Introduction
Acute leukemia (AL) is a malignant clonal disease of
hematopoietic stem cells. The incidence rate of leukemia in China
is 5.68/100,000, and it is the leading cause of cancer-related
mortality in children and adults under the age of 35 (1). Recent progress in medical sciences and
therapeutic approaches such as chemotherapy, radiotherapy,
biological regulation and hematopoietic stem cell transplantation
has resulted in significant advancements; however, leukemia
continues to be a significant health burden. An effective molecular
marker for early diagnosis, prognosis and treatment guidance, is
therefore, urgently required.
Numerous studies have shown that epigenetic
abnormalities play an important role in the development and
progression of AL (2). Non-coding
RNA has increasingly been shown to play an important role in
epigenetic regulation. Long non-coding RNAs (lncRNAs) are a class
of RNA more than 200 nucleotides in length that lack an open
reading frame and protein coding function, most of which are
transcribed by RNA polymerase II (3). lncRNAs can regulate gene expression at
the level of epigenetics, transcription and post-transcription,
participate in X chromosome inactivation, genomic imprinting,
chromatin modification, transcription activation, transcription
interference, nuclear transport and a variety of important
regulatory processes. lncRNAs are closely involved with the onset,
development and pathogenesis of human diseases (4–7).
In 2007, Rinn et al (8) were the first to identify an lncRNA
regulating gene expression in trans at the HOXC gene locus
on chromosome 12. The lncRNA is involved in regulation of HOX gene
clusters located on different chromosomes rather than
cis-regulation in the HOXC locus and was, therefore, named
HOX antisense intergenic RNA (HOTAIR). Gupta et al (9) subsequently found that the expression
level of HOTAIR in breast cancer metastases was significantly
higher than in primary breast cancer and normal breast tissues, and
that the high expression of HOTAIR was associated with metastasis
and a poor prognosis for patients. An additional two studies on
breast cancer reached similar conclusions (10,11).
Further studies showed that expression levels of HOTAIR were also
significantly increased in colorectal cancer (12), hepatocellular carcinoma (13), lung (14), pancreatic (15) and nasopharyngeal cancer (16), and other malignant tumors and
metastases. Furthermore, cancer patients with high HOTAIR
expression had lower survival rates and higher recurrence rates
(17). These studies suggested that
HOTAIR was involved in tumorigenesis, and had significant clinical
importance.
Studies have shown that HOTAIR epigenetically
regulates gene expression, acting as a scaffold for protein
complexes in both PRC2 and LSD1-mediated target gene silencing.
PRC2, a member of PcG family, consists of the core elements EZH2,
EED and SUZ12, as well as histone binding protein RbAp46 and PHFl.
EZH2 is an important subunit with catalytic activity, with a highly
conserved SET structural domain that methylates the 9th and 27th
lysine in the nucleosome histone H3, thereby inhibiting hundreds of
genes. These include genes involved in cell growth,
differentiation, tumor metastasis and expression of related genes.
SUZ12 and EED maintain the stability of the complex, and are
essential components in the catalysis of PRC2 complexes (18). The LSD1 complex is comprised of
LSD1, REST, CoREST, HDAC1-2, BHC80 and BRAF35 (19,20).
LSD1 removes the methyl groups on H3K4me1/2 and H3K9me1/2,
resulting in a single methyl group or no methyl group, thereby
regulating transcription of downstream genes (21). REST, as a DNA-binding protein, is
involved in localization of the LSD1 complex to the correct genomic
location. CoREST can bind with nucleosomes, and recruit LSD1 to
demethylate H3K4. Together, these proteins cooperatively inhibit
transcription.
Although HOTAIR has been implicated in the onset of
a variety of tumors, its role in hematological tumor formation
remains unclear. To date, its significance in terms of diagnosis
and prognosis in leukemia has not been extensively studied. HOTAIR
acts as a scaffold for histone modification complexes and is
involved in epigenetic gene regulation (22). The present study aimed to examine
whether expression of DNA methyltransferases and histone
methyltransferases in leukemia patients was modulated by HOTAIR,
and whether HOTAIR could act as a diagnostic/prognostic marker for
leukemia.
Materials and methods
Patients
Ninety-six bone marrow cell samples were collected
from patients diagnosed with leukemia and treated at the Affiliated
Union Hospital of Fujian Medical University between October 2011
and February 2015. Patients included 56 males and 40 females
between the ages of 14 and 84. Seventy-three cases were acute
myelogenous leukemia and 23 cases were acute lymphoblastic
leukemia. Eighty bone marrow samples from bone marrow donors and
patients with non-hematologic malignancies were studied as
controls. All specimens were obtained with approval from the
Medical Ethics Committee and the patients informed consent.
Inclusion criteria were as follows: i) diagnosis and classification
of AL according to the French-American-British (FAB) classification
criteria; ii) confirmation of the AL diagnosis by morphology,
immunophenotyping, cytogenetics, molecular cell biology (MICM) and
additional examinations. Cases of transformation of myelodysplastic
syndrome (MDS) and acute transformation of chronic myelogenous
leukemia (CML) were excluded; iii) patients had no other types of
malignant tumors or contraindications to bone marrow puncture.
Evaluation criteria for efficacy were based on ‘Diagnosis and
Treatment Standards of Blood Disease’, compiled by Zhang Zhinan.
Prognosis was based on standards recommended by the European
Leukemia Working Group (23).
Extraction of mononuclear cells from
bone marrow
Following bone marrow puncture, 5 ml of bone marrow
was harvested using sterile procedures and added to a tube
containing 0.2 ml heparin. Mononuclear cells in bone marrow were
isolated using lymphocyte separation medium (Hao Yang in Tianjin).
The cell pellet was processed for extraction of DNA, RNA and
proteins, or was stored at −80°C for later use.
Extraction of total cellular RNA and
cDNA synthesis
TRIzol reagent (Invitrogen, Carlsbad, CA, USA) was
used to extract RNA. Two microliters of RNA was used to measure
concentration and purity on a quantitation analyzer for nucleic
acids. Purity of RNA was confirmed by OD260/280 ratios between
1.8–2.0. RNA was immediately reverse transcribed (Thermo Fisher
Scientific, Inc., Waltham, MA, USA) to cDNA. A PCR tube was used,
RNA (1 µg) was added, random primer 1 µl, diethylpyrocarbonate
(DEPC) water to 12 µl; 65°C water bath for 5 min and quickly
returned to ice for cooling; then 5X reverse transcriptase buffer 4
µl was added, 10 mM dNTP mix 2 µl, reverse transcriptase 1 µl,
RNase inhibitor 1 µl to the total volume was 20 µl. The reaction
conditions were: 25°C for 5 min, 42°C for 60 min, 70°C for 5 min
and the temperature was then decreased to 4°C. The cDNA was either
immediately used as a template for PCR or stored at −20°C for later
use.
Detection of changes in mRNA levels
using real-time PCR
Real-time PCR (ABI 7500) was performed to detect
expression of the lncRNA HOTAIR, PRC2 complex (EZH2, SUZ12 and
EED), LSD1 complex (LSD1, REST and CoREST), and methyltransferase
(DNMT3A and DNMT3B) genes. The total reaction volume was 25 µl, and
included 12.5 µl SYBR-Green qPCR Mix (Roche), 1 µl cDNA, 0.75 µl
each of forward and reverse primers at 10 µmol/l (Table I), and DEPC water to 25 µl. Cycling
parameters were as follows: 50°C for 2 min for 1 cycle; 95°C for 2
min for 1 cycle; 95°C for 15 sec, then 60°C for 30 sec for 40
cycles; melting curve analysis at 95°C for l5 sec, 60°C for l min,
95°C for l5 sec, 60°C for l5 sec for 1 cycle. GAPDH was used as an
internal reference. Quantification of mRNA expression levels was
performed by comparison of Ct values. Copy numbers were normalized
to GAPDH and the ΔCt of target genes calculated as the average Ct
value of target genes - average Ct value of reference genes. The
mean was obtained and the relative expression values calculated
using the 2−ΔCt method and differences compared.
 | Table I.Primer sequences for qRT-PCR. |
Table I.
Primer sequences for qRT-PCR.
| Gene | Primer (5′-3′) | Product size
(bp) | Gene accession
no. |
|---|
| GAPDH | Forward |
CCCCTTCATTGACCTCAACTACAT | 135 | NM_002046.4 |
|
| Reverse |
CGCTCCTGGAAGATGGTGA |
|
|
| HOTAIR | Forward |
GGTAGAAAAAGCAACCACGAAGC | 169 | NR_047517.1 |
|
| Reverse |
ACATAAACCTCTGTCTGTGAGTGCC |
|
|
| EZH2 | Forward |
GGAACAACGCGAGTCGG | 101 | NM_004456.4 |
|
| Reverse |
CTGATTTTACACGCTTCCGC |
|
|
| Suz12 | Forward |
GCATTGCCCTTGGTGTACTC | 214 | NM_015355.2 |
|
| Reverse |
TGGTCCGTTGCGACTAAAA |
|
|
| EED | Forward |
GTGTGCGATGGTTAGGCG | 120 | NM_003797.3 |
|
| Reverse |
GTCACATTAGATTCACTGGGTTT |
|
|
| LSD1 | Forward |
ACCACAACAGACCCAGAAGG | 116 | NM_015013.3 |
|
| Reverse |
GGTGCTTCTAATTGTTGGAGAG |
| REST | Forward |
GGCACGGAAGGAGCAAGT | 97 | NM_005612.4 |
|
| Reverse |
GGTGAGAGATCCTCTGTGC |
|
|
| CoREST | Forward |
CGAGGACTAAAACTAGTGTGATGG | 86 | NM_015156.3 |
|
| Reverse |
TGCCTCTTCCAGTTCATCCT |
|
|
| DNMT3A | Forward |
TATTGATGAGCGCACAAGAGAGC | 111 | NM_015355.2 |
|
| Reverse |
GGGTGTTCCAGGGTAACATTGAG |
|
|
| DNMT3B | Forward |
GACTCGAAGACGCACAGCTG | 98 | NM_006892.3 |
|
| Reverse |
CTCGGTCTTTGCCGTTGTTATAG |
|
|
Statistical analysis
Statistical software including SPSS 22.0 and
GraphPad Prism 6.0 was used for data analysis. Quantitative data
are expressed as mean ± standard deviation or median ±
interquartile range, and qualitative data as the number of cases
and percentages. The two groups were compared using the
Mann-Whitney U test, while multiple groups were compared using the
Kruskal-Wallis H test, and rates were compared and analyzed by the
Chi-square or Fisher's exact tests. The Kaplan-Meier method and
log-rank test was adopted to compare differences in survival time.
Variables influencing survival times with significant differences
in univariate analysis were analyzed using a Cox proportional
hazards model. P<0.05 was considered statistically significant.
Overall survival (OS) was measured from the beginning of the trial
to the death of the patient (by any cause) or to the date of last
follow-up for surviving patients. Event-free survival (EFS) was
measured from the beginning of the trial to treatment failure,
relapse or death (from any cause). For patients not experiencing
these events, EFS was calculated to the date of the last follow-up.
For patients who did not reach complete remission, EFS was
calculated from the date of the beginning of clinical trials to the
date of disease progression or death.
Results
Expression of HOTAIR in AL patients,
and analysis of clinical features and prognosis
The expression of HOTAIR genes in mononuclear cells
from 96 patients with AL and 80 controls was examined by RT-qPCR.
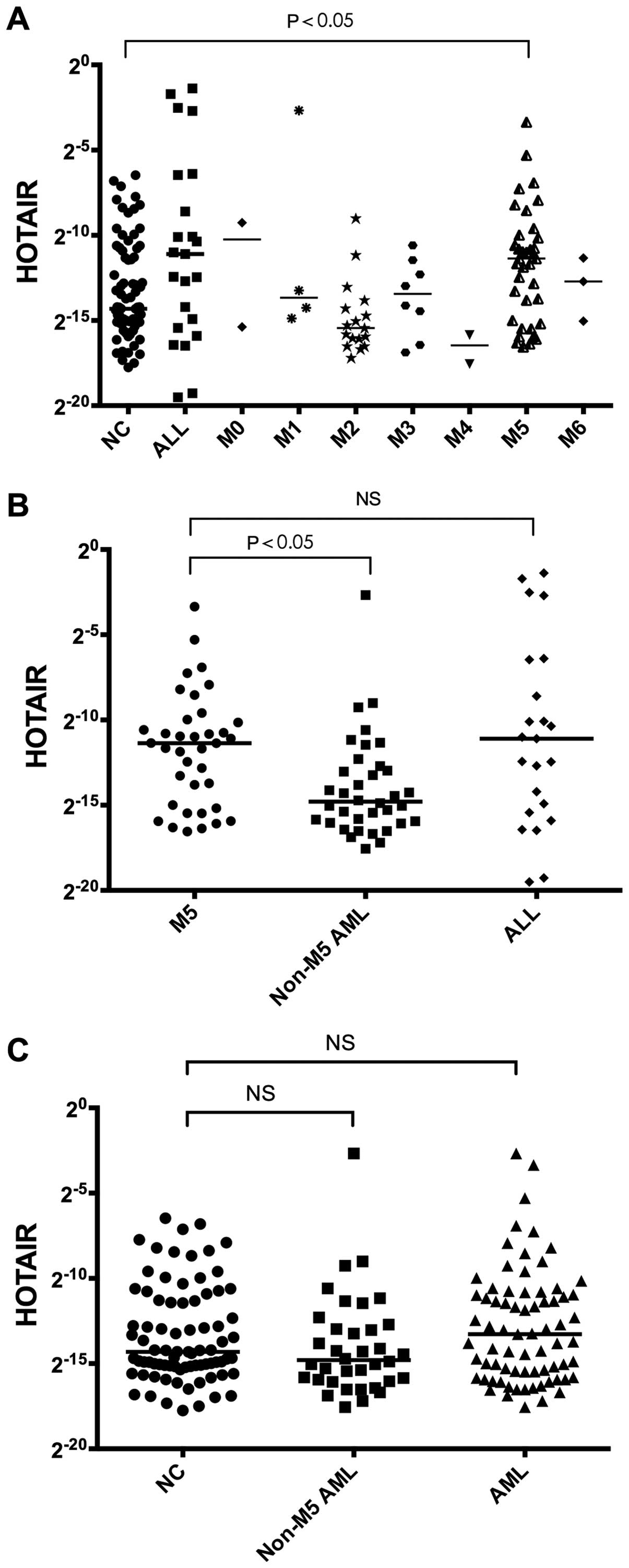
Our results show that the expression of HOTAIR in acute monocytic
leukemia (M5) patients increased relative to controls (P=0.0289;
Fig. 1A), and the increase is
greater than in patients with other types of acute myelogenous
leukemia (AML; P=0.0019; Fig. 1B).
No significant difference is observed when expression is compared
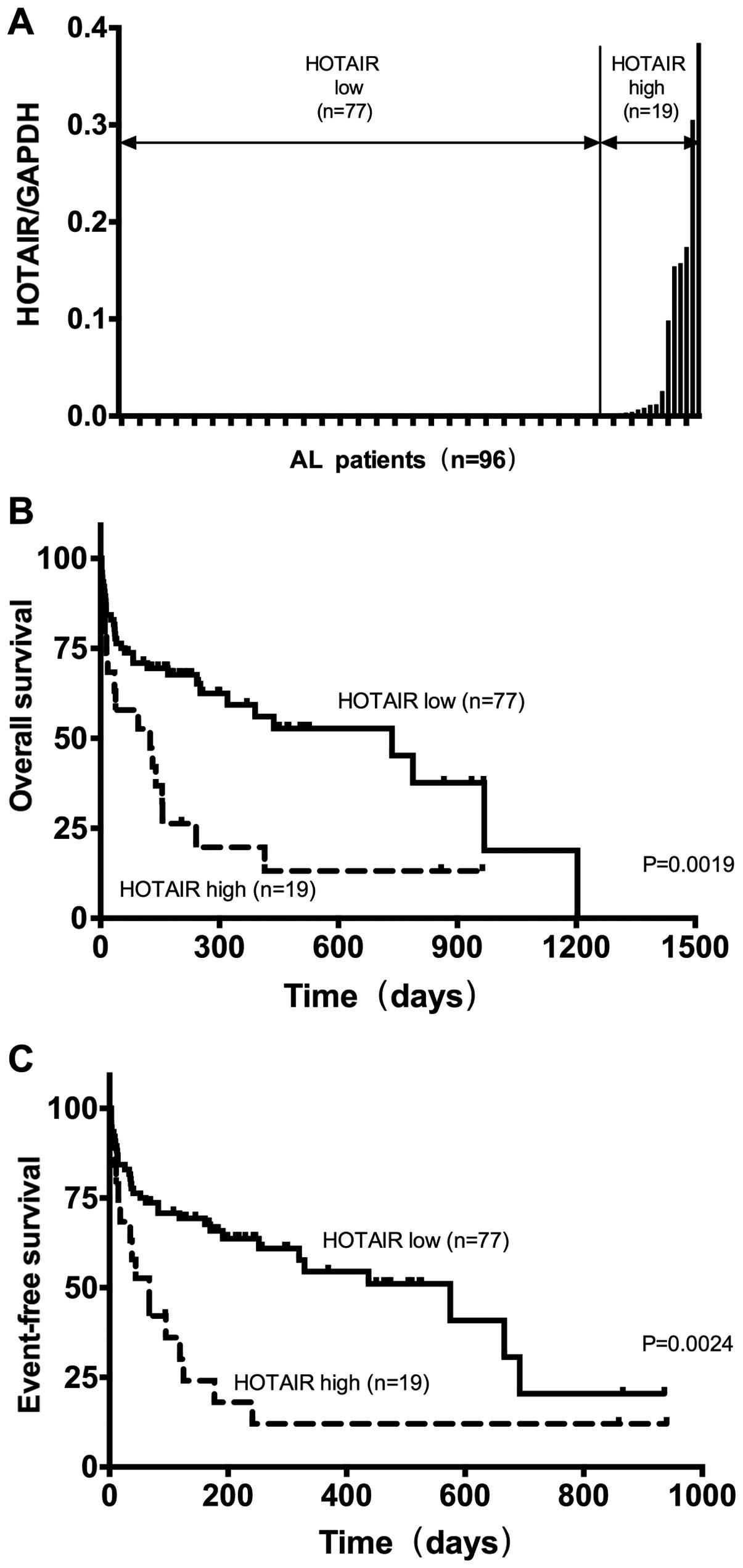
to ALL, AML, non-M5 AML patients (P=0.7041; Fig. 1B; P=0.1597, P=0.6209; Fig. 1C). Based on a HOTAIR/GAPDH
ratio=0.00098, patients were divided into a high HOTAIR expression
group (n=19) and a low expression group (n=77; Fig. 2A) (24), and the clinical features of the two
groups were analyzed. Patients from the two groups show no
statistically significant differences in gender, age, FAB type
(French-American-British Classification: A standardized
classification system for acute leukemias based on morphology),
white blood cell count, hemoglobin, platelets, proportion of
juvenile cells in the bone marrow, or risk stratification based on
karyotype (Table II). After
obtaining consent from the patients families, a telephone follow-up
was conducted in combination with inpatient and/or outpatient
medical record examination. Follow-up was completed May 30, 2015.
The Kaplan-Meier method was used to analyze the association of
expression levels of HOTAIR with OS and EFS in AL patients. The
results show that, compared with patients with low HOTAIR
expression, OS and EFS of patients with high HOTAIR expression
significantly decreased (P<0.05; Fig. 2B and C). Clinical prognostic
indicators were also analyzed. Univariate analysis of OS and EFS
shows that age, peripheral blood leucocyte count and the relative
level of HOTAIR expression are prognostic indicators, and although
the remaining the clinical features were not found to be prognostic
indicators, it could be due to an insufficient number of cases
(Table III). The characteristics
where P<0.05 were subjected to multivariable analysis showed
that, for OS and EFS in AL patients, age, peripheral blood
leucocyte count and HOTAIR expression are independent prognostic
factors (Table IV).
 | Table II.HOTAIR expression and
clinicopathological characteristics of patients. |
Table II.
HOTAIR expression and
clinicopathological characteristics of patients.
| Factors | HOTAIR high
expression (n=19) N (%) | HOTAIR low
expression (n=77) N (%) | P-value |
|---|
| Gender
(range/median) |
|
|
|
|
Male | 14 (73.7) | 42 (54.5) | 0.194 |
|
Female | 5 (26.3) | 35 (45.5) |
|
| Age (years,
range/median) |
|
|
|
|
<60 | 17 (89.5) | 63 (81.8) | 0.731 |
|
≥60 | 2 (10.5) | 14 (18.2) |
|
| FAB
classification |
|
|
|
|
AML | 13 (68.4) | 63 (81.8) | 0.216 |
|
ALL | 6 (31.6) | 14 (18.2) |
|
| WBC
(x109/l) |
|
|
|
|
<100 | 13 (68.4) | 59 (76.6) | 0.555 |
|
≥100 | 6 (31.6) | 18 (23.4) |
|
| HB (g/l) |
|
|
|
|
<60 | 4 (21.1) | 22 (28.6) | 0.579 |
|
≥60 | 15 (78.9) | 55 (71.4) |
|
| PLT
(x109/l) |
|
|
|
|
<30 | 10 (52.6) | 31 (40.3) | 0.438 |
|
≥30 | 9 (47.4) | 46 (59.7) |
|
| Bone marrow blasts
(%) |
|
|
|
|
<60 | 3 (15.8) | 13 (16.9) | 1 |
|
≥60 | 16 (84.2) | 64 (83.1) |
|
| Risk stratification
based on karyotypea |
|
|
|
|
Better-risk | 0 | 11 (14.3) | 0.075 |
|
Intermediate-risk | 7 (36.8) | 31 (40.3) |
|
|
Poor-risk | 2 (10.5) | 15 (19.5) |
|
|
Unknown | 10 (52.6) | 20 (26.0) |
 | Table III.Univariate analysis of OS and EFS of
patients with AL. |
Table III.
Univariate analysis of OS and EFS of
patients with AL.
| Characteristic | Group | n= | P-value (OS) | P-value (EFS) |
|---|
| Gender | Male/female | 56/40 | 0.238 | 0.171 |
| Age (years) | <60/≥60 | 79/17 | 0.003 | 0.005 |
| FAB
classification | AML/ALL | 73/23 | 0.615 | 0.523 |
| WBC
(x109/l) | <100/≥100 | 70/26 | <0.001 | <0.001 |
| HB (g/l) | <60/≥60 | 25/71 | 0.653 | 0.438 |
| PLT
(x109/l) | <30/≥30 | 40/56 | 0.251 | 0.265 |
| Bone marrow
blasts | <60/≥60% | 15/81 | 0.750 | 0.995 |
| HOTAIR | High/low | 19/77 | 0.003 | 0.003 |
 | Table IV.Multivariate analysis of OS and EFS
of patients with AL. |
Table IV.
Multivariate analysis of OS and EFS
of patients with AL.
|
|
|
| OS | EFS |
|---|
|
|
|
|
|
|
|---|
| Characteristic | Group | No. | HR | 95% CI | P-value | HR | 95% CI | P-value |
|---|
| Age (years) | <60/≥60 | 79/17 | 0.354 | 0.166–0.758 | 0.004 | 0.364 | 0.172–0.771 | 0.006 |
| WBC
(x109/l) | <100/≥100 | 70/26 | 0.379 | 0.186–0.773 | 0.001 | 0.438 | 0.219–1.877 | 0.002 |
| HOTAIR | High/low | 19/77 | 2.407 | 1.253–4.624 | 0.005 | 2.388 | 1.248–4.568 | 0.005 |
Expression levels of downstream genes
in the HOTAIR signaling pathway
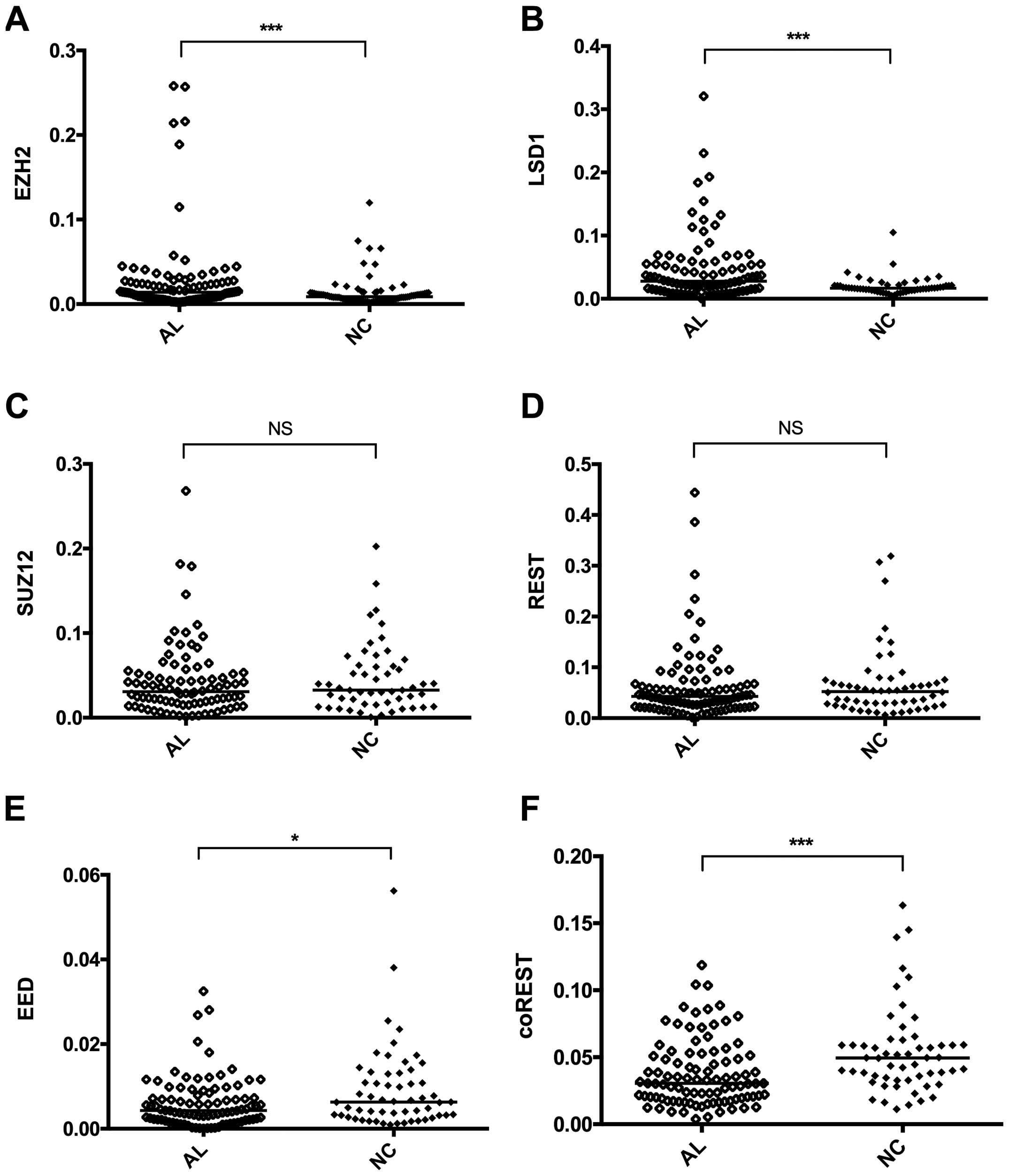
Expression of the PRC2 complex (EZH2, SUZ12 and EED)
and the LSD1 complex (LSD1, REST and CoREST), and downstream genes
in the HOTAIR signaling pathway was measured by RT-qPCR. Our
results show that, compared with normal controls, the expression of
EZH2 and LSD1 in AL patients significantly increased (P=0.0009 and
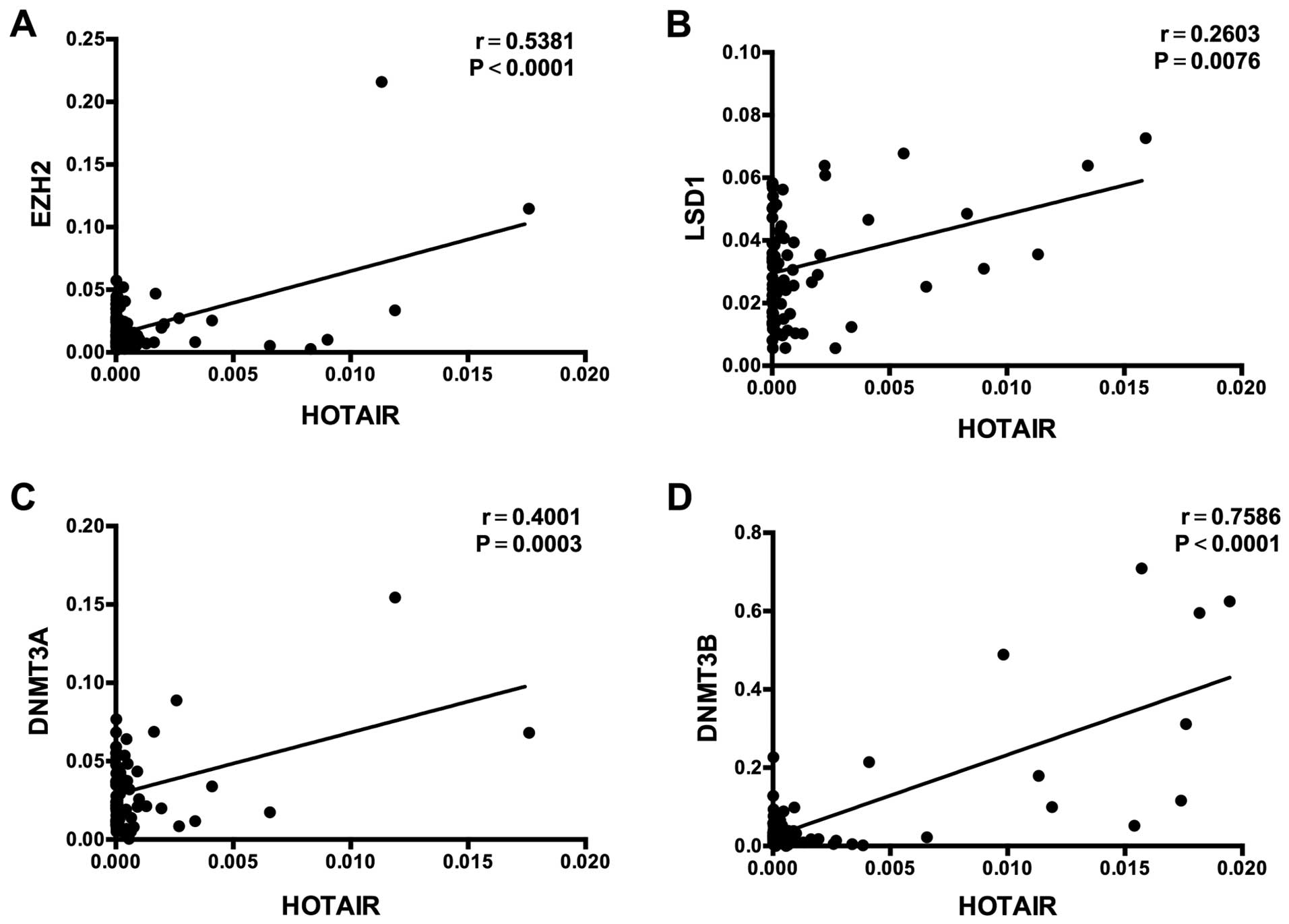
P=0.0003, respectively; Fig. 3A and
B), and showed a significant correlation with the high
expression of HOTAIR (EZH2, r=0.5381, P<0.0001; LSD1, r=0.2603,
P=0.0076; Fig. 5A and B).
Expression of SUZ12 and REST showed no significant differences
(P=0.5437 and P=0.4265, respectively; Fig. 3C and D), while expression of EED and
CoREST decreased (P=0.0213 and P=0.0003, respectively; Fig. 3E and F).
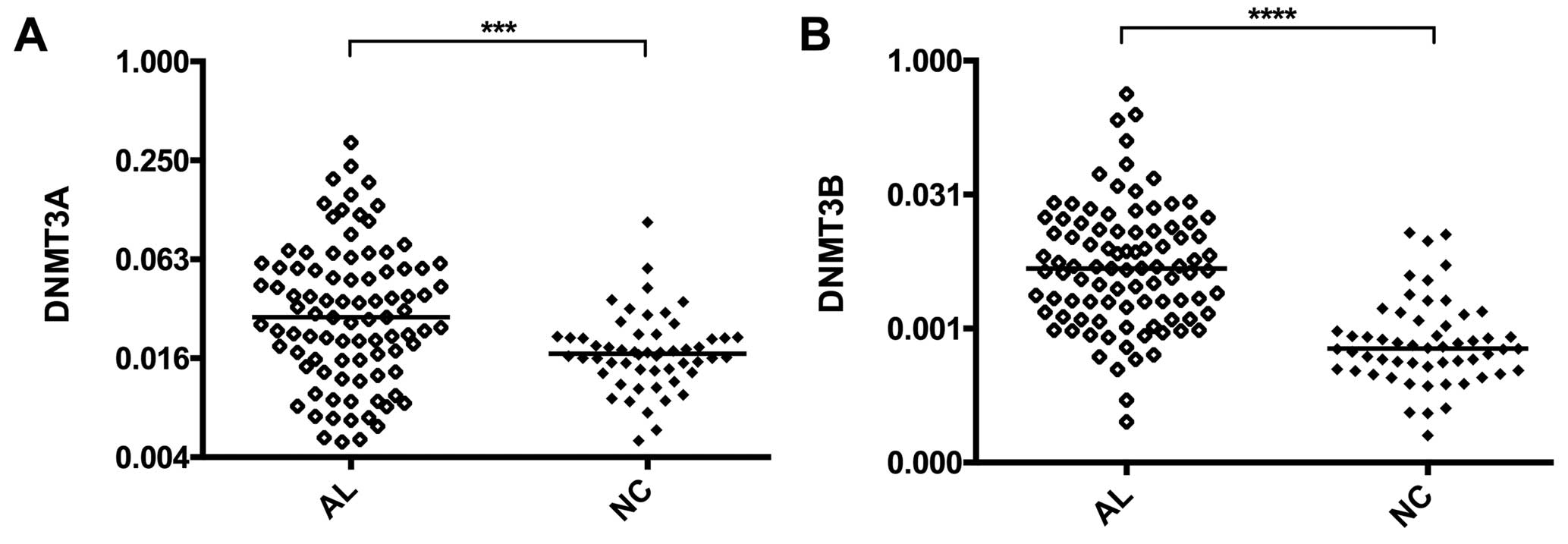
Expression levels of DNA
methyltransferase genes
DNA methylation plays an important role in the
occurrence and progression of leukemia, and expression of DNA
methyltransferase genes directly influences the functions of
certain oncogenes and tumor suppressor genes. DNA
methyltransferases are divided into two types: DNMT1 maintains DNA
methylation while DNMT3A, and DNMT3B remethylate demethylated CpG
sites. We used RT-qPCR to detect expression of DNMT3A and DNMT3B.
The results show that, compared with normal controls, the
expression of DNMT3A in AL patients significantly increases
(P=0.0002; Fig. 4A), as does DNMT3B
(P<0.0001, Fig. 4B). Expression
of these genes shows a significant positive correlation with high
expression of HOTAIR (DNMT3A, r=0.4001, P=0.0003; DNMT3B, r=0.7586,
P<0.0001; Fig. 5C and D).
Discussion
Previous studies have assigned an oncogenic function
to HOTAIR in a variety of tumors, findings supported in the present
study of AL. Compared with controls, the expression of HOTAIR in M5
patients increased, while the difference in other types of leukemia
patients did not reach statistical significance. Compared to
patients with low expression, high expression of HOTAIR was closely
associated with a poor prognosis in AL patients. Multivariate
analysis showed that age, peripheral blood leucocyte count and high
expression of HOTAIR were independent prognostic indicators for OS
and EFS. In addition, the examination of downstream genes in the
HOTAIR signaling pathway showed that the expression of EZH2, LSD1,
DNMT3A and DNMT3B in AL patients significantly increased, and
showed a significant positive correlation with high expression of
HOTAIR. Based on these observations, we speculated that HOTAIR may
participate in the development and progression of leukemia by
mediating the methylation of DNA and histones.
Although multiple groups have examined the function
of HOTAIR in tumorigenesis, its clinical significance and molecular
mechanisms in AL patients were not previously fully elucidated. Wu
et al (25) and Hao and Shao
(26) analyzed the clinical
features between HOTAIR and acute myeloid leukemia. Our research
further strengthens the evidence linking high HOTAIR expression
with poor prognosis in AL patients, and analyzed the relationship
between HOTAIR and downstream effector genes. The present study
found that, compared with normal controls, the expression of HOTAIR
in M5 patients increased, while in other types of leukemia patients
the difference did not reach statistical significance. We
speculated that, with an increase in sample size, the expression of
HOTAIR probably would increase in other types of AL. This will
require further examination and an increased study size. In the
present study we did not take into account the heterogeneity among
different types of AL. Differences in expression of HOTAIR could be
due to a different mechanism of action in different types of AL. We
followed up the AL patients and conducted clinical observations,
and found that the OS and EFS of patients with high expression of
HOTAIR were significantly reduced. Multivariate analysis showed
that high expression of HOTAIR is one of the risk factors
predicting OS and EFS, suggesting that HOTAIR could be used as a
molecular marker to predict leukemia and determine its
prognosis.
Studies focused on PRC2 complexes found that
transcription and activation of the complex in tumors was mediated
by EZH2, indicating that the EZH2 plays an important role in
function of the PRC2 complex. Overexpression of EZH2 was first
documented by gene chip analysis in prostate and breast cancer
(27,28). Overexpression of EZH2 was closely
related to invasiveness, eventual metastasis, poor clinical outcome
and poor prognosis in a variety of tumors (28–30). A
number of studies, however, have shown that the mechanisms involved
in the overexpression of EZH2 are cell type-dependent. EZH2 plays
an oncogenic role in most solid tumors through a variety of
molecular mechanisms. Increased EZH2 leads to an increase in
H3K27me3, thereby modifying and silencing the expression of tumor
suppressor genes in tumor cells epigenetically, resulting in
tumorigenesis. Increased EZH2 expression is involved in tumor
development through multiple signaling pathways. For example,
overexpression of the EZH2, MEK-ERK, and pRB-E2F signaling pathway
is associated with the occurrence of breast cancer (31,32).
MYC and ETS transcription factors can directly regulate the
transcription of EZH2 in prostate cancer (33,34).
In Ewing's sarcoma, the fusion oncoprotein EWS-FLI1 can induce the
expression of EZH2, and plays a key role in the differentiation of
the endothelium/neuroectoderm and in tumor growth (35). Paradoxically, recent studies have
shown that mutations or deletions in PRC2 complex components may
occur in some myelodysplastic syndromes (MDS) and
myeloproliferative neoplasms. Altered expression of EZH2 in cancers
may occur through complex mechanisms; the cellular environment
influences activation of oncogenic pathways leading to epigenetic
modifications promoting tumorigenesis (36,37).
The LSD1 subunit, which has a major catalytic function in the LSD1
complex, was the first identified histone demethylase. It is highly
expressed in a variety of tumor cells and promotes the growth,
metastasis and invasiveness of tumors, including prostate (38), breast (39), lung (40) and gastrointestinal cancer (41), and retinoblastoma (42). RNAi or small molecule inhibitors can
inhibit the expression and activity of LSD1, leading to inhibition
of tumor growth and metastasis. We examined the expression of the
PRC2 complex (EZH2, SUZ12 and EED) and LSD1 complex (LSD1, REST and
CoREST) in AL patients compared to normal controls in an attempt to
understand their significance in leukemia. Our results showed that,
compared with normal controls, the expression levels of EZH2 and
LSD1 in AL patients increased, those of SUZ12 and REST showed no
significant differences, and EED and CoREST decreased. We
speculated that the EHZ2 and LSD1 subunits play important roles in
the PRC2 and LSD1 complexes, respectively. HOTAIR could be involved
in the development of leukemia through regulation of the PRC2 and
LSD1 complexes and through mediating regulation of histone
methylation. The reduced expression of EED and CoREST could be due
to negative feedback or their participation in other regulatory
processes. Elucidation of specific mechanisms will be examined in
future studies by our group.
Methylation of genomic DNA is catalyzed and
sustained by DNA methyltransferase (DNMT). In mammals, DNMT can be
divided into three types based on structural and functional
differences: DNMT1, DNMT2 and DNMT3 (43–45).
The DNMT3 family includes DNMT3A, DNMT3B and the regulatory factor
DNMT3L which participate in the process of remethylation of
demethylated DNA (46). DNMT1
maintains methylation during DNA replication and repair, while the
function of DNMT2 is not yet clear (47). Several studies have shown that
abnormal DNA methylation is closely related to tumorigenesis and
that the expression of DNMTs in cancerous tissues is increased
(48,49). However, histone methylation and DNA
methylation were initially thought to be two separate gene
silencing systems. Viré et al (50) found that PRC2 complexes
co-immunoprecipitated with three transmethylases, and that
silencing of specific genes required the joint participation of
both EZH2 and DNA transmethylases. These findings revealed a direct
relationship between the two key epigenetic systems. LSD1 also
plays an important role in coordinating histones and DNA
methylation by acting directly on histones and DNMT1 (51). Understanding the role of DNMTs and
histone methylation in leukemia and exploring whether DNA
methylation and histone methylation are jointly involved in the
occurrence of the disease is particularly important. We examined
the expression of DNMT3A and DNMT3B, and found that, compared with
normal controls, the expression of DNMT3A and DNMT3B in AL patients
significantly increased. This suggests that interactions between
DNMTs and the PRC complex may occur in the process of gene
silencing by DNA methylation, resulting in the development and
progression of AL.
In conclusion, the present study examined the
expression levels of HOTAIR, DNA methyltransferase and histone
(de)methylase genes downstream of HOTAIR in AL patients. We found
that the expression of HOTAIR in M5 patients increased and was
closely related with a poor prognosis. In addition, the expression
of EZH2, LSD1, DNMT3A and DNMT3B in AL patients significantly
increased, and showed a significant positive correlation with the
high expression of HOTAIR. Based on our findings, we suggest that
HOTAIR may be involved in the development and progression of
leukemia by mediating the methylation of DNA and histones. The
present study provides a new perspective for the further
exploration of biological functions of lncRNAs, and identifies
novel targets involved in the development, progression and
treatment of certain types of AL.
Acknowledgements
The present study was supported by the National
Natural Science Foundation of China (nos. 81370629 and 81300428),
the Major Research of Fujian Medical University (09-ZD021), the
Science and Technology Department of Fujian Province (2011Y4002),
the Education Department of Fujian Province (JA10129), and the
National and Fujian Provincial Key Clinical Specialty Discipline
Construction Program (China).
Glossary
Abbreviations
Abbreviations:
|
AL
|
acute leukemia
|
|
AML
|
acute myelogenous leukemia
|
|
ALL
|
acute lymphoblastic leukemia
|
|
M0
|
acute myeloid leukemia with minimal
differentiation
|
|
M1
|
AML with partial differentiation
|
|
M2
|
AML with maturation
|
|
M3
|
acute promyelocytic leukemia
|
|
M4
|
acute myelomonocytic leukemia with
granules
|
|
M5
|
acute monocytic leukemia
|
|
M6
|
acute erythroleukemia
|
|
NC
|
normal control
|
|
lncRNA
|
long non-coding RNA
|
|
HOTAIR
|
HOX antisense intergenic RNA
|
|
DNMTs
|
DNA methyltransferases
|
|
PRC2
|
polycomb repressive complex 2
|
|
LSD1
|
lysine-specific demethylase 1
|
|
RT-qPCR
|
reverse transcription-quantitative
PCR
|
References
|
1
|
Liu YQ, Zhao FJ, Chen WQ, et al: An
analysis of incidence and mortality of leukemia in China, 2009.
China Cancer. 7:528–534. 2013.
|
|
2
|
Muto T, Sashida G, Oshima M and Iwama A:
Mutations of epigenetic regulator genes and myeloid malignancies.
Rinsho Ketsueki. 56:2287–2294. 2015.(In Japanese). PubMed/NCBI
|
|
3
|
Chalei V, Sansom SN, Kong L, Lee S,
Montiel JF, Vance KW and Ponting CP: The long non-coding RNA Dali
is an epigenetic regulator of neural differentiation. eLife.
3:e045302014. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Lee DH, Singh P, Tsai SY, Oates N, Spalla
A, Spalla C, Brown L, Rivas G, Larson G, Rauch TA, et al:
CTCF-dependent chromatin bias constitutes transient epigenetic
memory of the mother at the H19-Igf2 imprinting control region in
prospermatogonia. PLoS Genet. 6:e10012242010. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Haemmerle M and Gutschner T: Long
non-coding RNAs in cancer and development: Where do we go from
here? Int J Mol Sci. 16:1395–1405. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Guttman M, Donaghey J, Carey BW, Garber M,
Grenier JK, Munson G, Young G, Lucas AB, Ach R, Bruhn L, et al:
lincRNAs act in the circuitry controlling pluripotency and
differentiation. Nature. 477:295–300. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Li X, Wu Z, Fu X and Han W: lncRNAs:
Insights into their function and mechanics in underlying disorders.
Mutat Res Rev Mutat Res. 762:1–21. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Rinn JL, Kertesz M, Wang JK, Squazzo SL,
Xu X, Brugmann SA, Goodnough LH, Helms JA, Farnham PJ, Segal E, et
al: Functional demarcation of active and silent chromatin domains
in human HOX loci by noncoding RNAs. Cell. 129:1311–1323. 2007.
View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Gupta RA, Shah N, Wang KC, Kim J, Horlings
HM, Wong DJ, Tsai MC, Hung T, Argani P, Rinn JL, et al: Long
non-coding RNA HOTAIR reprograms chromatin state to promote cancer
metastasis. Nature. 464:1071–1076. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Bhan A, Hussain I, Ansari KI, Kasiri S,
Bashyal A and Mandal SS: Antisense transcript long noncoding RNA
(lncRNA) HOTAIR is transcriptionally induced by estradiol. J Mol
Biol. 425:3707–3722. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Chisholm KM, Wan Y, Li R, Montgomery KD,
Chang HY and West RB: Detection of long non-coding RNA in archival
tissue: Correlation with polycomb protein expression in primary and
metastatic breast carcinoma. PLoS One. 7:e479982012. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Wu ZH, Wang XL, Tang HM, Jiang T, Chen J,
Lu S, Qiu GQ, Peng ZH and Yan DW: Long non-coding RNA HOTAIR is a
powerful predictor of metastasis and poor prognosis and is
associated with epithelial-mesenchymal transition in colon cancer.
Oncol Rep. 32:395–402. 2014.PubMed/NCBI
|
|
13
|
Ye P, Wang T, Liu WH, Li XC, Tang LJ and
Tian FZ: Enhancing HOTAIR/MiR-10b drives normal liver stem cells
toward a tendency to malignant transformation through inducing
epithelial-to-mesenchymal transition. Rejuvenation Res. 18:332–340.
2015. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Zhuang Y, Wang X, Nguyen HT, Zhuo Y, Cui
X, Fewell C, Flemington EK and Shan B: Induction of long intergenic
non-coding RNA HOTAIR in lung cancer cells by type I collagen. J
Hematol Oncol. 6:352013. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Kim K, Jutooru I, Chadalapaka G, Johnson
G, Frank J, Burghardt R, Kim S and Safe S: HOTAIR is a negative
prognostic factor and exhibits pro-oncogenic activity in pancreatic
cancer. Oncogene. 32:1616–1625. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Nie Y, Liu X, Qu S, Song E, Zou H and Gong
C: Long non-coding RNA HOTAIR is an independent prognostic marker
for nasopharyngeal carcinoma progression and survival. Cancer Sci.
104:458–464. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Nakagawa T, Endo H, Yokoyama M, Abe J,
Tamai K, Tanaka N, Sato I, Takahashi S, Kondo T and Satoh K: Large
noncoding RNA HOTAIR enhances aggressive biological behavior and is
associated with short disease-free survival in human non-small cell
lung cancer. Biochem Biophys Res Commun. 436:319–324. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Simon JA and Lange CA: Roles of the EZH2
histone methyltransferase in cancer epigenetics. Mutat Res.
647:21–29. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Lee MG, Wynder C, Cooch N and Shiekhattar
R: An essential role for CoREST in nucleosomal histone 3 lysine 4
demethylation. Nature. 437:432–435. 2005.PubMed/NCBI
|
|
20
|
Shi YJ, Matson C, Lan F, Iwase S, Baba T
and Shi Y: Regulation of LSD1 histone demethylase activity by its
associated factors. Mol Cell. 19:857–864. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Shi Y, Lan F, Matson C, Mulligan P,
Whetstine JR, Cole PA, Casero RA and Shi Y: Histone demethylation
mediated by the nuclear amine oxidase homolog LSD1. Cell.
119:941–953. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Tsai MC, Manor O, Wan Y, Mosammaparast N,
Wang JK, Lan F, Shi Y, Segal E and Chang HY: Long noncoding RNA as
modular scaffold of histone modification complexes. Science.
329:689–693. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Döhner H, Estey EH, Amadori S, Appelbaum
FR, Büchner T, Burnett AK, Dombret H, Fenaux P, Grimwade D, Larson
RA, et al: European LeukemiaNet: Diagnosis and management of acute
myeloid leukemia in adults: Recommendations from an international
expert panel, on behalf of the European LeukemiaNet. Blood.
115:453–474. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Kogo R, Shimamura T, Mimori K, Kawahara K,
Imoto S, Sudo T, Tanaka F, Shibata K, Suzuki A, Komune S, et al:
Long noncoding RNA HOTAIR regulates polycomb-dependent chromatin
modification and is associated with poor prognosis in colorectal
cancers. Cancer Res. 71:6320–6326. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Wu S, Zheng C, Chen S, Cai X, Shi Y, Lin B
and Chen Y: Overexpression of long non-coding RNA HOTAIR predicts a
poor prognosis in patients with acute myeloid leukemia. Oncol Lett.
10:2410–2414. 2015.PubMed/NCBI
|
|
26
|
Hao S and Shao Z: HOTAIR is upregulated in
acute myeloid leukemia and that indicates a poor prognosis. Int J
Clin Exp Pathol. 8:7223–7228. 2015.PubMed/NCBI
|
|
27
|
Ku M, Koche RP, Rheinbay E, Mendenhall EM,
Endoh M, Mikkelsen TS, Presser A, Nusbaum C, Xie X, Chi AS, et al:
Genomewide analysis of PRC1 and PRC2 occupancy identifies two
classes of bivalent domains. PLoS Genet. 4:e10002422008. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Varambally S, Dhanasekaran SM, Zhou M,
Barrette TR, Kumar-Sinha C, Sanda MG, Ghosh D, Pienta KJ, Sewalt
RG, Otte AP, et al: The polycomb group protein EZH2 is involved in
progression of prostate cancer. Nature. 419:624–629. 2002.
View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Kleer CG, Cao Q, Varambally S, Shen R, Ota
I, Tomlins SA, Ghosh D, Sewalt RG, Otte AP, Hayes DF, et al: EZH2
is a marker of aggressive breast cancer and promotes neoplastic
transformation of breast epithelial cells. Proc Natl Acad Sci USA.
100:11606–11611. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Collett K, Eide GE, Arnes J, Stefansson
IM, Eide J, Braaten A, Aas T, Otte AP and Akslen LA: Expression of
enhancer of zeste homologue 2 is significantly associated with
increased tumor cell proliferation and is a marker of aggressive
breast cancer. Clin Cancer Res. 12:1168–1174. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Fujii S, Tokita K, Wada N, Ito K, Yamauchi
C, Ito Y and Ochiai A: MEK-ERK pathway regulates EZH2
overexpression in association with aggressive breast cancer
subtypes. Oncogene. 30:4118–4128. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Bracken AP, Pasini D, Capra M, Prosperini
E and Colli E K: EZH2 is downstream of the pRB-E2F pathway,
essential for proliferation and amplified in cancer. EMBO J.
22:5323–5335. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Kunderfranco P, Mello-Grand M, Cangemi R,
Pellini S, Mensah A, Albertini V, Malek A, Chiorino G, Catapano CV
and Carbone GM: ETS transcription factors control transcription of
EZH2 and epigenetic silencing of the tumor suppressor gene Nkx3.1
in prostate cancer. PLoS One. 5:e105472010. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Koh CM, Iwata T, Zheng Q, Bethel C,
Yegnasubramanian S and De Marzo AM: Myc enforces overexpression of
EZH2 in early prostatic neoplasia via transcriptional and
post-transcriptional mechanisms. Oncotarget. 2:669–683. 2011.
View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Richter GH, Plehm S, Fasan A, Rössler S,
Unland R, Bennani-Baiti IM, Hotfilder M, Löwel D, von Luettichau I,
Mossbrugger I, et al: EZH2 is a mediator of EWS/FLI1 driven tumor
growth and metastasis blocking endothelial and neuro-ectodermal
differentiation. Proc Natl Acad Sci USA. 106:5324–5329. 2009.
View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Ntziachristos P, Tsirigos A, Van
Vlierberghe P, Nedjic J, Trimarchi T, Flaherty MS, Ferres-Marco D,
da Ros V, Tang Z, Siegle J, et al: Genetic inactivation of the
polycomb repressive complex 2 in T cell acute lymphoblastic
leukemia. Nat Med. 18:298–301. 2012. View
Article : Google Scholar : PubMed/NCBI
|
|
37
|
Zhang J, Ding L, Holmfeldt L, Wu G,
Heatley SL, Payne-Turner D, Easton J, Chen X, Wang J, Rusch M, et
al: The genetic basis of early T-cell precursor acute lymphoblastic
leukaemia. Nature. 481:157–163. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Metzger E, Wissmann M, Yin N, Müller JM,
Schneider R, Peters AH, Günther T, Buettner R and Schüle R: LSD1
demethylates repressive histone marks to promote
androgen-receptor-dependent transcription. Nature. 437:436–439.
2005.PubMed/NCBI
|
|
39
|
Lim S, Janzer A, Becker A, Zimmer A,
Schüle R, Buettner R and Kirfel J: Lysine-specific demethylase 1
(LSD1) is highly expressed in ER-negative breast cancers and a
biomarker predicting aggressive biology. Carcinogenesis.
31:512–520. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Lv T, Yuan D, Miao X, Lv Y, Zhan P, Shen X
and Song Y: Over-expression of LSD1 promotes proliferation,
migration and invasion in non-small cell lung cancer. PLoS One.
7:e350652012. View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Chen C, Zhao M, Yin N, He B, Wang B, Yuan
Y, Yu F, Hu J, Yin B and Lu Q: Abnormal histone acetylation and
methylation levels in esophageal squamous cell carcinomas. Cancer
Invest. 29:548–556. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Yokoyama A, Takezawa S, Schüle R, Kitagawa
H and Kato S: Transrepressive function of TLX requires the histone
demethylase LSD1. Mol Cell Biol. 28:3995–4003. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
43
|
Bestor TH: The DNA methyltransferases of
mammals. Hum Mol Genet. 9:2395–2402. 2000. View Article : Google Scholar : PubMed/NCBI
|
|
44
|
Chedin F, Lieber MR and Hsieh CL: The DNA
methyltransferase-like protein DNMT3L stimulates de novo
methylation by Dnmt3a. Proc Natl Acad Sci USA. 99:16916–16921.
2002. View Article : Google Scholar : PubMed/NCBI
|
|
45
|
Chen T and Li E: Establishment and
maintenance of DNA methylation patterns in mammals. Curr Top
Microbiol Immunol. 301:179–201. 2006.PubMed/NCBI
|
|
46
|
Gowher H, Liebert K, Hermann A, Xu G and
Jeltsch A: Mechanism of stimulation of catalytic activity of Dnmt3A
and Dnmt3B DNA-(cytosine-C5)-methyltransferases by Dnmt3L. J Biol
Chem. 280:13341–13348. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
47
|
Margot JB, Cardoso MC and Leonhardt H:
Mammalian DNA methyltransferases show different subnuclear
distributions. J Cell Biochem. 83:373–379. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
48
|
Robaina MC, Mazzoccoli L, Arruda VO, Reis
FR, Apa AG, de Rezende LM and Klumb CE: Deregulation of DNMT1,
DNMT3B and miR-29s in Burkitt lymphoma suggests novel contribution
for disease pathogenesis. Exp Mol Pathol. 98:200–207. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
49
|
Pathania R, Ramachandran S, Elangovan S,
Padia R, Yang P, Cinghu S, Veeranan-Karmegam R, Arjunan P,
Gnana-Prakasam JP, Sadanand F, et al: DNMT1 is essential for
mammary and cancer stem cell maintenance and tumorigenesis. Nat
Commun. 6:69102015. View Article : Google Scholar : PubMed/NCBI
|
|
50
|
Viré E, Brenner C, Deplus R, Blanchon L,
Fraga M, Didelot C, Morey L, Van Eynde A, Bernard D, Vanderwinden
JM, et al: The Polycomb group protein EZH2 directly controls DNA
methylation. Nature. 439:871–874. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
51
|
Wang J, Hevi S, Kurash JK, Lei H, Gay F,
Bajko J, Su H, Sun W, Chang H, Xu G, et al: The lysine demethylase
LSD1 (KDM1) is required for maintenance of global DNA methylation.
Nat Genet. 41:125–129. 2009. View
Article : Google Scholar : PubMed/NCBI
|