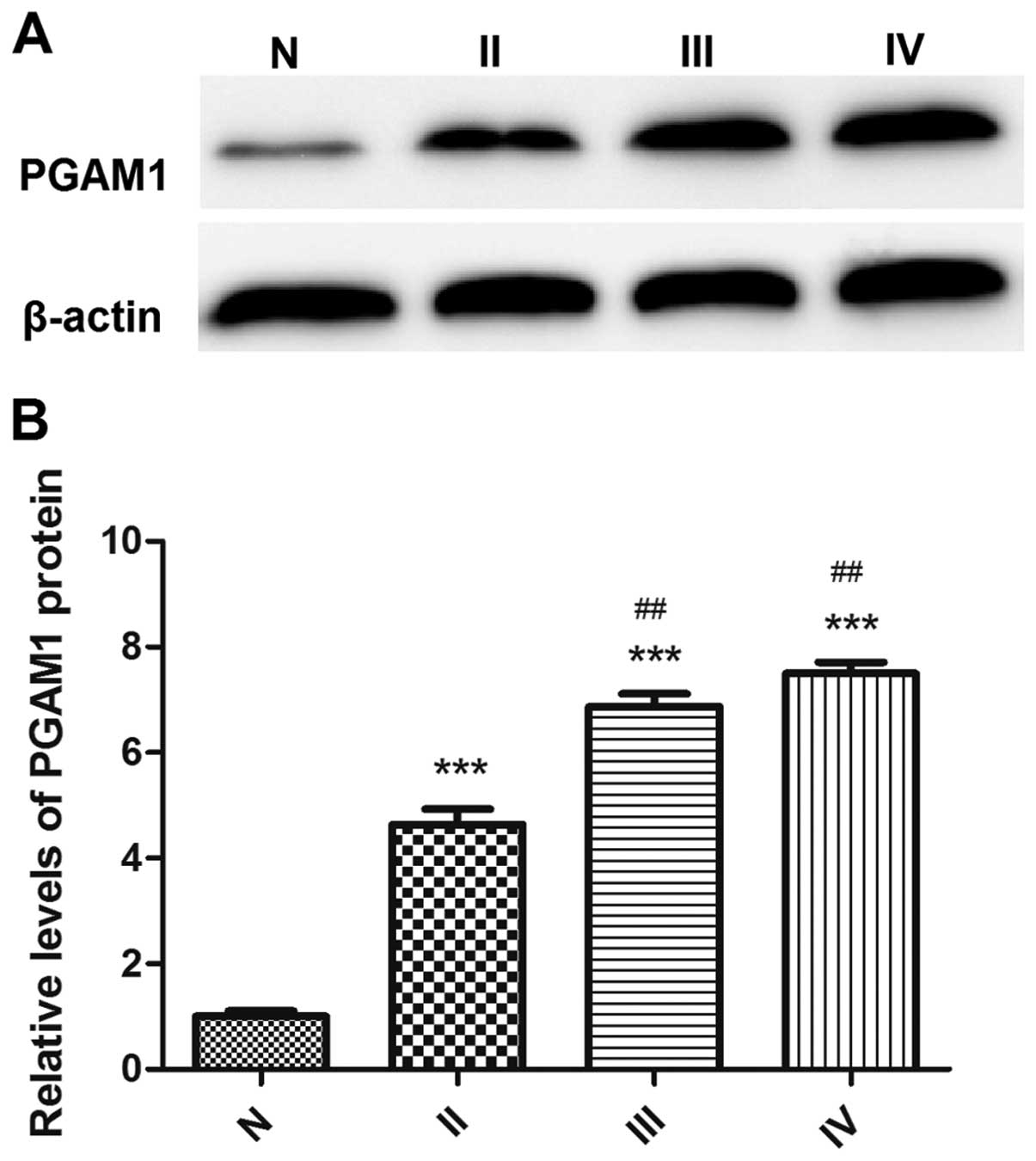
The protein expression of PGAM1 in tissue
by glioma grade
PGAM1 protein expression was associated with glioma
grade (Fig. 3). The expression of
PGAM1 was 56.3% in grade II glioma, 79.0% in grade III glioma and
81.5% in grade IV glioma on immunohistochemistry (Table II). PGAM1 protein expression was
higher in high-grade glioma than low-grade glioma tissue. The
significant upregulation of PGAM1 in glioma tissues indicated that
PGAM1 may function as a tumor promoter in glioma.
 | Table IIAssociation between PGAM1 protein
level and clinicopathological features of glioma patients. |
Table II
Association between PGAM1 protein
level and clinicopathological features of glioma patients.
| Variables | Total | −/+ (%) | ++/+++ (%) | P-value |
|---|
| Age (years) |
| <50 | 67 | 36 (53.7) | 31 (46.3) | |
| ≥50 | 57 | 29 (50.9) | 28 (49.1) | 0.751 >0.05 |
| Gender |
| Male | 69 | 35 (50.7) | 34 (49.3) | |
| Female | 55 | 30 (54.6) | 25 (45.4) | 0.672 >0.05 |
| WHO grade |
| II | 32 | 14 (43.7) | 18 (56.3) | |
| III | 38 | 8 (21.0) | 30 (79.0) | <0.05 III vs.
II |
| IV | 54 | 10 (18.5) | 44 (81.5) | <0.05 IV vs.
II |
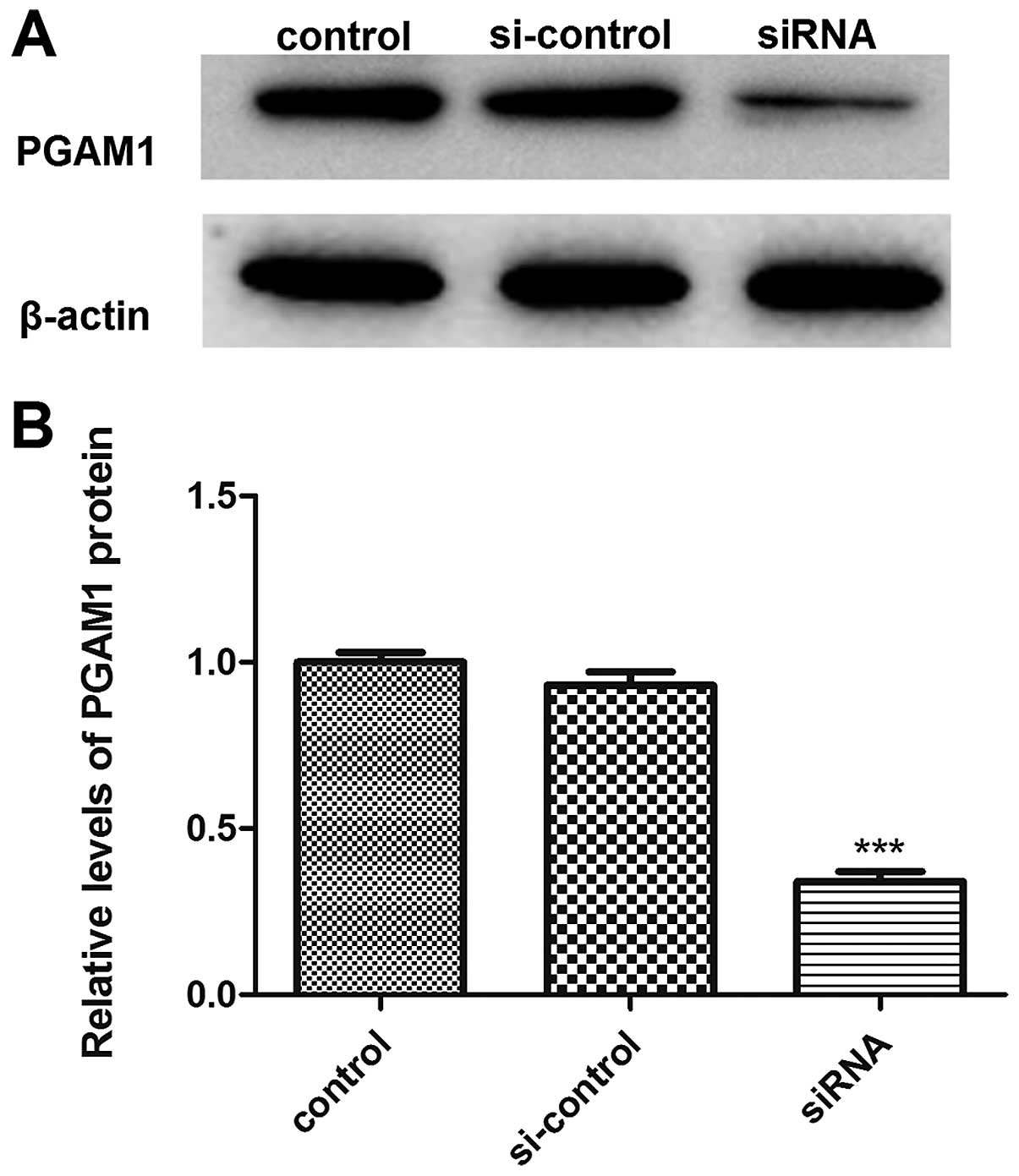
Downregulation of PGAM1 inhibits glioma
cell proliferation in vitro
We transiently transfected PGAM1 siRNA into U87
glioma cells to investigate the potential function of PGAM1 and
confirmed the downregulation of PGAM1 protein level with PGAM1
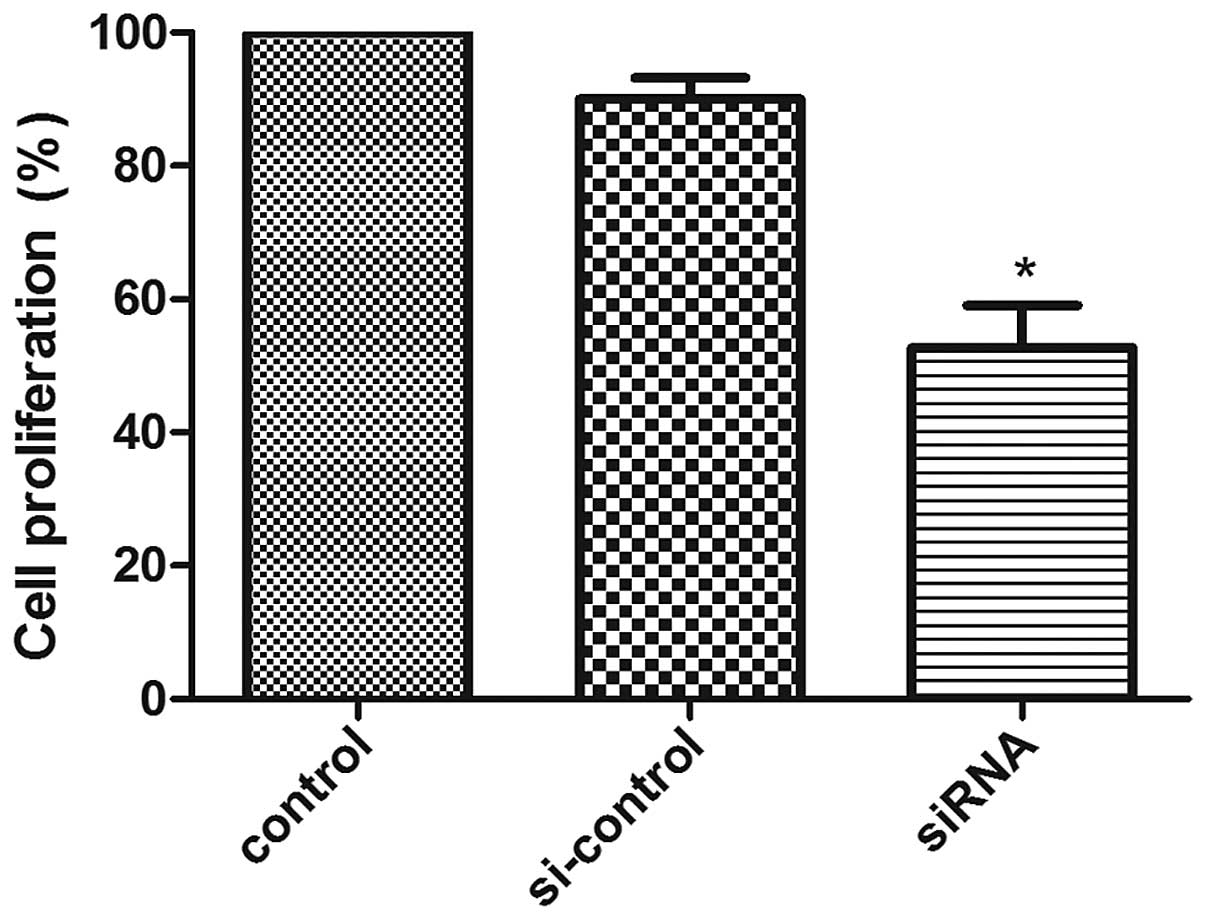
siRNA transfection as compared with controls (Fig. 4). Cell proliferation ability was
analyzed by MTT assay at 48 h after transfection in U87 cells. Cell
proliferation was reduced with PGAM1 siRNA transfection as compared
with negative control transfection in U87 cells (Fig. 5). Therefore, downregulation of PGAM1
inhibited glioma cell growth in vitro.
Knockdown of PGAM1 induces apoptosis in
U87 cells
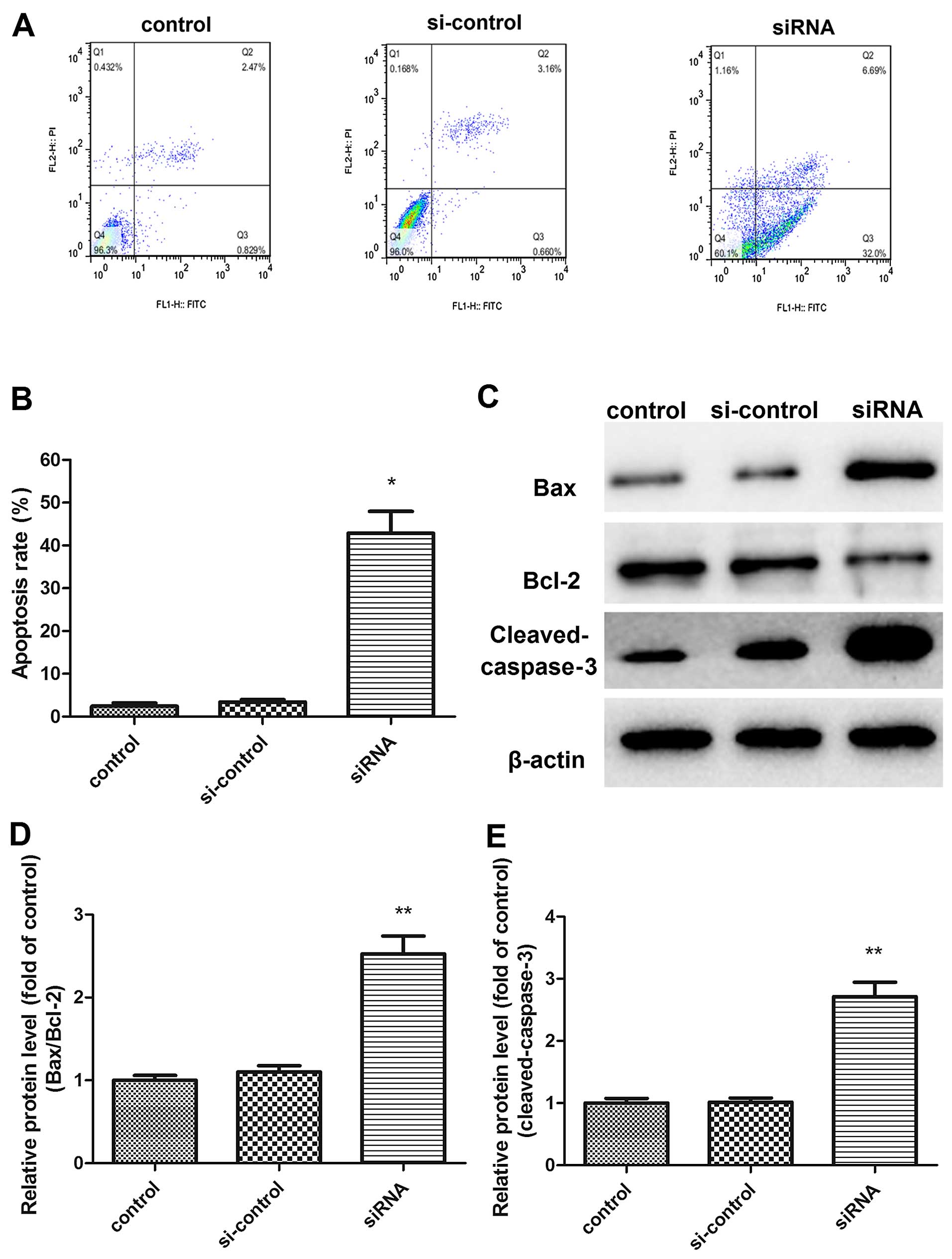
Cell apoptosis was measured by Annexin V FITC/PI
staining to further explore the mechanism of PGAM1 modulating
glioma cell growth (Fig. 6A). siRNA
knockdown of PGAM1 significantly increased cell apoptosis as
compared with controls. After transfection for 48 h, the early
apoptotic cells (right lower domain of the fluorocytogram) and late
apoptotic cells (right upper domain of the fluorocytogram)
increased from 0.83±0.67 and 2.47±0.39 to 32.0±4.26 and 6.69±3.11%,
respectively; thus, the total mean apoptotic rate reached 42.9%
with PGAM1 siRNA transfection (Fig.
6B). Apoptosis regulators were further examined (Fig. 6C). The ratio of Bax to Bcl-2
expression increased in U87 cells with PGAM1 siRNA knockdown
(Fig. 6D). Additionally, cleaved
caspase-3 activity was significantly upregulated in PGAM1
siRNA-transfected glioma cells compared with the control (Fig. 6E).
Silencing of PGAM1 induces cell cycle
arrest in U87 cells
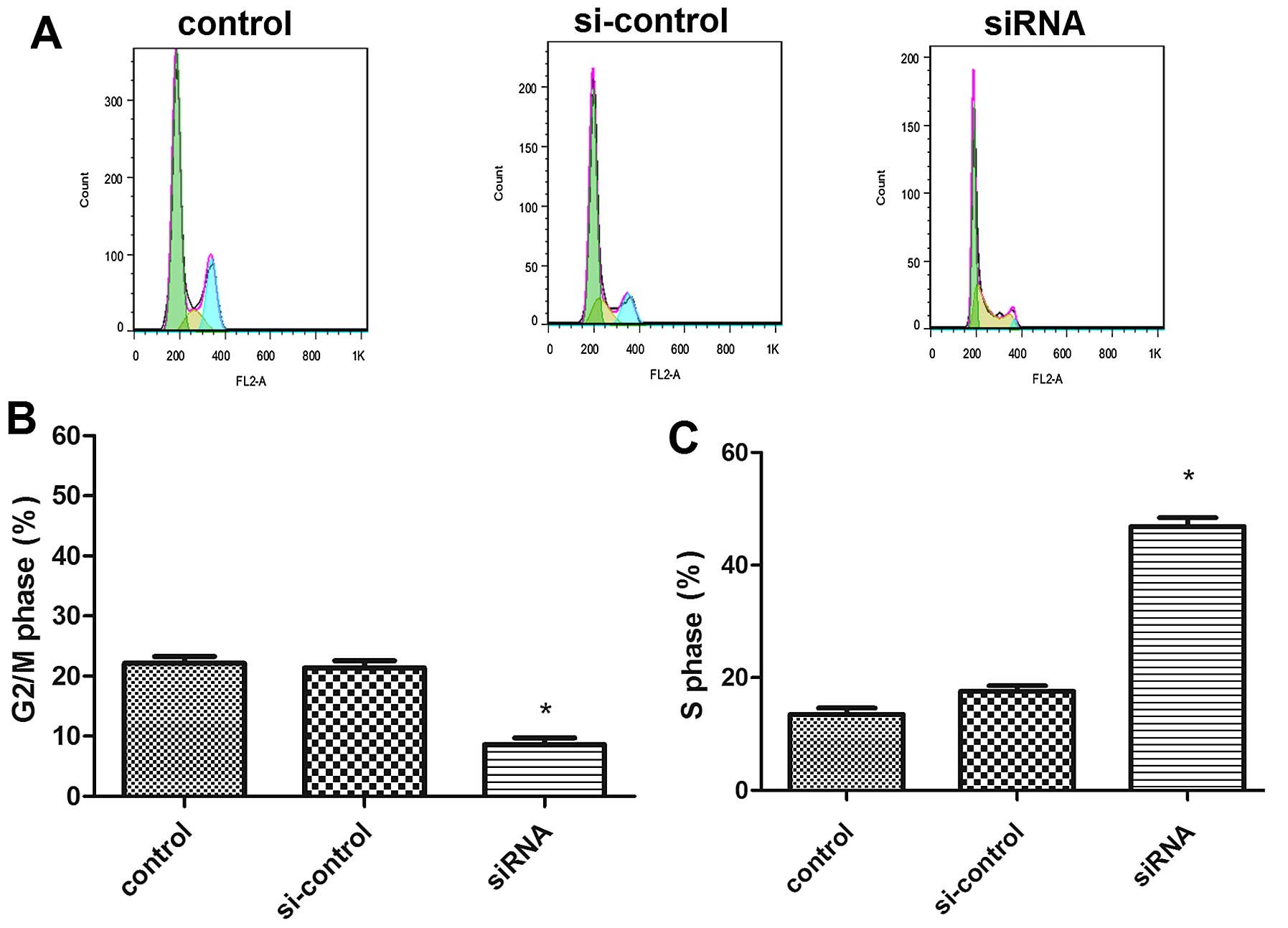
Knockdown of PGAM1 significantly increased the cell
population in the S phase and decreased that in the G2/M phase
(Fig. 7A). The controls had a mean
of 13.48±2.03% cells in the S phase; after siRNA transfection for
48 h, the rates increased to 46.86±2.73% (Fig. 7C), and the rates of cells in the
G2/M phase decreased from 22.16±0.95 to 8.61±0.37% (Fig. 7B). Therefore, PGAM1 siRNA knockdown
induced S-phase cell cycle arrest in U87 cells.
PGAM1 knockdown inhibited glioma cell
migration and invasion
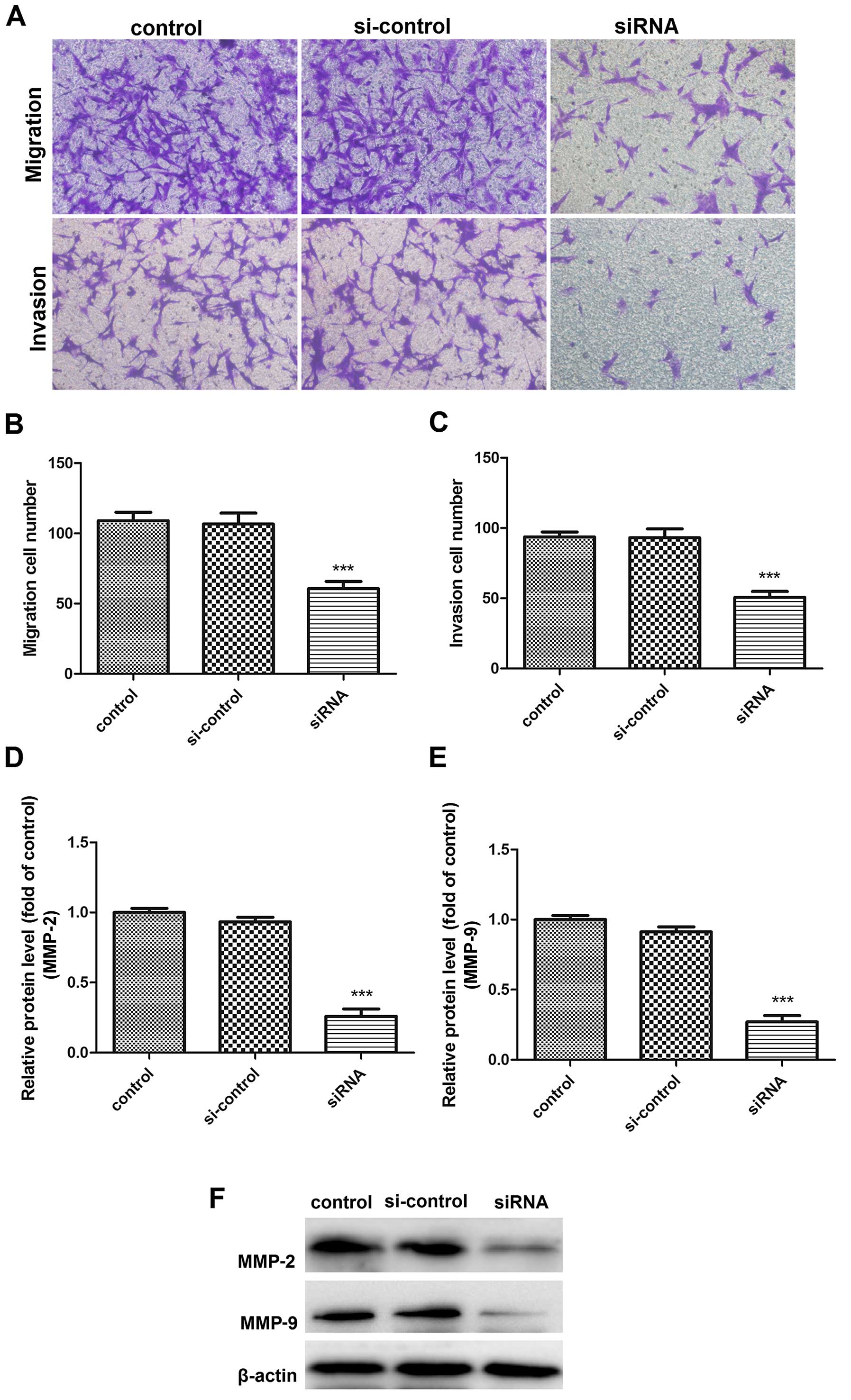
To test the effect of PGAM1 on glioma cell mobility,
we performed Transwell migration and invasion assays in U87 cells
at 48 h after PGAM1 siRNA transfection. The migration ability of
transfected U87 cells was decreased (Fig. 8A and B). Furthermore, the invasion
ability of transfected cells was inhibited as compared with
controls (Fig. 8A and C). Since
MMP2/9 play important roles in tumor cell migration and invasion,
we examined the effect of PGAM1 on their expression. PGAM1 siRNA
knockdown downregulated MMP2/9 protein levels in U87 cells
(Fig. 8D–F). PGAM1 knockdown may
significantly inhibit glioma cell migration and invasion in
vitro.
Discussion
Glioma is one of the most common adult primary
central nervous system tumors, with a median survival of only 14.6
months after diagnosis (14,15).
Gliomas can be classified into WHO grades I–IV based on malignant
behavior (14). Standard treatment
includes surgical operation followed by radiotherapy and
chemotherapy (16). Glioma is
rarely curable despite these treatments. Since the major barrier to
effective treatment of glioma is their highly invasive nature,
special attention needs to be paid to the molecular determinants
regulating their malignant behavior. In this study, we demonstrated
upregulated mRNA and protein levels of PGAM1 in glioma tissues.
Importantly, the expression of PGAM1 was significantly associated
with WHO grade of glioma. siRNA knockdown of PGAM1 inhibited glioma
cell growth, migration and invasion, and induced cell apoptosis and
cell cycle arrest. PGAM1 might be a novel therapeutic target in
glioma.
The Warburg effect is a property of cancer cells
whereby cancer cells maintain a high rate of aerobic glycolysis
even under the high-oxygen (20%) conditions of normal tissue
culture (17). The effect helps
cancer cells generate more ATP more quickly than normal cells,
which mainly rely on oxidative phosphorylation. As well, tumor
tissue accumulates more glucose than does normal tissue, because
cancer cells make use of large amounts of glucose as a molecular
source for anabolic biosynthesis of macromolecules, which are
necessary for cancer cell proliferation (18). The idea that tumors have a
particular metabolic phenotype that is associated with increased
glycolysis is supported by molecular and functional data (19). Microarray datasets collected from
several studies have consistently shown most of the genes involved
in glucose transport and glycolysis are upregulated in different
types of tumors (20,21). Glucose transporters are
overexpressed in hepatocarcinomas, breast cancer, neuroendocrine
carcinomas, lymphoblastic leukemia and others (22–25).
In mesenchymal glioma stem cells, the glycolysis pathway is highly
upregulated, involving aldehydedehydrogenase 1A3, which creates
heterogeneous cellular populations (26). A recent study demonstrated that the
activity of oxidative phosphorylation complexes and citrate
synthase was gradually decreased by tumor grade in glioma as
compared with normal brain tissue (27). This shift in cellular energy
production from oxidative phosphorylation to glycolysis is a result
of the glioma adapting to the surrounding environment. Several
mechanisms were involved in the downregulation of oxidative
phosphorylation in tumor cells. Lack of vascularization in tumors
causes severe hypoxia, which leads to a compensatory upregulation
of glycolysis in tumors. Genetic inactivation of p53 gene, a
regulator of oxidative phosphorylation, or activation of oncogenes
can result in a secondary decrease in oxidative phosphorylation
(28–31). Loss of function of components of
oxidative phosphorylation has been demonstrated in
pheochromocytomas and paragangliomas (32). In this context, glycolysis plays a
significant role in survival and growth of cancer cells (33). Therefore, intervening in glycolysis
could result in remarkable inhibition of cell growth and induction
of cell death (13,34).
PGAM1 is involved in the Warburg effect and
catalyzes the conversion of 3-PG to 2-PG, a crucial step in
glycolysis (35). PGAM1 controls
intracellular 3-PG and 2-PG levels and is important for anabolic
biosynthesis in cells. PGAM1 knockdown increases 3-PG level and
reduces 2-PG level. PGAM1 enzyme activity strikes a balance between
3-PG and 2-PG levels, which coordinates glycolysis and biosynthesis
to promote cancer cell proliferation (36).
PGAM1 activity is upregulated in many cancers.
Targeting PGAM1 by a small molecule inhibitor inhibited cancer cell
proliferation and tumor growth and altered 3-PG and 2-PG levels in
primary leukemia cells from human patients, thereby leading to
attenuated leukemia cell proliferation (4,13,36). A
recent study revealed that Y26 phosphorylation of PGAM1 enhanced
PGAM1 activity by stabilising the active conformation of PGAM1,
which promoted tumor growth (37).
Protein expression profile analysis revealed that overexpression of
PGAM1 was associated with poor survival of patients with lung
adenocarcinoma (5). Another study
suggested that overexpression of PGAM1 was strongly correlated with
poor differentiation and decreased survival rates in hepatocellular
carcinoma (7). A recent study also
demonstrated that PGAM1 knockdown had a marked survival benefit in
mice with intracranial xenografts (38). These studies suggest that PGAM1
probably participates in gliomagenesis. Our results show that PGAM1
expression was significantly upregulated in glioma tissues compared
with human brain tissues. Furthermore, PGAM1 expression increased
with increasing pathological glioma grade. Our result is consistent
with a recent report of protein expression profiles in gliomas
(39). Taken together, these
results indicate that PGAM1 might function as a tumor promoter in
glioma.
We confirmed the function of PGAM1 in glioma cells.
MTT assay showed that cell proliferation was inhibited with
transfection of PGAM1 siRNA into U87 cells. Moreover, siRNA
knockdown of PGAM1 expression significantly induced glioma cell
apoptosis in vitro by upregulating Bax expression,
downregulating Bcl-2 expression, and activating the caspase-3
signal in glioma cells. In addition, siRNA knockdown of PGAM1
induced S-phase cell cycle arrest and decreased the rate of cells
in the G2/M phase in U87 cells. The highly invasive nature of
glioma cells renders them incurable by localized therapy including
surgery and radiotherapy, thereby leading to poor patient survival
(40,41). Thus, using RNA interference, we
defined a specific role for PGAM1 in glioma migration and invasion.
After transfection with PGAM1 siRNA for 48 h, the rates of cell
migration and invasion were significantly reduced compared with
those in control cells, which suggests that PGAM1 knockdown is
deleterious for U87 cell migration and invasion. Importantly, PGAM1
knockdown suppressed the protein levels of MMP2/9, markers for cell
invasion, which may be important for inhibiting the invasive
potential of U87 cells. Our data strongly suggest that PGAM1 plays
an important role in glioma development and progression. Additional
molecular and functional studies of PGAM1 in glioma cells are
needed.
In conclusion, previous studies suggest that
targeting PGAM1 by siRNA or with a small molecule inhibitor
PGMI-004A attenuates cancer cell proliferation (7,36). In
agreement with these observations, we found that the expression of
PGAM1 is important in glioma cell proliferation. PGAM1 knockdown
efficiently inhibited glioma cell migration and invasion and
induced cell cycle arrest and apotosis in vitro. PGAM1 might
be a promising therapy in clinical treatment of glioma, which
heavily relies on aerobic glycolysis.
Acknowledgments
The present study was supported by the Natural
Scientific Foundation of China (no. 81141088), the Promotive
Research Fund for Excellent Young and Middle-aged Scientists of
Shandong Province (no. 2004BS02010), the Jinan Youth Technology
Star Plan (20120137), the Shandong Excellent Youth Scientist
Research Fund (BS2012YY022), and the Shandong University Innovation
Fund (2012TS135). Thanks to Laura Smales for the English
editing.
References
|
1
|
Furnari FB, Fenton T, Bachoo RM, Mukasa A,
Stommel JM, Stegh A, Hahn WC, Ligon KL, Louis DN, Brennan C, et al:
Malignant astrocytic glioma: Genetics, biology, and paths to
treatment. Genes Dev. 21:2683–2710. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Ostrom QT, Gittleman H, Farah P, Ondracek
A, Chen Y, Wolinsky Y, Stroup NE, Kruchko C and Barnholtz-Sloan JS:
CBTRUS statistical report: Primary brain and central nervous system
tumors diagnosed in the United States in 2006–2010. Neuro Oncol.
15(Suppl 2): ii1–ii56. 2013. View Article : Google Scholar :
|
|
3
|
Sant M, Minicozzi P, Lagorio S, Børge
Johannesen T, Marcos-Gragera R and Francisci S; EUROCARE Working
Group: Survival of European patients with central nervous system
tumors. Int J Cancer. 131:173–185. 2012. View Article : Google Scholar
|
|
4
|
Evans MJ, Saghatelian A, Sorensen EJ and
Cravatt BF: Target discovery in small-molecule cell-based screens
by in situ proteome reactivity profiling. Nat Biotechnol.
23:1303–1307. 2005. View
Article : Google Scholar : PubMed/NCBI
|
|
5
|
Chen G, Gharib TG, Wang H, Huang CC, Kuick
R, Thomas DG, Shedden KA, Misek DE, Taylor JM, Giordano TJ, et al:
Protein profiles associated with survival in lung adenocarcinoma.
Proc Natl Acad Sci USA. 100:13537–13542. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Narayanan NK, Narayanan BA and Nixon DW:
Resveratrol-induced cell growth inhibition and apoptosis is
associated with modulation of phosphoglycerate mutase B in human
prostate cancer cells: Two-dimensional sodium dodecyl
sulfate-polyacrylamide gel electrophoresis and mass spectrometry
evaluation. Cancer Detect Prev. 28:443–452. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Ren F, Wu H, Lei Y, Zhang H, Liu R, Zhao
Y, Chen X, Zeng D, Tong A, Chen L, et al: Quantitative proteomics
identification of phosphoglycerate mutase 1 as a novel therapeutic
target in hepatocellular carcinoma. Mol Cancer. 9:812010.
View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Liu L, Wang S, Zhang Q and Ding Y:
Identification of potential genes/proteins regulated by Tiam1 in
colorectal cancer by microarray analysis and proteome analysis.
Cell Biol Int. 32:1215–1222. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Turhani D, Krapfenbauer K, Thurnher D,
Langen H and Fountoulakis M: Identification of differentially
expressed, tumor-associated proteins in oral squamous cell
carcinoma by proteomic analysis. Electrophoresis. 27:1417–1423.
2006. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Fang MZ, Liu C, Song Y, Yang GY, Nie Y,
Liao J, Zhao X, Shimada Y, Wang LD and Yang CS: Overexpression of
gastrin-releasing peptide in human esophageal squamous cell
carcinomas. Carcinogenesis. 25:865–871. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Tong A, Wu L, Lin Q, Lau QC, Zhao X, Li J,
Chen P, Chen L, Tang H, Huang C, et al: Proteomic analysis of
cellular protein alterations using a hepatitis B virus-producing
cellular model. Proteomics. 8:2012–2023. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Zhang S, Zhao Y, Lei B, Li C and Mao X:
PGAM1 is involved in spermatogenic dysfunction and affects cell
proliferation, apoptosis, and migration. Reprod Sci. 22:1236–1242.
2015. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Engel M, Mazurek S, Eigenbrodt E and
Welter C: Phosphoglycerate mutase-derived polypeptide inhibits
glycolytic flux and induces cell growth arrest in tumor cell lines.
J Biol Chem. 279:35803–35812. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Louis DN, Ohgaki H, Wiestler OD, Cavenee
WK, Burger PC, Jouvet A, Scheithauer BW and Kleihues P: The 2007
WHO classification of tumours of the central nervous system (vol
114, pg 97, 2007). Acta Neuropathol. 114:547. 2007. View Article : Google Scholar
|
|
15
|
Wen PY and Kesari S: Malignant gliomas in
adults. N Engl J Med. 359:492–507. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Weller M, van den Bent M, Hopkins K, Tonn
JC, Stupp R, Falini A, Cohen-Jonathan-Moyal E, Frappaz D,
Henriksson R, Balana C, et al European Association for
Neuro-Oncology (EANO) Task Force on Malignant Glioma: EANO
guideline for the diagnosis and treatment of anaplastic gliomas and
glioblastoma. Lancet Oncol. 15:e395–e403. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Warburg O: On the origin of cancer cells.
Science. 123:309–314. 1956. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Kroemer G and Pouyssegur J: Tumor cell
metabolism: Cancer's Achilles' heel. Cancer Cell. 13:472–482. 2008.
View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Ortega AD, Sánchez-Aragó M, Giner-Sánchez
D, Sánchez-Cenizo L, Willers I and Cuezva JM: Glucose avidity of
carcinomas. Cancer Lett. 276:125–135. 2009. View Article : Google Scholar
|
|
20
|
Moreno-Sánchez R, Rodríguez-Enríquez S,
Marín-Hernández A and Saavedra E: Energy metabolism in tumor cells.
FEBS J. 274:1393–1418. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Altenberg B and Greulich KO: Genes of
glycolysis are ubiquitously overexpressed in 24 cancer classes.
Genomics. 84:1014–1020. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Amann T, Maegdefrau U, Hartmann A, Agaimy
A, Marienhagen J, Weiss TS, Stoeltzing O, Warnecke C, Schölmerich
J, Oefner PJ, et al: GLUT1 expression is increased in
hepatocellular carcinoma and promotes tumorigenesis. Am J Pathol.
174:1544–1552. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Meadows AL, Kong B, Berdichevsky M, Roy S,
Rosiva R, Blanch HW and Clark DS: Metabolic and morphological
differences between rapidly proliferating cancerous and normal
breast epithelial cells. Biotechnol Prog. 24:334–341. 2008.
View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Ozbudak IH, Shilo K, Tavora F, Rassaei N,
Chu WS, Fukuoka J, Jen J, Travis WD and Franks TJ: Glucose
transporter-1 in pulmonary neuroendocrine carcinomas: Expression
and survival analysis. Mod Pathol. 22:633–638. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Boag JM, Beesley AH, Firth MJ, Freitas JR,
Ford J, Hoffmann K, Cummings AJ, de Klerk NH and Kees UR: Altered
glucose metabolism in childhood pre-B acute lymphoblastic
leukaemia. Leukemia. 20:1731–1737. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Mao P, Joshi K, Li J, Kim SH, Li P,
Santana-Santos L, Luthra S, Chandran UR, Benos PV, Smith L, et al:
Mesenchymal glioma stem cells are maintained by activated
glycolytic metabolism involving aldehyde dehydrogenase 1A3. Proc
Natl Acad Sci USA. 110:8644–8649. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Feichtinger RG, Weis S, Mayr JA,
Zimmermann F, Geilberger R, Sperl W and Kofler B: Alterations of
oxidative phosphorylation complexes in astrocytomas. Glia.
62:514–525. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Bögler O, Huang HJ, Kleihues P and Cavenee
WK: The p53 gene and its role in human brain tumors. Glia.
15:308–327. 1995. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Ma W, Sung HJ, Park JY, Matoba S and Hwang
PM: A pivotal role for p53: Balancing aerobic respiration and
glycolysis. J Bioenerg Biomembr. 39:243–246. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Matoba S, Kang JG, Patino WD, Wragg A,
Boehm M, Gavrilova O, Hurley PJ, Bunz F and Hwang PM: p53 regulates
mitochondrial respiration. Science. 312:1650–1653. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Zhang H, Gao P, Fukuda R, Kumar G,
Krishnamachary B, Zeller KI, Dang CV and Semenza GL: HIF-1 inhibits
mitochondrial biogenesis and cellular respiration in VHL-deficient
renal cell carcinoma by repression of C-MYC activity. Cancer Cell.
11:407–420. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Astuti D, Latif F, Dallol A, Dahia PL,
Douglas F, George E, Sköldberg F, Husebye ES, Eng C and Maher ER:
Gene mutations in the succinate dehydrogenase subunit SDHB cause
susceptibility to familial pheochromocytoma and to familial
paraganglioma. Am J Hum Genet. 69:49–54. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Pelicano H, Martin DS, Xu RH and Huang P:
Glycolysis inhibition for anticancer treatment. Oncogene.
25:4633–4646. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Inoki K, Zhu T and Guan KL: TSC2 mediates
cellular energy response to control cell growth and survival. Cell.
115:577–590. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Grisolia S and Cleland WW: Influence of
salt, substrate, and cofactor concentrations on the kinetic and
mechanistic behavior of phosphoglycerate mutase. Biochemistry.
7:1115–1121. 1968. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Hitosugi T, Zhou L, Elf S, Fan J, Kang HB,
Seo JH, Shan C, Dai Q, Zhang L, Xie J, et al: Phosphoglycerate
mutase 1 coordinates glycolysis and biosynthesis to promote tumor
growth. Cancer Cell. 22:585–600. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Hitosugi T, Zhou L, Fan J, Elf S, Zhang L,
Xie J, Wang Y, Gu TL, Alečković M, LeRoy G, et al: Tyr26
phosphorylation of PGAM1 provides a metabolic advantage to tumours
by stabilizing the active conformation. Nat Commun. 4:17902013.
View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Sanzey M, Abdul Rahim SA, Oudin A, Dirkse
A, Kaoma T, Vallar L, Herold-Mende C, Bjerkvig R, Golebiewska A and
Niclou SP: Comprehensive analysis of glycolytic enzymes as
therapeutic targets in the treatment of glioblastoma. PLoS One.
10:1020152015. View Article : Google Scholar
|
|
39
|
Gao H, Yu B, Yan Y, Shen J, Zhao S, Zhu J,
Qin W and Gao Y: Correlation of expression levels of ANXA2, PGAM1,
and CALR with glioma grade and prognosis. J Neurosurg. 118:846–853.
2013. View Article : Google Scholar
|
|
40
|
Louis DN: Molecular pathology of malignant
gliomas. Annu Rev Pathol. 1:97–117. 2006. View Article : Google Scholar
|
|
41
|
Giese A, Bjerkvig R, Berens ME and
Westphal M: Cost of migration: Invasion of malignant gliomas and
implications for treatment. J Clin Oncol. 21:1624–1636. 2003.
View Article : Google Scholar : PubMed/NCBI
|