Introduction
MicroRNAs (miRNAs) are a class of non-coding highly
conserved RNAs that suppress protein expression through binding to
the 3′-untranslated region (3-UTR) of target mRNAs, and function as
post-transcriptional regulators of gene expression (1). Increasing evidence has confirmed that
miRNAs participate in diverse biological events such as
differentiation, disease, cell proliferation, apoptosis, migration
and invasion (2,3). Previous studies revealed that miRNAs
play important roles in human types of cancer and suggest that the
aberrant behavior of miRNAs contribute to the development and
progression of human types of cancer by modulating the expression
of oncogenes or tumor suppressors (4–6).
miR-455 has been shown to be significantly
dysregulated in different human tumors (7–11). For
instance, it was markedly downregulated in basal cell, glioblastoma
and colorectal carcinoma and in gastric and non-small cell lung
cancer (12–17). It inhibited the proliferation,
migration and invasion of lung cancer by directly modulating ZEB1
(18). Moreover, miR-455 inhibited
proliferation and invasion of colorectal carcinoma cells by
adjusting RAF proto-oncogene serine/threonine-protein kinase (RAF1)
and could be a prognostic marker for colorectal cancer patients
(19). Furthermore, miR-455 was a
predictor of chemoresistance in colon adenocarcinoma and diffuse
large B-cell lymphoma patients (20). These studies revealed that miR-455
serves as a suppressor miRNA in tumors. However, the functional
role of miR-455 and the underlying signaling mechanism by which
miR-455 regulates the development and progression of HCC have not
been established.
The present research aimed to identify the
biological function of miR-455 in HCC. We demonstrated that miR-455
was downregulated in HCC samples with an aggressive phenotype.
Downregulation of miR-455 was found to be correlated with adverse
prognostic characteristics and reduced 5-year overall and
disease-free survival of HCC patients. Gain- and loss-of-function
experiments demonstrated that miR-455 inhibited HCC migration and
invasion in vitro. Moreover, runt-related transcription
factor 2 (Runx2) was confirmed as a direct target of miR-455. Our
results elucidate the underlying mechanism involved through which
miR-455 suppresses HCC and propose the use of miR-455 as a
potential therapeutic strategy for HCC.
Materials and methods
Clinical specimens and data
One-hundred and four HCC specimens were obtained
from patients undergoing curative resection of their primary HCC at
the First and Second Affiliated Hospital of Wenzhou Medical
University, from January 2006 to December 2008, with a median
follow-up time of 40 months. The clinicopathological data were
collected and are summarized in Table
I. Informed consent was obtained and signed by all patients.
Patients did not receive preoperative chemotherapy or
embolization.
 | Table I.Correlation between the
clinicopathological characteristics and miR-455 expression in the
HCC cases (n=104). |
Table I.
Correlation between the
clinicopathological characteristics and miR-455 expression in the
HCC cases (n=104).
|
|
| Expression level |
|
|---|
|
|
|
|
|
|---|
| Clinical
parameters | Cases (n) |
miR-455high (n=48) | miR-455low
(n=56) | P-value
(aP<0.05) |
|---|
| Age (years) |
|
|
| 0.451 |
|
<65 | 32 | 13 | 19 |
|
| ≥65 | 72 | 35 | 37 |
|
| Gender |
|
|
| 0.834 |
| Male | 88 | 41 | 47 |
|
|
Female | 16 | 7 | 9 |
|
| Tumor size (cm) |
|
|
| 0.716 |
|
<5 | 50 | 24 | 26 |
|
| ≥5 | 54 | 24 | 30 |
|
| Tumor no. |
|
|
| 0.025a |
|
Solitary | 86 | 44 | 42 |
|
|
Multiple | 18 | 4 | 14 |
|
| Edmondson |
|
|
| 0.001a |
|
I+II | 35 | 24 | 11 |
|
|
III+IV | 69 | 24 | 45 |
|
| TNM stage |
|
|
| 0.008a |
|
I+II | 81 | 43 | 38 |
|
|
III+IV | 23 | 5 | 18 |
| Venous
infiltration |
|
|
| 0.021a |
|
Present | 21 | 5 | 16 |
|
|
Absent | 83 | 43 | 40 |
|
| AFP (ng/ml) |
|
|
| 0.544 |
|
<400 | 29 | 12 | 17 |
|
|
≥400 | 75 | 36 | 39 |
|
| HBsAg |
|
|
| 0.966 |
|
Positive | 89 | 41 | 48 |
|
|
Negative | 15 | 7 | 8 |
|
Cell line culture and treatment
The HCC cell lines (HepG2, Hep3B, SMMC-7721,
MHCC-97H and Bel-7402) and the normal hepatocyte cell line L02 were
purchased from the Institute of Biochemistry and Cell Biology
(Chinese Academy of Sciences, Shanghai, China) and were maintained
in Dulbeccos modified Eagles medium (DMEM) supplemented with 10%
fetal bovine serum (FBS; both from Gibco, Grand Island, NY, USA) in
a 5% CO2 humidified atmosphere at 37°C.
The miRNA expression vectors, including the miR-455
expression vector (HmiR0148-MR03), the control vector
(CmiR0001-MR04), miR-455 inhibitor (HmiR-AN0516) and the negative
control (CmiR-AN0001-AM04) and Runx2 overexpression plasmid
(LPP-H5215-lv101) were obtained from Genecopoeia (Guangzhou,
China). The specific siRNA against Runx2, with sequence
5′-UAACAGCAGAGGCAUUUCGUAGCUC-3′ and a scramble siRNA were produced
by Sangon Biotech Co., Ltd. (Shanghai, China). Cells were
transfected with the aforementioned vectors using Lipofectamine
2000 reagent (Invitrogen Life Technologies) in accordance with the
manufacturer's instructions.
Quantitative real-time polymerase
chain reaction (qRT-PCR)
Total cellular RNA was isolated from the cultured
cells or clinical tissues using TRIzol reagent (Invitrogen Life
Technologies). RNA was reverse-transcribed using a TaqMan human
miRNA assay kit (Applied Biosystems, Foster City, CA, USA) and cDNA
was then amplified with a SYBR® Premix Ex Taq™ II
(Perfect Real-Time) kit (Takara Bio Inc., Shiga, Japan).
Hsa-miR-455 primer (HmiRQP0516), snRNA U6 qPCR Primer (HmiRQP9001),
Runx2 (HQP016478) and GAPDH (HQP006940) were purchased from
Genecopoeia.
Western blot analysis
Cultured cells and clinical tissues were collected
and lysed and then the protein concentration was determined using a
BCA Protein Assay kit (Thermo Fisher Scientific, Rockford, IL,
USA). Thirty micrograms of denatured protein was subjected to 10%
SDS-PAGE and transferred to a polyvinylidene fluoride (PVDF)
membrane (Millipore, Billerica, MA, USA). The membranes were
incubated with respective primary antibodies: Runx2 and GAPDH
(1:1,000; Cell Signaling Technology, Inc.) at 4°C overnight. Then
the membranes were washed three times with TBST and incubated with
the appropriate HRP-conjugated secondary antibody for 2 h at room
temperature (ZSGB-BIO Co., Ltd., Beijing, China). Protein bands
were developed using an enhanced chemiluminescence kit (Amersham/GE
Healthcare Life Sciences, Little Chalfont, UK).
Immunohistochemistry
Paraffin-embedded sections were subjected to
immunohistochemical staining. Runx2 (1:300; Cell Signaling
Technology, Inc.) antibody was applied as the primary antibody
using a streptavidin peroxidase-conjugated (SP-IHC) method. The
percentage of positive cells was expressed as: 0 for <10%; 1 for
10–30%; 2 for 31–50%; 3 for >50%.
Transwell assays
Transwell migration and invasion assays were carried
out using 8-µm pore-sized Transwell inserts (Nalge Nunc
International Corp., Naperville, IL, USA). A cell suspension with a
density of 2.5×105 cells/ml in serum-free DMEM was
prepared and 200 µl of this cell suspension was loaded into the top
chamber, and 750 µl DMEM with 10% FBS was placed in the bottom
chamber. After a 24-h incubation, cells were fixed in 4%
paraformaldehyde for 15 min and stained with 0.3% crystal violet
dye for 15 min. The cells remaining in the top layer were swabbed
carefully. The BioCoat Matrigel invasion chamber (Becton Dickinson
Labware) was used to perform the Transwell invasion assay as
previously described above.
Dual-luciferase reporter gene
assay
The predicted Runx2 3′-UTR sequence and the
corresponding mutated sequence of the sites were produced and
embedded into the pRL-TK control vector (Promega, Corp., Madison,
WI, USA). MHCC-97H cells which achieved 90% confluency were seeded
in a 96-well plate. For the reporter assay, 120 ng miR-455
inhibitor or negative control was co-transfected with 30 ng of the
wild-type (wt) or mutant (mt) 3′-UTR of Runx2 mRNA using 0.45 µl of
FuGene® (Promega) and then were transfected into the
MHCC-97H cells. Following 48 h of transfection, the luciferase
activity was measured according to the manufacturers instructions
(Dual-Luciferase assay system; Promega). The assays were repeated
independently in triplicate.
Statistical analysis
Data are presented as the mean ± SD and performed
with at least three independent replicates. SPSS software, 16.0
(SPSS, Inc., Chicago, IL, USA) and GraphPad Prism 6.0 (GraphPad
Software, Inc., La Jolla, CA, USA) were used with a two-tailed
Students t-test, Pearson's correlation analysis, the Kaplan-Meier
method and the log-rank test to evaluate the statistical
significance. Significant differences were defined as
P<0.05.
Results
miR-455 expression is downregulated in
HCC tissues and cell lines
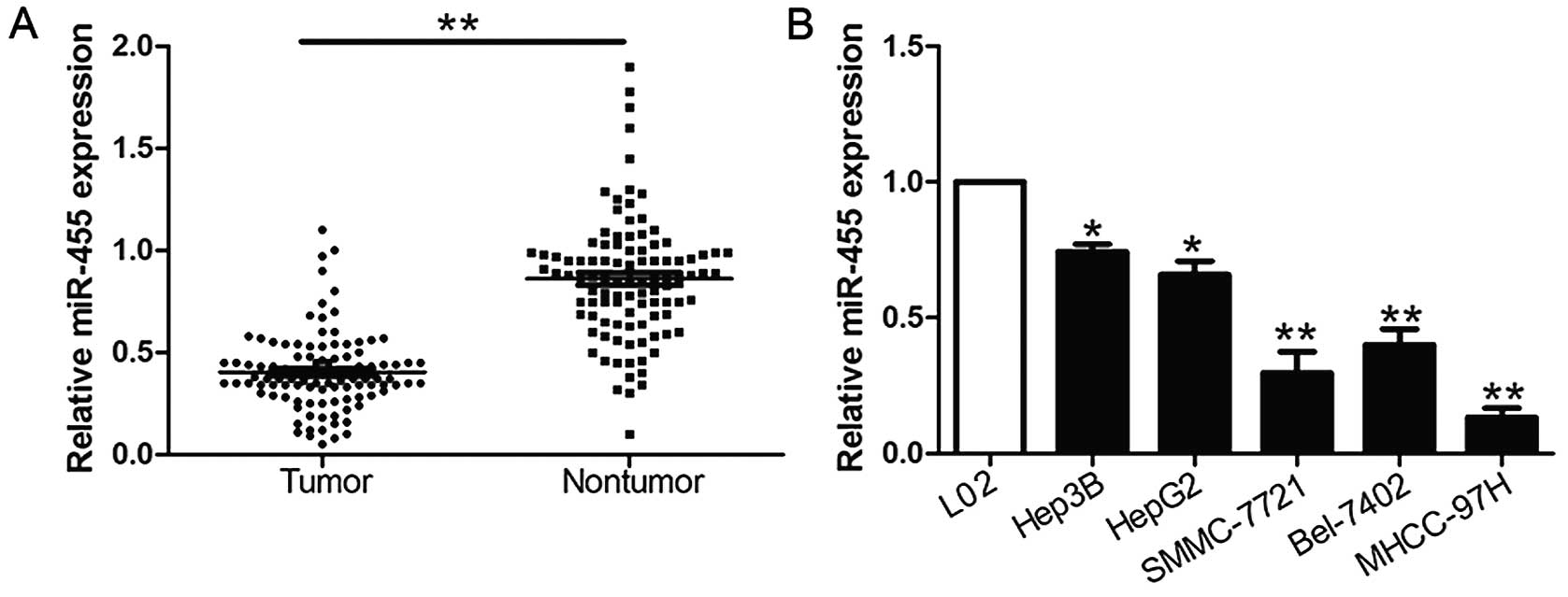
We investigated the expression of miR-455 compared
to an endogenous control (U6 RNA) in 104 pairs of HCC tissues and
the corresponding non-tumor tissues. We found that miR-455
expression in HCC tissues was significantly lower than levels noted
in the corresponding adjacent non-tumor tissues (P<0.05,
Fig. 1A). Furthermore, we analyzed
miR-455 expression in a non-transformed hepatic cell line (L02) and
five HCC cell lines (HepG2, Hep3B, SMCC-7721, Bel-7402 and
MHCC-97H). Notably, our results revealed that the relative
expression of miR-455 was evidently downregulated in the HCC cell
lines when compared with the L02 cells (P<0.05, Fig. 1B). These data suggest that the
downregulated expression of miR-455 potentially correlates with the
development and progression of HCC.
Clinical significance of the
downregulation of miR-455 expression in HCC samples
We defined two different miR-455 expression groups
according to the mean expression level of miR-455 in order to
investigate the association between miR-455 expression levels and
the clinical characteristics and prognosis of the HCC patients. As
shown in Table I, low expression of
miR-455 was clearly correlated with multiple tumor nodes (P=0.025),
venous infiltration (P=0.021), high Edmondson-Steiner grading
(P=0.001) and advanced tumor-node-metastasis (TNM) tumor stage
(P=0.008). Hence, these findings revealed that the reduced
expression of miR-455 was correlated with adverse prognostic
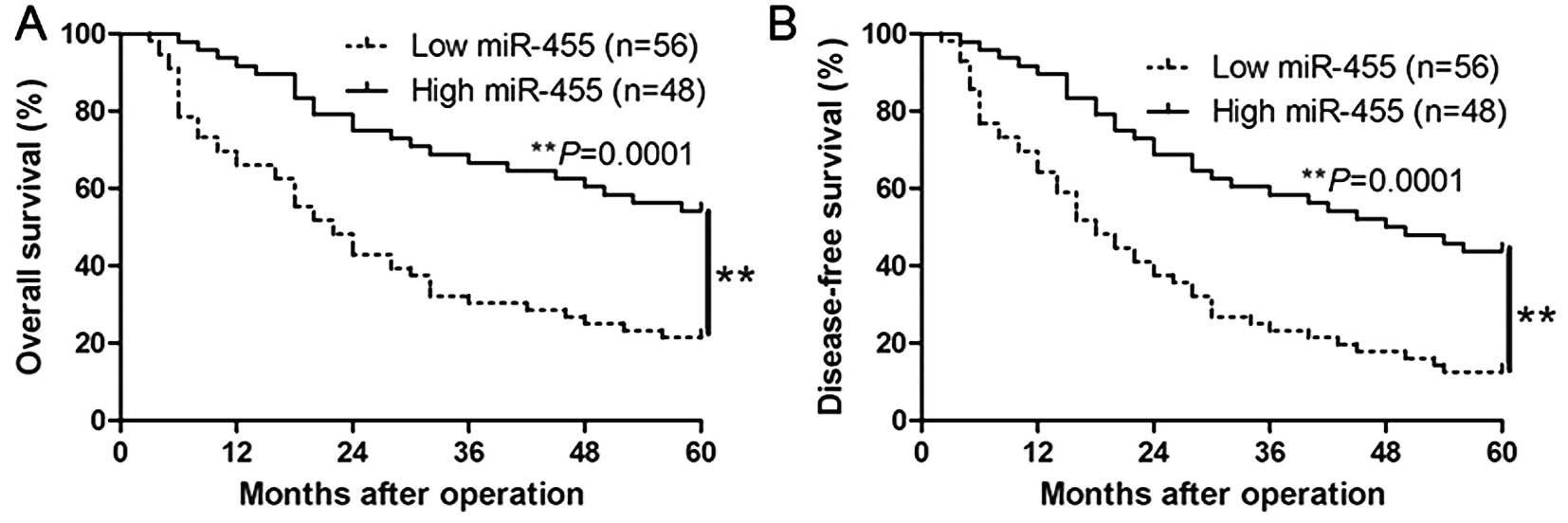
characteristics of HCC. Furthermore, Kaplan-Meier analysis revealed
that high expression of miR-455 was significantly correlated with a
prolonged overall survival (P=0.0001, Fig. 2A) and disease-free survival of the
HCC patients (P=0.0001, Fig. 2B).
Moreover, miR-455 expression was found to be a novel independent
factor for predicting both the 5-year overall and disease-free
survival in HCC patients (P=0.018 and 0.011, respectively; Table II). Our results suggest that
miR-455 could serve as a valuable biomarker for predicting the
outcome of HCC patients.
 | Table II.Multivariate Cox regression analysis
of the 5-year overall and disease-free survival of the 104 HCC
patients. |
Table II.
Multivariate Cox regression analysis
of the 5-year overall and disease-free survival of the 104 HCC
patients.
|
| Overall
survival | Disease-free
survival |
|---|
|
|
|
|
|---|
| Variables | HR | 95% CI | P-value | HR | 95% CI | P-value |
|---|
| miR-455
expression | 2.179 | 1.113–4.249 | 0.018a | 3.602 | 1.113–4.794 | 0.011a |
| Edmondson
grade | 1.213 | 0.713–1.983 | 0.230 | 1.123 | 0.694–1.869 | 0.347 |
| TNM stage | 2.812 | 1.355–5.813 | 0.007a | 2.194 | 1.020–4.649 | 0.009a |
| No. of tumor
nodules | 1.694 | 0.894–3.024 | 0.068 | 1.674 | 0.999–2.815 | 0.056 |
| Venous
infiltration | 3.253 | 2.108–5.983 | 0.001a | 3.521 | 2.238–6.372 | 0.001a |
miR-455 suppresses HCC cell migration
and invasion
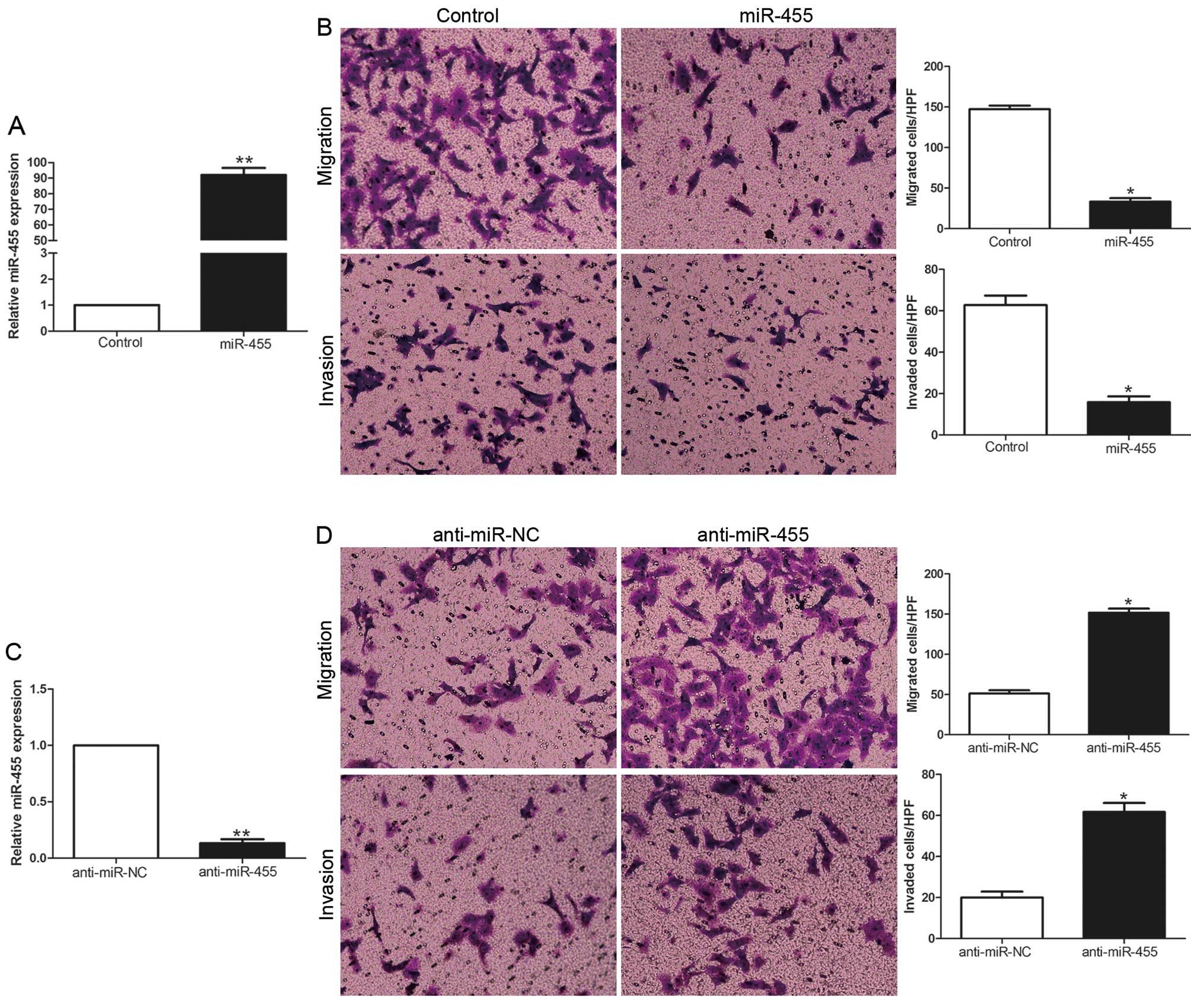
To identify the biological function of miR-455 in
HCC, we transduced HCC cell lines with the miR-455 expression
vector or anti-miR-455 vector with different endogenous miR-455
levels. As determined by qRT-PCR, the miR-455 vector obviously
increased the expression level of miR-455 in the MHCC-97H cells
(P<0.01, Fig. 3A), while the
anti-miR-455 vector significantly reduced the expression level of
miR-455 in the Hep3B cells (P<0.01, Fig. 3C). Transwell migration and invasion
assays showed that increased expression of miR-455 in the MHCC-97H
cells (MHCC-97H-miR-455) resulted in an obvious decrease in cell
migration and invasion (P<0.05, Fig.
3B), while downregulation of miR-455 in Hep3B cells
(Hep3B-anti-miR-455) showed a marked increase in the number of
migrated and invaded cells (P<0.05, Fig. 3D). Collectively, miR-455 may
function as an anti-metastatic molecule in HCC.
Runx2 is a direct target of miR-455 in
HCC
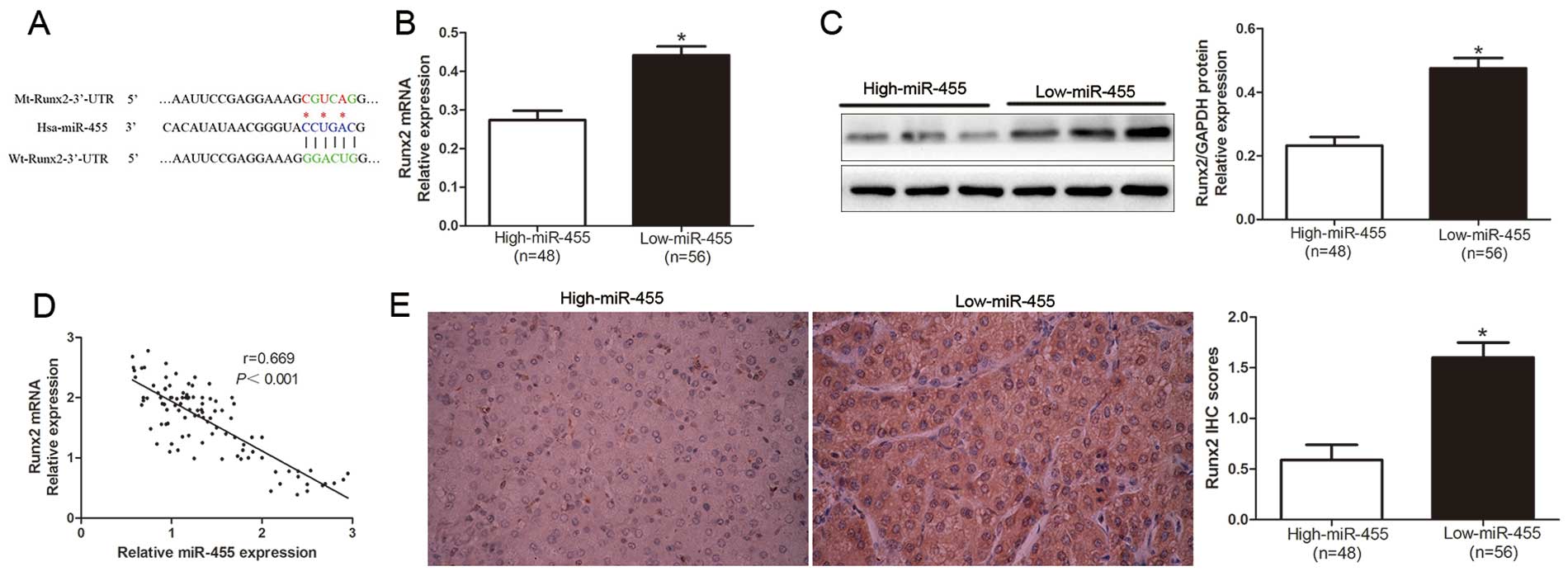
To further confirm the mechanism involved in the
miR-455 regulation in HCC cells, we explored the candidate target
genes of miR-455 by searching the publically available databases
TargetScan 6.2 and miRanda. As shown in Fig. 4A, we demonstrated that 3′-UTR of
Runx2 contained the highly conserved complementary sequence of
miR-455. Moreover, Zhang et al reported that Runx2 is a
target of miR-455 in chondrogenic differentiation (13). To confirm this prospect, we first
investigated the relationship between miR-455 and Runx2 in the HCC
cases. Our data revealed that both the Runx2 mRNA and protein
levels in the miR-455 high-expressing tumor tissues were obviously
lower than those in the miR-455 low-expressing tumors (P<0.05,
Fig. 4B and C). In addition, the
expression level of miR-455 was inversely associated with the
expression level of Runx2 mRNA in the HCC tissues (r=0.669,
P<0.0001, Fig. 4D). Similarily,
as determined by the IHC results, the protein expression level of
Runx2 was significantly decreased in patients with a high level of
miR-455 (P<0.05, Fig. 4E).
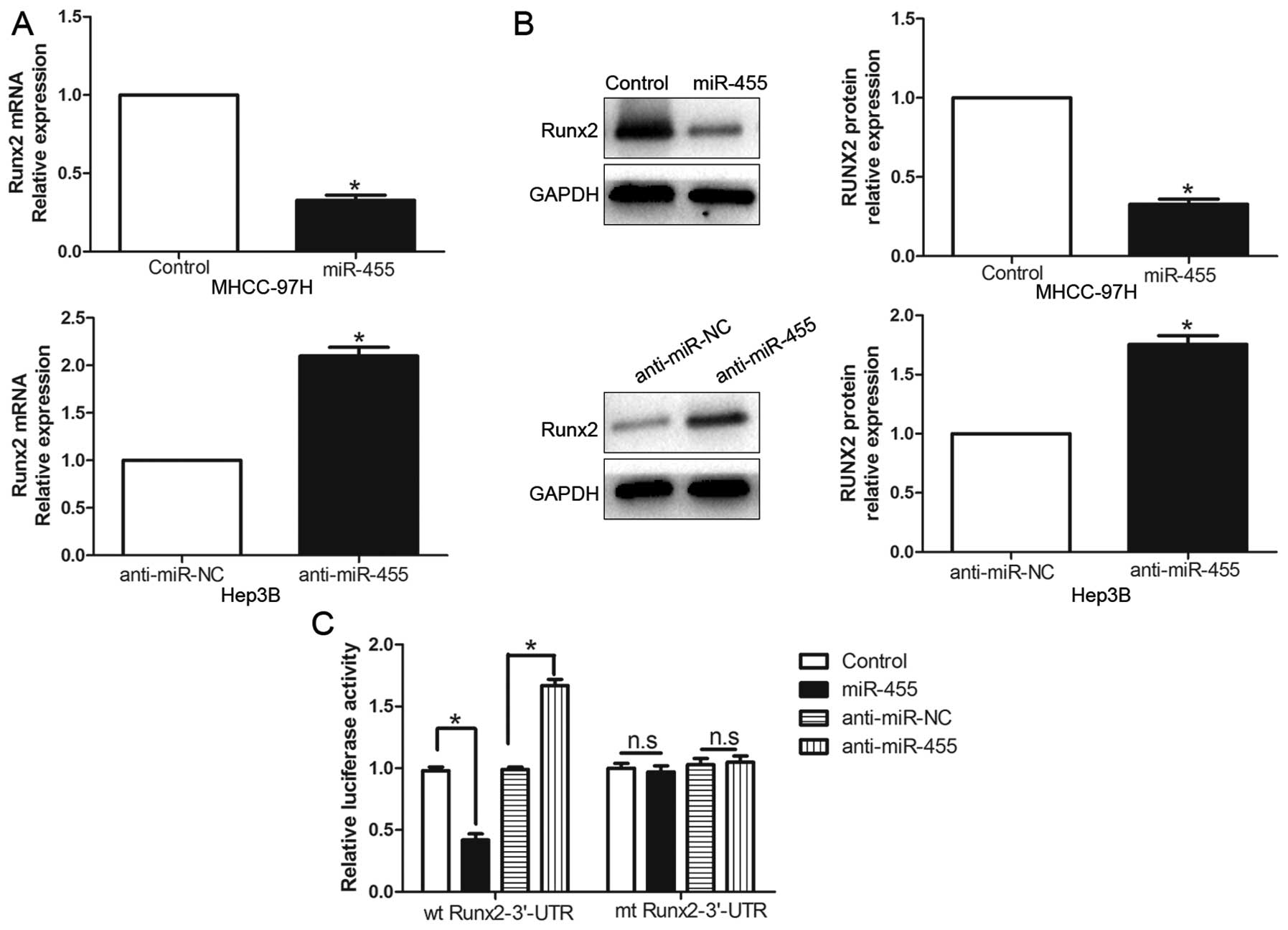
Furthermore, the expression levels of the Runx2 mRNA and protein
were prominently inhibited by the overexpression of miR-455 in the
MHCC-97H cells (P<0.05, respectively, Fig. 5A and B). By contrast, the
downregulation of miR-455 prominently promoted the mRNA and protein
expression levels of Runx2 in the Hep3B cells (P<0.05,
respectively, Fig. 5A and B). In
addition, the overexpression of miR-455 clearly suppressed the
luciferase activity of Runx2 containing a wt 3′-UTR but did not
inhibit the activity of mt Runx2 (P<0.001, Fig. 5C). Suppression of miR-455 increased
the luciferase activity of wt Runx2-3′-UTR (P<0.001, Fig. 5C). However, with the mt Runx2-3′-UTR
vectors, there was no relative increase in activity. Collectively,
these data strongly suggest that Runx2 is a downstream target of
miR-455 in HCC.
Alteration of Runx2 expression levels
affect the functional effects of miR-455 in HCC cells
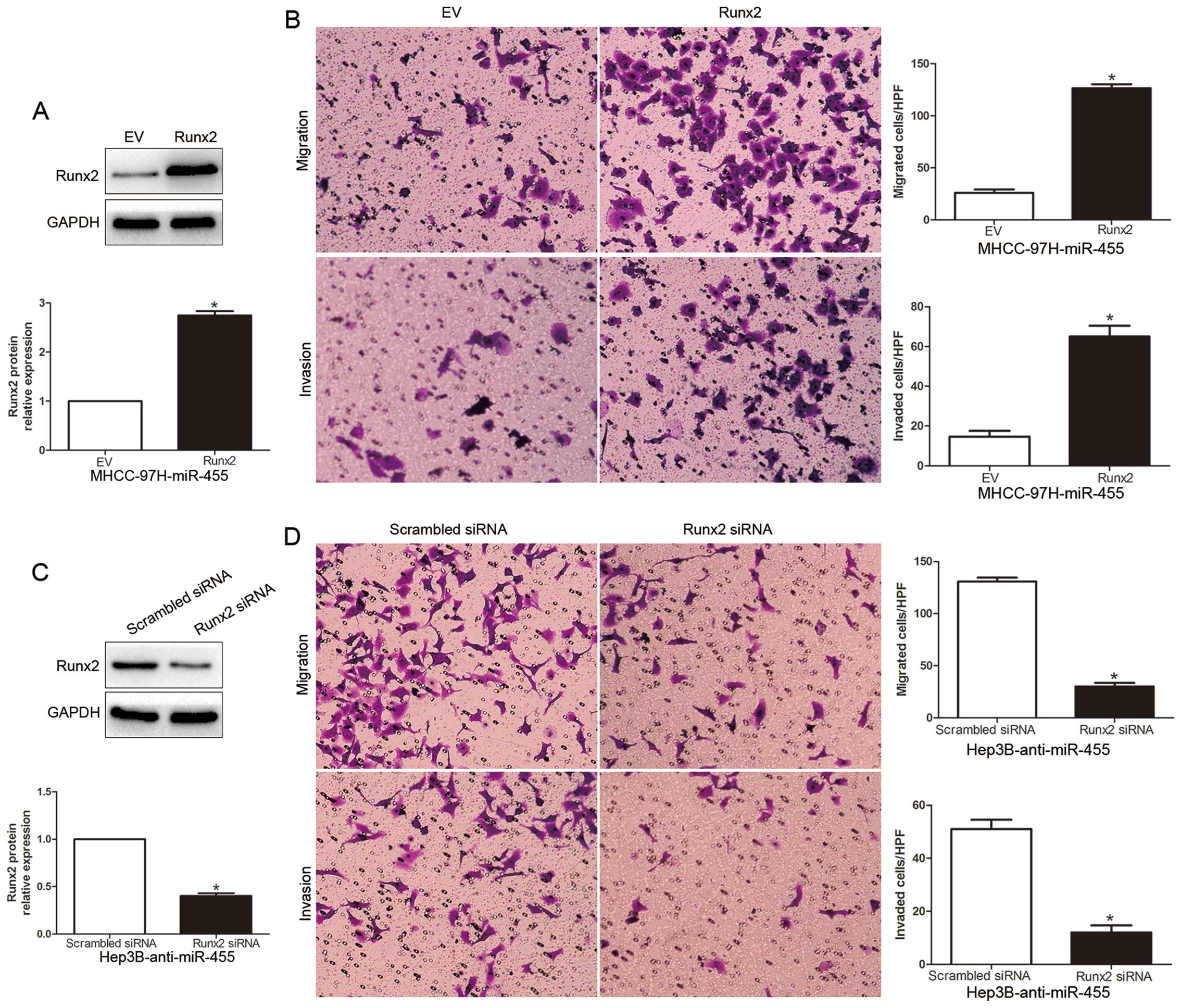
To explore that Runx2 is a functional target of
miR-455, we restored the Runx2 expression in MHCC-97H-miR-455 cells
by transfecting them with a Runx2 plasmid (P<0.05, Fig. 6A). We found that restoration of
Runx2 expression in the MHCC-97H-miR-455 cells partially suppressed
the effect of exogenous miR-455, resulting in a prominent increase
in cell migration and invasion (P<0.05, Fig. 6B). Similarly, silencing of Runx2 in
Hep3B-anti-miR-455 cells partially suppressed the effect of
anti-miR-455 on migration and invasion (P<0.05, Fig. 6C and D). These data demonstrate that
Runx2 is a downstream mediator in the function of miR-455 in
HCC.
Discussion
Increasing research has reported the critical roles
of miRNAs in the initiation and progression of various human types
of cancer (5). Exploration of
cancer-specific miRNAs and their functional molecular mechanism
contributes to the identification of novel biomarkers and
therapeutic strategies for human types of cancer (6). In this study, we identified that the
mean expression level of miR-455 was obviously decreased in HCC
tissues compared with that observed in the corresponding
tumor-adjacent tissues. Consistently, we also observed that miR-455
was decreased in HCC cell lines vs. a normal hepatic cell line.
These data suggest that miR-455 may be a novel tumor suppressor in
HCC. Furthermore, low expression of miR-455 was significantly
correlated with multiple tumor nodes, venous infiltration, high
Edmondson-Steiner grading and advanced TNM tumor stage. These data
indicate that the downregulated expression of miR-455 is associated
with adverse prognostic characteristics in HCC. Moreover, we
confirmed that increased expression of miR-455 predicted a
significant better 5-year survival for HCC patients. Multivariate
Cox repression analysis demonstrated that miR-455 is a novel
independent prognostic indicator for predicting the survival of HCC
patients. Collectively, these data reveal that miR-455 is critical
for the prognostic outcome in HCC patients.
Previous studies have confirmed miR-455 as a
suppressor in human types of cancer. Functionally, miR-455 exerts
anticancer functions by inhibiting tumor growth and metastasis
(21). Moreover, miR-455 suppressed
epithelial-mesenchymal transition (EMT) in NSCLC by targeting ZEB1
(18). These results were
consistent with the observations we performed in our experiments.
In the present study, we demonstrated that miR-455 overexpression
clearly reduced the number of migrated and invaded MHCC-97H cells
and miR-455 knockdown significantly promoted Hep3B cell migration
and invasion. Furthermore, we identified that miR-455 suppressed
migration and invasion, at least in part, by targeting Runx2.
miR-455 was inversely associated with the expression levels of both
Runx2 mRNA and protein in HCC tissues. miR-455 negatively regulated
Runx2 accumulation in HCC cells. Moreover, the complementary
sequence of miR-455 was confirmed in the 3′-UTR of Runx2 mRNA.
Knockdown of miR-455 increased the luciferase reporter activity of
wt 3′-UTR but not mt 3′-UTR of Runx2. Conversely, the
overexpression of miR-455 inhibited the luciferase activity of wt
3′-UTR but not mt 3′-UTR of Runx2. The function of miR-455
alteration on migration and invasion of HCC cells was also
dissipated by Runx2 modulation. In addition, Runx2 has been
reported to be a direct downstream target of miR-455 in early
chondrogenic differentiation (13).
Collectively, our results support Runx2 as a downstream mediator of
miR-455 function in HCC.
Previous studies have demonstrated that Runx2
regulates tumor progression, invasion and metastasis and its
function in migration and invasion has been documented in different
tumor cell types (22–26). Furthermore, Runx2 overexpression
upregulated transcription factors (SOX9, SNAI2 and SMAD3) that
participate in the process of EMT (27–29)
and whose features include increased motility and invasion
potential. Moreover, Runx2 was found to directly regulate the
expression levels of MMP9 and MMP13 and mediate the invasion of
human breast cancer cell lines (30). It has been shown that Runx2 plays a
critical role in hepatocarcinogenesis. This mechanism may account
for the effects of miR-455 in HCC.
In conclusion, the data revealed that the expression
of miR-455 was downregulated in HCC tissues and cell lines and its
low expression is related to malignant clinicopathological
characteristics. We confirmed that miR-455 is an independent
prognostic marker for predicting the 5-year survival of HCC
patients. In vitro, studies demonstrated that miR-455
suppressed HCC cell migration and invasion. Mechanistically, its
tumor-suppressive effects are mediated by Runx2. Collectively, the
downregulation of miR-455 may play a critical role in tumor
migration and invasion and may be a novel prognostic factor and of
potential use in therapeutic strategies for HCC.
References
|
1
|
Mendell JT: MicroRNAs: Critical regulators
of development, cellular physiology and malignancy. Cell Cycle.
4:1179–1184. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Bartel DP: MicroRNAs: Genomics,
biogenesis, mechanism, and function. Cell. 116:281–297. 2004.
View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Szabo G and Bala S: MicroRNAs in liver
disease. Nat Rev Gastroenterol Hepatol. 10:542–552. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Lu J, Getz G, Miska EA, Alvarez-Saavedra
E, Lamb J, Peck D, Sweet-Cordero A, Ebert BL, Mak RH, Ferrando AA,
et al: MicroRNA expression profiles classify human cancers. Nature.
435:834–838. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Calin GA and Croce CM: MicroRNA signatures
in human cancers. Nat Rev Cancer. 6:857–866. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Jansson MD and Lund AH: MicroRNA and
cancer. Mol Oncol. 6:590–610. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Liu C, Iqbal J, Teruya-Feldstein J, Shen
Y, Dabrowska MJ, Dybkaer K, Lim MS, Piva R, Barreca A, Pellegrino
E, et al: MicroRNA expression profiling identifies molecular
signatures associated with anaplastic large cell lymphoma. Blood.
122:2083–2092. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Hummel R, Wang T, Watson DI, Michael MZ,
Van der Hoek M, Haier J and Hussey DJ: Chemotherapy-induced
modification of microRNA expression in esophageal cancer. Oncol
Rep. 26:1011–1017. 2011.PubMed/NCBI
|
|
9
|
Pathak S, Meng WJ, Nandy SK, Ping J,
Bisgin A, Helmfors L, Waldmann P and Sun XF: Radiation and SN38
treatments modulate the expression of microRNAs, cytokines and
chemokines in colon cancer cells in a p53-directed manner.
Oncotarget. 6:44758–44780. 2015.PubMed/NCBI
|
|
10
|
Shoshan E, Mobley AK, Braeuer RR, Kamiya
T, Huang L, Vasquez ME, Salameh A, Lee HJ, Kim SJ, Ivan C, et al:
Reduced adenosine-to-inosine miR-455-5p editing promotes melanoma
growth and metastasis. Nat Cell Biol. 17:311–321. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Hudson J, Duncavage E, Tamburrino A,
Salerno P, Xi L, Raffeld M, Moley J and Chernock RD: Overexpression
of miR-10a and miR-375 and downregulation of YAP1 in medullary
thyroid carcinoma. Exp Mol Pathol. 95:62–67. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Sand M, Skrygan M, Sand D, Georgas D, Hahn
SA, Gambichler T, Altmeyer P and Bechara FG: Expression of
microRNAs in basal cell carcinoma. Br J Dermatol. 167:847–855.
2012. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Zhang Z, Hou C, Meng F, Zhao X, Zhang Z,
Huang G, Chen W, Fu M and Liao W: MiR-455-3p regulates early
chondrogenic differentiation via inhibiting Runx2. FEBS Lett.
589:3671–3678. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Boisen MK, Dehlendorff C, Linnemann D,
Nielsen BS, Larsen JS, Osterlind K, Nielsen SE, Tarpgaard LS,
Qvortrup C, Pfeiffer P, et al: Tissue microRNAs as predictors of
outcome in patients with metastatic colorectal cancer treated with
first line capecitabine and oxaliplatin with or without
bevacizumab. PLoS One. 9:e1094302014. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Bera A, VenkataSubbaRao K, Manoharan MS,
Hill P and Freeman JW: A miRNA signature of chemoresistant
mesenchymal phenotype identifies novel molecular targets associated
with advanced pancreatic cancer. PLoS One. 9:e1063432014.
View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Hamilton MP, Rajapakshe K, Hartig SM, Reva
B, McLellan MD, Kandoth C, Ding L, Zack TI, Gunaratne PH, Wheeler
DA, et al: Identification of a pan-cancer oncogenic microRNA
superfamily anchored by a central core seed motif. Nat Commun.
4:27302013. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Hiroki E, Akahira J, Suzuki F, Nagase S,
Ito K, Suzuki T, Sasano H and Yaegashi N: Changes in microRNA
expression levels correlate with clinicopathological features and
prognoses in endometrial serous adenocarcinomas. Cancer Sci.
101:241–249. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Li YJ, Ping C, Tang J and Zhang W:
MicroRNA-455 suppresses non-small cell lung cancer through
targeting ZEB1. Cell Biol Int. 40:621–628. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Chai J, Wang S, Han D, Dong W, Xie C and
Guo H: MicroRNA-455 inhibits proliferation and invasion of
colorectal cancer by targeting RAF proto-oncogene
serine/threonine-protein kinase. Tumour Biol. 36:1313–1321. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Song G, Gu L, Li J, Tang Z, Liu H, Chen B,
Sun X, He B, Pan Y, Wang S, et al: Serum microRNA expression
profiling predict response to R-CHOP treatment in diffuse large B
cell lymphoma patients. Ann Hematol. 93:1735–1743. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Yang H and Wang Y: Five miRNAs considered
as molecular targets for predicting neuroglioma. Tumour Biol.
37:1051–1059. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Baniwal SK, Khalid O, Gabet Y, Shah RR,
Purcell DJ, Mav D, Kohn-Gabet AE, Shi Y, Coetzee GA and Frenkel B:
Runx2 transcriptome of prostate cancer cells: Insights into
invasiveness and bone metastasis. Mol Cancer. 9:2582010. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Akech J, Wixted JJ, Bedard K, van der Deen
M, Hussain S, Guise TA, van Wijnen AJ, Stein JL, Languino LR,
Altieri DC, et al: Runx2 association with progression of prostate
cancer in patients: Mechanisms mediating bone osteolysis and
osteoblastic metastatic lesions. Oncogene. 29:811–821. 2010.
View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Brusgard JL, Choe M, Chumsri S, Renoud K,
MacKerell AD Jr, Sudol M and Passaniti A: RUNX2 and TAZ-dependent
signaling pathways regulate soluble E-cadherin levels and
tumorsphere formation in breast cancer cells. Oncotarget.
6:28132–28150. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Tandon M, Chen Z and Pratap J: Runx2
activates PI3K/Akt signaling via mTORC2 regulation in invasive
breast cancer cells. Breast Cancer Res. 16:R162014. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
van der Deen M, Akech J, Wang T, Fitz
Gerald TJ, Altieri DC, Languino LR, Lian JB, van Wijnen AJ, Stein
JL and Stein GS: The cancer-related Runx2 protein enhances cell
growth and responses to androgen and TGFβ in prostate cancer cells.
J Cell Biochem. 109:828–837. 2010.PubMed/NCBI
|
|
27
|
Cohen-Solal KA, Boregowda RK and Lasfar A:
RUNX2 and the PI3K/AKT axis reciprocal activation as a driving
force for tumor progression. Mol Cancer. 14:1372015. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Niu DF, Kondo T, Nakazawa T, Oishi N,
Kawasaki T, Mochizuki K, Yamane T and Katoh R: Transcription factor
Runx2 is a regulator of epithelial-mesenchymal transition and
invasion in thyroid carcinomas. Lab Invest. 92:1181–1190. 2012.
View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Tandon M, Chen Z, Othman AH and Pratap J:
Role of Runx2 in IGF-1Rβ/Akt- and AMPK/Erk-dependent growth,
survival and sensitivity towards metformin in breast cancer bone
metastasis. Oncogene. 35:4730–4740. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Wysokinski D, Blasiak J and Pawlowska E:
Role of RUNX2 in breast carcinogenesis. Int J Mol Sci.
16:20969–20993. 2015. View Article : Google Scholar : PubMed/NCBI
|